Abstract
Fragments of mature tRNAs have long been considered as mere degradation products without physiological function. However, recent reports show that tRNA-derived small RNAs (tsRNAs) play prominent roles in diverse cellular processes across a wide spectrum of species. Contrasting the situation in other small RNA pathways the mechanisms behind these effects appear more diverse, more complex, and are generally less well understood. In addition, surprisingly little is known about the expression profiles of tsRNAs across different tissues and species. Here, we provide an initial overview of tsRNA expression in different species and tissues, revealing very high levels of 5′ tRNA halves (5′ tRHs) particularly in the primate hippocampus. We further modulated the regulation capacity of selected 5′ tRHs in human cells by transfecting synthetic tsRNA mimics (“overexpression”) or antisense-RNAs (“inhibition”) and identified differentially expressed transcripts based on RNA-seq. We then used a novel k-mer mapping approach to dissect the underlying targeting rules, suggesting that 5′ tRHs silence genes in a sequence-specific manner, while the most efficient target sites align to the mid-region of the 5′ tRH and are located within the CDS or 3′ UTR of the target. This amends previous observations that tsRNAs guide Argonaute proteins to silence their targets via a miRNA-like 5′ seed match and suggests a yet unknown mechanism of regulation. Finally, our data suggest that some 5′ tRHs that are also able to sequence-specifically stabilize mRNAs as up-regulated mRNAs are also significantly enriched for 5′ tRH target sites.
Keywords: tRNA fragments, small noncoding RNAs, gene regulation, target prediction, target identification, k-mer mapping
INTRODUCTION
tRNAs are well known for their conserved role in protein biosynthesis. However, mature tRNAs and their precursors also give rise to a class of small noncoding RNAs, the tRNA-derived small RNAs (tsRNAs). While trailer tsRNAs (15–22 nt) are generated by clipping off the trailer sequence from the tRNA precursor molecule, 5′ tsRNAs and 3′ tsRNAs (18–35 nt) stem from the respective end of mature tRNAs (70–90 nt). Primarily considered as degradation products, tsRNAs are now recognized as additional players in small RNA-mediated gene regulation that act in a variety of cellular processes across species from all domains of life.
tsRNAs were found to play a role in fundamental physiological processes such as proliferation (Lee et al. 2009) and protein translation control (Ivanov et al. 2011; Gebetsberger et al. 2017; Keam et al. 2017; Guzzi et al. 2018). Moreover, tsRNAs are implicated in defense mechanisms of Escherichia coli against bacteriophages (Levitz et al. 1990) and human cells against trypanosoma or viruses (Garcia-Silva et al. 2014; Deng et al. 2015). Additionally, tsRNAs were found to prime the reverse transcription or inhibit the promotion of retroviruses and retrotransposons (Yeung et al. 2009; Ruggero et al. 2014; Martinez et al. 2017; Schorn et al. 2017). Furthermore, tsRNAs are associated with several diseases such as cancer (Lee et al. 2009; Goodarzi et al. 2015; Huang et al. 2017) and amyotrophic lateral sclerosis (Greenway et al. 2006; Ivanov et al. 2014) and were lately revealed to act as transgenerational transmitters that induce metabolic disorders and addictive behavior in mice (Chen et al. 2016; Sharma et al. 2016; Short et al. 2017; Sarker et al. 2019). While some of the described effects seem to base on similar mechanisms, tsRNAs from the same class were shown to trigger completely converse functions in other cases (Jehn and Rosenkranz 2019), suggesting that the regulatory potential of tsRNAs is more complex than observed for the well-studied major small RNA classes.
So far, two superordinate concepts that aim to explain how tsRNAs regulate gene expression have been identified. While some studies show that tsRNAs globally repress translation by inhibiting the assembly of the translation initiation machinery (Ivanov et al. 2011; Gebetsberger et al. 2017; Keam et al. 2017; Guzzi et al. 2018), others report a sequence-specific gene regulation (Haussecker et al. 2010; Deng et al. 2015; Zhou et al. 2017; Kuscu et al. 2018; Luo et al. 2018). As tsRNAs were found to associate with Argonaute proteins, a miRNA-like gene regulation mechanism seems apparent (Maute et al. 2013; Kumar et al. 2014; Kuscu et al. 2018). Indeed, studies could show that tsRNA-mediated transcript silencing is dependent on a 5′ seed, which is complementary to target sites within the 3′ UTR (Haussecker et al. 2010; Kuscu et al. 2018). Contrasting this, other studies observed sequence-specific gene silencing effects, where complementarity of the 5′ seed was neglectable in favor of other tsRNA portions (Deng et al. 2015; Zhou et al. 2017; Luo et al. 2018). Since Argonaute structure coerces the 5′ end of small RNAs for target recognition (Boland et al. 2011), the results of these studies suggest that tsRNAs interact with other effector proteins or act protein-independent to regulate genes. Indications from recent studies where tsRNAs were shown to interact with different proteins depending on the differentiation state of the cell or the modification status of the tsRNA further erode the AGO-centric view (Guzzi et al. 2018; Krishna et al. 2019) illustrating that we still lack a sufficiently deep understanding of how tsRNAs regulate gene expression mechanistically.
Surprisingly, although it has been noted that tsRNAs are much more abundant in tissues than in cultured cells (Torres et al. 2019), their expression profile across tissues and species has not yet been investigated. We therefore initially analyzed several available and our own small RNA sequencing data sets to provide a first overview on tsRNA expression profiles. Subsequently, we tested if 5′ tRHs regulate genes by modulating the level of 5′ tRHs-Glu-CTC and 5′ tRH-Gly-GCC in cultured cells by transfecting either synthetic RNA mimics or inhibiting antisense RNAs. By RNA sequencing, in silico target predictions and a novel k-mer mapping-based approach, we then dissected the targeting rules of the respective 5′ tRHs. Finally, applying the k-mer analysis on similar RNA sequencing data sets of cultured fly cells, we examined whether the identified targeting patterns of individual tsRNAs are conserved across species.
