Abstract
DNA origami is a promising molecular delivery system for a variety of therapeutic applications including cancer therapy, given its capability to fabricate homogeneous nanostructures whose physicochemical properties (size, shape, surface chemistry) can be precisely tailored. However, the correlation between DNA-origami design and internalization efficiency in different cancer cell lines remains elusive. We investigated the cellular uptake of four DNA-origami nanostructures (DONs) with programmed sizes and shapes in multiple human cancer cell lines. The cellular uptake efficiency of DONs was influenced by size, shape, and cell line. Scavenger receptors were responsible for the internalization of DONs into cancer cells. We observed distinct stages of the internalization process of a gold nanoparticle (AuNP)-tagged rod-shape DON, using high-resolution transmission electron microscopy. This study provides detailed understanding of cellular uptake and intracellular trafficking of DONs in cancer cells, and offers new insights for future optimization of DON-based drug delivery systems for cancer treatment.
Graphical Abstract
INTRODUCTION
Most approved drugs rely on passive delivery for uptake into cells, which largely undermines their therapeutic efficacy and raises off-target-associated toxicity issues. Effective and safe delivery of therapeutics into targeted cells has remained a challenging issue in modern medicine. One strategy to address this challenge is to use nanostructures as delivery vehicles to enable active transportation of drugs.1 Various materials have been explored to formulate nanoparticles for delivery, including liposomes, inorganic materials, and organic polymers.2–5 However, the ability to precisely control nanoparticle size, shape and surface chemistry using these materials is limited, which hinders the systematic investigation of cellular internalization and optimization of delivery performance.
Structural DNA nanotechnology,6 especially DNA origami,7 has demonstrated unprecedented abilities to construct uniform DNA nanostructures with prescribed size, shape, and surface functionality.8–14 DNA origami nanostructures (DONs) have served as robust platforms for a large variety of applications, including nanofabrication,15–20 biosensing,21,22 and drug delivery.23–29 In previous studies, DONs with varied sizes and shapes have been used to efficiently deliver small molecules (e.g., doxorubicin),23–27 proteins (e.g., antibody),28 and therapeutic nucleic acids (e.g., CpG oligonucleotides) into cells,29 but no systematic investigation has been undertaken to determine how DON design (i.e., size and shape) affects cellular uptake in different cell lines. In addition, mechanistic understanding of DONs’ cellular uptake process including the endocytosis pathway and intracellular trafficking remains elusive. Studies addressing these questions may offer critical insights for the design and optimization of DONs to achieve potent delivery of various therapeutics.
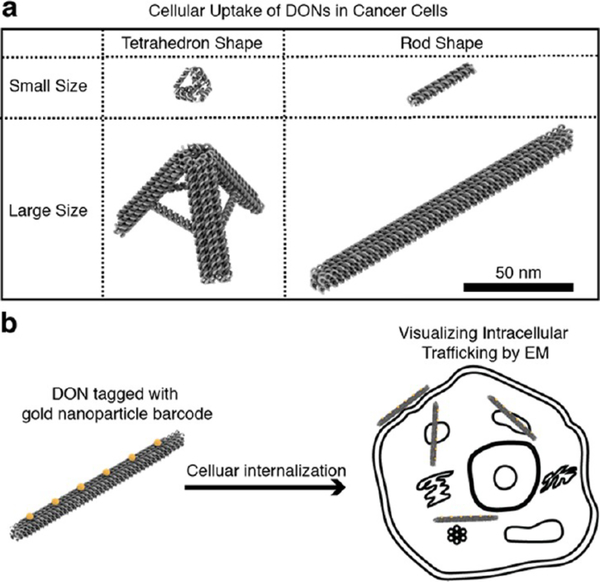
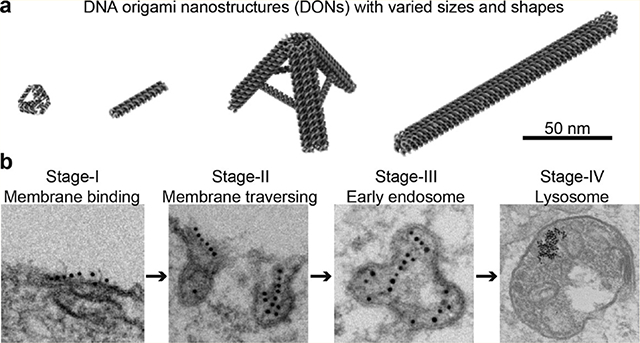
We studied the cellular uptake of a group of tetrahedron and rod shaped DONs with various sizes in two human lung cancer cell lines, H1299 (nonsmall cell lung cancer) and DMS53 (small cell lung cancer), as illustrated in Figure 1a. Lung cancer is the leading cause of cancer-related mortality in the United States,30 which includes nonsmall cell lung cancer and small cell lung cancer.31 Studying the uptake of DONs in both lung cancer cells may facilitate the translation of DNA origami nanotechnology for lung cancer treatment. Our results suggested a clear correlation between cellular uptake efficiency and DON design. Scavenger receptors were found to play an essential role in the uptake of DONs into both lung cell lines. Finally, we demonstrated that transmission electron microcopy (TEM) could be utilized to visualize the DON internalization process in detail by using small gold nanoparticles (AuNPs) to form barcodes on DONs (Figure 1b).
