Abstract
Caspases play central roles in mediating both cell death and inflammation. It has more recently become evident that caspases also drive other biological processes. Most prominently, caspases have been shown to be involved in differentiation. Several stem and progenitor cell types rely on caspases to initiate and execute their differentiation processes. These range from neural and glial cells, to skeletal myoblasts and osteoblasts, and several cell types of the hematopoietic system. Beyond differentiation, caspases have also been shown to play roles in other “noncanonical” processes, including cell proliferation, arrest, and senescence, thereby contributing to the mechanisms that regulate tissue homeostasis at multiple levels. Remarkably, caspases directly influence the course of the cell cycle in both a positive and negative manner. Caspases both cleave elements of the cell-cycle machinery and are themselves substrates of cell-cycle kinases. Here we aim to summarize the breadth of interactions between caspases and cell-cycle regulators. We also highlight recent developments in this area.
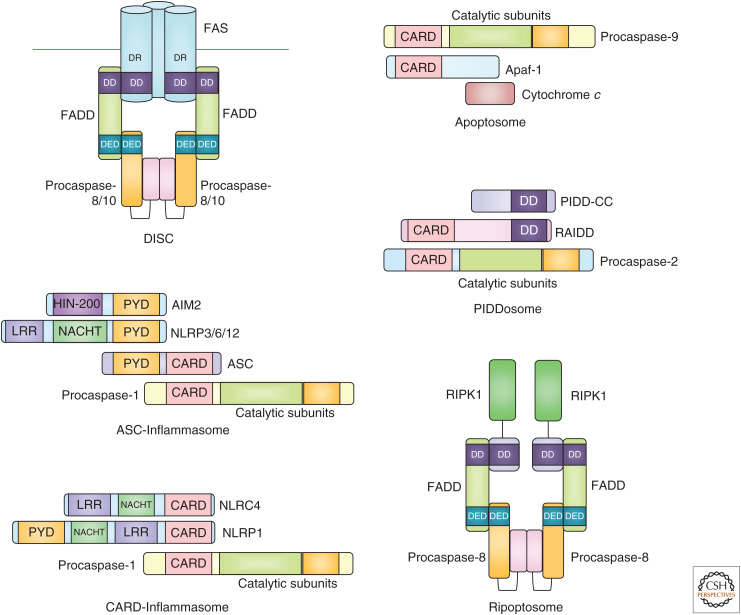
Caspases are a class of aspartic acid–specific endopeptidases that are best known for their roles in cell death and inflammation. Activation of caspases typically occurs as the result of complex intracellular signaling cascades, with multiple “checks and balances” to prevent their inappropriate activation. Cytoplasmic adaptor molecules recruit apical caspases into high molecular weight complexes that facilitate self-processing for autoactivation (Fig. 1). Once activated, apical caspase-8 or -9 processes and activates downstream effector caspases by proteolysis to induce apoptotic cell death. Others have roles in directing inflammatory responses, for example, caspase-1 or mouse caspase-11 (caspase-4 and -5 in humans) are activated by different types of inflammasomes for cytokine processing and executing a lytic type of cell death called pyroptosis (Bratton and Salvesen 2010; Julien and Wells 2017; Van Opdenbosch and Lamkanfi 2019).
Figure 1.
Schematic representations of different high-molecular weight caspase activation complexes. The inflammasome is represented in both ASC-dependent and -independent configurations.
Along with their traditional roles in apoptosis, caspases have more recently been shown to be crucial mediators of a wider range of biological processes. The most well studied of these has been cellular differentiation. Caspases have been shown to be essential to the differentiation of neural and glial cells, skeletal myoblasts and osteoblasts, as well as several cell types of the hematopoietic system, including erythrocytes (for review, see Crawford and Wells 2011; Connolly et al. 2014). In these systems, caspases are prevented from unrestrained runaway activation, which would normally lead to apoptotic cell death. Caspases cleave a limited set of substrates to affect the morphological and transcriptional changes that are characteristic of differentiation. Indeed, it has been argued that apoptosis itself is an extreme form of terminal differentiation, in analogy to the cornification of keratinocytes that causes their death (Fernando and Megeney 2007).
It may seem paradoxical to speak of activation of apoptotic initiator and effector caspases in the absence of cell death, but there are several well-studied mechanisms through which this can occur. One such mechanism is minority mitochondrial outer membrane permeabilization (miMOMP) (Ichim et al. 2015), a process in which only a fraction of a cell's mitochondria undergo permeabilization. This produces a low level of caspase activation, which is insufficient to trigger apoptosis. It has been shown that this process is used to trigger cytokine secretion in epithelial cells in response to viral, bacterial, and protozoal infection (Brokatzky et al. 2019). Moreover, it has even been reported that cells can survive and recover even from full apoptotic caspase activity. In a process termed “anastasis,” cells can reverse activation of caspases if a proapoptotic stimulus is removed, even at late stages of apoptosis, when morphological transformation of the cell has already occurred (Tang et al. 2012). This suggests that there exist mechanisms that can suppress “full-blown” caspase activation, apparently through a transcriptional mechanism that rapidly induces expression of antiapoptotic factors.
Another process in which caspases have shown “noncanonical” roles is in cell-cycle control and proliferation. This is admittedly a less well-studied area; however, a number of in vitro and in vivo studies have laid out a case for a potential role for caspases in regulation of the cell cycle. During both apoptosis and many forms of differentiation, caspases effect a terminal withdrawal from the cell cycle into a postmitotic state. In both cases, this is mediated both directly by proteolytic degradation of cell-cycle components, such as p21, p27, CDK11, or the retinoblastoma protein (Jänicke et al. 1996; Beyaert et al. 1997; Levkau et al. 1998; Podmirseg et al. 2016) and indirectly through cleavage of other targets, such as CHK1 or PAK2 that control proliferation rates in response to stress (Rudel and Bokoch 1997; Matsuura et al. 2008; Larsen et al. 2010). However, this is not the fullest extent to which caspases influence the cell cycle. There is also evidence for roles for caspases both in promoting normal cell-cycle progression, and in the response to perturbations in the cell cycle, in which caspases help mediate cell fate decisions. Along this line it is worth mentioning that when cells slip out of extended mitotic arrest, caspase-9 and -7 become activated (Orth et al. 2012). This leads to cleavage of the inhibitor of caspase-activated DNase, iCAD, releasing the endonuclease CAD, which causes limited DNA damage, as well as a p53 response. Hain et al. (2016) report a similar mechanism, tracing the caspase-induced DNA damage to telomeres, driving a DNA-PK-dependent cell-cycle arrest.
In this review, we summarize the evidence for interactions between caspases that act as initiators or effectors of apoptosis and the cell-cycle machinery. These interactions can be divided into two groups: modification of caspases by cell-cycle proteins, chiefly kinases such as CDK1, and (selective) proteolytic processing of cell-cycle proteins or their regulators by caspases. This area of research has undergone a reemergence in the recent past, particularly with respect to in vivo studies. It should be noted up front that the data outlined below often rely on measurement of cell proliferation in response to stimulation. Much of this work was completed before the discovery of necroptotic cell death as a response to caspase-8 inhibition (Holler et al. 2000; Degterev et al. 2005). As such, it is possible that necroptosis induction also contributes to the observed reduction in cell proliferation. Clearly, these phenomena should be reappraised in light of our current knowledge on the role of caspase-8 as an inhibitor of necroptosis. Finally, we highlight some of the most recent discoveries related to caspase-cell-cycle cross talk in the final section and advocate to reinvestigate the role of caspase substrates controlling cell-cycle progression in nonapoptotic settings.
LINKING CASPASES TO CELL-CYCLE REENTRY AND CELL-CYCLE PROGRESSION
Entry into the Cell Cycle from Quiescence
Many cell types, particularly stem and progenitor cells, remain in a quiescent but metabolically active state, termed G0. Upon tissue damage, infection or extensive cell loss, stem and progenitor cells reenter the cell cycle. There are several older examples in which caspase activity has been proposed to influence this transition into the cell cycle, particularly in the hematopoietic system.
An interesting example of this was shown in B cells where costimulation through CD40 and CD180 induced the cleavage and activation of both caspase-6 and -8 in a time-dependent manner, peaking at 12 and 24 h poststimulation, respectively (Olson et al. 2003). Chemical inhibition of either caspase greatly reduced the rate of proliferation triggered by either CD40/CD180 or LPS stimulation. Mechanistically, it was proposed that caspase-6-mediated cleavage of the transcriptional repressor SATB1 derepresses the transcription of cyclin D1/2 and CDK4, facilitating exit from G0 (Fig. 2). A caveat here is that the observed failure to proliferate could also be partly the result of non-cell-cycle effects, such as induction of necroptosis upon prolonged caspase-8 inhibition. Similarly, Watanabe et al. showed that upon stimulation of wild-type B cells with the CpG PAMP, activating TLR9, caspase-6 undergoes proteolytic cleavage to its active form, and that chemical inhibition of caspase-6 inhibited the resulting B-cell proliferation (Watanabe et al. 2008). Although the selectivity of pharmacological caspase-inhibitors is limited and caspase-8 may have been inhibited in parallel, Casp6 knockout mice showed a greatly increased fraction of splenic B cells in G1 phase. Knockout B cells differentiated more readily into plasma cells, and secreted IgG and IgM levels were higher. Hyperphosphorylation of the retinoblastoma protein was also observed in LPS-stimulated B cells lacking caspase-6 (Watanabe et al. 2008). Hence, caspase-6 might be a negative regulator of B-cell proliferation, and its absence appears to promote both proliferation and differentiation of this lineage. It remains to be clarified how caspase-6 may be activated in these settings, if not by its upstream sibling, caspase-3 that itself needs to be processed by caspase-9 (Slee et al. 1999). It deserves to be mentioned here that caspase-6 has been shown to cleave RIPK1 (van Raam et al. 2013). It is therefore possible that loss of caspase-6 may simply allow for greater RIPK1-mediated NF-κB activation upon stimulation, which would promote downstream differentiation signals.
Figure 2.
