Graphical abstract
Abbreviations: CD, Circular dichroism; CG, coarse-grained; HMGB1, High Mobility Group Box 1; MD, molecular dynamics; PACE, proteins with atomic details in a coarse-grained environment; SASA, solvent accessible surface area
Keywords: HMGB1, Acidic tail, Circular dichroism, Coarse-grained molecular dynamics simulations, PACE
Abstract
High mobility group box 1 (HMGB1) is a damage-associated molecular pattern (DAMP) molecule that triggers the progression of several pro-inflammatory diseases such as diabetes, Alzheimer’s disease and cancer, by inducing signals upon interaction with the receptors such as the receptor for advanced glycation end-products (RAGE) and toll-like receptors (TLRs). The acidic C-terminal tail of HMGB1 is an intrinsically disordered region of the protein which is known to determine the interaction of HMGB1 to DNA and histones. This study characterizes its structural properties using a combination of circular dichroism (CD) and molecular dynamics (MD) simulations. The full-length and tail-less forms of HMGB1 were compared to rationalise the role of the acidic tail in maintaining the stability of the entire structure of HMGB1 in atomistic detail. Consistent with experimental data, the acidic tail was predicted to adopt an extended conformation that allows it to make a range of hydrogen-bonding and electrostatic interactions with the box-like domains that stabilize the overall structure of HMGB1. Absence of the acidic tail was predicted to increase structural fluctuations of all amino acids, leading to changes in secondary structure from α-helical to more hydrophilic turns along with increased exposure of multiple amino acids to the surrounding solvent. These structural changes reveal the intrinsic conformational dynamics of HMGB1 that are likely to affect the accessibility of its receptors.
1. Introduction
Disordered regions in proteins frequently play significant biological roles which are often accompanied by large scale conformational shifts [1]. A disordered region is often characterized by, among other features, the simplicity of its amino acid sequence, which causes the protein to be dynamically disordered or unable to fold spontaneously [2]. Interestingly, about 50% of all human proteins contain intrinsically disordered regions, consisting of 40 amino acids or more [3]. Some intrinsically disordered regions only become ordered upon binding to other biomolecules, whereas others remain partially disordered even during interactions with other biomolecules [4]. Investigation of intrinsically disordered proteins by molecular dynamics (MD) simulation is now an area of increasing research interest [5] due to the challenges of capturing the unique folding and interaction kinetics of these proteins caused by their disordered regions [6]. High Mobility Group Box 1 (HMGB1) is a protein that contains an intrinsically disordered region [7]. Intracellular HMGB1 binds DNA and helps its transcription, whilst extracellular HMGB1 interacts with some pattern recognition receptors, mainly toll-like receptors (TLRs) and receptor of advanced glycation end products (RAGE), triggering diseases that involves tissue inflammation such as cancer and chronic sepsis [8].
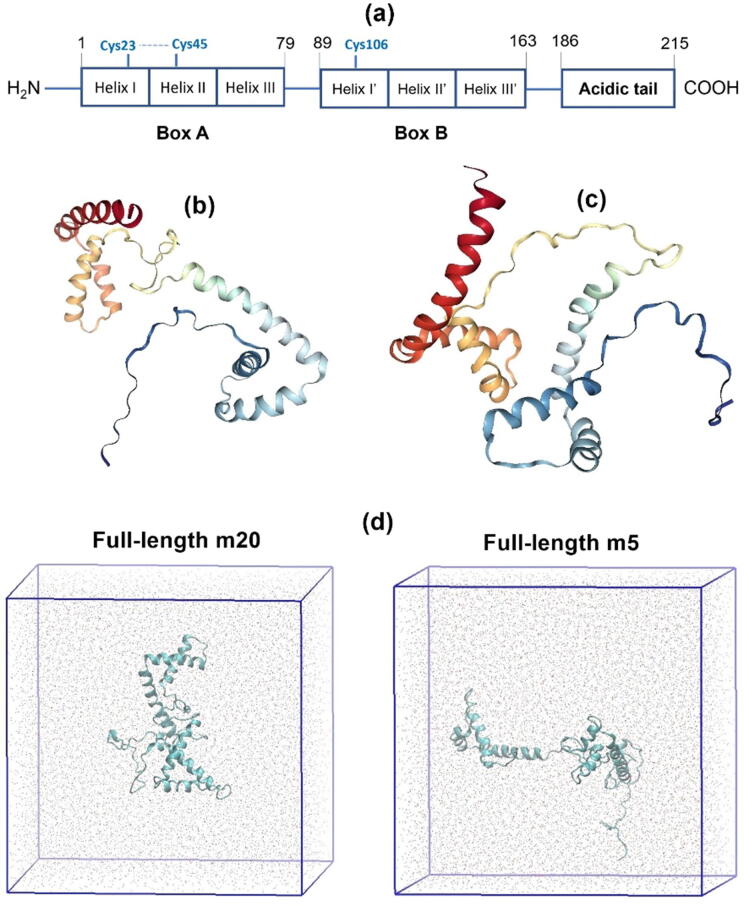
HMGB1 is composed of two tandem box-like domains, Box A and Box B, each consisting of three helices, as shown in Fig. 1. The predominant secondary structure of HMGB1 is reported to be α-helix [7], [9], [10], [11]. The acidic C-terminal tail preceding the boxes is an intrinsically disordered region that plays an important role in preserving the structural integrity of the entire protein by maintaining its stability against chemical denaturation and low pH [7]. A previous CD study put forward that, without the acidic tail, HMGB1 has lower thermal stability than full-length HMGB1 but upon interaction with the tail peptide, truncated HMGB1 has similar stability to that of full-length HMGB1 [9]. The acidic tail has a highly negative charge due to its 30 consecutive Asp and Glu residues [12], from residue 186 to 215 (Fig. 1a). The acidic tail in the HMG protein family is strongly associated with conserved domains within the HMG boxes and the basic linker region [13], thus playing a role in the interaction with binding partners. For example, the tail in HMGB1 can interact with the DNA binding regions of the two predominantly basic boxes and linker region, obstructing their DNA binding region and diminishing the ability of Box B to bend DNA [13], [14]. The interaction of the tail with the boxes and linker regions causes the whole structure of HMGB1 to collapse in an auto-inhibited conformation. This conformation exists in an equilibrium with the more open form, which is competent for binding DNA or other proteins [13], [15]. The tail was reported to bind to Box B with higher affinity than Box A by Stott et al. [15]. Their SAXS studies revealed that there is an ensemble of closed (collapsed) and open conformations, with the former being preferred [15]. The end portion of the tail was determined to protrude away from the rest of the protein [15].
Fig. 1.
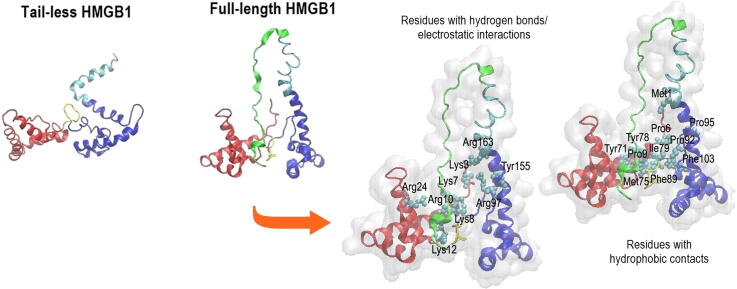
(a) Representation of the domain structure of full-length HMGB1. Beneath: structural models of full-length and tail-less HMGB1. These models were taken from the Protein Data Bank (PDB) entry 2YRQ: (b) m20 and (c) m5. In each structure, red regions represent Helix I, orange parts represent Helix II, and amber portions represents Helix III of Box A (residues 1–79). Yellow regions represent the linker residues 80–88. Meanwhile, cyan parts represent Helix I’, light blue regions represent Helix II’, and blue portions represent Helix III’ of Box B (residues 89–163). The navy-blue regions represent the linker between Box B and the acidic tail (residues 164–185). (d) Snapshots of the corresponding full-length structures of these models after MD simulation equilibration. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Previous NMR studies have indicated that a number of specific residues within the boxes interact with the acidic tail. The amino acids reported in common are Trp49, Thr77, Ala94, Ile159 and Arg163 [9], [10], [15]. More specifically, Knapp et al. reported residues Thr77, Ile79, Asp93, Ala94, Ile159 and Arg163 [9], Watson et al. reported residues Trp49, Lys50, Thr77, Lys112, Ala137 and Ile159 [10], whereas Stott et al. [15] reported residues Gln21, Lys43, Trp49, Asp62, Thr77, Tyr78, Ala94, Arg110, Ile159 and Ala160. This last study also suggested that different conformations of HMGB1 determine which amino acids of the boxes are likely to interact with the acidic tail [15].
