Functional protein microarray is a crucial tool in the study of proteins in native, unbiased, and high-throughput manner. There is a wide variety of applications, including the study of proteome-wide molecular interactions, analysis of post-translational modifications, identification of novel drug targets, and examination of pathogen-host interactions. Functional protein microarray is also useful in profiling antibody specificity, as well as in the discovery of novel biomarkers, especially for autoimmune diseases, infectious diseases, and cancers. Recently, the virion display method has been applied to produce functional GPCR array for various research and pharmaceutical applications.
Keywords: Protein array, protein-protein interactions, biofluids, diagnostic, pathogens
Graphical Abstract
Highlights
Summarize the development of functional protein microarray.
Application of functional proteome microarray in basic research.
Application of functional proteome microarray in translational research.
Fabrication of functional membrane protein array using virion display method.
Abstract
Protein microarrays are crucial tools in the study of proteins in an unbiased, high-throughput manner, as they allow for characterization of up to thousands of individually purified proteins in parallel. The adaptability of this technology has enabled its use in a wide variety of applications, including the study of proteome-wide molecular interactions, analysis of post-translational modifications, identification of novel drug targets, and examination of pathogen-host interactions. In addition, the technology has also been shown to be useful in profiling antibody specificity, as well as in the discovery of novel biomarkers, especially for autoimmune diseases and cancers. In this review, we will summarize the developments that have been made in protein microarray technology in both in basic and translational research over the past decade. We will also introduce a novel membrane protein array, the GPCR-VirD array, and discuss the future directions of functional protein microarrays.
Proteins are diverse biomolecules with a wide variety of structures and functions, and as such, it is a challenge to study them in a high-throughput fashion. There are three major types of protein microarrays: functional, analytical, and reverse phase. Functional protein microarrays are constructed with proteins purified/synthesized in a high-throughput fashion, enabling hundreds, and even thousands of different proteins to be probed for their biochemical properties in parallel. Analytical protein microarrays use affinity reagents that are immobilized on the array to detect or quantify complex biological samples. Finally, reverse phase protein microarrays utilize complex biological samples immobilized on the array and use affinity reagents for detection (1). In this short review, we focus on functional protein microarrays, summarize the recent developments in functional microarray technology, and discuss potential future applications.
Compared with other methods, such as mass spectrometry, functional protein microarrays are more capable of detecting weak interactions, more flexible with low abundancy proteins, and more amenable to analyzing crude samples such as serum. However, there are still some limitations for protein microarrays. The binding events observed during microarray experiments may not reflect the binding events that occur in the context of a cellular environment. Also, most of the detection methods involve labels and thus require proper controls (2). To date, many different types of functional protein microarrays have been developed in terms of differences in proteome coverage, protein lengths, and production pipelines. Some notable examples of the different categories of protein microarrays include purified proteome microarrays for full-length proteins, purified protein family microarrays for different protein categories, purified protein domain microarrays for user-defined domains/epitopes, and cell-free protein/peptide microarrays for in vitro translation from cDNA or in vitro synthesis. In Table I, we summarize the current developments in functional protein microarrays and divide them into these four categories. We also introduce a new concept, a membrane protein microarray (i.e. VirD1 array), and discuss the possible future directions of VirD array technology.
Table I. Summary of high-content functional protein microarrays.
| Organism/Protein classification | Protein No. | Coverage | Expression system | Refs. |
|---|---|---|---|---|
| Purified proteome microarray | ||||
| Homo sapiensa | 21,000 | 81% | S. cerevisiae | (11, 12) |
| S. cerevisiae | 5800 | 80% | S. cerevisiae | (3) |
| E. coli K12 | 4256 | 90% | E. coli | (8) |
| Arabidopsis thaliana | 15,000 | 56% | N. benthamiana | (9, 10) |
| M. tuberculosis | 4262 | 95% | S. cerevisiae | (7) |
| Coronavirus | 82 | 75% | S. cerevisiae | (4) |
| Epstein-Barr virus | 60 | 66% | S. cerevisiae | (6) |
| Zika and dengue viruses | 48 | 86% | S. cerevisiae | (5) |
| Herpes simplex virus-1/2 | 72 | 50% | E. coli | (94) |
| Purified protein family microarray | ||||
| G protein-coupled receptors | 315 | 84% | Mammalian cell lines | (14) |
| Membrane and secrete proteinsb | 505/1121 | <19% | E. coli | (15) |
| Influenza (HA antigens) | 283 | NA | Baculovirus or human cell | (16, 17) |
| HIV (gp120 and gp140) | 10 | NA | Mammalian or insect cell | (18) |
| Purified protein domain microarray | ||||
| Protein domains | ∼400 | NA | E. coli | (20, 111) |
| Protein epitope signature tags | 21,120 | NA | E. coli | (21, 22) |
| Consensus sequencec | 44 | NA | E. coli | (23) |
| Cell-free protein/peptide microarray | ||||
| Various pathogen antigensd | 100–7500 | 10–90% | In vitro expression | (24, 25) |
| Nucleic acid programmable | ∼10,000 | NA | In vitro expression | (26–28) |
aCommercialized with the trademark HuProtTM by CDI Laboratories and expended to >21,000 proteins in version 4.
bThe authors select proteins for expression based on a secretion signal peptide or at least one transmembrane domain. They express 505 proteins in full-length and 1121 protein fragments.
cDesigned 44 consensus coding sequences from 3,604 different dengue strains.
dPathogens included: Borrelia burgdorferi, Coxiella burnetiid, Burkholderia pseudomallei, Schistosoma japonicum, Chlamydia trachomatis, Bartonella henselae, Brucella melitensis, Hookworm Necator americanus, Leptospira interrogans, Plasmodium vivax, Schistosoma mansoni, Francisella tularensis, Toxoplasma gondii, Cytauxzoon felis, Plasmodium falciparum, Candida albicans, Mycobacterium tuberculosis, Salmonella enterica Typhi, Human papillomaviruses, and herpes simplex viruses 1&2.
Development of the Functional Protein Microarray
The proteome is the entire set of proteins that can be expressed by a genome. The development of a purified proteome microarray usually requires assembly of a genome-wide collection of open reading frames (ORFs) cloned into an expression vector, expression of the encoded proteins in cells, individual protein purification in a high-throughput fashion, and immobilization of the proteins on a microarray. Advances in purified proteome microarrays for model organisms, such as S. cerevisiae, E. coli, humans, and Arabidopsis thaliana, have propelled functional and biochemical studies of proteins to a proteomic level. The first of its kind is the S. cerevisiae (budding yeast) proteome array, developed by the Snyder group in 2001 and containing 5,800 full length yeast proteins (3). Currently, there are many purified proteome microarrays covering a wide variety of model systems, including coronaviruses (4), flaviviruses (5), human herpesviruses (6), M. tuberculosis (7), E. coli K12 (8), S. cerevisiae (3), Arabidopsis thaliana (9, 10), and humans (11, 12). Because of the coverage of ORF collections and the efficiency of protein expression/purification, the proteome coverage on such arrays ranges from 56% to 95% (Table I). The choice of protein expression system greatly influences post-translational modifications and can affect the success rate of protein purification. For example, because of a lack of eukaryotic posttranslational modifications and chaperones, proteins encoded by C. elegans were poorly expressed in E. coli, with an expression rate of 48%. Of this 48%, only 15% were soluble (13). Therefore, homologous expression systems are generally preferred to obtain the highest protein activity and expression efficiency. The S. cerevisiae, E. coli, and Arabidopsis thaliana proteome arrays are three of the best examples for use as homologues expression systems. In some cases, especially with mammalian cells, it is difficult and expensive to transfect cells, and thus one can use an alternative expression system, such as budding yeast, to accommodate protein production pipelines. Indeed, the human proteome microarray (i.e. HuProt) is one of the best examples to use a heterologous expression system, as it exhibits the most comprehensive human proteome collection purified from yeast (81% proteome coverage). Another commercial human proteome microarray, called ProtoArray, contained >9,000 human proteins purified from insect cells (43% proteome coverage), but was discontinued in 2018.
A protein family microarray is designed to interrogate specialized groups of proteins for their biochemical functions. Today, there are many different protein family microarrays, each used for different purposes. For example, one can utilize a G-protein coupled receptor (GPCR) array for pharmaceutical applications (14), a membrane/secreted protein array for profiling autoantibodies (15), a hemagglutinin antigen array for investigating influenza vaccines (16, 17), and a gp120/140 array from HIV for analyzing immune responses (18). Because most protein family microarrays have a relatively small number of proteins, the expression system can be tailored for desired qualities and quantities. For example, the GPCR array is developed using Virion Display (VirD) technology (19) to maintain the seven transmembrane structure and to obtain the best GPCR expression in several mammalian cell lines, including Vero, HEL, HeLa, and 293T cells (14).
Alternatively, protein domain microarrays can be designed to analyze certain regions, domains, or epitopes within the proteins. These arrays often involve the careful design of desired gene sequences before entering the protein production pipeline. Protein domain arrays, Protein Epitope Signature Tag (PrEST) arrays, and consensus sequence protein arrays are the three best examples of this sort. The protein domain arrays reported by Jones et al. contain all the human Src homology 2 and phosphotyrosine binding domains to profile the interaction networks for tyrosine phosphorylation on ErbB receptors (20). PrEST arrays contain the unique signature in the human proteome developed by the Human Protein Atlas Consortium for identifying multiple sclerosis autoantibodies (21) or for validating antibody specificity (22). In a consensus sequence protein array, Qi et al. summarize 44 consensus serotype sequences out of 3604 different dengue strains and construct a protein array accordingly for dengue serotyping (23). Overall, both purified proteome, protein family, and protein domain arrays have a wide variety of applications in basic and translational research, as well as pharmaceutical industry.