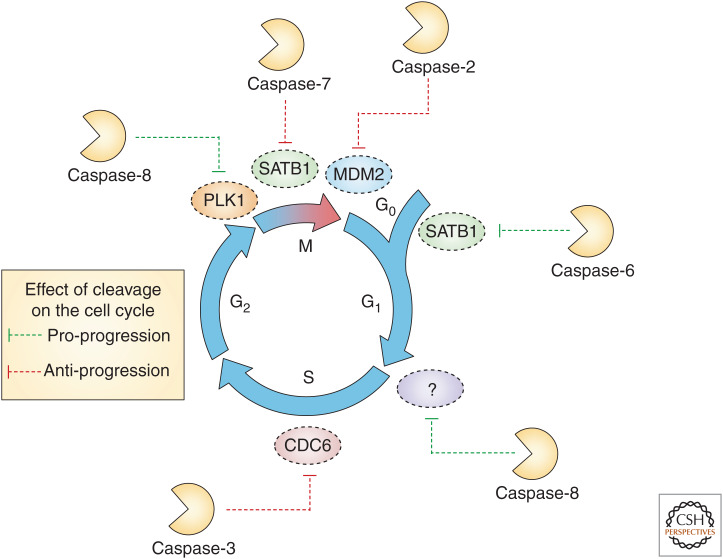
Substrates of caspases reported to be cleaved at different phases of the cell cycle in nonapoptotic contexts. Cleavage events are represented as either promoting or inhibiting cell-cycle progression.
In earlier work, Olson and colleagues demonstrated that unstimulated B cells show basal activation of caspase-3, as well as cleaved PARP1, potentially reflecting low level apoptosis in situ, that both disappear upon stimulation in tissue culture (Olson et al. 2003). This phenomenon coincides with another report demonstrating caspase-3 to be a negative regulator of B-cell proliferation (Woo et al. 2003), similar to caspase-6 (Watanabe et al. 2008). Caspase-3-deficient mice show splenomegaly and lymphadenopathy, showing both higher B-cell numbers and increased proliferation. Isolated B cells were also hyperresponsive to mitogenic stimuli, showing increased CDK2/4 activity and higher PCNA levels. Strangely, this hyperproliferation phenotype is mediated through the cell-cycle inhibitor CDKN1A/(p21), as Casp3 knockout mice show higher levels of p21, and codeletion of both Casp3 and p21 abolished the phenotype. Although this seems paradoxical, there is evidence that, under certain circumstances, p21 can actually promote the formation of active cyclin B/CDK1 complexes (LaBaer et al. 1997), suggesting that a caspase-processed form of p21 may be less potent in doing so. Interestingly, caspase-3 knockout mice also show an increased number of T cells, sufficient to maintain a normal B:T ratio, suggesting that T-cell activation or proliferation may also be negatively regulated by caspase-3 (Woo et al. 2003). This would suggest that processing of p21 by caspase-3 is needed for proper cell-cycle control in lymphocytes. One may also speculate that in the absence of caspase-3, homeostatic apoptosis may be inefficient and mitochondrial DNA release (McArthur et al. 2018; Riley et al. 2018) may cause inflammation-driven expansion of lymphocytes. Hence, conditional deletion of effector caspases in leukocytes may reveal interesting new insights about their noncanonical roles in signaling.
Transition into S Phase
The decision to transition from G1 into S phase is governed largely by CDK4/6-mediated phosphorylation of Rb, removing its inhibition of E2F family transcription factors. It was suggested early on that caspase-8 is a regulator of S-phase entry both in vitro and in vivo.
It has been shown that CD3-mediated stimulation of resting human T cells causes rapid cleavage of caspase-8 (Kennedy et al. 1999). No concomitant caspase-3 activation was noted, suggesting this phenomenon is distinct from any stimulus-induced apoptosis that can drive caspase-8 processing as a secondary consequence. Treatment with the caspase-8 inhibitor IETD-fmk prevented the induction of DNA synthesis and interleukin (IL)-2 secretion. Similarly, Arechiga et al. (2007) showed that caspase-8 knockout T cells are defective in S-phase entry upon CD3/CD28 costimulation. In another study, it was shown that caspase-inhibited T cells remain as a G0/G1 population upon CD3 stimulation, and fail to undergo CDK1/2 or cyclin A up-regulation or phosphorylation of Rb (Arechiga et al. 2007). Addition of exogenous IL-2 was able to restore T-cell proliferation under conditions of caspase-8 inhibition (Falk et al. 2004). Again, a role for caspase-8 in suppressing necroptosis upon T-cell stimulation needs to be considered here. How IL-2 signaling would counteract necrosome formation and cell death remains to be investigated. An attractive alternative explanation, however, would be that efficient T-cell activation-induced proliferation depends also on the formation of a heterodimer of caspase-8 with the paracaspase MALT1, that, similar to interaction with cFLIPL, alters substrate specificity (Oberst et al. 2011) and thereby facilitates BCL10-driven NF-κB activation and IL-2 transcription (Kawadler et al. 2008).
Beyond T cells, others reported similar findings in primary hepatocytes. Stimulation of primary rat hepatocytes with epidermal growth factor (EGF) stimulates a robust proliferation response (Gilot et al. 2005). During this time, caspase-8 cleavage is observed. In the absence of caspase-8, this proliferation response is blunted. The influence of caspase-8 upon S-phase entry is thought to be mediated through a FAS/FADD/cFLIPL complex and leads to increased S6 kinase activity through cyclin E/CDK2 activation. Salmena et al. (2003) suggest that caspase-8 stimulates proliferation through proteolysis of an unidentified cyclin E/CDK2 inhibitor. Both p21 and p27, the key inhibitors of cyclin E/CDK2 complexes, would qualify as they are known to be caspase substrates (Table 1).
Table 1.
Cell-cycle regulators that are caspase substrates
| Gene name | Protein name | Species | Cleavage sequence | References |
|---|---|---|---|---|
| ATP2B4 | PMCA4 | H | DEID (1080) | Pászty et al. 2002 |
| BUB1 | BUB1 | H | Undefined | Baek et al. 2005 |
| BUB1B | BUB1 beta/BUBR1 | H | DTCD (610), DVCD (579) | Kim et al. 2005 |
| CABLES2 | CDK5 and ABL1 enzyme substrate 2 | H | ISLDGRPP | Mahrus et al. 2008 |
| CCNA1 | Cyclin A1/A2 | X | DEPD (90) | Stack and Newport 1997 |
| CCNE1 | Cyclin E | H | LDVD (275) | Mazumder et al. 2002 |
| CCNT2 | Cyclin T2 | H | DVRDHYIA | Mahrus et al. 2008 |
| CDC27 | Cdc27 | H | Undefined | Zhou et al. 1998 |
| CDC42 | Cdc42 | H | DLRD (121) | Tu and Cerione 2001 |
| CDC5L | Cell division cycle 5–like protein | H | HESDFSGV | Mahrus et al. 2008 |
| CDC6 | Cdc6 | H | LVRD (99), SEVD (422) | Pelizon et al. 2002 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1 | H | DHVD (112) | Gervais et al. 1998 |
| CDKN1B | p27Kip1 | H | DPSD (139), ESQD (108) | Levkau et al. 1998 |
| CENPF | Centromere protein F | H | Undefined | Dix et al. 2008 |
| CHAF1A | Chromatin assembly factor 1 sub A | H | EQPDSLVD | Mahrus et al. 2008 |
| CKAP5 | Cytoskeleton-associated protein 5 | H | IEND (1495) | Dix et al. 2008 |
| EIF4EBP1 | eIF4E-BP1 | H | VLGD (25) | Bushell et al. 2000 |
| EIF4G1 | eIF4G 1 | H | DLLD (492), DRLD (1136) | Marissen et al. 2000 |
| EIF4G2 | eIF4G 2 | H | DETD (790) | Marissen et al. 2000 |
| KIF11 | Kinesin-like protein KIF23 | H | Undefined | Dix et al. 2008 |
| MAX | Max | H | IEVE (10), SAFD (135) | Krippner-Heidenreich et al. 2001 |
| MCM2 | MCM2 | H | Undefined | Schwab et al. 1998 |
| MCM3 | MCM3 | H | Undefined | Schwab et al. 1998 |
| MCM4 | MCM4 | H | Undefined | Schwab et al. 1998 |
| MCM5 | DNA RLF MCM5 | H | FYSDSFGG | Mahrus et al. 2008 |
| MCM6 | DNA RLF MCM6 | H | SGVDGYET | Mahrus et al. 2008 |
| MDM2 | MDM2 | H | DVPD (359) | Erhardt et al. 1997 |
| MDM4 | MDMX | M | DVPD (361) | Gentiletti et al. 2002 |
| MKI67 | Antigen KI-67 | H | THTDKVPG | Mahrus et al. 2008 |
| PCM1 | Pericentriolar material 1 protein | H | EDGDGAGA | Mahrus et al. 2008 |
| POLA1 | DNA polymerase α catalytic subunit | H | Undefined | Dix et al. 2008 |
| POLE | DNA polymerase ε | H | DQLD (216), DMED (1214) | Liu and Linn 2000 |
| RB1 | Rb | H | DEAD (886) | Jänicke et al. 1996 |
| TMEM30A | Cell-cycle control protein 50A | H | DEVDGGPP | Mahrus et al. 2008 |
| TP53 | p53 | H | Undefined | Sayan et al. 2006 |
| WEE1 | Wee1 | H | Undefined | Zhou et al. 1998 |
(H) human, (X) Xenopus, (M) mouse.
G2/Mitosis
Progression through mitosis is regulated by a series of checkpoints to monitor completeness of DNA replication, and ensure chromosomal integrity and proper spindle attachment. Caspases have been found to regulate several of these processes. One of the major regulators of the spindle assembly checkpoint (SAC) is the kinase BUB1b/BUBR1, an integral part of the mitotic checkpoint complex (MCC). Upon extended mitotic arrest, induced, for example, by microtubule targeting agents (MTAs) such as paclitaxel, caspases cleave BUBR1 at two separate sites (Baek et al. 2005; Kim et al. 2005). This cleavage removes the mitotic “brake,” allowing cells to exit from the forced mitotic arrest. Transfection with an uncleavable BUBR1 mutant greatly prolongs the duration of mitotic arrest, and leads to a polyploid or aneuploid phenotype. This raises the question of whether caspase activity noted in cells during normal mitosis (Hashimoto et al. 2008) exerts regulatory roles that contribute to faithful chromosome segregation and genomic stability (see below).