HMGB1 has three conserved Cys residues (Fig. 1a), which are sensitive towards the redox environment. The change of redox status modifies these residues and the conformation of HMGB1, which can determine its interactions with other proteins or molecules [16]. To enable extracellular HMGB1 to promote cytokine activity, these Cys residues must exist in specific reduced states. When Cys23 and Cys45 in Box A form disulfide bond and the Cys106 in Box B is reduced, HMGB1 is reported to interact with myeloid differentiation factor 2 (MD2) in the TLR4/MD2 complex [16]. When all Cys residues are reduced, HMGB1 interacts with CXC motif ligand 12 (CXCL12) [17].
In this study, the contribution of the acidic tail to the overall structure of HMGB1 was characterized using circular dichroism (CD) and MD simulation. A coarse-grained (CG) representation was selected to facilitate the simulation of this system for long enough time to observe biologically-relevant changes in molecular conformations [18]. Various MD force fields have been reported to not possess the correct balance between protein–protein and protein–water dispersion interactions, which are necessary to accurately model intrinsically disordered proteins [6]. We thus chose the united atom ‘proteins with atomic details in a coarse-grained environment’ (PACE) force field, which combines the simulation of a protein in atomistic detail and a coarse-grained description of the solvent to accurately describe protein folding [18] to describe its thermodynamic and kinetic properties [19].
Our simulations characterise the interactions of the acidic tail with the boxes and their relationship with the overall structural stability of HMGB1, which is discussed in the context of published experimental data. Absence of the acidic tail was observed to lead to significant changes in the secondary structure of HMGB1, from α-helical to more solvent-exposed turns as a result of increased fluctuations and solvent exposure of multiple amino acids in the protein. This suggests that the acidic tail may play a key role in determining HMGB1 function and its propensity for interaction with certain partners.
2. Materials and methods
2.1. Circular dichroism of HMGB1 constructs
The HMGB1 constructs, with and without the acidic tail, were isolated from crude protein lysate obtained from a culture of E. coli T7 Shuffle K transformed with plasmids containing HMGB1 gene. Protein purification was done in two-step chromatography, using HiTrap Heparin HP (GE Healthcare) and Superdex 200 10/300 GL (GE Healthcare) columns in a ÄKTA Purifier 100 system (GE Healthcare), as previously reported [11], [20]. These protein constructs were then dialyzed against a 10 mM KH2PO4 buffer at a pH of 7.4 [11].
CD measurements were conducted using a JASCO J-815 spectropolarimeter with a 1.0 mm path length rectangular Spectrosil Quartz cuvette (Starna). All measurements were performed with standard sensitivity (100 mdeg), a 1 nm data pitch and bandwidth, a speed of 100 nm/min in continuous scanning mode, and 1 s response. Absorbance was monitored over the range 260–185 nm. All experiments were initiated by blank measurements over five accumulated scans using 400 μl of the above buffer.
The reducing agent β-mercaptoethanol was used to disrupt the disulfide bond between Cys23 and Cys45 in HMGB1. The effect of adding β-mercaptoethanol to the HMGB1 constructs was determined by observing changes to their CD spectra. Three aliquots of each form of HMGB1 were prepared and β-mercaptoethanol was added to each one to the final working concentrations of 0 mM, 1 mM, 5 mM and 10 mM. The samples were incubated for 1 h on ice before measurements in the spectropolarimeter. The CD spectra obtained for each concentration was then blank-subtracted with the above described buffer containing the respective β-mercaptoethanol concentration. A similar procedure was performed for samples treated with different concentrations of the free cysteine alkylating agent, iodoacetamide. The final working concentrations for the iodoacetamide assay were 0.5 mM, 1 mM and 5 mM.
2.2. Secondary structure determination by CD
Determination of the secondary structure content in HMGB1 by CD was carried out as described previously [11]. CD measurements of the secondary structure of both full-length and tail-less HMGB1 at a concentration of 20 μM were conducted using a 10 mM KH2PO4 buffer (at a pH of 7.4 buffer) as the diluent, and the CD absorption spectra were accumulated over five scans. To obtain the CD spectra and secondary structure percentages, the raw data obtained were submitted to DichroWeb [21] and processed with reference set 6, which was specifically optimized for a wavelength range of 185–240 nm [22]. The CONTIN algorithm was used to fit the data and predict the secondary structure contents [23].
2.3. MD simulations
The reference structure for HMGB1 without its acidic C-terminal tail was taken from the solution NMR structure of the tandem HMG box domain of HMGB1 (PDB file 2YRQ) [24], [25], which consists of 173 residues. The Cys residues were kept in their reduced form. To obtain the complete structure of HMGB1, the 30 residue-sequence of the tail and its preceding Val174-Lys185 region was modelled using MODELLER [26]. Henceforth, these two structures will be referred to as full-length and tail-less HMGB1. As shown in Fig. 1b and c, two different structural models of HMGB1 (5 and 20, henceforth referred to as m5 and m20) were selected from the total of 20 models in the conformational ensemble determined by NMR spectroscopy. The m20 model was selected from amongst the three models that have the largest distance between the box domains, namely models 9, 14, and 20. Model m20 has the lowest atom-based root mean square deviation (RMSD) with respect to model 1, which was taken as the reference structure since it is the lowest-energy model in the NMR ensemble. Model 5 was selected as it exhibits the largest RMSD with respect to model m20. Models 5 and 20 thus represent the two most distinct conformations determined experimentally, which may exhibit different conformational behavior and interactions with the acidic tail.
The PACE united-atom protein force field [18] was applied to both forms of HMGB1 using the MD simulation program GROMACS [27]. The PACE force field was optimized for residue-specific conformational potentials and the interactions between side chains and the protein backbone, ensuring a balanced representation of structurally defined and disordered regions [19]. Cubic simulation cells were constructed with a 12 Å distance between the protein and the sides of the cell (Fig. 1d). In each system the protein was solvated using the MARTINI water model [28]. The net charge of the protein was neutralized, and an ionic strength of 0.15 M was simulated by adding 161 Na+ and 156 Cl− ions for full-length and 156 Na+ and 181 Cl− ions for tail-less HMGB1.
The production runs were performed using GROMACS 4.5.5 [29]. Electrostatic interactions between polar groups were calculated using Coulomb potentials. For consistency with the PACE force field, switching functions were used with Coulomb potentials from 0 to 1.2 nm and with Lennard-Jones 6–12 potentials from 0.9 to 1.2 nm [30]. All production runs were performed in the NPT ensemble. The system temperature was maintained at 323 K using the Berendsen thermostat [31]. The diffusion constant of water in the original MARTINI model (with which the PACE force field was developed) agrees with the experimental value at this temperature, which also prevents the freezing of water, a problem known to occur with the MARTINI water model at lower temperatures [32]. A constant isotropic pressure was maintained at 0.1 MPa with the Parrinello-Rahman barostat [33]. Before commencing the production run, an initial run of 0.4 µs was performed with an elongated structure of HMGB1, where the tail was located further away from the boxes. All production runs (from which all analyses were conducted) were carried out for 10 µs with a time step of 20 ps. The trajectory data was recorded every 10 ns. VMD was used for all visualization tasks [34].
Structural analyses were performed for the region encompassing residues 1–185, whereas computation of hydrophobic contacts and hydrogen bonds included the tail region encompassing residues 186–215. The secondary structure content of the protein was measured using the Dictionary of Secondary Structures of Protein (DSSP) [35]. Analysis of the average root-mean-square deviation (RMSD) of main chain Cα atoms was performed to assess the impact of the C-terminal acidic tail on the structural stability of HMGB1 by comparing between structures with and without the tail. The RMSD was measured with respect to the initial model chosen from the NMR structure (2YRQ). Clustering of conformations was carried using the g_cluster module in GROMACS [29] using a RMSD cut-off of 0.4 nm to identify unique conformations of HMGB1. Analysis of hydrophobic contacts between boxes A and B with the g_mindist module and of hydrogen bonds and/or electrostatic interactions between each box and the acidic tail with the g_hbond module were conducted for the two full-length forms of HMGB1, in both cases using a conservative cut-off of 0.5 nm. The average root-mean-square fluctuation (RMSF) [29] of each amino acid in the protein was also computed in the presence and absence of the tail. Lastly, computation of the solvent accessible surface area (SASA) [29] was done to determine the extent of surface area exposure towards the solvent, which is affected by conformational changes.