The cell-free protein/peptide microarray is designed to display a short peptide or full-length protein using a cell-free system. Cell-free expression is designed to bypass the expensive and often tedious work of cell-based protein production. To construct protein assays with an in vitro expression, many expression systems, including expression lysate from E. coli, insect cells, wheat germs, and human cells, are commercially available. For instance, the Felgner Lab established various pathogen arrays ranging from viruses to bacteria and yeasts by using an in vitro transcription and translation (IVTT) system adopted from E. coli (Table I and footnote) (24, 25). On the other hand, the LaBaer group utilized a DNA array, dubbed as the Nucleic Acid Programmable Protein Array (NAPPA), to construct human proteome arrays using in vitro transcription/translation system (26–28). Because cell-free expression lacks regulated protein folding, segregated cellular compartments, and coordinated post-translational modifications (PTMs), the protein functions are not guaranteed (27). The IVTT system also suffers from a lower yield of larger proteins (e.g. >50 kDa), potential contamination by other proteins presented in the lysates, and low array density (e.g. ∼2,000 features per array) (27). Nevertheless, protein arrays produced by cell-free expression are quite useful to analyze immune responses (24–26).
Application of Yeast Proteome Microarrays in Basic Research
Functional protein microarrays, especially purified proteome microarrays, are useful for profiling proteome-wide molecular interactions and allow for a comprehensive, unbiased screening. In basic research, researchers have been using functional protein microarrays to study protein-protein interactions, protein-lipid interactions, protein-cell/lysates, protein-DNA interactions, protein-RNA interactions, small molecule binding, and PTMs, such as glycosylation, ubiquitylation, SUMOylation, acetylation, phosphorylation, and methylation (Fig. 1A–1G). In Table II, we summarize representative studies based on the research applications illustrated in Fig. 1. Here, we review research studies based on the proteomes immobilized on microarrays.
Fig. 1.
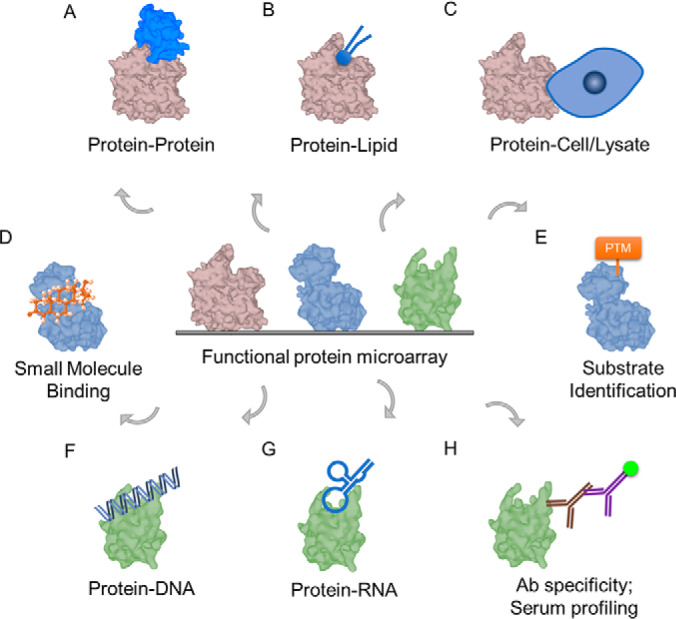
Application of Functional Protein Microarray. Applications of functional protein microarray for interrogating protein-binding property include A, Protein-protein interactions; B, Protein-lipid interactions; C, Protein-cell/lysate interactions; D, Small molecule binding; E, Enzyme-substrate relationships; F, Protein-DNA interactions; G, Protein-RNA interactions; H, Antibody specificity/Serum profiling. PTM = post-translational modification.
Table II. Representative studies using functional protein microarrays.
| Classification/Research | Tools | Major findings | Refs. |
|---|---|---|---|
| Protein-Protein | |||
| Calmodulin | Yeast proteome array | Identified 30 new targets | (3) |
| 4 antimicrobial peptides | E. coli proteome array | Identified many intracellular targets | (35) |
| 2-oxohistidine peptide | E. coli proteome array | Identified 9 redox targets | (112) |
| NS5A | HuProt | Identified 90 targets and validated Pim1 | (44) |
| PknG | HuProt | Identified 125 targets and validated CypA | (47) |
| MTB proteome array | Identified 59 targets | (7) | |
| ROP18 | HuProt | Identified 68 targets and validated 4 bindings | (46) |
| SidM, LidA, and AnkX | Human NAPPA | Identified 18, 20, and 8 host targets | (48, 49) |
| 61 ErbB peptides | SH2 PBD array | Profiled interaction networks | (20) |
| Protein-Lipid | |||
| 5 Phospholipids | Yeast proteome array | Identified 150 targets | (3) |
| Protein-Cell/Lysate | |||
| HBMEC | E. coli proteome array | Identified 23 targets and validated YojI | (38) |
| Macrophage lysate | MTB proteome array | Identified 26 targets | (113) |
| Small Molecule Binding | |||
| 2 inhibitors of rapamycin | E. coli proteome array | Identified 39 targets and validated Tep1p and Nir1p | (29) |
| Arsenic | HuProt | Identified 360 targets and validated hexokinase | (53) |
| 6-O-angeloylplenolin | HuProt | Identified 99 targets and validated STAT3 | (54) |
| Cyclic di-GMP | MTB proteome array | Identified 30 targets | (7) |
| E. coli proteome array | Identified 8 targets and validated CobB | (37) | |
| Substrate Identification | |||
| Six SUMO E3 ligases | HuProt | Identified 250 substrates and validated PYK2 | (114) |
| 289 kinases | HuProt ver. I | Constructed a high resolution kinase-substrate network | (115) |
| Four herpesvirus kinases | HuProt | Identified a conserved host pathway for viral replication | (116) |
| ppGalNAc-Ts | HuProt | Identified 128 common substrates for glycosylation | (51) |
| VopS and IbpAFic2 | Human NAPPA | Identified 21 AMPylation substrates | (52) |
| 87 yeast kinases | Yeast proteome array | Constructed a kinase-substrate network | (117) |
| Ubiquitin E3 Rsp5 | Yeast proteome array | Identified 84 substrates and validated Rnr2 | (33) |
| NuA4 | Yeast proteome array | Discovered two yeast ageing pathways involving Pck1p and Sip2 | (32, 118) |
| Tyrosine sulfation | E. coli proteome array | Identified 875 substrates | (40) |
| 11 MTB kinases | MTB proteome array | Identified 1,027 interaction network | (58) |
| Protein-DNA | |||
| Yeast genomic DNA | Yeast proteome array | Identified 200 targets and validated Arg5,6 | (30) |
| Mismatch and abasic site | E. coli proteome array | Validated YbaZ and YbcN | (8) |
| Promoter DNA of fimS | E. coli proteome array | Identified 19 targets and validated Spr | (34) |
| 460 DNA motifs | 4,191 human array | Discovered many unconventional DNA-binding proteins and showed Erk2 as a transcriptional repressor | (12) |
| Protein-RNA | |||
| BMV viral RNA | Yeast proteome array | Identified and validated Pus4 and App1's role in preventing viral spreading in tobacco | (31) |
| 13 IncRNAs | HuProt | Found many unconventional RNA-binding proteins and validated IDH1 | (42) |
| miR-122 | HuProt | Identified 40 targets and validated hnRNP K | (43) |
| Antibody specificity | |||
| mAbs against TFs | HuProt | Demonstrated the use of HuProt for specificity test of mAbs | (57) |
| 400 Abs against SH2 | SH2 PrESTs array | Verified Abs specificity | (111) |
mAbs = monoclonal antibodies.
Zhu et al. constructed the very first proteome microarray, the yeast proteome microarray, and utilized it to investigate protein-protein interactions and protein-lipid interactions. The array was probed with biotinylated calmodulin and 33 new calmodulin binding proteins with new common motifs were identified (3). In the same study, the yeast proteome array was probed with fluorescently labeled liposomes carrying various phosphatidyl-inositides and more than 150 phospholipid binding proteins were identified (3). Huang et al. used the same microarray to identify binding proteins for two small molecule inhibitors of rapamycin, SMIR3 and SMIR4, and identified 8 and 30 protein targets, respectively. Most target proteins were involved in PI3,4P2 signaling (29). Hall et al. used the yeast proteome microarray to profile DNA binding proteins and revealed a mitochondrial enzyme, Arg5,6, can regulate both nuclear and mitochondrial gene expression (30). Similarly, Zhu et al. used the yeast proteome microarray to profile RNA hairpin binding proteins and identified two proteins: Pus4 and App1. Their antiviral activity against the spread of brome mosaic virus was demonstrated in tobacco (31). The Zhu lab further demonstrated the utility of proteome arrays by performing covalent enzymatic reactions on the arrays. They were the first to establish the protein acetylation reactions using the yeast NuA4 complex, and two parallel signaling pathways in yeast aging were discovered (32). It has also been applied to determine the substrates of a HECT domain ubiquitin E3 ligase Rsp5 (33). These studies demonstrate the usefulness of the yeast proteome microarray in basic research.
