Abstract
Runt-related transcription factor 2 (Runx2) is an essential transcription factor for osteoblast differentiation, and is activated by mechanical stress to promote osteoblast function. Cleidocranial dysplasia (CCD) is caused by mutations of RUNX2, and CCD patients exhibit malocclusion and often need orthodontic treatment. However, treatment is difficult because of impaired tooth movement, the reason of which has not been clarified. We examined the amount of experimental tooth movement in Runx2+/− mice, the animal model of CCD, and investigated bone formation on the tension side of experimental tooth movement in vivo. Continuous stretch was conducted to bone marrow stromal cells (BMSCs) as an in vitro model of the tension side of tooth movement. Compared to wild-type littermates the Runx2+/− mice exhibited delayed experimental tooth movement, and osteoid formation and osteocalcin (OSC) mRNA expression were impaired in osteoblasts on the tension side of tooth movement. Runx2 heterozygous deficiency delayed stretch-induced increase of DNA content in BMSCs, and also delayed and reduced stretch-induced alkaline phosphatase (ALP) activity, OSC mRNA expression, and calcium content of BMSCs in osteogenic medium. Furthermore Runx2+/− mice exhibited delayed and suppressed expression of mammalian target of rapamycin (mTOR) and rapamycin-insensitive companion of mTOR (Rictor), essential factors of mTORC2, which is regulated by Runx2 to phosphorylate Akt to regulate cell proliferation and differentiation, in osteoblasts on the tension side of tooth movement in vivo and in vitro. Loss of half Runx2 gene dosage inhibited stretch-induced PI3K dependent mTORC2/Akt activity to promote BMSCs proliferation. Furthermore, Runx2+/− BMSCs in osteogenic medium exhibited delayed and suppressed stretch-induced expression of mTOR and Rictor. mTORC2 regulated stretch-elevated Runx2 and ALP mRNA expression in BMSCs in osteogenic medium. We conclude that Runx2+/− mice present a useful model of CCD patients for elucidation of the molecular mechanisms in bone remodeling during tooth movement, and that Runx2 plays a role in stretch-induced proliferation and osteogenesis in BMSCs via mTORC2 activation.
Keywords: Runx2+/− mice, BMSCs, Osteoblast, Mechanical stress, mTORC2, Experimental tooth movement
Highlights
-
•
Experimental tooth movement is delayed in Runx2+/− mice compared with wild-type mice.
-
•
Runx2 plays a role in stretch-induced proliferation and differentiation of BMSCs via mTORC2 activation.
-
•
Runx2+/− mice are useful model to clarify the mechanical stress-induced bone remodeling in CCD patients.
1. Introduction
Runx2 (Runt-related transcription factor 2 earlier called as Core binding factor α-1 (Cbfa1) is an essential transcription factor for osteoblast differentiation from mesenchymal stem cells (MSCs) and bone formation (Otto et al., 1997; Ducy et al., 1997; Komori et al., 1997). Mice with a homozygous mutation in Runx2 show arrested embryonic tooth development, complete absence of both intramembranous and endochondral ossification through a lack of osteoblast differentiation, and are embryonic lethal (Otto et al., 1997; Ducy et al., 1997; Komori et al., 1997; Mundlos, 1999; Aberg et al., 2004). Runx2 is activated by fluid shear stress in mouse cortical bone in vivo, and mechanical stresses such as stretching and fluid shear stress were reported to enhance Runx2 expression in osteoblastic lineage cells and promote its osteoblast differentiation in vitro (Liu et al., 2011a; Ziros et al., 2002; Kanno et al., 2007; Li et al., 2012). These findings suggest that Runx2 regulates mechanotransduction in osteoblastic cells for bone formation. However, underlying mechanism in biological function of Runx2 in mechanical stress-induced bone formation has not been fully clarified.
Runx2 heterozygous (Runx2+/−) mice show clavicular hypoplasia and delayed closer fontanelles, as regarded an animal model of an autosomal-dominant disorder of Cleidocranial dysplasia (CCD) caused by mutations of Runx2 in humans (Otto et al., 1997; Komori et al., 1997; Mundlos, 1999; Salingcarnboriboon et al., 2006; Tsuji et al., 2004). Orthodontic treatment is often necessary for CCD patients to recover masticatory function and esthetics because of the dental phenotypes such as delayed eruption of permanent teeth, multiple supernumerary teeth and malocclusion (Mundlos, 1999). The orthodontic treatment is difficult because of impaired tooth movement in CCD patients (Becker et al., 1997a; Becker et al., 1997b).
Orthodontic force acts as mechanical stress to influence the periodontal tissues such as periodontal ligament (PDL), alveolar bone, and gingiva, which support the tooth root and comprise cementum (Davidovitch, 1991). The PDL is a multifunctional fibrous tissue that connects the cementum covering the tooth root and the alveolar bone, contains a variety of cell populations including fibroblasts, osteoblasts, osteoclasts, endothelial cells, and MSCs, and senses orthodontic force (Davidovitch, 1991; Pavlin and Gluhak-Heinrich, 2001; Lekic and McCulloch, 1996; Beertsen et al., 1997). When force is loaded onto a tooth, osteoclastic activity is promoted on the pressure side of the tooth, and alveolar bone becomes selectively resorbed by osteoclasts, while bone formation is enhanced on the tension side by osteoblasts after proliferation and differentiation of PDL fibroblast and MSCs. As a result the tooth moves in the specified direction and a balance of bone apposition and resorption maintains the width of the PDL (Pavlin and Gluhak-Heinrich, 2001; Lekic and McCulloch, 1996; Takano-Yamamoto et al., 1994; Terai et al., 1999; Takimoto et al., 2015). It is likely that mutations of RUNX2 are associated with impaired orthodontic loading-induced bone remodeling during tooth movement in CCD patients. Therefore, it is hypothesized that mechanical loading-induced bone remodeling might be impaired in Runx2+/− mice.
Mammalian target of rapamycin (mTOR) is a catalytic subunit in mammals of two distinct complexes, namely mTOR complex 1 (mTORC1) and mTORC2 (Bhaskar and Hay, 2007). The defining subunits of mTORC1 and mTORC2 are regulatory-associated protein of mTOR (Raptor) and rapamycin-insensitive companion of mTOR (Rictor), respectively (Bhaskar and Hay, 2007). mTORC2 phosphorylates and activates Akt at serine 473, which regulates cell cycle progression, differentiation, apoptosis, and cell migration, and mTORC2 signaling is considered a key role in those biological process (Bhaskar and Hay, 2007; Zoncu et al., 2011). It has been reported that Rictor deficient mice exhibited impaired bone formation and showed reduced mechanical stress-induced bone formation in vivo (Chen et al., 2015). mTORC2 activation was induced by mechanical stretch in osteoblast lineage cells in vitro (Sen et al., 2014). mTOR expression is induced by recruitment of Runx2 to its promoter and mTORC2 signal is promoted (Tandon et al., 2014). Therefore, in the present study, we highlight mTORC2 signal for investigation of orthodontic force-induced bone formation in Runx2+/− mice, and hypothesized that Runx2 is associated with mTORC2 in mechanical loading-induced biological cellular response for bone formation, especially proliferation and osteoblast differentiation of bone marrow stromal cells (BMSCs).
In the present study, we investigated Runx2 function in mechanical stretch-induced bone remodeling by loading orthodontic force on teeth in Runx2+/− mice, an animal model of CCD. We examined proliferation and osteoblast differentiation in Runx2+/− mice on tension side of experimental tooth movement, and in stretched BMSCs derived from Runx2+/− mice. Finally, we examined mTORC2 activation in mechanical stretch-induced proliferation and osteoblast differentiation of BMSCs in Runx2+/− mice.
2. Materials and methods
2.1. Mice
Runx2+/− mice in NMRI background were a gift from Michael Owen (Imperial Cancer Research Fund, London, UK) (Aberg et al., 2004; Takano-Yamamoto et al., 1994). Mice were housed 5–6 animals per cage at the Facility for care and management with a 12-h/12-h light/dark cycle, and maintained by the animal technicians according to the guidelines of the Regulations for Animal Experiments and Related Activities of Okayama University and Tohoku University. Mice were allowed unlimited free range of food (Labo MR Stock, Nosan Corporation Life-Tech Department, Yokohama, Japan) and water. All experiments were approved by the Animal Committee of Okayama University and Tohoku University.
2.2. Reagents
We obtained SYBR premix Ex Taq from Takara Bio. Inc. (Shiga, Japan). Antibodies for cyclinD, p21, p27, and βactin for western blotting and mTOR, Rictor for immunohistochemistry were purchased from Santa Cruz Biotechnology Inc. (CA, USA). Antibodies for mTOR, Rictor, pAkt, Akt were purchased from Cell Signaling Technology (Beverly MA, USA). Fibronectin and p-nitrophenol phosphate (pNPP) were obtained from Sigma-Aldrich (St. Louis, MO, USA). We purchased mTOR inhibitor (KU63794, a specific inhibitor against phosphorylation on Ser2448/2481 of mTOR resulted in mTORC1/2 and its downstream, such as Akt (García-Martínez et al., 2009; Malagu et al., 2009)) from EMD Millipore (Darmstadt, German), PI3K inhibitor (LY294002, completely and specifically abolished PI3-kinase activity, and its one of downstream, Akt (Vlahos et al., 1994)) from Sigma-Aldrich, and Akt inhibitor (MK-2206 dihydrochloride, MK-2206 inhibits Akt1, 2 and 3 kinase activity and also inhibited auto-phosphorylation of both Akt T308 and S473 (Yan, 2009)) from Selleck (Houston, USA).