RESULTS AND DISCUSSION
5′ tRHs are highly abundant in the hippocampus of primates
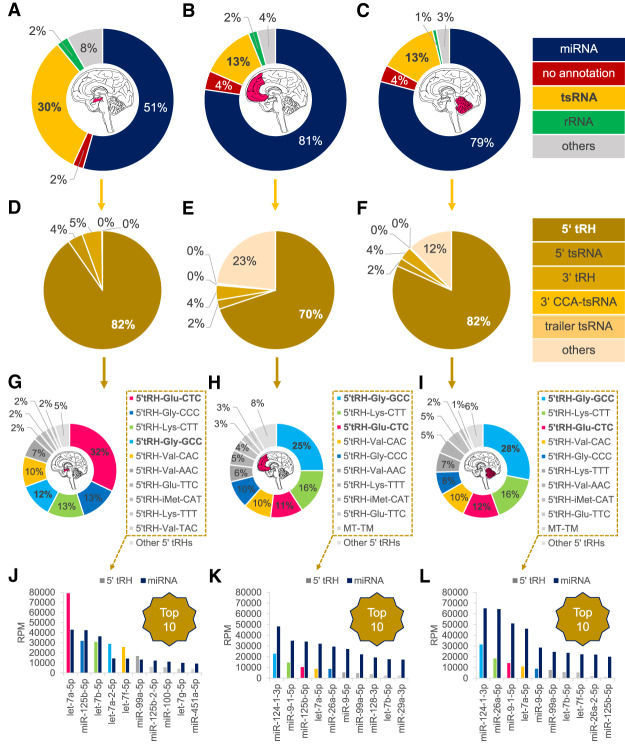
To investigate the expression levels of tsRNAs across different tissues, we annotated available human small RNA sequencing data sets complemented by our own small RNA sequencing data and found tsRNAs to be predominantly expressed in the hippocampus (Supplemental Fig. S1A). Here, 30% of all mapped reads were classified as tRNA-fragments with 82% of these tsRNA-annotated sequences being 5′ tRHs (Fig. 1A,D). According to this annotation, the most abundant 5′ tRHs have similar expression levels as the most abundant miRNAs suggesting a functional role in this brain region (Fig. 1J). In comparison, the overall tsRNA level was only 13% in both the frontal cortex and the cerebellum (Fig. 1B,C). Among the predominant tRNA-derived sequences in the hippocampus were the 5′ tRNA-halves 5′ tRH-Glu-CTC (26%) and 5′ tRH-Gly-GCC (10%) (Fig. 1D,G). Notably, these 5′ tRHs were also among the most abundant 5′ tRHs in the analyzed small RNA sequencing libraries of other tissues (Fig. 1H,I; Supplemental Fig. S1B). Both, 5′ tRH-Glu-CTC and 5′ tRH-Gly-GCC have been previously shown to play major roles in various cellular functions. They were among the sperm small RNAs that are involved in the epigenetic inheritance of paternal diet-induced metabolic disorders and addictive-like behavior in mice (Chen et al. 2016; Sharma et al. 2016; Zhang et al. 2018b; Sarker et al. 2019). Additionally, they were found among a group of 5′ tRHs to be up-regulated in cells upon infection by the respiratory syncytial virus (Wang et al. 2013). 5′ tRH-Gly-GCC was furthermore found to be part of a specific subset of 5′ tRHs that is dynamically expressed during stem cell differentiation (Krishna et al. 2019), while 5′ tRH-Glu-CTC was found to be highly expressed in human monocytes, where it triggers the transcriptional suppression of the surface glycoprotein CD1 (Zhang et al. 2016).
FIGURE 1.
Small RNA annotation of mapped reads from small RNA sequencing libraries of samples from the human hippocampus (left), frontal cortex (middle), and cerebellum (right). (A–C) Proportion of small RNA classes. (D–F) Proportion of tsRNA classes. (G–I) Proportion of 5′ tRH species. (J–L) Relative abundances of top 5′ tRH species in comparison to top miRNA species as a measure of reads per million (RPM).
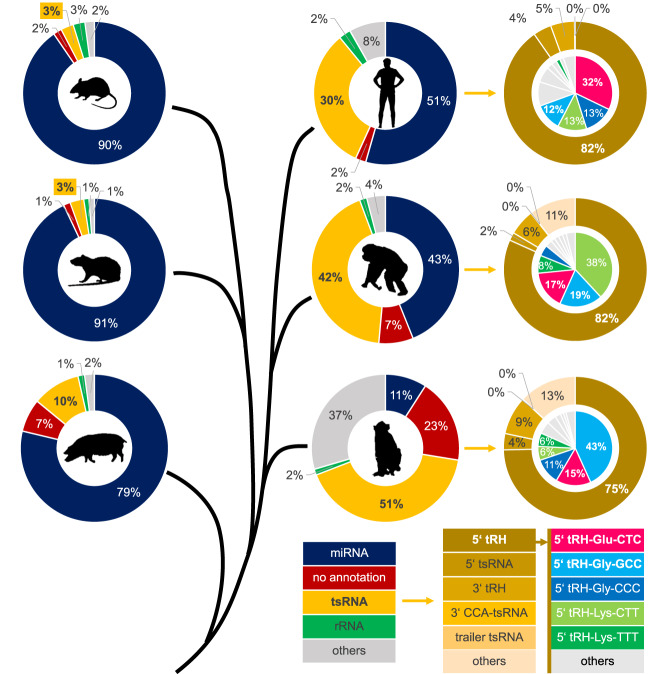
In order to investigate whether high 5′ tRH levels in the hippocampus are a common feature in mammals, we generated small RNA sequencing libraries of chimpanzee and macaque hippocampus samples, and additionally analyzed publicly available small RNA sequencing data sets from pig, rat, and mouse hippocampus. Interestingly, we found even higher proportions of reads being annotated as tsRNAs in the hippocampal libraries of the two primates (42% in the chimpanzee/51% in the macaque; Fig. 2) compared to the human hippocampus libraries (30%). Again, 5′ tRHs made up the majority of the tsRNA-annotated reads (82%/75%; Fig. 2) and 5′ tRH-Gly-GCC (15%/33%) and 5′ tRH-Glu-CTC (14%/11%) were among the most abundantly expressed tsRNAs (Fig. 2). In contrast, even though 5′ tRHs are also the predominant tsRNA class (represented mostly by 5′ tRH-Gly-GCC) in the analyzed sequencing libraries of the other mammals (Supplemental Fig. S2), only 10% of the pig and 3% of the rodent mapped reads were classified as tsRNAs (Fig. 2). This suggests that high levels of 5′ tRHs in the hippocampus are a primate-specific trait. As a subset of four 5′ tRHs that is shared between the three primate species (although with a different abundance ranking) is making up about two thirds of the reads assigned as 5′ tRHs in the hippocampus libraries (Fig. 2), this raises the question if a conserved subset of 5′ tRHs specifically fine-tunes hippocampal gene expression in primates.
FIGURE 2.
Phylogenetic tree and small RNA annotation of mapped reads from small RNA sequencing libraries of hippocampal samples from mouse, rat, pig, human, chimpanzee, and macaque.
miRNA- and piRNA-like targeting rules scarcely identify targets of the 5′ tRNA-halves Glu-CTC and Gly-GCC in HEK293T
In order to identify genes that are regulated by these 5′ tRHs, we overexpressed 5′ tRH-Glu-CTC and 5′ tRH-Gly-GCC in HEK293T cells by transfecting 50 nM synthetic 5′ tRH mimics. HEK293T cells were chosen as 5′ tRHs are barely expressed in this cell line: From the 9% of all mapped reads that were assigned as tRNA-fragments, only 1% are 5′ tRHs (Supplemental Fig. S3A). As validated by quantitative RT-PCR (qPCR), transfection of the RNA mimics successfully increased the number of 5′ tRH copies in comparison to a non-target-siRNA transfection control (Supplemental Fig. S4A).
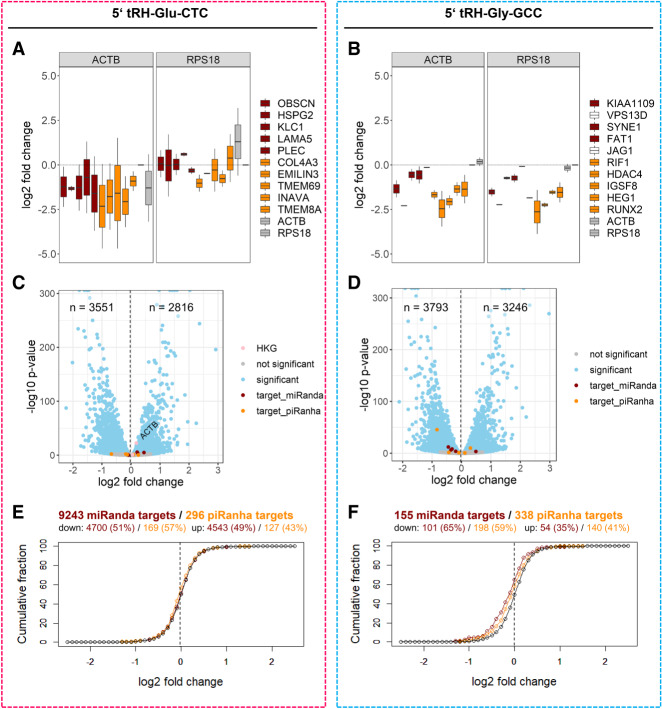
Given the lack of tsRNA target prediction algorithms, we examined whether existing target prediction algorithms designed for miRNAs or piRNAs might perform similarly well for tsRNAs. As a first test, we quantified the expression change of 20 abundantly expressed transcripts, which were predicted to represent targets of the respective tsRNAs according to miRNA (miRanda) or piRNA targeting rules, via qPCR. For piRNA-like target prediction, we developed a software named piRanha that by default applies targeting rules empirically verified by Zhang et al. (2018a). All 10 tested potential targets of 5′ tRH-Gly-GCC were lower expressed in the overexpression cells compared to the control cells (Fig. 3B). In contrast, only one miRanda- and three piRanha-predicted 5′ tRH-Glu-CTC-target transcripts were differentially expressed upon overexpression of the 5′ tRH-Glu-CTC (Fig. 3A), suggesting that target rules might differ between different tsRNAs.