Application of E. coli Proteome Microarrays in Basic Research
Chen et al. established a purified E. coli proteome microarray in 2008, comprising of 4256 unique proteins and applied it to identify potential new players in the DNA damage response. The E. coli proteome microarray was probed with several short DNA probes containing mismatched base pairs or abasic sites, and two DNA repair proteins were identified: YbaZ and YbcN (8). In another study the same array was used to detect DNA binding proteins to the promoter of type 1 fimbriae and identified Spr as a phase switch for type 1 fimbria expression (34). Ho et al. probed several antimicrobial peptides using the E. coli proteome array and identified many intracellular targets. Among the four antimicrobial peptides, they identified some shared and unique targets and suggested a synergistic effect on LfcinB and Bac7, as well as LfcinB and PR-39 (35). Hsiao et al. probed the E. coli proteome array with four glycosaminoglycans that are common on host cells and identified a hundred protein targets. They further validated ycbS as a bacterial factor for cell entry (36). Xu et al. probed the E. coli proteome array with an important bacterial second messenger, cyclic di-GMP, and identified CobB as a strong binder. Because CobB is a deacetylation enzyme, they subsequently found that cyclic di-GMP inhibits the enzymatic activity and forms a novel feedback loop to the cyclic di-GMP production (37). Feng et al. used E. coli proteome microarray to investigate protein-cell interactions by probing the human brain microvascular endothelial cells (HBMEC) on the array. They identified 23 target proteins and validated YojI as a protein for E. coli invasion. Moreover, they purified Yojl, probed using HuProt, and further identified interferon-alpha receptor as a host receptor for Yojl (38). Besides various binding assays, the E. coli proteome microarray has also been applied to identify substrates, including substrates of glycoproteins (39), tyrosine sulfation (40), and ClpYQ protease (41). As demonstrated by these representative works, the E. coli proteome microarray is widely used to study bacterial physiology as well as host-microbial interactions.
Application of Human Proteome Microarrays in Basic Research
The human proteome microarray is the most widely used array in basic research, translational research, and in the pharmaceutical industry. There are three popular human proteome microarrays: HuProt, ProtoArray, and NAPPA. HuProt contains ∼21,000 individual purified human proteins in full-length, which is by far the most comprehensive human proteome collection. ProtoArray contained ∼9000 human proteins purified from insect cells, but was discontinued commercially in 2018. NAPPA is an in vitro expression system that has been applied to express 10,000 human proteins.
The HuProt array was not made overnight. In its early stages, it contained 4191 unique human proteins, mostly transcription factors and co-factors. Hu et al. performed a large scale DNA-binding assay with 460 DNA motifs on this array and found 17,718 protein-DNA interactions. Not only were numerous known protein-DNA interactions recovered, but they also found many unconventional DNA-binding proteins, including a mitogen-activated protein kinase (MAPK), Erk2. In-depth mutagenesis studies and cell-based assays demonstrated that Erk2 acts as a transcriptional repressor in the regulation of interferon-gamma signaling (12). In 2012, the Zhu lab published the construction of HuProt version I, which contained 16,368 individual purified human proteins in full-length and demonstrated that it could serve as a useful tool to identify highly specific monoclonal antibodies (11). The work laid the foundation for the NIH-funded Protein Capture Reagents Program (PCRP; https://commonfund.nih.gov/proteincapture).
The birth of HuProt arrays expanded researchers' arsenal for interrogation of a great fraction of the entire human proteome for specific biochemical properties. For example, Liu et al. profiled the binding specificities of 13 long noncoding RNAs (lncRNAs) on HuProt to determine potential players in lncRNA-mediated biological processes. Ultimately, 671 lncRNA-binding proteins were found, 525 of which lacked any known RNA-binding domains. A novel RNA binding protein, IDH1, was further validated in cells and shown to bind thousands of RNA transcripts (42). Similarly, Fan et al. probed HuProt with miR-122 and identified 40 target proteins. Because miR-122 is required for hepatitis C virus (HCV) replication, they further validated the target hnRNP K as a repressor for HCV replication (43). Therefore, the human proteome microarray is a valuable tool to study the complex regulatory networks of protein-DNA and -RNA interactions (Fig. 1F and 1G).
The human proteome microarray is also useful for the analysis of protein-protein interactions, especially for determining players involved in pathogen-host interactions (Fig. 1A). Park et al. probed the nonstructural 5A protein from HCV on ProtoArray and identified 90 proteins. They further validated one of these proteins, Pim1, as a factor involved in HCV cell entry (44). Yoon et al. constructed a Zika virus-host protein-protein interaction network using a similar approach and compared its dengue virus counterparts to determine Zika virus-specific interactions (45). Further orthogonal large-scale screenings allowed them to pinpoint drug targets in the host involved in Zika virus replication. Yang et al. investigated the binding events of T. gondii virulence factor ROP18 using HuProt and identified 68 targets. They subsequently validated the crucial role of ROP18 on p53, p38, UBE2N, and SMAD1 through phosphorylation-dependent degradation (46). Wu et al. investigated the binding events of PknG, an important kinase in M. tuberculosis (MTB), using HuProt and identified 128 binding proteins. They further validated that one of these binding proteins, CypA, is degraded upon phosphorylation and subsequently inhibits inflammatory responses (47). Using human NAPPA, Yu et al. identified 18, 20, and 8 host proteins that interact with L. pneumophila effectors SidM, LidA, and AnkX, respectively (48, 49).
Human proteome microarrays have also been widely used to study PTMs (Fig. 1E). Song et al. established methods to detect global tyrosine phosphorylation, lysine acetylation, ubiquitylation, and SUMOylation on HuProt. The HuProt arrays were incubated with cell lysates diluted in different PTM reaction buffers to perform covalent protein modifications, and the modified proteins on the array were visualized using the corresponding PTM antibodies. Among the complex regulation of PTMs in cancers, they validated the hyperactivities of PTK2 and PTK2B kinases in ovarian cancer (50). Xu et al. surveyed the substrate for ppGalNAc-Ts using HuProt and identified 128 common substrates for O-GalNAc glycosylation (51). Yu et al. used human NAPPA to identify the 20 and 21 AMPylation substrates for VopS and IbpAFic2, respectively (52). Overall, the human proteome microarray serves as an unbiased platform for studying many kinds of binding events and enzyme-substrate relationships.
The two major pharmaceutical applications of the human proteome microarray are drug target identification (Fig. 1D) and specificity tests for monoclonal antibodies (mAbs) (Fig. 1H). HuProt was used to identify the targets of arsenic, a cancer drug, and 360 potential binders were identified. Hexokinase was validated to bind arsenic, and this binding event was further shown to result in the inhibition of glycolysis (53). With a similar strategy, Cheng et al. screened the targets of 6-O-angeloylplenolin, a drug that induces cell cycle arrest, and identified 99 proteins. The proteins Skp1 and STAT3 were further validated to show involvement in cell cycle arrest (54). Because the mAb-based biologicals are one of the fastest growing therapeutic modalities, quality control is extremely important. Many commercial mAbs, however, exhibit poor quality and have wasted $350 million annually in the United States alone (55). HuProt arrays are an ideal platform to screen mAbs for mono-specificity (56). As such, Venkataraman et al. established a production pipeline for the mAbs against transcription factors and adapted HuProt as a primary validation tool for specificity tests (57). Of the 5882 mAbs tested on HuProt arrays, 2000 passed the specificity tests, 1462 of which eventually passed the secondary cell-based validation for their ability to perform Western blot analysis and/or immunoprecipitation.
Proteome microarrays other than those already mentioned can also be quite useful in basic research, such as the Arabidopsis proteome array (9) and the MTB proteome array (7) to name a few. Popescu et al. established the Arabidopsis proteome microarray and profiled the binding of calmodulin and calmodulin-like proteins (9). Deng et al. developed the MTB proteome microarray and used it to identify the binding partners of PknG and the protein interactions of second messenger cyclic di-GMP (7). Wu et al. probed 11 serine/threonine protein kinases on the MTB proteome array and identified 492 binding proteins with 1027 network interactions (58).
Application of Functional Protein Microarrays in Translational Research
Serological biomarkers are valuable tools for diagnosis, prognosis and companion diagnosis in various autoimmune diseases, cancers, and infectious diseases (59, 60). One of the early applications of functional protein microarrays was to discover new serological biomarkers for autoimmune diseases because they can serve as antigen surveying platforms to detect subtle changes in antibody composition. In a dysregulated immune system, the antibodies that are generated by humoral immunity and react with self-antigens are referred to as autoantibodies (AAbs). When a functional protein array covers most of the human proteome (e.g. HuProt), a specific AAb signature can be readily detected by probing the array with a diluted patient serum/plasma sample. When this approach is used to profile AAb signatures for a large cohort, subsequent statistical analysis can reveal potential biomarkers associated with a disease of interest (Table III). This approach has three major advantages. First, patient samples are easy to obtain and store because they are mostly in the forms of serum, plasma or body fluid. Second, detection of AAbs on a human proteome array is very sensitive and quantitative, only requiring several microliters of samples. Finally, the presence of AAbs is detectable before symptoms can be identified, making early diagnosis possible.
