Abstract
Chronological aging as well as biological aging accelerated by various pathologies such as diabetes and obesity contribute to cardiovascular aging, and structural and functional tissue damage of the heart and vasculature. Cardiovascular aging in humans is characterized by structural pathologic remodeling including cardiac and vascular fibrosis, hypertrophy, stiffness, micro- and macro-circulatory impairment, left ventricular diastolic dysfunction precipitating heart failure with either reduced or preserved ejection fraction, and cardiovascular cell death. Cellular senescence, an important hallmark of aging, is a critical factor that impairs repair and regeneration of damaged cells in cardiovascular tissues whereas autophagy, an intracellular catabolic process is an essential inherent mechanism that removes senescent cells throughout life time in all tissues. Several recent reviews have highlighted the fact that all longevity treatment paradigms to mitigate progression of aging-related pathologies converge in induction of autophagy, activation of AMP kinase (AMPK) and Sirtuin pathway, and inhibition of mechanistic target of rapamycin (mTOR). These longevity treatments include health style changes such as caloric restriction, and drug treatments using rapamycin, the first FDA-approved longevity drug, as well as other experimental longevity drugs such as metformin, rapamycin, aspirin, and resveratrol. However, in the heart tissue, autophagy induction has to be tightly regulated since evidence show excessive autophagy results in cardiomyopathy and heart failure. Here we discuss emerging evidence for microRNA-mediated tight regulation of autophagy in the heart in response to treatment with rapamycin, and novel approaches to monitor autophagy progression in a temporal manner to diagnose and regulate autophagy induction by longevity treatments.
Keywords: cardiovascular, aging, senescence, autophagy, microRNA, longevity drugs, caloric restriction
Introduction
Global deaths from cardiovascular disease increased by 41% between 1990 and 2013 and population ageing alone contributed to an estimated 52.5% increase in these deaths (Roth et al., 2015; Roth et al., 2018). The 2019 United Nations statement predicts that by 2050, one in six people in the world will be over the age of 65, up from one in 11 in 2019 (United Nations, 2019). Older adults are particularly susceptible to cardiovascular diseases (CVD), a major contributor to frailty and mortality. The incidence of CVD is ~75% in those at the age of 60–79 years, and increases to ~86% in those above the age of 80 (Appelman et al., 2015; Childs et al., 2018; Roth et al., 2018; Rodgers et al., 2019; Noale et al., 2020; Stojanovic et al., 2020). Therefore, aging is both a driver and an independent risk factor for CVD.
Conversely, CVDs resulting from a variety of metabolic and environmental stressors or infections promote cellular senescence and accelerate cellular, organ-level, and organismal aging. Two well-established and preventable factors that accelerate biological aging independent of chronological age are obesity and type 2 diabetes. Currently 1.9 billion adults are overweight and 650 million people are obese (World Health Organization, 2018). Age-adjusted prevalence of obesity is 42.4% in the US and obesity affects 13.7 million children and adolescents (Hales et al., 2020). Obesity promotes diabetes—globally, an estimated 463 million adults have diabetes, a prevalence rate of one in 11 (International Diabetes Association, 2019). Moreover, newly diagnosed type 1 and type 2 diabetes cases have increased among the youth in US. Diabetes is an independent risk factor for CVD (National Diabetes Statistics Report, 2020). A report using the Swedish National Diabetes Registry comprised 318,083 patients with T2DM (1998–2012) and age-, sex-, and country-matched healthy <1.6 million controls shows that patients diagnosed of type 2 diabetes mellitus (T2DM) at ≤40 years had the highest excess risk for CVD and death. Thus, the younger obese and/or diabetic patients have increased cardiovascular risk and accelerated cardiovascular and biological aging despite their chronological young age.
Pro-Senescence Mechanisms of Cardiovascular Aging
Cardiovascular aging in humans is characterized by structural pathologic remodeling including fibrosis, hypertrophy, loss of capillary density, macro- and micro-circulatory impairment, and cardiomyocyte death. Left ventricular diastolic dysfunction that precipitates heart failure with reduced or preserved ejection fraction, vascular fibrosis, stiffness, and impairment of synthesis and/or secretion of endothelium-derived vasoactive molecules that underlie atherosclerosis and other vasculopathies mark cardiovascular aging. These cardiovascular pathologies are common to both chronological aging and accelerated biological aging induced by metabolic diseases such as diabetes and obesity despite the differences in the stressors in these different conditions (Ghebre et al., 2016; Upadhya and Kitzman, 2017; Ungvari et al., 2018; Jin, 2019; Wagner and Dimmeler, 2020). Cellular senescence, an important hallmark of aging, is a critical factor that impairs repair and regeneration of damaged cells in cardiovascular tissues (Childs et al., 2018; Lewis-McDougall et al., 2019).Cellular senescence is a stress-responsive process that is activated by multiple stressors including elevation of reactive oxygen species, pro-inflammatory cytokines, and metabolic, mechanical, and chemical toxicity such as glucotoxicity, lipotoxicity, and chemotoxicity (Colavitti and Finkel, 2005; Matsui-Hirai et al., 2011; Childs et al., 2018; Olivieri et al., 2018; Trayssac et al., 2018; Lewis-McDougall et al., 2019). Senescent cells release senescence-associated secretory phenotype (SASP) comprised of pro-inflammatory cytokines, chemokines, extracellular matrix degrading proteins, and other deleterious factors that disrupt tissue structure and function. Importantly, a relatively low abundance of senescent cells (10%) is sufficient to spread cellular senescence to surrounding cells in a tissue as well as to other tissues via SASP, and in conditions of diabetes, activation of DNA damage response (DDR) in cells induce senescence and thus activate a ‘malignant positive feedback loop' in which diabetes-DDR-cellular senescence further increases tissue damage and spreading of senescence (Tchkonia et al., 2013; Palmer et al., 2015; McHugh and Gil, 2018). Cellular senescence is also intrinsically linked to other hallmarks of aging such as telomere attrition, genomic instability, mitochondrial dysfunction, de-regulated nutrient sensing, stem cell exhaustion, loss of proteostasis, epigenetic alterations, and impaired intracellular communication (Lopez-Otin et al., 2013; Lidzbarsky et al., 2018). Therefore, extensive research is currently focused on understanding natural cellular processes that attenuate cellular senescence and remove senescent cells to prevent spreading of cellular senescence, and increasing these processes to mitigate aging-associated cardiovascular diseases via life style changes and/or longevity drugs.
Anti-Senescence Treatments for Cardiovascular Aging
Pre-clinical and clinical evidence show that caloric restriction (CR) is an effective method to ameliorate cardiovascular pathologic remodeling and to improve cardiovascular function (Ravussin et al., 2015; de Lucia et al., 2018; Han et al., 2018; Hansen et al., 2018; Sandesara and Sperling, 2018). For example, in a rat model for myocardial infarction and post-ischemic heart failure, 1-year long CR mitigated pathologic left ventricular remodeling and improved cardiac function and inotropic reserve. CR also improved sympathetic cardiac innervation and β-adrenergic receptor levels (de Lucia et al., 2018). An average 11% CR for a 2-year period reduced cardiometabolic risk factors and increased predictors of health span and longevity in a healthy human clinical trial (Ravussin et al., 2015). CR-induced increase in β-hydroxybutyrate (β-HB) is implicated in preventing vascular senescence (Han et al., 2018). However, CR is a life style change and it is difficult to achieve patient compliance.