2.3. Experimental tooth movement
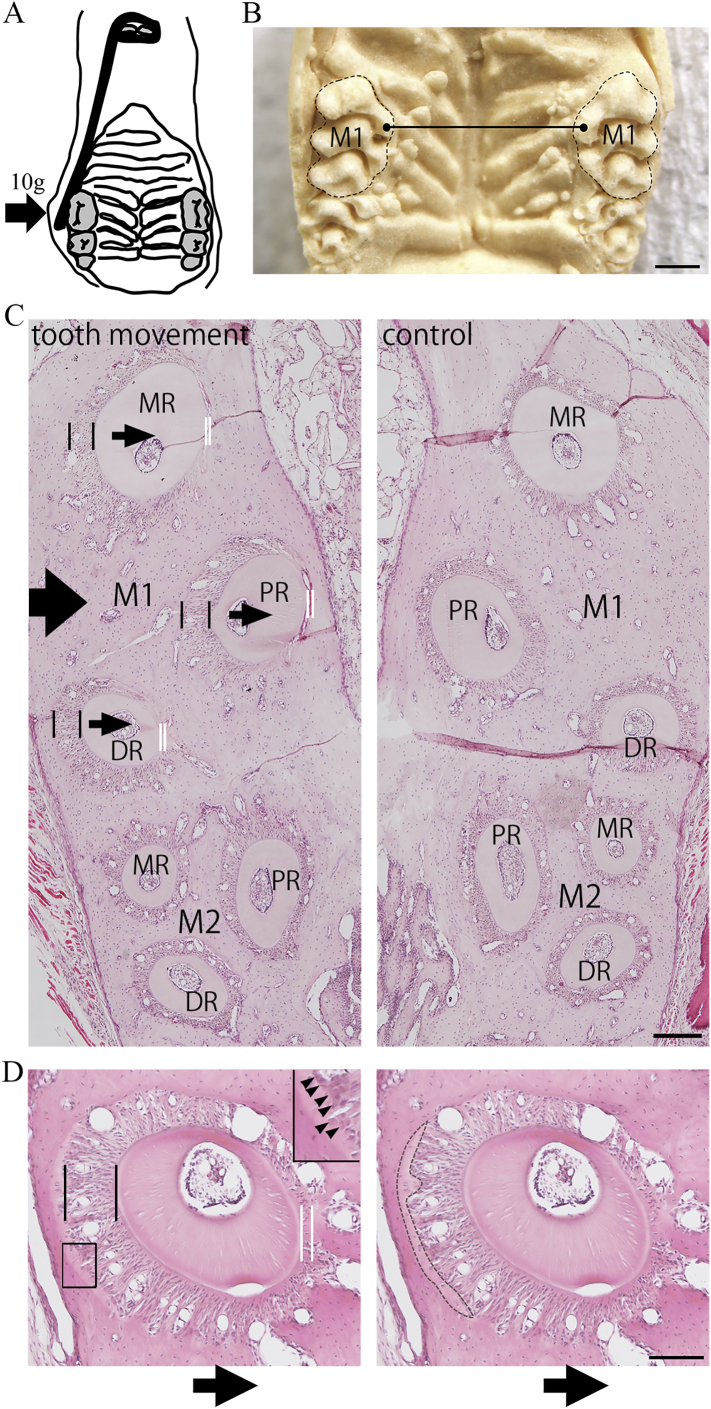
For measurement the amount of experimental tooth movement, three each of 6-week-old male wild-type and Runx2+/− mice were used. Orthodontic force was applied according to the method of Sakai et al. (2009). Briefly, nickel-titanium (Ni—Ti) wire, 0.012 in. in a diameter, was fixed to the maxillary incisor using composite resin for dental filling, and then the right maxillary first molar was moved toward the palatal side by the Ni—Ti wire at a continuous 10-g load for 21 days (Fig. 1 A). For measurement of the amount of tooth movement, maxillary impressions were taken with a silicone impression material under anesthesia at 0, 3, 7, 10, 14, and 21 days after the initiation of experimental tooth movement. For making plaster model of maxillay alveolar bone including the both sides of 1st molars, impressions were filled with dental plaster (New Fujirock GC Corp.). The distance between the tips of mesial palatal cusps of right and left maxillary 1st molars was measured using digital calipers (Fig. 1B). For each mouse, the measurement was taken four times, and then value was used for the amount of tooth movement of each mouse.
Fig. 1.
(A) Schematic drawing of experimental tooth movement in mice. Ni—Ti wire was fixed to the maxillary incisors with a composite resin for dental filling, and to load a continuous 10-g force (arrow) for movement of the right maxillary 1st molar toward the palatal side. (B) A representative image of plaster model of maxilla of mice for measurement of the amount of experimental tooth movement. The distance between the tips of mesial palatal cusps (Black dots) of right and left maxillary 1st molars was measured (Black line). Both sides of maxillary first molars were surrounded by dotted line. M1, maxillary 1st molar. Scale Bar, 500 μm. (C) Representative images of horizontal sections of maxillary alveolar bone during experimental tooth movement. Left panel represented right side of alveolar bone (tooth movement side), and right panel represented left side of alveolar bone (control side). Arrow indicated the direction of the force applied. Small narrow arrows indicated the direction of the tooth movement. Spaces between black lines indicated periodontal space of tension side, and the spaces between white lines represented that of compression sides. M1, maxillary 1st molar; M2, maxillary 2nd molar; MR, mesial root; PR, palatal root, DR, distal root. Scale Bar, 200 μm. (D) Representative images for identification of osteoid area. The distal root of the 1st molar during experimental tooth movement were shown. Arrow indicated the direction of the force applied. Spaces between black lines indicated periodontal space of tension side, and the spaces between white lines represented that of compression sides. Areas, identified as uniformly pink area staining with HE and covered with tall and cuboidal osteoblasts in tension side, were surrounded by dotted line as newly formed osteoid. Rectangle in left panel indicated the area enlarged in the inset. Arrowheads indicated the tall and cuboidal osteoblasts. Scale Bar, 100 μm.
2.4. Tissue preparation and histology
For histological examination, 15 of Runx2+/− and 15 of wild-type 6-week-old male mice were used. Samples of the maxillary alveolar bone region were prepared from wild-type and Runx2+/− mice at 0, 1, 3, 5, and 7 days after the initiation of experimental tooth movement. Under anesthesia, the animals were perfused with 4% paraformaldehyde (pH 7.4) and the maxillary bones and surrounding tissues were dissected and fixed in the above solution for approximately 12 h at 4 °C. The specimens, decalcified in 20% ethylenediaminetetraacetate (pH 7.4) for 14 days at 4 °C, were dehydrated and embedded in paraffin. The tissue blocks were cut into 7 μm thick of sections parallel to the occlusal plane, and the level of the sections from the furcation of the teeth were calculated by the counted number of the serially cut sections. In this study, the sections of between 175 and 245 μm from the furcation of the teeth were used. The representative image of these sections was shown in Fig. 1C. In these sections, the buccal sides of the mesial, palatal and distal roots of 1st molar were regarded as tension side with expansion of periodontal space and elongation of periodontal ligament fibers in the experimental tooth movement. The palatal sides of those were regarded as compression side with narrow periodontal space. The periodontal ligament fibers of the buccal side were tensioned and its direction was almost parallel to that of tooth movement in those areas (Fig. 1C arrowheads). In contrast, the periodontal space and periodontal ligament fibers of 2nd molar in tooth movement and molars in control had little change and a normal appearance (Fig. 1C).
The sections were stained with hematoxylin and eosin (HE) for histological examination (Meyer, 1956). New osteoid areas having wider periodontal spaces were identified as uniformly pink area staining with HE and covered with tall and cuboidal osteoblasts (Meyer, 1956; Pavlin et al., 2000). The osteoid areas in tension side of distal root of right maxillary 1st molars were measured in images obtained from bright-field microscope at 10× magnification of objective lens using software (Winroof; Mitani Corporation, Fukui, Japan) (Fig. 1D).
2.5. In situ hybridization
Digoxigenin (DIG)-11-UTP-labeled single-stranded RNA probes were prepared in a DIG RNA Labeling kit (Roche Diagnostics, Mannheirrir Germany) according to the manufacturer's instructions. A 0.95-kb fragment of mouse OSC was used to generate sense and anti-sense probes (Desbois et al., 1994). In situ hybridization was performed as described previously (Takano-Yamamoto et al., 1994; Terai et al., 1999). Controls were hybridized with sense probes.
2.6. Immunohistochemistry
Serial sections were incubated with 3% H2O2 at room temperature for 15 min after deparaffinization. Then, sections were incubated over night at 4 °C with antibodies for mTOR (1:50) and Rictor (1:100) in Can Get Signal Solution B (Toyobo, Osaka, Japan). As negative control, rabbit IgG (Sigma) was substituted for primary antibody. Then, sections were incubated with Histofine® Simple Stain Mouse MAX-PO (Nichirei Bioscience, Tokyo, Japan) for 30 min at room temperature. For visualization of the reaction products, 3, 3′-diaminobenzidine tetrachloride (DAB: Nichirei Bioscience) was used as substrate. We used wild-type mice on day 3 after the initiation of tooth movement as negative controls, rabbit IgG (Sigma, St. Louis, MO, USA) was substituted for the primary antibodies.
2.7. Cell culture
After euthanasia, femurs and tibiae of 6–9 week-old wild-type and Runx2+/− mice were carefully cleaned from adherent soft tissue and bone marrow cells were harvested. Collected cells were seeded at a density of 4 × 107 cells per 3.5 cm tissue culture dish (BD Falcon) and cultured in growth medium: Dulbecco's modified Eagle's medium-low glucose (DMEM; Sigma) containing 10% heat-inactivated fetal bovine serum (FBS; Nichirei) and penicillin/streptomycin (100 IU/ml and 100 μg/ml; Sigma), at 37 °C in 5% CO2 atmosphere. After 4 days of culture, nonadherent cells were removed and adherent cells were cultured 3 more days until 90% confluence to use as BMSCs in this study. For promoting osteogenesis, BMSCs were cultured in osteogenic medium: growth medium supplemented with 10 nM dexamethasone (Sigma), 82 μg/ml l-ascorbic acid (Wako) and 10 mM β-glycerophosphate (Sigma), at 37 °C in 5% CO2 atmosphere. Osteogenic medium was changed every three days.
2.8. Application of mechanical stretch on cells
BMSCs were seeded on 10 cm2 polydimethylsiloxane or 4-well-chambers (Strex Inc., Osaka, Japan) coated with 0.05 mg/ml fibronectin (Sigma) and cultured for 12 h at a density of 1 × 105 cells/cm2. After the cells had reached subconfluency, they were stretched by using a specially designed device that produced a 12% uni-axial increase in width of chamber. The stretch value was decided from the data of stretch-induced proliferation of BMSCs (Suppl. Fig. 1). In case of using inhibitor, cells were stretched after incubation with inhibitor for 1 h. Unstretched cells were incubated in the same conditions and used as controls.
2.9. Cell proliferation analysis
DNA content was measured based on previous report (Zhang et al., 2010). Briefly, BMSCs were washed with saline twice, collected with 625 mM Tris-HCl buffer (PH 9.0) containing 0.075% Triton X-100 (Wako Pure Chemical Industries, Ltd., Osaka, Japan), and sonicated on ice for 5 s. After centrifugation, DNA content was measured using Quant-iT PicoGreen (Invitrogen). Fluorescence measurement was carried out at 485/535 nm using infinite F200 (Tecan, Männedorf, Swiss) according to the manufacturer's instructions. DNA content was measured in amount per each well in 4-well-chamber. We also evaluated cell proliferation of BMSCs using Cell Counting Kit-8 reagent (Dojindo Laboratories, Kumamoto, Japan). Briefly, the reagents were added into the medium, followed by further incubation for 2 h. The optical density (OD) values were detected at 450 nm using microplate reader (Remote Sunrise; Tecan, Japan).
2.10. Alkaline phosphatase (ALP) activity and calcium content evaluation
ALP activity was evaluated based on previous report (Zhang et al., 2010). BMSCs were treated as described in “2.9. cell proliferation analysis” section. The supernatants were used for DNA content measurement and ALP activity evaluation. The cell pellet was used for measurement of calcium content. For measurement of ALP activity, 10-fold diluted supernatants with 0.5 mM MgCl2 (Sigma) and 0.5 mM p-nitrophenol phosphate (pNPP) were incubated at 37 °C for 30 min, stopping reaction by 0.1 N NaOH (Wako) and lysates were measured at 405 nm using Infinite F200. pNPP was normalized to the DNA concentration. For calcium content measurement, the cell pellet was washed twice with PBS, incubated at 37 °C for 16 h with 0.5 M HCl (Wako) and centrifuged at 13,000 ×g for 15 min. The supernatants were used for measuring calcium content by QuantiChrom Calcium Assay Kit (BioAssay Systems, Hayward, CA). The solution was read at 572 nm and calcium content was also normalized to the DNA content.
