Significance
Developmental genes can be coopted to generate evolutionary novelties by changing their spatial regulation. However, developmental genes seldom act independently, but rather work in a gene regulatory network (GRN). How is it possible to recruit a single gene from a whole GRN? What are the properties that allow parallel cooptions of the same genes during evolution? Here, we show that a novel engrailed gene expression underlies a novel wing color pattern in flies. We show that cooption is facilitated 1) because of GRN flexibility over development and 2) because every single gene of the GRN has its own functional time window. We suggest these two temporal properties could explain why the same gene can be independently recruited several times during evolution.
Keywords: cooption, novelty, gene network, drosophilids, pigmentation
Abstract
Organisms have evolved endless morphological, physiological, and behavioral novel traits during the course of evolution. Novel traits were proposed to evolve mainly by orchestration of preexisting genes. Over the past two decades, biologists have shown that cooption of gene regulatory networks (GRNs) indeed underlies numerous evolutionary novelties. However, very little is known about the actual GRN properties that allow such redeployment. Here we have investigated the generation and evolution of the complex wing pattern of the fly Samoaia leonensis. We show that the transcription factor Engrailed is recruited independently from the other players of the anterior–posterior specification network to generate a new wing pattern. We argue that partial cooption is made possible because 1) the anterior–posterior specification GRN is flexible over time in the developing wing and 2) this flexibility results from the fact that every single gene of the GRN possesses its own functional time window. We propose that the temporal flexibility of a GRN is a general prerequisite for its possible cooption during the course of evolution.
Organisms have evolved endless morphological, physiological, and behavioral novel traits during the course of evolution (1, 2). Novel traits are defined here as characters that are qualitatively novel features of an organism (3). Theory predicts that such innovations unlock ecological opportunity and allow organisms to invade new ecological niches (4, 5). Birds have evolved feathers, which allowed them to fly (6), plants evolved flowers and subsequent reproductive success (7), turtles evolved shells (8), and beetles have grown horns to fight over females (9). The way that most of these and other adaptations first evolved, however, is still largely unknown.
Even if such innovations can originate through the emergence of de novo genes (10–13), cooption of preexisting genes or GRNs has been emerging as a widespread evolutionarily relevant mechanism. Just to cite a few examples, gene cooption underpins butterfly wing pattern variation (14), wasp venom diversification (15), convergent evolution of caffeine in plants (16), and the emergence of numerous morphological traits (reviewed in refs. 17–20).
The Drosophila model system has contributed significantly to our understanding of how the cooption of key developmental (or toolkit) genes underlies the emergence of evolutionary novelties. For instance, the diffusible morphogen Wingless was coopted to generate discrete black dots on Drosophila guttifera wings (21), and the homeodomain protein Distal-less was coopted to make the male wing black spot in Drosophila biarmipes (22). Other examples include not only the redeployment of a single gene, but the reuse of a network of a larval breathing structure to evolve an adult genital lobe (23) and the redeployment of the EGFR and Dpp pathways to evolve respiratory appendages of the Drosophila eggshell (24). The cooption of toolkit genes or GRNs leads to the idea that such redeployment is possible through changes in cis-regulation (19, 25). Indeed, the spatiotemporal expression of toolkit genes is often controlled by modular and independent enhancers (26–28). This regulatory logic is thought to facilitate the gain or loss of enhancers over evolutionary time (25).
Nevertheless, it does not explain how the new expression of the coopted toolkit genes does not interfere with the development of the tissue. Some authors have suggested that the reuse of toolkit genes might only happen during late development after completion of the early function of the redeployed genes (21, 22, 29). However, little is known about the properties of a GRN that allow the cooption of one or several of its components/genes without impairing the development of the tissue.
In this study, we use the complex wing pigmentation pattern of the fly species Samoaia leonensis as a model to address how the temporal flexibility of GRN underlies the cooption of toolkit genes to make a complex color pattern. We argue that the traditional view of a GRN overlooks the temporal nature of development. We show that 1) the anterior–posterior specification GRN is flexible over time in the developing Drosophila wing and 2) this flexibility results from the fact that every single gene of the GRN possesses its own functional time window. We hypothesize that this flexibility allows the transcription factor Engrailed to be individually coopted to generate a novel wing color pattern during evolution. We propose that the temporal flexibility of a GRN is a general prerequisite for gene cooption during the course of evolution.
Materials and Methods
Animal Collection and Rearing.
Samoaia attenuata, Samoaia ocellaris, and Samoaia hirta were collected by net sweeping in American Samoa. The Samoaia species were grown on Wheeler–Clayton food in the laboratory at room temperature. Paper was added along the side of the vial and wetted with an antifungal agent. It helped to maintain a moist environment and to facilitate pupation.
Fly Stocks.
Chymomyza amoena, Drosophila deflecta, Drosophila funebris, D. guttifera, Drosophila quadrilineata, S. leonensis, and Zaprionus ghesquierei were obtained from Drosophila Species Stock Center (http://blogs.cornell.edu/drosophila/). Additional S. leonensis individuals were obtained from Masayoshi Watada (Ehime University, Matsuyama, Japan), Drosophila grimshawi, Drosophila hawaiiensis, and Drosophila silvestris were obtained from Kenneth Kaneshiro (University of Hawai‘i, Mānoa, Hawaii).
The following transgenic lines were used: UAS-ci (30), UAS-ciRNAi (TRiP JF01272), UAS-Dl (BDSC no. 5612), UAS-DlRNAi (TRiP HMS01309), UAS-Dll (31), UAS-dpp (BDSC no. 1486), UAS-dppRNAi (TRiP JF02455), UAS-en (32), UAS-enRNAi (VDRC GD35695), UAS-hh (33), UAS-hhRNAi (TRiP 25794), UAS-NRNAi (TRiP JF01356), UAS-ptc (BDSC no. 5817), UAS-ptcRNAi (TRiP JF03223), UAS-vvlRNAi (TRiP JF02126), UAS-wg (BDSC no. 5919), UAS-wgRNAi (TRiP HMS00794), hh-lacZ (34), nabNP3537-Gal4 (35), and tub-GAL80ts (BDSC no. 7018).
Data Collection and Phylogenetic Analysis.