Figure 1.
Study of the cellular uptake and intracellular trafficking of DONs in cancer cells. (a) DONs of different sizes and shapes were studied. (b) Intracellular trafficking of DONs tagged with gold nanoparticle barcode was visualized by electron microscopy.
RESULTS AND DISCUSSIONS
Design, Construction, and Characterization of DONs
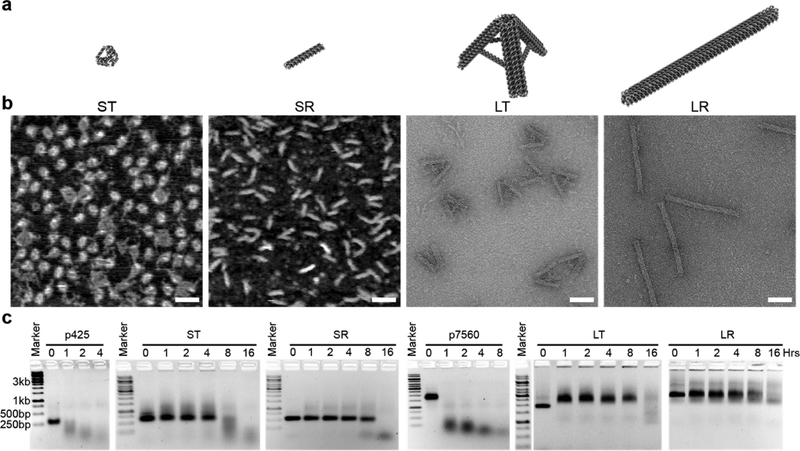
A group of four DONs were designed and constructed, including a three-dimensional (3D) small tetrahedron (ST), a one-dimensional (1D) small rod (SR), a large tripod that closely resembles a tetrahedron (LT), and a large rod (LR), as illustrated in Figure 2a (refer to Table S1 and Figures S1–S5 for design details). The ST and SR had similar molecular weights of ~250 kDa, and the LT and LR had similar molecular weights of ~4.5 megadalton. The LT and LR were constructed from a 7560-nucleotide-long M13 bacteriophage genome DNA (p7560), while the ST and SR were assembled from a custom 425-nucleotide-long single strand (p425) scaffold that was cut from p7560 by restriction enzyme digestion (BsaAI, Figure S6). We estimated the dimensions of each DON by using the parameters of B-form double-helix DNA (2 nm diameter, and 0.33 nm per base in the direction of the helical axis). Each edge of the hollow ST was 11 nm in length with a cross-section of 4 nm by 2 nm. The LT resembled a tripod shape with interarm angle of 60 degrees.17 Each arm of the LT was 47 nm in length with a cross-section of 7.2 nm by 12 nm. Both the SR and LR were solid rod shapes. The SR was 32 nm in length with a cross-section of 4 nm by 4 nm, and the LR was 127 nm in length with a cross-section of 8 nm by 8 nm.
Figure 2.
Design, construction, and characterization of DONs. (a) Schematic illustrations of small tetrahedron (ST), small rod (SR), large tetrahedron (LT), and large rod (LR). (b) AFM images of ST and SR, and TEM images of LT and LR confirmed the prescribed sizes and shapes of DONs agreed well with the designs. Scale bars: 50 nm. (c) Nuclease resistance study of DONs in 10% FBS. DONs were incubated in 10% FBS at 37 °C for 0, 1, 2, 4, 8, or 16 h. After incubation, the integrity of DONs was examined by 1% native agarose gel electrophoresis. All DONs exhibited significantly enhanced resistance to nuclease digestion, in comparison to the single-stranded p425 scaffold and p7560 scaffold.
DONs were prepared following a conventional thermal annealing protocol in aqueous buffer containing 10 mM of Mg2+. After assembly, DONs were initially characterized by native agarose gel electrophoresis. Discrete bands with expected mobility were observed for DONs, suggesting the formation of designed structures (Figure S7). The corresponding bands of DONs were then extracted from the gel for imaging characterization by atomic force microscopy (AFM) or TEM, as shown in Figure 2b and Figure S8, which unambiguously confirmed the formation of the DONs with expected sizes and shapes.
Nucleases are abundant in physiological environments, and are natural predators of nucleic acids that may hinder DONs’ potency as delivery vehicles. Therefore, we examined DONs’ resistance to nuclease digestion prior to cellular experiments. A time-course study (0, 1, 2, 4, 8, 16 h) was conducted by incubating DONs in 10% fetal bovine serum (FBS) at 37 °C. Native agarose gel electrophoresis was employed to examine the integrity of DONs immediately after incubation with FBS (Figure 2c). All DONs demonstrated enhanced resistance to nuclease digestion in comparison to p425 and p7560 scaffold DNAs, indicating that the densely packed DNA bundles in DONs provided protection against nuclease digestion. Scaffold DNAs underwent full degradation within 1 h of incubation within 10% FBS, while DONs remained intact even after 4–8 h. Interestingly, SR, LT, and LR exhibited higher resistance to nuclease digestion than ST, probably due to the fact that each edge of the ST contains only two bundled DNA duplexes, which are more exposed to nuclease attack.
