Abstract
Transcription–replication (T–R) conflicts are profound threats to genome integrity. However, whilst much is known about the existence of T–R conflicts, our understanding of the genetic and temporal nature of how cells respond to them is poorly established. Here, we address this by characterizing the early cellular response to transient T–R conflicts (TRe). This response specifically requires the DNA recombination repair proteins BLM and BRCA2 as well as a non-canonical monoubiquitylation-independent function of FANCD2. A hallmark of the TRe response is the rapid co-localization of these three DNA repair factors at sites of T–R collisions. We find that the TRe response relies on basal activity of the ATR kinase, yet it does not lead to hyperactivation of this key checkpoint protein. Furthermore, specific abrogation of the TRe response leads to DNA damage in mitosis, and promotes chromosome instability and cell death. Collectively our findings identify a new role for these well-established tumor suppressor proteins at an early stage of the cellular response to conflicts between DNA transcription and replication.
INTRODUCTION
Faithful replication of the genome is of utmost importance to sustain life and prevent genetic diseases like cancer. During replication, DNA polymerases meet numerous challenges including DNA damage and collision with RNA polymerases. Failure to successfully overcome these inevitable challenges during replication can manifest as genomic instability—a hallmark of cancer (1,2). To deal with disruption of DNA replication, cells may initiate a so-called replication stress response (3), which is characterized by activation of the ATR checkpoint kinase and subsequent cell cycle arrest. Whilst cell cycle arrest may be a desired response to various challenges, each type of replication impediment also requires a distinct action to be overcome. Yet, our current knowledge of pathway choice at stalled replication forks is limited. This is in part because fork stalling may lead to fork collapse, which is accompanied by a DNA damage response that masks the initial response to stalled forks (4). In particular the early cellular response to transcription–replication (T–R) conflicts has been difficult to study due to a lack of methods to rapidly and specifically induce endogenous T–R collisions.
Normally, transcription and replication are coordinated to minimize T–R conflicts (5). However, cancer cells are characterized by deregulated replication (4), rapid cell division (1) and widespread transcriptional activation collectively laying the grounds for frequent T–R collision (6). Moreover, T–R conflicts are inevitable at the largest genes in the genome because it takes more than one cell cycle to complete transcription of these genes (7). Under conditions of replication stress, transcription of large genes results in breaks at these specific regions on metaphase chromosomes known as common chromosomal fragile sites (CFSs) (8–10). It is likely that T–R conflicts that persist into mitosis contribute substantially to mutagenesis in cancer since regions of the genome that face common T–R conflicts including CFSs are hotspots for large deletions in a broad range of cancer genomes (7,11–15). However, it is unclear how T–R conflicts can go unnoticed into mitosis without activating cell cycle checkpoints.
Mechanistically, T–R conflicts probably occur via the formation of so-called transcriptional RNA–DNA hybrids, where nascent RNA hybridizes back to the complementary DNA template forming an RNA–DNA hybrid that displaces the non-coding strand of the DNA duplex. This structure is often referred to as an R loop. Specifically, RNA–DNA hybrids can cause replication stress, DNA breaks, chromosomal rearrangements, and chromatin alterations (16–18). Several cellular pathways keep levels of RNA–DNA hybrids in check. Firstly, RNase H1 and helicases actively degrade or remove RNA–DNA hybrids, respectively (19). Secondly, RNA maturation and splicing factors as well as topoisomerase I prevent accumulation of RNA–DNA hybrids (19). Moreover, disruption of DNA repair factors, BRCA1, BRCA2, FANCA, FANCM, BLM and RECQL5 leads to accumulation of RNA–DNA hybrids but it is unclear how these factors prevent nuclear buildup of RNA–DNA hybrids (17,20–23).
Investigation of specific CFSs showed accumulation of RNA–DNA hybrids in the absence of FANCD2 suggesting that FANCD2 may have a role at T–R conflicts (24–27). Moreover, purified chicken FANCD2 has high affinity for RNA–DNA hybrids (28), whereas human FANCD2 together with its binding partner FANCI binds the single-stranded DNA that forms as part of the R loop (29). The FANCD2 gene is one of 23 genes that when mutated give rise to the recessive genetic disorder Fanconi Anemia (FA). At the cellular level FA is characterized by hypersensitivity to chemotherapeutic DNA crosslinking agents and aldehydes (30). The role of FANCD2 in DNA interstrand crosslink repair is well characterized. It involves FANCD2 monoubiquitylation by a large E3 ubiquitin ligase complex where FANCL is the catalytic subunit (31–33). Many FA genes directly take part in the crosslink repair pathway, but others seem to act in parallel or downstream. This includes the tumor suppressor protein BRCA2 (also known as FANCD1) (34,35), which plays an important role during homologous recombination (36,37) and also works as a fork stabilizer (38). FANCD2 works together with the helicases BLM and FANCJ as well as BRCA2 to promote fork restart after hydroxyurea- or aphidicolin-mediated fork stalling (39,40). BLM is a tumor suppressor, which is mutated in a rare recessive genetic disorder called Bloom's syndrome, which is characterized by dramatic hyper-susceptibility to a wide range of cancers (41).
mRNA in eukaryotes is synthesized by RNA polymerase II (RNAP II). RNAP II pauses 20–100 base pairs downstream of the transcription start site (42–44). Here, P-TEFb (45), which is composed of cyclin T and Cdk9, phosphorylates Serine 2 (Ser2) of the carboxy-terminal domain repeat (CTD) of RNAP II to trigger the initiation-to-elongation transition (43,46). Pausing of RNAP II causes R-loop formation (47), which in turn may explain why promoter/5′UTR regions are genomic hotspots for R loops (48,49).
The early cellular response to T–R conflicts has been difficult to study due to a lack of methods to rapidly and specifically induce endogenous T–R collisions. Here we take advantage of Cdk9 inhibitors to investigate the cellular response to T–R conflicts. Using this approach, we find that T–R conflicts induce early colocalization of the recombination factors BRCA2, FANCD2 and BLM into nuclear foci and each of these factors are required for survival after transient induction of T–R conflicts, because they prevent T–R conflicts from escalating into genomic instability in mitotic cells. We furthermore provide insight into how T–R conflicts escape cell cycle arrest, as we find that they do not lead to activation of the ATR checkpoint kinase.
MATERIALS AND METHODS
Cell culture and transient transfection
DT40 cells were cultured in RPMI 1640 medium GlutaMAX (ThermoFisher Scientific) supplemented with 2% chicken serum (Sigma-Aldrich), 8% fetal bovine serum (ThermoFisher Scientific), 50 μM β-mercaptoethanol, 50 U/ml penicillin, and 50 μg/ml streptomycin at 39°C with 5% CO2.
U2OS cells were maintained in McCoy's 5A modified medium (ThermoFisher Scientific) supplemented with 10% fetal bovine serum, 50 U/ml penicillin and 50 μg/ml streptomycin (15070063, ThermoFisher Scientific) at 37°C with 5% CO2. For transient transfection, the Amaxa system (Nucleofector, Lonza) was used as previously described (50). PD20 (FA-D2) (51) and PD20 complemented cells described in (52) were maintained in DMEM+GlutaMAX medium (ThermoFisher Scientific) supplemented with 15% fetal bovine serum, 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 μg/ml Blasticidin S HCl (ThermoFisher Scientific) at 37°C with 5% CO2.
Live cell and immunofluorescence microscopy
Live cell microscopy was performed as previously described (53) using a wide-field microscope (Deltavision Elite, GE Healthcare) equipped with a 100× oil objective lens with a numerical aperture of 1.35 (GE Healthcare). Briefly, images were taken with 8 optical sections separated by 0.5 μm. Exposure times and excitation light intensities were optimized for each fluorophore. Fifteen minutes before imaging, 40 μl of DT40 cell culture were transferred to a μ-slide microscopy chamber (1 μ-slide Angiogenesis ibiTreat, Ibidi). As indicated in the figures, the following drugs were added for the indicated time before imaging: 1 μM flavopiridol (S1230, Selleckchem), 100 nM AZ20 (S7050, Selleckchem), ATMi 1 μM (S1570, Selleckchem), 50 μM DRB (D1916, Sigma-Aldrich), 100 nM Cisplatin (Sigma-Aldrich), 1 μM PladB (445493-23-2, Santa Cruz), 200 nM CHIR-124 (S2683, Selleckchem), 1μM MC-180295 (PC-35808, Probechem) or 100 nM CPT (7689-03-4, Sigma-Aldrich). Immunofluorescence microscopy of DT40 cells was performed at room temperature using the wide-field microscope (Deltavision Elite) as described above. For immunofluorescence, DT40 cells were allowed to set on poly-lysine covered coverslip for 10 min before fixation with 4% paraformaldehyde. Cells were blocked in PBS-T (PBS with 0.1% Tween 20) with 3% Bovine Serum Albumin (A0281, Sigma-Aldrich) for 1 h before overnight incubation with primary γH2AX antibody (1:500, 05-636, Millipore) at 4°C. Unbound primary antibodies were removed by washing three times for 5 min in PBS followed by incubation with secondary antibody (1:500, Alexa555 anti-mouse IgG A21422, Life Technologies) for 50 min. Coverslips were then washed three times for 5 min in PBS before mounting with mounting medium containing DAPI (4% n-propyl gallate, 80% glycerol, 1 μg/ml DAPI). For quantification of the S9.6 mean fluorescence intensity, cells were extracted with 100% ice-cold methanol for 10 min, followed by 100% ice-cold acetone for 1 min, and then RNases III (1:200, AM2290, ThermoFisher Scientific) and T1 (1:200, EN0541, ThermoFisher Scientific) was added followed by 5% bovine serum albumin (BSA) and 0.2% milk in PBS for 1 hour blocking. Samples were incubated with the S9.6 antibody (1:500, ENH001, Kerafast) in blocking buffer overnight at 4°C, and washed by PBS three times for 5 min. Secondary antibody staining and mounting was done as described above.
