Abstract
Centromeres pose an evolutionary paradox: strongly conserved in function but rapidly changing in sequence and structure. However, in the absence of damage, centromere locations are usually conserved within a species. We report here that isolates of the pathogenic yeast species Candida parapsilosis show within-species polymorphism for the location of centromeres on two of its eight chromosomes. Its old centromeres have an inverted-repeat (IR) structure, whereas its new centromeres have no obvious structural features but are located within 30 kb of the old site. Centromeres can therefore move naturally from one chromosomal site to another, apparently spontaneously and in the absence of any significant changes in DNA sequence. Our observations are consistent with a model in which all centromeres are genetically determined, such as by the presence of short or long IRs or by the ability to form cruciforms. We also find that centromeres have been hotspots for genomic rearrangements in the C. parapsilosis clade.
Centromeres are the point of assembly of the kinetochore, the position at which the spindle microtubules are connected to the chromosomes, enabling efficient and accurate separation of chromosome/chromatid pairs during cell division. Most eukaryotes have large “regional” centromeres that have been proposed to be epigenetically determined. They are specified by arrays of chromatin, compacted by di- or trimethylation at lysine 9 of histone H3 (H3K9me2/3). The position of the centromere in most species is determined by the presence of a variant of histone H3, called CENPA in mammals or Cse4 in yeast.
Centromere repositioning occurs on an evolutionary timescale, leading to the formation of evolutionarily new centromeres (ENCs). ENCs have played an important role in speciation, including in many mammals (Stanyon et al. 2008; Rocchi et al. 2012). An ancient ENC at one chromosome in orangutans is polymorphic; individuals can be homozygous for either the old or the new centromere or can be heterozygous for both (Locke et al. 2011; Rocchi et al. 2012). The new centromere location lacks the repetitive alpha satellites observed at other centromeres. In addition, damage to, or loss of, existing centromeres can be rescued by the formation of new (neo) centromeres at different locations. Neocentromere formation following damage has been observed in human clinical samples, as well as in other primates, in Equidae, marsupials, plants, and yeasts (for reviews, see Burrack and Berman 2012; Rocchi et al. 2012; Schubert 2018). Movement of centromeres among individuals within a species in a nonclinical context is much more rarely described. A small number of neocentromeres formed in human cells that have no obvious clinical effect have been reported; these were usually observed during routine amniocentesis (for review, see Rocchi et al. 2012). In addition, the location of one centromere in the horse (devoid of satellite DNA) varies among individuals (Wade et al. 2009; Purgato et al. 2015). The mechanisms underlying the formation of new centromeres are not fully understood, although many are likely to be associated with chromosomal inversion and translocation (Schubert 2018). The formation of neocentromeres following damage is particularly well studied in the yeast Candida albicans (Burrack and Berman 2012). Koren et al. (2010) suggested that, in this species, centromeres are associated with the presence of early origins of replication and that the formation of neocentromeres changes the activity of nearby origins.
Basic centromere organization is conserved in many fungi, including the basidiomycetes and the filamentous ascomycetes (Friedman and Freitag 2017). Centromeres in the budding yeasts (the Saccharomycotina) have undergone substantial changes associated with the loss of the lysine methylation machinery (Malik and Henikoff 2009). Within Saccharomycotina, the Saccharomycetaceae clade, containing the model yeast Saccharomyces cerevisiae, is by far the best studied. These species have small “point” centromeres, in which function is determined by sequence. The S. cerevisiae centromere consists of three conserved regions called centromere-determining elements (CDEs): CDEI, CDEII, and CDEIII (Schulman and Bloom 1991). Cse4 is present in one nucleosome at the centromere (Meluh et al. 1998; Furuyama and Biggins 2007; Henikoff and Henikoff 2012). Similar point centromeres are found in other Saccharomycetaceae species (Kitada et al. 1997; Mattei et al. 2002; Gordon et al. 2011). In Naumovozyma species, the sequences of the CDEs are different, but they still act as point centromeres (Kobayashi et al. 2015). The point centromeres in S. cerevisiae are among the fastest evolving sequences in the genome (Bensasson et al. 2008). However, point centromeres are not present in most fungal genomes (Malik and Henikoff 2009).
Centromere structure has also been investigated in other families in the Saccharomycotina, including the Pichiaceae and the CUG-Ser1 clade. Within the Pichiaceae, centromere structure is known in Kuraishia capsulata and Komagataella phaffii. In K. capsulata, centromeres lie in 2- to 6-kb regions with low GC content, and a 200-bp motif is conserved across some chromosomes (Morales et al. 2013). In K. phaffii, the centromeres consist of a 1-kb central (mid) region, flanked by a 2-kb inverted repeat (IR) (Coughlan et al. 2016). There is no conservation in sequence among the four centromeres in K. phaffii, and Cse4 localizes across the mid region and the IR.
The CUG-Ser1 clade within the Saccharomycotina contains many Candida and other species, characterized by translating CUG as serine rather than leucine (Ohama et al. 1993). The centromeres of C. albicans and Candida dubliniensis are described as “small regional”; they are characterized by gene-free regions of 4–18 kb, with 3–5 kb occupied by Cse4 (Sanyal et al. 2004; Padmanabhan et al. 2008; Roy and Sanyal 2011). The flanking compact chromatin extends up to 25 kb for C. albicans CEN7 (centromere of Chromosome 7) (Sreekumar et al. 2019). There is no sequence conservation between centromeres of different chromosomes. There are short unique IRs surrounding C. albicans CEN1, CEN4, and CENR, as well as longer repeats surrounding CEN5 (Sanyal et al. 2004). In the related species Candida tropicalis, the centromere cores are all flanked by IRs, and there is significant sequence conservation between different centromeres (Chatterjee et al. 2016). Centromeres in the more distantly related Clavispora lusitaniae have 4-kb regions occupied by Cse4, with no sequence conservation (Kapoor et al. 2015). The C. lusitaniae centromeres lie in regions with low GC content, which has also been proposed to mark centromeres in the CUG-Ser1 clade species Debaryomyces hansenii and Scheffersomyces stipitis (Lynch et al. 2010). The putative centromeres in these latter species contain clusters of retrotransposons (Lynch et al. 2010; Coughlan et al. 2016).
In this study, we aimed to determine the locations of centromeres in Candida parapsilosis using chromatin immunoprecipitation (IP) with DNA sequencing (ChIP-seq) and to use comparative genomics to study centromere evolution in the C. parapsilosis clade. Unexpectedly, we found that the locations are different in two different C. parapsilosis isolates that we examined.
Results
Identification of centromeres in C. parapsilosis
Many fungal centromeres are located in large intergenic regions and may be flanked by IR sequences. When we looked for regions that matched these criteria in the genome of C. parapsilosis CDC317 (the sequenced reference genome) (Butler et al. 2009), we identified one candidate centromere per chromosome (Fig. 1A,B). These regions range from 5.8 to 7.1 kb and lack genes. Each contains an IR sequence (shown in red in the dot matrix plot Fig. 1A), flanking a middle (mid) sequence. The IRs vary in size. Some are relatively short (e.g., 443 bp on Chromosome 6), and in others, the repeat region is broken into several sections (e.g., Chromosome 1, total size ∼1600 bp). The similarity between IRs ranges from 85%–96.7%. The sequences of the IRs are conserved among chromosomes, and the conservation extends beyond the IRs (Fig. 1B, black boxes). All IRs are predicted to form large secondary structures using RNAfold (Lorenz et al. 2011). However, there is no conservation among the mid regions that lie between the IRs on different chromosomes.
Figure 1.