Size, Shape, and Cell Line Dependent Uptake of DONs
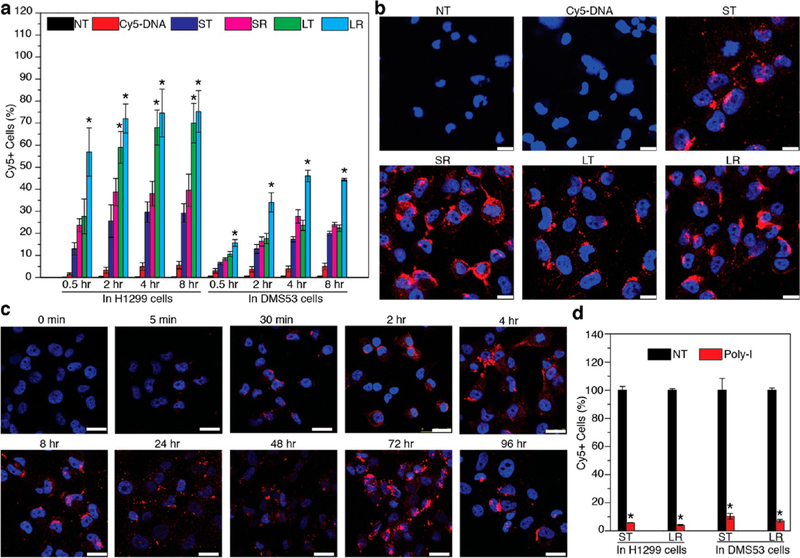
The cellular uptake of DONs was studied in H1299 and DMS53 lung cancer cell lines. A time-course study (0.5, 2, 4, 8 h) was conducted in parallel to examine and compare DONs cellular uptake over time in both cell lines, which were characterized qualitatively and quantitatively by confocal microscopy and flow cytometry, respectively (Figure 3). Along with DONs, nontreated (NT) and Cy5-DNA treated cells were studied as negative controls aiming to validate the delivery capability of DONs. To enable tracking of cellular uptake, multiple copies of Cy5-conjugated DNA (Cy5-DNA) were loaded onto DONs by hybridizing to complementary single-stranded handles on DONs (Table S1, Figure S9). Cy5-DNA was maintained at the same concentration (250 nM) across all DONs for cellular uptake experiments. It was reported that Cy5-conjugated oligonucleotides exhibited strong cellular uptake in multiple mammalian cells, possibly due to the positive charge of cyanine dyes,32,33 suggesting that it may interfere the cellular uptake of DONs. However, Cy5-DNA showed neglectable cellular uptake under our current experimental settings (i.e., 250 nM) as verified both by flow cytometry and confocal microscopy (Figure 3a,b), in agreement to the findings of Mirkin and collegues.34,35 Nevertheless, the cellular uptake of Cy5-DNA became prominent at a concentration of 1 μM (Figure S10), suggesting that a low concentration must be utilized to minimize the interfering effect of Cy5-DNA (e.g., 250 nM in the current study). Cellular uptake experiments were also independently conducted using a noncyanine dye (Alexa-488), and produced similar results (Figure S11).
Figure 3.
Cellular uptake study of DONs in H1299 and DMS53 cells. (a) Flow cytometry analysis of the uptake of DONs into H1299 and DMS53 cells after 0.5, 2, 4, and 8 h. Nontreated (NT) and Cy5-DNA treated cells were included as negative controls. Each column represents the average percentage of cells with Cy-5 positive fluorescence from three independent experiments. Error bars represent standard deviations. *P < 0.05 compared with NT, Cy5-DNA, ST, and SR. (b) Representative confocal microscopy images of DONs in H1299 cells after 8 h of incubation. Strong Cy5 fluorescence in the cytoplasm was observed for cells treated with DONs but not NT and Cy5-DNA controls. Blue: DAPI, red: Cy5. Scale bars: 25 μm. (c) Time-course study of ST uptake into H1299 cells by confocal microscopy. ST was found inside cells at as early as 5 min. Enhanced accumulation of Cy-5 fluorescence in cells was observed with increasing incubation time up to 72 h. Fluorescence was decreased after 96 h of incubation. Scale bars: 25 μm. (d) Uptake of ST and LR in both cell lines with inhibition of scavenger receptors. Significant inhibition (90% reduction) of ST and LR uptake into both H1299 and DMS53 cells was observed for cells treated with Poly-I, which binds and saturates scavenger receptors to block their accessibility to DONs. Each column represents the average percentage of cells with Cy-5 positive fluorescence from three independent experiments. Error bars represent standard deviation. *P < 0.05 compared against NT.