3. Results
3.1. Secondary structure and stability of full-length and tail-less HMGB1
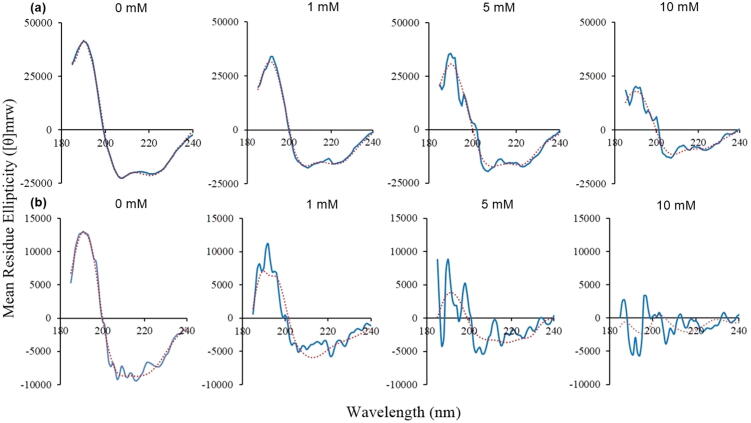
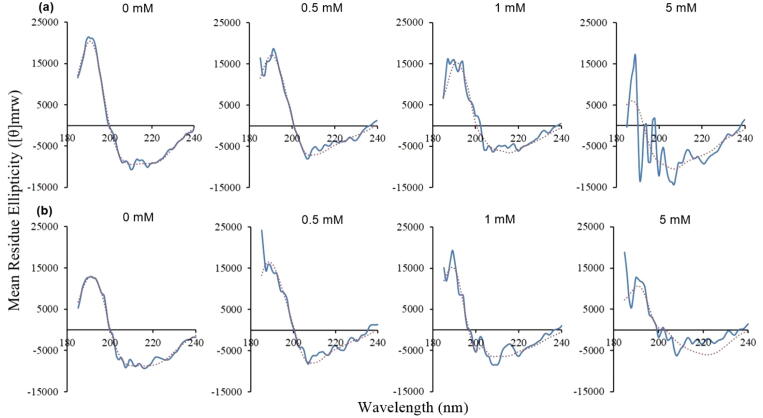
Comparison of the secondary structure content of full-length and tail-less HMGB1 in the absence of β-mercaptoethanol (Fig. 2) reveals that full-length HMGB1 exhibits a substantially higher proportion of helical content than the tail-less form. This is confirmed by the CONTIN fit (Table S1 in the Supporting Information). Fig. 2 also reveals that the amount of helical content in both forms decreases upon reduction with β-mercaptoethanol. The predicted amounts of secondary structure content are less reliable with higher concentrations of β-mercaptoethanol because the CONTIN fit is poorer, and this is especially the case for tail-less HMGB1 across all concentrations of β-mercaptoethanol (Fig. 2). Both observations suggest that full-length HMGB1 has a more stable structure upon reduction.
Fig. 2.
Circular dichroism spectra for (a) full-length HMGB1 and (b) tail-less HMGB1, upon addition of different concentrations of β-mercaptoethanol. The blue line is the experimental data and the red dotted line is the CONTIN fit. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Analysis of secondary structure with DSSP of the MD simulations of both the full-length m20 and m5 demonstrates that Box A (residues 1–79) and Box B (residues 89–163) are dominated by the presence of α-helical conformations (shown as blue areas in Fig. 3a and c). Other prominent conformations are turn (shown as yellow areas), which are especially apparent in the regions between residues 75–100 and 165–185, corresponding to the linker regions between Box A and Box B, and between Box B and the acidic tail, respectively (see Fig. 1).
Fig. 3.
Time evolution of the predicted secondary structure of (a) full-length m20; (b) tail-less m20; (c) full-length m5; and (d) tail-less m5. Secondary structure was determined with the DSSP method. Red arrows highlight regions in the sequence of tail-less m20 and m5 that exhibit shifts in conformation between α-helical and turn. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The corresponding analysis of secondary structure for both the tail-less m20 and m5 (Fig. 3b and d) suggests the occurrence of instabilities with respect to the full-length counterparts, which appear as frequent shifts between α-helical conformations and turns. The structural shift between α-helix to turn in tail-less m20 is indicated with red arrows which point to amino acids near positions Glu40, Phe60, Arg110, Leu120, and Asn135 (Fig. 3b). Meanwhile, the red arrows tail-less m5 indicate shifts in tail-less m5 around amino acids near positions Ser35, Phe60, Thr85, Asn135, and Ala160 (Fig. 3d).
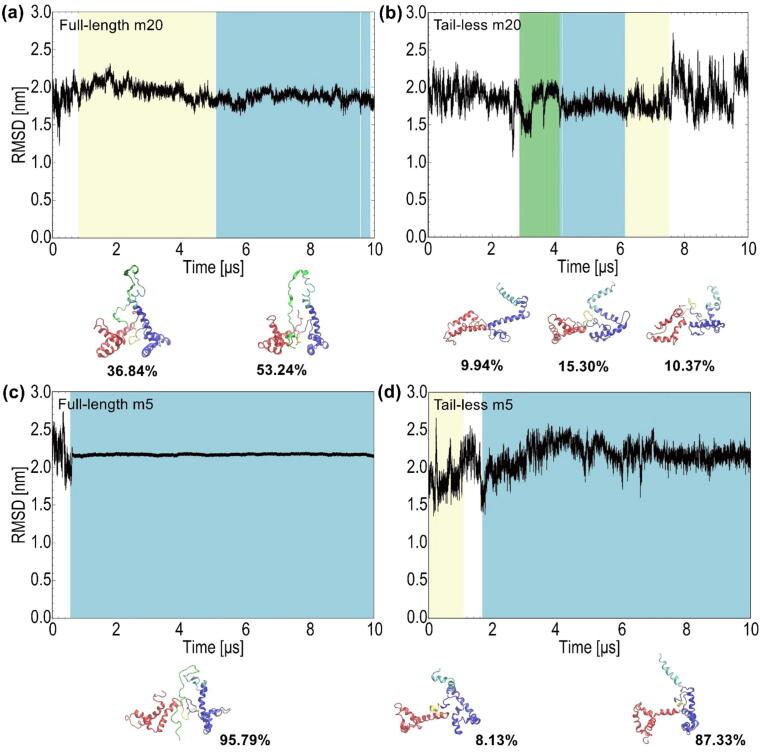
Analysis of the Cα-RMSD for full-length m20 revealed that the RMSD remained nearly constant around an average value of 1.84 nm (Fig. 4a). During the simulation the acidic tail adopted an extended conformation between the boxes, as shown in the representative conformations at the bottom of Fig. 4a. By contrast, the Cα-RMSD of tail-less m20 exhibited larger oscillations with an average value of 2.06 nm (Fig. 4b), demonstrating that the structure is not as stable as its full-length counterpart. In the case of full-length m5, analysis of the Cα-RMSD revealed that it rapidly reached a very stable value, as evidenced by a small fluctuation around an average value of 2.15 nm (Fig. 4c). The corresponding values of Cα-RMSD for tail-less m5 (Fig. 4d) reveal a similar degree of oscillations around an average of 2.07 nm to those seen for its tail-less m20 counterpart (Fig. 4b). This suggests that the structure of tail-less m5 is also not as stable as its full-length counterpart.
Fig. 4.