In addition, data from high content transcriptomics suggest that caspase-3 is the only caspase enriched during mitosis (Hsu et al. 2006). Notably, pretreatment of hepatoma cell lines with the caspase inhibitor z-DEVD-FMK prevented the efficient induction of mitotic arrest by the MTA nocodazole, suggesting a putative role for caspase-3 in mitotic entry. This study is contradicted, however, by another report (Lee et al. 2011), which showed that inhibition or depletion of multiple caspases, either alone or in combination, affected neither the duration nor magnitude of the mitotic checkpoint in MTA-treated cells. Unexpectedly, the authors noted that caspase inhibition increased the proportion of cells that died in MTA-induced mitotic arrest. This mechanism has only been demonstrated under the scenario of extended mitotic arrest. It remains to be seen whether caspase-mediated proteolysis plays a role in normal SAC disassembly and which type of cell death ensues upon chronic SAC activation when caspases are inhibited in hepatocellular carcinoma cells. It is worth mentioning that other cell types, including HeLa cervical carcinoma, A549 lung adenocarcinoma or colon carcinoma cell lines (Gascoigne and Taylor 2008; Haschka et al. 2015) mainly respond with mitotic slippage when caspases are blocked during mitotic arrest, in favor of the competing network hypothesis (see below).
Hashimoto et al. (2008) showed by immunofluorescence colocalization an accumulation of the active form of caspase-3 during mitosis in HepG2 cells. The authors also showed a partial activation of caspase-7, -8, and -9, although this may be accounted for by cells that fail to complete mitosis and undergo cell death. Knockdown of caspase-7 also appears to induce a delay or arrest in mitosis progression, which leads to cell death in a subset of HepG2 cells. This effect was reversed by transfection with an shRNA-resistant caspase-7. Caspase-7 cleaves the chromosomal passenger proteins CENP-C and INCENP, although in this context cleavage induced their mislocalization and the displacement of Aurora B from the centromeres. Although the effect on mitotic progression overall was not investigated, one would expect that inhibition of the normal function of Aurora B would lead to defects in chromosome condensation, spindle orientation, and cytokinesis, favoring cell death induction (Kelly and Funabiki 2009).
CONNECTING CELL-CYCLE DEFECTS AND APOPTOSIS
When cells experience severe genotoxic stress, they initiate apoptosis from within a certain phase of the cell cycle. This is often preceded by a cell-cycle arrest response (after DNA damage–induced activation of the ATM/CHK2/p53 axis), arresting cells mostly in G1 phase, or replication stress–associated activation of ATR/CHK1 in S-phase and after experiencing subsequent DNA damage at the G2/M boundary (Burgess and Misteli 2015). Errors in spindle to kinetochore attachments trigger activation of the SAC in M phase, preventing activation of the APC/C E3–ligase complex needed for mitotic exit (Etemad and Kops 2016).
During the last decades, significant resources have been directed to develop therapeutic strategies that interfere with mitotic events to eliminate rapidly proliferating cancer cells. A wide variety of cancers are commonly treated with cytotoxic drugs, such as taxanes and vinka alkaloids, which interfere with microtubules and disturb the mitotic spindle dynamics (Chan et al. 2012; Mitchison 2012). Despite the considerable clinical efficacy of antimitotic drugs, toxicity and drug-resistance phenotypes have posed a challenge for the application of these therapies. The effect of antimitotic drugs in nondividing cells such as neurons, causing neuropathies, has become a pivotal issue in the field and a crucial factor affecting patient quality of life (Hagiwara and Sunada 2004; McGrogan et al. 2008; Ho and Mackey 2014). To address these problems, extensive efforts have been made to develop a second generation of antimitotic agents targeting kinases that also play a role in spindle formation, like PLK1 (Polo-like kinase1) or Aurora kinases. However, the efficacy of these new compounds seems to be less promising than the classic microtubule poisons (Komlodi-Pasztor et al. 2012; Gutteridge et al. 2016).
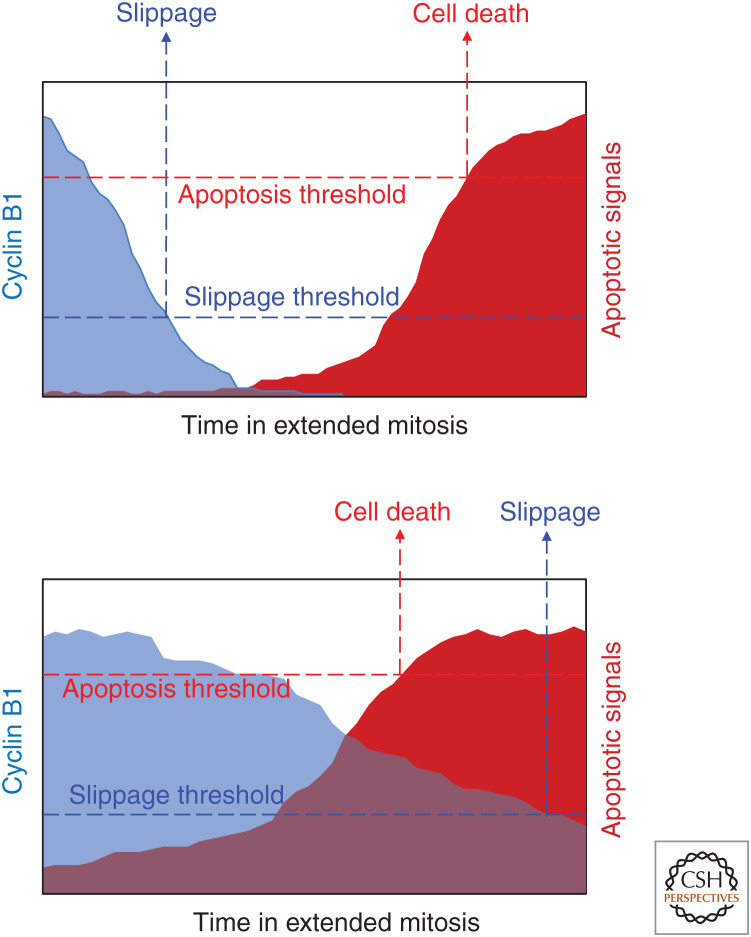
It is still unclear whether the success of antimitotic drugs to reduce cancer cells relies on their ability to promote cell death during prolonged mitotic arrest or not (Haschka et al. 2018). This idea is challenged by recent data indicating that division failures caused by antimitotic drugs can lead to micronuclei formation and an inflammatory response that seems to be key for the clinical efficacy of these drugs (Shi and Mitchison 2017). Of note, taxol concentrations used to treat primary breast tumors do not produce mitotic arrest of these cells ex vivo. Instead, the cells advance through mitosis and exhibit multipolar cell divisions that result in chromosome missegregation prior to cell death (Zasadil et al. 2014). Thus, it remains unclear how antimitotic drugs promote tumor clearance. However, in experimental settings, cells arrested in mitosis typically have one of two fates: they die during mitosis through the activation of mechanisms that resemble caspase-dependent apoptosis, or they exit mitosis, often without cell division, in a process known as “slippage” or “mitotic checkpoint adaptation.” Cells that undergo slippage can arrest in the next G1 phase, reenter the cell cycle, or die in this new, often tetraploid, G1 state (Rieder and Maiato 2004). Continued proliferation of genomically unstable cells that failed mitosis can lead to aneuploidy or chromotrypsis, both drivers of tumorigenesis and the development of drug resistance (Santaguida and Amon 2015). To explain the different fates of cells after extended mitotic arrest, Gascoigne and Taylor introduced the “competing network” hypothesis (Gascoigne and Taylor 2008; Topham and Taylor 2013). In this model, cell fate during mitotic arrest is regulated by two distinct signaling networks and the threshold that is crossed first decides cell fate (Figs. 3 and 4).
Figure 3.
The competing networks model of mitotic cell fate. During extended mitosis, cyclin B levels are gradually depleted by the APC/C E3 ligase complex that controls mitotic exit, while proapoptotic signaling gradually accumulates due to changes in the BCL2 network. Depending on which threshold is reached first, cells may undergo slippage (top) or cell death (bottom).
Figure 4.
Contributions of cell death proteins to abnormal mitotic fates. In response to mitotic abnormalities such as unattached chromosomes, cells enter into a prolonged mitotic arrest. This arrest is resolved through either mitotic cell death or slippage. Cells that have undergone slippage can die through apoptotic cell death, undergo arrest and senescence, or continue to cycle in a polyploid or aneuploidy state.
Activation of the SAC is required for effective cell death induction during mitosis. Drugs that interfere with microtubules or that cause mitotic spindle defects, such as reduced tension at kinetochores, activate the SAC and cause prometaphase arrest. The SAC inhibits the anaphase-promoting complex APC/C, preventing cyclin-dependent kinase 1 (CDK1) inactivation and mitotic exit, and then the apoptotic threshold is crossed (Lara-Gonzalez et al. 2012). Different kinases such as PLK1, BUB1/BUBR1, and Aurora B play critical roles in SAC signaling, including activation and amplification of the MCC, the main effector of SAC. MCC assembly inhibits APC/C activation and prevents proteolytic degradation of the separase inhibitor, securin, and the CDK1 activator, cyclin B1. Despite continued SAC activation, cyclin B1 levels are slowly degraded during prolonged mitosis by the APC/C in a noncanonical fashion, making it impossible for the cell to maintain high CDK1 activity. The gradual decline of cyclin B1 defines the period of time to mitotic exit (Brito and Rieder 2006).