Phylogenetic markers were identified in several complete genomes by BLASTN using Drosophila melanogaster sequences as a probe. D. grimshawi, D. melanogaster, Drosophila mojavensis, Drosophila pseudoobscura, Drosophila virilis, and Drosophila willistoni genomes were retrieved from FlyBase (http://flybase.org/). Alternatively, markers were amplified by PCR using degenerate primers (SI Appendix, Table S1) in species for which genomic data were not available. The sequences reported in this paper have been deposited in European Molecular Biology Laboratory under specific accession numbers (SI Appendix, Table S2).
Nucleotide sequences for individual markers were aligned with MUSCLE (36) and manually adjusted, and selected blocks obtained for each marker were concatenated and used for phylogenetic reconstruction. Maximum-likelihood (ML) searches were performed using PhyML v3.1 under the GTR+Γ4 model (37). One hundred bootstrap replicates were conducted for support estimation. Bayesian phylogenetic analyses were performed using MrBayes 3.2 under the GTR+Γ+I model (38). We ran four independent chains for at least 100,000 generations and discarded the first 25,000 generations as burn-in. The nucleotide sequence alignments and tree files are downloadable from the Dryad Digital Repository at http://dx.doi.org/10.5061/dryad.prr4xgxhs.
In Situ Hybridization and Immunostaining.
In situ hybridization was carried out as previously described (39), using digoxigenin (DIG)-labeled RNA probes at a hybridization temperature of 65 °C. Stained tissues were mounted in 80% glycerol-PBS and photographed using an Olympus SZX16 stereo microscope equipped with an Olympus DP71 digital camera. The PCR primers used to generate riboprobes are listed in SI Appendix, Table S1.
Anti-Dll was previously generated in S.B. Carroll’s lab and was used at a 1:100 dilution (40). Anti-En (4D9) came from Developmental Studies Hybridoma Bank (DSHB) and was used at a 1:50 dilution (41). Anti-LacZ (40-1a) came from DSHB and was used at a 1:2,000 dilution. Anti-Nub was used at a 1:100 dilution (42). Anti-Ptc (Apa1) came from DSHB and was used at a 1:50 dilution (43). Anti-pMad was used at a 1:100 dilution (44). Alexa 488- and 647-conjugated secondary antibodies were from Molecular Probes. Samples were mounted in Vectashield (Vector Labs) and examined using an Olympus Fluoview FV1000 confocal microscope.
Time Course Experiments.
We induced the silencing or overexpression of developmental genes at different time points during the wing development in D. melanogaster. UAS lines were crossed to the nabNP3537-Gal4;tub-GAL80ts line. Embryos developed until third-instar larvae at 18 °C, conditions for which GAL80TS inhibits GAL4 and the UAS line is not expressed. Wandering third-instar larvae were collected (t = 0) and sequentially moved to 30 °C at different time points. For a given genetic combination, reciprocal crosses were used as biological replicates. Some vials underwent the thermal shift as above while some vials remained at 18 °C to serve as control. Collected larvae developed to adulthood, and pharate wings were mounted in 80% glycerol and then photographed using an Olympus SZX16 stereo microscope equipped with an Olympus DP71 digital camera
Results and Discussion
The Samoaia Clade: A Model to Study Evolution of Pigmentation.
Samoaia is a small genus of seven described species endemic to the Samoan Islands in the central South Pacific (45, 46). This genus is undoubtedly embedded within the family Drosophilidae, although its exact phylogenetic position remains debated (47). Moreover, the relationships remain unresolved within the genus Samoaia.
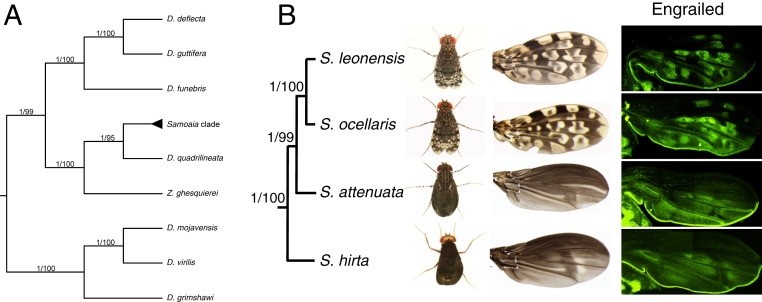
We employed a phylogenomic approach relying on a set of phylogenetic markers we developed in this study. We assembled a data set of 12 nuclear genes from 12 species of drosophilids, including four Samoaia species. Phylogenetic analyses support the Samoaia clade as sister to D. quadrilineata (Fig. 1A), confirming the topology previously found by Yassin and colleagues (48). Our phylogeny also provides insights into the internal structure of the Samoaia clade and the evolution of wing coloration. The black-wing species S. hirta and S. attenuata display a more primitive wing pattern, whereas the spotted-wing species S. ocellaris and S. leonensis display a more derived pattern (Fig. 1B). This finding suggests that spotted wings derive from the addition of white spots to uniformly black wings. Furthermore, ancestral reconstruction infers that wings were likely translucent in the last common ancestor of Drosophilidae (49). We hypothesize that an initial evolutionary change consisted in producing black pigment uniformly through the wing of the last common ancestor of the Samoaia species. Thus, the Samoaia clade represents a unique case study for the step-wise evolution of wing pigmentation. Our study primary aims at understanding how the spotted wing pattern is generated in the species S. leonensis.
Fig. 1.
Cooption of engrailed underlies wing pigmentation pattern in the genus Samoaia. (A) Phylogenetic reconstruction shows that the genus Samoaia is monophyletic and belongs to the Drosophilidae. (B) The black wing species represent the earliest-diverging lineages within the genus Samoaia. (C) The expression domain of the protein En prefigures the localization of the white spots in adult wings in S. leonensis and S. ocellaris. For each wing, two overlapping high-magnification clichés were acquired and manually stitched to cover the whole wing.
Cooption of Engrailed for Making a Complex Wing Pigmentation Pattern.
The S. leonensis wing pigmentation consists of a complex white and black spot pattern (Fig. 1B). White color results from the absence of melanin and the presence of a translucent wing membrane. We first focused on the generation of the black spots of the wing. In insects, black color very often results from the cuticular accumulation of melanin. The protein Yellow is required for the biosynthesis of melanin on fly wings (50). Similarly, we found that the spatial expression pattern of yellow prefigures the adult melanin wing pattern (SI Appendix, Fig. S1A). Then, we tested for putative candidate genes that could be involved in making the white spots. In pupal wings, the S. leonensis engrailed (en) transcript and protein are expressed where the adult white spots will appear (Fig. 1B and SI Appendix, Fig. S1B). The white spots are located both on the anterior and posterior sides of the wing, meaning that the expression of En is not restricted to the posterior compartment in the S. leonensis pupal wing. This finding is remarkable since en has been so far described as specifying posterior identity of embryonic segments (51) and wing discs (52) in Drosophila early development.