2.11. ALP and alizarin staining
BMSCs, stretched for 7 days, were washed with PBS twice and fixed with methanol for 10 min, and were applied substrate at 37 °C for 10 min. After fixation, the cell layers were stained with the reagents from ALP stain kit (Wako) according to the manufacturer's instructions. For alizarin staining, cells stretched for 21 days were washed with PBS twice, fixed with methanol for 10 min, and were stained with alizarin solution (Sigma) at 37 °C for 5 min.
2.12. Western blotting
Whole cell lysates were prepared with modified radio immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 1% Na-deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM NaF, pH 7.5) containing protease inhibitor cocktail (Sigma aldrich, St, Louis, MO). Protein (30 μg) was loaded to Tris-Glycine SDS-PAGE gel (Bio-Rad Laboratories Inc., CA, USA) and transferred to nitrocellurose or polyvinylidene difluoride membrane using Trans-Blot® Turbo™ Transfer System (Bio-Rad Laboratories Inc.). The membranes were incubated with primary antibody (1:1000) at 4 °C overnight, followed by incubation with secondary antibody conjugated with horseradish peroxidase (1:5000). Proteins were detected with SuperSignal West Femto Chemiluminescent Substrate (Pierce Chemical Co, CA, USA). The images were acquired using VersaDoc5000MP (Bio-Rad Laboratories Inc.).
2.13. RNA interference
For knockdown of the expression of Rictor, we employed an RNA interference method (Sen et al., 2014). siRictor against Rictor and its control siRNAs were purchased from Invitrogen (Carlsbad, CA). Cells were transfected with specific siRNA or control siRNA (10 nM) using PepMute Plus (SignaGen Lab, Rockville, MD) at 18 h after plating cells. Mechanical stretch was applied at 24 h after transfection. The siRNA sequences for Rictor were 5′-UCAUCUUUCUGACUAAGCGAAGGGC and for the control (nucleotide change within same sequence) were 5′-GCCCUCGUUGACUGAAAGAAUCUGA (Sen et al., 2014).
2.14. Real-time RT-PCR
The total RNA was extracted from BMSCs using TRIzol reagent® (Invitrogen) according to the manufacturer's instructions. Total RNA (1 μg) was reverse transcribed to generate complementary DNA (cDNA). Real-time PCR was performed using Thermal Cycler Dice Real Time System (Takara Bio. Inc., Shiga, Japan) with SYBR premix Ex Taq (Takara). Amplification reactions were conducted with thermal denaturation 95 °C for 10 min followed by 40 cycles at annealing 95 °C for 15 s and extension reaction at 60 °C for 60 s using the primer sets (Suppl. Table 1). Each sample was subjected to triplicate PCR reactions and the threshold cycle value of samples was normalized by expression of GAPDH as an endogenous housekeeping gene.
2.15. Statistical evaluations
The results were presented as means ± SD. Statistical analysis was performed using two-way ANOVA in vivo and two-way ANOVA in vitro followed by Sheffe's test. P values 0.05 were considered significant. All experiments were performed at least two times.
3. Results
3.1. Small amount of experimental tooth movement in Runx2+/− mice
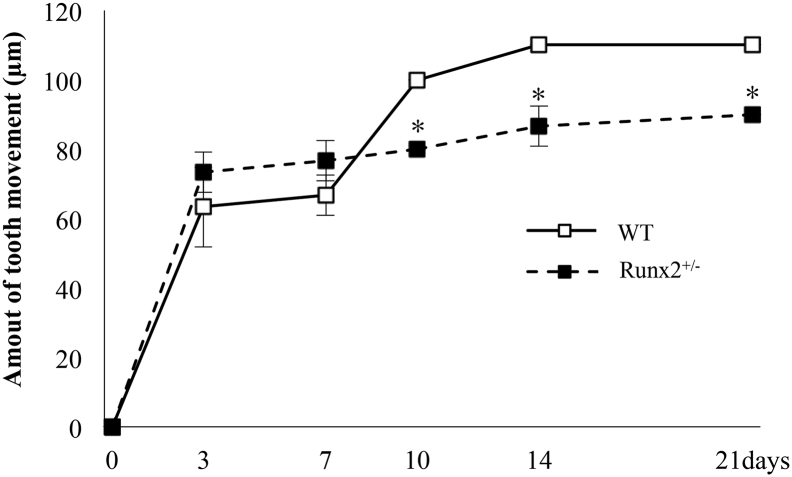
We examined the time course changes in the amount of experimental tooth movement in wild-type and Runx2+/− mice (Fig. 1C). No differences in daily behavior were seen in all mice during experiment period. In wild-type mice, the amount of tooth movement was 63.3 ± 11.5 μm at 3 days after the initiation of tooth movement and significantly increased from days 0 to 3, but not from days 3 to 7 (66.6 ± 5.7 μm). The distance significantly increased from days 7 to 10, but showed no significant difference between days 10 and 21. In Runx2+/− mice, the amount of tooth movement was 73.3 ± 5.7 μm on day 3, with no remarkable increases thereafter (Fig. 2).
Fig. 2.
Time course changes of amount of experimental tooth movement in wild-type and Runx2+/− mice. Time course changes of tooth movement at 3, 7, 10, 14, and 21 days after the initiation of tooth movement. Values are averages ± SD (n = 3). * significantly different from wild-type mice at corresponding time point.
There were no significant differences in the amount of tooth movement between wild-type and Runx2+/− mice until 7 days after the wire attachment. However, the distances in Runx2+/− mice on days 10, 14, and 21 were decreased compared with those in wild-type littermates (wild-type mice; 10 days, 100 ± 0 μm, 14 days, 110 ± 0 μm, 21 days, 110 ± 0 μm, Runx2+/− mice; 10 days, 80 ± 0 μm, 14 days, 86.6 ± 5.7 μm, 21 days, 90 ± 0 μm).
3.2. Osteoid area and OSC mRNA expression in osteoblasts on the tension side of tooth movement
To assess bone formation on the tension side of delayed experimental tooth movement in Runx2+/− mice, we evaluated the time course changes in osteoid area and osteogenesis in osteoblasts along the alveolar bone on the tension side of the upper first molar in mice.
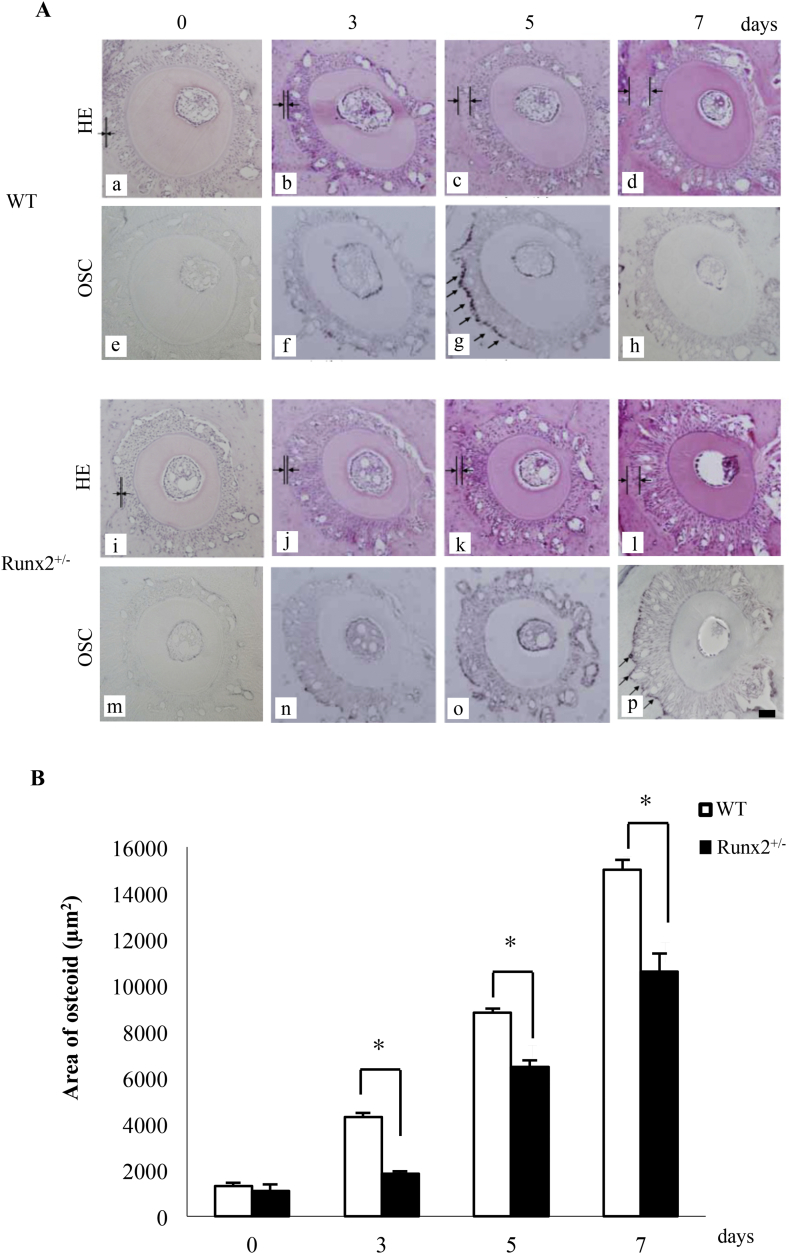
Using histological sections stained with HE stain, we found thin osteoid along the alveolar bone surface on the tension side of tooth movement in wild-type and Runx2+/− mice on day 3 after the initiation of tooth movement. The osteoid layers in both wild-type and Runx2+/− mice became thicker from days 5 to 7 (Fig. 3A a-d, and i-l). On day 3, both wild-type and Runx2+/− mice showed a significantly increased osteoid area (wild-type mice; 4187 ± 269 μm2, Runx2+/− mice; 1823 ± 95 μm2) compared with day 0 (wild-type mice; 1204 ± 239 μm2, Runx2+/− mice; 926 ± 393 μm2), with further osteoid deposition thereafter (wild-type mice; 5 days, 8663 ± 368 μm2, 7 days, 14,871 ± 457 μm2, Runx2+/− mice; 5 days, 6357 ± 323 μm2, 7 days, 10,367 ± 795 μm2). The osteoid areas did not alter between the two genotypes on day 0, but Runx2+/− mice exhibited a significantly smaller osteoid area than wild-type littermates on days 3, 5, and 7 (Fig. 3B).
Fig. 3.
Time course changes of osteoid area and OSC mRNA expression pattern in osteoblasts on tension side of alveolar bone in wild-type and Runx2+/− mice. (A) Time course changes of the osteoid area (a-d, and i-l) and OSC mRNA expression (e-h, and m-p) in osteoblasts on tension side of upper first molar in wild-type (a-h) and Runx2+/− mice (i-p). HE staining and in situ hybridization for OSC mRNA were performed on 0 (a, e, i, and m), 3 (b, f, j, and n), 5 (c, g, k, and o), and 7 (d, h, l, and p) days after the initiation of tooth movement. Arrows indicated osteoid layers in HE staining and arrows indicated OSC mRNA expression in osteoblasts in in situ hybridization. Scale bar, 50 μm. (B) Time course changes of the osteoid area on tension side of tooth movement on 0, 3, 5, and 7 days after the initiation of tooth movement. Values are averages ± SD (n = 3). * significantly different from wild-type mice at corresponding time point.