Time evolution of the Cα-RMSD and dominant conformations obtained from cluster analysis of (a) full-length m20; (b) tail-less m20; (c) full-length m5; and, (d) tail-less m5. The color scheme in the RMSD plots indicates the change of conformations, with cyan indicating the time frame for the most favored conformation, followed by crème and green as the area of the least preferred conformation, which are shown at the bottom of each RMSD plot. In each structure the red regions represent Box A, blue regions represent Box B, yellow portions represent the linker between boxes, cyan regions represent the linker between Box B and the acidic tail, and light green portions represent the acidic tail. Each conformation is shown with its corresponding percentage occupancy. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Clustering of conformations in m20 and m5 forms
A RMSD-based cluster analysis was performed to identify true representative and unique conformations sampled during the simulation. Each simulation trajectory of the full-length and tail-less forms of m20 and m5 was clustered separately. Dominant clusters were identified for each form of HMGB1, as shown at the bottom of the Cα-RMSD plots in Fig. 4. There were more clusters that could be identified; however, since their percentages were less than 2%, only clusters exhibiting percentages near 10% or more are presented. The dominant conformations of both tail-less m20 and m5 (Fig. 4b and d) show that the boxes fluctuate more than their full-length counterparts. Consequently, the distance between the boxes also differs significantly between conformations. On the other hand, the lack of variation in the position and distance between the boxes in the dominant conformations of full-length m20 and m5 (Fig. 4a and c) may be influenced by the position of the tail. The position of the acidic tail between the boxes seems to be the most favoured conformation of both full-length forms, whereby the tail inserts itself into a deeper position between the boxes in the dominant conformations of full-length m5 (Fig. 4c), whereas the tail in the dominant conformations of full-length m20 is located higher above the boxes (Fig. 4a).
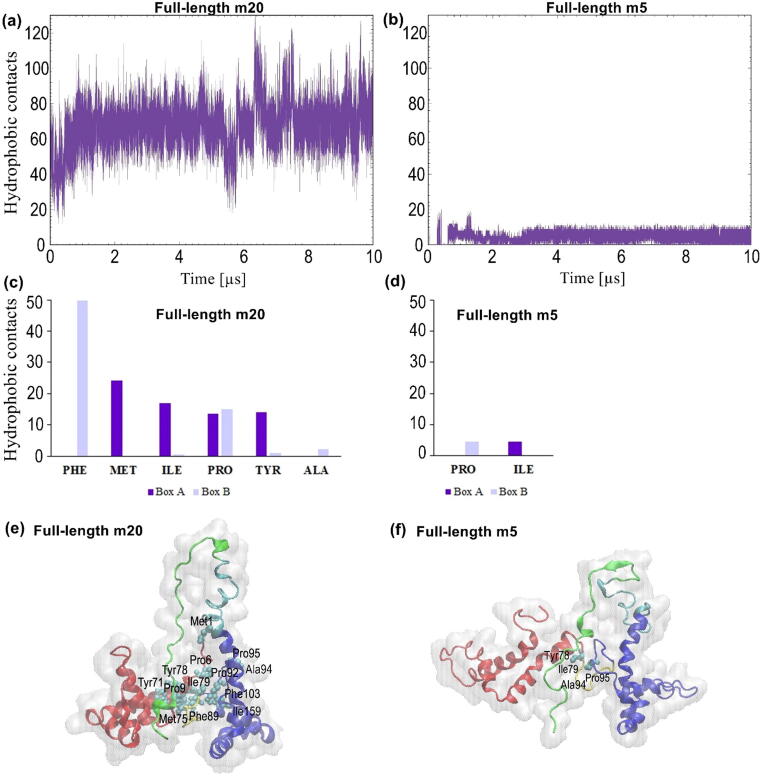
3.3. Hydrophobic contacts between Box a and Box B
Due to the position of the tail in the full-length form of HMGB1, the boxes appear to stay at a relatively fixed distance relative to each other, as revealed by the dominant conformations shown at the bottom of Fig. 4a and c. Both boxes have a net positive charge and, therefore, electrostatic interactions are not likely to dominate. Instead, any contacts between the boxes are likely to involve hydrophobic interactions. In order to characterize these interactions between the boxes, the number of hydrophobic contacts formed between Box A and Box B were calculated between carbon atoms of the side chains of nonpolar, hydrophobic amino acids. In full-length m20, an average of ~29 hydrophobic contacts were formed between the boxes (Fig. 5a), substantially higher than the average of ~7 hydrophobic contacts formed between the boxes in full-length m5, which were observed only after 0.6 µs in the simulation (Fig. 5b), at which point the acidic tail had occupied a deep, stable position between the boxes (Fig. 4c). The higher number of hydrophobic contacts in full-length m20 were found to be made by Phe, Met, Ile, Pro and Tyr residues (Fig. 5c), whereas in full-length m5 the lower number of hydrophobic contacts observed were made by Ile and Pro residues (Fig. 5d). The positions of specific residues mediating these interactions are shown within each box in Fig. 5e and f. Phe89 (Box B) forms the largest number of hydrophobic contacts with ~42. Met75 (Box A) establishes ~24 contacts and Pro95 (Box B) with ~15 contacts. Additionally, Tyr78 from Box A as well as Ala94 and Ile159 from Box B were predicted to form hydrophobic contacts with the boxes, each forming ~8, ~4, and ~0.4 contacts, respectively. These residues had been reported to interact with either the acidic tail or the linker residue between Box B and the tail [9], [10], [15].
Fig. 5.
Hydrophobic contacts formed between Box A and Box B. Time evolution of the average number of hydrophobic contacts between Box A and Box B in (a) full-length m20 and (b) full-length m5. Amino acid residues with the largest number of hydrophobic contacts in (c) full-length m20 and (d) full-length m5. Snapshots show residues forming hydrophobic contacts in (e) full-length m20 and (f) full-length m5. In each figure (e) and (f), the red regions represent Box A, blue regions represent Box B, yellow portions represent the linker between boxes, cyan regions represent the linker between Box B and the acidic tail, and light green portions represent the acidic tail. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
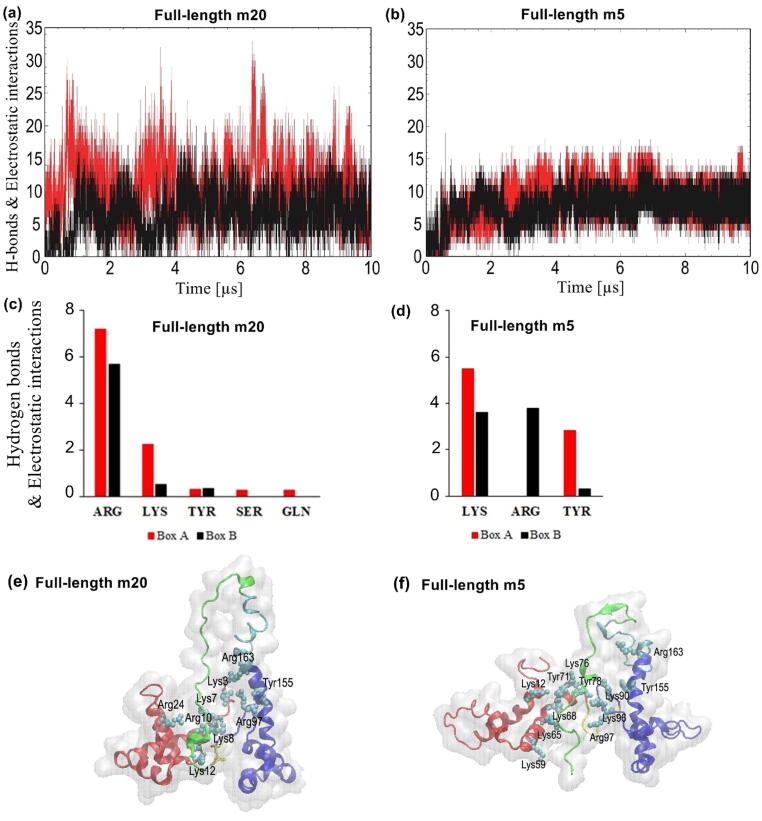
3.4. Hydrogen bonds and electrostatic interactions between the boxes and the acidic tail
The acidic tail carries a substantial negative charge and is thus expected to make a significant number of electrostatic and hydrogen-bonding interactions with the boxes. As seen at the bottom of Fig. 4a and c, the acidic tail seems to favor an elongated conformation positioned between the boxes in both full-length m20 and m5, thus forming multiple interactions with the boxes. Because the PACE coarse grained force field does not explicitly model hydrogen bonds, a distance-based analysis was conducted to compute hydrogen bonds and/or electrostatic interactions between the N and O atoms of the side chains of the acidic tail residues and the charged or neutral-polar amino acids in either Box A or Box B. Overall, the acidic tail was determined to form slightly more hydrogen bonds and electrostatic interactions with Box A. In full-length m20, there is an average of ~10 hydrogen bonds/electrostatic interactions between the tail and Box A and ~7 with Box B (Fig. 6a). Similarly, ~9 hydrogen bonds/electrostatic interactions were computed between Box A and the tail in full-length m5, whereas Box B has an average of ~8 with the tail (Fig. 6b). As expected, most of the interactions in full-length m20 are formed by the positively charged residues Arg and Lys, and to a much lesser extent by the polar residues Tyr, Ser, and Gln (Fig. 6c). Meanwhile, these interactions are formed almost equally by Lys, Arg and Tyr in full-length m5 (Fig. 6d). The larger fluctuations in the number of interactions between the acidic tail and the boxes in full-length m20 reflect the larger conformational changes (Fig. 4) compared to m5. Fig. 6e and f show the type of residues that mediate the interaction in each box. Amino acids Tyr155 and Arg163 were found to interact with the acidic tail with ~0.3 and ~5 hydrogen bonds/electrostatic interactions, respectively, along with Tyr78 with ~2 hydrogen bonds/electrostatic interactions. However, Fig. 6e and f also show that other amino acids, particularly Arg10 and Lys12, were predicted to form ~7 and ~2 hydrogen bonds/electrostatic interactions, respectively.