Flow cytometry revealed that all DONs had significantly higher cellular uptake efficiency, in terms of both percentage of Cy5-positive cells (Figure 3a) and fluorescence intensity (Figure S12), than NT and Cy5-DNA at all time points in both cell lines (detailed pairwise statistical analysis are included in Table S2), indicating that DONs served as delivery vehicles that transported Cy5-DNA into cells. This also suggested that Cy5-DNA remained conjugated to DONs before entering cells, since Cy5-DNA alone showed limited cell internalization. Similar cellular uptake was observed while introducing lower or higher amount of DONs to cells, with the exception that Cy5- DNA alone had strong uptake at high concentration (Figure S10). DONs exhibited different internalization behaviors in H1299 and DMS53 cells (Figure 3a, Table S2), with significantly higher cellular uptake efficiency in H1299 cells relative to DMS53 cells over all time points studied.
The influence of size and shape of DONs on cellular uptake was then examined (Figure 3a, Table S2). In H1299 cells, larger DONs (LR and LT) exhibited significantly higher cellular uptake efficiency than smaller DONs (ST and SR). Further comparison indicated that rod-shaped DONs performed better than tetrahedron-shaped DONs of similar sizes. A similar trend was observed in DMS53 cells except that LT was not internalized as well as observed in H1299 cells. These results suggested that both the size and shape of DONs influence cellular uptake. DONs of larger size may induce stronger interactions with the cell membrane that lead to higher cellular uptake efficiency. Tetrahedron-shaped DONs may not interact as strongly with cells as rod-shaped DONs due to their 3D configuration. Interestingly, though without well-defined 3D shape, p7560 scaffold DNA showed comparable cellular uptake as its DON products, while p425 scaffold DNA had minimal uptake (Figure S11 and Figure S13). This maybe attributed to the long and circular nature of p7560 scaffold DNA that may randomly coil into certain structures enabling cell uptake.
Quantitative polymerase chain reaction (qPCR) was utilized to quantify the intracellular DON numbers, following a methodology recently established by Kjems and colleagues.36,37 qPCR revealed similar cellular uptake trend to flow cytometry (Figure S14) with larger and rod-shaped DONs had higher numbers per cell than their smaller and tetrahedron-shaped counterparts, respectively. Although qPCR was quite sensitive and accurate on detecting DNAs, the acquired DON numbers within cells were underestimated given the loss of DNA prior to qPCR reactions, including loss due to intracellular nuclease degradation, cellular DNA extraction, and ethanol precipitation.
Confocal microscopy was used to visualize the cellular uptake of DONs (Figure 3b, Figure S15). Intense Cy5 fluorescence was observed for all DONs after 8 h of incubation in both cell lines. In contrast, cells with no treatment or treated with Cy5- DNA had no observable fluorescence, confirming that Cy5- DNA did not enter cells as well as DONs, in good agreement to flow cytometry studies. Observed fluorescent signals were predominantly localized in the cytoplasm, as confirmed by costaining of nuclei with DAPI (Figure 3b, Figure S15, Figure S16). The nonuniform distribution of Cy5 fluorescence in the cytoplasm may be attributed to DONs aggregation or segregation within certain cellular organelles (e.g., endosome or lysosome). A colocalization study of SR in H1299 cells revealed partial intracellular overlap between Cy5 and the lysosome after 8 h of incubation (Figure S17). In contrast, previous reports found the lysosome to be the primary compartment in which DONs were localized.23,24,38 The difference in observations may be attributed to differences in cell line and fluorophore. An extended time-course study of ST in H1299 cells between 5 min and 96 h revealed that the cellular uptake of ST began within 5 min, and accumulation gradually increased up to 72 h (Figure 3c). The decrease in Cy- 5 fluorescence observed at 96 h may result from increased cell proliferation or from exocytosis of the Cy-5 fluorophore.
Scavenger Receptor-Mediated Cellular Uptake of DONs
Scavenger receptors are known to mediate the endocytosis of certain polyanionic ligands, including nucleic acids.39,40 Mirkin et al. reported that scavenger receptors play important roles in the cellular uptake of spherical nucleic acids in multiple mammalian cell lines.34,40 Biocca et al. reported that scavenger receptors are essential for COS fibroblast cells to internalize octahedral DNA nanocages.41 Therefore, a mechanistic study was performed to assess the involvement of scavenger receptors in the uptake of DONs into H1299 and DMS53 cells. Polyinosine (Poly-I) was used to competitively bind and saturate scavenger receptors and thus minimize their accessibility to DONs. For both DONs studied (ST and LR), flow cytometry showed 90% reduction in uptake by cells treated with Poly-I compared to nontreated cells (Figure 3d), indicating that scavenger receptors are responsible for the uptake of DONs by both H1299 and DMS53 cells.