As the impaired osteoid formation on the tension side of experimental tooth movement in Runx2+/− mice, we examined the time course changes in OSC mRNA expression, which promoter regions have the Cbfa1 (Runx2)-binding motifs/osteoblast-specific cis-acting elements, Runx2 bind to the promoter to induce its expression as late osteoblast differentiation marker, in osteoblasts (Ducy et al., 1997). Wild-type mice revealed the strongest OSC mRNA expression on day 5 of experimental tooth movement and then the expression became weaker on day 7 (Fig. 3A-e-h). Runx2+/− mice showed the maximum OSC mRNA expression on day 7 (Fig. 3A-m-p), which was lower than the strongest expression in wild-type mice (Fig. 3A-g, and p).
3.3. Effect of stretch on proliferation of BMSCs from wild-type and Runx2+/− mice
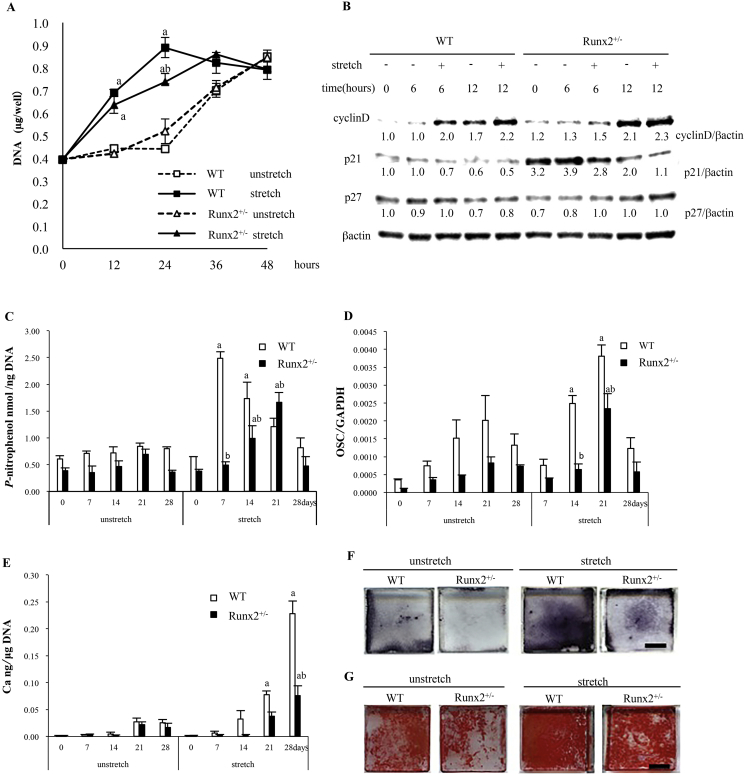
On the tension side of tooth movement, recruited MSCs in PDLs proliferate rapidly on the surfaces of alveolar bone, and initiate differentiation for osteoblast (Pavlin and Gluhak-Heinrich, 2001). Mineralization of the extracellular matrix then occurs and bone formation is promoted (Pavlin and Gluhak-Heinrich, 2001). To elucidate the mechanism for the reduced bone formation on the tension side of tooth movement in Runx2+/− mice, we stretched mouse BMSCs and examined stretch-induced osteoblast function in vitro. We examined the DNA content in wild-type and Runx2+/− BMSCs after stretching to evaluate whether Runx2 is associated with proliferation of osteoblast lineage cells. The DNA content in unstretched (control) wild-type and Runx2+/− BMSCs increased and reached a peak at 48 h after the initiation of mechanical loading (Fig. 4A). There were no significant differences in DNA content between unloaded wild-type and Runx2+/− BMSCs from 0 to 48 h (Fig. 4A). In wild-type BMSCs, mechanical stimulation significantly increased the DNA content at 12 and 24 h of stretch, with a peak at 24 h (Fig. 4A). Meanwhile, the DNA content from Runx2+/− BMSCs was significantly increased by stretching at 12 and 24 h, and a peak was observed at 36 h (Fig. 4A). Runx2+/− BMSCs exhibited a significantly reduced stretch-induced increase in DNA content compared with wild-type BMSCs at 24 h, while stretched wild-type and Runx2+/− BMSCs did not differ at 12, 36, and 48 h (Fig. 4A). In addition, we further quantitatively evaluated the cell proliferation of BMSCs using CCK-8. Twenty-four hours after the initiation of mechanical loading, mechanical stretch increased the proliferation of both wild-type and Runx2+/− BMSCs, and more importantly stretched Runx2+/− BMSCs significantly proliferated compared with wild-type BMSCs (Suppl. Fig. 2).
Fig. 4.
Stretch-induced proliferation and osteogenesis in wild-type and Runx2+/− BMSCs. (A) Change of DNA content of wild-type and Runx2+/− BMSCs at 0, 12, 24, 36, and 48 h after the initiation of mechanical stretch. (B) Expression of cell cycle regulation factors protein in BMSCs at 0, 6, and 12 h after the initiation of mechanical stretch. ALP activity (C), OSC mRNA (D) and calcium content (E) in BMSCs in osteogenic media were examined on day 0, 7, 14, 21, and 28. ALP staining on day 7 (F) and Alizarin red staining on day 21 (G) after the initiation of mechanical stretch were investigated. Values are averages ± SD (n = 3). a, significantly different between corresponding unstretched and stretched wild-type and Runx2+/− BMSCs. b, significantly different between stretched wild-type and Runx2+/− BMSCs. Scale bar, 1 cm. A quantification of normalized relative protein levels against respective βactin and corresponding unloaded wild-type mice cells is given below lane.
Cyclins and cyclin-dependent kinases (cdks) are integrators of growth factor-mediated signals that drive the cell cycle (Baldin et al., 1993; Lim and Kaldis, 2013). Growth factors promote the G1 phase of the cell cycle and D-type cyclins act primarily as growth factor sensors (Baldin et al., 1993; Lim and Kaldis, 2013). p21 and p27 are tight-binding inhibitors of cyclin D-cdk4 complex and inhibit G1 progression (Harper et al., 1993; Polyak et al., 1994). We evaluated the expression of cell cycle regulatory proteins in wild-type and Runx2+/− BMSCs at 6 and 12 h after the initiation of stretch to examine the association of cell cycle progression with stretch. Cyclin D was enhanced in unstretched wild-type and Runx2+/− BMSCs at 12 h compared with 0 h (Fig. 4B). Stretching remarkably induced cyclin D in wild-type BMSCs at 6 h, and in Runx2+/− BMSCs at 12 h (Fig. 4B). p21 protein expression in unstretched wild-type and Runx2+/− BMSCs did not change from 0 to 6 h, but was reduced from 6 to 12 h, although Runx2+/− BMSCs showed remarkably higher p21 expression from 0 to 12 h than wild-type BMSCs regardless of mechanical loading (Fig. 4B). In wild-type BMSCs, stretching attenuated p21 expression at 6 h, but not at 12 h, while in Runx2+/− BMSCs, stretching attenuated p21 expression at both 6 and 12 h (Fig. 4B). There was little difference in p27 expression in BMSCs from 0 to 12 h, regardless of loading and Runx2 gene dosage (Fig. 4B).
These findings indicated that stretch increased the proliferation of BMSCs and regulated cell cycle progression, and that the reduction of Runx2 gene dosage delayed mechanical stimulation-induced proliferation of BMSCs.
3.4. Effect of stretch on osteogenesis of BMSCs from wild-type and Runx2+/− mice
Next, we loaded stretch onto wild-type and Runx2+/− BMSCs in osteogenic medium to investigate the effect of Runx2 on stretch-induced osteoblast differentiation. In unstretched wild-type and Runx2+/− BMSCs, ALP activity, an early osteogenesis marker, gradually increased from 0 to 21 days after the initiation of stretching and decreased thereafter, with no significant differences between the two genotypes until day 28 (Fig. 4C). In wild-type BMSCs, stretching significantly increased ALP activity on days 7 and 14, and the peak of ALP activity occurred on day 7 (Fig. 4C). In Runx2+/− BMSCs, mechanical stimulation significantly enhanced ALP activity on day 14, but not on day 7, and the peak of ALP activity was observed on day 21. Furthermore, the heterozygous Runx2 deficiency significantly reduced stretch-induced ALP activity in BMSCs on day 14 (Fig. 4C). Stretch-induced peak of ALP activity in Runx2+/− BMSCs was significantly lower than that in wild-type BMSCs (Fig. 4C).
Mechanical loading significantly increased OSC mRNA expression in wild-type BMSCs on day 14 after the initiation of stretch, and the stretch-induced OSC mRNA expression peaked on day 21 and decreased thereafter (Fig. 4D). In contrast, stretch significantly induced OSC mRNA expression in Runx2+/− BMSCs on day 21, but not on day 14 (Fig. 4D). Runx2+/− BMSCs showed a significantly lower peak of stretch-induced OSC mRNA expression than wild-type BMSCs (Fig. 4D).
We further examined stretch-induced matrix-deposited calcium, a late marker of osteogenesis, in BMSCs from wild-type and Runx2+/− mice (Fig. 4E). In unloaded wild-type and Runx2+/− BMSCs, the calcium content increased constantly until day 28 after the initiation of stretch, but there was no significant difference between wild-type and Runx2+/− (Fig. 4E). Mechanical loading significantly enhanced the calcium content in wild-type BMSCs on days 21 and 28, and in Runx2+/− BMSCs on day 28, but not on day 21 (Fig. 4E). On day 28, the stretch-induced calcium content in Runx2+/− BMSCs was significantly lower than in wild-type BMSCs (Fig. 4E).
In addition, wild-type and Runx2+/− BMSCs were stained for ALP on day 7 and alizarin red on day 21 after the initiation of mechanical stimulation (Fig. 4F and G). Stretching caused remarkable staining of ALP and alizarin red in both genotypes (Fig. 4F and G). Unstretched and stretched Runx2+/− BMSCs showed less staining than the corresponding wild-type BMSCs (Fig. 4F and G).
Taken together, we found that stretch enhanced osteogenesis in both wild-type and Runx2+/− BMSCs, but Runx2+/− BMSCs showed reduced and delayed stretch-induced osteoblast differentiation compared with wild-type BMSCs.