Table III. Functional protein microarrays for biomarker idenitification.
| Diseases/Classifications | Tools | Major findings | Refs. |
|---|---|---|---|
| Autoimmune diseases | |||
| Autoimmune hepatitis | Purified 5,011 human array | Validate 3 AAbs | (61) |
| Purified 1,626 human array | Validate 6 AAbs | (15) | |
| Ankylosing spondylitis | NAPPA 3,498 human | Identify a set of AAbs | (62) |
| Multiple sclerosis | 11,520 PrESTs array | Validate 51 AAbs | (21) |
| ProtoArray | Validate CSF AAbs against RBPJ | (63) | |
| Type 1 diabetes | NAPPA 10,000 human | Validate 5 AAbs | (64) |
| Alzheimer's disease | ProtoArray | Validate 10 AAbs | (65) |
| Rheumatoid arthritis | ProtoArray | Validate 2 AAbs | (66) |
| Sjögren's syndrome | ProtoArray | Validate 4 saliva AAbs | (67) |
| Primary biliary cirrhosis | HuProt | Validate 6 AAbs | (68) |
| Amyotrophic lateral sclerosis | ProtoArray | Validate 20 AAbs | (69) |
| Male subfertility | ProtoArray | Validate AAbs against TGM4 in prostate | (70) |
| Juvenile idiopathic arthritis | NAPPA 768 human | Identify 18 AAbs | (71) |
| Behcet's disease | HuProt | Validate AAbs against CTDP1 | (72) |
| Sarcoidosis | 3,072 PrESTs array | Validate 4 AAbs | (73) |
| Cancers | |||
| Ovarian cancer | Purified 5,005 human array | Validate 4 AAbs | (74) |
| NAPPA 5,177 tumor antigens | Validate 3 AAbs | (75) | |
| Glioma | HuProt | Identify a set of AAbs | (76) |
| Lung cancer | HuProt | Validate 3 AAbs | (77) |
| Gastric cancer | HuProt | Validate 4 AAbs | (78) |
| Bladder cancer | ProtoArray | Validate 2 AAbs | (79) |
| Prostate cancer | Purified 123 antigen | Validate 3 AAbs | (80) |
| Colon cancer | ProtoArray | Validate 3 AAbs | (81) |
| Breast cancer | NAPPA 4,988 tumor antigens | Validate 28 AAbs | (82) |
| Myelodysplastic syndromes | HuProt | Validate 3 AAbs | (83) |
| Meningiomas | HuProt | Identify a set of AAbs | (84) |
| Infectious diseases | |||
| Coronaviruses | Coronaviruses proteome array | Identify a set of Abs | (4) |
| Flaviviruses | Zika/Dengue proteome array | Validate a set of Abs | (5) |
| M. tuberculosis | Purified MTB proteome array | Identify 14 Abs | (7) |
| NAPPA 4,045 MTB | Identify 8 Abs | (87) | |
| Varicella zoster virus | NAPPA 69 VZV | Identify 19 Abs | (86) |
| P. aeruginosa | NAPPA 262 P. aeruginosa | Identify 12 Abs | (85) |
| Herpes simplex virus | HSV-1&2 proteome array | Validate 2 Abs | (94) |
| L. interrogans | IVTT 3,359 L. interrogans array | Identify 191 Abs | (88) |
| S. Typhi | IVTT 2,724 S. Typhi array | Identify 93 Abs | (89) |
| B. melitensis | IVTT 3,046 B. melitensis array | Identify 33 Abs | (90) |
| Human papillomavirus | IVTT 104 HPV array | Identify E7 Ab in cancer | (91) |
| C. albicans | IVTT 451 C. albicans array | Identify 13 Abs | (92) |
| F. Tularensis | IVTT 1,741 F. Tularensis array | Identify 15 Abs | (93) |
| Other diseases | |||
| Asthma | ProtoArray | Validate 4 AAbs | (95) |
| Kawasaki disease | E. coli proteome array | Validate a set of AAbs | (96) |
| Preeclampsia | E. coli proteome array | Validate 5 AAbs | (97) |
| Bipolar disorder | E. coli proteome array | Validate 6 AAbs | (98) |
| Parkinson's disease | ProtoArray | Validate 10 AAbs | (99) |
| Osteoarthritis | 3,840 PrESTs array | Validate a set of AAbs | (100) |
| Chronic renal disease | ProtoArray | Validate 4 AAbs | (101) |
| Inflammatory bowel disease | ProtoArray | Validate AAbs against FAM84A | (102) |
| E. coli proteome array | Identify a set of AAbs | (103) | |
| Meniere's disease | ProtoArray | Identify 18 AAbs | (104) |
| Chronic humoral rejection | ProtoArray | No common AAbs | (119) |
AAbs = autoantibodies. AAbs if not specified, they are from blood. IVTT = in vitro transcription and translation.
Human proteome microarrays have been used to identify diagnostic AAbs for more than 13 autoimmune diseases, including autoimmune hepatitis (15, 61), ankylosing spondylitis (62), multiple sclerosis (21, 63), type 1 diabetes (64), Alzheimer's disease (65), rheumatoid arthritis (66), Sjögren's syndrome in saliva (67), primary biliary cirrhosis (68), amyotrophic lateral sclerosis (69), male subfertility (70), juvenile idiopathic arthritis (71), Behcet's disease (72), and sarcoidosis (73). For biomarker identification, it is necessary to include the most comprehensive human proteome collection for unbiased screening, and to validate candidate biomarkers using additional cohort to avoid overfitting. These requirements often result in a high price tag for biomarker research. Song et al. developed a strategy to overcome this issue by dividing the process into two phases. In phase I, also known as the biomarker discovery or screening step, they used the HuProt array to survey AAbs in a smaller cohort of serums from 22 autoimmune hepatitis (AIH) patients and 30 healthy controls. In this phase, they narrowed down thousands of human proteins to 11 candidate autoantigens. In phase II, also known as the biomarker verification or validation step, they fabricated a focused antigen array with the 11 candidate antigens to survey AAbs in a much larger cohort composed of sera from 44 AIH patients, 50 healthy controls, and 184 patients suffering from other autoimmune diseases as a disease comparison group. With this two-phase strategy, they identified and validated three new antigens, RPS20, Alba-like, and dUTPase as highly specific biomarkers for AIH (61).
In translational cancer research, it is important to identify early diagnosis markers to allow for earlier treatment and intervention. Human proteome arrays are widely used to profile the AAbs in 10 cancer types, including ovarian cancer (74, 75), glioma (76), lung cancer (77), gastric cancer (78), bladder cancer (79), prostate cancer (80), colon cancer (81), breast cancer (82), myelodysplastic syndromes (83), and meningiomas (84). Orenes-Pinero et al. performed serum profiling on ProtoArray and identified 171 autoantigens related to bladder cancer. They validated selected candidates by using a cancer tissue array and confirmed that dynamin is not only an autoantigen biomarker, but it is also associated with poor survival (79).
Regarding infectious disease, the purpose of using protein microarrays is quite different from autoimmune diseases or cancer because the serum antibodies in infectious diseases are a part of the normal immune response. Ways in which the protein microarray can be used to study infectious disease include serotyping, identifying markers for prognosis, and identifying immunogenic proteins for vaccine development. To serve these purposes, the protein array must be tailored according to the pathogens being studied. NAPPA techniques have been applied to vaccine development by profiling serum antibodies against P. aeruginosa in a varicella-zoster virus proteome array (85, 86). Because in vitro expression arrays are more flexible, most of the pathogen-protein arrays are built with either IVTT or NAPPA. Such arrays include MTB (87), varicella zoster virus (86), P. aeruginosa (85), L. interrogans (88), S. Typhi (89), B. melitensis (90), human papillomavirus (91), C. albicans (92), and F. Tularensis (93). A few pathogen-protein arrays are purified from yeast, including MTB (7), flaviviruses (5), and herpes simplex virus (94).
Other diseases with altered immune responses can also be examined using protein microarrays in order to identify AAbs relevant to disease. To date, there are nine inflammatory diseases with biomarkers that have been discovered using protein microarrays, including asthma (95), Kawasaki disease (96), preeclampsia (97), bipolar disorder (98), Parkinson's disease (99), osteoarthritis (100), chronic renal disease (101), inflammatory bowel disease (102, 103), and Meniere's disease (104).
GPCR-VirD Microarray
GPCRs form the largest transmembrane protein family in humans, consisting of seven transmembrane domains. This complex structure allows GPCRs to bind to a variety of ligands, ranging from protons, ATP, amino acids, peptides, proteins, and to many other unidentified ligands. To date, ∼40% of the FDA-approved drugs target GPCRs. Because the lipid bilayer is required to maintain the conformation of GPCRs, purification attempts often disrupt the GPCR conformation. To overcome this hurdle, Hu et al. developed VirD technology by replacing a viral envelope gene in herpes simplex virus-1 (HSV-1) with an ORF encoding a human transmembrane protein. The production of this recombinant virus from mammalian cells allowed the human receptor to be embedded in the viral envelope with correct conformation and function (19). More importantly, these recombinant viruses were arrayed on a glass slide to facilitate high-throughput screenings. Syu et al. expended the VirD technology to cover most of the non-odorant GPCRs (e.g. 315) for further biochemical interrogation (14). We demonstrated that the GPCR-VirD array is useful to profile specificity of mAbs (Fig. 2A). Among the 20 commercial mAbs tested, only 10 mAbs were determined to be ultra-specific. The rest either failed to show specificity entirely, or at least had several off-targets. Interestingly, all four mAbs with reported neutralization activity were shown to be ultra-specific on the GPCR-VirD array. Next, we performed specificity tests with known ligands (Fig. 2B) and revealed several off-targets for a peptide hormone, somatostatin-14. Two selected off-targets along with the canonical GPCR were validated with virion nano-oscillators for real-time and label-free detection (105) and showed significant binding affinities. Lastly, we probed the GPCR-VirD array with a clinical strain involved in neonatal meningitis (Group B Streptococcus K79) and identified five potential GPCR targets (Fig. 2C). CysLTR1 was further validated in vitro and in vivo as a host receptor for K79 invasion. We believe that the VirD array is a robust platform to profile many kinds of membrane protein interactions.
Fig. 2.
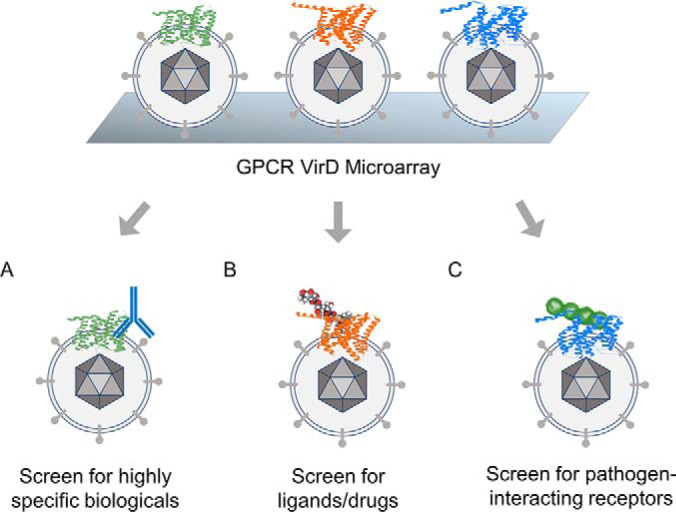
Application of GPCR-Virion Display (VirD) Microarray. 315 non-odorant GPCRs are displayed on the HSV-1 envelope to maintain the native conformation and form the world's largest functional GPCR-VirD array. The GPCR-VirD array is useful to screen for highly specific biologicals (A), ligands (B), small molecule drugs (B), and pathogen receptors (C).