Images were processed and quantitative measurements of fluorescence intensities and foci were performed using Volocity software (PerkinElmer). Images were pseudocolored according to the approximate emission wavelength of the fluorophores. Fluorescent proteins used in DT40 were TFP (pmTurquoise2-N1 (54)), eYFP (enhanced YFP, Takara Bio Inc.), Venus (55), and mCherry (56).
For measuring the percentage of H3 phospho-Ser10-positive cells in DT40 cell, cells are harvested and washed with cold PBS, fixed with 70% EtOH at −20°C for overnight, followed by two PBS washes, and then incubation of the pellet in pS10H3 antibody solution (1:40, 06-570, Millipore) at room temperature for 2 h. Secondary antibody staining and mounting was done followed by pipetting 40 μl of stained cell culture into Xcyto 2-chamber slide. Images were processed and quantitative measurement of fluorescence intensities was performed using Xcyto 10 (ChemoMetec).
For immunofluorescence microscopy of U2OS cells, asynchronous cultures were grown on coverslips. Cells were blocked in PBS-T (PBS with 0.1% Tween20) with 3% BSA (A0281, Sigma-Aldrich) for 1 h before overnight incubation with primary FANCD2 antibody (NB100-182SS, Novus Biologicals) at 4°C. Unbound primary antibodies were removed by washing three times for 5 min in PBS followed by incubation with Alexa 568 goat anti-rabbit IgG secondary antibody (1:500, A11011, Life Technologies) for 50 min. Coverslips were then washed three times for 5 min in PBS before mounting with mounting medium containing DAPI (4% n-propyl gallate, 80% glycerol, 1 μg/ml DAPI). In situ proximity ligation assay (PLA) was performed following instructions from Duolink PLA technology (Sigma-Aldrich) with the minor change that cells were treated with pre-extraction buffer (10 mM Tris–HCl pH 6.8, 2.5 mM MgCl2, 0.5% NP-40 and 1 mM PMSF) prior to blocking. 5-Ethynyl uridine (EU) incorporation was performed with reagents from Click-iT EdU kit (C10637, ThermoFisher Scientific) in accordance with the manufacturer's instructions. 120 mM of EU (E10345, ThermoFisher Scientific) was used for nascent transcripts detection. Images were acquired using the wide-field microscope (Deltavision Elite) as described above.
DT40 cell lines
DT40 cell lines used in this study are listed in supplemental table S1. For stable transfections, constructs were electroporated into DT40 cells (Gene Pulser Xcell, BioRad). Transfectants harbouring the PURO, BSR, NEO and HYG resistance genes were selected in the presence of 0.5 μg/ml puromycin (A11138-03, Life Technologies), 20 μg/ml blasticidin S (A11139-03, Life Technologies), 2 mg/ml G418 (CP11.3, Roth) or 2.0 mg/ml Hygromycin B (10687010, Thermo Fisher Science), respectively. Dilution cloning was used for selecting positive single clones. Clones were tested by fluorescence microscopy, and positive clones were cultured in DT40 medium with relevant drug selection. All resistance cassettes were flanked by LoxP sites and floxed as described previously (57). In brief, cell lines were transiently transfected with cDNA encoding the Cre recombinase and subsequently dilution cloned to obtain single colonies. Loss of selection markers was tested by treating the resulting cell lines with puromycin, G418, or blasticidin. In this study, Gallus gallus genes were endogenously tagged at their 3′ terminal except for FANCD2, which was tagged at the 5′ terminus.
Plasmids
Plasmids and primers used in this study are listed in Supplemental Table S2 and S3, respectively. The plasmid for overexpression of RNase H1 was generated by PCR amplifying the cDNA with BglII or SalI adapted primers VO317 and VO318 for directional cloning into pmCherry-C1 (Clontech, Takara). Human RNase H1 cDNA in pENTR221 plasmid (Ultimate ORF clones, IOH4870, ThermoFisher Scientific) was used as a template. Correct PCR amplification was verified by sequencing.
The plasmid pmCherry-BLM was generated by lifting the G. gallus BLM cDNA as a BamHI fragment from EGFP-gallusBLM (58) into the BamHI site in pmCherry-C1 (Clontech, Takara). Quikchange II Site-Directed Mutagenesis (#200523, Agilent) was used to generate Walker A motif mutant of G. gallus BLM cDNA K663T in accordance with the Instruction manual (primers for BLM K663T: VO336 and VO337). All constructs were verified by sequencing.
The targeting vector for endogenous C-terminal tagging of BRCA2 (Gene ID: 418915) was constructed by first PCR amplifying the 5′ homology arm using genomic DT40 DNA as a template with a KpnI adapted forward primer (LJ01) and a XhoI adapted reverse primer (LJ02). The 3′ homology arm was then amplified by PCR using a BcuI adapted forward primer (LJ03) and a NotI adapted reverse primer (LJ04). eYFP or TFP was amplified using peYFP-C1 (Clontech, Takara) or pmTurquoise2-N1 (54) as template, respectively, with a SalI adapted forward primer (LJ05) and a SmaI adapted reverse primer (LJ06). Fragments were subcloned into pBlueScript SK+ (Fermentas) by directional cloning. The puromycin or blasticidin (BSR) resistance cassettes, flanked by lox sequences were inserted into the BamHI restriction site.
Colony survival assay
DT40 cells were treated with drugs before seeding in methyl cellulose medium (59). The plates were then incubated in a humid box in a CO2 incubator at 37°C until the colonies were clearly visible and countable. Survival is plotted as a percentage of the colonies forming in the untreated control. Experiments were repeated three times.
PD20 and complemented cells were plated in 6-well plates and incubated in a CO2 incubator at 37°C for 24 h. The media was replaced by DMSO media or 1 μM flavo media. After treatment for the indicated time, the drug-containing media was removed and fresh warm media was added. Cells were incubated for 8–10 days, washed in PBS, stained with 0.5% crystal violet in 25% methanol for 2 min. Plates were rinsed with ddH2O. The plates were scanned and resulting images analysed by calculating the colony area according the published protocol (60). Survival is plotted as a percentage of the colonies area in the DMSO treated control. Experiments were repeated three times.
Metaphase spreads
Cell cultures were treated with 1 μM flavo for 1 h after which cells were washed and resuspended in prewarmed medium. Colcemid (Life Technologies) at a concentration of 0.1 μg/ml was added 150 min before harvest. Next, cells were swelled in 8 ml hypotonic buffer (20% FBS [v/v], 15 mM KCl) for 15 min. Next, 10 ml fixation buffer (25% acetone, 75% methanol) was added drop by drop. Cells were harvested by centrifugation at 1000 rpm for 5 min, followed by resuspension in 10 ml fixation buffer. Cells were stored at −20°C for 2 days. Finally, the cells were splatted onto the slides and mounted with cover slips using 5 μl of mounting medium containing DAPI (4% n-propyl gallate, 80% glycerol, 1 μg/ml DAPI). Metaphase chromosomes were visualized on a widefield microscope (AxioImager Z1; Carl Zeiss) equipped with a 100× objective lens (Plan Apochromat, NA 1.4; Carl Zeiss), a cooled CCD camera (Orca-ER; Hamamatsu Photonics), differential interference contrast (DIC), and an illumination source (HXP120C; Carl Zeiss);
FACS sorting
DT40 cells were stained by Vybrant Ruby Stain (5 μM, V10309, ThermoFisher Scientific) at 37°C for 15 min, protected from light according to manufacturer's protocol. 20 000 cells from G1, S and G2/M phase were sorted using FACSJazz (BD Biosciences) system with 561 nm excitation and >670 nm emission. Sorted cells were then incubated in pre-warmed RPMI medium for further treatment.
Dot blot
Genomic DNA was extracted using Genomic DNA extraction kit (51306, Qiagen). Next, genomic DNA was precipitated with isopropanol, washed with 70% ethanol, air-dried, and resuspended in Tris-EDTA (TE) buffer. Half of the DNA was treated with RNases III and T1 and half was treated with RNases III, T1 and H1 (M0297L, NEB), at 37°C for 6 h. Equal amounts of DNA were blotted onto a Hybond H+ membrane (RPN203B, GE Healthcare) using a dot blot apparatus (Bio-Rad). The membrane was dried overnight at room temperature and DNA was cross-linked to the membrane using UV. The membrane was blocked with 5% skimmed milk in PBS-T and incubated with S9.6 antibody (1:500, ENH001, Kerafast) and dsDNA antibody (1:5000, ab27156, Abcam), followed by incubation with secondary antibody (1:2000, P0161, Dako). The membrane was incubated with ECL reagents (RPN 2109, GE) for 3 min. The ECL signal was recorded using an ImageQuant LAS 4000 (GE Healthcare). ImageJ (NIH) was used for picture processing, and quantification of S9.6 mean intensity.