C. parapsilosis centromeres consist of unique mid regions surrounded by partially conserved inverted repeats (IRs). (A) Dot matrix plot comparing the putative centromere sequences in C. parapsilosis. Centromere regions (see Supplemental Table S2) were concatenated and are delineated by dark blue lines. IRs (right, IRR; left, IRL) are separated with cyan lines. Each dot represents a 25-bp window. Inverted sequences are shown in red; direct repeats, in black. (B) Diagrammatic representation of the information in A. Regions that are conserved among chromosomes are shown in black. Locations of IRs (>75% DNA sequence identity) are shown with white arrows. The mid regions are illustrated in different colors that indicate that each of them has a unique sequence. Adjacent genes are shown in gray. Each region shown is ∼10 kb in length. (C) Three copies of an HA tag were introduced into both alleles of the endogenous CSE4 gene in C. parapsilosis CLIB214 and 90-137 using CRISPR-Cas9 editing. The gene was cut between glycine 69 and glycine 70, and a repair template containing the HA tags was inserted by homologous recombination. The construct was confirmed by sequencing. (D) Visualization of the ChIP-seq signal across all chromosomes (Chr) in Cse4-tagged derivatives of C. parapsilosis CLIB214 and 90-137. (In) Input (before immunoprecipitation); IP1 and IP2 show two independent immunoprecipitation replicates from each strain. Strains derived from C. parapsilosis CLIB214 are shown in blue; from 90-137, in purple. There is one signal per chromosome in the IP samples, identifying the centromere, except for Chromosome 7, in which the rDNA locus (black asterisk) also generates a signal. The x-axis in each plot is the chromosome coordinates, and the y-axis is the number of reads mapping to a position. The maximum scale for C. parapsilosis CLIB214 is restricted to reduce the signal from the rDNA. Data are visualized using Integrative Genomics Viewer (IGV) (Thorvaldsdóttir et al. 2013).
To validate these predictions, we determined the location of the variant histone H3, Cse4, by ChIP. C. parapsilosis has a diploid genome. We introduced three copies of a nine-amino-acid epitope from human influenza hemagglutinin (HA), near the N terminus of both Cse4 alleles using CRISPR-Cas9 editing together with a synthetic repair template (Fig. 1C; Lombardi et al. 2017). The epitope was introduced into Cse4 twice independently in two different strains: C. parapsilosis CLIB214, which is the type strain, and C. parapsilosis 90-137, which was originally isolated from orbital tissue (Tavanti et al. 2005) and which can be efficiently edited using CRISPR-Cas9 (Lombardi et al. 2017, 2019a). We confirmed that the tagged protein is expressed and that it does not interfere with growth of the tagged strains, and we used ChIP-PCR to show that Cse4 binding is enriched at the predicted CEN1 sequence (Supplemental Fig. S1).
To identify all the regions in the genome where Cse4 binds, we used ChIP-seq. We obtained one very strong ChIP-seq signal per chromosome that was present in only the immunoprecipitated Cse4-HA strains and not in the input chromatin (Fig. 1D). We also identified a signal from the ribosomal DNA on Chromosome 7, an artifact owing to the high copy number that is also present in the control sample. More detailed analysis shows that the Cse4 signals from C. parapsilosis CLIB214 correspond with the regions that were bioinformatically identified as centromeres (Fig. 2). The centromeres are in regions that are devoid of open reading frames and are generally low in transcription (Fig. 2). Unlike C. tropicalis (Chatterjee et al. 2016) but similar to K. phaffii (Coughlan et al. 2016), Cse4 binding extends beyond the mid regions into the IRs, reducing in frequency toward the ends of the repeats.
Figure 2.
Natural polymorphisms for centromere location in C. parapsilosis. The ChIP-seq data from Figure 1D is shown in more detail, and the neocentromeres are highlighted with black boxes. The order of the tracks is the same in each panel but is labeled for CEN1 only. The top track shows the location of C. parapsilosis protein coding genes. The second track shows the IR sequences only (red), with an arrow indicating the direction of the repeat. The extent of the regions conserved between chromosomes is not shown. ChIP-seq read coverage is plotted in blue for C. parapsilosis CLIB214 and in purple for C. parapsilosis 90-137. Two independent immunoprecipitation experiments were performed per strain (IP1 and IP2). Only one control is shown; the total chromatin from C. parapsilosis CLIB214 (input). The equivalent data for C. parapsilosis 90-137, and for an experiment with no tagged Cse4, are available at GEO, accession number GSE136854. The bottom track (gray) shows gene expression measured by RNA-seq during growth in YPD (taken from SRR6458364 from Turner et al. 2018). The read depth scale is indicated in brackets; the total number of reads varied in each experiment. The maximum scale for C. parapsilosis CLIB214 is restricted to 500 to reduce the signal from the rDNA. The RNA expression data are plotted on a log scale. The apparent dips in coverage at the centromeres in the input data are likely to be an artifact of the mapping procedure because reads that map to more than one site in the genome were discarded. Some reads are also incorrectly mapped to nonidentical repeat sequences, resulting in a small Cse4 signal at CEN5 in 90-137. All data are visualized using IGV.
Polymorphic centromere locations in C. parapsilosis
The Cse4 signal in C. parapsilosis 90-137 is very similar to C. parapsilosis CLIB214 (Fig. 1D). Closer examination shows the pattern is almost identical for six of the eight chromosomes (Fig. 2). However, there are surprising differences at CEN1 and CEN5. For Chromosome 1, there is a signal at the expected centromere in C. parapsilosis 90-137, similar to C. parapsilosis CLIB214. However, there is an additional signal, ∼17 kb away in 90-137 (Fig. 2). This second signal, or neocentromere, partially overlaps two open reading frames, CPAR2_101630 and CPAR2_101640, which are transcribed in C. parapsilosis CLIB214 (RNA track in Fig. 2). The difference is even more striking on Chromosome 5. Here, C. parapsilosis 90-137 has no obvious Cse4 signal at the expected position of CEN5 (the small number of reads shown is an artifact of the mapping process, resulting from the presence of repeat sequences). Instead, the Cse4 signal is localized ∼29 kb away, again overlapping transcribed ORFs, CPAR2_502960 and CPAR2_502970. There are no IRs surrounding the new centromeres, and there is no sequence relationship with other centromeric regions.
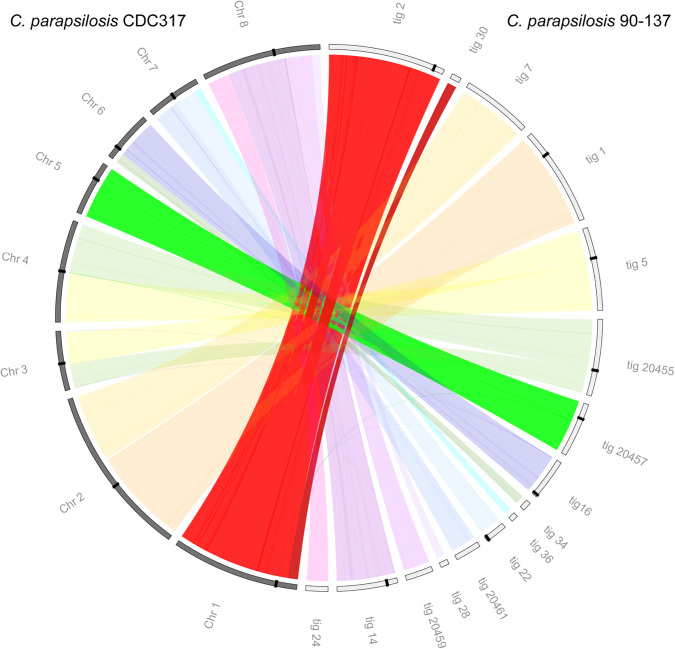
We considered that the occurrence of neocentromeres in C. parapsilosis 90-137 might coincide with possible rearrangements of the chromosomes in this isolate. We therefore determined the genome structure of the Cse4-HA tagged strain using long-read sequencing (Oxford Nanopore Technologies). The nuclear genome was assembled into 12 to 16 scaffolds >100 kb using Flye (Kolmogorov et al. 2019) or Canu (Koren et al. 2017), respectively (Fig. 3). The assemblies failed at some centromeric regions. However, Figure 3 shows that Chromosomes 1 and 5 are collinear between C. parapsilosis 90-137/Cse4-HA and the reference genome, including around the centromere regions. The IR structures and mid region at the original CEN1 and CEN5 locations are intact in C. parapsilosis 90-137, and in the Flye assembly, they are 99% identical to the reference genome.
Figure 3.
Lack of rearrangements at CEN1 and CEN5 in C. parapsilosis 90-137/Cse4-HA. The Circos plot compares the eight chromosomes of the reference strain C. parapsilosis CDC317 (gray; left) to the 16 largest minION scaffolds from the Canu assembly of C. parapsilosis 90-137/Cse4-HA (white; right). Centromeres are marked by black bands. Most chromosomes are collinear, including Chromosome 1 (assembled in two contigs in 90-137, contig 2 and contig 30) and Chromosome 5 (contig 20457). There is an apparent translocation between Chromosomes 3 and 4 (contig 5 and contig 20455) at a repetitive gene that is near (but not at) the centromere. This may represent an error in the reference assembly or represent a natural structural polymorphism. Some zeros have been removed from the contig (tig) names for clarity.