Several lines of evidence suggest that prolonged SAC activation triggers the intrinsic mitochondrial apoptosis pathway (Topham and Taylor 2013). Interactions between antiapoptotic proteins (for example, BCL2 and MCL1) and proapoptotic proteins (for example, BID, NOXA, and BIM) control activation of the apoptotic effectors BAX and BAK. For an extensive review of BCL2 family proteins modified during mitosis and their role in mitotic cell death, see Haschka et al. (2018). CDK1 plays a pivotal role during mitotic arrest. During an unperturbed cell division, CDK1 phosphorylates caspase-8 and -9 and inactivates the caspase cascade (Allan and Clarke 2007; Matthess et al. 2010, 2014). However, during prolonged mitosis, CDK1 also phosphorylates antiapoptotic proteins such as BCL2 or BCLX to reduce their antiapoptotic potential. Together with MCL1 degradation by noncanonical APC/C activity, this primes cells for apoptosis (Eichhorn et al. 2013; Allan et al. 2018). If apoptotic signals reach the death threshold first, MOMP is activated, culminating in activation of caspase-9, -3, and -7 and cell death.
Diverse studies have shown a direct correlation between caspase activation and cell death during mitotic arrest. Nevertheless, an open question here is whether these two networks are truly independent (Fava and Villunger 2014). Caspase activity is reported to be dispensable for mitotic slippage and inhibition of caspases in mitosis does not alter the degradation kinetics of endogenous cyclin B1 (Huang et al. 2010; Lee et al. 2011). However, the expression levels of some caspases as well as their activation are reportedly regulated in a cell-cycle, phase-specific manner, including in mitosis. Thus, there may be a tighter connection between both pathways (Hsu et al. 2006; Hashimoto et al. 2008). A correlation between antimitotic drug sensitivity and XIAP levels has also been established (Shi et al. 2008). In addition, preventing slippage by depleting CDC20 by siRNA triggers cell death (Huang et al. 2009). Allan and Clarke (2007) showed that caspase-9 is phosphorylated and inactivated by cyclin B/CDK1 in mitosis, proposing that gradual degradation of cyclin B1 results in gradual activation of caspase-9. Also, Kim et al. (2005) proposed that the apoptosis pathway directly promotes slippage by caspase-dependent proteolysis of the SAC protein BUBR1. Moreover, depletion of BAX, BAK and the mitochondrial fission protein DRP1 from U2OS cells delayed mitotic adaptation, which strongly indicates cross talk between mediators of mitotic apoptosis and SAC adaptation (Díaz-Martínez et al. 2014). More recently, it has been reported that a mitotic delay promotes DNA damage that depends on subapoptotic activation of the classical caspase-9/ -3/-7 pathway. Cleavage of iCAD liberates the caspase-activated DNase responsible for the DNA damage (Hain et al. 2016). This CAD activity may drive mutagenesis and genomic instability in cells surviving limited caspase activation after minority MOMP, as suggested by others (Ichim et al. 2015).
Altogether, it seems clear that prolonged mitotic arrest promotes caspase activation downstream of BCL2 family-regulated MOMP. What is less clear at present is whether caspases are activated at low levels (e.g., by miMOMP) (Ichim et al. 2015) to help override the SAC and avoid death in mitosis, or whether lack of caspase activation under such conditions favors alternative cell death pathways that are usually more inflammatory, particularly ferroptosis and necroptosis. We propose that caspase inhibition would be a potential strategy to induce inflammatory cell death upon antimitotic drug therapy, and this might promote antitumor immunity.
CELL-CYCLE REGULATORS AS CASPASE SUBSTRATES
It has long been noted that cell-cycle proteins are subjected to caspase-mediated proteolysis during apoptosis (Table 1). For example, the CDK inhibitors p21 and p27 (Levkau et al. 1998), as well as WEE1 kinase and the APC/C subunit CDC27 (Zhou et al. 1998) are cleaved. Paradoxically, cleavage of the latter causes futile activation of cell-cycle-promoting cyclin complexes involving CDK2 and CDK1 while caspase-processed p27 can no longer promote cell migration and invasion (Podmirseg et al. 2016).
There are several additional examples of caspase-mediated cleavage of cell-cycle proteins inducing their activation and/or translocation. Rb is cleaved by caspase-3 and -7 (Fattman et al. 2001). This cleaved form binds to cyclin D3 and represses transcriptional activity of E2F1 (Jänicke et al. 1996). Cleavage of the checkpoint kinase CHK1 by caspase-3 and -7 produces a truncated protein with hyperactive kinase activity (Matsuura et al. 2008; Okita et al. 2013). Interestingly, ectopic expression of this truncated CHK1 produces chromatin condensation and disruptions of nuclear architecture, reminiscent of changes seen during apoptosis. Precisely how this phenotype is induced remains unclear.
The cell-cycle driver CDC25A also undergoes caspase cleavage during apoptosis (Mazars et al. 2009). The resulting carboxy-terminal fragment is catalytically active and localizes to the nucleus, where it tightly associates with and dephosphorylates CDK2. Expression of the cleavage product is sufficient to induce apoptosis owing to excessive cyclin B/CDK1 activity (Chou et al. 2010), perhaps through CDK1 phosphorylation of BCL2 family members (Zhou et al. 2014).
Cleavage of the DNA replication regulator CDC6 by caspase-3 during apoptosis generates an amino-terminal fragment that lacks the nuclear export sequence while retaining the ATPase domain. Consequently, it is unable to undergo cytoplasmic translocation upon phosphorylation by CDK2 (Yim et al. 2003; Schories et al. 2004). Expression of this truncated form of CDC6 blocks MCM2 loading onto chromatin, promoting an ATM/ATR response, p53 accumulation, and BAX-mediated apoptosis (Yim et al. 2006).
A key question is whether all these cleavage events are relevant to apoptotic cell death. Arguably not, because caspases are activated during apoptosis at the point of no return (i.e., MOMP, has been passed already). It has been noted in other contexts involving caspase activity, such as cell differentiation, that cleavage of caspase substrates can produce entities with novel activities and functions. This phenomenon is not unique to the execution of apoptosis. For example, caspase-3 cleaves and activates the kinase MST1 during myogenic differentiation (Fernando et al. 2002). Hence it is possible that the apoptotic cleavage events described above may support other functions in nonapoptotic contexts.
CASPASES AS SUBSTRATES OF CELL-CYCLE REGULATORS
Modulation of caspase function by phosphorylation has been noted repeatedly. For example, the oncogenic nonreceptor tyrosine kinases SRC and c-ABL phosphorylate and inactivate caspase-8 and -9, respectively (Cursi et al. 2006; Serrano et al. 2017). Phosphorylation of caspase-3 and -7 by CK2 and PAK2, respectively, also appears to be inhibitory. Phosphorylation most likely affects dimer stability and/or substrate accessibility (Duncan et al. 2011; Li et al. 2011). Cyclin B/CDK1 has been shown to phosphorylate caspase-9 on Thr125, which is located between the CARD prodomain and large catalytic subunit. This site is modified by ERK in S phase, and during normal mitosis and mitotic arrest, when ERK activity is usually low. This suggests that caspase-9 is modified by different kinases depending on cell-cycle stage. Expression of caspase-9(T125A) was shown to accelerate cell death in response to nocodazole treatment but otherwise had no effect on mitotic progression (Allan and Clarke 2007). How phosphorylation of caspase-9 Thr125 affects recruitment into or autoactivation in the apoptosome remains to be addressed. Similarly, cyclin B/CDK1 is reported to phosphorylate caspase-2 at Ser340 during mitosis and in mitotically arrested cells, while in interphase this modification is removed by PP1 phosphatase (Andersen et al. 2009; Nutt et al. 2009). Here, the phospho site is located between the large and small subunits of caspase-2 and it may interfere with autoprocessing, which is needed for full activation. Caspase-2(S340A) sensitizes U2OS cells to death in mitosis in response to nocodazole (Andersen et al. 2009). Notably, caspase-8 was also shown to be phosphorylated in mitosis as a mechanism to limit the sensitivity of mitotic cells to FAS or TNFR ligation (Powley et al. 2016).
Collectively, these studies suggest that the apoptosis effector machinery is inactivated to avoid precocious activation of the caspase cascade during normal mitosis. This mechanism may also act as a fail-safe against partial activation of the apoptotic cascade when mitotic timing errors occur in response to spindle poisons (Orth et al. 2012) and during normal mitosis (e.g., because of chromosome alignment defects or lagging chromosomes) (Soto et al. 2019). It is interesting that phosphates are found in or near the linker region of all human apical caspases, including those controlling inflammation and pyroptosis, but are not in effector caspases (Andersen et al. 2009). Lytic cell death and inflammatory cytokine release might be regulated by proline-directed kinases, other than CDKs, because innate immune cells that are most susceptible to pyroptosis are usually terminally differentiated. However, homeostatic proliferation of tissue resident macrophages has been noted and epithelial cells also express caspase-1 and can undergo lytic cell death (Van Opdenbosch and Lamkanfi 2019).
RECENT DEVELOPMENTS LINKING CASPASES TO GENOMIC STABILITY AND (STERILE) INFLAMMATION
Although apoptotic caspases can modulate cell-cycle progression and cell fate by processing a set of substrates directly involved in cell-cycle control, they can also affect cell fate indirectly by promoting apoptosis and thereby repressing inflammatory responses. For example, hematopoietic stem cells (HSCs) deficient in caspase-9 exhaust prematurely but clear viral infections more effectively. Caspase-9 maintains stem cell dormancy by reducing interferon (IFN)-mediated stem cell mobilization, which is required to satisfy blood cell demand during viral infection. Specifically, activation of caspase-9-dependent apoptosis by BAK and BAX not only kills the cell, it also prevents mitochondrial DNA released into the cytosol by BAX and BAK from stimulating IFN production by the cGAS/STING/IRF3 pathway (Rongvaux et al. 2014; White et al. 2014; McArthur et al. 2018; Riley et al. 2018). Caspase-3 and -7, the key downstream effectors of caspase-9, have been reported to cull IFN signaling under normal conditions by cleaving cGAS, IRF3, as well as an alternative viral sensor that promotes IFN production called MAVS (Ning et al. 2019).
