Abstract
Emerging infectious diseases, the persistent potential for destabilising pandemics, remain a global threat leading to excessive morbidity and mortality. The current outbreak of pneumonia caused by 2019 novel coronavirus (COVID-19) illustrated difficulties in lack of effective drugs for treatment. Accurate and rapid diagnostic tools are essential for early recognition and treatment of infectious diseases, allowing timely implementation of infection control, improved clinical care and other public health measures to stop the spread of the disease. CRISPR-Cas technology speed up the development of infectious disease diagnostics with high rapid and accurate. In this review, we summarise current advance regarding diverse CRISPR-Cas systems, including CRISPR-Cas9, CRISPR-Cas12 and CRISPR-Cas13, in the development of fast, accurate and portable diagnostic tests and highlight the potential of CRISPR-Cas13 in COVID-19 Pneumonia and other emerging infectious diseases diagnosis.
Keywords: CRISPR-Cas, diagnostics, infectious diseases, COVID-19
Introduction
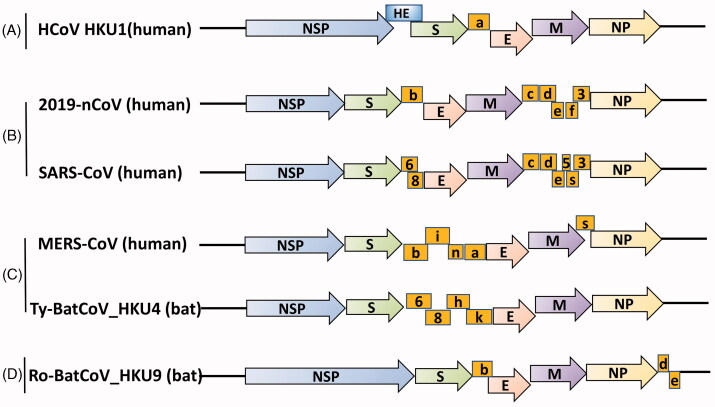
Coronaviruses are frequent causes of respiratory and neurological diseases in human, six major species are known to cause respiratory and neurological diseases in human [1]. These species include severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and less virulent species HKU1, OC43, NL63 and 229E [2]. An unknown pneumonia widely broke out in the word. Genetic sequencing of isolates obtained from patients with pneumonia identified a previously unknown coronavirus (2019-nCoV) [3], which has been recently denoted ‘COVID-19’ by WHO [4]. Based on genomic characteristics of coronavirus (Figure 1), the current diagnostic tests for coronavirus include RT‐PCR, rRT‐PCR (real‐time reverse transcription PCR), RT‐LAMP (reverse transcription loop‐mediated isothermal amplification), as well as real‐time RT‐LAMP [5–9].
Figure 1.
The genomic characteristics of coronavirus.
The epidemiology, aetiology and clinical characteristics of COVID-19 have recently been described in detail [3,10–13]. The current diagnostic of COVID-19 includes detection of virus by genomic techniques using either PCR-based method or deep sequencing [11,14,15]. Within days of Chinese researchers releasing the sequence of the virus on 11 January, scientists developed PCR-based tests to detect novel coronaviruses COVID-19 from patients [16]. Although diagnostic test kits have been approved, the low efficiency and low accuracy of these test kits makes severe situation that is difficult to keep up with the increasing number of patients. Up to April 14, 2020, over 1.92 million confirmed cases and 119000 death worldwide. However, these detection methods heavily rely on the presence of viral genome in sufficient amounts at the site of sample collection that can be amplified. Missing the time-window of viral replication can provide false negative results. Similarly, an incorrect sample collection can limit the usefulness of qPCR-based assay. A false negative diagnosis can have grave consequences, specially at this stage of the pandemic by allowing infected patients to spread the infection and hampering the efforts to contain the spread of virus [17]. Additional screening methods that can detect the presence of infection despite lower viral titres can be beneficial to ensure timely diagnosis of all infected patients. Therefore, it’s urgently needs to develop a diagnostic method with simple, quick and high accuracy to diagnose emerging pathogen infectious.