The species C. parapsilosis is therefore polymorphic for centromere location on two chromosomes. The centromere relocations are associated with a transition from a structured (IR) format to a format with no obvious structure or sequence dependence, within a single species. On Chromosome 5, it is likely that the centromeres on both copies of this chromosome have moved to a new location. It is possible that C. parapsilosis 90-137 is heterozygous at CEN1, with Cse4 at the expected location on one copy of Chromosome 1 and at a new location on the other copy.
Genomic rearrangements in C. orthopsilosis coincide with centromere locations
C. parapsilosis is closely related to Candida orthopsilosis and Candida metapsilosis; they are all members of the C. parapsilosis sensu lato clade (Tavanti et al. 2005). We surmised that the centromeres in these other species may have a similar structure to C. parapsilosis. The C. orthopsilosis 90-125 reference assembly (Riccombeni et al. 2012; Schröder et al. 2016) is not fully assembled at putative centromeres, so we used a minION assembly of this strain from Lombardi et al. (2019b). We identified one large region per chromosome likely to represent the centromere. The size of the regions ranges from 4.9–7.1 kb (Fig. 4A). Candidates on Chromosomes 1, 2, 5, 6, and 7 have a similar structure to C. parapsilosis centromeres. A pair of IR sequences, varying in size from 788 bp on Chromosome 5 to 2.2 kb on Chromosome 6, flank a core region of ∼3 kb. The similarity between IRs ranges from 91.0% to 99.8%, the sequences are conserved among chromosomes, and for Chromosomes 5, 6, and 7, the conservation among chromosomes extends beyond the IRs. The remaining inferred centromeres (CEN3, -4, -8) do not contain IR sequences. However, 135 bp to 2.2 kb of the flanking regions surrounding the 2.6- to 3.4-kb mid regions are conserved with other centromeres. Like in C. parapsilosis, there is no conservation between the mid regions identified on different chromosomes. In addition, none of the C. orthopsilosis CEN regions (not just the IR-less ones) share significant sequence similarity with any of the C. parapsilosis CEN regions.
Figure 4.
Identification of centromeres and centromere-proximal rearrangements in C. orthopsilosis and C. metapsilosis. (A) Cartoon of centromere structure in C. orthopsilosis 90-125 (Lombardi et al. 2019b). All mid regions are unique and are shown in different colors. Sequences in black are conserved among chromosomes. IRs are shown with white arrows, and adjacent genes are shown with gray boxes. Putative transposases with DDE domains are indicated. More detail is provided in Supplemental Figure S2 and Supplemental Table S2. (B,C) Synteny relationship between C. parapsilosis and C. orthopsilosis. SynChro (Drillon et al. 2014) was used (delta value of two) to identify potential orthologs (reciprocal best hits [RBHs]), represented by colored lines in the two species, and to generate synteny maps. (B) Location of RBHs on C. orthopsilosis chromosomes. The approximate location of the putative centromeres is indicated with a gray polygon. (C) C. parapsilosis chromosomes, colored with respect to the RBH from C. orthopsilosis. The location of the C. parapsilosis centromeres are indicated with an offset white circle. The location of syntenic C. orthopsilosis centromeres is shown in more detail in Figure 5. (D) Cartoon of centromere structure in C. metapsilosis. Sequences in black are conserved among chromosomes. IRs are shown with white arrows, which are sometimes fragmented and overlapping. Mid-core regions from some CENs are similar in sequence (>60%) and are shown in the same color. Adjacent genes are shown with gray boxes. More detail is provided in Supplemental Figure S2 and Supplemental Table S2. (E,F) Synteny relationship between C. parapsilosis and C. metapsilosis. (E) Location of RBHs on C. metapsilosis chromosomes. The approximate location of the putative C. metapsilosis centromeres are indicated with a gray star (centromeres were not identified on scaffolds 4 and 8). (F) C. parapsilosis chromosomes, colored with respect to the RBH from C. metapsilosis. The location of the C. parapsilosis centromeres are indicated with a white circle. The approximate location of syntenic C. metapsilosis centromeres are shown by name and with gray stars. The same colors are used for C. orthopsilosis (B) and C. metapsilosis (E). This does not indicate that synteny is completely conserved between these species; it is a feature of SynChro, which carries out pairwise comparisons.
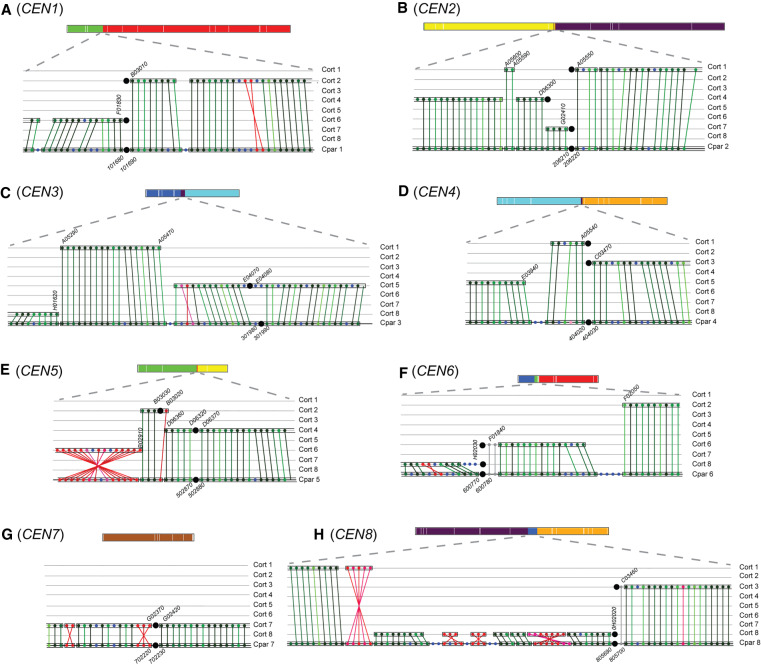
We compared the conservation of centromere position and gene order between C. parapsilosis and C. orthopsilosis using SynChro, a tool designed to visualize synteny blocks in eukaryotic genomes (Drillon et al. 2014). Putative orthologs between the two species were assigned by identifying reciprocal best hits (RBHs). Figure 4B shows the locations of genes in C. orthopsilosis that have a RBH in C. parapsilosis. Each chromosome is assigned a specific color. Figure 4C shows the locations of the same RBHs on the C. parapsilosis chromosomes, colored with respect to C. orthopsilosis chromosomes. It is immediately obvious that there is strong conservation of synteny between C. orthopsilosis and C. parapsilosis, as we have described previously (Riccombeni et al. 2012). One chromosome pair (Chromosome 7 in each species) is essentially collinear, as shown by the brown color (Fig. 4B,C). Most of the other chromosomes are represented by two major colors in C. parapsilosis, indicating that there has been one major translocation per chromosome between C. parapsilosis and C. orthopsilosis.
Overlaying the position of the mapped centromeres shows that most of the evolutionary rearrangements between C. parapsilosis and C. orthopsilosis involve breakpoints at or near the C. parapsilosis centromeres (Fig. 4C). For some chromosomes, there is a single breakpoint (e.g., Chromosome 1). For others, whereas most of the two arms of the C. parapsilosis chromosome matches two C. orthopsilosis chromosomes, the junction near the centromere includes short sections from a third chromosome (e.g., on Chromosome 8). These relationships are explored in Figure 5, which shows the gene order around each C. parapsilosis centromere in more detail. Individual RBHs (identified and visualized using SynChro) (Drillon et al. 2014) are shown. Each C. parapsilosis centromere is compared with all C. orthopsilosis chromosomes, and syntenic blocks are highlighted.
Figure 5.