Western blot
For Western blot analysis, cells were lysed in RIPA buffer (1% NP40, 0.5% Na deoxycholate, 0.1% SDS in 1× PBS) in the presence of protease inhibitor cocktail (Roche) by syringing eight times through a 25 G needle. Lysates were cleared by centrifugation at 10 000 g for 10 min. Samples were separated by SDS-PAGE on 8% polyacrylamide Tris-glycine gels for FANCD2 or 4–20% Tris-glycine gels (mini-Protean TGX, #456-1093, Bio-Rad) for all other western blots. For analysis of monoubiquitylation of Venus-FANCD2, mouse anti-GFP monoclonal (1:500, 11814460001, Roche) and anti-mouse IgG conjugated to HRP (1:2000, P0161, Dako) were used as primary and secondary antibodies, respectively. For analysis of CHK1-Ser345P, rabbit anti-phospho Ser345 -CHK1 (1:1000, #2348, Cell Signaling), and anti-rabbit IgG conjugated to HRP (1:2000, P0217, Dako) were used as primary and secondary antibodies, respectively. For analysis of CHK1, mouse anti-CHK1 (1:2000, sc-8408, Santa Cruz), and anti-mouse IgG conjugated to HRP (1:2000, P0161, Dako) were used as primary and secondary antibodies, respectively. For analysis of MCM2-Ser108P, rabbit anti-phospho MCM2 S108 antibody (1:1000, A300-094A, Bethyl), and anti-rabbit IgG conjugated to HRP (1:2000, P0217, Dako) were used as primary and secondary antibodies, respectively. For analysis of MCM2, mouse anti-MCM2 (1:1000, sc-373702, Santa Cruz), and anti-mouse IgG conjugated to HRP (1:2000, P0161, Dako) were used as primary and secondary antibodies, respectively. For analysis of ATM-pS1981, rabbit anti-phospho-ATM (1:1000, Ser1981, ab81292, Abcam), and anti-rabbit IgG conjugated to HRP (1:2000, P0217, Dako) were used as primary and secondary antibodies, respectively. For analysis of ATM, mouse anti-ATM (1:1000, sc-135663, Santa Cruz), and anti-mouse IgG conjugated to HRP (1:2000, P0161, Dako) were used as primary and secondary antibodies, respectively. For analysis of CHK2-Thr68P, rabbit anti-phospho Thr68 -CHK2 (1:1000, cst2197t, BioNordika), and anti-rabbit IgG conjugated to HRP (1:2000, P0217, Dako) were used as primary and secondary antibodies, respectively. For analysis of RNAP ll CTD Ser2 phosphorylation and RNAP ll CTD Ser5 phosphorylation, RNAP ll CTD Ser2 antibody (1:1000, MABE953, Merckmillipore), RNA pol ll CTD Ser5 antibody (1:1000, 04-1572, Merckmillipore), and anti-Rat IgG conjugated to HRP (1:2000, P0450, Dako) were used as primary and secondary antibodies, respectively.
Chromatin immunoprecipitation (ChIP)-qPCR
DT40 cultures (5 × 107 cells) were treated as indicated (1 μM flavo or DMSO for 40 min) and fixed for 10 min with 1× Crosslink Buffer (0.75% paraformaldehyde, 10 mM NaCl, 50 μM EGTA and 5 mM HEPES pH 8.0). Then fixation was stopped by addition of glycine to a final concentration of 125 mM and incubate for 5 min. Cells were washed with 1 X PBS twice and resuspended in 4 ml of SDS buffer (100 mM NaCl, 50 mM Tris-Cl pH 8.0, 5 mM EDTA, 0.1% SDS,10 mM glycerol-phosphate, protease inhibitor cocktail (11697498001, Merck) and 2 mM PMSF). Lysates were sonicated for 2 min using a Sonicator (Misonix) and centrifuged at 20 000 rpm for 10 min at 4°C. At this point, 1% of supernatant was saved for ‘Input’, the rest of sample was diluted with 1 ml Triton dilution buffer (100 mM Tris–Cl pH 8.6, 100 mM NaCl, 5 mM EDTA pH 8.0, 5% Triton X-100, 1 mM Na3VO4 and protease inhibitor cocktail). Diluted lysates were incubated with antibodies (RNAP II CTD Ser2P or RNAP II CDT Ser5P) rotating overnight at 4°C. Dynabeads Protein G (10004D, ThermoFisher Scientific) were added to antibody-chromatin mixture, and incubated on a rotating wheel for 2 h at 4°C. Beads were washed twice with Wash Buffer I (1% Triton X-100, 0.1% SDS, 150 mM NaCl, 2 mM EDTA pH 8.0 and 20 mM Tris–Cl pH8.0), followed by one wash with Wash Buffer II (1% Triton X-100, 0.1% SDS, 500 mM NaCl, 2 mM EDTA pH 8.0 and 20 mM Tris–Cl pH 8.0). Beads were resuspended in 150 μl freshly prepared Elution buffer (1% SDS and 0.1 M NaHCO3 pH 8.0) at 65°C for 30 min. Input samples and eluates were incubated with 2 M NaCl and 150 μg/ml RNase A at 65°C overnight, followed by addition of 0.1 mg/ml Protease K at 60°C for 1 h. DNA was purified using the GeneJET PCR Purification kit (K0702, ThermoFisher Scientific). Sequences of the primers used for qPCR are shown in supplementary Table S3, GAPDH amplicon 1 primers (VO355 and VO356), amplicon 2 primers (VO357 and VO358), amplicon 3 primers (VO359 and VO360); SMC2 amplicon 1 primers (VO361 and VO362), amplicon 2 primers (VO363 and VO364), amplicon 3 primers (VO365 and VO366). Reactions were run using Maxima SYBR Green/ROX qPCR Master Mix (K0222, ThermoFisher Scientific). All qPCR was performed by using a CFX96 Real-Time PCR Detection system (Bio-Rad). The dilution factor was adjusted and the percentage of the input was calculated.
DNA–RNA immunoprecipitation (DRIP)
DRIP-qPCR experiments were performed based on the published protocol (61). Briefly, genomic DNA was extracted using phenol–chloroform extraction. Then, DNA was fragmented using a restriction enzyme cocktail (EcoRI, HindIII, XbaI, SspI and BSrGI (NEB)). Half of the sample was digested with RNase H1 (M0297L, NEB) and the other half was treated with RNase III (AM2290, ThermoFisher Scientific) for 6 h at 37°C. Fragmented DNA was purified with phenol–chloroform extraction and re-suspended in TE buffer (10 mM Tris pH 8.0, 1 mM EDTA). RNA–DNA hybrids were immunoprecipitated from total nucleic acids using 5 μg of the S9.6 antibody in 1× binding buffer (10 mM NaPO4 pH 7.0, 140 mM NaCl, 0.05% Triton X-100) overnight rotating incubation at 4°C. 40 μl pre-washed Dynabeads conjugated to Protein G (10003D, ThermoFisher Scientific) were added to the hybrids and antibody mixture and incubated for 2 h at 4°C on a rotator. Isolated complexes were washed three times with 1× binding buffer before elution with 120 μl of elution buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS). Proteinase K digestion was performed at 55°C for 45 min, followed by PCR purification kit (K0702, ThermoFisher Scientific) extraction, and eluted in 150 μl TE buffer. The relative occupancy of the immunoprecipitated DNA–RNA hybrid at the specified sites in the SAE1 gene was determined by qPCR with the primers described in Table S3, SAE1 amplicon 1 primers (XS69 and XS70), amplicon 2 primers (XS71 and XS72), amplicon 3 primers (XS79 and XS80), amplicon 4 primers (XS81 and XS82). The results were normalized against input DNA.
Statistical methods
For microscopy experiments, the significance of the differences between cell populations was determined by a two-tailed unpaired t-test. P-values were defined as significant if P < 0.05. Error bars representing 95% confidence intervals were calculated by Prism 7 (GraphPad Software). For colony survival assay, the significance of the differences between groups was determined by two-tailed unpaired t-test. P-values were considered significant if P < 0.05. Error bars represent standard deviations. Three independent experiments were carried out to generate each data set.
RESULTS
FANCD2 is important for survival after transient block of RNAP II promoter proximal release
The drugs flavopiridol (flavo), DRB and MC180295 (MC295) are Cdk9 inhibitors that prevent phosphorylation of Ser2 in RNAP II CTD, thereby blocking the transition from transcription initiation to transcription elongation (62,63). As a consequence, Cdk9 inhibitors trap RNAP II near the promoter (64), which can potentially block an advancing replication fork. We confirmed the efficiency of Cdk9 inhibition by flavo and MC295 in our model systems, DT40 and U2OS, by western blot of the Cdk9 substrate RNAP II CTD Ser2P (Supplementary Figure S1A,B). As expected, Cdk9 inhibition also resulted in overall transcription inhibition as measured by decreased EU incorporation (Supplementary Figure S1C). It was previously shown that the promoter proximal paused RNAP II is stabilized with flavo or DRB treatment (65). Phosphorylation of Serine 5 (Ser5P) of the CTD of RNAP II precedes phosphorylation of RNAP II CTD Ser2 (43). We confirmed by ChIP of RNAP ll CTD Ser5P followed by qPCR that a large proportion of paused RNAP II (at amplicon 1) was still retained at the promoter regions of GAPDH or SMC2 gene after 40 min flavo treatment, whereas RNAP II CTD Ser5P at the gene body disappeared as expected (Supplementary Figure S1D, right). Also, as expected, RNAP II CTD Ser2P disappeared at both promoter region and gene body (Supplementary Figure S1D, left). The mild decrease of RNAP II CTD Ser5P at the first amplicon upon flavo addition may be a result of suppression of new transcriptional initiation by the paused RNAP II (65,66). To characterize the early cellular effects of Cdk9 inhibition we investigated whether known markers of CFSs, DNA double-strand breaks (DSBs) and replication stress (FANCD2 (25,26) and TopBP1 (53,67,68) change localization pattern in response to flavo, DRB and MC295. Intriguingly, all drugs induced a small but significant increase in FANCD2 focus formation that peaks around 30 or 60 min, after which the number of foci gradually decrease to levels below that observed in the untreated cell line (Figure 1A). Importantly, the overall level of Venus-FANCD2 does not change within the two-hour flavo treatment (Supplementary Figure S1E). The level of TopBP1 foci is not affected by flavo or DRB (Supplementary Figure S2A,B), indicating that no immediate DSBs or extensive single-stranded DNA (ssDNA) are formed (69,70). 53BP1 is a DSB repair factor and a G1 marker of replication stress-induced DNA damage inherited from the previous S phase (71). Consistently, flavo-induced FANCD2 foci do not colocalize with 53BP1 (Supplementary Figure S2C, D). Flavo-mediated induction of FANCD2 foci was also observed by immunostaining in human U2OS cells (Figure 1B).
Figure 1.