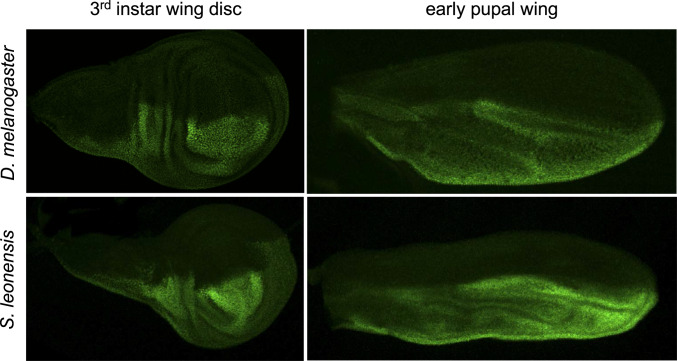
To test whether en has a totally different developmental function in S. leonensis, we investigated the dynamics of en expression (both at the transcript and protein level) during wing development in this species. In wing discs, the expression of En is homogeneous and restricted to the posterior compartment (Fig. 2), similarly to what is known in D. melanogaster (52). In early pupal wings (stage P5ii), the expression fades away in some regions, which leads to a nonuniform expression of the transcripts en (SI Appendix, Fig. S1 B and C) and the protein En (Fig. 2) in the posterior compartment. Expression in spots shows up in the posterior region first (stage P6), and then in the anterior part of the developing pupal wing (stage P7; SI Appendix, Fig. S1 D–G). Taken together, our data suggest that the early function of En as a posterior identity gene is conserved between D. melanogaster and S. leonensis. As for the novel function of En as a putative repressor of Yellow (53), it takes place during late pupal stages (stages P6/7). In other words, the cooption of the identity gene en to make pigmentation is possible during late pupal wing development without affecting the overall morphology of the wing.
Fig. 2.
Early expression pattern of the protein En. Immunostainings of En reveal conserved expression pattern between D. melanogaster and S. leonensis in both the third-instar wing disc and early pupal wing. At those stages, En is restricted to the posterior compartment. The anterior expression of En only shows up in the late pupal wing in S. leonensis.
We hypothesize that there is a critical time point beyond which en is no longer required in maintaining the posterior identity of a developing wing. We functionally tested this hypothesis in D. melanogaster because of the genetic tools available in this model.
Late Redeployment of Engrailed Does Not Affect Wing Morphology in D. melanogaster.
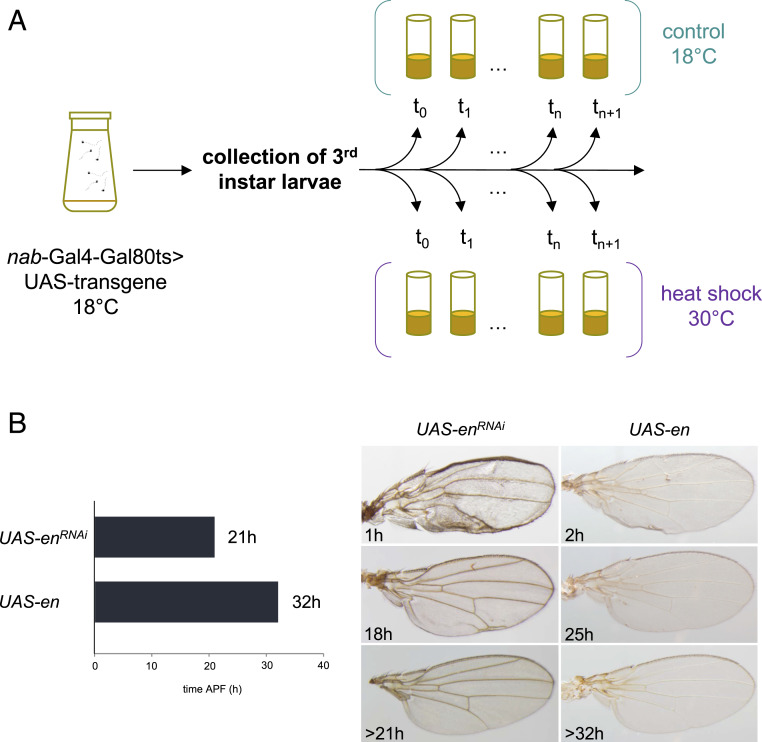
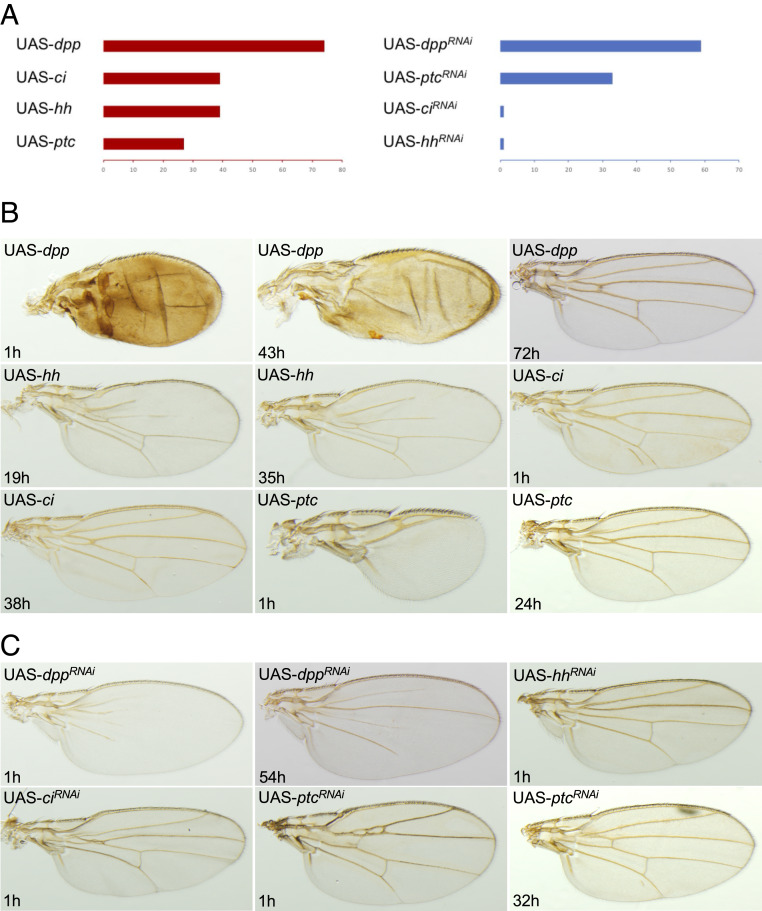
First, we used RNAi to deplete En function in the wing disc and the pupal wing strictly. The inducible expression system GAL4/GAL80 allowed us to control the time of the RNAi hairpin expression (Fig. 3A). Silencing en 1 h after puparium formation (APF) led to a drastic effect on the overall morphology of the adult wing (Fig. 3B). The wing tended to be symmetrical on either side of the first row of posterior cells, and the posterior cross-vein was absent. This phenotype is very similar to the one obtained when using the wing driver MS1096-Gal4 (54), validating the use of the nabNP3537-Gal4 driver in our study. When en knockdown was triggered between 18 and 21 h APF, this led to milder effects in the posterior margin (Fig. 3B). This time-course experiment identified 21 h APF as a critical time point that corresponds to the end of posterior identity function of En in D. melanogaster. Following the same approach, we overexpressed en throughout the whole larval wing disc or pupal wing. Overexpressing en until 32 h APF led to defaults in the adult wings, such as modified cross-veins and weak pigmentation (Fig. 3B). After 32 h APF, the overexpression of en did not affect wing morphology. Our data show that there is a critical time point during D. melanogaster wing development after which tinkering with en expression, by both loss or gain of function, does not affect wing morphology. We hypothesize that the developmental and evolutionary cooption of en for a new function in wing pigmentation is more likely to occur after this critical time because it does not impair its old function. In D. melanogaster, the gene en does not act independently but rather as a component of the Hedgehog circuit. We therefore investigated the role of the other genes of the Hedgehog circuit in S. leonensis.