At the moment, it remains unclear under which physiological or pathological conditions caspase activation is compromised to the degree that mitochondrial DNA actively drives IFN signaling, or, potentially even MLKL-dependent necrotic cell death via activation of the DNA sensor DAI/ZBP1. Considering that many viruses encode proteins that inhibit caspases, viral infections seem a plausible scenario. Cells and tissues that silence expression of the caspase-9 adaptor APAF1 may also be affected. Of note, the cGAS/STING/IRF3 signaling axis also drives inflammation in response to chromosome missegregation and the subsequent formation of micronuclei. Here, breakdown of the brittle nuclear envelope formed around chromosomes that were not included into the main nucleus exposes nuclear dsDNA that is recognized by cGAS (Harding et al. 2017; Mackenzie et al. 2017). Whether caspases suppress inflammatory signaling in cells presenting with micronuclei remains to be investigated.
For a long time, the role of caspase-2, the caspase most closely related to the prototypical Caenorhabditis elegans caspase Ced-3, remained mysterious (Kumar 2009; Fava et al. 2012). Most studies pointed to a role for caspase-2 in DNA damage–induced cell death, or potentially death-receptor-mediated apoptosis. Caspase-2, similar to caspase-8, can cleave the BCL2 family protein BID into tBID, which can then trigger activation of BAX and BAK (Upton et al. 2008; Fava et al. 2012). In the context of DNA damage–induced apoptosis, it was considered an effector of p53, because a component of the PIDDosome complex that activates caspase-2, PIDD1, is a transcriptional target of p53 (Lin et al. 2000; Tinel and Tschopp 2004). However, this issue remained controversial because multiple cell types derived from animals lacking PIDD1, caspase-2, or another PIDDosome component called RAIDD/CRADD exhibited normal p53-dependent apoptosis. Moreover, in contrast to animals with impaired p53 responses upon DNA damage (e.g., Puma/Bbc3, Atm, or Chk2-deficient mice), absence of the PIDDosome did not cause any obvious phenotypes (O'Reilly et al. 2002; Berube et al. 2005; Manzl et al. 2009). Thus, the idea that caspase-2 or the PIDDosome were needed for DNA damage–induced apoptosis became less palatable. However, loss of caspase-2 accelerates cancer in MYC-transgenic (Ho et al. 2009; Manzl et al. 2012), Her2-transgenic (Parsons et al. 2013), as well as ATM-deficient mice (Puccini et al. 2013), so there was residual interest in studying caspase-2 biology. A role for caspase-2 in cell-cycle control was postulated because Casp2−/− MEFs had difficulties arresting the cell cycle in response to DNA damage (Ho et al. 2008) and aneuploid cells were more frequent in MYC-driven lymphomas (Dorstyn et al. 2012) or in SV40 immortalized MEFs lacking caspase-2 (Peintner et al. 2015). A role for caspase-2 in killing aneuploid cells or cells arrested in mitosis by spindle poisons also was reported. Together, these findings pointed toward a role for caspase-2 in surveilling mitotic fidelity.
Looking into a potential link between cell-cycle progression and caspase-2, we were excited to find that caspase-2 becomes selectively activated in a PIDDosome-dependent manner when cells undergo cytokinesis failure (Fava et al. 2017). Specifically, the presence of supernumerary centrosomes once cells return to the G1 state drives a PIDDosome-dependent, p53-mediated cell-cycle arrest. Polyploidy in the presence of supernumerary centrosomes is considered to occur frequently and early on in tumor formation. Extra centrosomes, priming cells for multipolar division, have been discussed as a cause of cancer for more than 100 years (Holland and Cleveland 2009; Conduit et al. 2015).
Subsequently, we found that the presence of extra centrosomes, rather than the increased DNA content or an aneuploid karyotype, is the direct cue that activates caspase-2 in the PIDDosome (Fava et al. 2017). Active caspase-2 then processes and inactivates the ubiquitin ligase MDM2, which allows accumulation of p53 and p21-dependent cell-cycle arrest. Whether this function of caspase-2 contributes to tumor suppression in mouse models of cancer remains to be formally tested. Extra centrosomes owing to PLK4 overexpression has recently been shown to promote spontaneous carcinogenesis (Levine et al. 2017) and a PIDDosome-deficient background might accelerate tumor development.
These findings broaden our understanding of how extra centrosomes activate p53, and they open up new lines of research in the field of scheduled polyploidization during organogenesis or regeneration (e.g., in the naturally polyploid liver). Of note, PIDDosome mutant animals show increased liver ploidy and our unpublished findings suggest a tight control of PIDDosome expression during liver development and regeneration after partial hepatectomy involving E2F family proteins. Thus, E2F family members control the ability of hepatocytes to activate p53 in response to cytokinesis failure and extra centrosomes.
Finally, caspase-8, best known for its apoptosis-inducing function downstream of death receptors and as an inhibitor of necroptotic cell death, was linked to faithful chromosome alignment in mitosis. During normal mitosis, a ripoptosome-like complex (Fig. 1) recruited the mitotic kinase PLK1, where it was inactivated by caspase-8-mediated proteolysis. Cleavage of PLK1 limited phosphorylation of the SAC component BUBR1, fine-tuning chromosome segregation fidelity. It is possible that this role of caspase-8 is responsible for its tumor-suppressive activity, rather than its apoptosis-promoting function or its anti-inflammatory role in restraining necroptosis. Of note, low levels of RIPK1 together with high levels of PLK1 correlate with increased aneuploidy in breast, lung, and colorectal cancer (Liccardi et al. 2019). Somewhat at odds with these findings is the fact that mice lacking Casp8 together with the necroptosis executer Mlkl or its upstream activator, Ripk3, are viable. These animals do not seem to develop spontaneous tumors at a higher rate than normal. However, long-term observation is hampered by the fact that these knockouts develop lymphadenopathy and fatal autoimmune disease (Alvarez-Diaz et al. 2016; Zhang et al. 2016). Tissue-specific deletion of Casp8 and Mlkl outside the hematopoietic compartment may help to better address the role of caspase-8 in the maintenance of genomic stability and as a barrier against cancer. Of note, loss of Casp8 in B cells facilitates their transformation (Hakem et al. 2012) and CASP8 is found mutated in HCC, gastric, breast, as well as colorectal cancer (Kim et al. 2003; Soung et al. 2005a,b; Park et al. 2016).
Taken together, these studies suggest that caspases do more than their acknowledged roles in apoptosis and pathogen-driven inflammation. A reevaluation of the interaction of caspases with their substrates linked to cell-cycle control but originally described as targets for proteolysis under apoptotic conditions may open up exciting new lines of research.
OPEN QUESTIONS
Many of the findings discussed here were described in vitro. On the one hand, there are clear examples where these translate into in vivo phenotypes, for example, the B-cell proliferation defect in caspase-6 knockout mice. On the other hand, caspase-8 conditional knockout mice lack obvious cell-cycle defects. While cell-cycle transition and mitotic timing seem unaffected, loss of caspase-8 may facilitate the emergence of genomically unstable cells (Liccardi et al. 2019). If caspase-8 has cell-cycle functions beyond apoptosis promotion and necroptosis suppression, why do caspase-8 mutant mice lacking RIPK3 or MLKL not develop cancer spontaneously, nor have obvious cell-cycle defects (Alvarez-Diaz et al. 2016; Zhang et al. 2016)? Why does the increased ploidy and reported aneuploidy in Casp2-mutant mice not suffice to drive transformation (Dorstyn et al. 2012; Fava et al. 2017)? To what extent do compensatory mechanisms prevent the emergence of cell-cycle defects in animal models (Zheng et al. 2000)? Executioner caspase-3 and -7 show a measure of functional redundancy both in vitro and in vivo. It may be the case that multiple gene knockouts may be necessary for cell-cycle phenotypes to emerge. Combined loss of caspase-3 and -7 is lethal (Lakhani et al. 2006) and, so far, consequences of combined conditional ablation of both caspases have not been reported. Caspase-6 is mainly expressed in the nervous system, and therefore the analysis of caspase-3, -7 double-mutant animals may yield interesting new insights. Of note, even the loss of individual cyclin-dependent kinases, other than CDK1, can be well compensated in vivo (Barrière et al. 2007; Santamaria et al. 2007). Surprisingly, loss of any of the PIDDosome components is insufficient to drive spontaneous cancer, but can enhance tumor development driven by MYC overexpression (Manzl et al. 2012; Peintner et al. 2015). Consequently, there is much work ahead of us to define the role of caspase-2 within and potentially also outside the PIDDosome in tumorigenesis.
COMPETING INTEREST STATEMENT
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
We want to thank all members of our team for fruitful discussion. We apologize to all those whose valuable contributions were not cited because of space constrains. Work in our laboratories is supported by the County of South Tyrol, the Austrian Science Fund, FWF Grant Nos. P 26856, P 29499, PIR 3, and the ERC-AdG “POLICE.”