Interspecies synteny breakpoints occur at centromeres. Synteny between C. parapsilosis and C. orthopsilosis was visualized using SynChro (Drillon et al. 2014), with a delta value of two. Changing delta values had minor effects on predicted synteny. A diagrammatic representation of each C. parapsilosis chromosome, colored as in Figure 4C, is shown to scale at the top of each panel. The lower sections of each panel show the gene order around the centromere. (A–H) Gene order around the eight centromeres in C. parapsilosis compared with C. orthopsilosis. The bottom row in each panel shows gene order on the C. parapsilosis chromosome, and the eight C. orthopsilosis chromosomes are shown above. Each gene is indicated by a colored dot, and RBHs are joined by lines. Syntenic blocks are surrounded with a box. Centromeres are shown by large black circles. The chromosome number is indicated at the side of each panel. The names of some genes are shown for orientation purposes. We removed the prefix “CORT0” from C. orthopsilosis genes and “CPAR2_” from C. parapsilosis genes for brevity. The color of the dots indicates the similarity of the proteins. Noninverted RBHs are shown in green, ranging from darkest (>90% similarity) to lightest (<30% similarity), and inverted orthologs are shown in red. Genes without RBH orthologs are shown in blue. Genes in gray were not identified as RBHs by SynChro but were identified using CGOB (Fitzpatrick et al. 2010; Maguire et al. 2013).
Multiple rearrangements have occurred exactly at, or very close to, the centromere on almost all chromosomes (Fig. 5). For example, on C. parapsilosis Chromosome 1, genes to the right of the centromere are syntenic with genes on C. orthopsilosis Chromosome 2, and genes to the left of the centromere are syntenic with C. orthopsilosis Chromosome 6 (Fig. 5A). The break in synteny coincides exactly with the location of the predicted centromeres on the two C. orthopsilosis chromosomes and with C. parapsilosis CEN1. More complex rearrangements are seen at CEN2, CEN4, CEN6, and CEN8 (Fig. 5B,D,F,H). In each of these examples, there is a break in synteny at the C. parapsilosis centromere, so that the left and right flanks of the C. parapsilosis centromeres match two different C. orthopsilosis chromosomes, and the breakpoints in C. orthopsilosis also occur at or near its centromeres. However, in these four cases, there are also additional rearrangements nearby, at which at CEN2 (Fig. 5B) corresponds with a third centromere in C. orthopsilosis on Chromosome 4.
Even on Chromosome 7 (Fig. 5G), which is almost collinear between the two species, there has been an inversion beside the centromere. C. parapsilosis CEN3 is also collinear with C. orthopsilosis CEN5 (Fig. 5C). However, there have been two rearrangements on the left of C. parapsilosis CEN3, where a short block of genes on C. parapsilosis Chromosome 3 matches a region on C. orthopsilosis Chromosome 1. Most of the remainder of the left side of C. parapsilosis Chromosome 3 is syntenic with C. orthopsilosis Chromosome 8. Something similar is seen at C. parapsilosis Chromosome 5 (Fig. 5E), except here one rearrangement occurs at a second C. orthopsilosis centromere (CEN2). In summary, C. parapsilosis has synteny breakpoints relative to C. orthopsilosis at seven of its eight centromeres, and most of these breakpoints also map to C. orthopsilosis centromeres. We examined the sequences around each inter-chromosomal rearrangement site but did not find any sequence repeats that could have facilitated the rearrangements.
Genomic rearrangements in C. metapsilosis and L. elongisporus
C. metapsilosis originated from hybridization between two related species, generating a hybrid with a highly heterozygous diploid genome (Pryszcz et al. 2015). The best assembly of its genome is derived from Illumina sequencing only and is a consensus built from both haplotypes from two different isolates (Pryszcz et al. 2015). Of the nine largest C. metapsilosis scaffolds, we identified putative centromeres on seven (Fig. 4D,E). Scaffold 2 contained two candidate regions. Closer examination revealed that this scaffold contains a region (around CMET_4044) that is syntenic with two telomeres in C. parapsilosis (Chromosomes 5 and 6). We do not know if this represents a recent telomere-to-telomere fusion in C. metapsilosis or if it is an assembly error. We split scaffold 2 at CMET_4044, generating scaffolds 2A and 2B (Fig. 4E) and giving a total of eight centromeres. All the centromeres are surrounded by IRs, which have high levels of sequence similarity among chromosomes. The IRs on scaffolds 5, 6, 7, and 9 are relatively long (2.1–2.6 kb). IRs in conserved regions on scaffolds 6 and 7 are fragmented (Fig. 4D). IRs on scaffolds 1, 2A, 2B, and 3 are highly repetitive, with regions that sometimes overlap. The mid regions of C. metapsilosis centromeres vary in size from 1.2 to 2.2 kb, and unlike C. parapsilosis and C. orthopsilosis, there is sequence conservation among chromosomes. CEN2B, -5, -6, -7, and -9 share >75% identity, and CEN1 is ∼60% identical to these (Fig. 4D; Supplemental Fig. S2).
Figure 4F shows a pattern of interspecies chromosomal breakage at centromeres between C. metapsilosis and C. parapsilosis, similar to that seen with C. orthopsilosis, although the rearrangements are different and have therefore occurred independently. C. parapsilosis Chromosome 6 and C. metapsilosis scaffold 2B are collinear. Most other chromosomes have undergone a major rearrangement at points that correspond to the centromeres of both species. There have been complex rearrangements at these sites, similar to the C. orthopsilosis/C. parapsilosis comparisons. For example, the region around C. parapsilosis CEN2 is syntenic with regions near C. metapsilosis CEN2A, CEN5, and CEN7. Other apparent rearrangements may reflect gaps in the C. metapsilosis assembly (e.g., C. metapsilosis scaffold 8, which does not contain a centromere, maps to the end of C. parapsilosis Chromosome 8).
Lodderomyces elongisporus is an outgroup to the C. parapsilosis sensu lato species group (Fitzpatrick et al. 2006). We did not find any structures similar to the C. parapsilosis centromeres in the L. elongisporus genome (Butler et al. 2009). However, Koren et al. (2010) hypothesized that centromeres in L. elongisporus are adjacent to early-firing origins of replication, as in C. albicans. They identified putative regions by characterizing GC skew, which switches between strands at replication origins. Koren et al. (2010) identified nine candidate centromeres in the 11 largest L. elongisporus scaffolds that lie within intergenic regions and have a strong GC skew. Three may not represent true centromeres; one (on scaffold 9) is adjacent to the rDNA locus (Donovan et al. 2016), and two are in strongly transcribed regions (scaffold 7, scaffold 10) (Donovan et al. 2016) that are probably incorrectly annotated in the L. elongisporus genome. The most likely centromeres and a comparison of the synteny of C. parapsilosis with L. elongisporus are shown in Supplemental Figure S3. There are more rearrangements than observed between C. parapsilosis and C. orthopsilosis or C. metapsilosis. However, C. parapsilosis Chromosome 6 and L. elongisporus Chromosome 7 are collinear, and major rearrangements in the other chromosomes coincide with the location of the centromeres in C. parapsilosis and several of the remaining centromeres in L. elongisporus (Supplemental Fig. S3). It is therefore likely that six of the proposed centromere locations in L. elongisporus are correct and that centromeres are fragile sites in all four species. However, the centromere structure in L. elongisporus is very different to the C. parapsilosis sensu lato species. There are no IRs, and the sequences are mostly unique (Koren et al. 2010). They are therefore more similar to the epigenetic centromeres described in C. albicans and C. dubliniensis (Sanyal et al. 2004; Padmanabhan et al. 2008; Thakur and Sanyal 2013).
To identify the number of translocations that have occurred during the evolution of the C. parapsilosis clade, we inferred the most likely ancestral chromosomal structure using AnChro (Supplemental Fig. S4; Vakirlis et al. 2016). Some of the reference assemblies are quite fragmented, and the number of predicted chromosomes in the ancestral species are probably overestimated (13–15) (Supplemental Fig. S4). It is therefore difficult to fully resolve every rearrangement. However, the synteny comparisons identified 13 inter-chromosomal breaks between C. parapsilosis and C. orthopsilosis, and all are at or close to the centromeres as shown in Figure 5. Most rearrangements occurred on the branch leading to C. orthopsilosis (Supplemental Fig. S4). It is therefore clear that inter-chromosomal breaks are enriched at centromeres.