One of the underlying mechanisms for the anti-aging effect of CR is induction of autophagy, a process that removes senescent cells from tissues and thus prevent spreading of cellular senescence. It is now well established that autophagy is a converging point for the beneficial effects of longevity drugs such as rapamycin, other rapalogs, metformin, and resveratrol (Schiattarella and Hill, 2016; Hansen et al., 2018; Tian et al., 2019). Optimal levels of autophagy is an evolutionarily-conserved intracellular catabolism process essential to preserve cellular homeostasis in response to the same or similar stressors that induce cellular senescence. Therefore, therapeutic targeting of autophagy can be an effective approach to mitigate cardiovascular diseases (Schiattarella and Hill, 2016). In particular, cardiomyopathy caused by diabetes involves extensive deregulation of cardiac mitochondrial function and induction of mitochondrial autophagy (mitophagy) that may start as a survival mechanism, but can cause cell death when excessive (Lorenzo et al., 2013). Moreover, high glucose causes excessive mitochondrial fission that results in reactive oxygen bursts, increase in mitochondrial permeability transition pore (mPTP) opening that leads to apoptosis and Sirtuin 1 (Sirt1) inhibits process (Qin et al., 2019). Rapamycin is the first and only FDA-approved longevity drug. Metformin is an emerging longevity drug that was originally developed as an anti-glycemic drug. Resveratrol and aspirin are also drugs that are shown to have anti-aging properties. A common mode of action for all these drugs as well as CR involves inhibition of mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) and activation of AMP-dependent kinase (AMPK) and Sirt1 that result in the activation of autophagy process (Rubinsztein et al., 2011; Gelino et al., 2016; Golbidi et al., 2017). The mTOR can form either mTORC1 by partnering with Raptor that activates p70S6Kinase (s6K1) or mTOR Complex 2 (mTORC2) by partnering with Rictor in a Sin1 dependent manner that activates Akt (Saxton and Sabatini, 2017). Importantly, loss of mTORC2 enhances autophagy/mitophagy (Aspernig et al., 2019).
Autophagy Regulation and Dysregulation in CVD
Autophagy encompasses highly regulated cellular processes to maintain cellular homeostasis and proteostasis, and eliminates potentially harmful cellular stressors that induce cell death (Levine, 2005; Fernandez et al., 2018; Li et al., 2018). The highly-conserved autophagy machinery forms double-membraned autophagosomes to sequester portions of the cytoplasm and organelles, and trafficks these autophagosomes to lysosomes for degradation (Levine and Klionsky, 2004; Gatica et al., 2015; Miyamoto, 2019). Various forms of autophagy including macroautophagy, microautophagy, and chaperone-mediated autophagy all lead to turnover of intracellular components. Autophagy starts with the formation of phagophore comprised of several autophagy-related gene products (Atg1–Atg12) and other proteins that are organized in at least five molecular components: (1) the Atg1/unc-51-like kinase (ULK) complex; (2) the Beclin 1/class III phosphatidylinositol 3-kinase (PI3K) complex; (3) Atg9 and vacuole membrane protein 1 (VMP1); (4) two ubiquitin-like protein (Atg12 and Atg8/LC3) conjugation systems; and (5) proteins that mediate fusion between autophagosomes and lysosomes (Kroemer et al., 2010). Detailed mechanisms of autophagy process have been described in other reviews and not repeated here (Kroemer et al., 2010; Wang et al., 2019). While autophagy is a catabolic process that degrades damaged organelles, misfolded proteins and other harmful stressors, it also generates new building blocks (for example amino acids), energy for anabolism in conditions of nutrient deprivation, and promotes self-renewal and differentiation of pluripotent stem cells which is essential for repair of damaged tissue (Chen et al., 2018). Autophagy dysregulation tilts the balance from autophagy being the protective mechanism to exerting detrimental effects on cells leading to apoptosis, to whole-organ dysfunction, and organismal demise. Therefore, better understanding of the underlying molecular mechanisms of therapeutic induction of autophagy is of utmost importance, and the levels of autophagy need to be carefully monitored.
While basal level of autophagy is required to maintain normal cardiac function, autophagy becomes downregulated in the heart during the course of aging (Taneike et al., 2010). Age-induced impairment of autophagy further contributes to cardiac diseases (Madeo et al., 2015). For instance, mutations in ATG4C gene is associated with increased risk of heart disease in elderly patients and eventual death (Walter et al., 2011). Cardiomyocytes show age-related changes in the proteostasis pathways, resulting in calcium homeostasis impairment, reactive oxygen species induction, hypertrophy and fibrosis, and eventual structure damage and diminished cardiac function (Klionsky et al., 2016; Li et al., 2018). Cardiomyocyte-specific deletion of GSK-3α in mice accelerated cardiac aging and reduced basal autophagy levels (Zhou and Force, 2013; Zhou et al., 2013). Impaired autophagy with age slows the turnover of damaged proteasomes and contribute to age-associated CVDs and cardiomyocyte senescence (Korolchuk et al., 2009). Degradation of mitochondria by autophagy (mitophagy) is impaired in aged mice, and the induction of mitophagy improved mitochondrial function and reduced arterial wall stiffness (LaRocca et al., 2014; Palikaras et al., 2015).
Life-extending CR intervention modulates autophagy through multiple upstream regulators of autophagy, including Sirt1, AMPK, and mTOR (Rubinsztein et al., 2011; Gelino et al., 2016; Golbidi et al., 2017; Han et al., 2018; Hansen et al., 2018). The AMPK activator metformin has been shown to attenuate cardiomyocyte contractile defects in an aging-induced myocardial contractile dysfunction model (Burkewitz et al., 2016; Garg et al., 2017). Direct suppression of mTOR Complex 1 (mTORC1) via administration of rapamycin inhibits the adverse effects of aging, increases lifespan, and promotes autophagy in the heart as well as in many other cell types and organs, even when autophagy was suppressed by aging (Harrison et al., 2009; Bjedov et al., 2010). Supplementation with spermidine, a natural polyamine, has shown cardiac protective effects and lifespan extension by enhancing autophagy in aging-related skeletal muscle atrophy in young and old mice (Eisenberg et al., 2016; Fan et al., 2017).