Future Directions
Membrane proteins are one of the most important protein categories, as they play important roles in many biological processes, such as signal transduction, cell recognition, cell-cell communication, transport, and anchorage, to name a few. It is highly desirable to develop a high-content and high-throughput platform for functional membrane proteins to enable meaningful screening for ligands, biologicals and small molecule drugs. To date, many methods have been developed to maintain the native conformation of membrane proteins, including nanodiscs (106), macrodiscs (107), Salipro nanoparticales (108), virus-like particles (109), and VirD (14, 19). Unlike VirD, the other methods are not easy to scale up for multiplexed, highly parallel screening while maintaining the flexibility of massive production of the reagents from various mammalian cell lines. When the VirD array is coupled with nano-oscillator technology (105), the entire membrane protein family can be screened simultaneously with candidate drugs or biologicals in a label-free, real time fashion, and binding specificity and kinetics can be obtained in a single experiment. We envision that VirD array technology can expand to all kind of membrane protein families and holds promise for discovering biologicals, drugs, and receptor interactions. Besides VirD tailored for membrane proteins, all other human proteins need a proper expression system for the best folding and PTMs. For this reason, it would be desirable to use a mammalian expression system. In combination with transfection, transformation, and CRISPR knock-in technologies (110), it is possible to generate a human proteome microarray from human cells and accelerate research, potentially leading to the discovery of novel drugs or biologicals.
Footnotes
* This work was supported in part by the NIH/NCI IMAT R33-CA186790-01A1, MOST Taiwan fellowship 105-2917-I-564-078, and MOST Taiwan 108-2320-B-006-054-MY2. JD was supported in part by the CBI program (NIH 2T32GM080189-11) at The Johns Hopkins University. The authors declare that they have no conflicts of interest with the contents of this article.
1 The abbreviations used are:
- VirD
- Virion Display
- PrEST
- Protein Epitope Signature Tag
- IVTT
- in vitro transcription and translation
- NAPPA
- Nucleic Acid Programmable Protein Array
- PTM
- post-translational modifications
- HBMEC
- human brain microvascular endothelial cells.
REFERENCES
- 1. Moore C. D., Ajala O. Z., and Zhu H. (2016) Applications in high-content functional protein microarrays. Curr. Opin. Chem. Biol. 30, 21–27 [DOI] [PubMed] [Google Scholar]
- 2. Neiswinger J., Uzoma I., Cox E., Rho H., Song G., Paul C., Jeong J. S., Lu K. Y., Chen C. S., and Zhu H. (2016) Protein microarrays: flexible tools for scientific innovation. Cold Spring Harb. Protoc. 2016. [DOI] [PubMed] [Google Scholar]
- 3. Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., and Snyder M. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 4. Zhu H., Hu S., Jona G., Zhu X., Kreiswirth N., Willey B. M., Mazzulli T., Liu G., Song Q., Chen P., Cameron M., Tyler A., Wang J., Wen J., Chen W., Compton S., and Snyder M. (2006) Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. U.S.A. 103, 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song G., Rho H. S., Pan J., Ramos P., Yoon K. J., Medina F. A., Lee E. M., Eichinger D., Ming G. L., Munoz-Jordan J. L., Tang H., Pino I., Song H., Qian J., and Zhu H. (2018) Multiplexed biomarker panels discriminate Zika and Dengue virus infection in humans. Mol. Cell. Proteomics 17, 349–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu J., Liao G., Shan L., Zhang J., Chen M. R., Hayward G. S., Hayward S. D., Desai P., and Zhu H. (2009) Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J. Virol. 83, 5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng J., Bi L., Zhou L., Guo S. J., Fleming J., Jiang H. W., Zhou Y., Gu J., Zhong Q., Wang Z. X., Liu Z., Deng R. P., Gao J., Chen T., Li W., Wang J. F., Wang X., Li H., Ge F., Zhu G., Zhang H. N., Gu J., Wu F. L., Zhang Z., Wang D., Hang H., Li Y., Cheng L., He X., Tao S. C., and Zhang X. E. (2014) Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep. 9, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 8. Chen C. S., Korobkova E., Chen H., Zhu J., Jian X., Tao S. C., He C., and Zhu H. (2008) A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat. Methods 5, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Popescu S. C., Popescu G. V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., and Dinesh-Kumar S. P. (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 104, 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manohar M., Tian M., Moreau M., Park S. W., Choi H. W., Fei Z., Friso G., Asif M., Manosalva P., von Dahl C. C., Shi K., Ma S., Dinesh-Kumar S. P., O'Doherty I., Schroeder F. C., van Wijk K. J., and Klessig D. F. (2014) Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeong J. S., Jiang L., Albino E., Marrero J., Rho H. S., Hu J., Hu S., Vera C., Bayron-Poueymiroy D., Rivera-Pacheco Z. A., Ramos L., Torres-Castro C., Qian J., Bonaventura J., Boeke J. D., Yap W. Y., Pino I., Eichinger D. J., Zhu H., and Blackshaw S. (2012) Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics 11, O111 016253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H. S., Woodard C., Wang H., Jeong J. S., Long S., He X., Wade H., Blackshaw S., Qian J., and Zhu H. (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139, 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luan C. H., Qiu S., Finley J. B., Carson M., Gray R. J., Huang W., Johnson D., Tsao J., Reboul J., Vaglio P., Hill D. E., Vidal M., Delucas L. J., and Luo M. (2004) High-throughput expression of C. elegans proteins. Genome Res. 14, 2102–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Syu G. D., Wang S. C., Ma G., Liu S., Pearce D., Prakash A., Henson B., Weng L. C., Ghosh D., Ramos P., Eichinger D., Pino I., Dong X., Xiao J., Wang S., Tao N., Kim K. S., Desai P. J., and Zhu H. (2019) Development and application of a high-content virion display human GPCR array. Nat. Commun. 10, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zingaretti C., Arigo M., Cardaci A., Moro M., Crosti M., Sinisi A., Sugliano E., Cheroni C., Marabita F., Nogarotto R., Bonnal R. J., Marcatili P., Marconi M., Zignego A., Muratori P., Invernizzi P., Colombatto P., Brunetto M., Bonino F., De Francesco R., Geginat J., Pagani M., Muratori L., Abrignani S., and Bombaci M. (2012) Identification of new autoantigens by protein array indicates a role for IL4 neutralization in autoimmune hepatitis. Mol. Cell. Proteomics 11, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakajima R., Supnet M., Jasinskas A., Jain A., Taghavian O., Obiero J., Milton D. K., Chen W. H., Grantham M., Webby R., Krammer F., Carter D., Felgner P. L., and Davies D. H. (2018) Protein microarray analysis of the specificity and cross-reactivity of influenza virus hemagglutinin-specific antibodies. mSphere 3, pii: e00592–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desbien A. L., Van Hoeven N., Reed S. J., Casey A. C., Laurance J. D., Baldwin S. L., Duthie M. S., Reed S. G., and Carter D. (2013) Development of a high density hemagglutinin protein microarray to determine the breadth of influenza antibody responses. BioTechniques 54, 345–348 [DOI] [PubMed] [Google Scholar]
- 18. Dotsey E. Y., Gorlani A., Ingale S., Achenbach C. J., Forthal D. N., Felgner P. L., and Gach J. S. (2015) A high throughput protein microarray approach to classify HIV monoclonal antibodies and variant antigens. PLoS ONE 10, e0125581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu S., Feng Y., Henson B., Wang B., Huang X., Li M., Desai P., and Zhu H. (2013) VirD: a virion display array for profiling functional membrane proteins. Anal. Chem. 85, 8046–8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones R. B., Gordus A., Krall J. A., and MacBeath G. (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 [DOI] [PubMed] [Google Scholar]
- 21. Ayoglu B., Haggmark A., Khademi M., Olsson T., Uhlen M., Schwenk J. M., and Nilsson P. (2013) Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Mol. Cell. Proteomics 12, 2657–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sjoberg R., Mattsson C., Andersson E., Hellstrom C., Uhlen M., Schwenk J. M., Ayoglu B., and Nilsson P. (2016) Exploration of high-density protein microarrays for antibody validation and autoimmunity profiling. Nat. Biotechnol. 33, 582–592 [DOI] [PubMed] [Google Scholar]
- 23. Qi H., Zhou H., Czajkowsky D. M., Guo S., Li Y., Wang N., Shi Y., Lin L., Wang J., and Wu Tao S. C. (2017) Rapid production of virus protein microarray using protein microarray fabrication through gene synthesis (PAGES). Mol. Cell. Proteomics 16, 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang L., and Felgner P. L. (2015) A systems biology approach for diagnostic and vaccine antigen discovery in tropical infectious diseases. Curr. Opin. Infect. Dis. 28, 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vigil A., Davies D. H., and Felgner P. L. (2010) Defining the humoral immune response to infectious agents using high-density protein microarrays. Future Microbiol. 5, 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miersch S., Bian X., Wallstrom G., Sibani S., Logvinenko T., Wasserfall C. H., Schatz D., Atkinson M., Qiu J., and LaBaer J. (2013) Serological autoantibody profiling of type 1 diabetes by protein arrays. J. Proteomics 94, 486–496 [DOI] [PubMed] [Google Scholar]
