Abstract
Human CWC27 is an uncharacterized splicing factor and mutations in its gene are linked to retinal degeneration and other developmental defects. We identify the splicing factor CWC22 as the major CWC27 partner. Both CWC27 and CWC22 are present in published Bact spliceosome structures, but no interacting domains are visible. Here, the structure of a CWC27/CWC22 heterodimer bound to the exon junction complex (EJC) core component eIF4A3 is solved at 3Å-resolution. According to spliceosomal structures, the EJC is recruited in the C complex, once CWC27 has left. Our 3D structure of the eIF4A3/CWC22/CWC27 complex is compatible with the Bact spliceosome structure but not with that of the C complex, where a CWC27 loop would clash with the EJC core subunit Y14. A CWC27/CWC22 building block might thus form an intermediate landing platform for eIF4A3 onto the Bact complex prior to its conversion into C complex. Knock-down of either CWC27 or CWC22 in immortalized retinal pigment epithelial cells affects numerous common genes, indicating that these proteins cooperate, targeting the same pathways. As the most up-regulated genes encode factors involved in inflammation, our findings suggest a possible link to the retinal degeneration associated with CWC27 deficiencies.
INTRODUCTION
Splicing of pre-messenger RNA (pre-mRNA) is performed by a very large RNA protein complex: the spliceosome. The stepwise assembly of spliceosomes involves the recruitment of snRNP (small nuclear ribonucleoproteins) and numerous proteins (1). Extensive rearrangements in composition and conformation accompany the formation of successive complexes named: E (early), A (pre-spliceosome), B (pre-catalytic spliceosome), Bact (activated spliceosome), B* (catalytically activated spliceosome), C, C* (catalytic spliceosome), P (post-catalytic splicesome) and ILS (Intron Lariat Spliceosome). B* spliceosomes catalyse the first catalytic step generating cleaved 5′-exon and intron/3′-exon lariat intermediates while C* spliceosomes catalyse the second step yielding ligated exons and intron lariat (2).
The yeast CWC27 (Complexed with Cef1 27) interacts with Cef1 protein, an essential splicing factor. The human CWC27 homologue is also named NY-CO-10. In both human and yeast spliceosomes, CWC27 is part of the Bact complexes (3–5) and leaves before its conversion to B* (5,6). CWC27 comprises an inactive N-terminal peptidyl-prolyl isomerase (PPIase) domain that has been conserved throughout evolution from yeast to mammals, followed by an elongated, unstructured and solvent-exposed C-terminal domain (7). Mutations that are expected to generate truncations of CWC27 unstructured C-terminal domain have been identified in human patients with retinal degeneration with or without other developmental defects (8). In mouse models, CWC27 knock-out is lethal while a C-terminal protein-truncating mutation leads to retinal degeneration, suggesting that the N-terminal CWC27 PPIase domain is essential for viability (8). Despite being associated with the spliceosome at a specific step, the molecular function of CWC27 remains unknown.
To unravel its function, we investigated CWC27 co-immunoprecipitating proteins. We found CWC22 (Complexed with Cef1 22), another evolutionarily conserved splicing factor, to be the CWC27 major interaction partner. In both Saccharomyces cerevisiae and human spliceosomes, CWC22 borders the spliceosome ‘exon binding channel’ and stabilizes the 5′ exon before the first step of splicing (3,5). In humans, CWC22 has been proposed to escort eIF4A3, a core exon junction complex (EJC) subunit, to the spliceosome (9,10). The EJC is an RNA binding protein complex found in metazoans and deposited around 27 nt upstream exon–exon junctions (11,12). It is composed of four core subunits (eIF4A3, MAGOH, Y14 and MLN51) and interacts with various peripheral factors (13). The EJC is recruited by spliceosomes and accompanies spliced mRNAs from the nucleus to the cytoplasm where it is removed by the first translating ribosome. It participates to pre-mRNA splicing regulation and contributes to mature mRNA export, localization, translation and degradation (13,14). According to published cryo-EM spliceosome structures, the complete EJC is bound to the 5′ exon in the spliceosome C complex (15,16). However, how and when the four core EJC subunits are recruited and assembled onto mRNA remains largely unknown.
Using purified recombinant proteins, we reconstituted a CWC27/CWC22/eIF4A3 ternary complex and solved its 3D structure by X-ray crystallography. This structure possibly corresponds to eIF4A3 earliest contacts with the spliceosome. We propose that CWC22 and CWC27 in the Bact complex form a landing platform for eIF4A3 before the release of CWC27 and the assembly of a complete EJC core bound to CWC22. In addition, transcriptomic data of knock-downs of CWC27 and CWC22 in an immortalized retinal pigment epithelial cell line revealed that these proteins target the same pathways. Noteworthy, genes in the inflammation pathways are among the most strongly up-regulated, suggesting a link between retinal degeneration and CWC27 deficiency.
MATERIALS AND METHODS
Cells maintenance and transfections
Human HeLa and Hek293T cells were propagated at 37°C in a humidified 5% CO2 atmosphere in high glucose DMEM medium (31966-021, Life Technologies) supplemented with 10% fetal bovine serum and 100 U/ml Penicillin-Streptomycin (Life Technologies). For overexpression of CWC27 and eIF4A3 constructs, cells were transfected with JetPrime (Polyplus) according to manufacturer's instruction. Full-length human CWC27 cDNA was PCR amplified with Phusion DNA polymerase (New England Biolabs) from a HCT116 cDNA homemade library and cloned into p 3xFLAG-CMV-10 (Sigma). Truncated CWC27 versions were generated by inverted PCR from p 3xFLAG-CMV-10 CWC27. p3XFLAG-CMV eIF4A3 was obtained from (M.J. Moore). Point mutant D270G in eIF4A3 was generated by subjecting the p3XFLAG-CMV eIF4A3 plasmid to QuickChange Site-Directed Mutagenesis.
hTERT-RPE-1 cells were propagated at 37°C in a humidified 5% CO2 atmosphere in DMEM/F-12 GlutaMAX medium (31331-028, Life Technologies) supplemented with 10% FBS and 100 U/ml penicillin–streptomycin 1× (Life Technologies). HeLa and hTERT-RPE-1 cells were genotyped by Eurofins Forensik and routinely tested for mycoplasma by PCR.
Immunoprecipitation and Western Blotting
Cells were lysed with RIPA buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP40, 1% sodium deoxycholate, RQ1 DNase (Promega, 1:50) and Protease inhibitor (Sigma, 1:100)). RNase A+T1 (Thermo Scientific, 1:200) was added or not to the sample. IP was performed overnight with 1 mg of total protein and 40 μl of Anti-FLAG M2 Magnetic Beads (Sigma) or 40 μl of Dynabeads Protein A (Life Technologies) linked to the desired antibody. Washes were performed with IP150 buffer (10 mM Tris–HClL (pH 7.5), 150 mM NaCl, 2.5 mM MgCl2, 1% NP-40). After elution with SDS loading dye, samples were separated by electrophoresis in 4–12% Tris-glycine SDS/PAGE (Life Technologies) and were transferred onto 0.2-μm nitrocellulose membranes (Protan-BA83; GE Healthcare) using Thermofisher Transblot systems. Membranes were blocked in PBS with 10% (w/v) milk and 0.05% Tween-20 (Euromedex) before incubation with primary antibodies diluted 1:1000 in PBS 0,05% Tween for 1 h at RT or overnight at 4°C. Anti-CWC22, anti-eIF4A3, anti-MAGOH, anti-Y14 (9) and anti-CWC27 (Atlas, #HPA020344), anti-GAPDH (Cell signaling Technology, 2118S), anti-FLAG (Sigma, F7425) were used. After washing with 1× PBS, membranes were incubated with stabilized goat anti-rabbit secondary antibodies (1:10 000; Promega) and visualized using SuperSignal West Femto (Thermo Scientific) with LAS 4000 mini (GE Healthcare).
Immunofluorescence
Cells were seeded on coverslips coated with poly-lysine (Sigma, P1524) and fixed in 4% paraformaldehyde before permeabilization in PBS-Triton X (0.1%) for 2 min. After blocking, coverslips were incubated for 1 h at RT with the primary antibody diluted in PBS–BSA 1%. Nuclei were stained with Hoechst (diluted 1:400 in PBS-BSA 1%). Coverslips were then incubated for 1 h at room temperature with secondary antibodies (conjugated with Alexa Fluor 488 or Alexa Fluor 546 or Alexa Fluor 647 fluorochrome) diluted in PBS-BSA 1%. Coverslips were mounted in 5 μl of Fluoromount-G (Southern Biotech®) medium. Pictures were taken on Nikon Ti LGM. Images were processed and analyzed with Fiji software.