Above studies are testimonials for efficacy of longevity drugs and indicate that suppression of autophagy contributes to cellular senescence and enhancing autophagy renders anti-senescence effects and mitigate cardiovascular aging. However, emerging evidence indicate that CVD-promoting pathologies such as glucotoxicity and lipotoxicity that accelerate cardiovascular aging actually increase autophagy in cardiac tissue and cause cardiac damage. Diabetes causes progressive cardiomyopathy and excessive autophagy in human heart as evidenced by increase in the levels of autophagy marker LC3B-II and its mediator Beclin-1 and decreased expression of p62, which incorporates into autophagosomes to be efficiently degraded (Munasinghe et al., 2016). Moreover, palmitic acid-induced insulin resistance is accompanied by excessive autophagy that causes cardiomyocyte death in rat H9c2 cardiomyoblast model system (Li et al., 2017). Moreover, during severe ischemia, induction of excessive autophagy worsens cardiomyocyte death and heart failure (Li et al., 2018; Liu et al., 2018; Xiao et al., 2018). Studies on cancer cells have shown that autophagy can have a pro-senescence effect because autophagy-induced increase in availability of amino acids can contribute to protein synthesis for SASP that spreads senescence (Young et al., 2009). The TOR-autophagy spatial coupling compartment (TASCC) responsible for the protein synthesis of some SASP factors serves as the platform for coupling autophagy and senescence since at TASCC autophagy supplies amino acids to activate mTOR which is involved in the synthesis of interleukins 6 and 8 that are part of SASP (Narita et al., 2011).
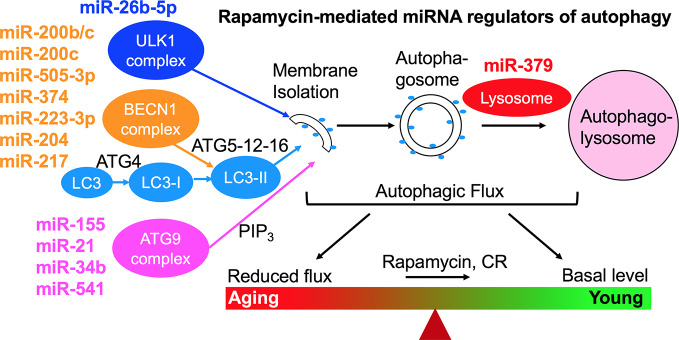
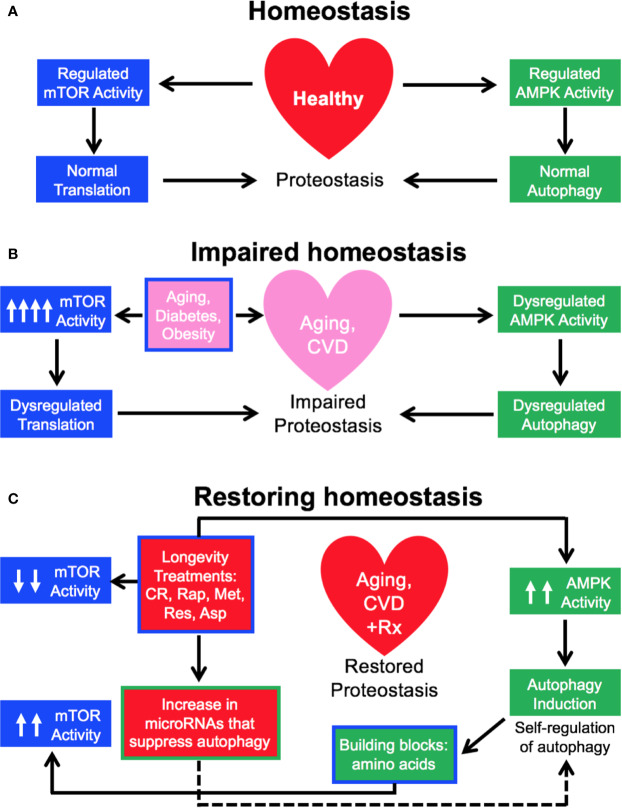
We have studied the effect of a 3-month treatment with a comparatively low dose of rapamycin (750 μg/kg/d delivered subcutaneously) on the heart tissues of male healthy Zucker lean (ZL) and obese, diabetic Zucker obese (ZO) rats that exhibit accelerated aging, and reported that while rapamycin treatment was beneficial in reducing body weight and suppressing fasting triglycerides, insulin, and uric acid in ZO rats, it also increased blood glucose levels. In ZO rats, rapamycin initially improved multiple diastolic parameters (E/E′, E′/A′, E/Vp), but by the end of treatment these beneficial effects were reversed. In ZL rats, rapamycin increased fibrosis and induced differential regulation of intracardiac cytokines (Luck et al., 2017). A cardiac microRNA transcriptome analysis (Narita et al., 2011) showed that 40 differentially expressed miRNAs by minimum 1.5-fold in saline treated ZO rats (ZO-C) versus saline treated ZL rats (ZL-C; p ≤ 0.05) were identical to the miRNAs that were differentially expressed between ZL-C and rapamycin-treated ZL (ZL-Rap) rats (Belenchia et al., 2018). Interestingly, most of these miRNAs that exhibited increased expression in both rapamycin-treated ZL rats and saline treated diabetic ZO rats are established suppressors of autophagy (Table 1). Since these miRNAs are increased in rapamycin-treated healthy ZL rats, induction of such a cardiac miRNA network indicates a pan-post-transcriptional regulation of autophagy promoting genes after induction of autophagy by an established autophagy inducer and longevity drug such as rapamycin. This can be a cardiac adaptive mechanism to modulate excessive autophagy that causes cardiomyopathy in response to continuous mTORC1 inhibition by 3-month rapamycin treatment. Alternatively, since these miRNAs (Table 1) show increased expression in saline-treated, obese and diabetic ZO rats compared to their age-matched healthy ZL rats, activation of this autophagy-inhibiting cardiac miRNA network could be a cardiac mechanism underlying the increasing cellular senescence induced by diabetes. Figure 1 shows the roles of autophagy regulating proteins in the autophagy machinery that are directly affected by cardiac miRNAs that are modulated similarly by Rapamycin treatment and diabetes. The fact that autophagy-inhibiting cardiac miRNA network (Figure 1) is activated in two independent cardiac autophagy-inducing conditions, namely diabetes and rapamycin treatment, suggest that induction of such a cardiac miRNA network is an inherent part of regulation of cardiac autophagy. Regulation of cardiac autophagy by miRNAs has been described in other CVDS as well (Sun et al., 2018), Figure 2 shows the complex interplay of mTOR and AMPK in health and disease that modulate autophagy, and the down-stream effects of autophagy or autophagy-inducing drugs (rapamycin) that further modulate autophagy. Specifically, autophagy generates amino acids that re-activate mTOR signaling whereas autophagy-regulating miRNAs suppress progression of autophagy (Figure 2). Induction of autophagy by either longevity promoting pharmacological treatment (rapamycin) or pathology (diabetes) in the heart is thus intrinsically opposed by other post-transcriptional mechanisms such as increase in thirteen autophagy-inhibiting microRNAs (Table 1). Only three of the miRNAs in this group were enhancers of autophagy. This observation highlights the fact that although rapamycin, a drug that is considered as CR mimetic, is an activator of autophagy (consistent with the increase in autophagy enhancing miRNAs as shown in Table 1), the exact progression of autophagy is highly regulated by post-transcriptional mechanisms such as autophagy-inhibiting cardiac miRNA networks (Table 1 and Figure 2). Since autophagy induction can be either protective or detrimental to cardiovascular tissues, effective methods to measure autophagic flux during disease progression in a temporal manner is critical.