Cdk9 inhibitors induce transient FANCD2 focus formation and FANCD2 is required for cellular survival in response to the inhibitors. (A) Left, quantification of FANCD2 foci at the indicated time points after addition of 1 μM flavo, 50 μM DRB or 1 μM MC295 using the DT40 cell line FANCD2+/Venus TopBP1+/+/mCherry PICH+/TFP. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). Right, representative images of Venus-FANCD2 in flavo-, DRB- and MC295-treated DT40 FANCD2+/Venus TopBP1+/+/mCherry PICH+/TFP cells at the indicated time points. Scale bar: 5 μm. (B) Left, quantification of FANCD2 foci detected by immunostaining of U2OS cells at the indicated time points after addition of 1 μM flavo or 100 nM cisplatin. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). Right, representative images of FANCD2-immunostaining of flavo-treated U2OS cells at the indicated time points. Scale bar: 5 μm. (C) Quantification of colony survival assay of DT40 WT or FANCD2−/− cells in response to the indicated drug treatments normalized to untreated. Results are mean of three independent experiments. Error bars are standard deviations. For all panels in this figure: ***P < 0.001, ****P < 0.0001. Ns, not significant (P > 0.05). Two-tailed t-test.
To address whether FANCD2 is important for cellular survival after transient treatment with Cdk9 inhibitors, we performed colony survival of DT40 FANCD2−/− cells after drug treatment for one hour. The interstrand crosslinker cisplatin was used as control. Whilst the DT40 parental cells (referred to as ‘wild-type’ (WT)) were not sensitive to the applied concentrations of the drugs, FANCD2−/− cells were indeed sensitive demonstrating that FANCD2 is required for survival after transient treatment with Cdk9 inhibitors (Figure 1C).
RNAP II stalling at promoter proximal sites leads to T–R conflicts
The important roles of FANCD2 at stalled replication forks (40,72,73) suggest that Cdk9-inhibitor-induced FANCD2 foci mark T–R conflicts. If that were the case, we would expect that flavo-mediated induction of FANCD2 foci occurs mainly in S phase. To test this idea, we FACS sorted cells from G1, S, and G2/M phases of the cell cycle before subjection to a one-hour flavo treatment (Supplementary Figure S2E). This revealed that flavo-mediated induction of FANCD2 foci indeed is restricted to S phase (Figure 2A). Moreover, we found that G1 cells were resistant to a one-hour treatment with flavo whereas S-phase cells and, surprisingly, also G2/M cells were sensitive to flavo (Figure 2B). This could indicate that induction of T–R conflicts at late stages of the replication process, which sometimes continues in G2 and even M (53,74,75), may be detrimental.
Figure 2.
Transient inhibition of Cdk9 leads to T–R conflicts. (A) Quantification of FANCD2 foci in DT40 population FACS-enriched for G1, S or G2/M phases as indicated. Cells were either untreated or treated with 1 μM flavo for 1 h. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). (B) Quantification of colony survival assay of DT40 WT or FANCD2−/− cell population FACS-enriched for G1, S or G2/M phases. Cells were either untreated or treated with 1 μM flavo for 1 h as indicated. Results are mean of three independent experiments. Error bars are standard deviations. Indications of significance (ns and *) refer to comparison with the untreated or WT. (C) Quantification of PLA foci between RNAP II CTD Ser5P and PCNA in U2OS cells treated with DMSO or flavo for the indicated time. Mean values and error bars representing 95% confidence intervals are indicated. Number of cells analysed is indicated (n). (D) Representative images of the PLA assay in U2OS cell with DMSO or flavo treatment for the indicated time. Scale bar: 6 μM. For all panels in this figure: *P < 0.05, ****P < 0.0001. Ns, not significant (P > 0.05). Two-tailed t-test.
To directly test whether Cdk9 inhibition triggers T–R conflicts we employed the proximity ligation assay (PLA) on fixed U2OS cells using anti-PCNA and anti-RNAP ll Ser5P antibodies. A striking increase of PLA foci at 30 min flavo treatment compared to DMSO treatment was evident, and this increase disappeared by 60 or 120 min flavo treatment (Figure 2C,D). Since RNAP II Ser5P is mainly found at the 5′ end of genes this strongly suggests an increase in T–R conflicts at promoter proximal pause sites shortly after Cdk9 inhibition. As expected, 60 min inhibition of Cdk9 by flavo leads to a severe reduction of RNAP II Ser2P and PCNA PLA signal (Supplementary Figure S2F) consistent with the flavo-induced reduction of RNAP II CTD Ser2P (Supplementary Figure S1A,B).
FANCD2 foci induced by Cdk9-inhibition depend on RNA–DNA hybrids
DRB has been shown to induce RNA–DNA hybrids at promoter proximal regions (64), which suggest that RNA–DNA hybrids underlie T–R conflicts induced by Cdk9 inhibition. Therefore, we asked whether flavo also induces RNA–DNA hybrids that may mediate its effect on FANCD2 localization. We used dot blotting and immunostaining with the S9.6 antibody that recognizes RNA–DNA hybrids (Figure 3A, B and Supplementary Figure S3A) (76). Both methods showed that DT40 WT and FANCD2−/− cells have increased levels of RNA–DNA hybrids when treated with flavo. Untreated FANCD2−/− cells displayed significantly higher levels than the untreated WT, consistent with previous work showing constitutive higher RNA–DNA hybrid levels in FANCD2-deficient cells (21). Moreover, we performed ChIP-qPCR at the SAE1 gene, which was previously reported to accumulate RNA–DNA hybrids at the transcription start site upon DRB treatment (64). This analysis showed that flavo induces RNA–DNA hybrids around the transcription start site of SAE1 (Figure 3C), suggesting that RNA–DNA hybrids are induced at promoter proximal regions by flavo similarly to DRB (64).
Figure 3.
RNA–DNA hybrids induced by transcription-stalling drugs, flavo and DRB, trigger FANCD2 focus formation. (A) Quantification of dot blot analysis. The level of RNA–DNA hybrids per input dsDNA is normalized against that of FANCD2−/− untreated. Results are mean of three independent experiments. Error bars represent mean with standard deviations. (B) Quantification of RNA–DNA hybrid immunostaining. Signal intensity in arbitrary units (a.u.) of RNA–DNA hybrids per cell in DT40 WT or FANCD2−/− cells before or after the indicated drug treatments. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). (C) DRIP-qPCR analysis of SAE1 gene in U2OS cells treated with DMSO or flavo. The amount of DRIP-qPCR product is plotted relative to qPCR for input (% input). Results are mean of three independent experiments. Error bars are standard deviations. Positions of the amplicons are shown as blue lines with numbers below. (D) FANCD2 foci in the DT40 cell line FANCD2Venus/Venus were quantified at the indicated time points after addition of 1 μM flavo in cells that were or were not transiently transfected with mCherry-RNase H1 expressing plasmid. Mean values and error bars representing 95% confidence intervals are indicated. Number of cells analysed is indicated (n). (E) Representative images of the DT40 cell line FANCD2Venus/Venus at the indicated time points after addition of 1 μM flavo in cells that were (bottom) or were not (top) transiently transfected with mCherry-RNase H1 expressing construct. Scale bar: 5 μm. For all panels in this figure: ***P < 0.001, ****P < 0.0001. Ns, not significant (P > 0.05). Two-tailed t-test.
To determine whether flavo-induced FANCD2 focus formation was dependent on RNA–DNA hybrids, we transiently overexpressed mCherry-tagged RNase H1 and quantified FANCD2 foci in transfected and untransfected cells (with or without red fluorescence, respectively). Unlike untransfected cells, a flavo-induced increase in FANCD2 foci was not observed in cells expressing RNase H1, suggesting that RNA–DNA hybrids lead to flavo-induced FANCD2 focus formation (Figure 3D, E).
The early cellular response to T–R conflicts does not involve FANCD2 monoubiquitylation or ATR activation
FANCD2 focus formation in response to DNA damage is normally accompanied by its monoubiquitylation, but we found that Cdk9 inhibitors did not induce FANCD2 monoubiquitylation in contrast to control treatment with the topoisomerase I poison, camptothecin (CPT) (Figure 4A). To firmly establish whether FANCD2 monoubiquitylation is important for the role of FANCD2 at T–R conflicts, we determined the sensitivity of FANCD2-K563R cell line to flavo. We also included a cell line with knockout of the FANCD2 binding partner FANCI. The interstrand crosslinking agent mitomycin C was used as a control. Neither FANCD2-K563R nor FANCI−/− cells were sensitive to flavo treatment (Figure 4B), FANCL is the catalytic subunit of the E3 ubiquitin ligase complex responsible for monoubiquitylation of FANCD2. In agreement with the resistance of the FANCD2 monoubiquitylation mutant, FANCL−/− cells were not sensitive to these drugs but showed the expected sensitivity to cisplatin (Supplementary Figure S3B). Moreover, the cell line PD20, which is a FANCD2 deficient cell line derived from a FA patient (51) was more sensitive to 2 or 4 h of flavo treatment, than PD20 cells in which the wildtype hFANCD2 or the monoubiquitylation defective hFANCD2-K561R had been reintroduced (Figure 4C).
Figure 4.
Monoubiquitylation of FANCD2 is dispensable for the TRe response, whereas basal activity of ATR is required. (A) Western blot to detect monoubiquitylated Venus-FANCD2 (ub-FANCD2) and non-ubiquitylated Venus-FANCD2 (FANCD2) in extracts from DT40 FANCD2+/Venus subjected to the indicated drug treatments before harvest. (B) Quantification of colony survival assay of DT40 WT, FANCD2−/−, FANCD2-K563R or FANCI−/− cells in response to the indicated drug treatments. Results are mean of three independent experiments. Error bars are standard deviations. (C) Quantification of colony survival assay of PD20 cells complemented with empty vector, hFANCD2 or hFANCD2-K561R in response to 1 μM flavo for the indicated duration. The experiment was done in triplicate. Error bars are standard deviations. (D) Western blot to assay ATR activation by detection of CHK1 phosphorylation at Ser345 (CHK1-Ser345P) and total CHK1 in extracts from FANCD2+/Venus cells subjected to the indicated drug treatments. Tubulin was used as a loading control. (E) Left, quantification of FANCD2 foci in FANCD2+/Venus with or without 30 min treatment with 1 μM flavo and/or 100 nM AZ20 as indicated. Mean values and error bars representing 95% confidence intervals are indicated. Number of cells analysed is indicated (n). Right, representative images of Venus-FANCD2 in flavo- or/and AZ20-treated DT40 FANCD2+/Venus cells at the indicated time points. Scale bar: 5 μm. (F) Western blot to assay ATM activation by detection of ATM autophosphorylation at Ser1981, CHK2 phosphorylation at Thr68 and total ATM in extracts from U2OS cells incubated with flavo for the indicated durations or subjected to irradiation (IR) with 10 Gray (Gy). Tubulin was used as a loading control. (G) Quantification of FANCD2 foci at the indicated time points after addition of 1 μM flavo and 200 nM CHIR-124 using the DT40 cell line FANCD2+/Venus TopBP1+/+/mCherry RPA+/CFP. Mean values and error bars representing 95% confidence interval are indicated. For all panels in this figure: *P < 0.05, **P < 0.01. Ns, not significant (P > 0.05). Two-tailed t-test.