Genome editing
HeLa cells were co-transfected with two plasmids. The first one derived from pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) (from E. Bertrand, IGMM Montpellier), expresses the nickase version of Streptococcus pyogenes Cas9 (Cas9n) and the two gRNAs (gRNA1 (5′-GGCCGCTCTCATCCCCCGTA-3′) and gRNA2 (5′-GCTCATCTTGGTCAGTACAA-3′). The other plasmid contains the repair sequence comprising the puromycin gene flanked by two lox sites. Puromycin-resistant colonies were isolated and expanded. Homozygous edited cell clones were identified by PCR on genomic DNA and by western blot with an anti-CWC27 antibody (Atlas, #HPA020344). To remove the puromycin gene, a FLAG-CWC27 expressing clone was transfected with a plasmid expressing both the Cre-recombinase and the Geneticin-resistance genes (X. Morin, IBENS, Paris) and maintained in Geneticin (G418, Thermo Scientific) containing medium for 40 hours to select transiently transfected cells. After clonal isolation and expansion, removal of the puromycin gene was checked by PCR on genomic DNA and CWC27 expression was analysed by Western Blot with an anti-CWC27 antibody.
Mass spectrometry
LC–MS/MS analysis of FLAG-CWC27
107 HeLa WT and HeLa FLAG-CWC27 cells were lysed in HKM300 buffer (10 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 300 mM NaCl, 0.2 mM EGTA, 0.5% NP-40). A nuclear fraction (P) was pelleted 10 min at 400 g. The P fraction was resuspended with 1 ml HKM300 digested by DNase RQ1 (1:50) 10 min on ice, sonicated and centrifuged at 10 000 g for 10 min at 4°C. The supernatant (1 mg of protein in a final volume of 1 ml) was incubated overnight at 4°C with 40 μl of Anti-FLAG M2 Magnetic Beads (Sigma). The beads were washed three times 5 min at 4°C in HKM300.
Spin-dried beads were digested overnight at 37°C by sequencing grade trypsin (12,5 μg/ml; Promega Madison, WI, USA) in 20 μl of 25 mM NH4HCO3. The digested peptide mixture was loaded on a Q-Exactive plus system coupled to a Nano-LC Proxeon 1000 column equipped with an EASY-Spray ion source (Thermo Scientific). Peptides were separated by chromatography on Acclaim PepMap100 C18 pre-column (2 cm, 75 μm i.d., 3 μm, 100 Å), Pepmap-RSLC Proxeon C18 column (50 cm, 75 μm i.d., 2 μm, 100 Å) with a gradient from 95% solvent A (water, 0.1% formic acid) to 35% solvent B (100% acetonitrile, 0.1% formic acid) over a period of 97 min at 300 nl/min flow rate. Peptides were analysed in the Orbitrap cell, in full ion scan mode, at a resolution of 120 000 (at m/z 200), with a mass range of m/z 350–1550 and an AGC target of 4 × 105. Fragments were obtained by high collision-induced dissociation (HCD) activation with a collisional energy of 30%, and a quadrupole isolation window of 1.6 Da. MS/MS data were acquired in the Orbitrap cell. Precursor priority was highest charge state, followed by most intense. Peptides with charge states from 2 to 8 were selected for MS/MS acquisition. The maximum ion accumulation times were set to 100 ms for MS acquisition and 60 ms for MS/MS acquisition.
LC–MS/MS analysis of CWC22 and eIF4A3 immunoprecipitates
Nuclei were prepared from HeLa cells essentially as previously described by A. Lamond (17). The clean pelleted nuclei were resuspended in 5 ml RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0,5% deoxycholate) with antiprotease cocktail, RQ1 RNAse-Free DNAse (1:50 volume, Promega), RNAse A (1:100 volume, Thermo Scientific) and RNAse T1 (1:100 volume, Thermo Scientific), sonicated at 4°C. The lysate was centrifuged at 2800 g for 10 min at 4°C. Supernatants were incubated for 2 h at 4°C with 100 μl protein A-coupled Dynabeads (Life Technologies) either or not (control) crosslinked with dimethylpimelidate to affinity-purified anti-eIF4A3 or anti-CWC22. The beads were next washed three times with IP buffer 300 (10 mM Tris–HCl, pH 7.5, 300 mM NaCl, 2.5 mM MgCl2, 1% NP-40, 1% protease-inhibitor mixture) and incubated 20 min at 25°C with 50 U RNase A and 25 U RNAse T1 in 200 μl of IP buffer 150. Following three washes with IP buffer 300. Proteins were eluted with 20 ng/μl of the appropriate immunogenic peptide.
A short SDS-PAGE (dye-front at 1 cm from the bottom of the well) was used as a cleanup step. Gel slices were washed in water and proteins were reduced with 10 mM DTT before alkylation with 55 mM iodoacetamide. After dehydration with 100% (v/v) acetonitrile, we performed in-gel digestion using trypsin/Lyc-C (Promega) overnight in 25 mM NH4HCO3 at 30°C. The peptide mixture was analyzed by LC-MS/MS using an RSLCnano system (Ultimate 3000, Thermo Scientific) coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific). Peptide separation was performed on a C18-reversed phase column (75 mm ID × 50 cm; C18 PepMapTM, Dionex) at a flow rate of 400 nl/min and an oven temperature of 40°C. The loading solution was 0.1% trifluoroacetic acid and 2% acetonitrile and for elution 100% water with 0.1% formic acid for channel A, and 0.085% formic acid and 100% acetonitrile for channel B. The peptides were eluted with a linear multi-step gradient of 1–6% solution B in 1 min, of 6–9% solution B in 11 min of 9–32% solution B in 82 min, and of 32–40% solution B in 6 min. We acquired Survey MS scans in the Orbitrap on the 400–1500 m/z range with the resolution set to a value of 120 000 and a 4 × 105 ion count target. Each scan was recalibrated in real time by co-injecting an internal standard from ambient air into the C-trap. Tandem MS was performed by isolation at 1.6 Th with the quadrupole, HCD fragmentation with normalized collision energy of 35, and rapid scan MS analysis in the ion trap. The MS2 ion count target was set to 104 and the max injection time was 100 ms. Only those precursors with charge state 2–7 were sampled for MS/MS. The dynamic exclusion duration was set to 60 s with a 10 ppm tolerance around the selected precursor and its isotopes. The instrument was run in top speed mode with 3 s cycles.
Mass spectrometry data analysis
The raw mass spectrometry data were analyzed by MaxQuant software (version 1.6.3.4) (18) using the embedded Andromeda search engine using the human protein database downloaded from Uniprot (20181204, 95146 entries) and completed with the contaminant list from MaxQuant. Up to two missed cleavages were allowed. The precursor mass tolerance was set to 4.5 ppm and the fragment mass tolerance to 0.5 Da. Carbamidomethylation of Cysteine residues was set as fixed modification and acetylation of protein N-terminus, oxidation of Methionine and deamidation of Asparagine and Glutamine were set as variable modifications. Minimal peptide length was set to seven amino acids. Second peptide option search was allowed. A false discovery rate (FDR) of 1% was independently applied for both peptide and protein identification. The ‘match between runs’ (MBR) option was allowed with a match time window of 1 min and an alignment time window of 20 min. In the case of identified peptides that are shared between two proteins, these were combined and reported as a single protein group. Label-free quantification (LFQ) option was enabled, with at least two peptides required for LFQ measurements. LFQ was done using both unique and razor peptides for each protein.
Bioinformatic analysis of the MaxQuant/Andromeda workflow output and the analysis of the abundances of the identified proteins was performed from the ‘proteinGroups.txt’ of MaxQuant output file with the Perseus software (version 1.6.2.3) (19). The lists of identified proteins were filtered to eliminate reverse hits and known contaminants. LFQ values were further transformed to a log2 scale. The missing values were imputed from normal distribution with a width of 0.3 and a down-shift of 1.8 to simulate signals from low abundant proteins. To distinguish specifically interacting proteins from the background, protein abundances were compared between sample and control groups, using the Student's t-test statistic (FDR ≤ 0.01, S0 = 2, n = 3 independent measurements), and results were visualized as volcano plots.
Recombinant proteins and in vitro interaction assays
Purification of recombinant proteins CBP-CWC22-S (pHL599), CBP-eIF4A3 (pHL241), eIF4A3 (pHL48) was previously described (9,20). For CBP-CWC22-MIF4G (pHL988), residues 117–406 were inserted in a variant of pET28a to fuse an N-terminal CBP tag and a C-terminal His6 Tag (Chamieh et al. 2008). For PTS-CWC27 (pHL1584), coding sequences of human CWC27 (1-472, Uniprot Q6UX04) were cloned between SalI and NotI in pET28a (Novagen) allowing for the fusion of N-terminal tags Protein A and TwinStrep between NheI and SalI and a C-terminal His6 Tag. For CWC27-C (pHL1553), residues 354–472 were PCR amplified and inserted between NheI and XhoI sites in pET28a allowing fusion of a C-terminal His6 Tag. The PTS-CWC27 protein fragments were successively purified on Nickel column (Ni-NTA, Clontech) and on StrepTactin affinity column (IBA). CWC27-iso2 (Uniprot Q6UX04-2) expressed as Sumo cleavable N-terminal His6 fusion protein was incubated with Senp2 protease overnight at 4°C and dialyzed against 50 mM Na2HPO4 pH 7.5, 150 mM NaCl. In vitro interaction assays were performed as previously described (20). For PTS pulldown, 12 μl of pre-blocked StrepTactin affinity beads (50% slurry, IBA) was used and precipitated proteins were eluted with 1× SDS loading buffer.