The roles of clathrin- and caveolin-dependent uptake pathways were also investigated, as they have previously been implicated to mediate the endocytosis of nanoparticles.42 Fan et al. reported the uptake of small DNA tetrahedral structures into HeLa cells through a caveolin-dependent endocytosis path- way.43 We examined the involvement of caveolin in the uptake of DONs by HeLa cells, using the above-reported small tetrahedral structure (ST-CONT) as a control. Pharmacological inhibition studies showed that ST, LR, and ST-CONT all exhibited caveolin-dependent internalization in HeLa cells since the treatment of cells with methyl-β-cyclodextrin (MβCD) resulted in ~50% reduction in uptake, whereas the blockade of other pathways (i.e., clathrin and phagocytosis) did not affect cellular uptake (Figure S18a). Poly-I treatment of HeLa cells resulted in ~50% decrease in the uptake of DONs and ST-CONT, indicating that HeLa cells also rely on scavenger receptors to internalize DONs. Similar studies were conducted in H1299 and DMS53 cells (Figure S18b). Surprisingly, considerable cytotoxicity was observed in H1299 and DMS53 cells treated with the pharmacological inhibitors at the recommended conditions (data not shown).34,40,43 Inhibitors were diluted significantly in order to maintain apparent cell viability, but no inhibition of cellular uptake of DONs was observed at such conditions (Figure S18c), possibly due to insufficient blockade of targeting pathways at the reduced concentrations.
Visualizing the Cell Internalization Process of DONs by TEM
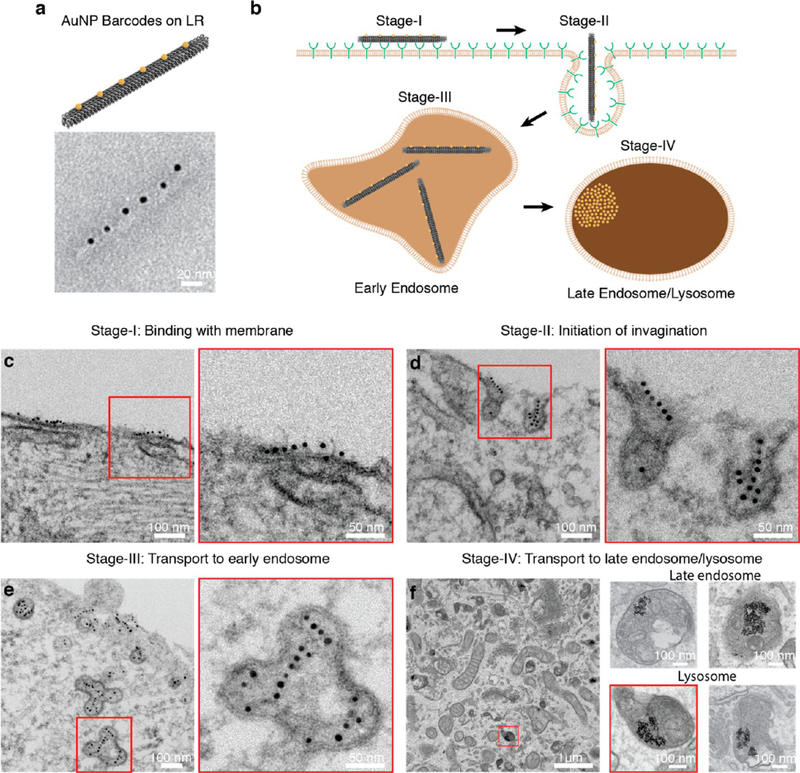
The cell internalization process of DONs was directly visualized at a single particle level using high-resolution TEM imaging techniques. Routinely used fluorescent microscopy imaging methods provide limited information regarding the behaviors of individual nanoparticles due to relatively low resolution. Herein, the visualization of individual DON under TEM was realized by tagging and barcoding a number of 5 nm (diameter) AuNPs onto DONs to form beads-on-a-string structures (Figure 4a). Considering that AuNPs may interfere the cellular uptake of DONs as DNA-conjugated AuNPs may be readily taken up by cells, we tagged small AuNPs (5 nm) onto the largest DON design (LR) aiming to minimize the interference from AuNPs. The majority of LRs had 6 AuNPs aligned on one side of the LR, as confirmed by gel electrophoresis and TEM imaging (Figure S19). The barcoded LRs were purified from the gel and then incubated with H1299 cells for varied periods of time (0.5, 2, 8, 12, 24, and 48 h) with an AuNP concentration of 2.5 nM. Internalization was terminated by removing the medium and fixing the cells. Fixed cells were sliced into thin sections (~80 nm) and stained for TEM imaging. Based on TEM observations, we proposed that it may involve four stages during the internalization of LR into H1299 cells, as illustrated in the schematic model (Figure 4b): Stage I: binding with membrane; Stage II: initiation of invagination; Stage III: transport to early endosome; and Stage IV: transport to late endosome/lysosome.
Figure 4.
Visualization of the LR cell internalization process in H1299 cells by transmission electron microscopy. (a) 5 nm (diameter) AuNPs were tagged onto LR to form a beads-on-a-string barcoded structure. (b) Proposed model illustrating the internalization process of LRs into H1299 cells based on TEM observations. Four distinct stages were identified in the internalization process of LRs into H1299 cells. (c) Stage I: LRs aligned onto cell membrane longitudinally. The longitudinal alignment may maximize the interaction between LRs and the cell membrane since more scavenger receptors are involved in this orientation. (d) Stage II: cell membrane invagination was initiated to internalize LRs with a transverse orientation. This transverse orientation may help to minimize energy expense by generating smaller membrane invaginations. (e) Stage III: LRs were transported into early endosome-like compartments. The nonspherical, tubular-vesicular like organelles closely resemble early endosomes. At this stage, AuNP barcodes stayed relatively intact, suggesting no severe degradation of LRs had occurred. (f) Stage IV: AuNPs accumulated in late endosome and lysosome-like organelles. At this stage, all AuNPs were clustered within cellular compartments that resemble late endosomes (right panel, top) and lysosomes (right panel, bottom). AuNP barcodes were not identifiable, suggesting that AuNPs may have detached from LRs. AuNPs were not found in other cellular compartments (e.g., golgi, mitochondria), cytosol, or nucleus.