FIGURE 3.
(A,B) qPCR quantified log2 fold change of potential 5′ tRH targets in 5′ tRH overexpression HEK293T cells compared to control cells. The selected transcripts were predicted by miRanda (red) and piRanha (orange) to be targets of the 5′ tRNA-halves Glu-CTC (A) or Gly-GCC (B). The housekeeping genes ACTB and RPS18 were used as normalizers. Note that normalization by ACTB is not suitable in case of the 5′ tRH-Glu-CTC overexpression (A), as the expression of ACTB is up-regulated in this condition (also revealed by RNA-seq; see C). (C,D) Volcano plot of differential expression analysis for protein-coding genes of 5′ tRH overexpression and control HEK293T cells (blue, adjusted P-value <0.01). The 10 previously tested putative target genes are highlighted in red (miRanda prediction) and orange (piRanda prediction). For the 5′ tRH-Glu-CTC analysis (C), the housekeeping genes ACTB and RPS18 are additionally highlighted in pink, showing that ACTB gets up-regulated upon 5′ tRH-Glu-CTC overexpression, while the RPS18 expression is not affected. (E,F) Cumulative fraction plots for log2-fold-change values of genes that were identified as potential targets of the 5′ tRHs Glu-CTC or Gly-GCC by miRanda (red; threshold: miRanda-score < −80) or piRanha (orange; threshold: piRanha-score < −30) and of genes not predicted as targets by the respective algorithm (black).
To get a global overview of 5′ tRH-regulated genes, we then sequenced the transcriptome of HEK293T 5′ tRH-overexpression and control cells and performed differential expression analysis. Upon overexpression of the respective 5′ tRH, more genes were significantly down-regulated than up-regulated (adjusted P-value <0.01; Fig. 3C,D). Even though most of the potential 5′ tRH-Gly-GCC targets previously quantified by qPCR were not significantly down-regulated according to the differential expression analysis, they still have the same tendency to be down-regulated apart from two (Fig. 3C,D).
When plotting the cumulative fraction of the log2 fold change values of all miRanda and piRanha predicted genes, only miRanda- and piRanha-targets of 5′ tRH-Gly-GCC were enriched for a higher degree of down-regulation over the remaining genes (Fig. 3E,F). Additionally, only in the 5′ tRH-Gly-GCC overexpression HEK293T cells more genes are down-regulated that were predicted by miRanda or piRanha compared to the pool of unpredicted genes (α < 0.001 for both algorithms; χ2 test). We therefore conclude that for 5′ tRNA-halves the targeting rules of piRNAs or miRNAs do not satisfyingly predict 5′ tRH targets in every case.
Non-miRNA-like targeting rules for 5′ tRHs
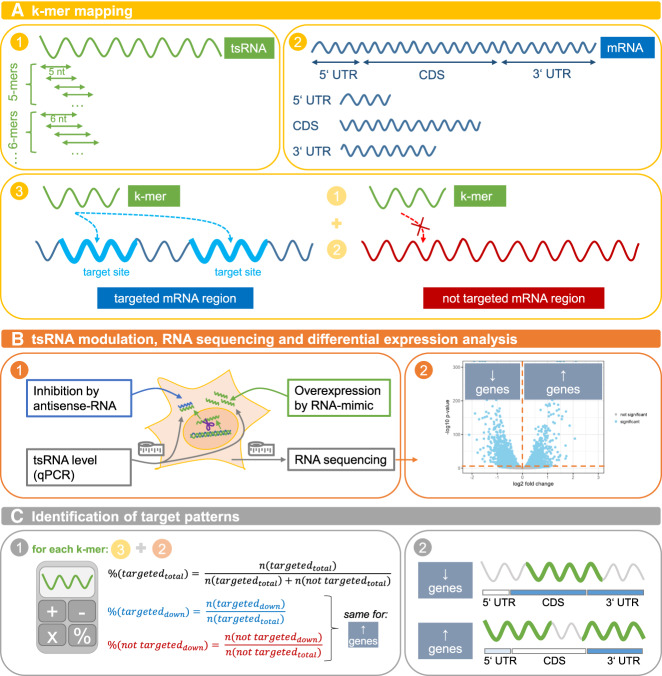
To unravel the targeting rules of 5′ tRHs, we developed a k-mer mapping approach, where k-mers of the 5′ tRH sequences with all possible lengths (k ≥ 5) and start positions within the tRH are mapped separately to the three major regions of each human transcript (5′ UTR, CDS and 3′ UTR). By calculating for each k-mer the fraction of “targeted” (k-mer alignment) or “not targeted” (no k-mer alignment) genes that get significantly down-regulated (adjusted P-value <0.01) we identify the portion within the tRH that is most likely to be important for target recognition and thus silencing. As we do this analysis for each mRNA region separately, we additionally get an indication in which transcript region effective targeting takes place. Figure 4 visualizes the underlying principles of the analysis.
FIGURE 4.
Graphical overview of identification of tsRNA targeting rules via a k-mer mapping approach.
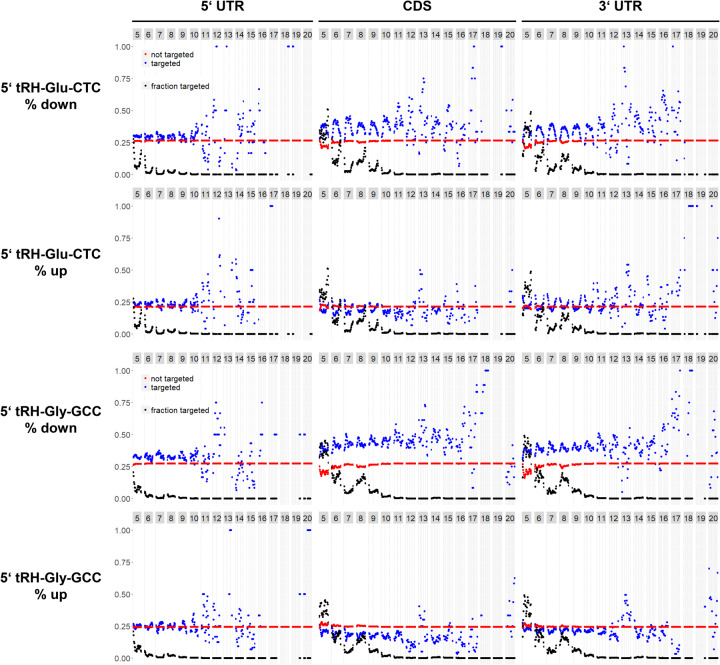
Our analysis revealed that down-regulation of transcripts with k-mer alignment (“targeted”) is significantly enriched over the number of down-regulated transcripts without k-mer alignment (“not targeted”) particularly when k-mers of 5–10 nt length with start positions in the middle of the 5′ tRH align to the 3′ UTR or the CDS of the target (Fig. 5). This suggests that 5′ tRNA-halves such as 5′ tRH-Glu-CTC and 5′ tRH-Gly-GCC, unlike miRNAs that bind to the 3′ UTR of their targets via a 7 nt long seed at the 5′ end, down-regulate transcripts mainly by binding with a 5–10 nt long stretch of their middle region to the 3′ UTR or the CDS of the target transcripts.