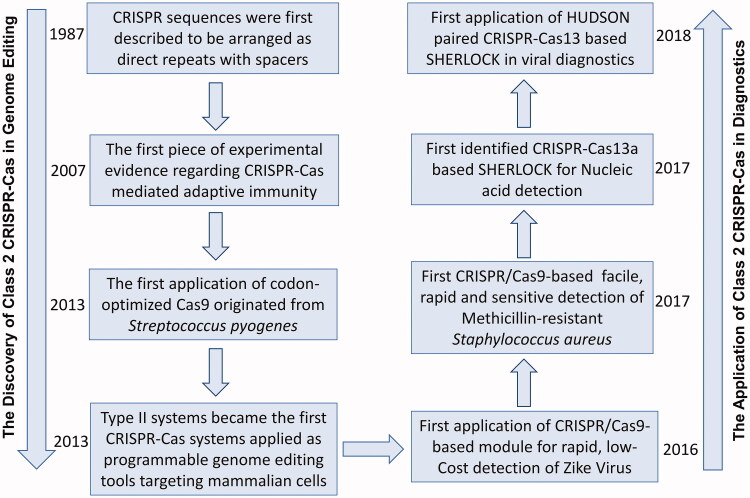
The discovery and recently advance in the biology of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated Cas proteins have led to rapidly research expansion and most recently molecular diagnostics (Figure 2). The development of CRISPR-Cas based systems for genome editing should acknowledge the efforts from different fields in the past of 30 years [18]. The CRISPR locus were first discovered in 1987 and described to be arranged as highly homologous repeats with spacers in Escherichia coli [19]. A subsequent sentinel discoveries were unveiled the functions and mechanisms of CRISPR-Cas system, setting up a new era of CRISPR-Cas mediated adaptive immunity in 2007 [20]. In 2013, the first application of CRISPR-Cas9 technology has revolutionised the field of gene editing targeting mammalian cells, accelerating the advance of novel applications in other CRISPR-Cas systems for basic sciences and clinical medicine [21,22]. The CRISPR/Cas9-based tools were firstly used to detect Zike virus in 2016 [23] and Methicillin-resistant Staphylococcus aureus in 2017 [24]. The discovery of RNA-guided, RNA-targeting CRISPR effector Cas13a [25] and subsequently founded Cas12a [26] set up a stage of CRISPR-Cas12 or/and -Cas13 based nucleic acid detection that used for clinical diagnosis [27]. Recently, CRISPR-Cas13 based SHERLOCK protocols described by Prof. Feng Zhang et al provides a rapid and accurate diagnostic assay for emerging 2019 Novel Coronavirus (COVID-19) Pneumonia. Therefore, CRISPR/Cas based technology have a good potential for application as a fast, accurate and portable diagnostic assay for emerging infectious disease.
Figure 2.
The advance of Class 2 CRISPR-Cas based genomic editing and application in diagnostics.
General aspects of CRISPR-cas systems
CRISPR-Cas systems, act as RNA-guided, DNA or RNA-targeting technique, which confer prokaryotes with heritable adaptive immunity against foreign genetic elements, including bacteriophages and plasmids from bacteria and archaea [20,28–34]. Excellent comprehensive reviews have summarised the main processes of CRISPR/Cas-mediated immunity, which include adaptation, crRNA maturation and interference [33]. Based on the composition of the interference complex, CRISPR-Cas systems are divided into Class 1 CRISPR-Cas systems (types I, III, and IV) and Class 2 CRISPR-Cas systems (II/Cas9, V/Cas12, and VI/Cas13) [34,35]. Class 1 CRISPR-Cas systems utilise crRNA together with multi-effector complex to recognise and cleave the target sequence, whereas Class 2 systems utilise single multi-domain Cas protein together with the crRNA for interference [34–37]. To date, Class 2 CRISPR-Cas systems have been widely used for genome editing and accurate and rapid diagnosis of infectious diseases [21,23,24,27]. CRISPR-Cas12a, -Cas13a and -Cas13b have been recently applied to develop practical and sensitive detection assays for human pathogens, including bacteria and virus [25–27,38].