Stage I: Binding with Membrane
LRs initially aligned longitudinally onto the cellular membrane (Figure 4c, Figure S20a). Considering our finding that scavenger receptors on the membrane are responsible for recognizing and capturing DONs from the environment, the longitudinal alignment of LRs may allow maximal interaction with the cells since more scavenger receptors are available for capturing LRs than in the transverse orientation. 44
Stage II: Initiation of Invagination
Once LRs were captured and aligned onto the membrane, the cell membrane started to invaginate to internalize LRs (Figure 4d, Figure S20b). Interestingly, LRs were found to rotate 90 deg to enter the membrane at a transverse orientation. This orientation may minimize energy expense by generating smaller invagination flasks. The same phenomenon was observed in the cell internalization process of gold nanorods.44 Both studies suggested that nanoparticles of high aspect ratio might enter through a similar process: maximizing capturing efficiency via longitudinal alignment, followed by minimizing energy expense via transverse entry. Membrane invaginations appeared flaskshaped, with sizes between 50 and 100 nm, closely resembling caveolae,34,42 suggesting that LRs may enter H1299 cells through a caveolin-dependent pathway.
Stage III: Transport to Early Endosome
After traversing the membrane, LRs were observed in nonspherical, tubular- vesicular organelles, which resembled early endosomes (Figure 4e, Figure S20c). Early endosomes are known as the first endocytic compartments to accept and sort incoming cargoes internalized through receptors.42 LRs remained relatively intact within early endosomes as AuNP barcoding patterns were still identifiable, although with a number of AuNPs missing from LRs. This suggests that severe degradation of LRs had not occurred at this stage, likely due to the relatively mild environment within early endosomes.
Stage IV: Transport to Late Endosome/Lysosome
LRs were transported to the late endosome and lysosome-like cellular compartments (Figure 4f,Figure S20d). The multivesicular body-like organelles containing internal vesicles (Figure 4f, right panel, top row) resemble late endosomes. Organelles with no identifiable vesicles in the lumen were assumed to be lysosomes. At this stage, no barcoding patterns of AuNPs were observed. Instead, AuNPs were found randomly clustered within organelles. Given the highly acidic and enzymatic environment within late endosomes and lysosomes, LRs were expected to undergo severe degradation over time and AuNPs would eventually detach, disrupting the beads-on-a- string pattern. No AuNPs were observed in the nucleus, cytosol, or other cellular compartments (e.g., Golgi, mitochondria), indicating that both free AuNPs and those tagged on LRs had limited ability to escape from endosomes.
The time-course study revealed that later internalization stages became increasingly prominent with extended incubation time (Figure S21). For instance, no barcoded LRs were observed internalizing or internalized into the cell at 0.5 h, except for a few individual AuNPs that may have dissociated from the LRs. In contrast, all AuNPs were located within late endosomes and lysosomes after 48 h of incubation, and no earlier stages of internalization were observed. However, at 24 h of incubation, AuNP-barcoded LRs were observed undergoing each stage of the internalization process. These representative images of four stages were primarily selected from cells incubated with LRs for 24 h. In contrast, no barcoded AuNP structures were observed when cells were incubated with individual AuNPs (Figure S22), which further confirmed that AuNP barcodes in the cells did not originate from random alignment of discrete AuNPs.