FIGURE 5.
k-mer analysis to elucidate the targeting rules of 5′ tRH-mediated transcript regulation in HEK293T cells. The y-axis refers to the fraction of up- or down-regulated genes. Blue dots refer to genes that have a corresponding k-mer alignment, red dots refer to genes that do not have a corresponding k-mer alignment. Black dots refer to the overall fraction of genes that align to the corresponding k-mer (declines with growing k-mer size). The numbers in the gray boxes above each plot indicate the respective k-mer length in nucleotides. That is, dots within the first column (with k-mer length = 5 nt) all refer to genes that align to k-mers with a length of 5 nt. Further, dots at the very left of a column refer to genes that align to k-mers from the very 5′ end of the parent tRH, while dots at the very right of a column refer to genes that align to k-mers from the very 3′ end of the parent tRH. Dots in between correspond to genes that align to k-mers derived from a corresponding position inside the parent tRH.
The resulting target pattern additionally suggests that 5′ tRH-Gly-GCC, but not 5′ tRH-Glu-CTC, is able to down-regulate targets by binding with its miRNA-seed like 5′ end to the transcript. This is in line with, the different target regulation behavior of 5′ tRH-Glu-CTC and 5′ tRH-Gly-GCC as revealed by qPCR quantification of predicted target transcripts and cumulative distribution analysis of the RNA sequencing data.
Interestingly, mRNA quantification by qPCR and RNA sequencing revealed that the housekeeping gene ACTB gets up-regulated upon 5′ tRH-Glu-CTC transfection (Fig. 3A,C). Unexpectedly, ACTB has several potential binding sites for 5′ tRH-Glu-CTC. Hence, we examined whether up-regulated genes might be enriched for 5′ tRHs target sites as well, which would indicate that 5′ tRH targeting might also have a protective effect. Indeed, our analysis revealed that transcripts that have target sites in their 3′ UTR for the 5′ and 3′ ends of 5′ tRH-Glu-CTC are more likely to be up-regulated than those that do not have a target site, while a corresponding up-regulating effect was not detectable for 5′ tRH-Gly-GCC (Fig. 5).
To test our k-mer mapping approach we repeated the analysis with k-mers of 5′ tRH-Glu-CTC that were randomly shuffled before they were mapped to the respective transcript regions. As shown exemplarily for the 3′ UTR, we obtain a similar enrichment for genes with alignments of these shuffled k-mers to be down-regulated (Supplemental Fig. S5B) like those observed for the original k-mer sequences (Supplemental Fig. S5A). This would suggest that the observed effects are unspecific. However, when selecting those genes that have target sites for k-mers that correspond to the proposed targeting rules (“k-mer targets,” e.g., genes with alignments in their 3′ UTR for 7-mers that start from position 10 to 16 of the 5′ tRH), only the k-mer targets of the original 7-mers are enriched to be down-regulated compared to genes that do not have such an alignment as seen in the cumulative fraction plots of log2 fold changes (Supplemental Fig. S5A.2). In contrast, the k-mer targets that were identified for randomly shuffled 7-mers (Supplemental Fig. S5B.2) as well as k-mers of the original sequence that do not follow the proposed targeting rule (e.g., 7-mers from the 5′ [start position <5] or the 3′ end [start position >23] of the 5′ tRH; Supplemental Fig. S5A.1+3) were not more likely to be down-regulated than genes that do not have such an alignment (Supplemental Fig. S5). Hence, we consider the targeting rules proposed for the two analyzed 5′ tRHs as appropriate but suggest to cross-check targeting patterns when applying the k-mer analysis on other data sets.
Concluding from the above outlined target pattern analyses we assume that tRNA-fragments, even from the same tsRNA-series, may recognize their targets via different parts of the tsRNA instead of a dominating 5′ seed match (Fig. 5). This suggests that an Argonaute-dependent regulation mechanism as proposed for tsRNAs previously (Haussecker et al. 2010; Kuscu et al. 2018) cannot fully explain the observed changes in gene expression. However, our results are in line with other studies identifying miRNA-untypical targeting (Deng et al. 2015; Zhou et al. 2017; Luo et al. 2018). Our analysis additionally suggests that some tsRNAs (e.g., 5′ tRH-Glu-CTC) trigger not only the down-regulation of target genes, but may also stabilize target transcripts (Fig. 5). This is a rather unexpected finding, since a stabilizing effect of small RNAs is rarely seen in eukaryotes, but rather a trait of prokaryotic small RNA pathways (Fröhlich and Vogel 2009). However, a recent study showed that sequence-specific binding of a 3′ tsRNA to the mRNAs of ribosomal proteins enhances the translation of the target transcript (Kim et al. 2017). It was suggested that structural changes induced by this interaction allow for higher transcription rates. Similar alterations in the secondary structure could also lead to a stabilization of the transcript.
Identification of genuine 5′ tRH targets by antisense-inhibition of 5′ tRH-lu-CTC suggests a role of 5′ tRHs in neurogenesis
Noteworthily, drawing conclusions regarding tsRNA targets only from overexpression experiments has several shortcomings. First, our synthetic RNA mimic does not carry post-transcriptional modifications as endogenous tsRNAs do. Thus, we might observe an artificial regulation behavior that does not reflect the physiological situation. Second, genes that were found to be differentially expressed upon tsRNA overexpression in cells that typically express these tsRNAs at very low levels must not necessarily be genuine targets of the modulated tsRNA under natural conditions. To circumvent these distorting effects and gain support for our conclusion, we additionally performed an inverse experiment where we inhibited the regulation capacity of 5′ tRH-Glu-CTC by transfecting an antisense RNA.