- 27. Ramachandran N., Raphael J. V., Hainsworth E., Demirkan G., Fuentes M. G., Rolfs A., Hu Y., and LaBaer J. (2008) Next-generation high-density self-assembling functional protein arrays. Nat. Methods 5, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramachandran N., Hainsworth E., Bhullar B., Eisenstein S., Rosen B., Lau A. Y., Walter J. C., and LaBaer J. (2004) Self-assembling protein microarrays. Science 305, 86–90 [DOI] [PubMed] [Google Scholar]
- 29. Huang J., Zhu H., Haggarty S. J., Spring D. R., Hwang H., Jin F., Snyder M., and Schreiber S. L. (2004) Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. U.S.A. 101, 16594–16599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hall D. A., Zhu H., Zhu X., Royce T., Gerstein M., and Snyder M. (2004) Regulation of gene expression by a metabolic enzyme. Science 306, 482–484 [DOI] [PubMed] [Google Scholar]
- 31. Zhu J., Gopinath K., Murali A., Yi G., Hayward S. D., Zhu H., and Kao C. (2007) RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. U.S.A. 104, 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., Wan J., Tao S. C., Qian J., Zhao Y., Boeke J. D., Berger S. L., and Zhu H. (2009) Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136, 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu J. Y., Lin Y. Y., Qian J., Tao S. C., Zhu J., Pickart C., and Zhu H. (2008) Functional dissection of a HECT ubiquitin E3 ligase. Mol. Cell. Proteomics 7, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y. W., Teng C. H., Ho Y. H., Jessica Ho T. Y., Huang W. C., Hashimoto M., Chiang I. Y., and Chen C. S. (2014) Identification of bacterial factors involved in type 1 fimbria expression using an Escherichia coli K12 proteome chip. Mol. Cell. Proteomics 13, 1485–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ho Y. H., Shah P., Chen Y. W., and Chen C. S. (2016) Systematic analysis of intracellular-targeting antimicrobial peptides, bactenecin 7, hybrid of pleurocidin and dermaseptin, proline-arginine-rich peptide, and lactoferricin B, by using Escherichia coli proteome microarrays. Mol. Cell. Proteomics 15, 1837–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hsiao F. S., Sutandy F. R., Syu G. D., Chen Y. W., Lin J. M., and Chen C. S. (2016) Systematic protein interactome analysis of glycosaminoglycans revealed YcbS as a novel bacterial virulence factor. Sci. Rep. 6, 28425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu Z., Zhang H., Zhang X., Jiang H., Liu C., Wu F., Qian L., Hao B., Czajkowsky D. M., Guo S., Xu Z., Bi L., Wang S., Li H., Tan M., Yan W., Feng L., Hou J., and Tao S. C. (2019) Interplay between the bacterial protein deacetylase CobB and the second messenger c-di-GMP. EMBO J. 38, e100948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feng Y., Chen C. S., Ho J., Pearce D., Hu S., Wang B., Desai P., Kim K. S., and Zhu H. (2018) High-throughput chip assay for investigating Escherichia coli interaction with the blood-brain barrier using microbial and human proteome microarrays (Dual-Microarray Technology). Anal. Chem. 90, 10958–10966 [DOI] [PubMed] [Google Scholar]
- 39. Wang Z. X., Deng R. P., Jiang H. W., Guo S. J., Le H. Y., Zhao X. D., Chen C. S., Zhang J. B., and Tao S. C. (2012) Global identification of prokaryotic glycoproteins based on an Escherichia coli proteome microarray. PLoS ONE 7, e49080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang B. Y., Chen P. C., Chen B. H., Wang C. C., Liu H. F., Chen Y. Z., Chen C. S., and Yang Y. S. (2017) High-throughput screening of sulfated proteins by using a genome-wide proteome microarray and protein tyrosine sulfation system. Anal. Chem. 89, 3278–3284 [DOI] [PubMed] [Google Scholar]
- 41. Tsai C. H., Ho Y. H., Sung T. C., Wu W. F., and Chen C. S. (2017) Escherichia coli proteome microarrays identified the substrates of ClpYQ protease. Mol. Cell. Proteomics 16, 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu L., Li T., Song G., He Q., Yin Y., Lu J. Y., Bi X., Wang K., Luo S., Chen Y. S., Yang Y., Sun B. F., Yang Y. G., Wu J., Zhu H., and Shen X. (2019) Insight into novel RNA-binding activities via large-scale analysis of lncRNA-bound proteome and IDH1-bound transcriptome. Nucleic Acids Res. 47, 2244–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fan B., Lu K. Y., Reymond Sutandy F. X., Chen Y. W., Konan K., Zhu H., Kao C. C., and Chen C. S. (2014) A human proteome microarray identifies that the heterogeneous nuclear ribonucleoprotein K (hnRNP K) recognizes the 5′ terminal sequence of the hepatitis C virus RNA. Mol. Cell. Proteomics 13, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park C., Min S., Park E. M., Lim Y. S., Kang S., Suzuki T., Shin E. C., and Hwang S. B. (2015) Pim kinase interacts with nonstructural 5A protein and regulates hepatitis C virus entry. J. Virol. 89, 10073–10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoon K. J., Song G., Qian X., Pan J., Xu D., Rho H. S., Kim N. S., Habela C., Zheng L., Jacob F., Zhang F., Lee E. M., Huang W. K., Ringeling F. R., Vissers C., Li C., Yuan L., Kang K., Kim S., Yeo J., Cheng Y., Liu S., Wen Z., Qin C. F., Wu Q., Christian K. M., Tang H., Jin P., Xu Z., Qian J., Zhu H., Song H., and Ming G. L. (2017) Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell 21, 349–358 e346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Z., Hou Y., Hao T., Rho H. S., Wan J., Luan Y., Gao X., Yao J., Pan A., Xie Z., Qian J., Liao W., Zhu H., and Zhou X. (2017) A human proteome array approach to identifying key host proteins targeted by toxoplasma kinase ROP18. Mol. Cell. Proteomics 16, 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu F. L., Liu Y., Zhang H. N., Jiang H. W., Cheng L., Guo S. J., Deng J. Y., Bi L. J., Zhang X. E., Gao H. F., and Tao S. C. (2018) Global profiling of PknG interactions using a human proteome microarray reveals novel connections with CypA. Proteomics 18, e1800265. [DOI] [PubMed] [Google Scholar]
- 48. Yu X., Decker K. B., Barker K., Neunuebel M. R., Saul J., Graves M., Westcott N., Hang H., LaBaer J., Qiu J., and Machner M. P. (2015) Host-pathogen interaction profiling using self-assembling human protein arrays. J Proteome Res 14, 1920–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu X., Noll R. R., Romero Duenas B. P., Allgood S. C., Barker K., Caplan J. L., Machner M. P., LaBaer J., Qiu J., and Neunuebel M. R. (2018) Legionella effector AnkX interacts with host nuclear protein PLEKHN1. BMC Microbiol 18, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song G., Chen L., Zhang B., Song Q., Yu Y., Moore C., Wang T. L., Shih I. M., Zhang H., Chan D. W., Zhang Z., and Zhu H. (2019) Proteome-wide tyrosine phosphorylation analysis reveals dysregulated signaling pathways in ovarian tumors. Mol. Cell. Proteomics 18, 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu Z., Li X., Zhou S., Xie W., Wang J., Cheng L., Wang S., Guo S., Xu Z., Cao X., Zhang M., Yu B., Narimatsu H., Tao S. C., and Zhang Y. (2017) Systematic identification of the protein substrates of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T1/T2/T3 using a human proteome microarray. Proteomics 17, doi: 10.1002/pmic.201600485 [DOI] [PubMed] [Google Scholar]
- 52. Yu X., Woolery A. R., Luong P., Hao Y. H., Grammel M., Westcott N., Park J., Wang J., Bian X., Demirkan G., Hang H. C., Orth K., and LaBaer J. (2014) Copper-catalyzed azide-alkyne cycloaddition (click chemistry)-based detection of global pathogen-host AMPylation on self-assembled human protein microarrays. Mol. Cell. Proteomics 13, 3164–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang H. N., Yang L., Ling J. Y., Czajkowsky D. M., Wang J. F., Zhang X. W., Zhou Y. M., Ge F., Yang M. K., Xiong Q., Guo S. J., Le H. Y., Wu S. F., Yan W., Liu B., Zhu H., Chen Z., and Tao S. C. (2015) Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc. Natl. Acad. Sci. U.S.A. 112, 15084–15089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheng X., Liu Y. Q., Wang G. Z., Yang L. N., Lu Y. Z., Li X. C., Zhou B., Qu L. W., Wang X. L., Cheng Y. X., Liu J., Tao S. C., and Zhou G. B. (2017) Proteomic identification of the oncoprotein STAT3 as a target of a novel Skp1 inhibitor. Oncotarget 8, 2681–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bradbury A., and Pluckthun A. (2015) Reproducibility: Standardize antibodies used in research. Nature 518, 27–29 [DOI] [PubMed] [Google Scholar]