Fig. 3.
Time course of inducible misexpression of en in D. melanogaster. (A) Detailed methodology. nab-Gal4-Gal80ts> UAS-transgene flies were kept at 18 °C to develop until the third-instar larva stage. Wandering larvae were then collected and placed back to 18 °C (control) or transferred to 30 °C to switch the expression of the transgene on. (B) Depletion of en before 21 h APF and overexpression of en before 32 h APF led to wing defects (Left). The phenotypes range from severe A–P polarity defects to weak vein malformation (Right).
Engrailed Is Partly Recruited Independently of the Hedgehog Circuit.
Hedgehog (Hh) is a major secreted morphogen involved in development (55). In Drosophila wing imaginal discs, hh is expressed exclusively in cells of the posterior compartment. The definition of this cellular boundary is initiated by the asymmetric expression of en in posterior cells (32), which induces, cell-autonomously, hh expression (reviewed in ref. 56). Hh produced in the posterior compartment diffuses into the anterior compartment (57, 58). On the contrary, En represses the expression of Cubitus interruptus (Ci) (59), an essential downstream component of Hh signaling (60, 61). The first steps in the reception and transduction of the HH signal are mediated by its receptor Patched (62) and the seven-transmembrane-domain protein Smoothened (63, 64). Hh controls the expression of the secreted signaling molecule Decapentaplegic (Dpp) in a thin stripe of anterior cells along the anterior–posterior boundary (65). Dpp acts as a symmetrical long-range morphogen to organize the growth and patterning of surrounding tissue (66).
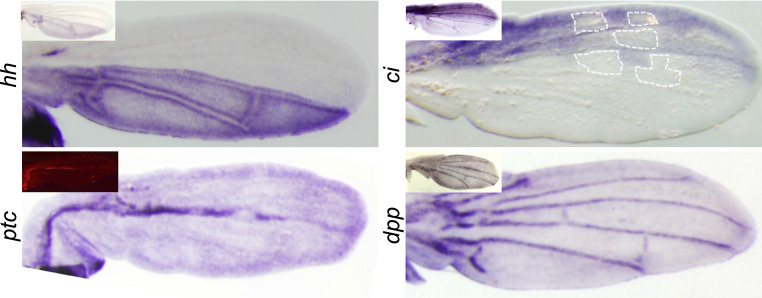
We investigated the expression of the main components of the Hedgehog circuit in the S. leonensis pupal wing. The expression of hh is restricted to the posterior compartment (Fig. 4), and does not follow the anterior en expression in spots. Moreover, in the posterior compartment, the down-regulation of en expression in discrete sites does not induce a similar effect on hh expression (Fig. 4). The gene ci is strictly expressed in the anterior compartment (Fig. 4). Its expression is not uniform and correlates with black regions where en is not expressed (Fig. 4). This observation suggests that En might still repress Ci in the anterior part of the wing, showing therefore that the Hedgehog circuit might be partly active. The gene ptc is strongly expressed along the anterior–posterior (AP) compartment border, and shows also a weak expression through the whole pupal wing (Fig. 4). The expression of ptc in S. leonensis pupal wing is reminiscent of ptc expression in D. melanogaster at similar developmental stages (Fig. 4, Inset). Thus, ptc is not involved in making the novel S. leonensis wing color pattern through the cooption of en. Finally, the gene dpp is expressed in the pupal veins like in D. melanogaster (Fig. 4). The expression of dpp is totally independent from en expression in the pupal wing, suggesting that dpp is not reused along with en to generate the novel color pattern in S. leonensis.
Fig. 4.
In situ hybridization of key genes of the Hedgehog circuit in late pupal wings. Large pictures show expression patterns in S. leonensis, and Insets depict pattern for homologs in D. melanogaster. The gene hh shows a similar expression pattern in both species, whereas en is expressed in spots at this stage in S. leonensis. This result suggests that hh is no longer activated by En in S. leonensis late pupal wing. On the contrary, ci seems to be still repressed by En in the anterior compartment. The localization of some of the future anterior white spots is indicated by the white dotted line.
Taken together, these results suggest that en is recruited independently of the Hedgehog circuit during S. leonensis pupal wing development to make the wing white spots. Contrary to the Hedgehog circuit in the wing disc, with its components interacting with each other, the same circuit might be more flexible during S. leonensis pupal development. We tested this hypothesis by comparing the function of the Hedgehog circuit components between wing disc and pupal development in D. melanogaster. We performed time-course silencing/overexpression of the Hedgehog circuit components and investigated the underlying molecular changes.
The Hedgehog Circuit Is Flexible over Time in D. melanogaster Wing.