Footnotes
Editors: Kim Newton, James M. Murphy, and Edward A. Miao
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
- Allan LA, Clarke PR. 2007. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell 26: 301–310. 10.1016/j.molcel.2007.03.019 [DOI] [PubMed] [Google Scholar]
- Allan LA, Skowyra A, Rogers KI, Zeller D, Clarke PR. 2018. Atypical APC/C-dependent degradation of Mcl-1 provides an apoptotic timer during mitotic arrest. EMBO J 37: e96831 10.15252/embj.201796831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Diaz S, Dillon CP, Lalaoui N, Tanzer MC, Rodriguez DA, Lin A, Lebois M, Hakem R, Josefsson EC, O'Reilly LA, et al. 2016. The pseudokinase MLKL and the kinase RIPK3 have distinct roles in autoimmune disease caused by loss of death-receptor-induced apoptosis. Immunity 45: 513–526. 10.1016/j.immuni.2016.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, Nutt LK, Thompson JW, Moseley MA, Kornbluth S. 2009. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J 28: 3216–3227. 10.1038/emboj.2009.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arechiga AF, Bell BD, Leverrier S, Weist BM, Porter M, Wu Z, Kanno Y, Ramos SJ, Ong ST, Siegel R, et al. 2007. A Fas-associated death domain protein/caspase-8-signaling axis promotes S-phase entry and maintains S6 kinase activity in T cells responding to IL-2. J Immunol 179: 5291–5300. 10.4049/jimmunol.179.8.5291 [DOI] [PubMed] [Google Scholar]
- Baek KH, Shin HJ, Jeong SJ, Park JW, McKeon F, Lee CW, Kim CM. 2005. Caspases-dependent cleavage of mitotic checkpoint proteins in response to microtubule inhibitor. Oncol Res 15: 161–168. 10.3727/096504005776367906 [DOI] [PubMed] [Google Scholar]
- Barrière C, Santamaría D, Cerqueira A, Galán J, Martín A, Ortega S, Malumbres M, Dubus P, Barbacid M. 2007. Mice thrive without Cdk4 and Cdk2. Mol Oncol 1: 72–83. 10.1016/j.molonc.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, Yeh WC, Mak TW, Benchimol S. 2005. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci 102: 14314–14320. 10.1073/pnas.0506475102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaert R, Kidd VJ, Cornelis S, Van de Craen M, Denecker G, Lahti JM, Gururajan R, Vandenabeele P, Fiers W. 1997. Cleavage of PITSLRE kinases by ICE/CASP-1 and CPP32/CASP-3 during apoptosis induced by tumor necrosis factor. J Biol Chem 272: 11694–11697. 10.1074/jbc.272.18.11694 [DOI] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. 2010. Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci 123: 3209–3214. 10.1242/jcs.073643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Rieder CL. 2006. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol 16: 1194–1200. 10.1016/j.cub.2006.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokatzky D, Dörflinger B, Haimovici A, Weber A, Kirschnek S, Vier J, Metz A, Henschel J, Steinfeldt T, Gentle IE, et al. 2019. A non-death function of the mitochondrial apoptosis apparatus in immunity. EMBO J 38: e100907 10.15252/embj.2018100907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Misteli T. 2015. Not all DDRs are created equal: non-canonical DNA damage responses. Cell 162: 944–947. 10.1016/j.cell.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Poncet D, Marissen WE, Flotow H, Lloyd RE, Clemens MJ, Morley SJ. 2000. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ 7: 628–636. 10.1038/sj.cdd.4400699 [DOI] [PubMed] [Google Scholar]
- Chan KS, Koh CG, Li HY. 2012. Mitosis-targeted anti-cancer therapies: where they stand. Cell Death Dis 3: e411 10.1038/cddis.2012.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Yen YC, Lee CM, Chen MS. 2010. Pro-apoptotic role of Cdc25A: activation of cyclin B1/Cdc2 by the Cdc25A C-terminal domain. J Biol Chem 285: 17833–17845. 10.1074/jbc.M109.078386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Wainman A, Raff JW. 2015. Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol 16: 611–624. 10.1038/nrm4062 [DOI] [PubMed] [Google Scholar]
- Connolly PF, Jäger R, Fearnhead HO. 2014. New roles for old enzymes: killer caspases as the engine of cell behavior changes. Front Physiol 5: 149 10.3389/fphys.2014.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. 2011. Caspase substrates and cellular remodeling. Annu Rev Biochem 80: 1055–1087. 10.1146/annurev-biochem-061809-121639 [DOI] [PubMed] [Google Scholar]
- Cursi S, Rufini A, Stagni V, Condò I, Matafora V, Bachi A, Bonifazi AP, Coppola L, Superti-Furga G, Testi R, et al. 2006. Src kinase phosphorylates Caspase-8 on Tyr380: a novel mechanism of apoptosis suppression. EMBO J 25: 1895–1905. 10.1038/sj.emboj.7601085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. 2005. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 1: 112–119. 10.1038/nchembio711 [DOI] [PubMed] [Google Scholar]
- Díaz-Martínez LA, Karamysheva ZN, Warrington R, Li B, Wei S, Xie XJ, Roth MG, Yu H. 2014. Genome-wide siRNA screen reveals coupling between mitotic apoptosis and adaptation. EMBO J 33: 1960–1976. 10.15252/embj.201487826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. 2008. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134: 679–691. 10.1016/j.cell.2008.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Puccini J, Wilson CH, Shalini S, Nicola M, Moore S, Kumar S. 2012. Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ 19: 1411 10.1038/cdd.2012.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Turowec JP, Duncan KE, Vilk G, Wu C, Luscher B, Li SS, Gloor GB, Litchfield DW. 2011. A peptide-based target screen implicates the protein kinase CK2 in the global regulation of caspase signaling. Sci Signal 4: ra30 10.1126/scisignal.2001682 [DOI] [PubMed] [Google Scholar]
- Eichhorn JM, Sakurikar N, Alford SE, Chu R, Chambers TC. 2013. Critical role of anti-apoptotic Bcl-2 protein phosphorylation in mitotic death. Cell Death Dis 4: e834 10.1038/cddis.2013.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt P, Tomaselli KJ, Cooper GM. 1997. Identification of the MDM2 oncoprotein as a substrate for CPP32-like apoptotic proteases. J Biol Chem 272: 15049–15052. 10.1074/jbc.272.24.15049 [DOI] [PubMed] [Google Scholar]
- Etemad B, Kops GJ. 2016. Attachment issues: kinetochore transformations and spindle checkpoint silencing. Curr Opin Cell Biol 39: 101–108. 10.1016/j.ceb.2016.02.016 [DOI] [PubMed] [Google Scholar]
- Falk M, Ussat S, Reiling N, Wesch D, Kabelitz D, Adam-Klages S. 2004. Caspase inhibition blocks human T cell proliferation by suppressing appropriate regulation of IL-2, CD25, and cell cycle-associated proteins. J Immunol 173: 5077–5085. 10.4049/jimmunol.173.8.5077 [DOI] [PubMed] [Google Scholar]
- Fattman CL, Delach SM, Dou QP, Johnson DE. 2001. Sequential two-step cleavage of the retinoblastoma protein by caspase-3/-7 during etoposide-induced apoptosis. Oncogene 20: 2918–2926. 10.1038/sj.onc.1204414 [DOI] [PubMed] [Google Scholar]
- Fava LL, Villunger A. 2014. Stop competing, start talking! EMBO J 33: 1849–1851. 10.15252/embj.201489466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava LL, Bock FJ, Geley S, Villunger A. 2012. Caspase-2 at a glance. J Cell Sci 125: 5911–5915. 10.1242/jcs.115105 [DOI] [PubMed] [Google Scholar]
- Fava LL, Schuler F, Sladky V, Haschka MD, Soratroi C, Eiterer L, Demetz E, Weiss G, Geley S, Nigg EA, et al. 2017. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev 31: 34–45. 10.1101/gad.289728.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando P, Megeney LA. 2007. Is caspase-dependent apoptosis only cell differentiation taken to the extreme? FASEB J 21: 8–17. 10.1096/fj.06-5912hyp [DOI] [PubMed] [Google Scholar]
- Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. 2002. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci 99: 11025–11030. 10.1073/pnas.162172899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS. 2008. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell 14: 111–122. 10.1016/j.ccr.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Gentiletti F, Mancini F, D'Angelo M, Sacchi A, Pontecorvi A, Jochemsen AG, Moretti F. 2002. MDMX stability is regulated by p53-induced caspase cleavage in NIH3T3 mouse fibroblasts. Oncogene 21: 867–877. 10.1038/sj.onc.1205137 [DOI] [PubMed] [Google Scholar]
- Gervais JL, Seth P, Zhang H. 1998. Cleavage of CDK inhibitor p21Cip1/Waf1 by caspases is an early event during DNA damage–induced apoptosis. J Biol Chem 273: 19207–19212. 10.1074/jbc.273.30.19207 [DOI] [PubMed] [Google Scholar]
- Gilot D, Serandour AL, Ilyin GP, Lagadic-Gossmann D, Loyer P, Corlu A, Coutant A, Baffet G, Peter ME, Fardel O, et al. 2005. A role for caspase-8 and c-FLIPL in proliferation and cell-cycle progression of primary hepatocytes. Carcinogenesis 26: 2086–2094. 10.1093/carcin/bgi187 [DOI] [PubMed] [Google Scholar]
- Gutteridge RE, Ndiaye MA, Liu X, Ahmad N. 2016. Plk1 inhibitors in cancer therapy: from laboratory to clinics. Mol Cancer Ther 15: 1427–1435. 10.1158/1535-7163.MCT-15-0897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara H, Sunada Y. 2004. Mechanism of taxane neurotoxicity. Breast Cancer 11: 82–85. 10.1007/BF02968008 [DOI] [PubMed] [Google Scholar]
- Hain KO, Colin DJ, Rastogi S, Allan LA, Clarke PR. 2016. Prolonged mitotic arrest induces a caspase-dependent DNA damage response at telomeres that determines cell survival. Sci Rep 6: 26766 10.1038/srep26766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem A, El Ghamrasni S, Maire G, Lemmers B, Karaskova J, Jurisicova A, Sanchez O, Squire J, Hakem R. 2012. Caspase-8 is essential for maintaining chromosomal stability and suppressing B-cell lymphomagenesis. Blood 119: 3495–3502. 10.1182/blood-2011-07-367532 [DOI] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548: 466–470. 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschka MD, Soratroi C, Kirschnek S, Häcker G, Hilbe R, Geley S, Villunger A, Fava LL. 2015. The NOXA-MCL1-BIM axis defines lifespan on extended mitotic arrest. Nat Commun 6: 6891 10.1038/ncomms7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschka M, Karbon G, Fava LL, Villunger A. 2018. Perturbing mitosis for anti-cancer therapy: is cell death the only answer? EMBO Rep 19: e45440 10.15252/embr.201745440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Yamauchi L, Hunter T, Kikkawa U, Kamada S. 2008. Possible involvement of caspase-7 in cell cycle progression at mitosis. Genes Cells 13: 609–621. 10.1111/j.1365-2443.2008.01192.x [DOI] [PubMed] [Google Scholar]