Fig. 6.
Hydrogen bonds and electrostatic interactions formed between the acidic tail and the boxes. Time evolution of the average number of hydrogen bonds and electrostatic interactions between the acidic tail and Box A (red) and Box B (black) in (a) full-length m20 and (b) full-length m5. Amino acid residues with the largest number of hydrogen bonds and electrostatic interactions from each box in (c) full-length m20 and (d) full-length m5. Snapshots show amino acids forming hydrogen bonds or electrostatic interactions in (e) full-length m20 and (f) full-length m5. In each figure (e) and (f), the red regions represent Box A, blue regions represent Box B, yellow portions represent the linker between boxes, cyan regions represent the linker between Box B and the acidic tail, and light green portions represent the acidic tail. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Effect of alkylation on structural stability
Iodoacetamide is often used to alkylate free Cys residues in a protein so that the Cys residues cannot form disulfide bonds [36]. The CD spectra shown in Fig. 7 demonstrate that alkylation of HMGB1 with an increasing concentration of iodoacetamide alters the secondary structure of HMGB1, both with and without its acidic tail. Treatment with as little as 0.5 mM iodoacetamide resulted in visible shifts in the CD spectra. Surprisingly, the predicted content of secondary structure by the CONTIN fit of the data (Table S2 in the Supporting Information) shows that α-helical content drastically decreases in full-length HMGB1 after extensive alkylation, with a similar but less pronounced effect in the tail-less form. The loss of α-helical structure in full-length HMGB1 was more significant after treatment with iodoacetamide than with β-mercaptoethanol (Table S1 in the Supporting Information).
Fig. 7.
Circular dichroism spectra for (a) full-length HMGB1 and (b) tail-less HMGB1, upon addition of different concentrations of iodoacetamide. The blue line is the experimental data and the red dotted line is the CONTIN fit. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Both Cys23 and Cys45 are surrounded by hydrophilic amino acids that are located at least three residues before and after them in the protein sequence. By contrast, Cys106 is mostly surrounded by hydrophobic or neutral amino acids. The three Cys residues were found to have relatively low average SASA values (Table 1). However, almost all surrounding amino acids have higher solvent exposure than these Cys residues.
Table 1.
Average SASA values of residues Cys23, Cys45 and Cys106, as well as of the three residues before and after them in the protein sequence. Hydrophilic residues are shown in bold.
| Residue | Number | Average SASA values (nm2) |
|||
|---|---|---|---|---|---|
| m20 |
m5 |
||||
| Full-length | Tail-less | Full-length | Tail-less | ||
| Val | 20 | 0.28 | 0.41 | 0.34 | 0.14 |
| Gln | 21 | 1.2 | 1.27 | 1.07 | 1.17 |
| Thr | 22 | 0.47 | 0.63 | 0.42 | 0.55 |
| Cys | 23 | 0.27 | 0.42 | 0.14 | 0.24 |
| Arg | 24 | 1.2 | 1.2 | 1.03 | 1.04 |
| Glu | 25 | 0.95 | 1.02 | 0.9 | 0.93 |
| Glu | 26 | 0.97 | 1.00 | 0.81 | 0.94 |
| Ser | 42 | 0.78 | 0.6 | 0.38 | 0.39 |
| Lys | 43 | 1.57 | 1.33 | 1.46 | 1.38 |
| Lys | 44 | 0.98 | 1.14 | 0.87 | 1.19 |
| Cys | 45 | 0.27 | 0.4 | 0.1 | 0.11 |
| Ser | 46 | 0.74 | 0.62 | 0.62 | 0.65 |
| Glu | 47 | 1.09 | 1.03 | 0.98 | 1.26 |
| Arg | 48 | 0.98 | 1.36 | 1.17 | 1.4 |
| Phe | 103 | 0.63 | 0.57 | 0.46 | 0.17 |
| Leu | 104 | 0.07 | 0.2 | 0.25 | 0.23 |
| Phe | 105 | 0.09 | 0.03 | 0.06 | 0.56 |
| Cys | 106 | 0.12 | 0.16 | 0.18 | 0.21 |
| Ser | 107 | 0.51 | 0.63 | 0.79 | 0.71 |
| Glu | 108 | 0.96 | 0.77 | 0.86 | 0.84 |
| Tyr | 109 | 0.77 | 0.32 | 0.5 | 0.51 |
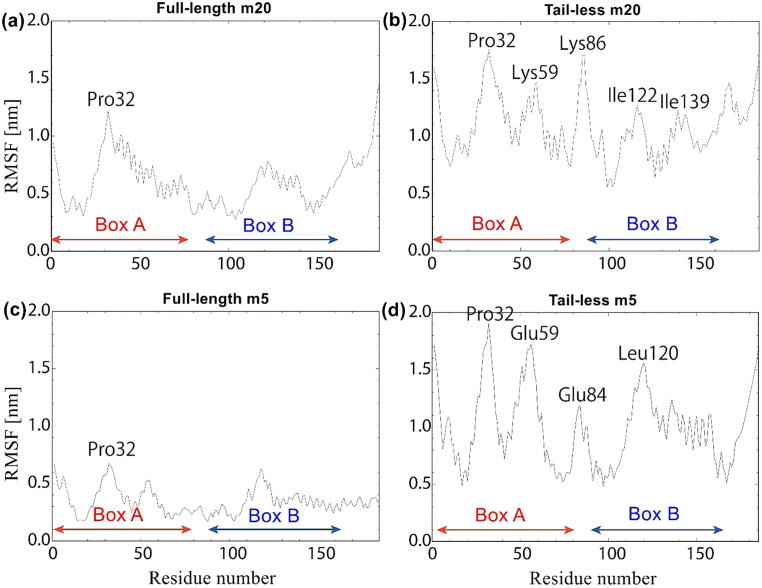
3.6. Residues involved in conformational fluctuations in full-length and tail-less HMGB1
Comparison of the computed RMSF values between the full-length and tail-less forms in the simulations of both m20 and m5 reveals that Pro32 exhibits a prominent peak compared to other residues in all four forms of HMGB1. Additionally, the residues within Box A display higher fluctuations than those in Box B, and the overall RMSF values of full-length m5 (Fig. 8c) were predicted to be considerably lower than those of its m20 counterpart (Fig. 8a). In the tail-less forms, there are residues other than Pro32 that show higher fluctuations in the m20 and m5 forms (Fig. 8b and d), as opposed to their significantly less noticeable full-length counterparts (Fig. 8a and c). In tail-less m20, the Lys59 in Box A and Ile122 and Ile139 in Box B display the largest fluctuations (Fig. 8b), whereas in tail-less m5 the residues with the largest fluctuations are Glu59 in Box A and Leu120 in Box B (Fig. 8d).
Fig. 8.
Average RMSF values for (a) full-length m20; (b) tail-less m20; (c) full-length m5; and (d) tail-less m5.