Before 59 h APF, depletion of dpp function generates incomplete vein development in the distal part of the wing (t = 1 and 54 h APF; Fig. 5), whereas proper wings are obtained after this critical time point. Early overexpression of dpp causes an overall change of the wing morphology and the differentiation of most wing tissue as vein (t = 1 h and 43 h APF; Fig. 5). Later overexpression leads to minor modifications of the veins (t = 72 h; Fig. 5), whereas no effects are detected from 74 h APF.
Fig. 5.
Time course experiments: genes of the Hedgehog circuit in D. melanogaster. (A) We identified critical time points from which loss or gain of function no longer has an effect on wing development for the genes ci, dpp, hh, and ptc. Remarkably, ci and hh show a similar window of activity. The phenotypes range from severe A–P polarity defects to weak vein malformation for gain-of-function (B) and loss-of-function (C) contexts.
In the case of hh depletion, we only obtained a phenotype showing an absence of the anterior cross-vein when the RNAi was induced 1 h APF (t = 1 h APF; Fig. 5). Up to 39 h APF, overexpression of hh causes incomplete vein formation in the anterior wing compartment (t = 19 h and 35 h APF; Fig. 5). Interestingly, the gene ci behaves similarly to hh. Prior to 39 h APF, overexpression of ci leads to minor defects in vein formation (t = 1 h and 38 h APF; Fig. 5), whereas ci depletion only shows vein defects when induced very early (t = 1 h APF; Fig. 5). These findings indicate that hh and ci lose their developmental function early during pupal wing development, whereas these two genes are key developmental players in the larval wing disc (reviewed in refs. 67 and 68).
Depletion of ptc transcripts causes severe vein defects (t = 1 h APF; Fig. 5) or mild ones (t = 32 h APF; Fig. 5) until a loss of phenotype from 33 h APF. Early overexpression of ptc results in an abnormal wing shape, reduction in size, and absence of veins (t = 1 h APF; Fig. 5). Later overexpression leads to minor defaults in the anterior cross-vein (t = 24 h APF; Fig. 5) and no visible defects from 27 h APF.
As for the gene en, we identified a critical time point during pupal development for key components of the hedgehog signaling, where their function is no longer needed to regulate vein and wing morphology. Whereas ci and hh lose their developmental function early during pupal wing development, ptc and dpp maintain a role in vein patterning until a later stage. Except for ci and hh, whose functions seem to remain synchronized in time, the different players of the Hh circuit have their own function and dynamics during pupal development.
In order to confirm the flexibility of the Hedgehog circuit over time, we also investigated the interaction of the different partners at the molecular level. First, we assessed the expression of En, Hh, Ptc, Ci, and Dpp proteins in third-instar wing discs. As previously described, the depletion of En protein affects the expression of the other partners in the wing disc (SI Appendix, Fig. S3, Top Right). The Ci and Ptc proteins are no longer repressed by En, resulting in their expression domains expanding to the posterior compartment (32). The Dpp protein becomes expressed at the border between en+ and en− cells (69, 70). Our study shows the repression of Ci by En is not active anymore in the pupal wing. Indeed, Ci expression is restrained in the anterior compartment of the late pupal wing in absence of En protein (SI Appendix, Fig. S3, Top Right). Interestingly, we found that Ci is no longer repressed by En in the S. leonensis pupal wing (Fig. 4).
We also studied the expression of these different proteins in a context of overexpression of En. As expected, the ectopic expression of En protein affects the expression of the other players in the wing disc (SI Appendix, Fig. S3, Bottom Right). Hh is now activated and expressed in both the anterior and posterior compartments (70); Dpp is expressed in newly en+ cells in the anterior compartment (70), and Ci is partly repressed in the anterior compartment (71). However, our study brings insights into the interaction between those proteins in the developing pupal wing. Whereas the protein En is still overexpressed throughout the whole wing, Hh remains expressed strictly in the posterior compartment (SI Appendix, Fig. S3, Bottom Right). Similarly, Ci remains only expressed in the anterior compartment (SI Appendix, Fig. S3, Bottom Right). Dpp is mainly expressed at the A–P boundary, with discrete expression sites along the anterior margin. Again, Ci and Hh do not respond to En in the late pupal wing, as they do in the wing disc.
Taken together, these results show that the Hedgehog circuit is flexible over time and emphasize the possibility that en is coopted partly independently from its wing disc network in S. leonensis to its novel expression pattern prefiguring the wing white spots. Examples of partial GRN recruitment are scarce, but such molecular events have been proposed to underlie the development of abdominal appendages in sepsid flies (72, 73), the formation of eyespots in butterflies (74, 75), and the origin of gin-trap in beetle pupae (76).
So far, and based on their expression pattern, we only identified y, en, and ci as putative genes involved in wing pigmentation in S. leonensis. Are there additional genes involved in making the wing spot pattern in S. leonensis?
Several Developmental Genes Are Redeployed in the S. leonensis Wing.
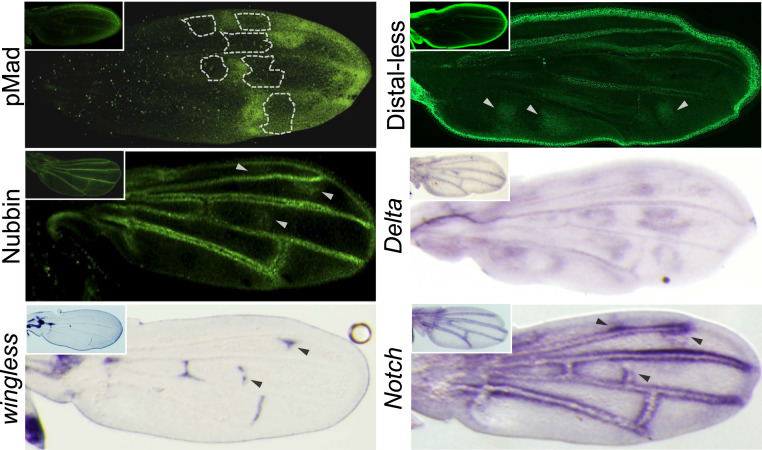
We tested for additional candidate genes that might be reused to make the wing color pattern in S. leonensis. In Drosophila, the protein Mothers against dpp (Mad) is required for both transduction of Dpp signals (77) and Wg signaling (78) during the formation of the wing anterior–posterior and proximo–distal (PD) axes, respectively. In S. leonensis, the phosphorylated form pMad is expressed in spots that prefigure some of the distal black spots on the adult wing (Fig. 6), whereas pMad is restricted to the wing margin in D. melanogaster (Fig. 6, Inset). This result suggests that pMad might also be involved in the color wing pattern in Samoaia.