- Ho MY, Mackey JR. 2014. Presentation and management of docetaxel-related adverse effects in patients with breast cancer. Cancer Manag Res 6: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LH, Read SH, Dorstyn L, Lambrusco L, Kumar S. 2008. Caspase-2 is required for cell death induced by cytoskeletal disruption. Oncogene 27: 3393–3404. 10.1038/sj.onc.1211005 [DOI] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. 2009. A tumor suppressor function for caspase-2. Proc Natl Acad Sci 106: 5336–5341. 10.1073/pnas.0811928106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487. 10.1038/nrm2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J. 2000. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 1: 489–495. 10.1038/82732 [DOI] [PubMed] [Google Scholar]
- Hsu SL, Yu CT, Yin SC, Tang MJ, Tien AC, Wu YM, Huang CY. 2006. Caspase 3, periodically expressed and activated at G2/M transition, is required for nocodazole-induced mitotic checkpoint. Apoptosis 11: 765–771. 10.1007/s10495-006-5880-x [DOI] [PubMed] [Google Scholar]
- Huang HC, Shi J, Orth JD, Mitchison TJ. 2009. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell 16: 347–358. 10.1016/j.ccr.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Mitchison TJ, Shi J. 2010. Stochastic competition between mechanistically independent slippage and death pathways determines cell fate during mitotic arrest. PLoS One 5: e15724 10.1371/journal.pone.0015724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichim G, Lopez J, Ahmed SU, Muthalagu N, Giampazolias E, Delgado ME, Haller M, Riley JS, Mason SM, Athineos D, et al. 2015. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol Cell 57: 860–872. 10.1016/j.molcel.2015.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänicke RU, Walker PA, Lin XY, Porter AG. 1996. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J 15: 6969–6978. 10.1002/j.1460-2075.1996.tb01089.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien O, Wells JA. 2017. Caspases and their substrates. Cell Death Differ 24: 1380–1389. 10.1038/cdd.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawadler H, Gantz MA, Riley JL, Yang X. 2008. The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol Cell 31: 415–421. 10.1016/j.molcel.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Funabiki H. 2009. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol 21: 51–58. 10.1016/j.ceb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy NJ, Kataoka T, Tschopp J, Budd RC. 1999. Caspase activation is required for T cell proliferation. J Exp Med 190: 1891–1896. 10.1084/jem.190.12.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Lee JW, Soung YH, Park WS, Kim SY, Lee JH, Park JY, Cho YG, Kim CJ, Jeong SW, et al. 2003. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology 125: 708–715. 10.1016/S0016-5085(03)01059-X [DOI] [PubMed] [Google Scholar]
- Kim M, Murphy K, Liu F, Parker SE, Dowling ML, Baff W, Kao GD. 2005. Caspase-mediated specific cleavage of BubR1 is a determinant of mitotic progression. Mol Cell Biol 25: 9232–9248. 10.1128/MCB.25.21.9232-9248.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komlodi-Pasztor E, Sackett DL, Fojo AT. 2012. Inhibitors targeting mitosis: tales of how great drugs against a promising target were brought down by a flawed rationale. Clin Cancer Res 18: 51–63. 10.1158/1078-0432.CCR-11-0999 [DOI] [PubMed] [Google Scholar]
- Krippner-Heidenreich A, Talanian RV, Sekul R, Kraft R, Thole H, Ottleben H, Lüscher B. 2001. Targeting of the transcription factor Max during apoptosis: phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid residue in position P1. Biochem J 358: 705–715. 10.1042/bj3580705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. 2009. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more? Nat Rev Cancer 9: 897–903. 10.1038/nrc2745 [DOI] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev 11: 847–862. 10.1101/gad.11.7.847 [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GJ, Booth CJ, Mehal WZ, Inayat I, Flavell RA. 2006. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 311: 847–851. 10.1126/science.1115035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. 2012. The spindle assembly checkpoint. Curr Biol 22: R966–980. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. 2010. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci 107: 4230–4235. 10.1073/pnas.0913089107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kenny AE, Rieder CL. 2011. Caspase activity is not required for the mitotic checkpoint or mitotic slippage in human cells. Mol Biol Cell 22: 2470–2479. 10.1091/mbc.e11-03-0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, et al. 2017. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell 40: 313–322.e5. 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkau B, Koyama H, Raines EW, Clurman BE, Herren B, Orth K, Roberts JM, Ross R. 1998. Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: role of a caspase cascade. Mol Cell 1: 553–563. 10.1016/S1097-2765(00)80055-6 [DOI] [PubMed] [Google Scholar]
- Li X, Wen W, Liu K, Zhu F, Malakhova M, Peng C, Li T, Kim HG, Ma W, Cho YY, et al. 2011. Phosphorylation of caspase-7 by p21-activated protein kinase (PAK) 2 inhibits chemotherapeutic drug-induced apoptosis of breast cancer cell lines. J Biol Chem 286: 22291–22299. 10.1074/jbc.M111.236596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liccardi G, Ramos Garcia L, Tenev T, Annibaldi A, Legrand AJ, Robertson D, Feltham R, Anderton H, Darding M, Peltzer N, et al. 2019. RIPK1 and caspase-8 ensure chromosome stability independently of their role in cell death and inflammation. Mol Cell 73: 413–428.e7. 10.1016/j.molcel.2018.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S. 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet 26: 124–127. [DOI] [PubMed] [Google Scholar]
- Liu W, Linn S. 2000. Proteolysis of the human DNA polymerase ε catalytic subunit by caspase-3 and calpain specifically during apoptosis. Nucleic Acids Res 28: 4180–4188. 10.1093/nar/28.21.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548: 461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. 2008. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134: 866–876. 10.1016/j.cell.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, Logette E, Tschopp J, Villunger A. 2009. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol 185: 291–303. 10.1083/jcb.200811105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Peintner L, Krumschnabel G, Bock F, Labi V, Drach M, Newbold A, Johnstone R, Villunger A. 2012. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ 19: 1722–1732. 10.1038/cdd.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissen WE, Guo Y, Thomas AA, Matts RL, Lloyd RE. 2000. Identification of caspase 3-mediated cleavage and functional alteration of eukaryotic initiation factor 2α in apoptosis. J Biol Chem 275: 9314–9323. 10.1074/jbc.275.13.9314 [DOI] [PubMed] [Google Scholar]
- Matsuura K, Wakasugi M, Yamashita K, Matsunaga T. 2008. Cleavage-mediated activation of Chk1 during apoptosis. J Biol Chem 283: 25485–25491. 10.1074/jbc.M803111200 [DOI] [PubMed] [Google Scholar]
- Matthess Y, Raab M, Sanhaji M, Lavrik IN, Strebhardt K. 2010. Cdk1/cyclin B1 controls Fas-mediated apoptosis by regulating caspase-8 activity. Mol Cell Biol 30: 5726–5740. 10.1128/MCB.00731-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthess Y, Raab M, Knecht R, Becker S, Strebhardt K. 2014. Sequential Cdk1 and Plk1 phosphorylation of caspase-8 triggers apoptotic cell death during mitosis. Mol Oncol 8: 596–608. 10.1016/j.molonc.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazars A, Fernandez-Vidal A, Mondesert O, Lorenzo C, Prévost G, Ducommun B, Payrastre B, Racaud-Sultan C, Manenti S. 2009. A caspase-dependent cleavage of CDC25A generates an active fragment activating cyclin-dependent kinase 2 during apoptosis. Cell Death Differ 16: 208–218. 10.1038/cdd.2008.142 [DOI] [PubMed] [Google Scholar]
- Mazumder S, Gong B, Chen Q, Drazba JA, Buchsbaum JC, Almasan A. 2002. Proteolytic cleavage of cyclin E leads to inactivation of associated kinase activity and amplification of apoptosis in hematopoietic cells. Mol Cell Biol 22: 2398–2409. 10.1128/MCB.22.7.2398-2409.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V, Geoghegan ND, Chappaz S, Davidson S, San Chin H, et al. 2018. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359: eaao6047 10.1126/science.aao6047 [DOI] [PubMed] [Google Scholar]
- McGrogan BT, Gilmartin B, Carney DN, McCann A. 2008. Taxanes, microtubules and chemoresistant breast cancer. Biochim Biophys Acta 1785: 96–132. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. 2012. The proliferation rate paradox in antimitotic chemotherapy. Mol Biol Cell 23: 1–6. 10.1091/mbc.e10-04-0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X, Wang Y, Jing M, Sha M, Lv M, Gao P, Zhang R, Huang X, Feng JM, Jiang Z. 2019. Apoptotic caspases suppress type I interferon production via the cleavage of cGAS, MAVS, and IRF3. Mol Cell 74: 19–31.e7. 10.1016/j.molcel.2019.02.013 [DOI] [PubMed] [Google Scholar]
- Nutt LK, Buchakjian MR, Gan E, Darbandi R, Yoon SY, Wu JQ, Miyamoto YJ, Gibbon JA, Andersen JL, Freel CD, et al. 2009. Metabolic control of oocyte apoptosis mediated by 14-3-3ζ-regulated dephosphorylation of caspase-2. Dev Cell 16: 856–866. 10.1016/j.devcel.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. 2011. Catalytic activity of the caspase-8-FLIPL complex inhibits RIPK3-dependent necrosis. Nature 471: 363–367. 10.1038/nature09852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita N, Yoshimura M, Watanabe K, Minato S, Kudo Y, Higami Y, Tanuma S. 2013. CHK1 cleavage in programmed cell death is intricately regulated by both caspase and non-caspase family proteases. Biochim Biophys Acta 1830: 2204–2213. 10.1016/j.bbagen.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Olson NE, Graves JD, Shu GL, Ryan EJ, Clark EA. 2003. Caspase activity is required for stimulated B lymphocytes to enter the cell cycle. J Immunol 170: 6065–6072. 10.4049/jimmunol.170.12.6065 [DOI] [PubMed] [Google Scholar]