X-ray structure
For X-ray structure, recombinant proteins CWC22 and CWC27 were co-expressed in E. coli BL21 (DE3) grown in TB-medium at 37°C, as GST-3C-N-terminal His6 fusion or Sumo cleavable N-terminal His6 fusion proteins respectively. EIF4A3 was expressed as a Sumo cleavable N-terminal His6 fusion. Overexpression was induced at 18°C with 0.5 mM IPTG. Cells were lysed by sonication in 50 mM Na2HPO4 pH 7.5, 250 mM NaCl, 10 mM imidazole, 1 mM PMSF, and 25 mg/ml DNaseI, and the extract was cleared by centrifugation (4°C, 75 000 g, 30 min). In a first step, proteins were purified via a Ni2+-NTA affinity column (5 ml, GE healthcare). In order to remove N-terminal His6-tags, proteins were incubated with 3C or Senp2 proteases overnight at 4°C and dialyzed against 50 mM Na2HPO4 pH 7.5, 150 mM NaCl for subsequent heparin chromatography (5 ml Heparin Q sepharose, GE Healthcare). Protein complexes were isolated by size exclusion chromatography (SEC) after concentrating to 20–30 mg/ml in a buffer containing 10 mM HEPES pH 7.5, 150 mM NaCl and 1 mM DTT using a HiLoad Superdex 75 column (GE Healthcare). The complex was stored at 80°C in SEC buffer.
Crystallization of CWC22-CWC27-EIF4A3(RecA2) complex
The complex was set up for crystallization at 20 mg/ml in SEC buffer by sitting-drop vapor diffusion in 0.2 ul drops obtained by mixture of equal volumes of protein and crystallization solution. Crystals appeared after 2 days at 4°C as monoclinic prism after mixing with 20% (w/v) PEG20000, 50 mM MES pH 6.5 and were cryoprotected in reservoir solution containing 33% (v/v) ethylene glycol prior to flash freezing in liquid nitrogen.
Data collection and crystal structure determination
Diffraction data were collected at the PXII beamline at the Swiss Light Source (SLS) in Villigen, Switzerland, and were processed with XDS (21) prior to scaling with Aimless of the CCP4 package (22). The structure of CWC22-CWC27-EIF4A3 (RecA2) was determined from selenomethionine substituted protein crystals. Single anomalous dispersion data were recorded at the Se peak wavelength, and AUTOSOL as part of the PHENIX package was used to locate Se sites. A combination of single anomalous dispersion and molecular replacement was used to solve the structure at 3.0 Å using known EIF4A3-CWC22 structure (PDB IDs: 4C9B) using the program Phaser (23). The asymmetric unit contained four molecules of the complex. The model was completed by iterative cycles of model building in COOT (24), followed by refinement in PHENIX (25) using NCS restraints.
mRNA-seq and data analysis
For mRNA-seq, h-TERT RPE-1 cells were transfected at 60–70% confluency with 9 μl of Lipofectamine and 1.5 μl of 20 μM DsiRNAs (Integrated DNA Technologies) CWC27, CWC22 or control. A mix of two different DsiRNA targeting different regions of the CWC27 and CWC22 genes was used. Transfections for replicates, were performed independently. Forty eight hours after transfection, RNAs were extracted using Monarch Total RNA Miniprep Kit (New England Biolabs). DsiRNA efficiency was checked by WB. RNA-seq was performed by Fasteris on paired-end libraries run on Illumina HiSeq using 2 × 150 bp.
Bioinformatic analysis
After trimming of the adapters using cutadapt (26), the reads were mapped on hg38 (Gencode GRCh38.p12.genome.fa) using STAR (version 020201,(27)) with default parameters, and adding the -quantMode GeneCounts option to generate gene count files. Gencode hg38 V29 gtf annotations were used. BamCoverage from deepTools (28) was used to generate bigwig file for quick visualization of the read counts on IGV (29). Principal component analysis was performed using the ‘pcaMethods’ R package directly on gene counts. Intron retention was assessed using iREAD 0.8.0 (30), adding a post-treatment to remove the regions where two genes overlap. Resulting intron count files of controls vs CWC22 or CWC27 siRNA triplicates were processed with DESeq2 to find significant changes associated with the Knock Downs (KDs). Introns with |log2(FC)| > 1 and P-value < 0.05 were considered as significantly retained. Jsplice (31) was run in junction mode to find differential splicing. Alternative splicing modules (ASMs) with |FC| > 1.5 and P-value < 0.05 were considered as significantly differentially spliced. We used JSplice classification for alternative splicing events. For differential expression, DESeq2 (32) was run on gene counts of controls versus CWC22 or CWC27 siRNA triplicates, with default parameters. Genes with |log2(FC)| > 1 and P-value < 0.05 were considered as significantly regulated. Gene Ontology (GO) analysis was performed with GOrilla (33,34) on genes upregulated with a P-value of 0.05. ‘Process’ was chosen as ontology and default parameters were used. Revigo (35) was used for summarizing GO categories and for generating the graphics. Default parameters were used. TreeMap was used as representation. Subcategories were removed from the original TreeMap graph but they are indicated in the table.
RESULTS
CWC27 associates with CWC22 and eIF4A3
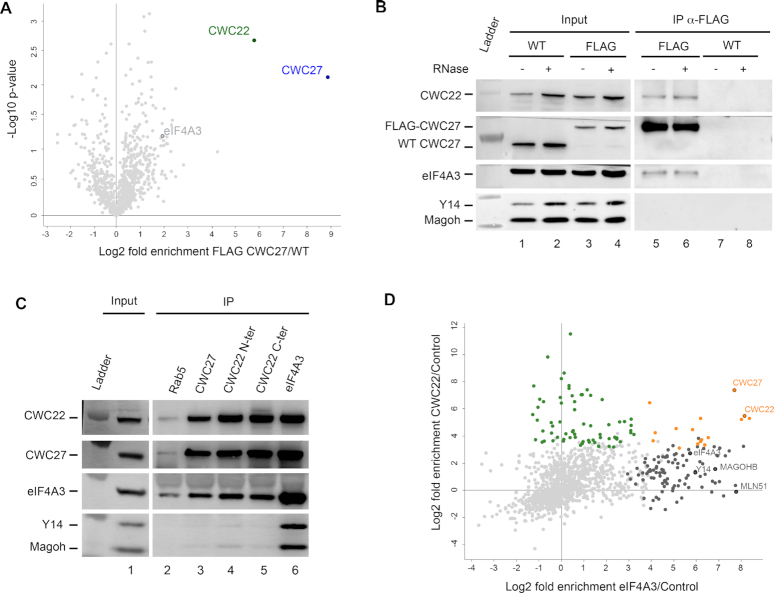
To better characterize CWC27 function, we first looked for its protein partners by immunoprecipitation. To maximize immunoprecipitation specificity and efficiency, CWC27 was epitope tagged with a N-terminal 3xFLAG peptide. To minimize artefacts due to overexpression, the FLAG was inserted by CRISPR–Cas9 editing of all CWC27 alleles in HeLa cells (Supplementary Figure S1A). Indeed, a Western Blot using affinity purified anti-CWC27 polyclonal antibodies confirmed that expression level of FLAG-CWC27 is similar to that of wild-type CWC27 in the parental cell line (Supplementary Figure S1B). Triplicates of FLAG immunoprecipitation from FLAG-CWC27 cells were analysed by label-free quantitative mass spectrometry (LC–MS/MS) using the parental HeLa cells as a negative control. The splicing factor CWC22 is found as the most significant interacting protein (Figure 1A). CWC22 is also detected by Western blotting after FLAG immunoprecipitation from FLAG-CWC27 cells but not from the parental ones (Figure 1B, lanes 5–8). The interaction between CWC27 and CWC22 is not RNA dependent since co-precipitation is unaffected by RNase treatment (Figure 1B, lane 6). As a further confirmation, CWC27-specific antibodies immunoprecipitated CWC22 (Figure 1C, lane 3) and two distinct CWC22-specific antibodies immunoprecipitated CWC27 (Figure 1C, lanes 4 and 5). CWC22 is bound to the EJC inside the spliceosome after the first step of splicing (36,37) and it is important for the recruitment of eIF4A3 into spliceosome (9,10,38). eIF4A3 is co-immunoprecipitated by FLAG antibodies from FLAG-CWC27 cell lysates with or without RNase treatment (Figure 1B) and by affinity purified CWC22 or CWC27 antibodies from HeLa cell lysates (Figure 1C, lanes 3–5). The other EJC subunits Y14 and MAGOH are weakly immunoprecipitated by CWC22 antibodies and hardly detected following CWC27 immunoprecipitation (Figure 1C, lanes 3–5).
Figure 1.