Fig. 6.
Expression of genes putatively involved in the wing pigmentation in S. leonensis. Large pictures show expression patterns in S. leonensis, and Insets depict pattern for homologs in D. melanogaster. The localization of some of the future anterior white spots is indicated by the white dotted line.
We have also identified relevant expression changes for the homeodomain protein Distal-less (Dll). In Drosophila, Dll plays a role in patterning the wing through the differentiation of the wing margin (79). The protein Dll is similarly expressed in the wing margin and along longitudinal veins in D. melanogaster (Fig. 6) and S. leonensis (Fig. 6, Inset). However, the S. leonensis Dll has extra expression domains in spots that correspond to the black patches on the adult wing (Fig. 6, white arrowheads). Interestingly, two previous studies have demonstrated that Dll is recruited to make the wing black spot in the D. biarmipes adult male (22) and dark pigmentation in Bicyclus anynana butterflies (80, 81). These observations raise the question of the evolutionary origin of the link between the melanin pathway protein Yellow and Dll. Has Dll been independently recruited several times during the course of evolution? We searched for developmental genes whose expression pattern prefigures wing black ornamentation outside the subgenus Sophophora. We uncovered a strict correlation between melanin and Dll expression patterns in the Hawaiian drosophilids, as well as in Chymomyza amoena (SI Appendix, Fig. S4), suggesting that Dll has been recruited independently several times to make black pigmentation during evolution.
The nubbin (nub) gene encodes a POU-domain protein required for proximal–distal patterning in the Drosophila wing (82). Here it is also interesting to note that indirect evidence suggests that nub might regulate the expression of en (83). Both S. leonensis and D. melanogaster Nub proteins are expressed in the wing margin, as well as along the veins of the pupal wing (Fig. 6). This vein expression is compatible with the putative role of Nub in wing vein formation in D. melanogaster (84, 85). The S. leonensis Nub protein is additionally expressed at the distal tip of the longitudinal vein L2 and in a few spots in the posterior compartment (Fig. 6, white arrowheads). Interestingly, the Nub expression domains specific to S. leonensis coincide with specific expression domains of the receptor Notch (N), and wingless to some extent, in the same species (Fig. 6, black arrowheads). It has been previously shown that Nub represses N-dependent target genes in the D. melanogaster wing disc (86). Our data suggest that the interaction between Nub and Notch activity is still functional in the S. leonensis pupal wing.
However, the ligand Delta (Dl) is the component of the Notch signaling pathway that shows a striking correlation between its expression and the location of white spots on the S. leonensis wing (Fig. 6). This expression totally differs from the Dl expression in D. melanogaster that is mainly restricted to the veins (Fig. 6, Inset), suggesting that Dl has been coopted along with en to make the white wing spots in S. leonensis. What do we know about a possible interaction between en and Dl in development? Very little is known except that Drosophila hindgut patterning requires the repression of the gene Dl by En (87, 88). Our study clearly unravels a case where Dl and En are coexpressed in the same populations of cells, suggesting that En does not repress the expression of Dl in the Samoaia pupal wing. This same coexpression is also seen in the center of eyespot patterns in multiple species of butterfly (89). Further investigation would be required to elucidate the nature of the direct/indirect interaction between En and the Notch signaling pathway in the present context.
By performing similar time course experiments, we identified specific critical time points for these additional investigated genes in D. melanogaster (SI Appendix, Fig. S5). Remarkably, we showed that the depletion of wg and Dll transcripts rapidly stops having a phenotypic effect during pupal wing development (SI Appendix, Fig. S5, Top). The early functional “relaxation” of wg and Dll during pupal development might have facilitated their cooption in the making of wing pigmentation in D. guttifera (21), and in D. biarmipes (22), the Hawaiian flies (SI Appendix, Fig. S4), the genus Chymomyza (SI Appendix, Fig. S4), respectively. It would be interesting to test whether this characteristic of early “relaxation” is a general theme in other cases of morphological parallel evolution (90–93).
Conclusion
Key developmental genes are organized in networks or circuits. Several studies have shown that the heterotopic redeployment of such GRNs underlies the emergence of evolutionary novelties. Remarkably, the expression of a coopted gene does not impair the overall development of the novel location (tissue). We propose that 1) temporal flexibility is a property of GRN that is fundamental to allow cooption during the emergence of novel traits and 2) this flexibility results from the fact that every single gene of the GRN possesses its own functional time window. Moreover, differences in the time of functional relaxation of toolkit genes might explain why some genes are more easily coopted than others (SI Appendix, Fig. S6). In other words, relaxation time as an intrinsic property of a given GRN could explain why the same genes have been independently coopted during the course of evolution.
Supplementary Material
Acknowledgments
We thank Jane Selegue for technical assistance; the Sean Carroll laboratory for discussions and financial support; Antónia Monteiro for comments on the manuscript; Anastasios Pavlopoulos (University of Cambridge) for the nab-Gal4NP3537 line; Thomas Kornberg (University of California, San Francisco) for the UAS-ci, UAS-en, and UAS-hh lines; and Richard Mann (Columbia University) for the UAS-Dll line.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All accession sequence numbers are indicated in SI Appendix, Table S2. The nucleotide sequence alignments and tree files are downloadable from the Dryad Digital Repository at http://dx.doi.org/10.5061/dryad.prr4xgxhs.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002092117/-/DCSupplemental.