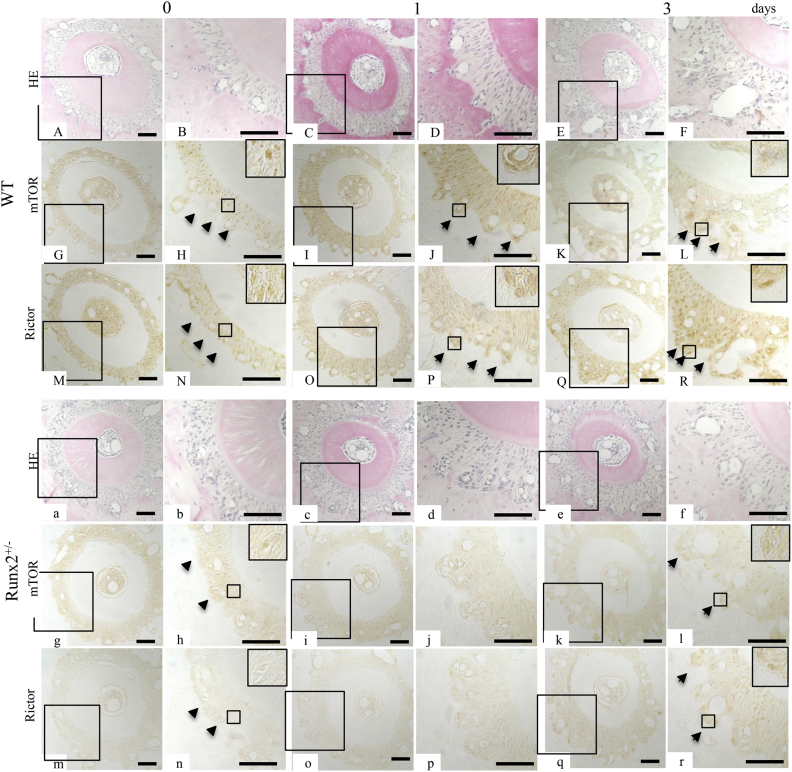
3.5. mTOR and Rictor protein expression in osteoblasts on the tension side of alveolar bone
To assess the association of mTORC2 with bone formation on the tension side of delayed tooth movement in Runx2+/− mice, we examined the protein expression of mTOR and Rictor in osteoblasts on the tension side of experimental tooth movement.
Before the initiation of experimental tooth movement (day 0), mTOR and Rictor were expressed in the PDL in both wild-type and Runx2+/− mice, although Runx2+/− mice showed weaker expression of both proteins compared with wild-type littermates (Fig. 5H, N, h, and n, arrowheads). Enhanced mTOR and Rictor expression in osteoblasts on the tension side of experimental tooth movement in wild-type mice was detected on days 1 and 3 of experimental tooth movement (Fig. 5J, P, L, and R, arrows), while Runx2+/− mice exhibited weaker expressions of these proteins on day 3 (Fig. 5j, p, l, and r, arrows). Expression of mTOR and Rictor was not detected in osteoblasts of Runx2+/− mice on day 1 (Fig. 5i, j, o, and p). We used wild-type mice on day 3 after the initiation of tooth movement as negative controls, and there was no expression (data not shown).
Fig. 5.
Time course changes of mTOR and Rictor protein expression pattern in osteoblasts on tension side of alveolar bone in wild-type and Runx2+/− mice. HE staining (A-F, and a-f) and immunohistochemistry for mTOR (G-L, and g-l) and Rictor (M-R, and m-r) expression in osteoblasts on tension side of distal root of upper right 1st molar in wild-type (A-R) and Runx2+/− (a-r) mice on 0 (A, B, G, H, M, N, a, b, g, h, m, and n) and 1 (C, D, I, J, O, P, c, d, i, j, o, and p), 3 (E, F, K, L, Q, R, e, f, k, l, q, and r) days after the wire attachment. Arrowheads indicate strong mTOR and Rictor protein expression in periodontal ligament.. B, D, F, H, J, L, N, P, and R are enlarged views of squares in A, C, E, G, I, K, M, O, and Q, and b, d, f, h, j, l, n, p, and r are enlarged views of a, c, e, g, i, k, m, o, and q. Squares in H, J, L, N, P, R, h, l, n, and r indicate the areas enlarged in the inset. Scale bar, 50 μm.
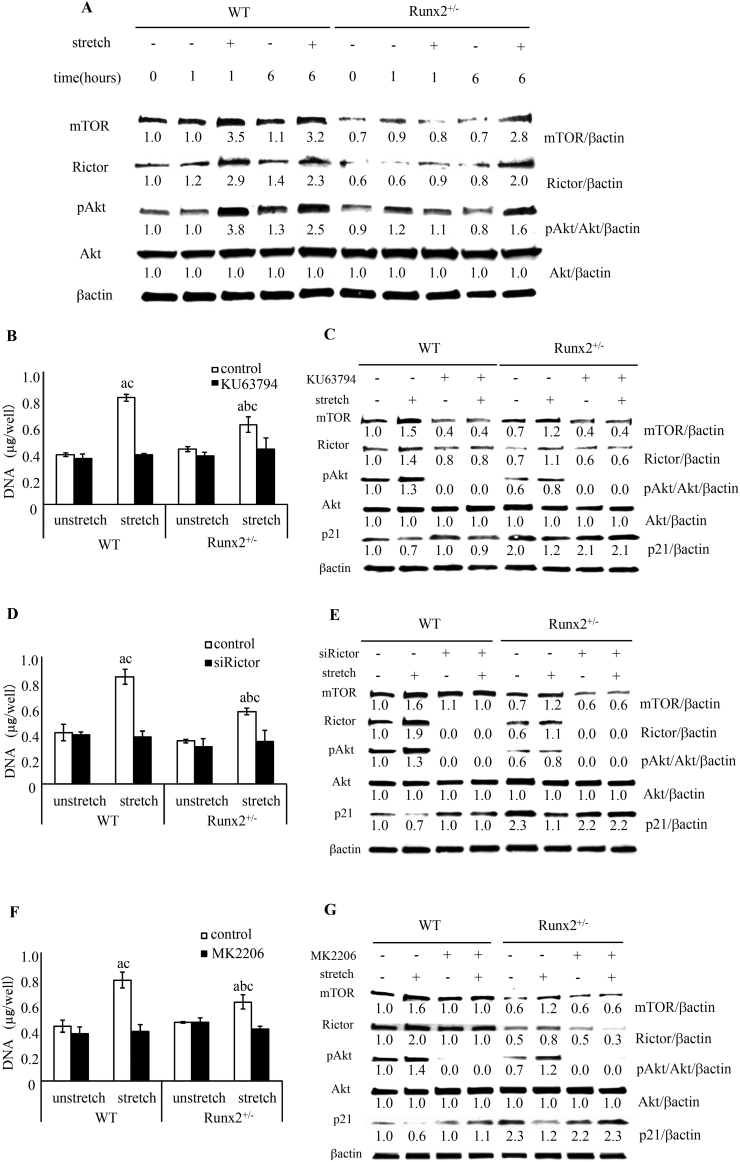
3.6. Association of mTORC2 activation with stretch-induced proliferation of BMSCs
To assess whether delayed stretch-induced proliferation of Runx2+/− BMSCs was caused by failure of mTORC2 activation, we evaluated mTORC2 activation in BMSCs after mechanical loading. In unstretched wild-type and Runx2+/− BMSCs, there was little change in mTOR protein expression from 0 to 6 h after the initiation of stretching (Fig. 6A). Mechanical stimulation remarkably increased mTOR protein expression in wild-type BMSCs at both 1 and 6 h, and in Runx2+/− BMSCs at 6 h, but not at 1 h (Fig. 6A). The Runx2 heterozygous deficiency reduced the expression of mTOR in BMSCs regardless of stretch (Fig. 6A). Similar results were confirmed for Rictor, an essential component factor of mTORC2, and Akt phosphorylation at serine 473, a target of mTORC2 (Fig. 6A).
Fig. 6.
Stretch-induced mTORC2/Akt signal and proliferation of wild-type and Runx2+/− BMSCs. (A) Change of mTOR and Rictor protein expression and Akt phosphorylation in BMSCs after 0, 1, and 6 h after the initiation of stretch. DNA content of BMSCs treated with KU63794 (5 μM) (B), siRictor (D), and MK2206 (5 μM), and stretched for 24 h (F). Values are averages ± SD (n = 3). a, significantly different between corresponding unstretched and stretched wild-type and Runx2+/− BMSCs. b, significantly different between stretched wild-type and Runx2+/− BMSCs. c significantly different between control and inhibitor or siRNA treated stretched wild-type and Runx2+/− BMSCs. p21 and mTORC2 related protein expression pattern in BMSCs treated with KU63794 (5 μM) (C), siRictor (E), and MK2206 (5 μM) (G) and stretched for 6 h. A quantification of normalized relative protein levels against respective βactin and corresponding unloaded wild-type mice cells is given below lane.
To examine the effect of mTORC2 on stretch-induced cell proliferation, we inhibited component factors of mTORC2 and Akt phosphorylation. KU63794, an inhibitor of mTOR activation, did not change the DNA content in unstretched wild-type and Runx2+/− BMSCs but dramatically inhibited the stretch induced-increase in DNA content (Fig. 6B). By immunoblot analysis, we found that KU63794 down-regulated mTOR expression and Akt phosphorylation in wild-type and Runx2+/− BMSCs regardless of stretching, and inhibited both stretch-induced Rictor expression and stretch-induced reduction of p21 expression (Fig. 6C). To confirm the involvement of mTORC2, but not mTORC1, in the stretch-induced cell proliferation, we silenced Rictor by short interfering RNA (siRNA). siRictor inhibited the stretch-induced DNA content in wild-type and Runx2+/− BMSCs (Fig. 6D). Immunoblot analysis revealed that siRictor silenced Rictor expression and Akt phosphorylation in both wild-type and Runx2+/− BMSCs and inhibited stretch-regulated mTOR and p21 expression (Fig. 6E). Furthermore, MK2206, an Akt phosphorylation inhibitor, down-regulated the increase in DNA content and weakened p21 expression in response to stretching (Fig. 6F and G). Interestingly, MK2206 also inhibited stretch-induced mTOR and Rictor expression (Fig. 6G).
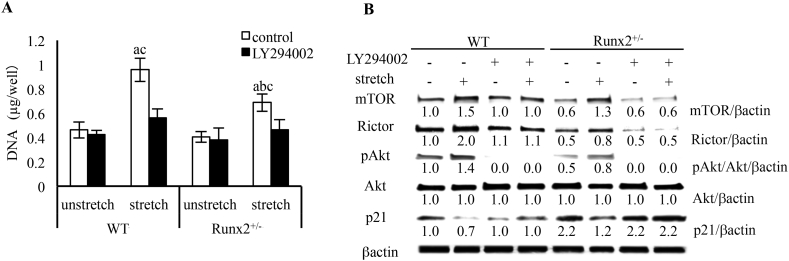
3.7. PI3K signaling on stretch-induced proliferation of BMSCs
Growth factors such as insulin-like growth factor (IGF), cytokines and stretch activate PI3K and phosphorylate Akt (Vanhaesebroeck et al., 2010; Watabe et al., 2011). PI3K/Akt signaling regulates cell proliferation and differentiation of bone-related cells such as MSCs, osteoblasts, and chondrocytes (Ghosh-Choudhury et al., 2002; Yun et al., 2008; Hidaka et al., 2001).
To examine the involvement of PI3K and mTORC2 signaling in stretch-induced cell proliferation, we stretched wild-type and Runx2+/− BMSCs after treatment with LY294002, a PI3K inhibitor. We found that LY294002 inhibited stretch-induced proliferation of wild-type and Runx2+/− BMSCs (Fig. 7A). Furthermore, wild-type and Runx2+/− BMSCs treated with LY294002 showed inhibition of stretch-regulated mTOR and Rictor expression, Akt phosphorylation (Fig. 7B). In addition, LY294002 inhibited stretch-induced reduction of p21 expression in both wild-type and Runx2+/− BMSCs (Fig. 7B).
Fig. 7.