CWC27 is associated with CWC22 and eIF4A3. (A) Volcano plot of protein enrichment for FLAG-CWC27 immunoprecipitation versus control purification. Not-significant proteins (FDR > 0.01) are indicated in light grey. (B) Western blots of proteins coimmunoprecipitated with FLAG-CWC27. Protein extracts from WT and FLAG-CWC27 (FLAG) cells were treated with (+) or without (−) RNase. Detected proteins are indicated on the left. (C) Same as (B), with affinity purified antibodies anti-Rab5, anti-CWC27, anti-CWC22 (N-terminus), anti-CWC22 (C-terminus) and anti-eIF4A3 used for immunoprecipitations with protein extracts from HeLa cells treated with RNase. (D) Enrichment plot of proteins from eIF4A3 and CWC22 purifications. These enrichments were computed using the same control purifications without antibodies during the immunoprecipitation step. Proteins significantly (FDR ≤ 0.01) enriched in anti-CWC22 immunoprecipitation (green), in anti-eIF4A3 (dark gray) or in both (orange) are indicated on the plot. Not-significant proteins are indicated in light gray.
We next wanted to explore the CWC22 and eIF4A3 interacting network, notably with nuclear splicing factors. For this, we used sucrose density centrifugation to isolate HeLa cell nuclei (see Materials and Methods) before immunoprecipitation. Then, we performed triplicate immunoprecipitations coupled to label-free quantitative mass spectrometry (LC–MS/MS) using this time affinity-purified polyclonal anti-CWC22 and anti-eIF4A3 antibodies. 88 and 107 statistically significant proteins were identified with CWC22 and eIF4A3 antibodies, respectively (Figure 1D and Supplementary Tables S1 and S2). Among the 17 statistically significant proteins common to both CWC22 and eIF4A3 immunoprecipitates, 10 are splicing-related factors and the remaining ones are linked to other mRNA maturation steps (Supplementary Table S3). Several splicing factors such as SLU7, CWC15, CWC22, CWC27 and CDC5L (the human orthologue of Cef1) belong to Bact spliceosome and subsequent complexes. Noteworthy, CWC27 is one of the most enriched proteins. Taken together these results indicate that CWC27, CWC22 and eIF4A3 interact with each other in spliceosomes.
A CWC22/CWC27/eIF4A3 ternary association requiring the CWC27 C-terminal domain
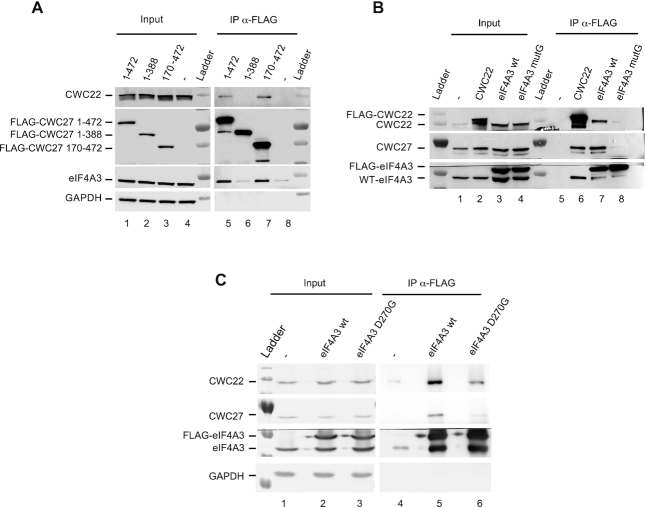
To map the interaction domains, we transiently expressed FLAG-tagged versions of the full-length (1–472) or truncated CWC27 proteins. All proteins were correctly expressed and localized in the nucleus (Figure 2A and Supplementary Figure S2). CWC22 and eIF4A3 both co-precipitated with the full-length and a truncation lacking the N-terminal PPIase domain (170–472) (Figure 2A, lanes 5 and 7) while they did not co-precipitate with truncations lacking fragments of the unstructured C-terminal domain (1–306) and (1–388) (Figure 2A, lane 6 and Supplementary Figure S2A, lanes 9 and 10), despite the truncated proteins remaining localized in the nucleus (Supplementary Figure S2B). These results indicate that the last 84 amino acids of CWC27 are required to interact with both CWC22 and eIF4A3.
Figure 2.
CWC27 C-terminal region mediates interaction with CWC22 and eIF4A3 (A) Western blots of proteins coimmunoprecipitated with FLAG-CWC27 isoforms or FLAG empty vector (−) transiently expressed in HeLa cells. Detected proteins are indicated on the left. (B) Same as (A), with transiently expressed FLAG-CWC22, FLAG-eIF4A3 WT or mutG. (C) Same as (A), with transiently expressed FLAG-eIF4A3 WT or the mutant FLAG-eIF4A3 D270G.
Conversely, transfected FLAG-CWC22 and FLAG-eIF4A3 proteins co-immunoprecipitate CWC27 (Figure 2B, lanes 6 and 7). We next investigated two mutated versions of eIF4A3 known to affect its binding to CWC22. A quadruple mutation of the 298–301 sequence at the surface of eIF4A3 (REAN>HARD), called eIF4A3-mutG mutation, had been shown to strongly reduce eIF4A3-CWC22 interaction in vitro (9). The eIF4A3 D270G mutation is associated with the Richieri Costa Pereira syndrome (39). eIF4A3 D270 directly contacts lysine K174 of CWC22 (40) but its impact on eIF4A3-CWC22 interaction had not been investigated. We observed that both mutations not only reduce the interaction between CWC22 and eIF4A3 but also their interaction with CWC27 (Figure 2B and C). These observations strongly suggest that in live cells CWC27 forms a ternary complex with CWC22 and eIF4A3, requiring an intact CWC22/eIF4A3 interaction.
In vitro reconstitution of a CWC27/CWC22/eIF4A3 ternary complex
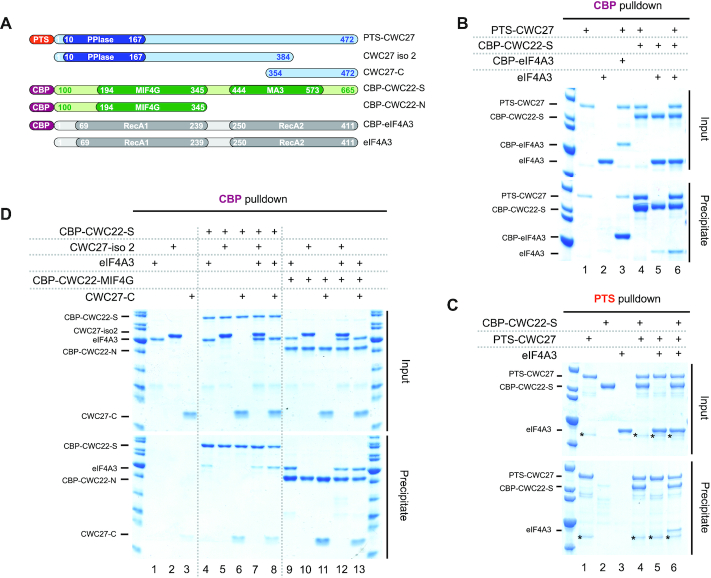
To better characterize the CWC27, CWC22 and eIF4A3 association, we used in vitro reconstitution experiments with recombinant proteins purified from bacteria (Figure 3A). All proteins were fused with a C-terminal His6 tag and when indicated, with a Calmodulin Binding Peptide (CBP) or a tandem Protein A-Twin Strep (PTS) N-terminal tag. Full-length CWC22 does not express well in bacteria, therefore we used a shorter version (CWC22-S; residues 100–665) more suitable for in vitro binding studies (9). The proteins were mixed, incubated with calmodulin beads, and after extensive washes, calmodulin bound protein(s) were fractionated by SDS-PAGE and visualized by Coomassie staining. CBP-CWC22-S co-retains PTS-CWC27 while CBP-eIF4A3 does not co-retain more PTS-CWC27 than control (Figure 3B, lanes 1, 3 and 4). As previously described (9), CBP-CWC22-S co-retains some eIF4A3 above control (Figure 3B, lanes 2 and 5). eIF4A3 co-retained with CWC22-S increases significantly in the presence of PTS-CWC27 (Figure 3B, lane 6). We next used a preformed CBP-CWC22-S/CWC27 heterodimer obtained by co-expression in bacteria and mixed it with eIF4A3. Again, the heterodimer co-retains more eIF4A3 than CBP-CWC22-S alone (Supplementary Figure S3). Conversely, PTS-CWC27 efficiently retains CBP-CWC22-S on StrepTactin beads whether eIF4A3 is added or not (Figure 3C, lanes 4 and 6). In contrast, addition of CBP-CWC22-S is an absolute requirement to retain eIF4A3 on the beads (Figure 3C, lanes 5 and 6). Taken together, these experiments suggest that eIF4A3 binds primarily to CWC22 and that CWC27 stabilizes this interaction.
Figure 3.
Direct interaction of CWC27 and eIF4A3 with CWC22 (A) Schematic representation of the different purified recombinant proteins (CWC27 in blue, CWC22 in green and eIF4A3 in gray) and the fused affinity tags (PTS in orange, CBP in purple). (B) Interaction of CWC27 with CWC22 and eIF4A3. Mixed recombinant proteins (input: 35% of total) and proteins retained with CBP-eIF4A3 (lane 3), CBP-CWC22-S (lanes 4 to 6) or not (lanes 1 and 2) on calmodulin beads (precipitate) were analyzed by SDS-PAGE. Proteins are indicated on the left. (C) Same as (B), with the indicated proteins retained with PTS-CWC27 (lanes 1, 4, 5 and 6) or not (lanes 2 and 3) on streptavidin beads. An unspecific contaminating protein co-purifying with PTS-CWC27 is marked (*). (D) Same as (b), with proteins retained with CBP-CWC22-S (lanes 4–8), with CBP-CWC22-MIF4G (lanes 9–13) or not (lanes 1–3) on calmodulin beads.