- O'Reilly LA, Ekert P, Harvey N, Marsden V, Cullen L, Vaux DL, Hacker G, Magnusson C, Pakusch M, Cecconi F, et al. 2002. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ 9: 832–841. 10.1038/sj.cdd.4401033 [DOI] [PubMed] [Google Scholar]
- Orth JD, Loewer A, Lahav G, Mitchison TJ. 2012. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol Biol Cell 23: 567–576. 10.1091/mbc.e11-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HL, Ziogas A, Chang J, Desai B, Bessonova L, Garner C, Lee E, Neuhausen SL, Wang SS, Ma H, et al. 2016. Novel polymorphisms in caspase-8 are associated with breast cancer risk in the California Teachers Study. BMC Cancer 16: 14 10.1186/s12885-015-2036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, McCormick L, Janke L, Howard A, Bouchier-Hayes L, Green DR. 2013. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ 20: 1174–1182. 10.1038/cdd.2013.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pászty K, Verma AK, Padányi R, Filoteo AG, Penniston JT, Enyedi A. 2002. Plasma membrane Ca2+ATPase isoform 4b is cleaved and activated by caspase-3 during the early phase of apoptosis. J Biol Chem 277: 6822–6829. 10.1074/jbc.M109548200 [DOI] [PubMed] [Google Scholar]
- Peintner L, Dorstyn L, Kumar S, Aneichyk T, Villunger A, Manzl C. 2015. The tumor-modulatory effects of caspase-2 and Pidd1 do not require the scaffold protein Raidd. Cell Death Differ 22: 1803–1811. 10.1038/cdd.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelizon C, d'Adda di Fagagna F, Farrace L, Laskey RA. 2002. Human replication protein Cdc6 is selectively cleaved by caspase 3 during apoptosis. EMBO Rep 3: 780–784. 10.1093/embo-reports/kvf161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podmirseg SR, Jäkel H, Ranches GD, Kullmann MK, Sohm B, Villunger A, Lindner H, Hengst L. 2016. Caspases uncouple p27Kip1 from cell cycle regulated degradation and abolish its ability to stimulate cell migration and invasion. Oncogene 35: 4580–4590. 10.1038/onc.2015.524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley IR, Hughes MA, Cain K, MacFarlane M. 2016. Caspase-8 tyrosine-380 phosphorylation inhibits CD95 DISC function by preventing procaspase-8 maturation and cycling within the complex. Oncogene 35: 5629–5640. 10.1038/onc.2016.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccini J, Shalini S, Voss AK, Gatei M, Wilson CH, Hiwase DK, Lavin MF, Dorstyn L, Kumar S. 2013. Loss of caspase-2 augments lymphomagenesis and enhances genomic instability in Atm-deficient mice. Proc Natl Acad Sci 110: 19920–19925. 10.1073/pnas.1311947110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Maiato H. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell 7: 637–651. 10.1016/j.devcel.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Riley JS, Quarato G, Cloix C, Lopez J, O'Prey J, Pearson M, Chapman J, Sesaki H, Carlin LM, Passos JF, et al. 2018. Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J 37: e99238 10.15252/embj.201899238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, et al. 2014. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159: 1563–1577. 10.1016/j.cell.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T, Bokoch GM. 1997. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science 276: 1571–1574. 10.1126/science.276.5318.1571 [DOI] [PubMed] [Google Scholar]
- Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY, Berry DM, Tamblyn L, Shehabeldin A, Migon E, et al. 2003. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev 17: 883–895. 10.1101/gad.1063703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16: 473–485. 10.1038/nrm4025 [DOI] [PubMed] [Google Scholar]
- Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Caceres JF, Dubus P, Malumbres M, Barbacid M. 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448: 811–815. 10.1038/nature06046 [DOI] [PubMed] [Google Scholar]
- Sayan BS, Sayan AE, Knight RA, Melino G, Cohen GM. 2006. p53 is cleaved by caspases generating fragments localizing to mitochondria. J Biol Chem 281: 13566–13573. 10.1074/jbc.M512467200 [DOI] [PubMed] [Google Scholar]
- Schories B, Engel K, Dörken B, Gossen M, Bommert K. 2004. Characterization of apoptosis-induced Mcm3 and Cdc6 cleavage reveals a proapoptotic effect for one Mcm3 fragment. Cell Death Differ 11: 940–942. 10.1038/sj.cdd.4401411 [DOI] [PubMed] [Google Scholar]
- Schwab BL, Leist M, Knippers R, Nicotera P. 1998. Selective proteolysis of the nuclear replication factor MCM3 in apoptosis. Exp Cell Res 238: 415–421. 10.1006/excr.1997.3850 [DOI] [PubMed] [Google Scholar]
- Serrano BP, Szydlo HS, Alfandari D, Hardy JA. 2017. Active site-adjacent phosphorylation at Tyr-397 by c-Abl kinase inactivates caspase-9. J Biol Chem 292: 21352–21365. 10.1074/jbc.M117.811976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Mitchison TJ. 2017. Cell death response to anti-mitotic drug treatment in cell culture, mouse tumor model and the clinic. Endocr Relat Cancer 24: T83–T96. 10.1530/ERC-17-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Orth JD, Mitchison T. 2008. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res 68: 3269–3276. 10.1158/0008-5472.CAN-07-6699 [DOI] [PubMed] [Google Scholar]
- Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, et al. 1999. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol 144: 281–292. 10.1083/jcb.144.2.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto M, Raaijmakers JA, Medema RH. 2019. Consequences of genomic diversification induced by segregation errors. Trends Genet 35: 279–291. 10.1016/j.tig.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim SY, Jang J, Park YG, Park WS, Nam SW, Lee JY, Yoo NJ, Lee SH. 2005a. CASPASE-8 gene is inactivated by somatic mutations in gastric carcinomas. Cancer Res 65: 815–821. [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim SY, Sung YJ, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. 2005b. Caspase-8 gene is frequently inactivated by the frameshift somatic mutation 1225_1226delTG in hepatocellular carcinomas. Oncogene 24: 141–147. 10.1038/sj.onc.1208244 [DOI] [PubMed] [Google Scholar]
- Stack JH, Newport JW. 1997. Developmentally regulated activation of apoptosis early in Xenopus gastrulation results in cyclin A degradation during interphase of the cell cycle. Development 124: 3185–3195. [DOI] [PubMed] [Google Scholar]
- Tang HL, Tang HM, Mak KH, Hu S, Wang SS, Wong KM, Wong CS, Wu HY, Law HT, Liu K, et al. 2012. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol Biol Cell 23: 2240–2252. 10.1091/mbc.e11-11-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304: 843–846. 10.1126/science.1095432 [DOI] [PubMed] [Google Scholar]
- Topham CH, Taylor SS. 2013. Mitosis and apoptosis: how is the balance set? Curr Opin Cell Biol 25: 780–785. 10.1016/j.ceb.2013.07.003 [DOI] [PubMed] [Google Scholar]
- Tu S, Cerione RA. 2001. Cdc42 is a substrate for caspases and influences Fas-induced apoptosis. J Biol Chem 276: 19656–19663. 10.1074/jbc.M009838200 [DOI] [PubMed] [Google Scholar]
- Upton JP, Austgen K, Nishino M, Coakley KM, Hagen A, Han D, Papa FR, Oakes SA. 2008. Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol Cell Biol 28: 3943–3951. 10.1128/MCB.00013-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opdenbosch N, Lamkanfi M. 2019. Caspases in cell death, inflammation, and disease. Immunity 50: 1352–1364. 10.1016/j.immuni.2019.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raam BJ, Ehrnhoefer DE, Hayden MR, Salvesen GS. 2013. Intrinsic cleavage of receptor-interacting protein kinase-1 by caspase-6. Cell Death Differ 20: 86–96. 10.1038/cdd.2012.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe C, Shu GL, Zheng TS, Flavell RA, Clark EA. 2008. Caspase 6 regulates B cell activation and differentiation into plasma cells. J Immunol 181: 6810–6819. 10.4049/jimmunol.181.10.6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, et al. 2014. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159: 1549–1562. 10.1016/j.cell.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M, Hakem R, Furlonger C, Hakem A, Duncan GS, Sasaki T, Bouchard D, Lu L, Wu GE, Paige CJ, et al. 2003. Caspase-3 regulates cell cycle in B cells: a consequence of substrate specificity. Nat Immunol 4: 1016–1022. 10.1038/ni976 [DOI] [PubMed] [Google Scholar]
- Yim H, Jin YH, Park BD, Choi HJ, Lee SK. 2003. Caspase-3-mediated cleavage of Cdc6 induces nuclear localization of p49-truncated Cdc6 and apoptosis. Mol Biol Cell 14: 4250–4259. 10.1091/mbc.e03-01-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H, Hwang IS, Choi JS, Chun KH, Jin YH, Ham YM, Lee KY, Lee SK. 2006. Cleavage of Cdc6 by caspase-3 promotes ATM/ATR kinase-mediated apoptosis of HeLa cells. J Cell Biol 174: 77–88. 10.1083/jcb.200509141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadil LM, Andersen KA, Yeum D, Rocque GB, Wilke LG, Tevaarwerk AJ, Raines RT, Burkard ME, Weaver BA. 2014. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci Transl Med 6: 229ra43 10.1126/scitranslmed.3007965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fan C, Zhang H, Zhao Q, Liu Y, Xu C, Xie Q, Wu X, Yu X, Zhang J, et al. 2016. MLKL and FADD are critical for suppressing progressive lymphoproliferative disease and activating the NLRP3 inflammasome. Cell Rep 16: 3247–3259. 10.1016/j.celrep.2016.06.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, Lazebnik Y, Flavell RA. 2000. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat Med 6: 1241–1247. 10.1038/81343 [DOI] [PubMed] [Google Scholar]
- Zhou BB, Li H, Yuan J, Kirschner MW. 1998. Caspase-dependent activation of cyclin-dependent kinases during Fas-induced apoptosis in Jurkat cells. Proc Natl Acad Sci 95: 6785–6790. 10.1073/pnas.95.12.6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Cai X, Han X, Xu N, Chang DC. 2014. CDK1 switches mitotic arrest to apoptosis by phosphorylating Bcl-2/Bax family proteins during treatment with microtubule interfering agents. Cell Biol Int 38: 737–746. 10.1002/cbin.10259 [DOI] [PubMed] [Google Scholar]