Type II: CRISPR-Cas9 based diagnostics
Type II CRISPR-Cas9 based technology has been reported by several groups for infectious disease diagnostics [23,24,39]. Pardee et al. developed a novel method that combine CRISPR-Cas9 with an isothermal amplification technique called NASBA (nucleic acid sequence-based amplification) to differentiate ZIKV strains in single‐base discrimination [23]. The investigators exploited the (ds)DNA, an intermediate of the NASBA amplification process, which serve as substrate for the Cas9 endonuclease. sgRNA-Cas9 complex cleave the resulting dsDNA, resulting in the truncated or full-length DNA fragments formed upon Cas9 cleavage with or without a strain-specific PAM, respectively. Full-length strands but not the truncated DNA fragments triggered the toehold switch, leading to a colour change to distinguish the different strains. Guk et al. developed a method to detect methicillin‐resistant Staphylococcus aureus (MRSA) via combination CRISPR-Cas9 with FISH(DNA fluorescent in situ hybridization) [24]. In this method, dCas9/sgRNA complex targets and recognises target mecA gene, which is associated with methicillin resistance in MRSA [40]. dCas9 does not induce DNA cleavage when the dCas9/sgRNA complex recognises the target DNA sequence, which can be detected by FISH and the corresponding fluorescence intensity reflects the concentration of MRSA. It’s easy to detect MRSA at a detection concentration of 10 CFU/ml and rapid distinguish between S. aureus isolates in the presence or absence of mecA gene. However, a new mecA homologue mecALGA251 shared 70% nucleotide homology with mecA [41], which results in possible false negative results by using this method. Meanwhile, the detection of the mecA gene is not specific to MRSA, because a small proportion of methicillin‐susceptible Staphylococcus aureus (MSSA) with mecA gene and a large proportion of MRSA strains lack of mecA [42,43]. Combination CRISPR-Cas9 with optical DNA mapping was also applied to identify bacterial antibiotic resistance genes [39]. In this assay, Cas9/gRNA complex specifically recognises and cleaves nucleic acid sequences of resistance genes. Then fluorescent dye (YOYO-1) and netropsin independently and selectively binds to the resulting DNA based on AT-rich regions, resulting in a difference emission intensity allows to detect different resistance genes.
Type V: CRISPR-Cas12 based diagnostics
Identification of CRISPR-Cas12 systems (Type V) have expand the CRISPR-Cas arsenal for genomic editing [44]. Cpf1 was characterised firstly and renamed as Cas12a [45], C2c1 (Cas12b) [46] and other type V members were identified subsequently [37,47,48]. Chen et al. reported a technique named DETECTR (DNA endonuclease-targeted CRISPR trans reporter), which provides a straightforward platform for molecular diagnostics [38]. DETECTR achieves attomolar sensitivity for DNA detection by combination the activation of non-specific single-stranded deoxyribonuclease of Cas12a ssDNase with isothermal amplification that enables fast and specific detection of virus from patient samples. In this assay, crRNA-Cas12a complex binds to target DNA and induces indiscriminate cleavage of ssDNA that is coupled to a fluorescent reporter. The author used DETECTR to differentiate between different types of human papillomavirus from cultured human cells and clinical samples within 1 h [38]. In addition, another technique combines CRISPR-Cas12 with a fluorescence-based point-of-care (POC) system is recently reported for rapid and detection for accurate African Swine Fever Virus (ASFV) [49]. In this method, Cas12a/crRNA detects and binds to targeting DNA, the Cas12a/crRNA/DNA complex becomes activated and degrades a fluorescent ssDNA reporter. The author used this method to detect ASFV at a detection limit of 1 mM within 2 h. A detection limit of 100 fM can be achieved after 24 h of incubation.