Next, we tested whether Cdk9 inhibitors activate ATR by examining CHK1 Ser345P. We found that none of the Cdk9 inhibitors activate ATR at any of the indicated time points (Figure 4D). Rather, flavo and MC295 reduced phosphorylation levels CHK1 Ser345P, whereas DRB did not lead to changes in CHK1 phosphorylation (Figure 4D). Thus, Cdk9 inhibition may lead to a decrease in CHK1 Ser345P by an unknown mechanism. Nevertheless, the lack of ATR activation by Cdk9 inhibitors is in stark contrast to the increase in CHK1 Ser345P observed after 15 min treatment with CPT (Figure 4D). Moreover, we analyzed the effect of flavo and MC295 on MCM2 Serine 108 phosphorylation. This residue is also a substrate for ATR and it has been suggested that MCM2 Serine 108 phosphorylation recruits FANCD2 to stalled replication forks (73). However, no effect on MCM2 phosphorylation was detected after flavo or MC295 treatment (Supplementary Figure S3C). The results indicate that early cellular response to T–R conflicts does not globally activate ATR, which is consistent with the finding that TOPBP1, which is an ATR activator, rarely colocalizes with flavo-induced FANCD2 foci (Supplementary Figure S2A, B). These results suggest that T–R conflicts trigger a distinct cellular response, which we name the T–R conflict early (TRe) response. To analyze whether ATR or CHK1 activity is dispensable for flavo-induced FANCD2 focus formation, we used the ATR inhibitor AZ20 and CHK1 inhibitor CHIR-124 (77) at concentrations that inhibit their activities (Supplementary Figure S3D, E). Surprisingly, AZ20, but not CHIR-124, inhibits flavo-induced FANCD2 focus formation, suggesting that even though ATR is not activated by flavo, basal ATR activity is required for the TRe response (Figure 4E,G). Moreover, a dose of flavo that induces FANCD2 foci, did not induce ATM autophosphorylation on Serine 1981 or phosphorylation of the ATM substrate Threonine 68 in CHK2 (Figure 4F). Treatment with ionizing radiation was used as a positive control for ATM autophosphorylation. Finally, ATR but not ATM inhibition also abolished flavo-induced FANCD2 focus formation in U2OS cells (Supplementary Figure S3F).
BLM and BRCA2 are involved in the early cellular response to T–R conflicts
The BLM and FANCJ helicases are physical and/or genetic interactors of FANCD2 (39,78,79). To address whether BLM or FANCJ work in the TRe response, we tested BLM−/− (80) and FANCJ−/− (81) cell lines for sensitivity to flavo and DRB. The results show that BLM is required for cellular survival after treatment with flavo or DRB as well as cisplatin (Figure 5A). In contrast, FANCJ−/− cells were not sensitive to transient treatment with the T–R conflict-inducing drugs (Figure 5A). To further confirm a role of BLM in the TRe response, we stably expressed mCherry-tagged BLM in the BLM−/− cell line and quantified focus formation in response to flavo treatment. BLM focus formation after addition of flavo is similar to that of FANCD2 with an initial increase in the number of foci followed by a drop at later time points (Supplementary Figure S3G). A BLM−/− cell line stably expressing mCherry-tagged BLM and endogenous Venus-tagged FANCD2 revealed that flavo-induced BLM and FANCD2 foci colocalize (Figure 5B, C) and that the total fluorescence intensity of mCherry-BLM was not affected by flavo (Supplementary Figure S1E). Next, we addressed the interdependency of BLM and FANCD2 for flavo-induced focus formation. The results shown in figure 5D reveal that BLM foci are not induced by flavo in FANCD2−/− cells and vice versa FANCD2 foci are not induced in BLM−/− cells (Supplementary Figure S3H, I). Thus, BLM and FANCD2 are interdependent for focus formation in response to flavo.
Figure 5.
BLM and FANCD2 work together in the cellular response to T–R conflicts. (A) Quantification of colony survival assay of DT40 WT, BLM−/− or FANCJ−/− cells in response to the indicated drug treatments. Results are mean of three independent experiments. Error bars are standard deviations. (B) Quantification of BLM and FANCD2 colocalizing foci in the DT40 cell line BLM−/− FANCD2+/Venus stably expressing mCherry-BLM at the indicated time points after treatment with flavo. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). (C) Representative images of Venus-FANCD2 and mCherry-BLM in flavo-treated DT40 BLM−/−FANCD2+/Venus cells stably expressing mCherry-BLM at the indicated time points. Scale bar: 5 μm. (D) Quantification of BLM foci in the DT40 cell line FANCD2−/− stably expressing mCherry-BLM (left) or FANCD2 foci in the DT40 cell line BLM−/− FANCD2+/Venus (right) at the indicated time points after addition of 1 μM flavo. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). For all panels in this figure: *P < 0.05, **P < 0.01. Ns, not significant (P > 0.05). Two-tailed t-test.
Previous studies have shown that mutation of the Walker A motif of the ATP-binding site of BLM compromises its helicase function (82,83). We investigated whether an equivalent point mutation in chicken BLM (K663T) could disrupt the TRe response. The number of BLM-K663T-mCherry foci were quantified in BLM−/− DT40 cells complemented with BLM-K663T-mCherry. We found that the level of BLM-K663T foci is not affected by flavo, while a reduction of FANCD2 foci was observed after flavo treatment (Supplementary Figure S4A). More importantly, the colocalization between BLM-K663T and FANCD2 was not changed by flavo (Supplementary Figure S4A). Furthermore, in contrast to cells complemented with wildtype BLM cDNA, BLM−/− cells expressing BLM-K663T were as sensitive as cells lacking BLM after transient exposure to either flavo or MC295 (Supplementary Figure S4B). Taken together these results suggest that the helicase function of BLM is required for the TRe response.
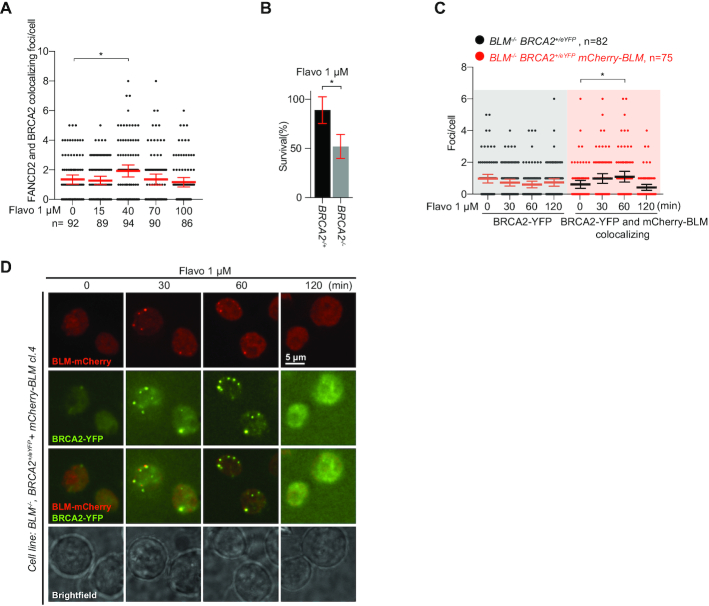
Similar to FANCD2 (21,22) and BLM (20), disruption of BRCA2 confers increased levels of RNA–DNA hybrids (17), suggesting that BRCA2 may also function in the TRe response. To test this, we first used a cell line with fluorescent tags on endogenous BRCA2 and FANCD2 and found that flavo induced the formation of colocalizing foci of FANCD2 and BRCA2 (Figure 6A and Supplementary Figure S4C). Moreover, BRCA2−/− cells were hypersensitive to flavo treatment compared to the heterozygous BRCA2+/− control cell line (Figure 6B). Finally, BRCA2 foci were not induced by flavo in a BLM−/− cell line but this phenotype was rescued by introducing mCherry-tagged BLM (Figure 6C and Supplementary Figure S4D), and it was evident that flavo-induced BRCA2 foci colocalize with BLM (Figure 6C,D). Total levels of BRCA2-YFP was not changed within 2 hours of flavo treatment (Supplementary Figure S1E). These results suggest that BRCA2 is part of the TRe response together with FANCD2 and BLM.
Figure 6.
BRCA2 acts in concert with FANCD2 and BLM in the TRe response. (A) Quantification of FANCD2 and BRCA2 colocalizing foci in the DT40 cell line BRCA2+/TFP FANCD2+/Venus at the indicated time points after addition of 1 μM flavo. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). (B) Quantification of colony survival assay of DT40 BRCA2+/− or BRCA2−/− cells in response to the indicated drug treatments. Results are mean of three independent experiments. Error bars are standard deviations. (C) Quantification of BRCA2 foci in the DT40 cell line BLM−/− BRCA2+/eYFP (left) or BRCA2 and BLM foci in the DT40 cell line BLM−/− BRCA2+/eYFP stably expressing mCherry-BLM (right) at the indicated time points after addition of 1 μM flavo. Mean values and error bars representing 95% confidence interval are indicated. Number of cells analysed is indicated (n). (D) Representative images of mCherry-BLM and BRCA2-YFP in flavo-treated DT40 BLM−/− BRCA2+/eYFP cells stably expressing mCherry-BLM at the indicated time points. Scale bar: 5 μm. For all panels in this figure: *P < 0.05. Two-tailed t-test.