Table 1.
Rapamycin-induced cardiac miRNAs in 19-wk old healthy rats that modulate autophagy.
| MicroRNA | Fold change in Rap-treated healthy rat heart | Effect on autophagy | Model system | Mechanism | Reference |
|---|---|---|---|---|---|
| miR-155 | 4.56 | Enhancer | Human umbilical vein endothelial cells; cervical cancer; human and mouse septic lung injury | Inhibition of Pdk1 or growth factor-β-activated kinase-1-binding protein 2 (Tab2) | (Liu et al., 2017; Zhang et al., 2017; Wang F. et al., 2018) |
| miR-200b/c | 4.99/2.83 | Suppressor | Suppressor in Human lung adenocarcinoma and breast cancer cells. | Suppression via inhibition of Atg12 conjugation. Other targets are ZEB1 and UBQLN1 | (Pan et al., 2015; Sun et al., 2015) |
| miR-200c | 2.83 | Enhancer | PC-3 cells | Increases microtubule-associated protein 1A/1B-LC3-II via endoplasmic reticulum stress mediator IRE1α | (Sohn, 2018) |
| miR-21 | 5.28 | Suppressor | NRK-52E cells, rat ischemia/reperfusion model, nucleus pulposus cells, non-small cell lung cancer cells | Inhibits Ras-related protein Rab11A and PTEN | (Liu et al., 2013; Liu et al., 2015; Wang W. et al., 2018) |
| miR-26b-5p | 5.13 | Suppressor | Rat model for exercise induced left ventricular hypertrophy and rat H9c2 cardiomyoblasts | Inhibits ULK1 | (Qi et al., 2020) |
| miR-411 | 7.16 | Suppressor | Chondrocytes | Inhibits HIF-1α | (Yang et al., 2020) |
| miR-301a | 4.86 | Suppressor | Prostate cancer cells in hypoxia | Inhibits NDRG2 | (Wang et al., 2016) |
| miR-505-3p | 4.92 | Suppressor | Mouse cortical neurons, mouse model for axonal development | Inhibits autophagy-related gene Atg12 | (Yang et al., 2017; Sun et al., 2018) |
| miR-140 | 4.00 | Enhancer | Human primary chondrocytes | Inhibits FUT1 | (Wang Z. et al., 2018) |
| miR-374 | 5.94 | Suppressor | HEK293T cells, Squamous cell carcinoma | Inhibits UV radiation resistance-associated gene UVRAG and autophagy-related gene ATG5 | (Huang et al., 2012; Jing et al., 2015) |
| miR-223-3p | 3.56 | Suppressor | Experimental autoimmune encephalomyelitis mouse model, bone marrow derived macrophages, BV2 microglial cells, hepatocellular carcinoma | Inhibits ATG16L1 and FOXO3 | (Li et al., 2019; Zhou et al., 2019) |
| miR-204 | 7.01 | Suppressor | Rat ischemia-reperfusion model | Inhibits microtubule-associated protein 1A/1B-LC3-II | (Jegga et al., 2011; Xiao et al., 2011) |
| miR-34b | 3.29 | Suppressor | Caenorhabditis elegans | Inhibits autophagy-related Atg9 | (Yang et al., 2013) |
| miR-217 | 2.98 | Suppressor | Mouse glomerular mesangial cells | Targets Atg1, LC3, and Becn1 | (Deshpande et al., 2018) |
| miR-541 | 3.18 | Suppressor | human hepatocellular carcinoma | ATG2A and Ras-related protein RAB1B | (Xu et al., 2019) |
| miR-379 | 2.91 | Suppressor | SHSY5Y cells | Lysosome-associated membrane protein 2A | (Alvarez-Erviti et al., 2013) |
Pdk1, phosphoinositide-dependent kinase-1; Atg12, autophagy-related gene 12; LC3, light chain 3; PTEN, phosphatase and tensin homolog; ULK1, Unc-51 like autophagy activating kinase 1; HIF-1α, hypoxia-inducible factor 1 alpha; NDRG2, N-myc downstream-regulated gene 2; FUT1, fucosyltransferase 1; ATG16L1, autophagy-related 16 like gene; FOXO3, Forkhead Box O3; RAB1B, Rab-1B.
Figure 1.
Rapamycin-induced cardiac miRNA regulation of autophagy machinery. Since these same miRNAs are also differentially expressed in response to diabetes, this autophagy-inhibiting miRNA network is an inherent cardiac mechanism in response to conditions that induce activation of autophagy. This autophagy-inhibiting miRNA network may be an adaptive mechanism to prevent excessive autophagy that leads to cardiomyopathy and heart failure, or a detrimental mechanism that prevents autophagy progression and promotes cardiac senescence. It is important to note that careful monitoring of this autophagy regulation by determining changes in cardiac autophagic flux is a critical step in evaluating whether a longevity drug- or life style-induced increase in induction of autophagy is progressing appropriately to protect the heart tissue. New non-invasive imaging approaches need to be developed to couple cardiac autophagy progression with cardiac function.
Figure 2.
Tight Regulation of cardiac autophagy—inherent mechanisms and pathways. (A, B) In healthy individuals, tightly regulated activity of mTOR and AMP kinase (AMPK) maintains normal proteostasis, however, factors that accelerate cardiovascular aging also dysregulates mTOR and AMPK activity and autophagy. (C) Longevity treatments induce autophagy in the heart via initial inhibition of mTOR and activation of AMPK. However, building blocks such as amino acids generated by autophagy can reactivate mTOR, whereas an increase in microRNAs that inhibit various component proteins of autophagy machinery (shown in Figure 1) can inhibit further progression of autophagy in the heart.
Autophagy Detection and Imaging Technology
Autophagic flux, the dynamic process of autophagy, refers to the autophagophore membrane isolation, autophagosome formation/maturation, and fusion with lysosome to form autophagolysosomes (Gottlieb et al., 2015; Klionsky et al., 2016). Fluorescent LC3 fusion reporters are well suited to track autophagic flux and LC3-mediated proteolysis (Kabeya et al., 2000; Kimura et al., 2007). In mCherry-LC3 transgenic mice expressing the fluorescent LC3 under the alpha myosin heavy chain promoter, in vivo imaging was performed by planar imaging at 4 h after the co-injection of rapamycin (autophagy induction) and chloroquine (blocked flux). The authors reported a 23% increase in cardiac fluorescence signal compared to the baseline levels prior to rapamycin/chloroquine injection (Gottlieb et al., 2015). Autophagic flux impairment is increasingly linked to aging-associated neurodegenerative disorders (Zhang et al., 2013). GFP-LC3 transgenic mice expressing LC3 systemically, previously shown to track autophagy activation in the brain after transient middle cerebral artery occlusion (Tian et al., 2010), are well suited for intravital microscopy imaging at single-neuron resolution. GFP-LC3 punctate was seen accumulated in neurons in the ischemic hemisphere, consistent with an increase in apoptosis. GFP-LC3 overexpression did not impact neuronal aggregate clearance thus potentially allowing proteostasis during aging to be studied in vivo. Autophagy readout can be further combined with functional medical imaging of cerebral blood flow, a key to maintaining increased oxygen and glucose supply during healthy aging (Jahrling et al., 2015), during lifespan extension therapy such as rapamycin.