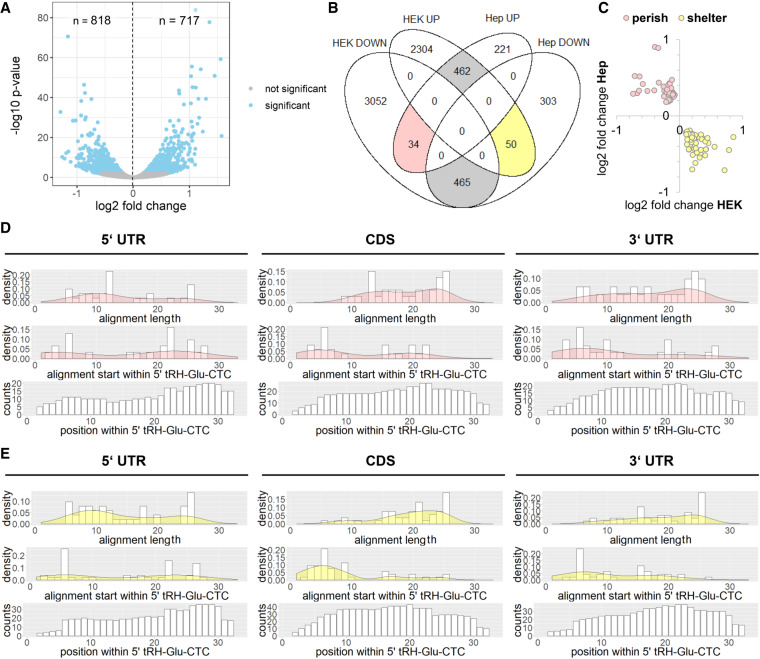
We chose to perform the experiment with HepG2 cells, in order to exclude cell line specific regulation effects. HepG2 cells have a similar overall tsRNA level compared to HEK293T cells (6% of the total reads compared to 9% in HEK293T cells; Supplemental Fig. S3B), but express a higher proportion of 5′ tRHs according to small RNA sequencing data (17% of the reads assigned to tRNAs compared to 1% in HEK293T cells; Supplemental Fig. S1B). Like in HEK293T cells, 5′ tRH-Glu-CTC and 5′ tRH-Gly-GCC are the most abundant 5′ tRHs (6% and 3% of the reads assigned to tRNAs) in the analyzed library. As quantified by qPCR, transfection of the antisense-RNA decreased the number of 5′ tRH-Glu-CTC copies in HepG2 cells by about 70% (Supplemental Fig. S4B). Even though the level of 5′ tRH-Glu-CTC was decreased, only a small amount of genes was significantly differentially expressed compared to the control state (818 down-regulated and 717 up-regulated genes, adjusted P-value <0.01; Fig. 6A). While the majority of significantly differentially expressed genes of the antisense-inhibited HepG2 was likewise regulated as in the overexpression HEK293T cells (Fig. 6B, gray areas), only a few genes were inversely regulated (Fig. 6B, rosy and lutescent area) and we assume these genes to represent the genuine targets of 5′ tRH-Glu-CTC. In the following we refer to the 34 genes that might get down-regulated by 5′ tRH-Glu-CTC as “perish targets” (Fig. 6B, rosy area) and name the subset of 50 genes that get up-regulated possibly due to stabilizing effects exerted by 5′ tRH-Glu-CTC “shelter targets” (Fig. 6B, lutescent area). It must be noted that the log2 fold changes of the perish and shelter targets all range below 1 (Fig. 6C). This may indicate that 5′ tRH-dependent gene regulation is rather fine-tuning cell processes than causing drastic changes. However, we observe these effects in cell lines where 5′ tRHs are rather low expressed and their impact in the hippocampus might be a different one. Additionally, it has to be considered that transcripts in the HepG2 antisense-inhibition experiment might get down-regulated not due to the loss of a stabilizing effect by 5′ tRH-Glu-CTC targeting but by silencing committed by Argonautes loaded with the antisense-RNA instead.
FIGURE 6.
(A) Volcano plot of differential expression analysis for protein-coding genes of 5′ tRH antisense-inhibition and control HepG2 cells (blue, adjusted P-value <0.01). (B) Venn diagram of significantly differentially expressed genes. Genes that are likewise regulated in HEK as in HepG2 cells are highlighted in gray. Genes that are inversely regulated in HEK overexpression and HepG2 inhibition cells are highlighted rosy (“potential perish targets”) and lutescent (“potential shelter targets”). (C) Log2 fold change values of perish and shelter transcripts in the HEK overexpression and HepG2 antisense-inhibition experiments. (D,E) Analysis of thermodynamically favored alignments for the major transcript regions of “potential perish targets” or “potential shelter targets” with 5′ tRH-Glu-CTC.
In order to characterize 5′ tRH-targeting further, we subsetted the three major transcript regions (5′ UTR, CDS, 3′ UTR) of the potential perish and shelter targets and computed the thermodynamics for RNA-duplex formation with the 5′ tRH-Glu-CTC. Considering the free energy needed to open intrinsic secondary structures, the free energy of the interaction and the so-called “dangling end” energies that noninteracting sequence stretches are causing, we identified the energetic optimal region of interaction. For the CDS and the 3′ UTR of both the potential perish targets and the potential shelter targets, these optimal binding sites were enriched for k-mer alignments with a length of around 20 bp (Fig. 6D,E) and alignments preferentially started around the sixth position from the 5′ end of the tRH. Thus, most thermodynamically favored interactions involved big stretches of the middle part of 5′ tRH-Glu-CTC. This is in line with the target pattern identified by the k-mer mapping approach in case of the potential perish targets, but not with the target pattern for the potential shelter targets. As stabilizing effects probably involve other protein interactors than silencing effects, this objection may be a result of the stabilizing interactor exposing only the ends of the tRH for target recognition.
As we found 5′ tRHs like 5′ tRH-Glu-CTC to be extremely abundant in small RNA libraries of primate hippocampal tissues and previous studies suggested a role in targeting genes involved in neural processes (Krishna et al. 2019; Sarker et al. 2019), we were interested whether the identified potential targets of 5′ tRH-Glu-CTC are implicated in neuronal processes. Indeed, we found 20% of the potential shelter targets (transcripts might be stabilized), but only 5% of the potential perish targets (transcripts might be degraded) to have an assigned neuronal function. In comparison only 7% and 8% of all expressed genes in HEK293T and HepG2 cells were assigned the gene ontology term “neurogenesis” (GO:0022008). Hence, there is a statistic correlation between genes that are potential targets of 5′ tRH-Glu-CTC and neurogenesis (α < 0.001 for shelter targets; χ2 test). While genes like MDK, VEGFA and EVL play a role in the regulation of axon outgrowth (Kurosawa et al. 2001; Drees and Gertler 2008; He et al. 2015), genes like NOTCH1 and NP1L1 are involved in neuronal differentiation (Patten et al. 2006; Qiao et al. 2018). Interestingly, decapitation studies with the planarian Dugesia japonica showed that 5′ tRH-Gly-GCC is not only up-regulated in regenerating animals, but is furthermore required for proper head regeneration (Z Cao, D Rosenkranz, S Wu, H Liu, Q Pang, B Liu, B Zhao, in prep.). Taking into consideration that planarians share many CNS genes with humans (Mineta et al. 2003), and that the hippocampus is one of the few brain regions known to have high rates of adult neurogenesis (Eriksson et al. 1998; Boldrini et al. 2018), it is tempting to speculate that 5′ tRHs may have critical functions in human neurogenesis as well.
Targeting patterns of 5′ tRHs/tsRNAs are unique for each fragment, but seem to be conserved across species
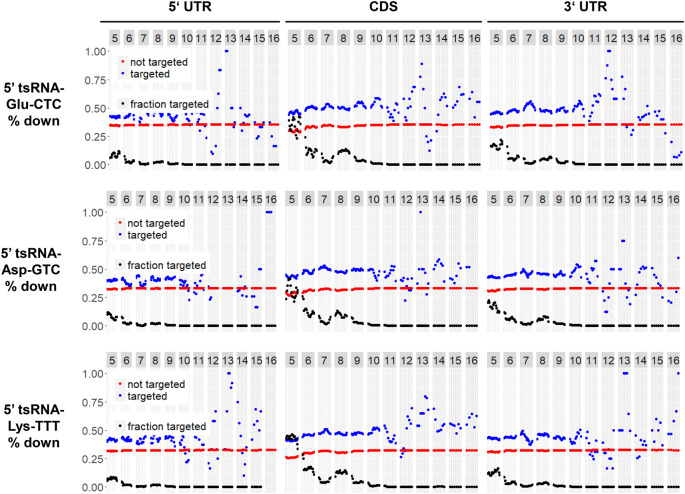
In order to gain further support for our assumptions, we researched whether the identified 5′ tRH target patterns are conserved among different species. We therefore analyzed published RNA sequencing data from Drosophila S2 cells that were transfected with a 20 nt long 5′ tsRNA-Glu-CTC, which exhibits high sequence homology with the human 5′ tRNA-half (Supplemental Fig. S6). As is the case for the human 5′ tRNA-half, the fly 5′ tRNA-fragment of the tRNA-Glu-CTC showed the strongest down-regulating effect on targets when its ∼7 nt long middle part binds to the 3′ UTR or CDS of the target transcript (Fig. 7). However, unlike the human 5′ tRNA-half, the both ends of the fly 5′ tsRNA did not seem to contribute to target up-regulation (data not shown), which is possibly due to its shorter sequence which lacks the corresponding nucleotides. Given the similar target regulation pattern, we suggest that 5′ tRNA-fragments can regulate their targets via conserved mechanisms across different species, while the sequence stretch being most important for target regulation can vary for fragments from different parental tRNAs.