Residues 79–89 in the linker region between Box A and Box B in the full-length m20 and m5 forms were predicted to have much lower fluctuations than residues in the boxes (Fig. 8a and c). By contrast, in the same region of the tail-less m20 and m5 forms, some residues exhibit significantly large RMSF values, peaking for Lys86 in m20 (Fig. 8b) and Glu84 in m5 (Fig. 8d). Nonetheless, the linker region in the m5 form exhibited lower RMSF values than in the m20 form.
3.7. Changes in solvent exposure of amino acids
Table 2 lists the amino acid residues of both m20 and m5 which have more than 0.5 nm2 increase or decrease in SASA values. All residues shown in Table 2 are either hydrophobic or hydrophilic. Interestingly, changes in secondary structure revealed by DSSP analysis (Fig. 3) appear to correspond to a number of amino acids listed in Table 2. The red arrows indicate areas around certain amino acids in the tail-less form m20 (Fig. 3b) that undergo structural switch between α-helix and turn, which correspond to Phe41, Arg110, Ile122 and Thr136. These residues are among the amino acids in the tail-less form of m20 that exhibit large increases or decreases in SASA values (Table 2). In the DSSP analysis of the tail-less form m5 (Fig. 3d), the red arrows correspond to Phe38, Lys57, Met63, Lys87, Lys88, Thr136 and Ala160. These residues are also among the amino acids with large increases or decreases in SASA values (Table 2). Furthermore, almost all of these amino acids correspond to regions with high RMSF values (Fig. 8a and b).
Table 2.
Average SASA values of amino acid residues in full-length (FL) and tail-less (TL) forms of both m20 and m5 that display the largest changes. Hydrophilic residues are shown in bold, while all others are hydrophobic residues. The residues that experience more than 0.5 nm2 increase in SASA value in the tail-less form compared to the full-length form are highlighted in grey.
| Residue | Number | Average SASA values (nm2) |
Residue | Number | Average SASA values (nm2) |
||
|---|---|---|---|---|---|---|---|
| FL m20 | TL m20 | FL m5 | TL m5 | ||||
| Arg | 10 | 1.48 | 0.95 | Tyr | 16 | 0.84 | 0.39 |
| Met | 13 | 0.93 | 0.34 | Phe | 38 | 1.05 | 0.23 |
| Phe | 41 | 0.18 | 0.67 | Lys | 55 | 1.65 | 1.15 |
| Trp | 49 | 0.58 | 1.09 | Lys | 57 | 0.93 | 1.63 |
| Lys | 57 | 1.63 | 1.08 | Met | 63 | 1.27 | 0.79 |
| Met | 75 | 0.29 | 0.73 | Glu | 72 | 0.36 | 0.93 |
| Lys | 76 | 1.69 | 1.04 | Lys | 82 | 1.85 | 1.14 |
| Tyr | 78 | 0.27 | 0.78 | Lys | 87 | 0.80 | 1.51 |
| Thr | 85 | 0.53 | 1.07 | Lys | 88 | 0.76 | 1.47 |
| Lys | 86 | 0.76 | 2.00 | Gly | 115 | 0.17 | 0.66 |
| Phe | 89 | 0.15 | 0.90 | Pro | 118 | 1.15 | 0.69 |
| Pro | 95 | 0.87 | 0.38 | Leu | 120 | 0.64 | 1.34 |
| Phe | 105 | 0.06 | 0.56 | Glu | 131 | 1.41 | 0.95 |
| Arg | 110 | 0.94 | 1.57 | Thr | 136 | 0.18 | 1.14 |
| Ile | 122 | 1.31 | 0.34 | Ala | 138 | 0.79 | 0.29 |
| Met | 132 | 0.46 | 1.01 | Ile | 159 | 0.39 | 0.90 |
| Thr | 136 | 0.29 | 0.80 | Ala | 160 | 0.61 | 0.06 |
| Tyr | 144 | 0.32 | 1.23 | Val | 175 | 0.39 | 0.85 |
| Val | 176 | 0.15 | 0.72 | ||||
The significant number of hydrophobic contacts shown in Fig. 6 is reflected in reductions of SASA values for a large number of residues within the boxes (Table 2). Several hydrophobic or bulky residues, particularly Phe, Ile, Met, and Tyr, have significantly lower values in full-length m20 compared to the tail-less form, and are found to contribute the most in forming hydrophobic contacts (Fig. 5c). By contrast, full-length m5 does not have as many residues with reduced values of SASA. This explains the higher number of hydrophobic contacts in full-length m20 compared to m5. Furthermore, formation of hydrogen bonds and/or electrostatic interactions between the boxes and the acidic tail (Fig. 6c and d) also corresponds to polar/charged amino acid residues (Lys, Arg and Tyr) that exhibit lower SASA values in the full-length forms compared to their tail-less counterparts (Table 2).
4. Discussion
Previous all-atom MD simulation studies of HMGB1 have only been performed in the submicrosecond timescale. A study of the stability and conformational changes caused by different redox states of the Cys residues in HMGB1 was conducted with simulations of up to 100 ns [16]. Another study of the aggregation of HMGB1 facilitated by the binding of Cys106 to (−)-epigallocatechin-3-gallate (EGCG) was performed with simulations of up to 300 ns [25]. Both studies were conducted using GROMACS with the AMBER99SB-ILDN and GROMOS96 53a6 force fields, respectively [16], [25]. However, large protein conformational changes can occur in the µs to ms time scale. In this study, a solvent coarse graining approach was adopted using the PACE force field to enable the first simulation of substantially longer times for the characterization of equilibrium properties of full-length and tail-less HMGB1.
In this study, the most abundant secondary structure in full-length HMGB1 is α-helical, as also shown by the previously published characterization of the structure of full-length HMGB1 using CD spectroscopy [7], [11]. The overall shape of the CD spectra (Fig. 2, 0 mM) with the dips in ellipticity at 208 and 222 nm, is similar to those previously reported [9], [10], indicative of the predominant α-helical content in both full-length and tail-less HMGB1, although it is lower in the latter form. Full-length HMGB1 was indeed predicted to retain its structural integrity better than the tail-less form. This was especially the case for the full-length m5 form, which rapidly attained a stable conformation, as shown in Fig. 4c and its dominant conformations. The role of the acidic tail in stabilizing the full-length HMGB1 is also confirmed by the dominant conformations, which show that the tail mostly favors occupying a position between Box A and Box B in both full-length m20 and m5 (Fig. 4a and c), leading to an observed increased stability of the full-length structures compared to the tail-less ones. We note that the CD experiments and MD simulations were done in the reduced state of HMGB1, which is expected to be the predominant form in the nuclear and intracellular environment. Our findings agree with the previous studies by Knapp et al. and Belgrano et al., who described the higher stability of full-length HMGB1 compared to the tail-less one [7], [9].
The difference of stability in full-length m5 and m20 appears to be related to the distance between Box A and Box B. This distance in m20 is significantly larger than in m5 (Fig. 1b and c), which was indeed one of the reasons for selecting these structures, apart from the fact that they have the largest difference in RMSD among all pairs of structural models. During the simulation, however, the distance between the boxes decreased, as shown by the dominant conformations of full-length m20 and m5 (Fig. 4a and c). The closer distance between the boxes seemed to promote the formation of a significant number of hydrophobic contacts between Box A and Box B (Fig. 5). Our simulations also observed the formation of a relatively constant number of hydrogen bonds and electrostatic interactions between each box and the acidic tail, involving primarily positively charged residues (Lys and Arg) as well as other polar residues (Fig. 6), and which are likely to help to stabilize both full-length forms. In particular, Tyr78 from Box A as well as Ala93, Ile159 and Arg163 from Box B had been reported to interact with either the acidic tail or the linker region [9], [10], [15] and were predicted to establish hydrogen bonds and/or electrostatic interactions with the tail region (Fig. 6e and f). Interestingly, since Arg163 was reported to interact with both RAGE [37] and the acidic tail [9], [10], it is likely that there is competition for binding between the tail and RAGE. It is important to highlight Phe103 of Box B, which is known to bind to DNA [14], and which was found to establish ~7 hydrophobic contacts with Box A, further highlighting the possibility of binding competition.