The translation of drug discovery from murine models to humans has been challenging, due to the lack of animal models that completely recapitulate the onset or the progression of CVDs. For instance, heart failure in murine models occurs acutely after surgical or pharmacological induction, whereas in human patients develops chronically over years (Strait and Lakatta, 2012; Valero-Munoz et al., 2017). The ability to carefully characterize systolic and diastolic dysfunction coupled with non-invasive imaging of autophagic flux in vivo thus warrants further investigation. A nanoparticle approach in apamycin-induced autophagic cardiomyocytes showed robust uptake of fluorescently labeled iron oxide Feraheme nanoparticle, due to enhanced endocytosis (Chen et al., 2013; Galluzzi and Green, 2019). Feraheme is FDA-approved thus with well-established biosafety profile in both animals and humans, possess superparamagnetic properties suitable for magnetic resonance imaging, and is amendable for further contrast agent development (Bashir et al., 2015; Yuan et al., 2018).
In conclusion, longevity treatments (pharmacological or lifestyle changes) may activate miRNA networks that can regulate progression of autophagy in the heart either as an adaptive mechanism to regulate excessive autophagy or a method for spreading of senescence. Temporal visualization of cardiac autophagy in the context of heart function is needed to ensure safety of longevity treatments.
Author Contributions
LP and HC wrote and edited manuscript and approved final version.
Funding
This work was supported by NIH NHLBI 1R01HL138988-01A1 (LP) and NIH K99/R00HL121152 (HC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alvarez-Erviti L., Seow Y., Schapira A. H., Rodriguez-Oroz M. C., Obeso J. A., Cooper J. M. (2013). Influence of microRNA deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in Parkinson's disease. Cell Death Dis. 4, e545. 10.1038/cddis.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelman Y., van Rijn B. B., Ten Haaf M. E., Boersma E., Peters S. A. (2015). Sex differences in cardiovascular risk factors and disease prevention. Atherosclerosis 241 (1), 211–218. 10.1016/j.atherosclerosis.2015.01.027 [DOI] [PubMed] [Google Scholar]
- Aspernig H., Heimbucher T., Qi W., Gangurde D., Curic S., Yan Y., et al. (2019). Mitochondrial Perturbations Couple mTORC2 to Autophagy in C. Elegans. Cell Rep. 29 1399-1409 (6), e1395. 10.1016/j.celrep.2019.09.072 [DOI] [PubMed] [Google Scholar]
- Bashir M. R., Bhatti L., Marin D., Nelson R. C. (2015). Emerging applications for ferumoxytol as a contrast agent in MRI. J. Magn. Reson. Imaging 41 (4), 884–898. 10.1002/jmri.24691 [DOI] [PubMed] [Google Scholar]
- Belenchia A. M., Gavini M. P., Toedebusch R. G., DeMarco V. G., Pulakat L. (2018). Comparison of Cardiac miRNA Transcriptomes Induced by Diabetes and Rapamycin Treatment and Identification of a Rapamycin-Associated Cardiac MicroRNA Signature. Oxid. Med. Cell Longev. 2018, 8364608. 10.1155/2018/8364608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I., Toivonen J. M., Kerr F., Slack C., Jacobson J., Foley A., et al. (2010). Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11 (1), 35–46. 10.1016/j.cmet.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K., Weir H. J., Mair W. B. (2016). AMPK as a Pro-longevity Target. Exp. Suppl. 107, 227–256. 10.1007/978-3-319-43589-3_10 [DOI] [PubMed] [Google Scholar]
- Chen H. H., Mekkaoui C., Cho H., Ngoy S., Marinelli B., Waterman P., et al. (2013). Fluorescence tomography of rapamycin-induced autophagy and cardioprotection in vivo. Circ. Cardiovasc. Imaging 6 (3), 441–447. 10.1161/CIRCIMAGING.112.000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., He Y., Lu F. (2018). Autophagy in Stem Cell Biology: A Perspective on Stem Cell Self-Renewal and Differentiation. Stem Cells Int. 2018, 9131397. 10.1155/2018/9131397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs B. G., Li H., van Deursen J. M. (2018). Senescent cells: a therapeutic target for cardiovascular disease. J. Clin. Invest. 128 (4), 1217–1228. 10.1172/JCI95146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavitti R., Finkel T. (2005). Reactive oxygen species as mediators of cellular senescence. IUBMB Life 57 (4-5), 277–281. 10.1080/15216540500091890 [DOI] [PubMed] [Google Scholar]
- de Lucia C., Gambino G., Petraglia L., Elia A., Komici K., Femminella G. D., et al. (2018). Long-Term Caloric Restriction Improves Cardiac Function, Remodeling, Adrenergic Responsiveness, and Sympathetic Innervation in a Model of Postischemic Heart Failure. Circ. Heart Fail 11 (3), e004153. 10.1161/CIRCHEARTFAILURE.117.004153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S., Abdollahi M., Wang M., Lanting L., Kato M., Natarajan R. (2018). Reduced Autophagy by a microRNA-mediated Signaling Cascade in Diabetes-induced Renal Glomerular Hypertrophy. Sci. Rep. 8 (1), 6954. 10.1038/s41598-018-25295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T., Abdellatif M., Schroeder S., Primessnig U., Stekovic S., Pendl T., et al. (2016). Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 22 (12), 1428–1438. 10.1038/nm.4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Yang X., Li J., Shu Z., Dai J., Liu X., et al. (2017). Spermidine coupled with exercise rescues skeletal muscle atrophy from D-gal-induced aging rats through enhanced autophagy and reduced apoptosis via AMPK-FOXO3a signal pathway. Oncotarget 8 (11), 17475–17490. 10.18632/oncotarget.15728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. F., Sebti S., Wei Y., Zou Z., Shi M., McMillan K. L., et al. (2018). Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558 (7708), 136–140. 10.1038/s41586-018-0162-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Green D. R. (2019). Autophagy-Independent Functions of the Autophagy Machinery. Cell 177 (7), 1682–1699. 10.1016/j.cell.2019.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G., Singh S., Singh A. K., Rizvi S. I. (2017). Antiaging Effect of Metformin on Brain in Naturally Aged and Accelerated Senescence Model of Rat. Rejuvenation Res. 20 (3), 173–182. 10.1089/rej.2016.1883 [DOI] [PubMed] [Google Scholar]
- Gatica D., Chiong M., Lavandero S., Klionsky D. J. (2015). Molecular mechanisms of autophagy in the cardiovascular system. Circ. Res. 116 (3), 456–467. 10.1161/CIRCRESAHA.114.303788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelino S., Chang J. T., Kumsta C., She X., Davis A., Nguyen C., et al. (2016). Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PloS Genet. 12 (7), e1006135. 10.1371/journal.pgen.1006135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebre Y. T., Yakubov E., Wong W. T., Krishnamurthy P., Sayed N., Sikora A. G., et al. (2016). Vascular Aging: Implications for Cardiovascular Disease and Therapy. Transl. Med. (Sunnyvale) 6 (4), 183. 10.4172/2161-1025.1000183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbidi S., Daiber A., Korac B., Li H., Essop M. F., Laher I. (2017). Health Benefits of Fasting and Caloric Restriction. Curr. Diabetes Rep. 17 (12), 123. 10.1007/s11892-017-0951-7 [DOI] [PubMed] [Google Scholar]
- Gottlieb R. A., Andres A. M., Sin J., Taylor D. P. (2015). Untangling autophagy measurements: all fluxed up. Circ. Res. 116 (3), 504–514. 10.1161/CIRCRESAHA.116.303787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales C. M., CD F., Ogden C. L. (2020). Prevalence of obesity and severe obesity among adults: United States, 2017–2018. NCHS Data Brief, no 360 (Hyattsville, MD: National Center for Health Statistics; ). https://www.cdc.gov/nchs/products/databriefs/db360.htm. [PubMed] [Google Scholar]