In an attempt to define interaction domains, we performed the reconstitution experiments with protein fragments. The above described transfection experiments indicated that the last 84 aa of CWC27 isoform 1 (Uniprot Q6UX04-1) are required to interact with both CWC22 and eIF4A3 in live cells. Therefore, we first repeated the in vitro binding assays using CWC27 isoform 2 (Uniprot Q6UX04-2) in which, due to an alternative splicing, the last 88 aa are replaced by 6 aa. Indeed, this isoform (CWC27-iso2) is not retained by CBP-CWC22-S whether or not eIF4A3 is added (Figure 3D, lanes 5 and 7). Conversely, a CWC27 C-terminal fragment (354–472) is retained by CBP-CWC22-S whether or not eIF4A3 is added (Figure 3D, lanes 6 and 8). This result demonstrates that CWC22 binds to the unstructured CWC27 C-terminal domain and not the N-terminal PPIase domain. eIF4A3 has been previously reported to interact with the MIF4G domain of CWC22 (9). A CWC22 fragment (119–431) containing this domain (CBP-CWC22-N) indeed retains eIF4A3 (Figure 3D, lane 9). This fragment also retains the CWC27 C-terminal fragment (Figure 3D, lane 11). It is thus possible to reconstitute a minimal complex (Figure 3D, lane 13) with the CWC22 MIF4G domain, the last 118 aa of CWC27 and eIF4A3 that might be suitable for structural studies.
3D structure of the CWC27/CWC22/eIF4A3 ternary complex
In order to obtain the 3D structure of the CWC27/CWC22/eIF4A3 ternary complex by X-ray crystallography, we expressed and purified recombinant MIF4G domain of CWC22 (residues 119–359), a fragment of the C-terminal region of CWC27 (320–431) and eIF4A3. No crystals were obtained upon large crystallization screening. A limited proteolysis experiment allowed us to identify a shorter CWC27 construct (378–431) still interacting with CWC22. A combinatory crystallization screening approach led us to crystallize the ternary complex CWC22 (119–359) / CWC27 (378–431)/eIF4A3 RecA2 domain (246–411). The crystal structure was solved by a combination of single-wavelength anomalous dispersion (SAD) using selenomethionine substitution (CWC22/CWC27) and molecular replacement (See methods). The structure is refined at 3.0 Å resolution, with a free R factor of 27%, a working R factor of 23% and good stereochemistry (Supplementary Table S4).
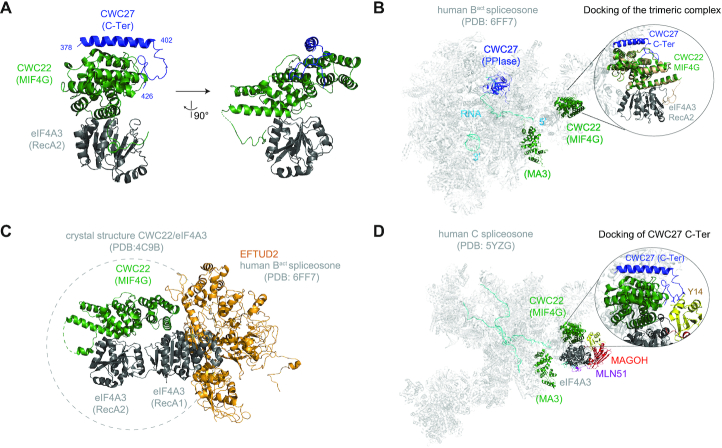
The final model encompasses the MIF4G domain of CWC22 (130–401), the RecA2 domain of eIF4A3 (246–411) and residues 378–426 of CWC27 (Figure 4A). The last five residues of CWC27 (427–431) as well as the N-terminal residues and a loop of CWC22 (116–122 and 142–148) are not visible in the electron density map. The RecA2 domain of eIF4A3 contacts CWC22 MIF4G domain, as observed in the previously published CWC22-eIF4A3 crystal structure. On the opposite side of the MIF4G domain, residues 378–402 of CWC27 form an extended helix that packs against a groove formed by a three alpha helices bundle of CWC22 (Figure 4A). The CWC27 C-terminal domain residues in contact with CWC22 are evolutionary conserved from yeast to mammals (Supplementary Figures S4A and S4b). The long CWC27 helix is followed by a loop (402–426) that folds around one side of MIF4G domain of CWC22 (Figure 4A). No direct contacts between eIF4A3 and CWC27 are detected.
Figure 4.
3D structure of the complex CWC27/CWC22/eIF4A3 (A) Cartoon representations of the complex, shown in two orientations. Human CWC27 is in blue, human CWC22 MIF4G domain is in green and human eIF4A3 RecA2 domain is in dark gray. This and all other cartoon drawings were generated using PyMOL (http://www.pymol.org/). (B) Cartoon representation of human Bact spliceosome (PDB: 6FF7). The PPIase domain of CWC27 is in blue, the MIF4G and MA3 domains of CWC22 in green, the spliceosome in light gray and pre-mRNA in cyan. Zoom-in shows the docking of CWC27/CWC22/eIF4A3 structure in which CWC22 MIF4G domain is in wheat, while CWC27 and eIF4A3 RecA2 domain are colored as in (A). (C) Cartoon representation of CWC22/eIF4A3 (PDB: 4C9B) docked onto the structure of the Bact spliceosome (PDB: 6FF7). eIF4A3 RecA1 domain clashes with the EFTUD2 spliceosomal protein. EFTUD2 is in orange, CWC22 in green, eIF4A3 in dark gray. (D) Cartoon representation of human C spliceosome (PDB: 5YZG). Zoom-in shows the docking of CWC27, highlighting the clash between CWC27 loop and the EJC core protein Y14. MLN51 is in purple, MAGOH in red, Y14 in yellow, eIF4A3 in dark gray, CWC27 in blue and CWC22 in green.
CWC22 MIF4G and MA3 domains, as well as CWC27 PPIase N-terminal domain are clearly observed in cryo-EM structures of human Bact spliceosomes (3,5) (Figure 4B). However, neither the C-terminal region of CWC27 (427–472) nor eIF4A3 are visible in these structures. The MIF4G domain in our new structure was perfectly aligned to the one in the Bact spliceosome structure (3), and docking of the entire CWC27/CWC22/eIF4A3 new structure shows no particular clashes (Figure 4B). We then wanted to investigate what conformation could assume eIF4A3 in Bact spliceosomes. We aligned the CWC22 MIF4G domain of the CWC22/eIF4A3 crystal structure (40) (PDB: 4C9B) to the one present in the Bact spliceosome cryo-EM structure (3) (PDB: 6FF7). The RecA1 domain of eIF4A3 clashes with the spliceosomal factor EFTUD2 (Figure 4C), indicating that eIF4A3 must adopt in the spliceosome a closer conformation than the open conformation observed in the crystal structure of CWC22/eIF4A3.
In the C complex, after the first catalytic splicing reaction, CWC27 is no longer present and the MIF4G domain of CWC22 contacts the EJC, which is assembled onto mRNAs around 27 nt upstream the exon-exon junction (41). We docked our structure to that of the C spliceosome (PDB: 5YZG) and found that the loop of CWC27 clashes with the Y14 EJC subunit (Figure 4D). This is consistent with the fact that CWC27 leaves the spliceosome before EJC assembly. Our new data indicate that CWC27 is another player in eIF4A3 recruitment, with our structure illustrating the early contacts of eIF4A3 with Bact spliceosomes.
CWC27 and CWC22 knock-down in retinal cells impact common pathways linked to inflammation
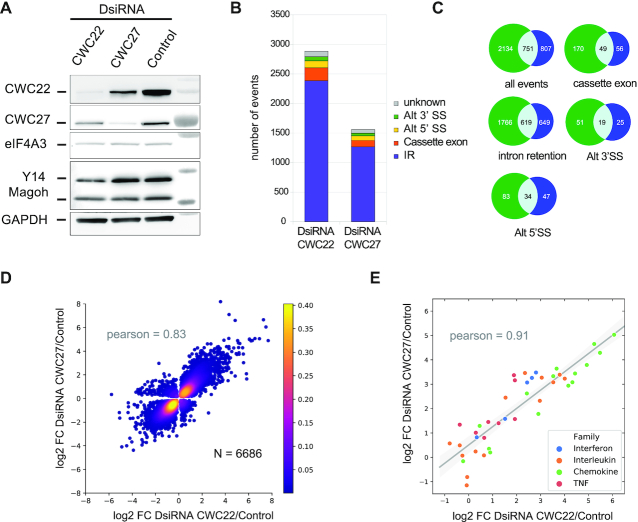
Patients with genetic mutations in CWC27 are prone to retinal degeneration (8). To explore the impact of CWC27 depletion on gene expression, we performed siRNA knock-down (KD) on immortalized hTERT RPE-1 cells from human retinal pigment epithelium. These cells express CWC22, CWC27 and eIF4A3 correctly, and both EJC and the ternary complex CWC22/CWC27/eIF4A3 are detected as shown by co-immunoprecipitation of endogenous proteins (Supplementary Figure S5A). We performed separate KD of CWC27 and CWC22 followed by large-scale sequencing of mRNAs. eIF4A3 KD was not investigated as it resulted in rapid cell death within a few hours of treatment. The efficiency of CWC27 and CWC22 down-regulation was checked by RT-qPCR (Supplementary Figure S5B) and Western Blot analysis (Figure 5A). Interestingly, CWC27 KD reduces CWC22 protein levels and vice versa, while their respective transcripts are not affected. This shows that each protein stabilizes its partner, further supporting that the two proteins interact together in vivo. We sequenced KDs and control samples in triplicate with paired-end Illumina sequencing and obtained between 33 and 40 million reads per sample. After read mapping on hg38 with STAR (27), principal component analysis on gene counts showed clustering of the samples according to their experimental conditions (Supplementary Figure S5C). This, as well as visual inspection of read counts on IGV (29), validated sample reproducibility as well as the quality of library preparation and sequencing.