CONCLUSION
In summary, a parallel study was performed to investigate the cellular uptake of a group of four DONs with varied sizes and shapes in multiple human cancer cell lines. All DONs demonstrated significantly higher cellular uptake efficiency than Cy5-DNA alone in both H1299 and DMS53 cells, confirming the potent delivery capability of these DONs. DONs exhibited drastically different uptake behaviors between cell lines, e.g., DONs had significantly higher uptake efficiency in H1299 cells than in DMS53 cells. The size and shape of DONs were relevant in determining cellular uptake efficiencies. In general, larger DONs exhibited higher cell uptake efficiency, presumably by involving a higher number of receptors on the cell membrane. Shape is also relevant as it may affect the interaction orientations between DONs and cells. Scavenger receptors were found critical in mediating the uptake of DONs. However, no specific endocytosis pathway (clathrin-dependent versus caveolin-dependent) was identified in either H1299 or DMS53 cells, possibly due to the cell lines studied being less tolerant to pharmacological inhibitors as severe cytotoxicity was observed under recommended experimental conditions. Nevertheless, DONs’ internalization into HeLa cells was found to be caveolin-dependent, in agreement with previous studies.43 We successfully barcoded AuNPs onto LRs to enable high-resolution visualization of cellular uptake and intracellular trafficking at a single particle level by TEM imaging, which may serve as a general methodology to track DONs in vitro or even in vivo. Four distinct stages of LR internalization were observed in H1299 cells using this methodology. LRs undergo a process of aligning onto the membrane longitudinally, traversing the membrane transversely by rotating 90°, transporting to early endosomes, and eventually transporting to late endosomes and lysosomes. No AuNPs were observed to escape from endosomes or lysosomes into the cytosol, which might present a challenge in utilizing DONs as delivery vehicles for cargoes that require cytosol or nuclei transportation. The findings from this study may provide mechanistic insight for the design and optimization of DONs and other nanoparticle vehicles for the delivery of drugs and other substrates for biomedical applications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the NCI R21EB022828-01A1 to D.M.S. and Y.K., and Winship Cancer Institute grant #IRG-14-188-0 from the American Cancer Society to Y.K. The EM data was collected on a Hitachi HT7700 120 kv TEM supported by the Georgia Clinical and Translational Science Alliance under award number UL1TR002378 to the Robert P. Apkarian Integrated Electron Microsocpy Core at the Emory College of Arts and Sciences and the Emory University School of Medicine. This research used an AFM supported by NIH grants GM084070 and 3R01GM084070-07S1 to Laura Finzi in the Department of Physics at Emory University. We thank Dr. Anthea Hammond for her assistance in critical reading and editing of the manuscript.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b09024.
Designs of DONs, experimental methods, additional data, and strand sequences (PDF)
REFERENCES
- (1).Petros RA; DeSimone JM Nat. Rev. Drug Discovery 2010, 9 (8), 615–627. [DOI] [PubMed] [Google Scholar]
- (2).Lv H; Zhang S; Wang B; Cui S; Yan JJ Controlled Release 2006, 114 (l), 100–109. [DOI] [PubMed] [Google Scholar]
- (3).Rosi NL; Giljohann DA; Thaxton CS; Lytton-Jean AK; Han MS; Mirkin CA Science 2006, 312 (5776), 1027–1030. [DOI] [PubMed] [Google Scholar]
- (4).Peer D; Karp JM; Hong S; FarokHzad OC; Margalit R; Langer R Nat. Nanotechnol. 2007, 2 (12), 751–760. [DOI] [PubMed] [Google Scholar]
- (5).Davis ME; Chen ZG; Shin DM Nat. Rev. Drug Discovery 2008, 7 (9), 771–782. [DOI] [PubMed] [Google Scholar]
- (6).Seeman NC Nature 2003, 421 (6921), 427–431. [DOI] [PubMed] [Google Scholar]
- (7).Rothemund PW Nature 2006, 440 (7082), 297–302. [DOI] [PubMed] [Google Scholar]
- (8).Wang P; M TA.; Pan V; Dutta PK; Ke Y Chem. 2017, 2 (3), 359–382. [Google Scholar]
- (9).Dietz H; Douglas SM; Shih WM Science 2009, 325 (5941), 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Douglas SM; Dietz H; Liedl T; Hogberg B; Graf F; Shih WM Nature 2009, 459 (7245), 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Han DR; Pal S; Nangreave J; Deng ZT; Liu Y; Yan H Science 2011, 332 (6027), 342–346. [DOI] [PubMed] [Google Scholar]
- (12).Benson E; Mohammed A; Gardell J; Masich S; Czeizler E; Orponen P; Hogberg B Nature 2015, 523 (7561), 441–444. [DOI] [PubMed] [Google Scholar]
- (13).Zhang F; Jiang SX; Wu SY; Li YL; Mao CD; Liu Y; Yan H Nat. Nanotechnol. 2015, 10 (9), 779–784. [DOI] [PubMed] [Google Scholar]