- Han Y. M., Bedarida T., Ding Y., Somba B. K., Lu Q., Wang Q., et al. (2018). beta-Hydroxybutyrate Prevents Vascular Senescence through hnRNP A1-Mediated Upregulation of Oct4. Mol. Cell 71 (6), 1064–1078 e1065. 10.1016/j.molcel.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Rubinsztein D. C., Walker D. W. (2018). Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19 (9), 579–593. 10.1038/s41580-018-0033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. E., Strong R., Sharp Z. D., Nelson J. F., Astle C. M., Flurkey K., et al. (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460 (7253), 392–395. 10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Guerrero-Preston R., Ratovitski E. A. (2012). Phospho-DeltaNp63alpha-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle 11 (6), 1247–1259. 10.4161/cc.11.6.19670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Diabetes Association (2019). IDF Diabetes Atlas, 9th edition, https://diabetesatlas.org/en/sections/demographic-and-geographic-outline.html [Accessed March 31, 2020].
- Jahrling N., Becker K., Wegenast-Braun B. M., Grathwohl S. A., Jucker M., Dodt H. U. (2015). Cerebral beta-Amyloidosis in Mice Investigated by Ultramicroscopy. PloS One 10 (5), e0125418. 10.1371/journal.pone.0125418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegga A. G., Schneider L., Ouyang X., Zhang J. (2011). Systems biology of the autophagy-lysosomal pathway. Autophagy 7 (5), 477–489. 10.4161/auto.7.5.14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K. (2019). A Microcirculatory Theory of Aging. Aging Dis. 10 (3), 676–683. 10.14336/AD.2019.0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Z., Han W., Sui X., Xie J., Pan H. (2015). Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 356 (2 Pt B), 332–338. 10.1016/j.canlet.2014.09.039 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19 (21), 5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3 (5), 452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Acevedo Arozena A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12 (1), 1–222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V. I., Menzies F. M., Rubinsztein D. C. (2009). A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 5 (6), 862–863. 10.4161/auto.8840 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Marino G., Levine B. (2010). Autophagy and the integrated stress response. Mol. Cell 40 (2), 280–293. 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca T. J., Hearon C. M., Jr., Henson G. D., Seals D. R. (2014). Mitochondrial quality control and age-associated arterial stiffening. Exp. Gerontol 58, 78–82. 10.1016/j.exger.2014.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6 (4), 463–477. 10.1016/s1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Levine B. (2005). Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120 (2), 159–162. 10.1016/j.cell.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Lewis-McDougall F. C., Ruchaya P. J., Domenjo-Vila E., Shin Teoh T., Prata L., Cottle B. J., et al. (2019). Aged-senescent cells contribute to impaired heart regeneration. Aging Cell 18 (3), e12931. 10.1111/acel.12931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li H., Yang D., Yu X., Irwin D. M., Niu G., et al. (2017). Excessive Autophagy Activation and Increased Apoptosis Are Associated with Palmitic Acid-Induced Cardiomyocyte Insulin Resistance. J. Diabetes Res. 2017, 2376893. 10.1155/2017/2376893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang D., Wiersma M., Brundel B. (2018). Role of Autophagy in Proteostasis: Friend and Foe in Cardiac Diseases. Cells 7 (12), 279. 10.3390/cells7120279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou D., Ren Y., Zhang Z., Guo X., Ma M., et al. (2019). Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy 15 (3), 478–492. 10.1080/15548627.2018.1522467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidzbarsky G., Gutman D., Shekhidem H. A., Sharvit L., Atzmon G. (2018). Genomic Instabilities, Cellular Senescence, and Aging: In Vitro, In Vivo and Aging-Like Human Syndromes. Front. Med. (Lausanne) 5, 104. 10.3389/fmed.2018.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. L., Wang H., Liu J., Wang Z. X. (2013). MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol. Cell Biochem. 372 (1-2), 35–45. 10.1007/s11010-012-1443-3 [DOI] [PubMed] [Google Scholar]
- Liu X., Hong Q., Wang Z., Yu Y., Zou X., Xu L. (2015). MiR-21 inhibits autophagy by targeting Rab11a in renal ischemia/reperfusion. Exp. Cell Res. 338 (1), 64–69. 10.1016/j.yexcr.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Liu F., Nie C., Zhao N., Wang Y., Liu Y., Li Y., et al. (2017). MiR-155 Alleviates Septic Lung Injury by Inducing Autophagy Via Inhibition of Transforming Growth Factor-beta-Activated Binding Protein 2. Shock 48 (1), 61–68. 10.1097/SHK.0000000000000839 [DOI] [PubMed] [Google Scholar]
- Liu C. Y., Zhang Y. H., Li R. B., Zhou L. Y., An T., Zhang R. C., et al. (2018). LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat. Commun. 9 (1), 29. 10.1038/s41467-017-02280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G. (2013). The hallmarks of aging. Cell 153 (6), 1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Ramirez E., Picatoste B., Egido J., Tunon J. (2013). Alteration of energy substrates and ROS production in diabetic cardiomyopathy. Mediators Inflammation 2013, 461967. 10.1155/2013/461967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck C., DeMarco V. G., Mahmood A., Gavini M. P., Pulakat L. (2017). Differential Regulation of Cardiac Function and Intracardiac Cytokines by Rapamycin in Healthy and Diabetic Rats. Oxid. Med. Cell Longev. 2017, 5724046. 10.1155/2017/5724046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Zimmermann A., Maiuri M. C., Kroemer G. (2015). Essential role for autophagy in life span extension. J. Clin. Invest. 125 (1), 85–93. 10.1172/JCI73946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui-Hirai H., Hayashi T., Yamamoto S., Ina K., Maeda M., Kotani H., et al. (2011). Dose-dependent modulatory effects of insulin on glucose-induced endothelial senescence in vitro and in vivo: a relationship between telomeres and nitric oxide. J. Pharmacol. Exp. Ther. 337 (3), 591–599. 10.1124/jpet.110.177584 [DOI] [PubMed] [Google Scholar]
- McHugh D., Gil J. (2018). Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 217 (1), 65–77. 10.1083/jcb.201708092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S. (2019). Autophagy and cardiac aging. Cell Death Differ. 26 (4), 653–664. 10.1038/s41418-019-0286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munasinghe P. E., Riu F., Dixit P., Edamatsu M., Saxena P., Hamer N. S., et al. (2016). Type-2 diabetes increases autophagy in the human heart through promotion of Beclin-1 mediated pathway. Int. J. Cardiol. 202, 13–20. 10.1016/j.ijcard.2015.08.111 [DOI] [PubMed] [Google Scholar]
- Narita M., Young A. R., Arakawa S., Samarajiwa S. A., Nakashima T., Yoshida S., et al. (2011). Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332 (6032), 966–970. 10.1126/science.1205407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Diabetes Statistics Report (2020). https://www.cdc.gov/diabetes/library/features/diabetes-stat-report.html [Accessed March 31, 2020].