Figure 5.
Impact of CWC27 and CWC22 KD on gene expression in RPE1 cells. (A) Western blots of CWC22 or CWC27 from hTERT RPE-1 cells treated with the corresponding DsiRNA. Detected proteins are indicated on the left. Molecular size markers are on the right. (B) Classification of alternative splicing events induced by CWC22 and CWC27 KDs. Intron retention (IR) events were identified with iREAD (30), using a minimum fold change of 2 (P-value < 0.05). Other splicing events were detected with JSplice (31), using a minimum fold change of 1.5 (P-value < 0.05). Alternative 3′- and 5′-splice sites: alt 3′ SS and alt 5′ SS, respectively. Unknown corresponds to complex splicing defects involving several junctions classified as unknown by JSplice. (C) Venn diagrams showing the overlap between significant splicing events changes in CWC22 (green) and CWC27 (blue) KDs. (D) Logarithmic fold change (log2 FC) of genes significantly up- or down- regulated (N = 6686, P-value < 0.05) in both CWC22 and CWC27 KD compared to control cells. Pearson correlation coefficient is 0.83 (P-value < 0.001). (E) Logarithmic fold change (log2 FC) of for expression of all detected cytokines in CWC22 and CWC27 KD compared to control cells. Linear regression (gray line) with 95% confidence interval of squared residuals (gray shade). Pearson correlation coefficient is 0.91 (P-value < 10−16).
Since CWC22 and CWC27 are spliceosomal proteins, we first examined the impact of KD on splicing. 2385 and 1268 introns were significantly (P-value < 0.05, fold change > 1.5) more retained in CWC22 and CWC27 KD respectively (Figure 5B). About half of the retained introns (619) after CWC27 KD were also retained after CWC22 KD (Figure 5C). CWC22 and CWC27 KD affected 500 and 290 alternative splicing events respectively (fold change > 1.5; figure 5B and C). Of these events, 40% (132) were common to both CWC27 and CWC22 KD (Figure 5C). These results show that both proteins are involved in common splicing events.
The expression of 2040 and 1701 genes increased, and that of 1176 and 1526 decreased significantly (P-value<0.05, fold change > 2) in CWC27 and CWC22 KD, respectively. Changes in gene expression in CWC27 KD were highly correlated to changes in CWC22 KD (Pearson = 0.83, P-value < 0.001) (Figure 5D). To annotate gene function, we performed a GO analysis on significantly up-regulated genes (P-value < 0.05) from both samples. Interestingly, they show enrichment in genes related to inflammation (Supplementary Table S5 and S6). Among the 10 most up-regulated genes in CWC27 KD (>30-fold), eight are linked to inflammation (Supplementary Table S7). Moreover, half of the 122 genes up-regulated >10-fold have a pro-inflammatory function, they correspond to cytokines (interferons, chemokines, members of the tumor necrosis factor super family), chemokine receptors, adhesion molecules, interferon-inducible transcripts, inflammasome components, inflammatory pathway modulators and actors of antigen presentation (Supplementary Table S7). The same genes are also up-regulated following CWC22 KD. For instance, all detected cytokines are up-regulated in both CWC27 and CWC22 KD with a fairly good correlation (Pearson coefficient of 0.91 and an associated P-value < 0.001) (Figure 5E). Among the 67 genes down-regulated >5-fold following CWC27 KD (Supplementary Table S8), some belong to the transforming growth factor beta signalling cascade, others are linked to the actin cytoskeleton, others are mitochondrial encoded transcripts for oxidative phosphorylation enzymes. The majority of these genes are also down-regulated following CWC22 KD (Supplementary Table S8). Together, our results show that CWC27 KD has a wide impact on gene expression. Moreover, down-regulated and up-regulated genes as well as alternative splicing events follow the same trend after CWC22 KD, indicating that common pathways are targeted by both proteins and that both proteins are physically and functionally linked.
DISCUSSION
In this study, we provide new structural and functional insights into the splicing factor CWC27. In both yeast and human, CWC27 is composed of an inactive PPIase domain followed by a long and disordered region. The PPIase domain of CWC27 is the only part of CWC27 visible in spliceosome cryo-EM structures. It plays a conserved role during spliceosome assembly, as it is positioned identically in yeast (4) and human (3,5) Bact spliceosomes, and it is released concomitantly with the RNF113A protein (CWC24 in yeast) during the B to B* spliceosome conversion (5,6). Prior to our work, little was known about the long C-terminal region of CWC27. In humans, truncations of this region are associated with retinitis pigmentosa and developmental defects (8). Here, we show that the C-terminus of human CWC27 directly contacts its splicing partner CWC22 and together, these proteins offer a landing platform for the EJC core component eIF4A3.
We find that CWC22 is the main protein interacting with CWC27 in cell lysates. By biochemical and structural approaches, we showed that the interaction between the two proteins is direct and mediated by the C-terminus of CWC27 and the MIF4G domain of CWC22. Our evidences strongly suggest that the CWC22/CWC27 heterodimer can be considered as a building block because it exists independently of the spliceosome: (i) CWC22 is by far the major protein enriched in CWC27 immunoprecipitations, (ii) each protein stabilizes the other and (iii) both proteins are stably integrated in the spliceosome during the transition from B to Bact (3,5). Thus, we propose that CWC22 and CWC27 are recruited to Bact spliceosomes as a heterodimer.
CWC22 had previously been proposed to bind the EJC subunit eIF4A3 and escort it to the spliceosome (9,10,38). Here, we show that a small proportion of cellular CWC27 is bound to both CWC22 and eIF4A3. We were able to identify the interacting domains and to reconstitute a ternary complex containing CWC22 MIF4G domain, CWC27 C-terminal domain and eIF4A3. We solved the 3D structure of this complex and found that contacts between the CWC22 MIF4G and the eIF4A3 RecA2 domains are kept almost identical to those previously seen in our CWC22/eIF4A3 structure (40). The CWC27 C-terminal sequence (378–402) folds into a helix that tightly binds the MIF4G domain on the side opposite to eIF4A3 RecA2 domain. This C-terminal helix is followed by a loop (402–426) that packs into a groove on the CWC22 surface. Both eIF4A3 RecA1 domain and the last C-terminal 46 amino acids of CWC27 are not present in our structure. In the absence of direct contact between CWC27 and eIF4A3, how CWC27/CWC22 stabilizes the binding of eIF4A3 remains an open question.
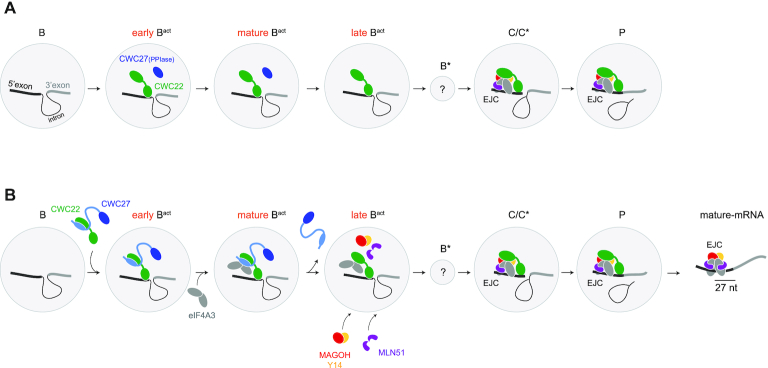
The cryo-EM structures of multiple splicing complexes have been solved and notably the human pre-B, B, early Bact, mature Bact, late Bact, C, C*, P and ILS complexes (2). Neither CWC22, CWC27 nor eIF4A3 are found in B complexes (42,43) (Figure 6A). CWC22 and CWC27 are found in ‘early’ and ‘mature’ (3,5) Bact complex structures. eIF4A3 is not visible in these structures but it was identified by mass spectrometry in Bact spliceosomes isolated by Haselbach and colleagues (Supplementary Figure S4 in Haselbach et al. (3)). Noteworthy, eIF4A3 is the sole EJC subunit copurifying with the Bact complex. Loose interactions with spliceosome complexes might prevent its co-detection in 3D structures. While CWC27 is released before conversion of the ‘mature’ Bact into ‘late’ Bact complex (5), CWC22 remains within the spliceosome from the ‘late’ Bact to the P complex at the end of splicing (Figure 6A). In the structure of C (41), C* (37) and P (44,45) complexes, CWC22 is bound to eIF4A3 within an assembled EJC on its target RNA (Figure 6A).