FIGURE 7.
Target pattern analysis of published RNA sequencing data of fly S2 cells, where the 20 nt long 5′ tsRNA Glu-CTC, 5′ tsRNAs Asp-GTC or Lys-TTT was overexpressed by tsRNA mimic transfection (Luo et al. 2018). Plotted is the percentage of genes with (blue) or without (red) k-mer alignment that get down-regulated.
To confirm this assumption, we analyzed additional published RNA sequencing data sets from fly S2 cells that were transfected with the 20 nt long 5′ tRNA-fragments Asp-GTC and Lys-TTT (Fig. 7). These analyses support our hypothesis, that 5′ tRNA-fragments from different parental tRNAs act via different regions to regulate their targets on a sequence-complementary basis. What they have in common, is that 3′ UTR and CDS targeting leads to the biggest regulatory effect.
Concluding remarks
Despite the rise of NGS techniques, exploring the mechanisms by which small RNAs target genes remains challenging. Even for miRNAs, where gene regulation mechanisms are well studied, correct target prediction is difficult since the in vivo accessibility of potential target sites is difficult to assess. RNA binding proteins may not only occupy the putative binding site but may also change the secondary structure of the target when binding elsewhere in the transcript. Predicting and identifying targets of rather unexplored small RNAs like tsRNAs is more complicated, as it is unknown if and to which extent mismatches, wobble base pairs and bulges are tolerated. Aggravatingly is that target interactions can be surprisingly variable (Backofen and Hess 2010).
Using a k-mer mapping approach, we sought to identify the target patterns of 5′ tRHs. Our analysis suggested that 5′ tRHs silence genes, which have complementary binding sites for long stretches of the tRNA half that not necessarily need to include the miRNA-typical 5′ region. This finding suggests that Argonaute proteins are not necessarily indispensable as effector proteins for tsRNA-dependent gene regulation as it had been suggested by other studies (Haussecker et al. 2010; Kuscu et al. 2018). Whether 5′ tRHs independently regulate targets or do so in association with other effector proteins than Argonautes remains to be investigated. As it was shown that specific subsets of tsRNAs bind to certain proteins (Ivanov et al. 2011; Saikia et al. 2014; Goodarzi et al. 2015; Krishna et al. 2019), it is likely that 5′ tRHs might interact with different proteins to regulate distinct targets. The tRNA 3′ processing endoribonuclease RNase Z might be such an effector protein, as it was shown to cleave transcripts, which form RNA hybrids with 5′ tRHs that have similar secondary structures like pre-tRNAs (Elbarbary et al. 2009). Identifying more proteins involved in tsRNA-mediated gene regulation and elucidating the underlying mechanism will greatly enhance our understanding of gene regulation.
Analyzing the expression profile of tsRNAs across tissues and species, we found 5′ tRHs to be particularly high expressed in the hippocampus of primates, while their expression was rather low in the hippocampus of the pig, the rat and the mouse. In the hippocampus of all three tested primate species (human, chimpanzee and macaque), 5′ tRH-Glu-CTC and 5′ tRH-Gly-GCC were among the most abundant 5′ tRHs suggesting that they have a conserved role in the primate hippocampus. As we find transcripts that are presumably stabilized by 5′ tRH-Glu-CTC targeting to be substantially enriched for a function in neuronal processes such as axon outgrowth and neuronal differentiation, we hypothesize that 5′ tRHs play a role in fine-tuning primate neurogenesis. Alternatively, since retrotransposons are highly active in hippocampal neurons (Upton et al. 2015) and as 5′ tRH-Gly-GCC was shown to down-regulate transcripts that are driven by the LTR-retrotransposon MERVL in the developing mouse embryo (Sharma et al. 2016), it is also possible that hippocampal 5′ tRH expression serves the purpose of regulating transposition-related events.
MATERIALS AND METHODS
Small RNA sequencing, data processing, and annotation
Small RNA sequencing data of various human tissues was downloaded from the SRA Database (for accession numbers see Supplemental Table S1) and quality controlled by FastQC (version 0.11.2). Adapter and quality trimming was performed using BBDuk (version 36.77; ktrim = r overwrite = true k = 20 mink = 9 ziplevel = 2 hdist = 1 qtrim = rl trimq = 10 minlen = 15 maxlen = 34; for Encode data additionally: forcetrimleft = 6) before the reads were examined via FastQC again and mapped to the human genome (version GCA_000001405.27_GRCh38.p12) using Bowtie 2 (version 2.3.0). Based on these map-files small RNA annotation was performed with unitas (version 1.7.3; Gebert et al. 2017). Using the custom Perl script annotationtable2RPM.pl the RPM values were calculated for the respective tsRNA species.
Total RNA from adult normal male human hippocampal tissue was obtained from AMS Biotechnology (Cat. Nr: R1234052-10). A small RNA library was prepared as described in Gebert et al. (2015). In brief, small RNAs (15–40 nt) were extracted from a denaturing polyacrylamide gel. Subsequently, a 3′ adapter (5′-rAppCTGTAGGCACCATCAATddC-3′) and a 5′ adapter (5′-GACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′) were ligated to the small RNAs. Following cDNA synthesis with the RT-Primer 5′-ATTGATGGTGCCTACAG-3′, the cDNA transcripts were PCR amplified using the forward primer 5′-ACATGGACTGAAGGAGTAGA-3′ and the index-containing reverse primer 5′-ggctcATTGATGGTGCCTACAG-3′. The generated library was high throughput sequenced in parallel with six other indexed samples by GENterprise (Mainz) on an Illumina HiSeq 2000 system. After converting the fastq file to fasta formate using the NGS toolbox Perl script fastq2fasta, the 5′ adapter sequence was clipped off the 120 nt long reads using the NGS toolbox Perl script clip (-m 5.AGTAGAAA). As reads may contain the reverse complementary sequence, the reverse complementary variant of the 5′ adapter sequences was also clipped off (-m TTTCTACT.3) and the remaining sequences were transcribed to the original sequence direction using the NGS toolbox Perl script rev-comp. Both outputs were then concatenated using the NGS toolbox Perl script concatenate. To extract only the reads with the right index and to remove the 3′ adapter sequence the NGS toolbox Perl script clip (-m CTGTA.GAGCC.3) was applied. Subsequent analysis was performed as described below.
Total RNA from tissue samples of human adult brain regions (hippocampus, cortex, cerebellum) was obtained from the BioChain Institute (Newark, CA, USA). Total RNA of hippocampal tissue samples of a female and a male chimpanzee brain was obtained from the National Chimpanzee Brain Resource (www.chimpanzeebrain.org, USA). A macaque brain provided by the Primate Brain Bank (The Netherlands Institute for Neuroscience, Amsterdam, Netherlands) was dissected and hippocampal tissue was homogenized in TRIzol (Thermo Fisher). Total RNA was isolated according to the manufacturers protocol. The total RNA was sent to LC Sciences (Houston, TX, USA) for small RNA library preparation and small RNA sequencing. The 3′ adapter sequences were clipped off using the NGS toolbox (version 2.1; http://www.smallrnagroup.uni-mainz.de/) Perl script clip (-m TGGAATTC.3).