Stretch-induced PI3K signal and cell proliferation in wild-type and Runx2+/− BMSCs. (A) DNA content from BMSCs treated with LY294002 (5 μM) and stretched for 24 h. Values are averages ± SD (n = 3). a, significantly different between corresponding unstretched and stretched wild-type and Runx2+/− BMSCs. b, significantly different between stretched wild-type and Runx2+/− BMSCs. c significantly different between control and inhibitor treated stretched wild-type and Runx2+/− BMSCs. (B) Immunoblot for p21 and mTORC2 related protein and expression pattern in BMSC treated with LY294002 (5 μM) and stretched for 6 h. A quantification of normalized relative protein levels against respective βactin and corresponding unloaded wild-type mice cells is given below lane.
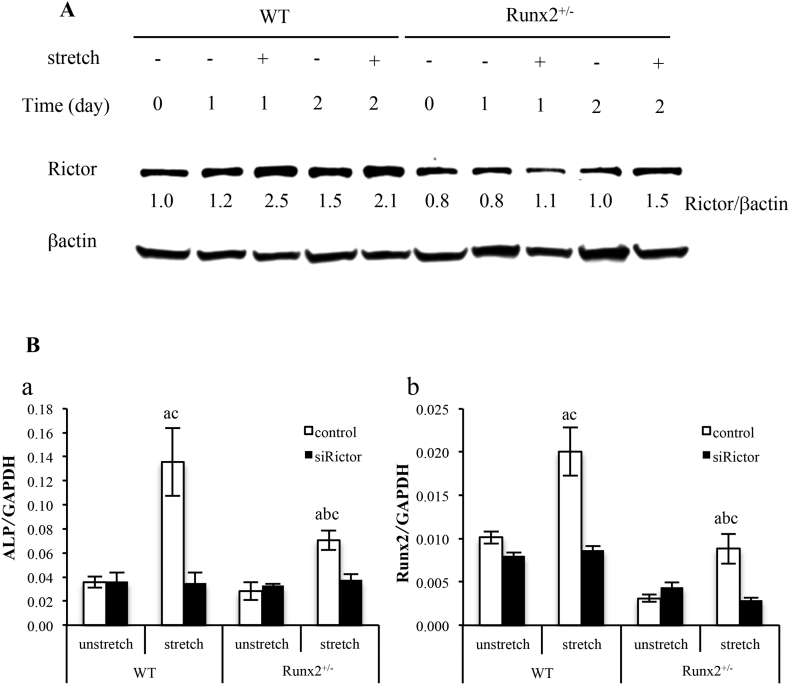
3.8. Effect of Rictor on stretch-induced osteoblast differentiation in BMSCs
To assess whether delayed stretch induced osteogenesis of BMSCs, we evaluated mTORC2 activation in BMSCs after mechanical loading in osteogenic medium. In unstretched wild-type BMSCs, Rictor protein expression was induced from 0 to 2 days after the initiation of stretching, while in Runx2+/− BMSCs, there was little change in Rictor protein expression (Fig. 8A). Mechanical stimulation remarkably increased Rictor protein expression in wild-type BMSCs on both days 1 and 2, and in Runx2+/− BMSCs on day 2, but not on day 1 (Fig. 8A). The Runx2 heterozygous deficiency reduced the expression of Rictor in BMSCs regardless of stretch (Fig. 8A). To investigate the association of mTORC2 with stretch-induced osteoblast differentiation, we stretched BMSCs in osteogenic medium after siRNA-mediated down-regulation of Rictor, and examined ALP and Runx2 mRNA expression by real-time PCR. After control siRNA treatment, wild-type and Runx2+/− BMSCs expressed similar level of ALP mRNA in unstretched condition (Fig. 8B-a). While, stretching significantly increased ALP mRNA expression in both genotypes, and Runx2+/− BMSCs exhibited significantly reduced stretch-induced ALP mRNA expression compared with wild-type BMSCs (Fig. 8B-a). siRictor did not alter ALP mRNA expression in unstretched cells, but inhibited stretch-induced ALP expression regardless of Runx2 gene dosage (Fig. 8B-a). In control siRNA-treated cells, stretch significantly enhanced Runx2 mRNA expression in both wild-type and Runx2+/− BMSCs, but Runx2 mRNA expression in Runx2+/− BMSCs showed significantly decresed compared with that in wild-type BMSCs (Fig. 8B-b). In siRictor-treated cells, there was no significant difference in Runx2 mRNA expression in unstretched wild-type and Runx2+/− BMSCs, while siRictor inhibited stretch-induced Runx2 mRNA expressions in both genotypes (Fig. 8B-b).
Fig. 8.
Stretch-induced osteoblast differentiation in wild-type and Runx2+/− BMSCs treated by siRictor. (A) BMSCs were down-regulated with siRictor, and stretched for 1 and 2 days after 1 h followed by change of osteogenic medium. (B) mRNA expression of ALP (a), and Runx2 (b) in BMSCs in osteogenic medium on 2 days after mechanical stretch. Values are averages ± SD (n = 3). a, significantly different between corresponding unstretched and stretched wild-type and Runx2+/− BMSCs. b, significantly different between stretched wild-type and Runx2+/− BMSCs. c, significantly different between control and inhibitor treated stretched wild-type and Runx2+/− BMSCs. A quantification of normalized relative protein levels against respective βactin and corresponding unloaded wild-type mice cells is given below lane.
4. Discussion
In the present study, we demonstrated for the first time that experimental tooth movement is delayed in Runx2+/− mice, the animal model of human CCD patients. We loaded a 10-g force to the upper first molars of 6-week-old Runx2+/− mice toward the palatal side using Ni—Ti wires for 21 days to induce experimental tooth movement according to Sakai et al. (2009). Tooth movement was initiated in Runx2+/− mice but it came to a halt after 3 days resulting in a significantly smaller amount of tooth movement compared to wild-type littermates between days 7 to 21. Meanwhile, Chung et al. (2004) reported no significant difference in the amount of experimental tooth movement between 13 and 22-week-old adult wild-type and Runx2+/− mice. They used a Ni—Ti coiled spring fixed to the maxillary incisor to load a mesial 10-g force on the maxillary first molar, and measured the distance of tooth movement by μCT (Chung et al., 2004). The reasons for the differences with our findings may include the differences in experimental design, such as age of mice, apparatus and direction of mechanical stretch, and measurement method for the amount of tooth movement. Tsuji et al. (2004) demonstrated that reduced osteogensis of bone marrow cells derived from aged adult Runx2+/− mice (7.5 ± 0.5 months old) compared with wild-type mice. The possible reason for the difference between Chung's result and ours might be the age of mice.
When orthodontic force is applied to teeth in rodents, three phases of tooth movement are induced (Wise and King, 2008; Yan et al., 2015; Takano-Yamamoto et al., 2017). The first phase involves initial tooth displacement by tooth inclination in the PDL space, followed by a lag phase, and finally a tooth movement phase accompanied by bone remodeling with bone resorption by osteoclasts and bone formation by osteoblasts (Wise and King, 2008; Yan et al., 2015; Takano-Yamamoto et al., 2017). Therefore, bone resorption and formation couple to remodeling in the alveolar bone around the root of the tooth during orthodontic tooth movement. In the present study, suppressed osteoid formation and reduced and delayed OSC mRNA expression were observed in osteoblasts on the tension side of tooth movement in Runx2+/− mice as compared with wild-type mice, indicating impaired stretch-induced osteoblast function and bone formation. These findings suggest that the delayed tooth movement is associated with impaired bone formation on the tension side in Runx2+/− mice. In contrast, Runx2 enhanced osteoclast differentiation by regulating production of receptor activator of NF-κB ligand (RANKL) and osteoprotegerin, both are important factors for osteoclastogenesis (Enomoto et al., 2003). In addition, RANKL promoter had Runx2 binding site (Kitazawa et al., 2003). Therefore, Runx2 would probably have some effects on bone resorption on the compression side of tooth movement. However, further investigations are needed to clarify the osteoclast function in Runx2+/− mice.
On the tension side of tooth movement, mechanical stress-induced proliferation and osteoblast differentiation of MSCs result in promotion of bone formation (Pavlin and Gluhak-Heinrich, 2001). In the present study, in order to elucidate the underlying mechanism of orthodontic force-induced impaired bone formation in Runx2+/− mice, we cultured mouse BMSCs in silicon chambers and loaded continuous uniaxial stretching to the chambers, which simulates the tension side of tooth movement in vivo, and investigated cell proliferation in growth medium and osteogenesis in osteogenic medium.
It has been reported that chondrocyte proliferation in the growth plates of tibia and femur is reduced in Runx2 knockout embryos compared with wild-type mice, and that Runx2 promotes chondrocyte proliferation through induction of Indian hedgehog and Tcf7 (Yoshida et al., 2004; Mikasa et al., 2011). In addition, recent studies found that Runx2 phosphorylation increased the proliferation of human bone marrow endothelial cells (hECs) (Qiao et al., 2006; Pierce et al., 2012). We found that the stretch-induced growth rate of BMSCs was significantly delayed in Runx2+/− mice compared with wild-type littermates, suggesting for the first time an association of Runx2 with stretch-induced proliferation of BMSCs. Contrary to our results, it was reported that Runx2 overexpression suppressed the proliferation of MC3T3 mouse preosteoblastic cells and C2C12 mouse mesenchymal cells, and that Runx2 overexpression in MC3T3 cells caused a delay in G1 phase progression (Galindo et al., 2005). On the contrary, calvarial osteoblastic cells from Runx2-deficient mice also exhibited an increased cell growth rate compared with those from wild-type littermates (Pratap et al., 2003). In the present study, stretch elevated cyclin D (G1) expression in wild-type and Runx2+/− BMSCs, it did not promote osteoblastic phenotypes (Suppl. Fig. 3), indicating that Runx2 response for mechanical stretch enhanced proliferation of BMSCs but did not promote osteoblast differentiation. Consistent with our findings, Hata et al. (2013) showed that uniaxial stretch increased proliferation of stem cells isolated from dental pulp of Sprague-Dawlay rats, while its osteogenic differentiation was inhibited. Therefore, the influences of Runx2 in cell proliferation may be dependent on osteoblastic differentiation-stages. It is suggested that Runx2 dosage might influence mechanical stress induced-proliferation of BMSCs at early stage of osteogenesis.
It was reported that Rictor-deficient mice show suppressed axial loading-induced mineral apposition on the periosteal surface of the tibia compared with wild-type littermates (Sen et al., 2014). A bi-axial cyclic stretch elevated mTOR and Rictor protein expression to mTORC2 activation for cytoskeletal reorganization of MSCs (Tandon et al., 2014). Tandon et al. (2014) revealed that recruitment of Runx2 to the promoter of mTOR activated PI3K/Akt signaling via mTORC2 to induce survival in a human breast cancer cell line. Therefore, Runx2 might be associated with mechanical loading-induced osteoblast function via mTORC2 activation. In the present study, for the first time, we found reduced and delayed expression of mTOR and Rictor in osteoblasts on the tension side of tooth movement in Runx2+/− mice compared with wild-type littermates. Therefore, we suggest the possible association between Runx2 and mTORC2 in bone formation on the tension side of tooth movement.
In the present study, reduced and delayed expression of mTOR and Rictor, were observed in osteoblasts on the tension side of tooth movement in Runx2+/− mice as compared with wild-type littermates in vivo and in vitro. In addition, we demonstrated that silenced mTOR and Rictor expression, and inhibition of Akt phosphorylation completely inhibited stretch-induced proliferation of BMSCs in vitro, indicating that stretch-induced mTORC2/Akt signaling promoted BMSC proliferation. Taken together, these findings suggest that mTORC2 activation was involved in delayed mechanical stress-induced tooth movement in RUNX2+/− mouse (CCD) model.