References
- 1.Pigliucci M., What, if anything, is an evolutionary novelty? Philos. Sci. 75, 887–898 (2008). [Google Scholar]
- 2.Wagner G. P., Lynch V. J., Evolutionary novelties. Curr. Biol. 20, R48–R52 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Peterson T., Müller G. B.. What is evolutionary novelty? Process versus character based definitions. J. Exp. Zoolog. B Mol. Dev. Evol. 320, 345–350 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Schluter D., The Ecology of Adaptive Radiation. Oxford Series in Ecology and Evolution (Oxford Univ Press, Oxford, United Kingdom, 2000). [Google Scholar]
- 5.Losos J. B., Lizards in an Evolutionary Tree: Ecology and Adaptive Radiation of Anoles (Univ California Press, Berkeley, CA, 2009), chap. 3. [Google Scholar]
- 6.Prum R. O., Evolution of the morphological innovations of feathers. J. Exp. Zoolog. B Mol. Dev. Evol. 304, 570–579 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Chanderbali A. S., Berger B. A., Howarth D. G., Soltis P. S., Soltis D. E., Evolving ideas on the origin and evolution of flowers: New perspectives in the genomic era. Genetics 202, 1255–1265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cebra-Thomas J., et al. , How the turtle forms its shell: A paracrine hypothesis of carapace formation. J. Exp. Zoolog. B Mol. Dev. Evol. 304, 558–569 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Emlen D. J., Marangelo J., Ball B., Cunningham C. W., Diversity in the weapons of sexual selection: Horn evolution in the beetle genus onthophagus (Coleoptera: Scarabaeidae). Evolution 59, 1060–1084 (2005). [PubMed] [Google Scholar]
- 10.Santos M. E., Le Bouquin A., Crumiere A. J. J., Khila A.. Taxon-restricted genes at the origin of a novel trait allowing access to a new environment. Science 358, 386–390 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Khalturin K., Hemmrich G., Fraune S., Augustin R., Bosch T. C. G., More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 25, 404–413 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Johnson B. R., Tsutsui N. D., Taxonomically restricted genes are associated with the evolution of sociality in the honey bee. BMC Genomics 12, 164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arendsee Z. W., Li L., Wurtele E. S., Coming of age: Orphan genes in plants. Trends Plant Sci. 19, 698–708 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Jiggins C. D., Wallbank R. W. R., Hanly J. J., Waiting in the wings: What can we learn about gene co-option from the diversification of butterfly wing patterns? Philos. Trans. R Soc. B Biol. Sci. 372, 20150485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinson E. O., Mrinalini Y. Kelkar D., Chang C. H., Werren J. H., The evolution of venom by co-option of single-copy genes. Curr. Biol. 27, 2007–2013.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang R., O’Donnell A. J., Barboline J. J., Barkman T. J., Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc. Natl. Acad. Sci. U.S.A. 113, 10613–10618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman J. L., Briginshaw L. N., Florent S. N., Evolution and co-option of developmental regulatory networks in early land plants. Curr. Top. Dev. Biol. 131, 35–53 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Das Gupta M., Tsiantis M., Gene networks and the evolution of plant morphology. Curr. Opin. Plant Biol. 45, 82–87 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Rebeiz M., Tsiantis M., Enhancer evolution and the origins of morphological novelty. Curr. Opin. Genet. Dev. 45, 115–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halfon M. S., Perspectives on gene regulatory network evolution. Trends Genet. 33, 436–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werner T., Koshikawa S., Williams T. M., Carroll S. B., Generation of a novel wing colour pattern by the Wingless morphogen. Nature 464, 1143–1148 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Arnoult L., et al. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339, 1423–1426 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Glassford W. J., et al. , Co-option of an ancestral hox-regulated network underlies a recently evolved morphological novelty. Dev. Cell 34, 520–531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vreede B. M. I., Lynch J. A., Roth S., Sucena E., Co-option of a coordinate system defined by the EGFr and Dpp pathways in the evolution of a morphological novelty. Evodevo 4, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Goto T., Macdonald P., Maniatis T., Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57, 413–422 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Gómez-Skarmeta J. L., et al. , Cis-regulation of achaete and scute: Shared enhancer-like elements drive their coexpression in proneural clusters of the imaginal discs. Genes Dev. 9, 1869–1882 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Adachi Y., et al. , Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech. Dev. 120, 1113–1126 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Koshikawa S., et al. , Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 112, 7524–7529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domínguez M., Brunner M., Hafen E., Basler K., Sending and receiving the hedgehog signal: Control by the Drosophila gli protein Cubitus interruptus. Science 272, 1621–1625 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Gorfinkiel N., Morata G., Guerrero I., The homeobox gene Distal-less induces ventral appendage development in Drosophila. Genes Dev. 11, 2259–2271 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabata T., Schwartz C., Gustavson E., Ali Z., Kornberg T. B., Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development 121, 3359–3369 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Capdevila J., Guerrero I., Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 13, 4459–4468 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J. J., von Kessler D. P., Parks S., Beachy P. A., Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71, 33–50 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Terriente Félix J., Magariños M., Díaz-Benjumea F. J., Nab controls the activity of the zinc-finger transcription factors Squeeze and Rotund in Drosophila development. Development 134, 1845–1852 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S., et al. , New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Ronquist F., et al. , MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturtevant M. A., Roark M., Bier E., The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev. 7, 961–973 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Panganiban G., Sebring A., Nagy L., Carroll S., The development of crustacean limbs and the evolution of arthropods. Science 270, 1363–1366 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Patel N. H., et al. , Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58, 955–968 . (1989). [DOI] [PubMed] [Google Scholar]
- 42.Ng M., Diaz-Benjumea F. J., Vincent J. P., Wu J., Cohen S. M., Specification of the wing by localized expression of wingless protein. Nature 381, 316–318 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Capdevila J., Pariente F., Sampedro J., Alonso J. L., Guerrero I., Subcellular localization of the segment polarity protein patched suggests an interaction with the wingless reception complex in Drosophila embryos. Development 120, 987–998 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Nahmad M., Stathopoulos A., Dynamic interpretation of hedgehog signaling in the Drosophila wing disc. PLoS Biol. 7, e1000202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malloch J. R., “Part VI. Diptera” in Insects of Samoa and other Samoan terrestrial arthropoda (British Museum Natural History, London, 1934), pp. 267–328. [Google Scholar]
- 46.Wheeler M. R., Kambysellis M. P., Notes on the Drosophilidae (Diptera) of Samoa. University of Texas Publication 6615 (Univ Texas Press, Austin, TX, 1966). [Google Scholar]