Figure 6.
Model of EJC assembly by spliceosome (A) Cartoon representation of published cryo-EM structures of human spliceosomal complexes. Spliceosome complexes are represented by gray disks. Proteins involved in EJC core and assembly are colored. CWC27 PPIase domain is in dark blue, CWC22 in green. (B) Model of stepwise EJC assembly onto mRNA. The CWC27 C-terminal region is in light blue, with an ellipse representing the CWC27 alpha-helix interacting with CWC22. See discussion for description.
Based on these data, we propose a new step-wise pathway for EJC core assembly by the splicing machinery (Figure 6B). CWC27 and CWC22 are bound together outside of the spliceosome before being integrated together in the Bact spliceosome in which the heterodimer forms a stable landing platform for eIF4A3. We propose that the ternary complex we found exists as part of the Bact complex because it is the only moment where CWC27, CWC22 and eIF4A3 are present according to the structures of different spliceosomal complexes. We suppose that in the Bact spliceosome eIF4A3 adopts a semi-closed conformation when bound to CWC27 and CWC22, since its open conformation is not compatible with the spliceosome and that the closed formation necessitates of the presence of the other EJC components (46,47). The structure of spliceosomes in which this entire ternary complex is visible, would tell us whether eIF4A3 directly contact both CWC27 and CWC22 and whether other spliceosome components participate to eIF4A3 attachment. When docked on the C spliceosome our structure shows that the loop of CWC27 clashes with the Y14 EJC subunit (Figure 4C). This finding accounts for the release of CWC27 during the transition between Bact to B*, thus allowing the concomitant association of MAGOH/Y14 and MLN51 with eIF4A3 already bound to the spliceosome through the CWC22 MIF4G domain. The binding of CWC22 MA3 domain to mRNA 5′ exon serves to position eIF4A3 clamping to RNA around 27 nt upstream 5′-exon extremity. This pre-EJC is maintained until spliceosome disassembly after P complex (44,45) during which the release of CWC22 allows the complete folding of MLN51 around eIF4A3 and binding to RNA. This overall picture of EJC assembly still contains loopholes as we ignore when exactly MAGOH/Y14 and MLN51 are recruited. Obtaining the missing cryo-EM structure of the B* complex may also help characterize the mechanistic aspects of EJC recruitment and assembly. A complete picture of the recruitment of EJC core subunits to the spliceosome is essential to understand the mechanisms potentially modulating EJC assembly and thus EJC-dependent mRNA destiny.
Changes in gene expression generated in hTERT RPE1 cells by CWC27 KD were highly correlated to those generated by CWC22 KD. These observations strongly support a functional link between the two proteins. hTERT RPE1 cells are immortalized retinal pigmented epithelium cells that are known to contribute to immune and inflammatory responses in the eye (48). The major changes in gene expression following CWC27 KD or CWC22 KD relate to the activation of a pro-inflammatory state. Half of the most up-regulated genes correspond to interleukins that activate all categories of leukocytes as well as adhesion molecules that mediate the migration and adhesion of leukocytes (Supplementary Table S7). Conversely, TGFβ2 (Transforming Growth Factor Beta), one of the most down-regulated genes (Supplementary Table S8), is known to prevent inflammation in the eye (49). Among the most down-regulated genes, we found genes coding for actin, myosin, tropomyosin and transgelin that are associated with the actin cytoskeletal network (Supplementary Table S8). The expression of many down-regulated genes, including some of those associated with the actin cytoskeleton relies upon TGFβ (50). Down-regulation of TGFβ2 can lead to disruption of the actin networks (51). Integrity of the actin cytoskeleton contributes to mitochondrial DNA (mtDNA) maintenance (52) and knocking-out β-actin results in mitochondrial dysfunction characterized by mtDNA accumulation and aggregation of TFAM, a nuclear encoded mitochondrial transcription factor (53). Remarkably, TFAM as well as all 13 mitochondrial-encoded proteins involved in oxidative phosphorylation are down-regulated as a consequence of CWC27 KD or CWC22 KD. A decrease in mitochondrial enzymes involved in oxidative phosphorylation most likely results in decreased ATP production, leading to a reduced protein synthesis capacity and thus a general decrease in ribosomal protein transcripts. Mitochondrial dysfunction can trigger a pro-inflammatory state (54). It generates reactive oxygen species (ROS) and mtDNA activates the AIM2 inflammasome. As a result, mitochondrial dysfunction activates genes in the oxidative stress and pro-inflammatory pathways. We speculate that TGFβ2, actin cytoskeleton associated and mitochondrial-transcript down-regulation contributes to the activation of a pro-inflammatory state.
Mutations in human CWC27 gene have been associated primarily to retinal degeneration and to a spectrum of other phenotypes with various degrees of severity, such as brachydactyly, craniofacial abnormalities, short stature, and neurological defects (8). TGFβ pathway deficiency in the retinal microglia induces inflammatory contributions to retinal degeneration (55). In particular, deficiencies in CTGF and GDF6 downstream the TGF cascade, are associated with retinal dystrophies (56,57). GDF6 is also involved in early mouse cranial development (58). A target knock-out of TFAM in mouse retinal pigment epithelia leads to retinal degeneration (59) and mutations in the MT-ATP6 gene cause the Neuropathy Ataxia Retinitis Pigmentosa (NARP) syndrome (60). Furthermore, patients with defective oxidative phosphorylation are subject to craniofacial anomalies and brachydactyly (61). Given the results of our knock-down experiments and the evidence mentioned above, we hypothesize that TGFβ2 and MT-gene down-regulation contributes to patient phenotypes associated with CWC27 deficiencies.
Spliceosomopathies are genetic disorders associated with mutations in constitutive splicing factors (8,62). Several of them share common phenotypes, including Retinitis Pigmentosa or craniofacial development defects. Our findings suggest inflammation as a possible link to the retinal degeneration associated with CWC27 deficiencies. Future work should investigate how the knock-down of spliceosomal proteins related to Retinitis Pigmentosa compared those related to craniofacial disorders impact transcriptomes. We can suppose that in some specific cell types, the transcriptome is more sensitive to constitutive splicing defects. Much remains to be done to identify precursor mRNAs which processing defects contribute to these pathologies.