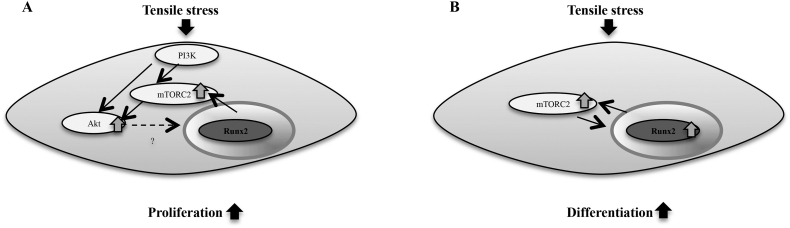
Next, we investigated cell cycle regulators to elucidate the mechanism of the delayed stretch-induced proliferation of Runx2+/− BMSCs. It was reported that glucose treatment facilitated Runx2 DNA-binding activity and cell cycle progression in hECs, and that targeted knockdown of Runx2 delayed progression in G1 phase (Pierce et al., 2012). Consistent with these findings, we revealed delayed stretch-elevated expression of cyclin D, a G1 cell cycle progression marker, in Runx2+/− BMSCs compared with that in wild-type BMSCs. Akt phosphorylation promotes cell proliferation through inhibition of p21 expression, an inhibitor of cdk and G1 transition (Gu et al., 2011). Phosphorylated Runx2 is translocated into the nucleus and recruited to the promoter of p21 to suppress its expression in hECs and mouse osteoblastic cell lines (Pierce et al., 2012; Westendorf et al., 2002). Furthermore, Runx2 is a direct substrate of Akt and is phosphorylated in human breast cancer cells (Pande et al., 2013). In the present study, stretch-induced mTORC2/Akt activation was inhibited in Runx2+/− BMSCs compared with that in wild-type BMSCs. In addition, suppression of p21 expression induced by stretch was abolished by inhibition of mTORC2 and Akt phosphorylation. These findings suggest that Runx2 is associated with stretch-regulated expression of cell cycle regulators in BMSCs, and affects stretch-induced cell proliferation. It is likely that down-regulation of p21 by stretching via Runx2/mTORC2/Akt axis regulated cell cycle progression. We propose that there may be two underlying mechanisms, one involving indirect inhibition by Runx2 for p21 expression via mTORC2/Akt activation, and the other involving direct inhibition by recruitment of Akt-phosphorylated Runx2 to the p21 promoter. PI3K is a kinase that phosphorylates Akt and induces mTORC2 activation (Masui et al., 2014). In the present study, inhibition of PI3K suppressed stretch-induced mTOR and Rictor expression and Akt phosphorylation, and also inhibited stretch-increased cell proliferation, suggesting that regulation of Runx2 via PI3K-dependent mTOR/Akt signaling plays a critical role in stretch-induced osteoblastic cell proliferation (Fig. 9A). Interestingly, we found that inhibition of Akt phosphorylation downstream of mTORC2 down-regulated stretch-enhanced mTOR and Rictor expression. It is suggested that another upstream mechanism of Akt and mTORC2 for stretch-induced mTORC2 activity may be involved, but this remains to be clarified.
Fig. 9.
Runx2 regulates stretch-induced osteoblast function via mTORC2. (A) Runx2 regulates PI3K/mTORC2/Akt signaling axis in stretch-induced proliferation of BMSCs. (B) Runx2 interacts with mTORC2 in stretch-induced osteoblast differentiation.
Several studies have revealed that mechanical stress activates Runx2 to promote osteogenesis in osteoblastic cells (Liu et al., 2011a; Ziros et al., 2002; Kanno et al., 2007; Li et al., 2012; Salingcarnboriboon et al., 2006; Liu et al., 2011b). Consistent with these past reports, we showed that Runx2+/− BMSCs in osteogenic medium exhibited delayed and reduced stretch-induced early and late osteoblast differentiation compared with wild-type BMSCs. Overexpression of Runx2 up-regulated Akt phosphorylation, a target of mTORC2, in MC3T3 cells (Fujita et al., 2004). However, there are no reports on the association of mTORC2 with osteoblast differentiation promoted by Runx2. Akt, a downstream of mTORC2, was phosphorylated by overexpression of Runx2 in MC3T3 cells (Fujita et al., 2004). In the present study, the expression of stretch-enhanced mTOR and Rictor in Runx2+/− BMSCs cultured in osteogenic medium was delayed and reduced compared with that in wild-type BMSCs. Akt phosphorylation promoted BMP-induced osteoblast differentiation and enhanced Runx2 mRNA and protein expression (Choi et al., 2014), and IGF phosphorylated Akt and elevated ALP activity during osteoblast differentiation (Fujita et al., 2004). We found that silencing of Rictor suppressed stretch-elevated mRNA expression of Runx2 and ALP in wild-type and Runx2+/− BMSCs in osteogenic medium in vitro, indicating stretch-promoted osteoblast differentiation in BMSCs via mTORC2 activation. Taken together, we suggest that the bidirectional interaction of Runx2 and mTORC2 affects stretch-induced osteoblast differentiation (Fig. 9B).
Resently, Dai et al. (2017) have reported that preosteoblast-specific knockout of mTOR impaired mouse osteoblast differentiation and cause CCD-like bone phenotype. They showed ablation of Raptor, specific factor of mTORC1, reduced Runx2 expression in osteoblasts and suppressed osteoblast differentiation. In our preliminary data, stretch induced Raptor protein expression in BMSCs and Runx2+/− BMSCs showed delayed and reduced stretch-induced expression, as is the case with Rictror (data not shown). There is a possibility association of mTORC1 as well as mTORC2 for mechanical stress-induced osteoblast function through Runx2 activation, but it remains to be studied. In addition, several studies have reported involvement of other signaling pathway, such as MAPK, BMP/Smad and Wnt signal, for mechanical stimuli-induced Runx2 activation and osteoblast function (Kanno et al., 2007; Liu et al., 2011b; Yan et al., 2016). Therefore, these signaling pathways would probably have some effects on bone formation on the tension side of experimental tooth movement in Runx2+/− mice. However, further investigations are needed to clarify the involvement of other signaling pathways in mechanical stress-induced bone formation in Runx2+/− mice.
5. Conclusions
We have shown for the first time that tooth movement is delayed in Runx2+/− mice compared with wild-type mice. This prompted us to use Runx2+/− mice as a model for delayed orthodontic tooth movement in CCD patients having RUNX2+/−. In addition, we have also shown that Runx2+/− BMSCs decreased stretch-induced proliferation, and differentiation to osteoblasts via mTORC2 activation. Our findings suggest that the association of Runx2 with mTORC2/Akt activation is one of critical role for stretch-induced proliferation and differentiation to osteoblasts in osteogenesis during tooth movement.
Author contribution
T.T.-Y. contributed to the conception and design of the study. N.Tamamura, Y.S., T.F., N.Takeshita, T.Y., I.T. and T.T.-Y. contributed to the in vivo studies. T.A. T.F., S.S. and T.T.-Y. contributed to the in vitro studies. All statistical analysis was carried out by T.A.. T.T.-Y. and T.A. prepared the manuscript. All authors read the manuscript and provided comments and revisions.
Declaration of competing interest
All authors declare that they have no competing interest.
Acknowledgments
We thank Dr. E. Fukumoto (Tohoku University), Dr. H. Kawaki (Asahi University), Dr. E. Ikeda (RIKEN Center for Developmental Biology), and Dr. Kitaura (Tohoku University) for excellent technical support, and Biomedical Research Unit of Tohoku University Hospital for technical equipment support.
This study was supported by a grant-in Aid for scientific Research from Ministry of Education, Culture, Sports, Science and Technology, Japan (15659491, 17209064, 20249081, 23249085, 15K11335 and 15H05048 to T.T.-Y., 15K11335 and 18K09827 to T.F., 24792270 and 26861768 to T.A.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bonr.2020.100285.
Contributor Information
Tomohiro Fukunaga, Email: tfukunaga@dent.tohoku.ac.jp.
Nobuo Takeshita, Email: nob@m.tohoku.ac.jp.
Takashi Yamashiro, Email: yamashiro@dent.osaka-u.ac.jp.
Irma Thesleff, Email: irma.thesleff@helsinki.fi.
Teruko Takano-Yamamoto, Email: t-yamamo@m.tohoku.ac.jp.