Small RNA sequencing data of hippocampal tissue from three male adult Wistar rats (Study: PRJEB24026, Run accessions: ERR2226477, ERR2226482 and ERR2226487) was downloaded from the European Nucleotide Archive. Subsequent analysis was performed as described below.
Small RNA sequencing data of hippocampal tissue from the pig (SRR3105507 and SRR3105508) and the mouse (SRR5144167, SRR5144168 and SRR5144169) was downloaded via the SRA toolkit tool fastq-dump (version 2.8.2). The 3′ adapter sequences were clipped off using the NGS toolbox (version 2.1) Perl script clip (-m TGGAATTC.3). Subsequent analysis was performed as described below.
Total RNA of HEK293T cells was extracted with TRI Reagent (Thermo Fisher) 24 h after transfection with 10 nM Silencer Select Negative Control No. 1 siRNA (4390843; Thermo Fisher) using Lipofectamine RNAiMAX (Thermo Fisher). The RNA was sent to BGI (Hongkong) for small RNA library preparation and 50 bp single-end sequencing. The reads were delivered adapter-trimmed. Subsequent analysis was performed as described below.
Small RNA sequencing data of HepG2 cells (SRR6823987) was downloaded via the SRA toolkit tool fastq-dump (version 2.8.2). 3′ adapter sequences were clipped off using the NGS toolbox (version 2.1) Perl script clip (-m AGATCGGA.3). Subsequent analysis was performed as described below.
Adapter-trimmed data was first quality controlled by FastQC (version 0.11.2), then converted to the fasta-format, length filtered (15–40 nt), collapsed to nonidentical reads and depleted for low complexity reads using the NGS toolbox Perl scripts length-filter, collapse and duster (version 2.1; http://www.smallrnagroup.uni-mainz.de/). The remaining sequences were mapped to the respective genome (versions GCA_000001405.27_GRCh38.p12, GCA_000001515.5_Pan_tro_3.0, GCA_000772875.3_Mmul_8.0.1, GCA_000003025.6_Sscrofa11.1, GCA_000001895.4_Rnor_6.0 and GCA_000001635.8_GRCm38.p6) using the Perl script sRNAmapper (version 1.0.5; -a best) that uses SeqMap (Jiang and Wong 2008) as mapping tool. The map-files were used for small RNA annotation with unitas (version 1.6.1; Gebert et al. 2017).
Transfection of tsRNA-mimics and tsRNA antisense 2′-OMe-RNAs
HEK293T (2.5E4 cells/well) and HepG2 (1E5 cells/well) cells were seeded in 24-well plates and cultured in 1× GlutaMAX-I DMEM supplemented with 10% FBS (Thermo Fisher). The next day the cells were transfected with 50 nM tsRNA-mimics or tsRNA antisense 2′ OMe-RNAs (biomers) using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer's protocol. Sequences of the transfected RNAs are available in Supplemental Table S2. As control, cells were transfected with Silencer Select Negative Control No. 1 siRNA (Thermo Fisher). Forty-eight hours after transfection, the RNA was isolated according to the TRI Reagent protocol (Thermo Fisher).
RT-PCR quantification of tsRNAs
In order to measure the tsRNA level after transfection, the 15–40 nt small RNA fraction was eluted from a denaturing polyacrylamide gel (see Gebert et al. 2015) and polyadenylated using the A-Plus Poly(A) Polymerase Tailing Kit (Cellscript). After ethanol precipitation the poly(A) tailed RNA was reversely transcribed with the SuperScript IV reverse transcriptase (Thermo Fisher) using the RT-primer 5′-CGAATTCTAGAGCTCGAGGCAGGCGACATGT25VN-3′. For qPCR 1 µL of cDNA was mixed with 0.5 µL 10 µM sequence-specific forward primer, 0.5 µL 10 µM RT-primer-specific reverse primer, 3 µL water and 5 µL 2× QuantiFast SYBR Green PCR Master Mix (Qiagen). Technical duplicates of this reaction mix were then analyzed on a Corbett Rotor-Gene 6000 real-time PCR cycler. Finally, the copy numbers of the respective tsRNAs were quantified by standard curves of the individual primer pair amplicons. As normalizers the miRNAs miR25, miR532 and miR99a were used. The boxplots were created with R using the R packages ggplot2 and Cairo (version 3.4.3). qPCR primer sequences are available in Supplemental Table S3.
RT-PCR quantification of miRanda/piRanha predicted targets
Potential target transcripts of the tsRNAs were predicted using the algorithm miRanda (version 3.3a) that bases on miRNA targeting rules (Enright et al. 2003) and a self-developed software named piRanha (version 0.0.0) that bases on piRNA targeting rules (Zhang et al. 2018a). Both tools were used with default settings, expected to yield best results for miRNAs or piRNAs, respectively. The reference transcriptome was downloaded from the Ensembl database (release 94). The custom Perl scripts MRscript.pl and PRscript.pl were applied to extract and sort the transcript IDs from the output files by the respective miRanda and piRanha score, which is the sum of binding energies of all alignments with the tsRNA seed. It is assumed that the lower the miRanda or piRanha score (i.e., free energy of the alignments) of a transcript, the more likely it is a target of the respective tsRNA. For further analysis, only transcripts that have a TPM value above 0.2 in HEK293 cells were considered. Therefore the RNA sequencing data sets SRR629569 and SRR629570 were downloaded with NCBI's fastq-dump (-I ‐‐split-files ‐‐gzip) and uploaded to the Galaxy server (usegalaxy.org), where they were mapped to the human genome (Galaxy hg38) using RNA STAR (Galaxy Tool Version 2.6.0b-1). Transcript wise counting was performed with featureCounts (Galaxy Tool Version 1.6.0.6) on the basis of an Ensemble GTF-file (Homo_sapiens.GRCh38.90), which had been converted to UCSC coordinates using the File Chameleon tool of Ensembl. The generated count tables and gene length files were used to calculate the mean TPM values and select the expressed transcripts with R using the R packages biomaRt and stats (version 3.4.3). For each tsRNA and algorithm, the five transcripts with the lowest miRanda or piRanha scores were chosen for RT-PCR quantification. Primers with the length of ∼20 nt were designed to be either exon-junction spanning or to include intronic regions that are bigger than 700 bp. Furthermore, only primers that exclusively amplify the same amplicon from the different splicing isoforms that are potential targets were taken into account. For each primer pair a test PCR with cDNA from untransfected HEK293T cells was performed to evaluate the amplicon quality and length on an agarose gel. In order to compare the relative copy number of the selected potential tsRNA targets in tsRNA-mimic and control transfected cells, the respective total RNA was extracted with TRI Reagent (Thermo Fisher), reversely transcribed and quantified as described for the poly(A) tailed RNA above. The housekeeping genes ACTB and RPS18 were used as normalizers to calculate the relative expression by means of the delta-delta-CT method. qPCR primer sequences are available in Supplemental Table S3. The boxplots were created with R using the R packages ggplot2 and Cairo (version 3.4.3).