Discussion
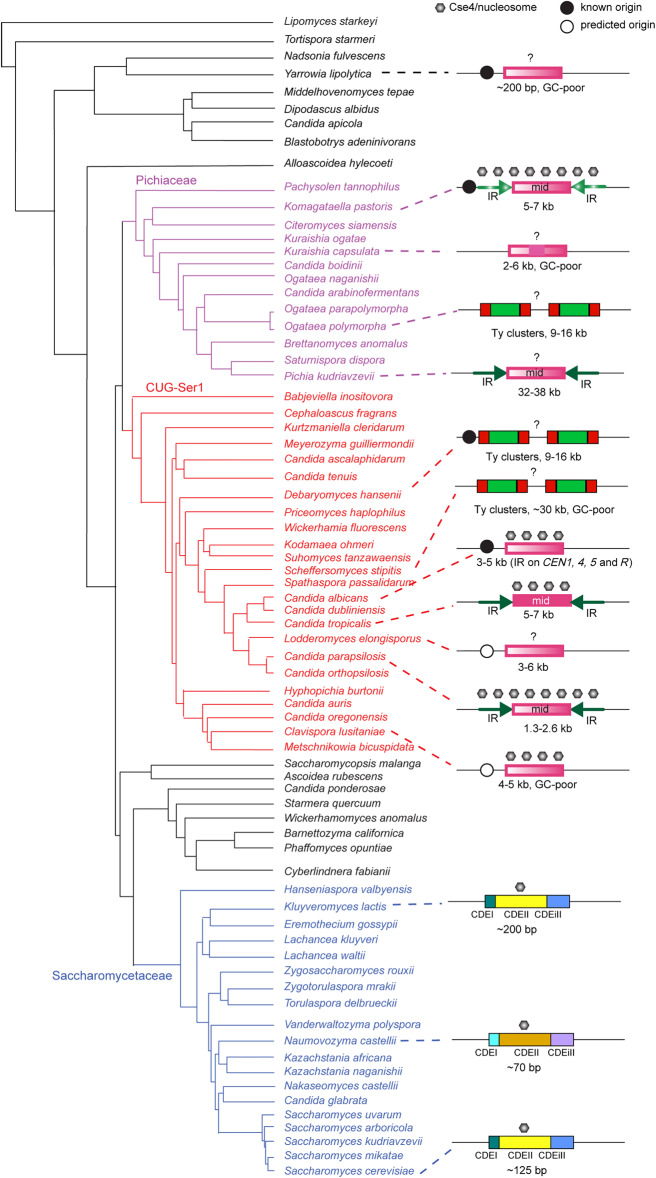
Centromeres evolve remarkably rapidly, considering their conserved function (Henikoff et al. 2001). Species in the CUG-Ser1 clade have a very wide range of centromere types (Fig. 6). Centromeres of C. albicans and C. dubliniensis have been proposed to be epigenetically determined and have little obvious sequence similarity and few IRs (Sanyal et al. 2004; Padmanabhan et al. 2008). We have shown that the centromeres in the C. parapsilosis sensu lato species group consist of a mid region that is mostly unique and is usually surrounded by IR sequences. The centromere structures in the C. parapsilosis sensu lato clade are most similar to those of C. tropicalis (Fig. 6; Padmanabhan et al. 2008; Chatterjee et al. 2016). However, in C. tropicalis, the mid regions of all centromeres are similar (∼80% identity), and the IRs are highly homogenized. Chatterjee et al. (2016) suggested that the ancestral centromere in Candida species consisted of an IR surrounding a core and that most of the IRs have been lost in C. albicans and C. dubliniensis. Orthology of the centromeres on each chromosome within the CUG-Ser1 clade, despite their structural variation, is supported by evidence that gene order is partially conserved around centromeres among C. albicans, C. dubliniensis, and C. tropicalis (Padmanabhan et al. 2008; Chatterjee et al. 2016). Synteny is conserved between C. albicans CEN3 and C. parapsilosis CEN5, and there is partial conservation of synteny around C. albicans CEN5 with centromeres in C. parapsilosis, Scheffersomyces stipitis, and C. lusitaniae, even though centromeres do not contain IRs in the latter two species (Lynch et al. 2010; Chatterjee et al. 2016).
Figure 6.
Organization of centromeres in Saccharomycotina species. The phylogeny is adapted from Shen et al. (2016). The size indicated on the centromeres refers to the region bound by Cse4 when known, or else when predicted bioinformatically, except for the Saccharomycetaceae, for which the size of the point centromere is shown. Solid color indicates conservation of sequence across centromeres in the same species, whereas a color gradient indicates unique sequences. IRs are shown with arrows; Ty clusters, as red and green boxes. Black circles show known (solid) or predicted (open) early-firing origins of replication (for details, see text). Point centromeres are conserved across the Saccharomycetaceae except for the Naumovozyma lineage, which has different sequences. Question marks indicate that localization of Cse4 nucleosomes has not been determined.
The IR structure of centromeres is likely to be old because it is also found in some species in the sister clade, the family Pichiaceae (Fig. 6). In Pichia kudriavzevii, the IRs at each CEN are very similar and they are conserved across centromeres. In addition, these IRs share some similarity with mid sequences on other chromosomes (Douglass et al. 2018). In K. phaffii (Pichia pastoris), both the IRs and the mid regions are unique at each CEN (Coughlan et al. 2016). The ancestor of the Pichiaceae and the CUG-Ser1 clade species therefore likely had an IR surrounding a mid region, with unique sequences at each centromere. The IRs have undergone homogenization in several species (P. kudriavzevii, C. tropicalis, and C. parapsilosis sensu lato), and the mid regions have been homogenized in C. tropicalis and to a lesser extent in C. metapsilosis. IRs have probably been lost in C. albicans, C. lusitaniae, and K. capsulata. In other species in the CUG-Ser1 clade (D. hansenii, S. stipitis) and in the Pichiaceae (Ogataea polymorpha), the CENs are associated with retrotransposons (Ty5-like elements). A retrotransposon (member of the Ty3/Gypsy family) is found at CEN7 in C. tropicalis, C. albicans, and C. dubliniensis (Padmanabhan et al. 2008; Chatterjee et al. 2016). DDE-type transposases are found adjacent to C. orthopsilosis CEN4 and CEN8, but these are likely to be DNA transposons (Nesmelova and Hackett 2010), more similar to CEN-associated transposons in the basidiomycete Cryptococcus neoformans (Janbon et al. 2014).
It is not clear what the ancestral centromere structure was in the subphylum Saccharomycotina because centromeres have been characterized in very few species outside the Pichiaceae and the CUG-Ser1 clade (Fig. 6). The point centromeres in the Saccharomycetaceae are unusual and probably represent a derived state (Malik and Henikoff 2009; Lefrançois et al. 2013; Kobayashi et al. 2015). Centromeric regions have been identified in Yarrowia lipolytica, an outgroup to the three clades (Fig. 6). These lie in regions of poor GC-content, adjacent to autonomously replicating sequences (Fournier et al. 1993; Lynch et al. 2010). Y. lipolytica centromeres may be small and have conserved short palindromic repeats of 17–21 bp (Yamane et al. 2008). However, the exact structure of the centromere and the location of CENPA (Cse4) in Y. lipolytica has never been determined. More experimental analysis of centromeres from other clades of the Saccharomycotina is therefore required before conclusions can be drawn about the ancestral centromere structure.
Kasinathan and Henikoff (2018) postulated that all centromeres, whether apparently epigenetic or sequence-dependent, share a common feature: They are at regions that can make non-B form DNA. This can be achieved via dyad symmetry (IRs) in the DNA or by the activity of specific DNA-binding proteins (such as binding of Cbf1 in the Saccharomycetaceae). IRs have the capacity to form cruciform structures, especially when associated with replication origins (Pearson et al. 1996). In particular, Kasinathan and Henikoff (2018) found that neocentromeres in vertebrates are particularly enriched in regions of short dyad symmetry.
The formation of “rescue” neocentromeres when the endogenous centromere is damaged has been well studied in C. albicans (for review, see Burrack and Berman 2012). When CEN5 or CEN7 is damaged, neocentromeres form, either adjacent to the original centromere or up to 450 kb away (Ketel et al. 2009; Thakur and Sanyal 2013). Koren et al. (2010) found that natural C. albicans CENs are near early-firing replication origins and that the formation of neocentromeres changes the timing of firing at adjacent origins. By characterizing the switches in base composition skew that occur at replication origins, they predicted that CENs are also near early-firing origins of replication in L. elongisporus, C. lusitaniae, and Y. lipolytica (experimentally confirmed for Y. lipolytica by Fournier et al. [1993]).