DATA AVAILABILITY
Sequencing raw data has been deposited in GEO. The accession number is GSE145872. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145872.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Bertrand, M.J. Moore and X. Morin for plasmids, E. Del Nery for RPE-1 cells and lab members for fruitful discussions. We thank D. Loew, F. Dingli and G. Arras of the Mass Spectrometry laboratory (Institut Curie, Paris, France) and, T. Léger, B. Morlet and C. Garcia of the Mass Spectrometry facility of Institut Jacques Monod (CNRS, Paris, France).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ANR differEnJCe grant [ANR-13-BSV8-0023]; ANR spEJCificity [ANR-17-CE12-0021 to H.L-H.] from the French Agence Nationale de la Recherche; program « Investissements d’Avenir » launched by the French Government and implemented by ANR [ANR-10-LABX-54 MEMOLIFE and ANR-10-IDEX-0001-02 PSL* Research University to V.B. and H.L.H.]; Labex Memolife and the Foundation LNCC (Ligue Nationale Contre le Cancer to V.B.); Centre National de Recherche Scientifique, the Ecole Normale Supérieure and the Institut National de la Santé et de la Recherche Médicale, France. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wahl M.C., Will C.L., Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 2. Wilkinson M.E., Charenton C., Nagai K.. RNA splicing by the spliceosome. Annu. Rev. Biochem. 2019; doi:10.1146/annurev-biochem-091719-064225. [DOI] [PubMed] [Google Scholar]
- 3. Haselbach D., Komarov I., Agafonov D.E., Hartmuth K., Graf B., Dybkov O., Urlaub H., Kastner B., Luhrmann R., Stark H.. Structure and conformational dynamics of the human spliceosomal B(act) complex. Cell. 2018; 172:454–464. [DOI] [PubMed] [Google Scholar]
- 4. Yan C., Wan R., Bai R., Huang G., Shi Y.. Structure of a yeast activated spliceosome at 3.5 A resolution. Science. 2016; 353:904–911. [DOI] [PubMed] [Google Scholar]
- 5. Zhang X., Yan C., Zhan X., Li L., Lei J., Shi Y.. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018; 28:307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wan R., Bai R., Yan C., Lei J., Shi Y.. Structures of the catalytically activated yeast spliceosome reveal the mechanism of branching. Cell. 2019; 177:339–351. [DOI] [PubMed] [Google Scholar]
- 7. Ulrich A., Wahl M.C.. Structure and evolution of the spliceosomal peptidyl-prolyl cis-trans isomerase Cwc27. Acta Crystallogr. D. Biol. Crystallogr. 2014; 70:3110–3123. [DOI] [PubMed] [Google Scholar]
- 8. Xu M., Xie Y.A., Abouzeid H., Gordon C.T., Fiorentino A., Sun Z., Lehman A., Osman I.S., Dharmat R., Riveiro-Alvarez R. et al.. Mutations in the spliceosome component CWC27 cause retinal degeneration with or without additional developmental anomalies. Am. J. Hum. Genet. 2017; 100:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barbosa I., Haque N., Fiorini F., Barrandon C., Tomasetto C., Blanchette M., Le Hir H.. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat. Struct. Mol. Biol. 2012; 19:983–990. [DOI] [PubMed] [Google Scholar]
- 10. Steckelberg A.L., Boehm V., Gromadzka A.M., Gehring N.H.. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep. 2012; 2:454–461. [DOI] [PubMed] [Google Scholar]
- 11. Hocq R., Paternina J., Alasseur Q., Genovesio A., Le Hir H.. Monitored eCLIP: high accuracy mapping of RNA-protein interactions. Nucleic Acids Res. 2018; 46:11553–11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le Hir H., Izaurralde E., Maquat L.E., Moore M.J.. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000; 19:6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le Hir H., Sauliere J., Wang Z.. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 2016; 17:41–54. [DOI] [PubMed] [Google Scholar]
- 14. Leung C.S., Johnson T.L.. The exon junction complex: a multitasking guardian of the transcriptome. Mol. Cell. 2018; 72:799–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Merz C., Urlaub H., Will C.L., Luhrmann R.. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007; 13:116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reichert V.L., Le Hir H., Jurica M.S., Moore M.J.. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002; 16:2778–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I.. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002; 12:1–11. [DOI] [PubMed] [Google Scholar]
- 18. Tyanova S., Temu T., Cox J.. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016; 11:2301–2319. [DOI] [PubMed] [Google Scholar]
- 19. Tyanova S., Cox J.. Perseus: A bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol. Biol. 2018; 1711:133–148. [DOI] [PubMed] [Google Scholar]
- 20. Ballut L., Marchadier B., Baguet A., Tomasetto C., Seraphin B., Le Hir H.. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005; 12:861–869. [DOI] [PubMed] [Google Scholar]
- 21. Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Storoni L.C., McCoy A.J., Read R.J.. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:432–438. [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:doi.org/10.14806/ej.17.1.200. [Google Scholar]
- 27. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H.D., Funk C.C., Price N.D.. iREAD: a tool for intron retention detection from RNA-seq data. BMC Genomics. 2020; 21:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christinat Y., Pawlowski R., Krek W.. jSplice: a high-performance method for accurate prediction of alternative splicing events and its application to large-scale renal cancer transcriptome data. Bioinformatics. 2016; 32:2111–2119. [DOI] [PubMed] [Google Scholar]
- 32. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eden E., Lipson D., Yogev S., Yakhini Z.. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007; 3:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Supek F., Bosnjak M., Skunca N., Smuc T.. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bertram K., Agafonov D.E., Liu W.T., Dybkov O., Will C.L., Hartmuth K., Urlaub H., Kastner B., Stark H., Luhrmann R.. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017; 542:318–323. [DOI] [PubMed] [Google Scholar]
- 37. Zhang X., Yan C., Hang J., Finci L.I., Lei J., Shi Y.. An atomic structure of the human spliceosome. Cell. 2017; 169:918–929. [DOI] [PubMed] [Google Scholar]
- 38. Alexandrov A., Colognori D., Shu M.D., Steitz J.A.. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. PNAS. 2012; 109:21313–21318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Favaro F.P., Alvizi L., Zechi-Ceide R.M., Bertola D., Felix T.M., de Souza J., Raskin S., Twigg S.R., Weiner A.M., Armas P. et al.. A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am. J. Hum. Genet. 2014; 94:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buchwald G., Schussler S., Basquin C., Le Hir H., Conti E.. Crystal structure of the human eIF4AIII-CWC22 complex shows how a DEAD-box protein is inhibited by a MIF4G domain. PNAS. 2013; 110:E4611–E4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhan X., Yan C., Zhang X., Lei J., Shi Y.. Structure of a human catalytic step I spliceosome. Science. 2018; 359:537–545. [DOI] [PubMed] [Google Scholar]
- 42. Bertram K., Agafonov D.E., Dybkov O., Haselbach D., Leelaram M.N., Will C.L., Urlaub H., Kastner B., Luhrmann R., Stark H.. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell. 2017; 170:701–713. [DOI] [PubMed] [Google Scholar]
- 43. Zhan X., Yan C., Zhang X., Lei J., Shi Y.. Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res. 2018; 28:1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fica S.M., Oubridge C., Wilkinson M.E., Newman A.J., Nagai K.. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science. 2019; 363:710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X., Zhan X., Yan C., Zhang W., Liu D., Lei J., Shi Y.. Structures of the human spliceosomes before and after release of the ligated exon. Cell Res. 2019; 29:274–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andersen C.B., Ballut L., Johansen J.S., Chamieh H., Nielsen K.H., Oliveira C.L., Pedersen J.S., Seraphin B., Le Hir H., Andersen G.R.. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006; 313:1968–1972. [DOI] [PubMed] [Google Scholar]
- 47. Bono F., Ebert J., Lorentzen E., Conti E.. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006; 126:713–725. [DOI] [PubMed] [Google Scholar]
- 48. Holtkamp G.M., Kijlstra A., Peek R., de Vos A.F.. Retinal pigment epithelium-immune system interactions: cytokine production and cytokine-induced changes. Prog. Retin. Eye Res. 2001; 20:29–48. [DOI] [PubMed] [Google Scholar]
- 49. Zamiri P., Sugita S., Streilein J.W.. Immunosuppressive properties of the pigmented epithelial cells and the subretinal space. Chem. Immunol. Allergy. 2007; 92:86–93. [DOI] [PubMed] [Google Scholar]
- 50. Kubo E., Shibata S., Shibata T., Kiyokawa E., Sasaki H., Singh D.P.. FGF2 antagonizes aberrant TGFbeta regulation of tropomyosin: role for posterior capsule opacity. J. Cell. Mol. Med. 2017; 21:916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Montecchi-Palmer M., Bermudez J.Y., Webber H.C., Patel G.C., Clark A.F., Mao W.. TGFbeta2 induces the formation of cross-linked actin networks (CLANs) in human trabecular meshwork cells through the smad and non-smad dependent pathways. Invest. Ophthalmol. Vis. Sci. 2017; 58:1288–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reyes A., He J., Mao C.C., Bailey L.J., Di Re M., Sembongi H., Kazak L., Dzionek K., Holmes J.B., Cluett T.J. et al.. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011; 39:5098–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kang I., Chu C.T., Kaufman B.A.. The mitochondrial transcription factor TFAM in neurodegeneration: emerging evidence and mechanisms. FEBS Lett. 2018; 592:793–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. West A.P. Mitochondrial dysfunction as a trigger of innate immune responses and inflammation. Toxicology. 2017; 391:54–63. [DOI] [PubMed] [Google Scholar]
- 55. Ma W., Silverman S.M., Zhao L., Villasmil R., Campos M.M., Amaral J., Wong W.T.. Absence of TGFbeta signaling in retinal microglia induces retinal degeneration and exacerbates choroidal neovascularization. Elife. 2019; 8:e42049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Asai-Coakwell M., March L., Dai X.H., Duval M., Lopez I., French C.R., Famulski J., De Baere E., Francis P.J., Sundaresan P. et al.. Contribution of growth differentiation factor 6-dependent cell survival to early-onset retinal dystrophies. Hum. Mol. Genet. 2013; 22:1432–1442. [DOI] [PubMed] [Google Scholar]
- 57. Ma T., Dong L.J., Du X.L., Niu R., Hu B.J.. Research progress on the role of connective tissue growth factor in fibrosis of diabetic retinopathy. Int. J. Ophthalmol. 2018; 11:1550–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Clendenning D.E., Mortlock D.P.. The BMP ligand Gdf6 prevents differentiation of coronal suture mesenchyme in early cranial development. PLoS One. 2012; 7:e36789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., Ahern K., Snyder M., Bok D., Dunaief J.L. et al.. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 2011; 121:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lefevere E., Toft-Kehler A.K., Vohra R., Kolko M., Moons L., Van Hove I.. Mitochondrial dysfunction underlying outer retinal diseases. Mitochondrion. 2017; 36:66–76. [DOI] [PubMed] [Google Scholar]
- 61. Cormier-Daire V., Rustin P., Rotig A., Chretien D., Le Merrer M., Belli D., Le Goff A., Hubert P., Ricour C., Munnich A.. Craniofacial anomalies and malformations in respiratory chain deficiency. Am. J. Med. Genet. 1996; 66:457–463. [DOI] [PubMed] [Google Scholar]
- 62. Lehalle D., Wieczorek D., Zechi-Ceide R.M., Passos-Bueno M.R., Lyonnet S., Amiel J., Gordon C.T.. A review of craniofacial disorders caused by spliceosomal defects. Clin. Genet. 2015; 88:405–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing raw data has been deposited in GEO. The accession number is GSE145872. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145872.