Disruption of the TRe response manifests as DNA damage in mitosis
Our data suggest that BLM, BRCA2 and FANCD2 work together to promote survival after induction of T–R conflicts. Assuming that T–R conflicts result in genomic instability in the absence of a functional TRe response, we examined breaks and gaps on metaphase chromosomes in flavo-treated WT and FANCD2−/− DT40 cells. Indeed, we found that FANCD2−/− cells have an increase in chromosomal aberrations after exposure to flavo for 1 h (Figure 7A). Consistently, flavo-treated FANCD2−/− interphase cells also displayed a small but significant increase in γH2AX foci—marking DNA damage in chromatin (84) (Figure 7B), indicating that T–R conflicts can result in DNA damage, when FANCD2 is deficient.
Figure 7.
FANCD2 prevents DNA damage and genomic instability after T–R conflicts. (A) Left, representative images of metaphase chromosomes from untreated or flavo-treated DT40 WT or FANCD2−/− cells. Close up image shows break indicated by red arrow. Scale bar: 7.8 μm. Right, quantification of chromosomal aberrations on metaphase macrochromosomes from DT40 WT or FANCD2−/− cells untreated or treated with 1 μM flavo for 1 h. Error bars represent standard deviations. (B) Quantification of γH2AX foci in the DT40 WT or FANCD2−/− cell lines subjected to flavo-treatment for 1 h followed by 1 h release before imaging or cisplatin treatment for 1 h. Mean values and error bars representing 95% confidence intervals are indicated. Number of cells analysed is indicated (n). (C) Quantification of γH2AX foci in prometaphase/metaphase (pro-meta) or in anaphase/telophase DT40 WT, FANCD2−/− or FANCL−/− cells untreated or subjected flavo-treatment for one hour followed by one hour release before fixation. Mean values and error bars representing 95% confidence intervals are indicated. Number of cells analysed is indicated (n). (D) Representative images of immunostained γH2AX foci in prometaphase/metaphase or in anaphase/telophase of WT, FANCD2−/− or FANCL−/− cells untreated or subjected flavo-treatment for one hour followed by one hour release before fixation. DAPI/γH2AX merge as well as brightfield images are also shown. Scale bar: 5 μm. (E) Model depicting how RNAP II transcription stalling at a promotor proximal site induces an RNA–DNA hybrid, which in turn can cause a T–R conflict (co-directional or head-on) with approaching DNA polymerases (DNAP). The T–R conflict can be resolved by activating the TRe response that involves BLM, FANCD2 and BRCA2. BLM and FANCD2 are interdependent and BRCA2 depends on BLM (indicated by red arrows). It is uncertain whether BRCA2 and FANCD2 are interdependent and whether BLM rely on BRCA2 in the TRe (white arrows). If the TRe is impaired (indicated by the X), the T–R conflict will cause DNA damage and genomic instability. For all panels in this figure: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Ns, not significant (P > 0.05). Two-tailed t-test.
Because flavo treatment does not trigger CHK1 phosphorylation and therefore might not activate the G2/M checkpoint, we wondered whether flavo-induced T–R conflicts arising in very late S phase may manifest as DNA damage in M phase. Thus, we analyzed the level of γH2AX foci in different phases of mitosis. In prometa- and metaphase, FANCD2−/− cells displayed a higher level of γH2AX foci compared to WT DT40 and this is further significantly increased by flavo treatment (Figure 7C, D). In ana- and telophase, γH2AX foci were rare in both untreated and in flavo-treated WT cells. In contrast, untreated and flavo-treated FANCD2−/− cells in ana-/telophase presented γH2AX foci, with the latter showing a highly significant increase in γH2AX foci compared to untreated ana-/telophase FANCD2−/− (Figure 7C, D). This demonstrates that in FANCD2-deficient cells, flavo treatment before M phase induces DNA damage in mitotic cells. This may explain why the G2-enriched population of FANCD2−/−, which probably also contains some late S phase cells, was sensitive to flavo (Figure 2B). FANCL−/− cells also present with high levels of γH2AX foci in prometaphase and metaphase but this is not further increased by flavo. In ana-/telophase, levels of γH2AX foci in FANCL−/− cells decreased similar to that observed in FANCD2−/− cells and, surprisingly, flavo treatment significantly decreases γH2AX levels in FANCL deficient cells (Figure 7C, D). Taken together these data indicate that disruption of the TRe response manifests as DNA damage in mitosis.
Splicing inhibition triggers the TRe response
DRB and flavo both induce RNA–DNA hybrids by inhibiting Cdk9 (Figure 3A–C) (64). However, RNA–DNA hybrids also form upon splicing inhibition (85,86). Thus, to address whether RNA–DNA hybrids generally induce the TRe response, we used the drug Pladienolide B (PladB), which inhibits splicing (87). Similar to flavo, DRB and MC295, PladB treatment induced transient FANCD2 foci, which were not accompanied by FANCD2 ubiquitylation or ATR activation (Supplementary Figure S4E), suggesting that the TRe response is activated by RNA–DNA hybrids.
DISCUSSION
Here, we delineate the early cellular response to T–R conflicts, the TRe response, which manifests as rapid accumulation of BRCA2, FANCD2 and BLM in colocalizing foci, and acts as the first defense to avoid DNA damage and genomic instability as a consequence of T–R conflicts (Figure 7E). In this study, we have used Cdk9 inhibitors to prolong pausing of RNAP II at promoter proximal pause sites. Whilst all three Cdk9 inhibitors used in this study are ATP competitive-drugs they are structurally diverse. Flavo is a flavonoid alkaloid and DRB is a nucleotide analog, whereas MC295 has a thiazole core and a bulky norbonane group that is thought to provide this drug specificity for Cdk9. Although all three Cdk9 inhibitors probably can inhibit other Cdks at high concentrations, it is highly unlikely that they have the same off targets.
BLM and FANCD2 are interdependent for focus formation in response to flavo, which is in line with the recent finding that FANCD2 and BLM are epistatic for suppression of RNA–DNA hybrids (20). Also, BLM depends on BRCA2 for focus formation, suggesting that BRCA2, FANCD2 and BLM work together in the TRe response. Cdk9 inhibition triggers an initial increase followed by a drop in BRCA2, FANCD2 and BLM foci. The decrease in FANCD2 foci after longer treatments with flavo or DRB is in agreement with previously observations (21) and is likely due to the block to new transcription initiation. Also, 1–2 h after flavo addition, RNAP II CTD Ser5P drops. Since RNAP II CTD Ser5P is mainly associated with promoter proximal RNAP II (88) this observation suggests that buildup of RNAP II at the promoters does not continue at later time points after flavo addition (Supplementary Figure S1A). The drop in FANCD2 foci after extended flavo treatment thus indicates that many spontaneous FANCD2 foci represent T–R conflicts.
Intriguingly, we found that ATR is not globally activated in response to drug-induced T–R conflicts. As a likely consequence, T–R conflicts cause damage in mitosis when the TRe response is disrupted. Thus, our results offer an explanation for the observation that T–R conflicts lead to the persistence of underreplicated regions into mitosis (7,9,24) by showing that T–R conflicts do not elicit immediate checkpoint activation. The lack of full ATR activation may have a physiological role by ensuring that replication continues globally, which increases the chance that stalled forks are rescued by adjacent forks.
In line with the lack of ATR activation, we also find that the TRe response does not involve FANCD2 monoubiquitylation. FANCD2 monoubiquitylation works to engage nucleases (89,90). Since T–R conflicts do not involve damage in the DNA template, it seems plausible that the cellular response avoids activation of DNA repair nucleases, because it may have adverse effects to activate nucleases, if the replication fork is stalled for reasons other than damage in the DNA template (91). Paradoxically however, hydroxyurea and aphidicolin trigger FANCD2 monoubiquitylation though these stresses do not directly damage the DNA template either (27). In vitro studies suggest that ssRNA and ssDNA can stimulate monoubiquitylation of FANCD2 (29), whereas R loops seem to be dispensable for aphidicolin-induced FANCD2 monoubiquitylation in cells (28), suggesting that aphidicolin-induced monoubiquitylation of FANCD2 in vivo is triggered by ssDNA formed in response to aphidicolin rather than T–R conflicts.
A recent study using an episomal system to dissect the cellular response to head-on and co-directional T–R collision found that head-on collisions trigger the ATR-CHK1 axis whereas co-directional collisions activate ATM (92). Moreover, co-directional collisions lead to clearance of RNA–DNA hybrids (92). We speculate that T–R collisions induced by Cdk9 inhibitors are mainly co-directional. This is based on the finding that replication is generally co-oriented with transcription in the human genome (93). Thus, forced transcription pausing around transcription start sites is likely to induce co-directional clashes. Our results show that the TRe response does not involve global activation of ATM or ATR. However, the previously reported responses may reflect the outcome of prolonged T–R conflicts (92,94), whereas we describe the early response, which may be required for clearance of RNA–DNA hybrids that occur upon co-directional collisions.
The involvement of FANCD2, BRCA2 and BLM in the TRe response may account for the observation that cells disrupted for either of these genes display elevated levels of RNA–DNA hybrids (17,20–22,95). Notably, FANCA-disrupted cells also accumulate RNA–DNA hybrids although our work suggests that FANCA, which is part of the FA core complex responsible for FANCD2 monoubiquitylation, is not part of the TRe response. Intriguingly, it has recently been suggested that RNA–DNA hybrids may be susceptible to interstrand crosslinks (22,96). Therefore, we speculate that the canonical FA pathway could be required for the repair of RNA–DNA crosslinks, whereas FANCD2 may be involved in the removal of both crosslinked and non-crosslinked RNA–DNA hybrids, potentially explaining why RNA–DNA hybrids also accumulate in FANCA deficient cells.