- (14).Veneziano R; Ratanalert S; Zhang KM; Zhang F; Yan H; Chiu W; Bathe M Science 2016, 352 (6293), 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kuzyk A; Schreiber R; Fan ZY; Pardatscher G; Roller EM; Hogele A; Simmel FC; Govorov AO; Liedl T Nature 2012, 483 (7389), 311–314. [DOI] [PubMed] [Google Scholar]
- (16).Knudsen JB; Liu L; Kodal ALB; Madsen M; Li Q; Song J; Woehrstein JB; Wickham SFJ; Strauss MT; Schueder F; Vinther J; Krissanaprasit A; Gudnason D; Smith AAA; Ogaki R; Zelikin AN; Besenbacher F; Birkedal V; Yin P; Shih WM; Jungmann R; Dong MD; Gothelf KV Nat. Nanotechnol. 2015, 10 (10), 892–898. [DOI] [PubMed] [Google Scholar]
- (17).Zhan P; Dutta PK; Wang P; Song G; Dai M; Zhao SX; Wang ZG; Yin P; Zhang W; Ding B; Ke Y ACS Nano 2017, 11 (2), 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sun W; Boulais E; Hakobyan Y; Wang WL; Guan A; Bathe M; Yin P Science 2014, 346 (6210), 1258361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Gopinath A; Miyazono E; Faraon A; Rothemund PW K. Nature 2016, 535 (7612), 401–405. [DOI] [PubMed] [Google Scholar]
- (20).Tian C; Kim H; Sun W; Kim Y; Yin P; Liu HT ACS Nano 2017, 11 (1), 227–238. [DOI] [PubMed] [Google Scholar]
- (21).Langecker M; Arnaut V; Martin TG; List J; Renner S; Mayer M; Dietz H; Simmel FC Science 2012, 338 (6109), 932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wei RS; Martin TG; Rant U; Dietz H Angew. Chem., Int. Ed. 2012, 51 (20), 4864–4867. [DOI] [PubMed] [Google Scholar]
- (23).Jiang Q; Song C; Nangreave J; Liu XW; Lin L; Qiu DL; Wang ZG; Zou GZ; Liang XJ; Yan H; Ding BQ J. Am. Chem. Soc. 2012, 134 (32), 13396–13403. [DOI] [PubMed] [Google Scholar]
- (24).Halley PD; Lucas CR; McWilliams EM; Webber MJ; Patton RA; Kural C; Lucas DM; Byrd JC; Castro CE Small 2016, 12 (3), 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhao YX; Shaw A; Zeng XH; Benson E; Nystrom AM; Hogberg B ACS Nano 2012, 6 (10), 8684–8691. [DOI] [PubMed] [Google Scholar]
- (26).Yan J; Hu C; Wang P; Zhao B; Ouyang X; Zhou J; Liu R; He D; Fan C; Song S Angew. Chem., Int. Ed. 2015, 54 (8), 2431–2435. [DOI] [PubMed] [Google Scholar]
- (27).Zhang Q; Jiang Q; Li N; Dai L; Liu Q; Song L; Wang J; Li Y; Tian J; Ding B; Du Y ACS Nano 2014, 8 (7), 6633–6643. [DOI] [PubMed] [Google Scholar]
- (28).Douglas SM; Bachelet I; Church GM Science 2012, 335 (6070), 831–834. [DOI] [PubMed] [Google Scholar]
- (29).Schuller VJ; Heidegger S; Sandholzer N; Nickels PC; Suhartha NA; Endres S; Bourquin C; Liedl T ACS Nano 2011, 5 (12), 9696–9702. [DOI] [PubMed] [Google Scholar]
- (30).Siegel RL; Miller KD; Jemal A Ca-Cancer J. Clin. 2018, 68 (1), 7–30. [DOI] [PubMed] [Google Scholar]
- (31).Han B; Park D; Li R; Xie M; Owonikoko TK; Zhang G; Sica GL; Ding C; Zhou J; Magis AT; Chen ZG; Shin DM; Ramalingam SS; Khuri FR; Curran WJ; Deng X Cancer Cell 2015, 27 (6), 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Rhee WJ; Bao G Nucleic Acids Res. 2010, 38 (9), e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lorenz S; Tomcin S; Mailander V Microsc. Microanal. 2011, 17 (3), 440–445. [DOI] [PubMed] [Google Scholar]
- (34).Choi CH; Hao L; Narayan SP; Auyeung E; Mirkin CA Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (19), 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wu XCA; Choi CHJ; Zhang C; Hao LL; Mirkin CA J. Am. Chem. Soc. 2014, 136 (21), 7726–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Okholm AH; Nielsen JS; Vinther M; Sorensen RS; Schaffert D; Kjems J Methods 2014, 67 (2), 193–197. [DOI] [PubMed] [Google Scholar]
- (37).Schaffert DH; Okholm AH; Sorensen RS; Nielsen JS; Torring T; Rosen CB; Kodal ALB; Mortensen MR; Gothelf KV; Kjems J Small 2016, 12 (19), 2634–2640. [DOI] [PubMed] [Google Scholar]
- (38).Fu M; Dai L; Jiang Q; Tang Y; Zhang X; Ding B; Li J Chem. Commun. 2016, 52 (59), 9240–9242. [DOI] [PubMed] [Google Scholar]
- (39).Greaves DR; Gordon SJ Lipid Res. 2005, 46 (1), 11–20. [DOI] [PubMed] [Google Scholar]
- (40).Patel PC; Giljohann DA; Daniel WL; Zheng D; Prigodich AE; Mirkin CA Bioconjugate Chem. 2010, 21 (12), 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Vindigni G; Raniolo S; Ottaviani A; Falconi M; Franch O; Knudsen BR; Desideri A; Biocca S ACS Nano 2016, 10 (6), 5971–5979. [DOI] [PubMed] [Google Scholar]
- (42).Doherty GJ; McMahon HT Annu. Rev. Biochem. 2009, 78, 857–902. [DOI] [PubMed] [Google Scholar]
- (43).Liang L; Li J; Li Q; Huang Q; Shi JY; Yan H; Fan CH Angew. Chem., Int. Ed. 2014, 53 (30), 7745–7750. [DOI] [PubMed] [Google Scholar]
- (44).Yang H; Chen Z; Zhang L; Yung WY; Leung KC; Chan HY; Choi CH Small 2016, 12 (37), 5178–5189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.