Examination of the known and predicted centromeres in CUG-Ser1 clade species shows that they all contain IRs (either long or short, including retrotransposon LTRs), and/or they are located near early-firing replication origins (known or predicted). All of these structures can form cruciforms, which may be necessary to recruit Cse4, as has been reported for Schizosaccharomyces pombe (Folco et al. 2008). The loss of the IRs at centromeres in L. elongisporus and C. lusitaniae, and from some centromeres in C. albicans and C. dubliniensis, may be compensated by the presence of a nearby early-firing replication origin (Fig. 6). Therefore, there may be no true “epigenetic” centromeres in this clade; as Kasinathan and Henikoff (2018) suggest, at least some part of centromere formation always requires cruciform or non-B form DNA, however it is made. The neocentromeres formed in C. parapsilosis 90-137 do not contain large IRs like the originals in this species. The hypothesis predicts that the neocentromeres form in regions capable of making cruciform structures, which may be facilitated by transcription. The C. parapsilosis neocentromeres are formed at regions that are transcribed, and transcription is known to facilitate centromere activity in S. cerevisiae (Ohkuni and Kitagawa 2011).
We found that the majority of chromosomal rearrangements between species in the C. parapsilosis/L. elongisporus clade involve breakpoints at or near centromeres and that, in several cases, multiple closely spaced breaks occurred near centromeres. Rearrangements between C. albicans and C. tropicalis also appear to be enriched around centromeres, which Chatterjee et al. (2016) suggested was facilitated by repeat sequences. However, rearrangements at centromeres in other species are unusual and, for example, were rarely seen in Saccharomycetaceae species (Dujon et al. 2004; Gordon et al. 2011; Vakirlis et al. 2016). It therefore appears that centromeres are hotspots for chromosome breakage in the CUG-Ser1 clade and particularly in species closely related to C. parapsilosis (e.g., CENs in C. albicans and C. dubliniensis are collinear) (Padmanabhan et al. 2008). Although fragility may be associated with the presence of repeats (IRs) at the centromeres and with the similarity of centromere sequences among chromosomes, even the centromeres of L. elongisporus, which have no IRs or other repeats, coincide with evolutionary breakpoints (Supplemental Fig. S3). Interspecies rearrangements of the karyotype by breakage at centromeres have also been reported in the basidiomycete yeast Cryptococcus (Sun et al. 2017).
There are many unanswered questions about how and why the centromere relocations in C. parapsilosis 90-137 occurred. We do not know how frequent centromere location polymorphism is in C. parapsilosis, but the fact that we observed it in one of only two strains tested, affecting two of eight chromosomes, suggests that it is not rare. There are some genome differences between C. parapsilosis strains. However, there is little evidence of substantial diversity, and heterozygosity levels are generally low (Butler et al. 2009; Pryszcz et al. 2013; Zhai et al. 2020). Such centromere sliding may also be frequent in other organisms (including humans) but has not been observed because of a lack of investigation (Rocchi et al. 2012). We also do not know what factors caused the original centromere sites to become disused in C. parapsilosis 90-137. The IR structure at the original sites appears to be intact, so it is unclear why neither allele of CEN5 binds Cse4. Similarly, we do not know what makes the new centromere sites, at both CEN1 and CEN5, attractive for Cse4 binding. They have no repeats and no obvious features such as strong base composition skew. However, they are both within 30 kb of the original site, which means that diploids heterozygous for Cse4 bound at old and new sites (like at CEN1 in C. parapsilosis 90-137) can still establish proper spindle tension. Similar heterozygous centromeric sites have been reported in orangutans (Locke et al. 2011), in horses (Wade et al. 2009; Purgato et al. 2015), and in C. albicans following damage at one allele (Thakur and Sanyal 2013). Lastly, we do not know why the new sites only bind Cse4 in C. parapsilosis 90-137 and not in C. parapsilosis CLIB214. Our discovery of “natural” neocentromeres in C. parapsilosis is one of the few known examples of within-species polymorphism for CEN locations and provides an ideal opportunity for further future investigation of how centromere location and function are determined (Wade et al. 2009; Locke et al. 2011; Rocchi et al. 2012).
Methods
Bioinformatic prediction of centromere location
Genomic sequences of intergenic regions >2 kb were extracted from the reference sequence of C. parapsilosis CDC317 (Butler et al. 2009), C. orthopsilosis 90-125 (Riccombeni et al. 2012; Schröder et al. 2016), and the chimeric reference assembly of C. metapsilosis strains PL429 (SZMC1548) and SZMC8094 (Pryszcz et al. 2015) using a custom script (Supplemental Code). Sequences were compared using BLASTN v 2.2.26 with default parameters and tabular alignment output (Altschul 1990). An IR pair was defined as a sequence identity >75% with a region in the opposite orientation (E-value cutoff 0.005). Candidate regions were selected for manual investigation. Predicted centromere locations in the C. orthopsilosis 90-125 reference assembly (Riccombeni et al. 2012; Schröder et al. 2016), available at CGOB (Fitzpatrick et al. 2010), had long regions of ambiguous bases, so we extracted equivalent regions from a minION assembly from Lombardi et al. (2019b; Supplemental Table S2). Dot matrix plots were constructed using DNAMAN (www.lynnon.com) with a criterion of 23 matches per 25-bp window. Synteny was visualized using SynChro with a delta value of two (Drillon et al. 2014), using genome assemblies and annotations from CGOB (Fitzpatrick et al. 2010; Maguire et al. 2013). To reconstruct ancestral genomes, SynChro was run using delta values between one and six. The ancestor of C. parapsilosis and C. orthopsilosis (A1) was reconstructed using AnChro (Vakirlis et al. 2016), varying delta values from one to six for each branch. C. metapsilosis and L. elongisporus were used as outgroups. The best A1 candidate, with the smallest number of chromosomes (13) and conflicts (six), was chosen as recommended by Vakirlis et al. (2016; Supplemental Fig. S4). The A1 reconstruction was then compared with the other genomes using SynChro (delta values one to six), and a second ancestral genome (A2) was constructed from A1 and C. metapsilosis, with L. elongisporus as an outgroup. The best A2 candidate, with the smallest number of chromosomes (15) and conflicts (one) and the highest number of genes (4409) was chosen (Supplemental Fig. S4). Inter-chromosomal breaks were identified using pairwise comparison of synteny maps.
Tagging Cse4
C. parapsilosis strains CLIB214 and 90-137 were edited using a tRNA plasmid based CRISPR-Cas9 gene editing system as described by Lombardi et al. (2017, 2019a). Primers gRNA_CSE4_TOP and gRNA_CSE4_BOT were annealed and cloned into pCP-tRNA, and 5 µg plasmid was transformed together with 5 µg of a 594-bp synthetic DNA fragment containing a section of the H3 histone variant Cse4 with a 3xHA tag inserted between amino acids 69 and 70, and 250 bp homology arms (Integrated DNA Technologies) (Supplemental Fig. S1). Transformants were selected on YPD agar supplemented with 200 µg/mL nourseothricin and screened by colony PCR using primers CSE4_N_RT_fw and CSE4_col_inTag_rv. The structure was confirmed using ChIP-seq and minION sequencing as described below (Supplemental Fig. S1). Loss of pCP-tRNA was induced by patching transformants onto YPD agar without nourseothricin. For western blots, protein extracts were prepared from 15 A600 units of C. parapsilosis 90-137 and two Cse4-HA tagged strains cultured overnight in YPD. Cell pellets were washed in 500 µL water, resuspended in 500 µL ice-cold extraction buffer (1× PBS, 0.1% Tween 20, 1 mM PMSF), and homogenized with glass beads. The protein extract was separated by centrifugation at 10,000 rpm at 4°C. Twenty microliters of protein extracts diluted 1:1 (v/v) with ice-cold 2× Laemmli sample buffer (Sigma-Aldrich) was separated by 12% SDS-PAGE, at 200 V constant voltage for 1 h, and electroblotted onto nitrocellulose membranes at 100 V for 45 min. Immunoblotting was performed using the mouse epitope tag antibody, Anti-HA.11 (BioLegend 901513), at a 1:1000 dilution in milk/TBS blocking buffer (5 g nonfat dry milk to 100 mL TBS–100 mM Tris-HCl at pH 7.5, 150 mM NaCl) and HRP-conjugated secondary antibody anti-mouse IgG (Cell Signaling Technology 7076P2) at 1:2000 dilution. Immunoblots were detected using the Pierce ECL western blotting substrate (Thermo Fisher Scientific) and enhanced chemiluminescence (G:BOX Chemi XRQ, Syngene).