- Noale M., Limongi F., Maggi S. (2020). Epidemiology of Cardiovascular Diseases in the Elderly. Adv. Exp. Med. Biol. 1216, 29–38. 10.1007/978-3-030-33330-0_4 [DOI] [PubMed] [Google Scholar]
- Olivieri F., Prattichizzo F., Grillari J., Balistreri C. R. (2018). Cellular Senescence and Inflammaging in Age-Related Diseases. Mediators Inflammation 2018, 9076485. 10.1155/2018/9076485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K., Lionaki E., Tavernarakis N. (2015). Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521 (7553), 525–528. 10.1038/nature14300 [DOI] [PubMed] [Google Scholar]
- Palmer A. K., Tchkonia T., LeBrasseur N. K., Chini E. N., Xu M., Kirkland J. L. (2015). Cellular Senescence in Type 2 Diabetes: A Therapeutic Opportunity. Diabetes 64 (7), 2289–2298. 10.2337/db14-1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B., Feng B., Chen Y., Huang G., Wang R., Chen L., et al. (2015). MiR-200b regulates autophagy associated with chemoresistance in human lung adenocarcinoma. Oncotarget 6 (32), 32805–32820. 10.18632/oncotarget.5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Luo X., Ma Z., Zhang B., Li S., Zhang J. (2020). Downregulation of miR-26b-5p, miR-204-5p, and miR-497-3p Expression Facilitates Exercise-Induced Physiological Cardiac Hypertrophy by Augmenting Autophagy in Rats. Front. Genet. 11, 78. 10.3389/fgene.2020.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R., Zhang L., Lin D., Xiao F., Guo L. (2019). Sirt1 inhibits HG-induced endothelial injury: Role of Mff-based mitochondrial fission and Factin homeostasis-mediated cellular migration. Int. J. Mol. Med. 44 (1), 89–102. 10.3892/ijmm.2019.4185 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ravussin E., Redman L. M., Rochon J., Das S. K., Fontana L., Kraus W. E., et al. (2015). A 2-Year Randomized Controlled Trial of Human Caloric Restriction: Feasibility and Effects on Predictors of Health Span and Longevity. J. Gerontol A Biol. Sci. Med. Sci. 70 (9), 1097–1104. 10.1093/gerona/glv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. L., Jones J., Bolleddu S. I., Vanthenapalli S., Rodgers L. E., Shah K., et al. (2019). Cardiovascular Risks Associated with Gender and Aging. J. Cardiovasc. Dev. Dis. 6 (2), 19. 10.3390/jcdd6020019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth G. A., Forouzanfar M. H., Moran A. E., Barber R., Nguyen G., Feigin V. L., et al. (2015). Demographic and epidemiologic drivers of global cardiovascular mortality. N. Engl. J. Med. 372 (14), 1333–1341. 10.1056/NEJMoa1406656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth G. A., Johnson C. O., Abate K. H., Abd-Allah F., Ahmed M., Alam K., et al. (2018). Global Burden of Cardiovascular Diseases Collaboration. The Burden of Cardiovascular Diseases Among US States 1990-2016. JAMA Cardiol. 3 (5), 375–389. 10.1001/jamacardio.2018.0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D. C., Marino G., Kroemer G. (2011). Autophagy and aging. Cell 146 (5), 682–695. 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Sandesara P. B., Sperling L. S. (2018). Caloric Restriction as a Therapeutic Approach to Heart Failure: Can Less Be More in (Mice) and Men? Circ. Heart Fail 11 (3), e004930. 10.1161/CIRCHEARTFAILURE.118.004930 [DOI] [PubMed] [Google Scholar]
- Saxton R. A., Sabatini D. M. (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 168 (6), 960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiattarella G. G., Hill J. A. (2016). Therapeutic targeting of autophagy in cardiovascular disease. J. Mol. Cell Cardiol. 95, 86–93. 10.1016/j.yjmcc.2015.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn E. J. (2018). (MicroRNA 200c-3p regulates autophagy via upregulation of endoplasmic reticulum stress in PC-3 cells). Cancer Cell Int. 18, 2. 10.1186/s12935-017-0500-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic S. D., Fiedler J., Bauersachs J., Thum T., Sedding D. G. (2020). Senescence-induced inflammation: an important player and key therapeutic target in atherosclerosis. Eur. Heart J. 0, 1–14. 10.1093/eurheartj/ehz919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strait J. B., Lakatta E. G. (2012). Aging-associated cardiovascular changes and their relationship to heart failure. Heart Fail Clin. 8 (1), 143–164. 10.1016/j.hfc.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Liu T., Yuan Y., Guo Z., Xie G., Du S., et al. (2015). MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1. Int. J. Cancer 136 (5), 1003–1012. 10.1002/ijc.29065 [DOI] [PubMed] [Google Scholar]
- Sun T., Li M. Y., Li P. F., Cao J. M. (2018). MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells 7 (8), 104. 10.3390/cells7080104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneike M., Yamaguchi O., Nakai A., Hikoso S., Takeda T., Mizote I., et al. (2010). Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 6 (5), 600–606. 10.4161/auto.6.5.11947 [DOI] [PubMed] [Google Scholar]
- Tchkonia T., Zhu Y., van Deursen J., Campisi J., Kirkland J. L. (2013). Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J. Clin. Invest. 123 (3), 966–972. 10.1172/JCI64098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Deguchi K., Yamashita T., Ohta Y., Morimoto N., Shang J., et al. (2010). In vivo imaging of autophagy in a mouse stroke model. Autophagy 6 (8), 1107–1114. 10.4161/auto.6.8.13427 [DOI] [PubMed] [Google Scholar]
- Tian Y., Song W., Li D., Cai L., Zhao Y. (2019). Resveratrol As A Natural Regulator Of Autophagy For Prevention And Treatment Of Cancer. Onco. Targets Ther. 12, 8601–8609. 10.2147/OTT.S213043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trayssac M., Hannun Y. A., Obeid L. M. (2018). Role of sphingolipids in senescence: implication in aging and age-related diseases. J. Clin. Invest. 128 (7), 2702–2712. 10.1172/JCI97949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungvari Z., Tarantini S., Kiss T., Wren J. D., Giles C. B., Griffin C. T., et al. (2018). Endothelial dysfunction and angiogenesis impairment in the ageing vasculature. Nat. Rev. Cardiol. 15 (9), 555–565. 10.1038/s41569-018-0030-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations (2019). Department of Economic and Social Affairs, Population Division, World Population Ageing 2019: Highlights (ST/ESA/SER.A/430).