It is known that the acidic tail regulates the interaction between HMGB1 boxes and DNA, ultimately facilitating the collapse of the structure of HMGB1 [13], [15]. Specific amino acid residues within the box domains in HMGB1 are paramount for interaction with the acidic tail and linker region between Box B and the tail. The NMR experiments by Knapp et al., Watson et al., and Stott et al. similarly highlighted the role of Thr77 in Box A and Ile159 in Box B in the interaction with the tail [9], [10]. In particular, Ile159 was proposed to specifically interact with residues 190–204 of the acidic tail [10]. The predicted SASA values in Table 2 show that Ile159 undergoes a prominent decrease in solvent exposure, confirming that it interacts with other parts of the protein. Stott et al. also demonstrated that certain hydrophobic residues, Phe89, Ala94, Phe103, Ile122 and Ile159 in Box B interact with the linker region between Box B and the tail [15]. In this study, two of the listed hydrophobic residues, namely Phe89 and Ile159, were found to experience a significant decrease in solvent exposure (Table 2). Additionally, Trp49, Tyr78 and Arg110, which had been reported to exhibit large shifts during salt titration (thus implying that they interact with the acidic tail) [15], were predicted to exhibit significant decreases in solvent exposure (Table 2). The reduction in solvent exposure of these amino acid residues can be directly associated with inter-domain interactions.
The movement and relative position of the boxes is related to the dynamics of the linker region. Panneerselvam et al. stated that flexible linkers, such as the one between Box A and Box B (Fig. 1), gives the connecting domains freedom to twist and rotate with respect to each other [16], The predicted RMSF values (Fig. 8) show that the linker region fluctuates more in the tail-less forms. This fluctuation can be associated with the shift of the boxes with respect to each other, as shown by the dominant conformations at the bottom of Fig. 4b and d. On the other hand, the RMSF values in the linker region are lower than those of the boxes in the full-length forms, particularly in m5. This is likely to correspond to the relatively fixed position of the boxes in the full-length forms (Fig. 4a and c) compared to the tail-less ones (Fig. 4b and d).
The addition of iodoacetamide equally disrupted both the full-length and tail-less forms of HMGB1, as shown in Fig. 7. Since iodoacetamide is known to attack free Cys residues, it was initially assumed that Cys106, as the free cysteine residue of HMGB1, is equally accessible in either the full-length or the tail-less structures, making it susceptible to alkylation, which in turn may lead to changes in the secondary structure of the protein. However, as shown by the SASA values reported in Table 1, all Cys residues and their surrounding amino acids in the protein sequence have similar exposure to the solvent regardless of the presence or absence of the acidic tail. Therefore, all the Cys residues may have the same potential to be alkylated by iodoacetamide, regardless of the presence of the tail. It is therefore possible to speculate that Cys106 is not the only “free” Cys residue, as thiol/disulfide exchange [36] may occur between Cys23, Cys45, and Cys106, making them all equally likely to be alkylated.
A significant decrease in the level of solvent exposure was observed in the hydrophobic residues in both tail-less m20 and m5 forms compared to their full-length counterparts (Table 2). The secondary structure analysis by DSSP for the tail-less forms revealed that regions in the protein near those hydrophobic residues appear to shift from a turn conformation in their full-length forms (Fig. 3a and c) to α-helix in their tail-less forms (Fig. 3b and d). Ile residues stabilize the hydrophobic core of globular proteins [38], so they are likely to be present in secondary structures that are less exposed to the solvent. Ala, Met and Phe are residues with a preference for forming α-helical structures [39], [40]. However, not all hydrophobic amino acids with predicted low SASA values in the tail-less forms will necessarily be involved in α-helix formation, as is the case, for example, of Pro95 in tail-less m20 and Pro118 in tail-less m5. Pro residues tend to be found in several types of turns, including loops, which is due to their rigid structure prevents formation of helical conformations [41]. As can be seen in Fig. 3b and d, these Pro residues occupy turn regions.
A number of hydrophilic residues in the tail-less m20 and m5 structures were predicted to have high SASA values compared to their full-length structures (Table 2). Arg and Lys are positively charged residues that have a preference for exposure to the solvent [41]. Negatively charged Glu residues have a higher degree of hydrophilicity than other amino acids [42]. Lastly, Thr residues have a tendency to break secondary structures [41]. The increased solvent exposure of these residues was likely to contribute to the change in secondary structure from α-helix to turn, as shown in Fig. 3b and d. Turns and loops are known to have a tendency to be more exposed to the solvent [43]. Therefore, in the absence of the acidic tail, some regions of HMGB1 can be predicted to adopt conformations with higher solvent exposure of their hydrophilic residues.
5. Conclusion and future directions
This paper reports a MD simulation study that describes the interactions of the acidic C-terminal tail within the structure of HMGB1. A key conclusion, consistent with experimental data, is that the acidic tail promotes the structural stability of HMGB1. The hydrogen bonding and electrostatic interactions of the acidic tail with the box domains of HMGB1 appear to be associated with a lesser degree of solvent exposure of hydrophilic residues in certain regions of full-length HMGB1. Additionally, the stability of the structure of HMGB1 is further enhanced by the hydrophobic interactions between Box A and Box B, possibly due to the role of acidic tail in bringing the boxes closer to each other. Some of the amino acid residues involved in these interactions are also known to bind DNA and other proteins. Therefore, it is important to examine the position of such residues to determine whether they are competent for binding to other proteins or are obstructed by other domains. The reduction in solvent exposure of hydrophobic residues is due to the interactions of those residues between the boxes. These findings shed light on the stability associated with different, dominant conformations of HMGB1.
Intrinsically disordered regions of proteins play a significant role in protein conformation and aggregation [2], which is also associated closely with disease progression [3]. For instance, the well-studied amyloid-β (Aβ) protein aggregates in the brains of patients with Alzheimer’s disease due to interactions involving its intrinsically disordered region [44]. In addition, the Cys residues in a disulfide bond can regulate the formation of protein aggregates. A study of the formation of hen egg white lysozyme (HEWL) aggregates indicated that preventing disulfide bond formation could weaken the interaction between protein monomers, reduce aggregate size, and cause the non-polar groups in aggregates of HEWL to be more solvent exposed [45]. Previous studies have also indicated that the redox status of Cys residues is crucial to understanding the role of the conformation of HMGB1 in its interactions with other proteins [46], [47]. Consequently, characterizing the contribution of the acidic tail and other amino acids, especially Cys residues, on oligomer formation in HMGB1 is important because these oligomers may be involved in the progression of inflammatory diseases, such as diabetes, Alzheimer’s disease and cancer [8]. This gives HMGB1 the potential to become part of the next generation of FDA-approved therapeutic targets and inflammatory biomarkers [24].
Author contributions
WLA, EH and RLM designed the experiments. WLA performed all experiments. EY and KY designed the simulations. KO performed all simulations. All authors contributed to the analysis and interpretation of the data, as well as the preparation of the manuscript.
Funding sources
WLA was supported by MEXT (Ministry of Education, Culture, Sports, Science and Technology, Japan) Grant-in-Aid for the “Program for Leading Graduate Schools”. EY was supported by MEXT Grant-in-Aid for the “Building of Consortia for the Development of Human Resources in Science and Technology”.
Acknowledgements
The authors gratefully acknowledge Dr. Rashmi Panigrahi (University of Western Australia) for her assistance with the CD experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.05.012.
Contributor Information
Wresti L. Anggayasti, Email: wl.anggayasti@ub.ac.id.
Kenji Yasuoka, Email: yasuoka@mech.keio.ac.jp.