One role of BRCA2, BLM and FANCD2 at T–R conflicts could be to stabilize the fork until it is rescued by a nearby fork. However, the observation that FANCD2 deletion leads to higher levels of RNA–DNA hybrids (Figure 2A,B) suggests that FANCD2 also plays an active role in removing RNA–DNA hybrids or solving T–R conflict. Similarly, cells depleted for BRCA2 accumulate RNA–DNA hybrids (17). Finally, BLM clearly has a role in the TRe response, even though it is not involved in protecting the fork from nucleolytic degradation (72), arguing that the TRe response is different from fork stabilization. We have identified a role for the helicase function of BLM in the TRe response, but it is uncertain whether BLM unwinds RNA–DNA hybrids or acts at the replication fork. Interestingly, BLM has previously been shown to unwind RNA–DNA hybrids in vitro and BLM also localizes RNA–DNA hybrids in human cells (20). The mechanistic action of BRCA2, BLM and FANCD2 in the TRe response is thus a subject for future investigations, but may relate to the role of FANCD2 interaction with RNA processing factors DDX47 and hnRNP U that was recently described (97). Given the reported roles in recombination of BRCA2, BLM and FANCD2 (98–100) as well as fork protection of BRCA2 and FANCD2 (38,72), the potential role in the TRe response of the essential key recombinase and fork protector RAD51 is also an important aspect that remains to be addressed.
FANCD2 has a well-characterized role in interstrand crosslink repair, which involves FANCD2 monoubiquitylation at lysine 561 (K563 in chicken) (33,101), but recent studies also indicated that FANCD2 has monoubiquitylation-independent functions (24,40,73). The non-canonical role of FANCD2 in the TRe response could explain these observations.
Biallelic mutations in BRCA2 (FANCD1) account for approximately 3% of FA cases (102). This subtype is associated with higher cancer risk than most other FA subtypes. Together with the dramatically increased cancer risk of Bloom's syndrome patients, this indicates that the TRe response is a crucial mechanism to avoid carcinogenesis, reflecting that T–R conflicts might underlie replication stress and genomic instability in early steps of carcinogenesis (6,103,104). The results presented here may have clinical implications given that flavo and PladB show promising anticancer activity (86,87,105–107). Our results suggest that the TRe-response status of cancers could be used to stratify drug sensitivities to flavo, PladB and other drugs with similar mechanisms.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Louise Juhl and Linea Busch for cloning and testing BRCA2-targeting constructs and to K.J. Patel, Shunichi Takeda, Minoru Takata and Jakob Nilsson for sharing reagents and to K.J. Patel for useful discussions. We are also thankful to Chemometec for sharing their Xcyto image cytometer.
Author contributions: X.S. performed all experiments and data analyses. A.M.J. assisted with some of the experiments shown in Figure 5. N.G.H. provided the PD20 cell lines and advice on their maintenance. V.H.O. conceived, supervised and coordinated the project and wrote the manuscript. M.L., X.S. and V.H.O. acquired funding. All authors commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese Scholarship Council [201506240152 to X.S.]; Novo Nordisk Foundation (to V.H.O.); Villum Foundation and the Danish Cancer Society (to M.L. and V.H.O.); Dagmar Marshalls Fond, Neye-Fonden, the Danish Agency for Science, Technology and Innovation, and A.P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til almene Formaal (to M.L.). Funding for open access charge: Novo Nordisk fonden [NNF18OC0052089].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 2. Dereli-Oz A., Versini G., Halazonetis T.D.. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol. 2011; 5:308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Techer H., Koundrioukoff S., Nicolas A., Debatisse M.. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet. 2017; 18:535–550. [DOI] [PubMed] [Google Scholar]
- 5. Wei X., Samarabandu J., Devdhar R.S., Siegel A.J., Acharya R., Berezney R.. Segregation of transcription and replication sites into higher order domains. Science. 1998; 281:1502–1506. [DOI] [PubMed] [Google Scholar]
- 6. Kotsantis P., Silva L.M., Irmscher S., Jones R.M., Folkes L., Gromak N., Petermann E.. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016; 7:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Helmrich A., Ballarino M., Tora L.. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011; 44:966–977. [DOI] [PubMed] [Google Scholar]
- 8. Wilson T.E., Arlt M.F., Park S.H., Rajendran S., Paulsen M., Ljungman M., Glover T.W.. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2014; 25:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pentzold C., Shah S.A., Hansen N.R., Le Tallec B., Seguin-Orlando A., Debatisse M., Lisby M., Oestergaard V.H.. FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells. Nucleic Acids Res. 2017; 46:1280–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glover T.W., Wilson T.E., Arlt M.F.. Fragile sites in cancer: more than meets the eye. Nat. Rev. Cancer. 2017; 17:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C. et al.. Signatures of mutation and selection in the cancer genome. Nature. 2010; 463:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M. et al.. The landscape of somatic copy-number alteration across human cancers. Nature. 2010; 463:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oestergaard V.H., Lisby M.. Transcription-replication conflicts at chromosomal fragile sites-consequences in M phase and beyond. Chromosoma. 2017; 126:213–222. [DOI] [PubMed] [Google Scholar]
- 14. Wilson T.E., Arlt M.F., Park S.H., Rajendran S., Paulsen M., Ljungman M., Glover T.W.. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015; 25:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Tallec B., Dutrillaux B., Lachages A.M., Millot G.A., Brison O., Debatisse M.. Molecular profiling of common fragile sites in human fibroblasts. Nat. Struct. Mol. Biol. 2011; 18:1421–1423. [DOI] [PubMed] [Google Scholar]
- 16. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 17. Bhatia V., Barroso S.I., Garcia-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A.. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 18. Gan W., Guan Z., Liu J., Gui T., Shen K., Manley J.L., Li X.. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011; 25:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 20. Chang E.Y., Novoa C.A., Aristizabal M.J., Coulombe Y., Segovia R., Chaturvedi R., Shen Y., Keong C., Tam A.S., Jones S.J.M. et al.. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J. Cell Biol. 2017; 216:3991–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwab R.A., Nieminuszczy J., Shah F., Langton J., Lopez Martinez D., Liang C.C., Cohn M.A., Gibbons R.J., Deans A.J., Niedzwiedz W.. The fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell. 2015; 60:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia-Rubio M.L., Perez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., Aguilera A.. The fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015; 11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L. et al.. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 2015; 57:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madireddy A., Kosiyatrakul S.T., Boisvert R.A., Herrera-Moyano E., Garcia-Rubio M.L., Gerhardt J., Vuono E.A., Owen N., Yan Z., Olson S. et al.. FANCD2 facilitates replication through common fragile sites. Mol. Cell. 2016; 64:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naim V., Rosselli F.. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009; 11:761–768. [DOI] [PubMed] [Google Scholar]
- 26. Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D.. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009; 11:753–760. [DOI] [PubMed] [Google Scholar]
- 27. Howlett N.G., Taniguchi T., Durkin S.G., D’Andrea A.D., Glover T.W.. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005; 14:693–701. [DOI] [PubMed] [Google Scholar]
- 28. Okamoto Y., Iwasaki W.M., Kugou K., Takahashi K.K., Oda A., Sato K., Kobayashi W., Kawai H., Sakasai R., Takaori-Kondo A. et al.. Replication stress induces accumulation of FANCD2 at central region of large fragile genes. Nucleic Acids Res. 2018; 46:2932–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang Z., Liang F., Teng Y., Chen X., Liu J., Longerich S., Rao T., Green A.M., Collins N.B., Xiong Y. et al.. Binding of FANCI-FANCD2 complex to RNA and R-loops stimulates robust FANCD2 Monoubiquitination. Cell Rep. 2019; 26:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garaycoechea J.I., Patel K.J.. Why does the bone marrow fail in Fanconi anemia. Blood. 2014; 123:26–34. [DOI] [PubMed] [Google Scholar]
- 31. Alpi A.F., Pace P.E., Babu M.M., Patel K.J.. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell. 2008; 32:767–777. [DOI] [PubMed] [Google Scholar]
- 32. Meetei A.R., de Winter J.P., Medhurst A.L., Wallisch M., Waisfisz Q., van de Vrugt H.J., Oostra A.B., Yan Z., Ling C., Bishop C.E. et al.. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003; 35:165–170. [DOI] [PubMed] [Google Scholar]
- 33. Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D’Andrea A.D.. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001; 7:249–262. [DOI] [PubMed] [Google Scholar]
- 34. Howlett N.G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G. et al.. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002; 297:606–609. [DOI] [PubMed] [Google Scholar]
- 35. Wooster R., Neuhausen S.L., Mangion J., Quirk Y., Ford D., Collins N., Nguyen K., Seal S., Tran T., Averill D. et al.. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994; 265:2088–2090. [DOI] [PubMed] [Google Scholar]
- 36. Prakash R., Zhang Y., Feng W., Jasin M.. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015; 7:a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patel K.J., Yu V.P., Lee H., Corcoran A., Thistlethwaite F.C., Evans M.J., Colledge W.H., Friedman L.S., Ponder B.A., Venkitaraman A.R.. Involvement of Brca2 in DNA repair. Mol. Cell. 1998; 1:347–357. [DOI] [PubMed] [Google Scholar]
- 38. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chaudhury I., Sareen A., Raghunandan M., Sobeck A.. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013; 41:6444–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raghunandan M., Chaudhury I., Kelich S.L., Hanenberg H., Sobeck A.. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle. 2015; 14:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cunniff C., Bassetti J.A., Ellis N.A.. Bloom's syndrome: clinical spectrum, molecular pathogenesis, and cancer predisposition. Mol Syndromol. 2017; 8:4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adelman K., Lis J.T.. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012; 13:720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harlen K.M., Churchman L.S.. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017; 18:263–273. [DOI] [PubMed] [Google Scholar]