Appendix A. Supplementary data
Supplementary material
References
- Aberg T., Cavender A., Gaikwad J.S. Phenotypic changes in dentition of Runx2 homozygote-null mutant mice. J. Histochem. Cytochem. 2004;52(1):131–139. doi: 10.1177/002215540405200113. [DOI] [PubMed] [Google Scholar]
- Baldin V., Lukas J., Marcote M.J. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7(5):812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Becker A., Lustmann J., Shteyer A. Cleidocranial dysplasia: part 1--general principles of the orthodontic and surgical treatment modality. Am. J. Orthod. Dentofac. Orthop. 1997;111(1):28–33. doi: 10.1016/s0889-5406(97)70298-1. [DOI] [PubMed] [Google Scholar]
- Becker A., Shteyer A., Bimstein E., Lustmann J. Cleidocranial dysplasia: part 2--treatment protocol for the orthodontic and surgical modality. Am. J. Orthod. Dentofac. Orthop. 1997;111(2):173–183. doi: 10.1016/s0889-5406(97)70213-0. [DOI] [PubMed] [Google Scholar]
- Beertsen W., McCulloch C.A., Sodek J. The periodontal ligament: unique, multifunctional connective tissue. Periodontol. 1997;2000(13):20–40. doi: 10.1111/j.1600-0757.1997.tb00094.x. [DOI] [PubMed] [Google Scholar]
- Bhaskar P.T., Hay N. The two TORCs and Akt. Dev. Cell. 2007;12(4):487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Chen J., Holguin N., Shi Y. mTORC2 signaling promotes skeletal growth and bone formation in mice. J. Bone Miner. Res. 2015;30(2):369–378. doi: 10.1002/jbmr.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.H., Kim Y.J., Jeong H.M. Akt enhances Runx2 protein stability by regulating Smurf2 function during osteoblast differentiation. FEBS J. 2014;281(16):3656–3666. doi: 10.1111/febs.12887. [DOI] [PubMed] [Google Scholar]
- Chung C.R., Tsuji K., Nifuji A. Micro-CT evaluation of tooth, calvaria and mechanical stress-induced tooth movement in adult Runx2/Cbfa1 heterozygous knock-out mice. J Med Dent Sci. 2004;51(1):105–113. [PubMed] [Google Scholar]
- Dai Q., Xu Z., Ma X. mTOR/raptor signaling is critical for skeletogenesis in mice through the regulation of Runx2 expression. Cell Death Differ. 2017;24(11):1886–1899. doi: 10.1038/cdd.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovitch Z. Tooth movement. Crit. Rev. Oral Biol. Med. 1991;2(24):411–450. doi: 10.1177/10454411910020040101. [DOI] [PubMed] [Google Scholar]
- Desbois C., Hogue D.A., Karsenty G. The mouse osteocalcin gene cluster contains three genes with two separate spatial and temporal patterns of expression. J. Biol. Chem. 1994;269(2):1183–1190. [PubMed] [Google Scholar]
- Ducy P., Zhang R., Geoffroy V. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Enomoto H., Shiojiri S., Hoshi K. Induction of osteoclast differentiation by Runx2 through receptor activator of nuclear factor-kappa B ligand (RANKL) and osteoprotegerin regulation and partial rescue of osteoclastogenesis in Runx2−/− mice by RANKL transgene. J. Biol. Chem. 2003;278(26):23971–23977. doi: 10.1074/jbc.M302457200. [DOI] [PubMed] [Google Scholar]
- Fujita T., Azuma Y., Fukuyama R. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J. Cell Biol. 2004;166(1):85–95. doi: 10.1083/jcb.200401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M., Pratap J., Young D.W. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 2005;280(21):20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez J.M., Moran J., Clarke R.G. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem. J. 2009;421(1):29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Choudhury N., Abboud S.L., Nishimura R. Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J. Biol. Chem. 2002;277(36):33361–33368. doi: 10.1074/jbc.M205053200. [DOI] [PubMed] [Google Scholar]
- Gu Y., Lindner J., Kumar A. Rictor/mTORC2 is essential fining a balance between beta-cell proliferation and cell size. Diabetes. 2011;60(3):827–837. doi: 10.2337/db10-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hata M., Naruse K., Ozawa S. Mechanical stretch increases the proliferation while inhibiting the osteogenic differentiation in dental pulp stem cells. Tissue Eng Part A. 2013;19(5–6):625–633. doi: 10.1089/ten.tea.2012.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka K., Kanematsu T., Takeuchi H. Involvement of the phosphoinositide 3-kinase/protein kinase B signaling pathway in insulin/IGF-I-induced chondrogenesis of the mouse embryonal carcinoma-derived cell line ATDC5. Int. J. Biochem. Cell Biol. 2001;33(11):1094–1103. doi: 10.1016/s1357-2725(01)00067-x. [DOI] [PubMed] [Google Scholar]
- Kanno T., Takahashi T., Tsujisawa T. Mechanical stress-mediated Runx2 activation is dependent on Ras/ERK1/2 MAPK signaling in osteoblasts. J. Cell. Biochem. 2007;101(5):1266–1277. doi: 10.1002/jcb.21249. [DOI] [PubMed] [Google Scholar]
- Kitazawa S., Kajimoto K., Kondo T. Vitamin D3 supports osteoclastogenesis via functional vitamin D response element of human RANKL gene promoter. J. Cell. Biochem. 2003;89(4):771–777. doi: 10.1002/jcb.10567. [DOI] [PubMed] [Google Scholar]
- Komori T., Yagi H., Nomura S., Yamaguchi A. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lekic P., McCulloch C.A. Periodontal ligament cell population: the central role of fibroblasts in creating a unique tissue. Anat. Rec. 1996;245(2):327–341. doi: 10.1002/(SICI)1097-0185(199606)245:2<327::AID-AR15>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Li Y., Ge C., Long J.P. Biomechanical stimulation of osteoblast gene expression requires phosphorylation of the Runx2 transcription factor. J. Bone Miner. Res. 2012;27(6):1263–1274. doi: 10.1002/jbmr.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- Liu L., Shao L., Li B., Zong C. Extracellular signal regulated kinase1/2 activated by fluid shear stress promotes osteogeneic differentiaiton of human bone marrow-derived mesenchymal stem cells through novel signaling pathways. Int. J. Biochem. Cell Biol. 2011;43(11):1591–1601. doi: 10.1016/j.biocel.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Liu L., Shao L., Li B., Zong C. Extracellular signal regulated kinase1/2 activated by fluid shear stress promotes osteogeneic differentiaiton of human bone marrow-derived mesenchymal stem cells through novel signaling pathways. Int. J. Biochem. Cell Biol. 2011;43(11):1591–1601. doi: 10.1016/j.biocel.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Malagu K., Duggan H., Menear K. The discovery and optimisation of pyrido[2,3-d]pyrimidine-2,4-diamines as potent and selective inhibitors of mTOR kinase. Bioorg. Med. Chem. Lett. 2009;19(20):5950–5953. doi: 10.1016/j.bmcl.2009.08.038. [DOI] [PubMed] [Google Scholar]
- Masui K., Cavenee W.K., Mischel P.S. mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol. Metab. 2014;25(6):364–373. doi: 10.1016/j.tem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P.C. The histological identification of osteoid tissue. J Pathol Bacteriol. 1956;71(2):325–333. doi: 10.1002/path.1700710207. [DOI] [PubMed] [Google Scholar]
- Mikasa M., Rokutanda S., Komori H. Regulation of Tcf7 by Runx2 in chondrocyte maturation and proliferation. J. Bone Miner. Metab. 2011;29(3):291–299. doi: 10.1007/s00774-010-0222-z. [DOI] [PubMed] [Google Scholar]
- Mundlos S. Cleidocranial dysplasia: clinical and molecular genetics. J. Med. Genet. 1999;36(3):177–182. [PMC free article] [PubMed] [Google Scholar]
- Otto F., Thornell A.P., Crompton T. Cbfa1, a candidate gene for Cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pande S., Browne G., Padmanabhan S. Oncogenic Cooperation Between PI3K/Akt Signaling and Transcription Factor Runx2 Promotes the Invasive Properties of Metastatic Breast Cancer Cells. J. Cell. Physiol. 2013;228(8):1784–1792. doi: 10.1002/jcp.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlin D., Gluhak-Heinrich J. Effect of mechanical loading on periodontal cells. Crit. Rev. Oral Biol. Med. 2001;12(5):414–424. doi: 10.1177/10454411010120050401. [DOI] [PubMed] [Google Scholar]
- Pavlin D., Goldman E.S., Gluhak-Heinrich J. Orthodontically stressed periodontium of transgenic mouse as a model for studying mechanical response in bone: the effect on the number of osteoblasts. Clin. Orthod. Res. 2000;3(2):55–66. [PubMed] [Google Scholar]
- Pierce A.D., Anglin I.E., Vitolo M.I. Glucose-activated RUNX2 phosphorylation promotes endothelial cell proliferation and an angiogenic phenotype. J. Cell. Biochem. 2012;113(1):282–292. doi: 10.1002/jcb.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Kato J.Y., Solomon M.J. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8(1):9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- Pratap J., Galindo M., Zaidi S.K. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003;63(17):5357–5362. [PubMed] [Google Scholar]
- Qiao M., Shapiro P., Fosbrink M. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J. Biol. Chem. 2006;2281(11):7118–7128. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Balam T.A., Kuroda S. CTGF and apoptosis in mouse osteocytes induced by tooth movement. J. Dent. Res. 2009;88(4):345–350. doi: 10.1177/0022034509334649. [DOI] [PubMed] [Google Scholar]
- Salingcarnboriboon R., Tsuji K., Komori T. Runx2 is a target of mechanical unloading to alter osteoblastic activity and bone formation in vivo. Endocrinology. 2006;147(5):2296–2305. doi: 10.1210/en.2005-1020. [DOI] [PubMed] [Google Scholar]
- Sen B., Xie Z., Case N. mTORC2 regulates mechanically induced cytoskeletal reorganization and lineage selection in marrow-derived mesenchymal stem cells. J. Bone Miner. Res. 2014;29(1):78–89. doi: 10.1002/jbmr.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Yamamoto T., Takemura T., Kitamura Y. Site-specific expression of mRNAs for osteonectin, osteocalcin, and osteopontin revealed by in situ hybridization in rat periodontal ligament during physiological tooth movement. J. Histochem. Cytochem. 1994;42(7):885–896. doi: 10.1177/42.7.8014472. [DOI] [PubMed] [Google Scholar]
- Takano-Yamamoto T., Sasaki K., Fatemeh G. Synergistic acceleration of experimental tooth movement by supplementary high-frequency vibration applied with a static force in rats. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-13541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto A., Kawatsu M., Yoshimoto Y. Scleraxis and osterix antagonistically regulate tensile force-responsive remodeling of the periodontal ligament and alveolar bone. Development. 2015;142(4):787–796. doi: 10.1242/dev.116228. [DOI] [PubMed] [Google Scholar]
- Tandon M., Chen Z., Pratap J. Runx2 activates PI3K/Akt signaling via mTORC2 regulation in invasive breast cancer cells. Breast Cancer Res. 2014;16(1):R16. doi: 10.1186/bcr3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai K., Takano-Yamamoto T., Ohba Y. Role of osteopontin in bone remodeling caused by mechanical stress. J. Bone Miner. Res. 1999;14(6):839–849. doi: 10.1359/jbmr.1999.14.6.839. [DOI] [PubMed] [Google Scholar]
- Tsuji K., Komori T., Noda M. Aged mice require full transcription factor, Runx2/Cbfa1, gene dosage for cancellous bone regeneration after bone marrow ablation. J. Bone Miner. Res. 2004;19(9) doi: 10.1359/JBMR.040601. (1481-1289) [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010;11(5):329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Vlahos C.J., Matter W.F., Hui K.Y. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J. Biol. Chem. 1994;269(7):5241–5248. [PubMed] [Google Scholar]
- Watabe H., Furuhama T., Tani-Ishii N., Mikuni-Takagaki Y. Mechanotransduction activates α₅β₁ integrin and PI3K/Akt signaling pathways in mandibular osteoblasts. Exp. Cell Res. 2011;317(18):2642–2649. doi: 10.1016/j.yexcr.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Westendorf J.J., Zaidi S.K., Cascino J.E. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol. Cell. Biol. 2002;22(22):7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise G.E., King G.J. Mechanisms of tooth eruption and orthodontic tooth movement. J. Dent. Res. 2008;87(5):414–434. doi: 10.1177/154405910808700509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. AACR Annual Meeting-Apr 18–22, 2009. May 2009. MK-2206: A potent oral allosteric AKT inhibitor. Denver, CO Published. [Google Scholar]
- Yan Y., Liu F., Kou X. T cells are required for orthodontic tooth movement. J. Dent. Res. 2015;94(10):1463–1470. doi: 10.1177/0022034515595003. [DOI] [PubMed] [Google Scholar]
- Yan Y., Sun H., Gong Y. Mechanical strain promotes osteoblastic differentiation through integrin-β1-mediated β-catenin signaling. Int. J. Mol. Med. 2016;38(2):594–600. doi: 10.3892/ijmm.2016.2636. [DOI] [PubMed] [Google Scholar]
- Yoshida C.A., Yamamoto H., Fujita T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18(8):952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.P., Lee M.Y., Ryu J.M. Role of HIF-1alpha and VEGF in human mesenchymal stem cell proliferation by 17beta-estradiol: involvement of PKC, PI3K/Akt, and MAPKs. Am. J. Phys. Cell Phys. 2008;296(2):C317–C326. doi: 10.1152/ajpcell.00415.2008. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Deng X., Scheller E.L. The effects of Runx2 immobilization on poly (epsilon-caprolactone) on osteoblast differentiation of bone marrow stromal cells in vitro. Biomaterials. 2010;31(12):3231–3236. doi: 10.1016/j.biomaterials.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziros P.G., Gil A.P., Georgakopoulos T. The bone-specific transcriptional regulator Cbfa1 is a target of mechanical signals in osteoblastic cells. J. Biol. Chem. 2002;277(26):23934–23941. doi: 10.1074/jbc.M109881200. [DOI] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material