ChIP-PCR and ChIP-seq
ChIP was performed as described by Coughlan et al. (2016) from log phase cultures in 200 mL YPD using EZview Red Anti-HA Affinity Gel from Sigma-Aldrich (E6779). Control IPs were performed in the absence of the anti-HA antibody (Mock-IP), and from C. parapsilosis 90-137 without a tagged Cse4 (CTRL). Dilutions of the protein extracts before IP (Input), and following IP and mock IP were used to assess binding to CEN1 by PCR amplification, using primers from five regions within the predicted CEN1 area, one pair from within the next largest intergenic region on Chromosome 1 (Chr 1: 1,948,277–1,955,373; to serve as negative control), and a region from within the actin gene ACT1 (Supplemental Fig. S1; Supplemental Table S1). ChIP sequencing was performed by Beijing Genomics Institute (BGI) on the BGISEQ500 platform. Approximately 20 million single-end reads (50 bases) were obtained per sample. ChIP-seq reads were mapped to the genome of C. parapsilosis CDC317 (Butler et al. 2009) using the aln/samse algorithm from BWA v0.7.17-r1188 (Li and Durbin 2010), with default parameters. Mapped reads were sorted and indexed with SAMtools v 1.9 (Li et al. 2009), and the read coverage across the genome was computed using BEDTools v2.27.1 (Quinlan and Hall 2010). Genome coverage files were changed into bigWig format using bedGraphToBigWig v4 (Kent et al. 2010) and loaded into IGV (Thorvaldsdóttir et al. 2013) for visualization.
minION sequencing
One derivative of C. parapsilosis 90-137 containing Cse4-HA was sequenced using the minION device from Oxford Nanopore Technologies (ONT). DNA was extracted using the MagJET genomic DNA kit K2721 from Thermo Fisher Scientific. Libraries were prepared with the rapid sequencing kit (RSK-SQK004) from ONT and sequenced on a minION flow cell (FLO-MIN106), yielding 30× coverage. Base-calling was performed using Guppy v2.3.7+e041753. Read length and quality were assessed using NanoPlot v1.23.1 (De Coster et al. 2018). NanoFilt v2.3.0 (De Coster et al. 2018) was used to remove reads with a quality score of less than seven. Assemblies were constructed using Canu v1.8 (Koren et al. 2017) with options genomeSize = 13030174 (to specify the genome size) and -nanopore-raw (for ONT data), generating 25 nuclear contigs, and using Flye v2.5 (Kolmogorov et al. 2019) with options ‐‐nano-raw (for ONT data), and -i 5 (five rounds of polishing), generating 14 nuclear contigs. Nanopolish v0.11.1 (Loman et al. 2015) was used to improve the consensus accuracy of the Canu assembly, and the sequence qualities of both assemblies were further improved by incorporating the BGISEQ data from the “input” sample of the ChIP-seq experiment using Pilon v1.23 (Walker et al. 2014). The assembly qualities were assessed with Quast v4.6.1 (Gurevich et al. 2013). Circoletto and Circos v0.69 (Krzywinski et al. 2009; Darzentas 2010) were used to visualize alignments between the C. parapsilosis CDC317 reference genome and the Canu C. parapsilosis 90-137/Cse4-HA assembly. There were some differences between the Canu and Flye assemblies, including some small deletions/insertions at the left IR of the original CEN1 location in the Canu assembly of C. parapsilosis 90-137/Cse4-HA. However, Chromosomes 1 and 5 are collinear in both.
Data access
The raw and processed ChIP-seq and minION data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA563885 with the Flye assembly at https://doi.org/10.6084/m9.figshare.12292850.v1.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by Science Foundation Ireland (12/IA/1343 to G.B. and 13/IA/1910 to K.H.W.; https://www.sfi.ie), the Wellcome Trust (102406/Z/13/Z and 109167/Z/15/Z; https://wellcome.ac.uk), and the European Research Council (789341, to K.H.W.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.257816.119.
Freely available online through the Genome Research Open Access option.
References
- Altschul S. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zarowiecki M, Burt A, Koufopanou V. 2008. Rapid evolution of yeast centromeres in the absence of drive. Genetics 178: 2161–2167. 10.1534/genetics.107.083980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Berman J. 2012. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res 20: 607–619. 10.1007/s10577-012-9296-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. 10.1038/nature08064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee G, Sankaranarayanan SR, Guin K, Thattikota Y, Padmanabhan S, Siddharthan R, Sanyal K. 2016. Repeat-associated fission yeast-like regional centromeres in the ascomycetous budding yeast Candida tropicalis. PLoS Genet 12: e1005839 10.1371/journal.pgen.1005839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan AY, Hanson SJ, Byrne KP, Wolfe KH. 2016. Centromeres of the yeast Komagataella phaffii (Pichia pastoris) have a simple inverted-repeat structure. Genome Biol Evol 8: 2482–2492. 10.1093/gbe/evw178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzentas N. 2010. Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26: 2620–2621. 10.1093/bioinformatics/btq484 [DOI] [PubMed] [Google Scholar]
- De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34: 2666–2669. 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan PD, Schröder MS, Higgins DG, Butler G. 2016. Identification of non-coding RNAs in the Candida parapsilosis species group. PLoS One 11: e0163235 10.1371/journal.pone.0163235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AP, Offei B, Braun-Galleani S, Coughlan AY, Martos AAR, Ortiz-Merino RA, Byrne KP, Wolfe KH. 2018. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: one species, four names. PLoS Pathog 14: e1007138 10.1371/journal.ppat.1007138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillon G, Carbone A, Fischer G. 2014. SynChro: a fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One 9: e92621 10.1371/journal.pone.0092621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, et al. 2004. Genome evolution in yeasts. Nature 430: 35–44. 10.1038/nature02579 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6: 99 10.1186/1471-2148-6-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, O'Gaora P, Byrne KP, Butler G. 2010. Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics 11: 290 10.1186/1471-2164-11-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97. 10.1126/science.1150944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P, Abbas A, Chasles M, Kudla B, Ogrydziak DM, Yaver D, Xuan JW, Peito A, Ribet AM, Feynerol C. 1993. Colocalization of centromeric and replicative functions on autonomously replicating sequences isolated from the yeast Yarrowia lipolytica. Proc Natl Acad Sci 90: 4912–4916. 10.1073/pnas.90.11.4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S, Freitag M. 2017. Centrochromatin of fungi. Prog Mol Subcell Biol 56: 85–109. 10.1007/978-3-319-58592-5_4 [DOI] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci 104: 14706–14711. 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH. 2011. Mechanisms of chromosome number evolution in yeast. PLoS Genet 7: e1002190 10.1371/journal.pgen.1002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. 2012. “Point” centromeres of Saccharomyces harbor single centromere-specific nucleosomes. Genetics 190: 1575–1577. 10.1534/genetics.111.137711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, et al. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10: e1004261 10.1371/journal.pgen.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Zhu L, Froyd C, Liu T, Rusche LN. 2015. Regional centromeres in the yeast Candida lusitaniae lack pericentromeric heterochromatin. Proc Natl Acad Sci 112: 12139–12144. 10.1073/pnas.1508749112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinathan S, Henikoff S. 2018. Non-B-form DNA is enriched at centromeres. Mol Biol Evol 35: 949–962. 10.1093/molbev/msy010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207. 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel C, Wang HSW, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J. 2009. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5: e1000400 10.1371/journal.pgen.1000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Yamaguchi E, Hamada K, Arisawa M. 1997. Structural analysis of a Candida glabrata centromere and its functional homology to the Saccharomyces cerevisiae centromere. Curr Genet 31: 122–127. 10.1007/s002940050185 [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Suzuki Y, Schoenfeld LW, Müller CA, Nieduszynski C, Wolfe KH, Tanaka TU. 2015. Discovery of an unconventional centromere in budding yeast redefines evolution of point centromeres. Curr Biol 25: 2026–2033. 10.1016/j.cub.2015.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37: 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Koren A, Tsai H-J, Tirosh I, Burrack LS, Barkai N, Berman J. 2010. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet 6: e1001068 10.1371/journal.pgen.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois P, Auerbach RK, Yellman CM, Roeder GS, Snyder M. 2013. Centromere-like regions in the budding yeast genome. PLoS Genet 9: e1003209 10.1371/journal.pgen.1003209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang S-P, Wang Z, Chinwalla AT, Minx P, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469: 529–533. 10.1038/nature09687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12: 733–735. 10.1038/nmeth.3444 [DOI] [PubMed] [Google Scholar]