Type VI: CRISPR-Cas13 based diagnostics
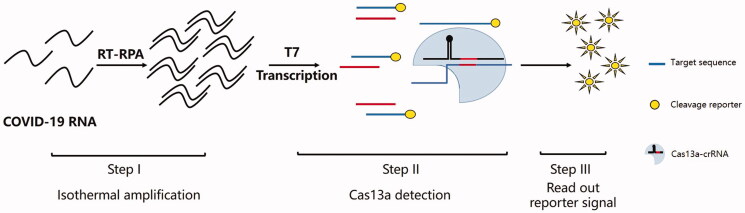
Several subtypes of Type VI CRISPR-Cas13 systems have been reported to serve as potential tools for RNA editing [50–52]. Generally, Type VI CRISPR-Cas13 systems contain a single ‘effector’ protein designated Cas13a, which combines with crRNA form a crRNA-guided RNA-targeting CRISPR effector complex to cleave ssRNA rather than DNA [34,50,53]. The first platforms based on Type VI CRISPR-Cas13 systems is termed SHERLOCK (specific high-sensitivity enzymatic reporter unlocking), which integrates isothermal RPA (recombinase polymerase amplification) or reverse transcription (RT)-RPA with nuclease activity of Cas13a [25]. A crRNA-Cas13a complex specifically binds and cleaves target nucleic acid as well as nontarget RNA coupled to a fluorescent reporter, which provides a fluorescent signal for rapid and real-time detection of pathogen even at low concentrations. Using SHERLOCK, Gootenberg et al. identified Escherichia coli and Pseudomonas aeruginosa and differentiated these bacteria with Mycobacterium tuberculosis, Klebsiella pneumoniae and Staphylococcus aureus. Clinical isolates of K. pneumoniae that possess different drug-resistant gene, including New Delhi metallo-beta-lactamase-1 resistance (NDM) or carbapenemase (KPC), can be distinguished. Moreover, the most powerful applications of SHERLOCK allowed for the discrimination of targets that differed in only a single base pair and successfully distinguished such targets as African and American strains of ZIKV, different serotypes of DENV, five healthrelated gene alleles from human saliva, and various cancer-related mutations in suspensions of cell-free DNA. Several important characteristics, including the estimated cost of the reagents and materials is less than $1 per test, a paper-based test can be used, reagents are stable and the detection can be performed within 2 h, which make it feasible for use. The revised method based on SHERLOCK, termed SHERLOCKv2, can simultaneously detect one DNA target and three ssRNA targets in a single reaction [26]. In this modified method, Cas13 together with Csm6, an auxiliary CRISPR-type III associated nuclease [54,55], which can increase about 3.5-fold signal sensitivity [26]. This method can be applied to detect specific nucleic acid with high sensitivity. It takes less than 90 min to accuratly detects Zika virus, Dengue virus and synthetic dsDNA [26]. Moreover, Myhrvold et al. developed a novel technique based on SHERLOCK together with HUDSON (heating unextracted diagnostic samples to obliterate nucleases) [27], which allows pathogen detection directly from bodily fluids of the patient without nucleic acid extraction. This method with high sensitivity detection that can identify dengue virus in the saliva, whole blood and serum of patient samples within 2 h. Recently, a protocol for detection of COVID-19 using CRISPR-based SHERLOCK described by Feng Zhang et al. (Figure 3). The author mentioned that the test can be performed starting with RNA isolated from patient samples, and can be read out using a dipstick in less than an hour. This protocol will provide reference for researchers interested in further advancing this diagnostics system for COVID-19 or other emerging infectious disease.
Figure 3.
Diagnostic applications of CRISPR-Cas13 based SHERLOCK technique for COVID-19. The protocol includes three steps: Step (I): isothermal amplification of the extracted nucleic acid sample using a commercially available recombinase polymerase amplification (RPA) kit (25 min incubation); Step (II): detection of pre-amplified viral RNA sequence using Cas13 (30 min incubation); Step (III): visual read out of the detection result by eye using a commercially-available paper dipstick (2 min incubation).
Conclusions and perspectives
The limitation in overall performance characteristics of traditional diagnostic methods makes novel molecular tools were quickly integrated into clinical use. Tremendous advances in novel methods associated with PCR and DNA sequencing allow rapidly identified the pathogen of emerging infectious diseases and facilitate timely treatment of infections. However, an ideal diagnostic reagent is characterised by rapid, reliable, inexpensive and easy-to-use. Perhaps non-specific DNA or cleavage observed in CRISPR-Cas type II (Cas9), V (Cas12), and VI (Cas13) systems provides a promising advance in CRISPR-based diagnostics for emerging infectious diseases. These three different techniques based on Class II CRISPR-Cas systems applied a combination of the CRISPR/Cas based system with other technique to achieve described applications in diagnosis. Although there is an urgent need for fast, accurate and inexpensive tests, and the possibility that the CRISPR-Cas based diagnosis may set up a novel stage in terms of the accuracy and the speed of detection. These emerging advanced techniques based diagnostic methods will require careful validation and field testing to guarantee their functionality.