- Upadhya B., Kitzman D. W. (2017). Heart Failure with Preserved Ejection Fraction in Older Adults. Heart Fail Clin. 13 (3), 485–502. 10.1016/j.hfc.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero-Munoz M., Backman W., Sam F. (2017). Murine Models of Heart Failure with Preserved Ejection Fraction: a “Fishing Expedition”. JACC Basic Transl. Sci. 2 (6), 770–789. 10.1016/j.jacbts.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. U. G., Dimmeler S. (2020). Cellular cross-talks in the diseased and aging heart. J. Mol. Cell Cardiol. 138, 136–146. 10.1016/j.yjmcc.2019.11.152 [DOI] [PubMed] [Google Scholar]
- Walter S., Atzmon G., Demerath E. W., Garcia M. E., Kaplan R. C., Kumari M., et al. (2011). A genome-wide association study of aging. Neurobiol. Aging 32 2109 (11), e2115–e2128. 10.1016/j.neurobiolaging.2011.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Liu M., Guan Y., Wu Q. (2016). Hypoxia-Responsive Mir-301a and Mir-301b Promote Radioresistance of Prostate Cancer Cells via Downregulating NDRG2. Med. Sci. Monit. 22, 2126–2132. 10.12659/msm.896832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Shan S., Huo Y., Xie Z., Fang Y., Qi Z., et al. (2018. a). MiR-155-5p inhibits PDK1 and promotes autophagy via the mTOR pathway in cervical cancer. Int. J. Biochem. Cell Biol. 99, 91–99. 10.1016/j.biocel.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Wang W. J., Yang W., Ouyang Z. H., Xue J. B., Li X. L., Zhang J., et al. (2018. b). MiR-21 promotes ECM degradation through inhibiting autophagy via the PTEN/akt/mTOR signaling pathway in human degenerated NP cells. BioMed. Pharmacother. 99, 725–734. 10.1016/j.biopha.2018.01.154 [DOI] [PubMed] [Google Scholar]
- Wang Z., Hu J., Pan Y., Shan Y., Jiang L., Qi X., et al. (2018. c). miR-140-5p/miR-149 Affects Chondrocyte Proliferation, Apoptosis, and Autophagy by Targeting FUT1 in Osteoarthritis. Inflammation 41 (3), 959–971. 10.1007/s10753-018-0750-6 [DOI] [PubMed] [Google Scholar]
- Wang L., Ye X., Zhao T. (2019). The physiological roles of autophagy in the mammalian life cycle. Biol. Rev. Camb. Philos. Soc. 94 (2), 503–516. 10.1111/brv.12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2018). Obesity and Overweight Key Facts, https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight [Accessed March 31, 2020].
- Xiao J., Zhu X., He B., Zhang Y., Kang B., Wang Z., et al. (2011). MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J. BioMed. Sci. 18, 35. 10.1186/1423-0127-18-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Wang K., Xu Y., Hu H., Zhang N., Wang Y., et al. (2018). Transplanted Mesenchymal Stem Cells Reduce Autophagic Flux in Infarcted Hearts via the Exosomal Transfer of miR-125b. Circ. Res. 123 (5), 564–578. 10.1161/CIRCRESAHA.118.312758 [DOI] [PubMed] [Google Scholar]
- Xu W. P., Liu J. P., Feng J. F., Zhu C. P., Yang Y., Zhou W. P., et al. (2019). miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut. 0, 1–13. 10.1136/gutjnl-2019-318830 [DOI] [PubMed] [Google Scholar]
- Yang J., Chen D., He Y., Melendez A., Feng Z., Hong Q., et al. (2013). MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr) 35 (1), 11–22. 10.1007/s11357-011-9324-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Yu B., Cheng C., Cheng T., Yuan B., Li K., et al. (2017). Mir505-3p regulates axonal development via inhibiting the autophagy pathway by targeting Atg12. Autophagy 13 (10), 1679–1696. 10.1080/15548627.2017.1353841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang R., Ma H., Zhao X., Wang G. (2020). miRNA-411 Regulates Chondrocyte Autophagy in Osteoarthritis by Targeting Hypoxia-Inducible Factor 1 alpha (HIF-1alpha). Med. Sci. Monit. 26, e921155. 10.12659/MSM.921155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. R., Narita M., Ferreira M., Kirschner K., Sadaie M., Darot J. F., et al. (2009). Autophagy mediates the mitotic senescence transition. Genes Dev. 23 (7), 798–803. 10.1101/gad.519709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Wilks M. Q., Normandin M. D., El Fakhri G., Kaittanis C., Josephson L. (2018). Heat-induced radiolabeling and fluorescence labeling of Feraheme nanoparticles for PET/SPECT imaging and flow cytometry. Nat. Protoc. 13 (2), 392–412. 10.1038/nprot.2017.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. J., Chen S., Huang K. X., Le W. D. (2013). Why should autophagic flux be assessed? Acta Pharmacol. Sin. 34 (5), 595–599. 10.1038/aps.2012.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Pan X., Yang S., Ma A., Wang K., Wang Y., et al. (2017). miR-155 Promotes ox-LDL-Induced Autophagy in Human Umbilical Vein Endothelial Cells. Mediators Inflammation 2017, 9174801. 10.1155/2017/9174801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Force T. (2013). Focusing the spotlight on GSK-3 in aging. Aging (Albany NY) 5 (6), 388–389. 10.18632/aging.100568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Freeman T. A., Ahmad F., Shang X., Mangano E., Gao E., et al. (2013). GSK-3alpha is a central regulator of age-related pathologies in mice. J. Clin. Invest. 123 (4), 1821–1832. 10.1172/JCI64398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Chen E., Tang Y., Mao J., Shen J., Zheng X., et al. (2019). miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 10 (11), 843. 10.1038/s41419-019-2053-8 [DOI] [PMC free article] [PubMed] [Google Scholar]