- Lombardi L, Turner SA, Zhao F, Butler G. 2017. Gene editing in clinical isolates of Candida parapsilosis using CRISPR/Cas9. Sci Rep 7: 8051 10.1038/s41598-017-08500-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L, Oliveira-Pacheco J, Butler G. 2019a. Plasmid-based CRISPR-Cas9 gene editing in multiple Candida species. mSphere 4: e00125-19 10.1128/mSphere.00125-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L, Zoppo M, Rizzato C, Bottai D, Hernandez AG, Hoyer LL, Tavanti A. 2019b. Characterization of the Candida orthopsilosis agglutinin-like sequence (ALS) genes. PLoS One 14: e0215912 10.1371/journal.pone.0215912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DB, Logue ME, Butler G, Wolfe KH. 2010. Chromosomal G+C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol Evol 2: 572–583. 10.1093/gbe/evq042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire SL, ÓhÉigeartaigh SS, Byrne KP, Schröder MS, O'Gaora P, Wolfe KH, Butler G. 2013. Comparative genome analysis and gene finding in Candida species using CGOB. Mol Biol Evol 30: 1281–1291. 10.1093/molbev/mst042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. 2009. Major evolutionary transitions in centromere complexity. Cell 138: 1067–1082. 10.1016/j.cell.2009.08.036 [DOI] [PubMed] [Google Scholar]
- Mattei S, Sampaolese B, De Santis P, Savino M. 2002. Nucleosome organization on Kluyveromyces lactis centromeric DNAs. Biophys Chem 97: 173–187. 10.1016/S0301-4622(02)00066-2 [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Mitchell Smith M. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613. 10.1016/S0092-8674(00)81602-5 [DOI] [PubMed] [Google Scholar]
- Morales L, Noel B, Porcel B, Marcet-Houben M, Hullo M-F, Sacerdot C, Tekaia F, Leh-Louis V, Despons L, Khanna V, et al. 2013. Complete DNA sequence of Kuraishia capsulata illustrates novel genomic features among budding yeasts (Saccharomycotina). Genome Biol Evol 5: 2524–2539. 10.1093/gbe/evt201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesmelova IV, Hackett PB. 2010. DDE transposases: structural similarity and diversity. Adv Drug Deliv Rev 62: 1187–1195. 10.1016/j.addr.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama T, Suzuki T, Mori M, Osawa S, Ueda T, Watanabe K, Nakase T. 1993. Non-universal decoding of the leucine codon CUG in several Candida species. Nucleic Acids Res 21: 4039–4045. 10.1093/nar/21.17.4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Kitagawa K. 2011. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr Biol 21: 1695–1703. 10.1016/j.cub.2011.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Thakur J, Siddharthan R, Sanyal K. 2008. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci 105: 19797–19802. 10.1073/pnas.0809770105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Zorbas H, Price GB, Zannis-Hadjopoulos M. 1996. Inverted repeats, stem-loops, and cruciforms: Significance for initiation of DNA replication. J Cell Biochem 63: 1–22. [DOI] [PubMed] [Google Scholar]
- Pryszcz LP, Németh T, Gácser A, Gabaldón T. 2013. Unexpected genomic variability in clinical and environmental strains of the pathogenic yeast Candida parapsilosis. Genome Biol Evol 5: 2382–2392. 10.1093/gbe/evt185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz LP, Németh T, Saus E, Ksiezopolska E, Hegedűsová E, Nosek J, Wolfe KH, Gacser A, Gabaldón T. 2015. The genomic aftermath of hybridization in the opportunistic pathogen Candida metapsilosis. PLoS Genet 11: e1005626 10.1371/journal.pgen.1005626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purgato S, Belloni E, Piras FM, Zoli M, Badiale C, Cerutti F, Mazzagatti A, Perini G, Della Valle G, Nergadze SG, et al. 2015. Centromere sliding on a mammalian chromosome. Chromosoma 124: 277–287. 10.1007/s00412-014-0493-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccombeni A, Vidanes G, Proux-Wéra E, Wolfe KH, Butler G. 2012. Sequence and analysis of the genome of the pathogenic yeast Candida orthopsilosis. PLoS One 7: e35750 10.1371/journal.pone.0035750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R. 2012. Centromere repositioning in mammals. Heredity (Edinb) 108: 59–67. 10.1038/hdy.2011.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Sanyal K. 2011. Diversity in requirement of genetic and epigenetic factors for centromere function in fungi. Eukaryot Cell 10: 1384–1395. 10.1128/EC.05165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Baum M, Carbon J. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Nat Acad Sci 101: 11374–11379. 10.1073/pnas.0404318101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder MS, Martinez de San Vicente K, Prandini THR, Hammel S, Higgins DG, Bagagli E, Wolfe KH, Butler G. 2016. Multiple origins of the pathogenic yeast Candida orthopsilosis by separate hybridizations between two parental species. PLoS Genet 12: e1006404 10.1371/journal.pgen.1006404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I. 2018. What is behind “centromere repositioning”? Chromosoma 127: 229–234. 10.1007/s00412-018-0672-y [DOI] [PubMed] [Google Scholar]
- Schulman I, Bloom KS. 1991. Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu Rev Cell Biol 7: 311–336. 10.1146/annurev.cb.07.110191.001523 [DOI] [PubMed] [Google Scholar]
- Shen XX, Zhou X, Kominek J, Kurtzman CP, Hittinger CT, Rokas A. 2016. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 6: 3927–3939. 10.1534/g3.116.034744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar L, Jaitly P, Chen Y, Thimmappa BC, Sanyal A, Sanyal K. 2019. Cis- and trans-chromosomal interactions define pericentric boundaries in the absence of conventional heterochromatin. Genetics 212: 1121–1132. 10.1534/genetics.119.302179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone MF, Bigoni F, Archidiacono N. 2008. Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res 16: 17–39. 10.1007/s10577-007-1209-z [DOI] [PubMed] [Google Scholar]
- Sun S, Yadav V, Billmyre RB, Cuomo CA, Nowrousian M, Wang L, Souciet J-L, Boekhout T, Porcel B, Wincker P, et al. 2017. Fungal genome and mating system transitions facilitated by chromosomal translocations involving intercentromeric recombination. PLoS Biol 15: e2002527 10.1371/journal.pbio.2002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanti A, Davidson AD, Gow NAR, Maiden MCJ, Odds FC. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J Clin Microbiol 43: 284–292. 10.1128/JCM.43.1.284-292.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Sanyal K. 2013. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res 23: 638–652. 10.1101/gr.141614.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SA, Ma Q, Ola M, Martinez de San Vicente K, Butler G. 2018. Dal81 regulates expression of arginine metabolism genes in Candida parapsilosis. mSphere 3: e00028-18 10.1128/mSphere.00028-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakirlis N, Sarilar V, Drillon G, Fleiss A, Agier N, Meyniel J-P, Blanpain L, Carbone A, Devillers H, Dubois K, et al. 2016. Reconstruction of ancestral chromosome architecture and gene repertoire reveals principles of genome evolution in a model yeast genus. Genome Res 26: 918–932. 10.1101/gr.204420.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326: 865–867. 10.1126/science.1178158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane T, Ogawa T, Matsuoka M. 2008. Derivation of consensus sequence for protein binding site in Yarrowia lipolytica centromere. J Biosci Bioeng 105: 671–674. 10.1263/jbb.105.671 [DOI] [PubMed] [Google Scholar]
- Zhai B, Ola M, Rolling T, Tosini NL, Joshowitz S, Littmann ER, Amoretti LA, Fontana E, Wright RJ, Miranda E, et al. 2020. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med 26: 59–64. 10.1038/s41591-019-0709-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.