- 47.O’Grady P. M., DeSalle R., Phylogeny of the genus Drosophila. Genetics 209, 1–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yassin A., et al. , Polyphyly of the Zaprionus genus group (Diptera: Drosophilidae). Mol. Phylogenet. Evol. 55, 335–339 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Wittkopp P. J., Carroll S. B., Kopp A., Evolution in black and white: Genetic control of pigment patterns in Drosophila. Trends Genet. 19, 495–504 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Wittkopp P. J., True J. R., Carroll S. B., Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development 129, 1849–1858 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Kornberg T., Sidén I., O’Farrell P., Simon M., The engrailed locus of Drosophila: In situ localization of transcripts reveals compartment-specific expression. Cell 40, 45–53 (1985). [DOI] [PubMed] [Google Scholar]
- 52.Brower D. L., Engrailed gene expression in Drosophila imaginal discs. EMBO J. 5, 2649–2656 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gompel N., Prud’homme B., Wittkopp P. J., Kassner V. A., Carroll S. B., Chance caught on the wing: Cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Layalle S., et al. , Engrailed homeoprotein acts as a signaling molecule in the developing fly. Development 138, 2315–2323 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Lee R. T. H., Zhao Z., Ingham P. W., Hedgehog signalling. Development 143, 367–372 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Basler K., Waiting periods, instructive signals and positional information. EMBO J. 19, 1168–1175 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basler K., Struhl G., Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368, 208–214 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Tabata T., Kornberg T. B., Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76, 89–102 (1994). [DOI] [PubMed] [Google Scholar]
- 59.Eaton S., Kornberg T. B., Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 4, 1068–1077 (1990). [DOI] [PubMed] [Google Scholar]
- 60.Aza-Blanc P., Ramírez-Weber F. A., Laget M. P., Schwartz C., Kornberg T. B., Proteolysis that is inhibited by hedgehog targets cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043–1053 (1997). [DOI] [PubMed] [Google Scholar]
- 61.Ohlmeyer J. T., Kalderon D., Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396, 749–753 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Nakano Y., et al. , A protein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature 341, 508–513 (1989). [DOI] [PubMed] [Google Scholar]
- 63.van den Heuvel M., Ingham P. W., Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature 382, 547–551 (1996). [DOI] [PubMed] [Google Scholar]
- 64.Alcedo J., Noll M., Hedgehog and its patched-smoothened receptor complex: A novel signalling mechanism at the cell surface. Biol. Chem. 378, 583–590 (1997). [DOI] [PubMed] [Google Scholar]
- 65.Méthot N., Basler K., Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96, 819–831 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Affolter M., Basler K., The decapentaplegic morphogen gradient: From pattern formation to growth regulation. Nat. Rev. Genet. 8, 663–674 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Blair S. S., Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 23, 293–319 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Hartl T. A., Scott M. P., Wing tips: The wing disc as a platform for studying Hedgehog signaling. Methods 68, 199–206 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Sanicola M., Sekelsky J., Elson S., Gelbart W. M., Drawing a stripe in Drosophila imaginal disks: Negative regulation of decapentaplegic and patched expression by engrailed. Genetics 139, 745–756 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zecca M., Basler K., Struhl G., Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development 121, 2265–2278 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Schwartz C., Locke J., Nishida C., Kornberg T. B., Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development 121, 1625–1635 (1995). [DOI] [PubMed] [Google Scholar]
- 72.Bowsher J. H., Nijhout H. F., Partial co-option of the appendage patterning pathway in the development of abdominal appendages in the sepsid fly Themira biloba. Dev. Genes Evol. 219, 577–587 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajaratnam G., Supeinthiran A., Meier R., Su K. F. Y., CRISPR/Cas9 deletions in a conserved exon of Distal-less generates gains and losses in a recently acquired morphological novelty in flies. iScience 10, 222–233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keys D. N., et al. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283, 532–534 (1999). [DOI] [PubMed] [Google Scholar]
- 75.Monteiro A., Gene regulatory networks reused to build novel traits: Co-option of an eye-related gene regulatory network in eye-like organs and red wing patches on insect wings is suggested by optix expression. BioEssays 34, 181–186 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Hu Y., et al. , A morphological novelty evolved by co-option of a reduced gene regulatory network and gene recruitment in a beetle. Proc. R Soc. B Biol. Sci., 10.1098/rspb.2018.1373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sekelsky J. J., Newfeld S. J., Raftery L. A., Chartoff E. H., Gelbart W. M., Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics 139, 1347–1358 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eivers E., et al. , Mad is required for wingless signaling in wing development and segment patterning in Drosophila. PLoS One 4, e6543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campbell G., Tomlinson A., The roles of the homeobox genes aristaless and Distal-less in patterning the legs and wings of Drosophila. Development 125, 4483–4493 (1998). [DOI] [PubMed] [Google Scholar]
- 80.Monteiro A., et al. , Distal-less regulates eyespot patterns and melanization in bicyclus butterflies. J. Exp. Zool. B Mol. Dev. Evol. 320, 321–331 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Connahs H., et al. , Activation of butterfly eyespots by Distal-less is consistent with a reaction-diffusion process. Development, 10.1242/dev.169367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ng M., Diaz-Benjumea F. J., Cohen S. M., Nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development 121, 589–599 (1995). [DOI] [PubMed] [Google Scholar]
- 83.Gibert J. M., et al. , Heterospecific transgenesis in Drosophila suggests that engrailed.a is regulated by POU proteins in the crustacean Sacculina carcini. Dev. Genes Evol. 212, 19–29 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Johannes B., Preiss A., Wing vein formation in Drosophila melanogaster: Hairless is involved in the cross-talk between Notch and EGF signaling pathways. Mech. Dev. 115, 3–14 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Kölzer S., Fuss B., Hoch M., Klein T., Defective proventriculus is required for pattern formation along the proximodistal axis, cell proliferation and formation of veins in the Drosophila wing. Development 130, 4135–4147 (2003). [DOI] [PubMed] [Google Scholar]
- 86.Neumann C. J., Cohen S. M., Boundary formation in Drosophila wing: Notch activity attenuated by the POU protein Nubbin. Science 281, 409–413 (1998). [DOI] [PubMed] [Google Scholar]
- 87.Fuss B., Hoch M., Notch signaling controls cell fate specification along the dorsoventral axis of the Drosophila gut. Curr. Biol. 12, 171–179 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Iwaki D. D., Lengyel J. A., A Delta-Notch signaling border regulated by Engrailed/Invected repression specifies boundary cells in the Drosophila hindgut. Mech. Dev. 114, 71–84 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Oliver J. C., Tong X. L., Gall L. F., Piel W. H., Monteiro A., A single origin for nymphalid butterfly eyespots followed by widespread loss of associated gene expression. PLoS Genet. 8, e1002893 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Colosimo P. F., et al. , Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 (2005). [DOI] [PubMed] [Google Scholar]
- 91.McGregor A. P., et al. , Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448, 587–590 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Liu Y., et al. , Convergent sequence evolution between echolocating bats and dolphins. Curr. Biol. 20, R53–R54 (2010). [DOI] [PubMed] [Google Scholar]
- 93.Kratochwil C. F., et al. , Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362, 457–460 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.