- 56. Michaud G. A., Salcius M., Zhou F., Bangham R., Bonin J., Guo H., Snyder M., Predki P. F., and Schweitzer B. I. (2003) Analyzing antibody specificity with whole proteome microarrays. Nat. Biotechnol. 21, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 57. Venkataraman A., Yang K., Irizarry J., Mackiewicz M., Mita P., Kuang Z., Xue L., Ghosh D., Liu S., Ramos P., Hu S., Bayron Kain D., Keegan S., Saul R., Colantonio S., Zhang H., Behn F. P., Song G., Albino E., Asencio L., Ramos L., Lugo L., Morell G., Rivera J., Ruiz K., Almodovar R., Nazario L., Murphy K., Vargas I., Rivera-Pacheco Z. A., Rosa C., Vargas M., McDade J., Clark B. S., Yoo S., Khambadkone S. G., de Melo J., Stevanovic M., Jiang L., Li Y., Yap W. Y., Jones B., Tandon A., Campbell E., Montelione G. T., Anderson S., Myers R. M., Boeke J. D., Fenyo D., Whiteley G., Bader J. S., Pino I., Eichinger D. J., Zhu H., and Blackshaw S. (2018) A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nat. Methods 15, 330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu F. L., Liu Y., Jiang H. W., Luan Y. Z., Zhang H. N., He X., Xu Z. W., Hou J. L., Ji L. Y., Xie Z., Czajkowsky D. M., Yan W., Deng J. Y., Bi L. J., Zhang X. E., and Tao S. C. (2017) The Ser/Thr protein kinase protein-protein interaction map of M. tuberculosis. Mol. Cell. Proteomics 16, 1491–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang Y., and Zhu H. (2017) Protein array-based approaches for biomarker discovery in cancer. Genomics Proteomics Bioinformatics 15, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu X., Petritis B., and LaBaer J. (2016) Advancing translational research with next-generation protein microarrays. Proteomics 16, 1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song Q., Liu G., Hu S., Zhang Y., Tao Y., Han Y., Zeng H., Huang W., Li F., Chen P., Zhu J., Hu C., Zhang S., Li Y., Zhu H., and Wu L. (2010) Novel autoimmune hepatitis-specific autoantigens identified using protein microarray technology. J. Proteome Res. 9, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wright C., Sibani S., Trudgian D., Fischer R., Kessler B., LaBaer J., and Bowness P. (2012) Detection of multiple autoantibodies in patients with ankylosing spondylitis using nucleic acid programmable protein arrays. Mol. Cell. Proteomics 11, M9 00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Querol L., Clark P. L., Bailey M. A., Cotsapas C., Cross A. H., Hafler D. A., Kleinstein S. H., Lee J. Y., Yaari G., Willis S. N., and O'Connor K. C. (2013) Protein array-based profiling of CSF identifies RBPJ as an autoantigen in multiple sclerosis. Neurology 81, 956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bian X., Wasserfall C., Wallstrom G., Wang J., Wang H., Barker K., Schatz D., Atkinson M., Qiu J., and LaBaer J. (2017) Tracking the antibody immunome in type 1 diabetes using protein arrays. J. Proteome Res. 16, 195–203 [DOI] [PubMed] [Google Scholar]
- 65. Nagele E., Han M., Demarshall C., Belinka B., and Nagele R. (2011) Diagnosis of Alzheimer's disease based on disease-specific autoantibody profiles in human sera. PLoS ONE 6, e23112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Auger I., Balandraud N., Rak J., Lambert N., Martin M., and Roudier J. (2009) New autoantigens in rheumatoid arthritis (RA): screening 8268 protein arrays with sera from patients with RA. Ann. Rheum. Dis. 68, 591–594 [DOI] [PubMed] [Google Scholar]
- 67. Hu S., Vissink A., Arellano M., Roozendaal C., Zhou H., Kallenberg C. G., and Wong D. T. (2011) Identification of autoantibody biomarkers for primary Sjogren's syndrome using protein microarrays. Proteomics 11, 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hu C. J., Song G., Huang W., Liu G. Z., Deng C. W., Zeng H. P., Wang L., Zhang F. C., Zhang X., Jeong J. S., Blackshaw S., Jiang L. Z., Zhu H., Wu L., and Li Y. Z. (2012) Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Mol. Cell. Proteomics 11, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. May C., Nordhoff E., Casjens S., Turewicz M., Eisenacher M., Gold R., Bruning T., Pesch B., Stephan C., Woitalla D., Penke B., Janaky T., Virok D., Siklos L., Engelhardt J. I., and Meyer H. E. (2014) Highly immunoreactive IgG antibodies directed against a set of twenty human proteins in the sera of patients with amyotrophic lateral sclerosis identified by protein array. PLoS ONE 9, e89596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Landegren N., Sharon D., Shum A. K., Khan I. S., Fasano K. J., Hallgren A., Kampf C., Freyhult E., Ardesjo-Lundgren B., Alimohammadi M., Rathsman S., Ludvigsson J. F., Lundh D., Motrich R., Rivero V., Fong L., Giwercman A., Gustafsson J., Perheentupa J., Husebye E. S., Anderson M. S., Snyder M., and Kampe O. (2015) Transglutaminase 4 as a prostate autoantigen in male subfertility. Sci. Transl. Med. 7, 292ra101. [DOI] [PubMed] [Google Scholar]
- 71. Gibson D. S., Qiu J., Mendoza E. A., Barker K., Rooney M. E., and LaBaer J. (2012) Circulating and synovial antibody profiling of juvenile arthritis patients by nucleic acid programmable protein arrays. Arthritis Res. Ther. 14, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hu C. J., Pan J. B., Song G., Wen X. T., Wu Z. Y., Chen S., Mo W. X., Zhang F. C., Qian J., Zhu H., and Li Y. Z. (2017) Identification of novel biomarkers for Behcet disease diagnosis using human proteome microarray approach. Mol. Cell. Proteomics 16, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Haggmark A., Hamsten C., Wiklundh E., Lindskog C., Mattsson C., Andersson E., Lundberg I. E., Gronlund H., Schwenk J. M., Eklund A., Grunewald J., and Nilsson P. (2015) Proteomic profiling reveals autoimmune targets in sarcoidosis. Am. J. Respir. Crit. Care Med. 191, 574–583 [DOI] [PubMed] [Google Scholar]
- 74. Hudson M. E., Pozdnyakova I., Haines K., Mor G., and Snyder M. (2007) Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 104, 17494–17499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Anderson K. S., Cramer D. W., Sibani S., Wallstrom G., Wong J., Park J., Qiu J., Vitonis A., and LaBaer J. (2015) Autoantibody signature for the serologic detection of ovarian cancer. J. Proteome Res. 14, 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Syed P., Gupta S., Choudhary S., Pandala N. G., Atak A., Richharia A., Zhu K. P. M. H., Epari S., Noronha S. B., Moiyadi A., and Srivastava S. (2015) Autoantibody profiling of glioma serum samples to identify biomarkers using human proteome arrays. Sci. Rep. 5, 13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pan J., Song G., Chen D., Li Y., Liu S., Hu S., Rosa C., Eichinger D., Pino I., Zhu H., Qian J., and Huang Y. (2017) Identification of serological biomarkers for early diagnosis of lung cancer using a protein array-based approach. Mol. Cell. Proteomics 16, 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang L., Wang J., Li J., Zhang H., Guo S., Yan M., Zhu Z., Lan B., Ding Y., Xu M., Li W., Gu X., Qi C., Zhu H., Shao Z., Liu B., and Tao S. C. (2016) Identification of serum biomarkers for gastric cancer diagnosis using a human proteome microarray. Mol. Cell. Proteomics 15, 614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Orenes-Pinero E., Barderas R., Rico D., Casal J. I., Gonzalez-Pisano D., Navajo J., Algaba F., Piulats J. M., and Sanchez-Carbayo M. (2010) Serum and tissue profiling in bladder cancer combining protein and tissue arrays. J. Proteome Res. 9, 164–173 [DOI] [PubMed] [Google Scholar]
- 80. Adeola H. A., Smith M., Kaestner L., Blackburn J. M., and Zerbini L. F. (2016) Novel potential serological prostate cancer biomarkers using CT100+ cancer antigen microarray platform in a multi-cultural South African cohort. Oncotarget 7, 13945–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Babel I., Barderas R., Diaz-Uriarte R., Martinez-Torrecuadrada J. L., Sanchez-Carbayo M., and Casal J. I. (2009) Identification of tumor-associated autoantigens for the diagnosis of colorectal cancer in serum using high density protein microarrays. Mol. Cell. Proteomics 8, 2382–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Anderson K. S., Sibani S., Wallstrom G., Qiu J., Mendoza E. A., Raphael J., Hainsworth E., Montor W. R., Wong J., Park J. G., Lokko N., Logvinenko T., Ramachandran N., Godwin A. K., Marks J., Engstrom P., and Labaer J. (2011) Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J. Proteome Res. 10, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mias G. I., Chen R., Zhang Y., Sridhar K., Sharon D., Xiao L., Im H., Snyder M. P., and Greenberg P. L. (2013) Specific plasma autoantibody reactivity in myelodysplastic syndromes. Sci. Rep. 3, 3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gupta S., Mukherjee S., Syed P., Pandala N. G., Choudhary S., Singh V. A., Singh N., Zhu H., Epari S., Noronha S. B., Moiyadi A., and Srivastava S. (2017) Evaluation of autoantibody signatures in meningioma patients using human proteome arrays. Oncotarget 8, 58443–58456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Montor W. R., Huang J., Hu Y., Hainsworth E., Lynch S., Kronish J. W., Ordonez C. L., Logvinenko T., Lory S., and LaBaer J. (2009) Genome-wide study of Pseudomonas aeruginosa outer membrane protein immunogenicity using self-assembling protein microarrays. Infect. Immun. 77, 4877–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ceroni A., Sibani S., Baiker A., Pothineni V. R., Bailer S. M., LaBaer J., Haas J., and Campbell C. J. (2010) Systematic analysis of the IgG antibody immune response against varicella zoster virus (VZV) using a self-assembled protein microarray. Mol. Biosyst. 6, 1604–1610 [DOI] [PubMed] [Google Scholar]