Funding Statement
This work was supported by Education Fund of Chongqing Medical and Pharmaceutical College [YGZJGWT1907], The Research Plan from the Nanan Health and Family Planning Commission [Grant No. 2017-38], The General Programme of Chongqing Science and Technology Commission [cstc2018jcyjAX0667].
Acknowledgements
This paper was conceived and designed by XXH and YZF. XXH, QKL, ZZ, LFY, XY, LY and YZF wrote the paper. All authors have read and approved the manuscript.
Disclosure statement
The authors declare that there is no conflict of interests.
References
- 1.Corman VM, Eckerle I, Bleicker T, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39):20285. [DOI] [PubMed] [Google Scholar]
- 2.Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020;92(4):401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang N, Luo C, Liu H, et al. Characterization of a new member of alphacoronavirus with unique genomic features in rhinolophus bats. Viruses. 2019;11(4):379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. [DOI] [PubMed] [Google Scholar]
- 7.Bhadra S, Jiang YS, Kumar MR, et al. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV). PLoS One. 2015;10(4):e0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JF, Choi GK, Tsang AK, et al. Development and evaluation of novel real-time reverse transcription-PCR assays with locked nucleic acid probes targeting leader sequences of human-pathogenic coronaviruses. J Clin Microbiol. 2015;53(8):2722–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X, Whitaker B, Sakthivel SK, et al. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52(1):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang D, Lin M, Wei L, et al. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA. 2020;323(11):1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of Novel Coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang N, Wang L, Deng X, et al. Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92(4):408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- 19.Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. [DOI] [PubMed] [Google Scholar]
- 21.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardee K, Green AA, Takahashi MK, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165(5):1255–1266. [DOI] [PubMed] [Google Scholar]
- 24.Guk K, Keem JO, Hwang SG, et al. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens Bioelectron. 2017;95:67–71. [DOI] [PubMed] [Google Scholar]
- 25.Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–182. [DOI] [PubMed] [Google Scholar]
- 29.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Oost J, Westra ER, Jackson RN, et al. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12(7):479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. [DOI] [PubMed] [Google Scholar]
- 32.Hale CR, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sternberg SH, Richter H, Charpentier E, et al. Adaptation in CRISPR-Cas systems. Mol Cell. 2016;61(6):797–808. [DOI] [PubMed] [Google Scholar]
- 34.O’Connell MR. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR-Cas systems. J Mol Biol. 2019;431(1):66–87. [DOI] [PubMed] [Google Scholar]
- 35.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller V, Rajer F, Frykholm K, et al. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci Rep. 2016;6:37938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers HF, Hartman BJ, Tomasz A. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J Clin Invest. 1985;76(1):325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Alvarez L, Holden MT, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11(8):595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elhassan MM, Ozbak HA, Hemeg HA, et al. Absence of the mecA gene in methicillin resistant staphylococcus aureus isolated from different clinical specimens in Shendi City, Sudan. Biomed Res Int. 2015;2015:895860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imani Fooladi AA, Ashrafi E, Tazandareh SG, et al. The distribution of pathogenic and toxigenic genes among MRSA and MSSA clinical isolates. Microb Pathog. 2015;81:60–66. [DOI] [PubMed] [Google Scholar]
- 44.Lewis KM, Ke A. Building the class 2 CRISPR-Cas arsenal. Mol Cell. 2017;65(3):377–379. [DOI] [PubMed] [Google Scholar]
- 45.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L, Chen P, Wang M, et al. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol Cell. 2017;65(2):310–322. [DOI] [PubMed] [Google Scholar]
- 47.Yan WX, Hunnewell P, Alfonse LE, et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363(6422):88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burstein D, Harrington LB, Strutt SC, et al. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542(7640):237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Q, Yu D, Bao M, et al. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens Bioelectron. 2020;154:112068. [DOI] [PubMed] [Google Scholar]
- 50.East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smargon AA, Cox DBT, Pyzocha NK, et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618.e7–630.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konermann S, Lotfy P, Brideau NJ, et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665.e14–676.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niewoehner O, Jinek M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA. 2016;22(3):318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staals RH, Zhu Y, Taylor DW, et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56(4):518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]