Ricardo L. Mancera, Email: R.Mancera@curtin.edu.au.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kmiecik S., Gront D., Kolinski M., Wieteska L., Dawid A.E., Kolinski A. Coarse-grained protein models and their applications. Chem Rev. 2016;116:7898–7936. doi: 10.1021/acs.chemrev.6b00163. [DOI] [PubMed] [Google Scholar]
- 2.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toth-Petroczy A, Palmedo P, Ingraham J, Hopf TA, Berger B, Sander C, Marks DS, Structured states of disordered proteins from genomic sequences, Cell 167 (2016) 158–170 e112. [DOI] [PMC free article] [PubMed]
- 4.Schafer N.P., Kim B.L., Zheng W., Wolynes P.G. Learning To Fold Proteins Using Energy Landscape Theory. Isr J Chem. 2014;54:1311–1337. doi: 10.1002/ijch.201300145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore S.J., Sonar K., Bharadwaj P., Deplazes E., Mancera R.L. Characterisation of the structure and oligomerisation of islet amyloid polypeptides (IAPP): a review of molecular dynamics simulation studies. Molecules. 2018;23 doi: 10.3390/molecules23092142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henriques J., Skepo M. Molecular dynamics simulations of intrinsically disordered proteins: on the accuracy of the TIP4P-D water model and the representativeness of protein disorder models. J Chem Theory Comput. 2016;12:3407–3415. doi: 10.1021/acs.jctc.6b00429. [DOI] [PubMed] [Google Scholar]
- 7.Belgrano F.S., de Abreu da Silva I.C., Bastos de Oliveira F.M., Fantappie M.R., Mohana-Borges R. Role of the acidic tail of high mobility group protein B1 (HMGB1) in protein stability and DNA bending. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0079572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anggayasti W.L., Mancera R.L., Bottomley S., Helmerhorst E. The self-association of HMGB1 and its possible role in the binding to DNA and cell membrane receptors. FEBS Lett. 2017;591:282–294. doi: 10.1002/1873-3468.12545. [DOI] [PubMed] [Google Scholar]
- 9.Knapp S., Muller S., Digilio G., Bonaldi T., Bianchi M.E., Musco G. The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry. 2004;43:11992–11997. doi: 10.1021/bi049364k. [DOI] [PubMed] [Google Scholar]
- 10.Watson M., Stott K., Thomas J.O. Mapping intramolecular interactions between domains in HMGB1 using a tail-truncation approach. J Mol Biol. 2007;374:1286–1297. doi: 10.1016/j.jmb.2007.09.075. [DOI] [PubMed] [Google Scholar]
- 11.Anggayasti W.L., Mancera R.L., Bottomley S., Helmerhorst E. The effect of physicochemical factors on the self-association of HMGB1: a surface plasmon resonance study. Biochim Biophys Acta. 1864;2016:1620–1629. doi: 10.1016/j.bbapap.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Stros M., Polanska E., Kucirek M., Pospisilova S. Histone H1 differentially inhibits DNA bending by reduced and oxidized HMGB1 protein. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0138774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stott K., Watson M., Bostock M.J., Mortensen S.A., Travers A., Grasser K.D. Structural insights into the mechanism of negative regulation of single-box high mobility group proteins by the acidic tail domain. J Biol Chem. 2014;289:29817–29826. doi: 10.1074/jbc.M114.591115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blair R.H., Horn A.E., Pazhani Y., Grado L., Goodrich J.A., Kugel J.F. The HMGB1 C-terminal tail regulates DNA bending. J Mol Biol. 2016;428:4060–4072. doi: 10.1016/j.jmb.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stott K., Watson M., Howe F.S., Grossmann J.G., Thomas J.O. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J Mol Biol. 2010;403:706–722. doi: 10.1016/j.jmb.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 16.Panneerselvam S., Durai P., Yesudhas D., Achek A., Kwon H.K., Choi S. Cysteine redox state plays a key role in the inter-domain movements of HMGB1: a molecular dynamics simulation study. Rsc Adv. 2016;6:100804–100819. [Google Scholar]
- 17.Yang H., Antoine D.J., Andersson U., Tracey K.J. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol. 2013;93:865–873. doi: 10.1189/jlb.1212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han W., Schulten K. Further optimization of a hybrid united-atom and coarse-grained force field for folding simulations: improved backbone hydration and interactions between charged side chains. J Chem Theory Comput. 2012;8:4413–4424. doi: 10.1021/ct300696c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H., Han W., Ma W., Schulten K. Transient beta-hairpin formation in alpha-synuclein monomer revealed by coarse-grained molecular dynamics simulation. J Chem Phys. 2015;143 doi: 10.1063/1.4936910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anggayasti W.L., Mancera R.L., Bottomley S., Helmerhorst E. Optimization of surface plasmon resonance experiments: case of high mobility group box 1 (HMGB1) interactions. Anal Biochem. 2016;499:43–50. doi: 10.1016/j.ab.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 23.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 24.VanPatten S., Al-Abed Y. High mobility group box-1 (HMGb1): current wisdom and advancement as a potential drug target. J Med Chem. 2018;61:5093–5107. doi: 10.1021/acs.jmedchem.7b01136. [DOI] [PubMed] [Google Scholar]
- 25.Meng X.Y., Li B., Liu S., Kang H., Zhao L., Zhou R. EGCG in green tea induces aggregation of HMGB1 protein through large conformational changes with polarized charge redistribution. Sci Rep. 2016;6:22128. doi: 10.1038/srep22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eswar N., Eramian D., Webb B., Shen M.Y., Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 27.Lindahl E., Hess B., van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- 28.Monticelli L., Kandasamy S.K., Periole X., Larson R.G., Tieleman D.P., Marrink S.J. The MARTINI coarse-grained force field: extension to proteins. J Chem Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 29.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 30.Ward M.D., Nangia S., May E.R. Evaluation of the hybrid resolution PACE model for the study of folding, insertion, and pore formation of membrane associated peptides. J Comput Chem. 2017;38:1462–1471. doi: 10.1002/jcc.24694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., Haak J.R. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 32.Yesylevskyy S.O., Schafer L.V., Sengupta D., Marrink S.J. Polarizable water model for the coarse-grained MARTINI force field. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parrinello M., Rahman A. Crystal-structure and pair potentials – a molecular-dynamics study. Phys Rev Lett. 1980;45:1196–1199. [Google Scholar]
- 34.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J Mol Graph Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 36.Winther J.R., Thorpe C. Quantification of thiols and disulfides. Biochim Biophys Acta. 1840;2014:838–846. doi: 10.1016/j.bbagen.2013.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauvala H., Rouhiainen A. Physiological and pathophysiological outcomes of the interactions of HMGB1 with cell surface receptors. Biochim Biophys Acta. 2010;1799:164–170. doi: 10.1016/j.bbagrm.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Kathuria S.V., Chan Y.H., Nobrega R.P., Ozen A., Matthews C.R. Clusters of isoleucine, leucine, and valine side chains define cores of stability in high-energy states of globular proteins: sequence determinants of structure and stability. Protein Sci. 2016;25:662–675. doi: 10.1002/pro.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malkov S.N., Zivkovic M.V., Beljanski M.V., Stojanovic S.D., Zaric S.D. A reexamination of correlations of amino acids with particular secondary structures. Protein J. 2009;28:74–86. doi: 10.1007/s10930-009-9166-3. [DOI] [PubMed] [Google Scholar]
- 40.Dougherty DA, Cation-pi interactions involving aromatic amino acids, J Nutr, 137 (2007) 1504S–1508S; discussion 1516S–1517S. [DOI] [PubMed]
- 41.Imai K., Mitaku S. Mechanisms of secondary structure breakers in soluble proteins. Biophysics (Nagoya-shi) 2005;1:55–65. doi: 10.2142/biophysics.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trevino S.R., Scholtz J.M., Pace C.N. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J Mol Biol. 2007;366:449–460. doi: 10.1016/j.jmb.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rauscher S., Baud S., Miao M., Keeley F.W., Pomes R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure. 2006;14:1667–1676. doi: 10.1016/j.str.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Zheng W., Tsai M.Y., Chen M., Wolynes P.G. Exploring the aggregation free energy landscape of the amyloid-beta protein (1–40) Proc Natl Acad Sci U S A. 2016;113:11835–11840. doi: 10.1073/pnas.1612362113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravi V.K., Goel M., Kotamarthi H.C., Ainavarapu S.R., Swaminathan R. Preventing disulfide bond formation weakens non-covalent forces among lysozyme aggregates. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0087012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fassi E.M.A., Sgrignani J., D'Agostino G., Cecchinato V., Garofalo M., Grazioso G. Oxidation state dependent conformational changes of HMGB1 regulate the formation of the CXCL12/HMGB1 heterocomplex. Comput Struct Biotechnol J. 2019;2019 doi: 10.1016/j.csbj.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S., He M., VanPatten S., Al-Abed Y. Mechanistic insights into high mobility group box-1 (HMGb1)-induced Toll-like receptor 4 (TLR4) dimer formation. J Biomol Struct Dyn. 2019;37:3721–3730. doi: 10.1080/07391102.2018.1526712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.