- 87. Song L., Wallstrom G., Yu X., Hopper M., Van Duine J., Steel J., Park J., Wiktor P., Kahn P., Brunner A., Wilson D., Jenny-Avital E. R., Qiu J., Labaer J., Magee D. M., and Achkar J. M. (2017) Identification of antibody targets for tuberculosis serology using high-density nucleic acid programmable protein arrays. Mol. Cell. Proteomics 16, S277–S289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lessa-Aquino C., Wunder E. A. Jr, Lindow J. C., Rodrigues C. B., Pablo J., Nakajima R., Jasinskas A., Liang L., Reis M. G., Ko A. I., Medeiros M. A., and Felgner P. L. (2015) Proteomic features predict seroreactivity against leptospiral antigens in leptospirosis patients. J. Proteome Res. 14, 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liang L., Juarez S., Nga T. V., Dunstan S., Nakajima-Sasaki R., Davies D. H., McSorley S., Baker S., and Felgner P. L. (2013) Immune profiling with a Salmonella Typhi antigen microarray identifies new diagnostic biomarkers of human typhoid. Sci. Rep. 3, 1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liang L., Tan X., Juarez S., Villaverde H., Pablo J., Nakajima-Sasaki R., Gotuzzo E., Saito M., Hermanson G., Molina D., Felgner S., Morrow W. J., Liang X., Gilman R. H., Davies D. H., Tsolis R. M., Vinetz J. M., and Felgner P. L. (2011) Systems biology approach predicts antibody signature associated with Brucella melitensis infection in humans. J. Proteome Res. 10, 4813–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Luevano M., Bernard H. U., Barrera-Saldana H. A., Trevino V., Garcia-Carranca A., Villa L. L., Monk B. J., Tan X., Davies D. H., Felgner P. L., and Kalantari M. (2010) High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays. Virology 405, 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mochon A. B., Jin Y., Kayala M. A., Wingard J. R., Clancy C. J., Nguyen M. H., Felgner P., Baldi P., and Liu H. (2010) Serological profiling of a Candida albicans protein microarray reveals permanent host-pathogen interplay and stage-specific responses during candidemia. PLoS Pathog. 6, e1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sundaresh S., Randall A., Unal B., Petersen J. M., Belisle J. T., Hartley M. G., Duffield M., Titball R. W., Davies D. H., Felgner P. L., and Baldi P. (2007) From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23, i508–i518 [DOI] [PubMed] [Google Scholar]
- 94. Kalantari-Dehaghi M., Chun S., Chentoufi A. A., Pablo J., Liang L., Dasgupta G., Molina D. M., Jasinskas A., Nakajima-Sasaki R., Felgner J., Hermanson G., BenMohamed L., Felgner P. L., and Davies D. H. (2012) Discovery of potential diagnostic and vaccine antigens in herpes simplex virus 1 and 2 by proteome-wide antibody profiling. J. Virol. 86, 4328–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu M., Subramanian V., Christie C., Castro M., and Mohanakumar T. (2012) Immune responses to self-antigens in asthma patients: clinical and immunopathological implications. Hum. Immunol. 73, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kuo H. C., Huang Y. H., Chung F. H., Chen P. C., Sung T. C., Chen Y. W., Hsieh K. S., Chen C. S., and Syu G. D. (2018) Antibody profiling of Kawasaki Disease using Escherichia coli proteome microarrays. Mol. Cell. Proteomics 17, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hsu T. Y., Lin J. M., Nguyen M. T., Chung F. H., Tsai C. C., Cheng H. H., Lai Y. J., Hung H. N., and Chen C. S. (2018) Antigen analysis of pre-eclamptic plasma antibodies using Escherichia coli proteome chips. Mol. Cell. Proteomics 17, 1457–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chen P. C., Syu G. D., Chung K. H., Ho Y. H., Chung F. H., Chen P. H., Lin J. M., Chen Y. W., Tsai S. Y., and Chen C. S. (2015) Antibody profiling of bipolar disorder using Escherichia coli proteome microarrays. Mol. Cell. Proteomics 14, 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Han M., Nagele E., DeMarshall C., Acharya N., and Nagele R. (2012) Diagnosis of Parkinson's disease based on disease-specific autoantibody profiles in human sera. PLoS ONE 7, e32383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Henjes F., Lourido L., Ruiz-Romero C., Fernandez-Tajes J., Schwenk J. M., Gonzalez-Gonzalez M., Blanco F. J., Nilsson P., and Fuentes M. (2014) Analysis of autoantibody profiles in osteoarthritis using comprehensive protein array concepts. J. Proteome Res. 13, 5218–5229 [DOI] [PubMed] [Google Scholar]
- 101. Butte A. J., Sigdel T. K., Wadia P. P., Miklos D. B., and Sarwal M. M. (2011) Protein microarrays discover angiotensinogen and PRKRIP1 as novel targets for autoantibodies in chronic renal disease. Mol. Cell. Proteomics 10, M110.000497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Vermeulen N., de Beeck K. O., Vermeire S., Van Steen K., Michiels G., Ballet V., Rutgeerts P., and Bossuyt X. (2011) Identification of a novel autoantigen in inflammatory bowel disease by protein microarray. Inflamm. Bowel Dis. 17, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 103. Chen C. S., Sullivan S., Anderson T., Tan A. C., Alex P. J., Brant S. R., Cuffari C., Bayless T. M., Talor M. V., Burek C. L., Wang H., Li R., Datta L. W., Wu Y., Winslow R. L., Zhu H., and Li X. (2009) Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Mol. Cell. Proteomics 8, 1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kim S. H., Kim J. Y., Lee H. J., Gi M., Kim B. G., and Choi J. Y. (2014) Autoimmunity as a candidate for the etiopathogenesis of Meniere's disease: detection of autoimmune reactions and diagnostic biomarker candidate. PLoS ONE 9, e111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ma G., Syu G. D., Shan X., Henson B., Wang S., Desai P. J., Zhu H., and Tao N. (2018) Measuring Ligand Binding Kinetics to Membrane Proteins Using Virion Nano-oscillators. J. Am. Chem. Soc. 140, 11495–11501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nath A., Atkins W. M., and Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 107. Park S. H., Berkamp S., Cook G. A., Chan M. K., Viadiu H., and Opella S. J. (2011) Nanodiscs versus macrodiscs for NMR of membrane proteins. Biochemistry 50, 8983–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Frauenfeld J., Loving R., Armache J. P., Sonnen A. F., Guettou F., Moberg P., Zhu L., Jegerschold C., Flayhan A., Briggs J. A., Garoff H., Low C., Cheng Y., and Nordlund P. (2016) A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods 13, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hirozane Y., Motoyaji T., Maru T., Okada K., and Tarui N. (2014) Generating thermostabilized agonist-bound GPR40/FFAR1 using virus-like particles and a label-free binding assay. Mol. Membr. Biol. 31, 168–175 [DOI] [PubMed] [Google Scholar]
- 110. Koch B., Nijmeijer B., Kueblbeck M., Cai Y., Walther N., and Ellenberg J. (2018) Generation and validation of homozygous fluorescent knock-in cells using CRISPR-Cas9 genome editing. Nat. Protoc. 13, 1465–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sjoberg R., Sundberg M., Gundberg A., Sivertsson A., Schwenk J. M., Uhlen M., and Nilsson P. (2012) Validation of affinity reagents using antigen microarrays. Nat. Biotechnol. 29, 555–563 [DOI] [PubMed] [Google Scholar]
- 112. Lin J. M., Tsai Y. T., Liu Y. H., Lin Y., Tai H. C., and Chen C. S. (2016) Identification of 2-oxohistidine interacting proteins using E. coli proteome chips. Mol. Cell. Proteomics 15, 3581–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. He X., Jiang H. W., Chen H., Zhang H. N., Liu Y., Xu Z. W., Wu F. L., Guo S. J., Hou J. L., Yang M. K., Yan W., Deng J. Y., Bi L. J., Zhang X. E., and Tao S. C. (2017) Systematic identification of Mycobacterium tuberculosis effectors reveals that BfrB suppresses innate immunity. Mol. Cell. Proteomics 16, 2243–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Uzoma I., Hu J., Cox E., Xia S., Zhou J., Rho H. S., Guzzo C., Paul C., Ajala O., Goodwin C. R., Jeong J., Moore C., Zhang H., Meluh P., Blackshaw S., Matunis M., Qian J., and Zhu H. (2018) Global identification of small ubiquitin-related modifier (SUMO) substrates reveals crosstalk between SUMOylation and phosphorylation promotes cell migration. Mol. Cell. Proteomics 17, 871–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Newman R. H., Hu J., Rho H. S., Xie Z., Woodard C., Neiswinger J., Cooper C., Shirley M., Clark H. M., Hu S., Hwang W., Jeong J. S., Wu G., Lin J., Gao X., Ni Q., Goel R., Xia S., Ji H., Dalby K. N., Birnbaum M. J., Cole P. A., Knapp S., Ryazanov A. G., Zack D. J., Blackshaw S., Pawson T., Gingras A. C., Desiderio S., Pandey A., Turk B. E., Zhang J., Zhu H., and Qian J. (2013) Construction of human activity-based phosphorylation networks. Mol. Syst. Biol. 9, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li R., Zhu J., Xie Z., Liao G., Liu J., Chen M. R., Hu S., Woodard C., Lin J., Taverna S. D., Desai P., Ambinder R. F., Hayward G. S., Qian J., Zhu H., and Hayward S. D. (2011) Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 10, 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., Meng L., Stark M. J., Stern D. F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P. F., and Snyder M. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 [DOI] [PubMed] [Google Scholar]
- 118. Lu J. Y., Lin Y. Y., Sheu J. C., Wu J. T., Lee F. J., Chen Y., Lin M. I., Chiang F. T., Tai T. Y., Berger S. L., Zhao Y., Tsai K. S., Zhu H., Chuang L. M., and Boeke J. D. (2011) Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 146, 969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Porcheray F., DeVito J., Yeap B. Y., Xue L., Dargon I., Paine R., Girouard T. C., Saidman S. L., Colvin R. B., Wong W., and Zorn E. (2010) Chronic humoral rejection of human kidney allografts associates with broad autoantibody responses. Transplantation 89, 1239–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]