- 44. Fuda N.J., Ardehali M.B., Lis J.T.. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009; 461:186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ni Z., Saunders A., Fuda N.J., Yao J., Suarez J.R., Webb W.W., Lis J.T.. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 2008; 28:1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larochelle S., Amat R., Glover-Cutter K., Sanso M., Zhang C., Allen J.J., Shokat K.M., Bentley D.L., Fisher R.P.. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 2012; 19:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zatreanu D., Han Z., Mitter R., Tumini E., Williams H., Gregersen L., Dirac-Svejstrup A.B., Roma S., Stewart A., Aguilera A. et al.. Elongation factor TFIIS prevents transcription stress and r-loop accumulation to maintain genome stability. Mol. Cell. 2019; 76:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Svikovic S., Crisp A., Tan-Wong S.M., Guilliam T.A., Doherty A.J., Proudfoot N.J., Guilbaud G., Sale J.E.. R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J. 2019; 38:e99793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chedin F.. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Franklin R., Sale J.E.. Transient transfection of DT40. Subcell. Biochem. 2006; 40:379–382. [DOI] [PubMed] [Google Scholar]
- 51. Timmers C., Taniguchi T., Hejna J., Reifsteck C., Lucas L., Bruun D., Thayer M., Cox B., Olson S., D’Andrea A.D. et al.. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell. 2001; 7:241–248. [DOI] [PubMed] [Google Scholar]
- 52. Rego M.A., Kolling F.W.t., Vuono E.A., Mauro M., Howlett N.G.. Regulation of the Fanconi anemia pathway by a CUE ubiquitin-binding domain in the FANCD2 protein. Blood. 2012; 120:2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pedersen R.T., Kruse T., Nilsson J., Oestergaard V.H., Lisby M.. TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells. J. Cell Biol. 2015; 210:565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goedhart J., van Weeren L., Hink M.A., Vischer N.O., Jalink K., Gadella T.W. Jr. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods. 2010; 7:137–139. [DOI] [PubMed] [Google Scholar]
- 55. Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A.. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002; 20:87–90. [DOI] [PubMed] [Google Scholar]
- 56. Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y.. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004; 22:1567–1572. [DOI] [PubMed] [Google Scholar]
- 57. Arakawa H., Lodygin D., Buerstedde J.M.. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001; 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hirano S., Yamamoto K., Ishiai M., Yamazoe M., Seki M., Matsushita N., Ohzeki M., Yamashita Y.M., Arakawa H., Buerstedde J.M. et al.. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005; 24:418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Simpson L.J., Sale J.E.. Colony survival assay. Subcell. Biochem. 2006; 40:387–391. [DOI] [PubMed] [Google Scholar]
- 60. Guzmán C., Bagga M., Kaur A., Westermarck J., Abankwa D.. Colonyarea: an imagej plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014; 9:e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanz L.A., Chédin F.. High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019; 14:1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. de Azevedo W.F. Jr, Canduri F., da Silveira N.J.. Structural basis for inhibition of cyclin-dependent kinase 9 by flavopiridol. Biochem. Biophys. Res. Commun. 2002; 293:566–571. [DOI] [PubMed] [Google Scholar]
- 63. Baumli S., Endicott J.A., Johnson L.N.. Halogen bonds form the basis for selective P-TEFb inhibition by DRB. Chem. Biol. 2010; 17:931–936. [DOI] [PubMed] [Google Scholar]
- 64. Chen L., Chen J.Y., Zhang X., Gu Y., Xiao R., Shao C., Tang P., Qian H., Luo D., Li H. et al.. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell. 2017; 68:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shao W., Zeitlinger J.. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017; 49:1045–1051. [DOI] [PubMed] [Google Scholar]
- 66. Gressel S., Schwalb B., Decker T.M., Qin W., Leonhardt H., Eick D., Cramer P.. CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife. 2017; 6:e29736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamane K., Wu X., Chen J.. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 2002; 22:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang J., Gong Z., Chen J.. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J. Cell Biol. 2011; 193:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Germann S.M., Oestergaard V.H., Haas C., Salis P., Motegi A., Lisby M.. Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair (Amst.). 2011; 10:210–224. [DOI] [PubMed] [Google Scholar]
- 70. Acevedo J., Yan S., Michael W.M.. Direct binding to replication protein A (RPA)-coated Single-stranded DNA Allows recruitment of the ATR activator TopBP1 to sites of DNA damage. J. Biol. Chem. 2016; 291:13124–13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grofte M., Chan K.L., Hickson I.D., Bartek J. et al.. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011; 13:243–253. [DOI] [PubMed] [Google Scholar]
- 72. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lossaint G., Larroque M., Ribeyre C., Bec N., Larroque C., Decaillet C., Gari K., Constantinou A.. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol. Cell. 2013; 51:678–690. [DOI] [PubMed] [Google Scholar]
- 74. Bergoglio V., Boyer A.S., Walsh E., Naim V., Legube G., Lee M.Y., Rey L., Rosselli F., Cazaux C., Eckert K.A. et al.. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. J. Cell Biol. 2013; 201:395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Minocherhomji S., Ying S., Bjerregaard V.A., Bursomanno S., Aleliunaite A., Wu W., Mankouri H.W., Shen H., Liu Y., Hickson I.D.. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015; 528:286–290. [DOI] [PubMed] [Google Scholar]
- 76. Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J.. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986; 89:123–130. [DOI] [PubMed] [Google Scholar]
- 77. Foote K.M., Blades K., Cronin A., Fillery S., Guichard S.S., Hassall L., Hickson I., Jacq X., Jewsbury P.J., McGuire T.M. et al.. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J. Med. Chem. 2013; 56:2125–2138. [DOI] [PubMed] [Google Scholar]
- 78. Pichierri P., Franchitto A., Rosselli F.. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004; 23:3154–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bridge W.L., Vandenberg C.J., Franklin R.J., Hiom K.. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005; 37:953–957. [DOI] [PubMed] [Google Scholar]
- 80. Wang W., Seki M., Narita Y., Sonoda E., Takeda S., Yamada K., Masuko T., Katada T., Enomoto T.. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000; 19:3428–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sarkies P., Murat P., Phillips L.G., Patel K.J., Balasubramanian S., Sale J.E.. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012; 40:1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Davies S.L., North P.S., Hickson I.D.. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007; 14:677. [DOI] [PubMed] [Google Scholar]
- 83. Wu L., Chan K.L., Ralf C., Bernstein D.A., Garcia P.L., Bohr V.A., Vindigni A., Janscak P., Keck J.L., Hickson I.D.. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005; 24:2679–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M.. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998; 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 85. Li X., Manley J.L.. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 86. Wan Y., Zheng X., Chen H., Guo Y., Jiang H., He X., Zhu X., Zheng Y.. Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J. Cell Biol. 2015; 209:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y.. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007; 3:570–575. [DOI] [PubMed] [Google Scholar]
- 88. Zaborowska J., Egloff S., Murphy S.. The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 2016; 23:771–777. [DOI] [PubMed] [Google Scholar]
- 89. Knipscheer P., Raschle M., Smogorzewska A., Enoiu M., Ho T.V., Scharer O.D., Elledge S.J., Walter J.C.. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009; 326:1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lachaud C., Moreno A., Marchesi F., Toth R., Blow J.J., Rouse J.. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science. 2016; 351:846–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hamperl S., Bocek M.J., Saldivar J.C., Swigut T., Cimprich K.A.. Transcription-replication conflict orientation modulates R-Loop levels and activates distinct DNA damage responses. Cell. 2017; 170:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Petryk N., Kahli M., d’Aubenton-Carafa Y., Jaszczyszyn Y., Shen Y., Silvain M., Thermes C., Chen C.L., Hyrien O.. Replication landscape of the human genome. Nat. Commun. 2016; 7:10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Matos D.A., Zhang J.M., Ouyang J., Nguyen H.D., Genois M.M., Zou L.. ATR protects the genome against R loops through a MUS81-Triggered feedback loop. Mol. Cell. 2020; 77:514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shivji M.K.K., Renaudin X., Williams C.H., Venkitaraman A.R.. BRCA2 regulates transcription elongation by RNA polymerase II to Prevent R-Loop accumulation. Cell Rep. 2018; 22:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tan S.L.W., Chadha S., Liu Y., Gabasova E., Perera D., Ahmed K., Constantinou S., Renaudin X., Lee M., Aebersold R. et al.. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017; 169:1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Okamoto Y., Abe M., Itaya A., Tomida J., Ishiai M., Takaori-Kondo A., Taoka M., Isobe T., Takata M.. FANCD2 protects genome stability by recruiting RNA processing enzymes to resolve R-loops during mild replication stress. FEBS J. 2019; 286:139–150. [DOI] [PubMed] [Google Scholar]
- 98. Nakanishi K., Yang Y.G., Pierce A.J., Taniguchi T., Digweed M., D’Andrea A.D., Wang Z.Q., Jasin M.. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:1110–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Moynahan M.E., Pierce A.J., Jasin M.. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001; 7:263–272. [DOI] [PubMed] [Google Scholar]
- 100. Wu L., Hickson I.D.. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003; 426:870–874. [DOI] [PubMed] [Google Scholar]
- 101. Yamamoto K., Hirano S., Ishiai M., Morishima K., Kitao H., Namikoshi K., Kimura M., Matsushita N., Arakawa H., Buerstedde J.M. et al.. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell. Biol. 2005; 25:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Alter B.P., Rosenberg P.S., Brody L.C.. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J. Med. Genet. 2007; 44:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A. Jr, Kastrinakis N.G., Levy B. et al.. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005; 434:907–913. [DOI] [PubMed] [Google Scholar]
- 104. Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C. et al.. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005; 434:864–870. [DOI] [PubMed] [Google Scholar]
- 105. Karp J.E., Blackford A., Smith B.D., Alino K., Seung A.H., Bolanos-Meade J., Greer J.M., Carraway H.E., Gore S.D., Jones R.J. et al.. Clinical activity of sequential flavopiridol, cytosine arabinoside, and mitoxantrone for adults with newly diagnosed, poor-risk acute myelogenous leukemia. Leuk. Res. 2010; 34:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zeidner J.F., Karp J.E.. Clinical activity of alvocidib (flavopiridol) in acute myeloid leukemia. Leuk. Res. 2015; 39:1312–1318. [DOI] [PubMed] [Google Scholar]
- 107. Zeidner J.F., Foster M.C., Blackford A.L., Litzow M.R., Morris L.E., Strickland S.A., Lancet J.E., Bose P., Levy M.Y., Tibes R. et al.. Final results of a randomized multicenter phase II study of alvocidib, cytarabine, and mitoxantrone versus cytarabine and daunorubicin (7 + 3) in newly diagnosed high-risk acute myeloid leukemia (AML). Leuk. Res. 2018; 72:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.