RNA sequencing, data processing, and differential expression analysis
Total RNA isolated from tsRNA-mimic, tsRNA-antisense and control transfected cells was sent for library construction and paired-end sequencing to BGI (Hong Kong). On average 35 million paired-end reads were obtained. Using the online platform Galaxy (usegalaxy.org) the reads were first mapped to the human genome (Galaxy hg38) by RNA STAR (Galaxy Tool Version 2.6.0b-1). Afterwards, gene wise counting was performed with featureCounts (Galaxy Tool Version 1.6.0.6) on the basis of the above mentioned GTF-file. Based on the generated count tables, DESeq2 (Galaxy Tool Version 2.11.40.2) was used to identify differentially expressed genes (adjusted P-value <0.01). Analysis of the Drosophila data sets (Study: PRJNA378597, Run accessions: SRR6930617, SRR6930619, SRR6930621; Luo et al. 2018) was likewise conducted using the organism-specific files. Volcano plots based on the DESeq2 result file and cumulative fraction plots based on the DESeq2 result file with thresholds of -80 for miRanda scores and -30 for piRanha scores were created with R using the R packages ggplot2, ggrepel and Cairo (version 3.4.3).
Identification of targeting patterns using a k-mer mapping approach
tsRNA sequences with length n were split into all possible k-mers with k = 5…n. All k-mers were mapped individually to the 5′ UTR, the CDS, and the 3′ UTR of the transcripts of the corresponding organism in reverse complementarity to identify putative target sites. Only the longest transcript per annotated gene was taken into account. The transcriptomes and respective genome annotation files were downloaded from Ensembl database (release 94). Splitting the transcripts into 5′ UTR, CDS and 3′ UTR was performed using the custom Perl scripts select + split_dmel.pl and select + split_hsap.pl. The following numbers of total mismatches (mm, including insertions/deletions) within a k-mer alignment were allowed: For k ≥ 6 1 mm, for k ≥ 12 2 mm, for k ≥ 18 3 mm, for k ≥ 22 4 mm, for k ≥ 26 5 mm. The numbers of allowed insertions/deletions (indel) within a k-mer alignment were: For k ≥ 12 1 indel, for k ≥ 18 2 indels, for k ≥ 22 3 indels. Mismatches were not allowed in the first two or last two positions of the alignment. Nested k-mer alignments, i.e., alignments that completely overlapped with larger k-mer alignments, were ignored. k-mer mapping and filtering was performed using the custom Perl script map_kmers.pl.
Gene expression values (fpkm) were calculated based on the featureCounts count tables and genes with an average expression ratio below 1 fpkm were depleted from the DESeq2 result file for further analysis using the R script DESeq2-Analysis-for-get_targets.R. The custom Perl script get_targets_for_DESeq2.pl was then used to count for each tsRNA and its k-mers defined by start position and length the number of significantly up-/down-regulated genes (adjusted P-value <0.01) that have a corresponding k-mer alignment, and the number of significantly up-/down-regulated genes that do not have a corresponding k-mer alignment. The R script get_targets_visualization.R was used to visualize for each transcript region the fraction of targeted as well as not targeted genes that are up-/down-regulated. For the individual plots, the script calculates the average values of a 5 nt sliding window for the start position with sliding window increment of 1 nt.
Analysis of potential targets regarding thermodynamically favored alignments 5′ tRH-Glu-CTC and GO term annotation
Based on the DESeq2 result tables, a Venn diagram of significantly differentially expressed genes (adjusted P-value <0.01) was generated using the R packages VennDiagram and polyclip (version 3.4.3). Genes that were inversely regulated in the HEK overexpression and the HepG2 inhibition cells were assigned to the groups “potential perish targets” (HEK: log2FC < 0; HepG2: log2FC > 0) and “potential shelter targets” (HEK: log2FC > 0; HepG2: log2FC < 0).
Input files for the program RNAup were generated using the custom Perl script RNAup_input.pl, which prints for each potential target gene the sequence of each transcript region (5′ UTR, CDS, 3′ UTR) together with the tsRNA sequence as fasta-format. RNAup (version 2.4.13) from the ViennaRNA Package (Lorenz et al. 2011) was executed for each input file with the parameters -b -d2 ‐‐noLP -c “S.” Using the custom Perl script merge_RNAupOutput.pl the RNAup output information was merged per transcript region. Using the R script visualize_RNAup.R the respective merged RNAup output files were visualized as a histogram displaying the alignment length, a histogram displaying the alignment start within the tsRNA and a bar plot displaying the alignment count per position within the tsRNA.
The list of human protein-coding genes with the gene ontology term “neurogenesis” (GO:0022008) was downloaded from AmiGO 2 (http://amigo.geneontology.org/amigo). Corresponding Ensembl gene identifier for the UniProt identifiers were retrieved from UniProt (https://www.uniprot.org/uploadlists/). To evaluate a potential statistical correlation between the perish or shelter targets of 5′ tRH-Glu-CTC and the GO term “neurogenesis” the χ2 test was applied.
Code availability and data deposition
All above mentioned custom Perl and R scripts are freely available at GitHub (github.com/jjehn/tRH-targeting). Sequencing data sets are accessible at NCBI's sequence read archive (SRA) under the accession numbers SRR10091207 (first sRNA-seq human hippocampus), SRR10091206 (second sRNA-seq human hippocampus), SRR10091205 (sRNA-seq human cortex), SRR10091204 (sRNA-seq human cerebellum), SRR10092006 (sRNA-seq female chimpanzee hippocampus), SRR10092005 (sRNA-seq male chimpanzee hippocampus), SRR10092004 (sRN-Aseq macaque hippocampus), SRR10082984 (sRNA-seq HEK293T cells), and SRR10085693 to SRR10085704 (RNA-seq of the HEK293T and HepG2 experiments).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Wolfrum and Strand laboratory for kindly providing HEK and HepG2 cells. We further thank René Ketting, Susanne Strand, Daniel Gebert, Christine Barbara Kiefer, and Lena Mazzariello for helpful comments and discussion. Chet C. Sherwood and Cheryl Stimpson from the National Chimpanzee Brain Resource (www.chimpanzeebrain.org, USA) are acknowledged for their great assistance in preparing chimpanzee RNA samples. The chimpanzee samples were supported by the National Institutes of Health (NIH) grant NS092988. This work was supported by the International PhD Program (IPP) coordinated by the Institute of Molecular Biology IMB, Mainz, Germany, funded by the Boehringer Ingelheim Foundation.
Author contributions: D.R. and J.J. designed the study. V.E. and C.H. analyzed the published small RNA sequencing data sets of human tissues. L.W. and I.F. generated and analyzed the small RNA libraries of the human hippocampus. R.N.K. dissected the macaque brain and homogenized the hippocampal tissue sample. I.F. isolated the total RNA from the macaque sample and coordinated the procurement and sequencing of all primate brain samples. J.J. analyzed the small RNA sequencing data sets of the cell lines, the primate brain samples, and the pig, rat, and mouse hippocampus. J.J., S.W., and B.O. performed the laboratory experiments. J.J., J.T., and D.R. analyzed the RNA sequencing data. J.J. and D.R. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.073395.119.
Freely available online through the RNA Open Access option.
REFERENCES
- Backofen R, Hess WR. 2010. Computational prediction of sRNAs and their targets in bacteria. RNA Biol 7: 33–42. 10.4161/rna.7.1.10655 [DOI] [PubMed] [Google Scholar]
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. 2011. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci 108: 10466–10471. 10.1073/pnas.1103946108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V, Rosoklija GB, Stankov A, Arango V, Dwork AJ, et al. 2018. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell 22: 589–599.e5. 10.1016/j.stem.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng G-h, Peng H, Zhang X, Zhang Y, et al. 2016. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351: 397–400. 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, Deng X, Zhou J, Lee I, Lee YS, et al. 2015. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther 23: 1622–1629. 10.1038/mt.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F, Gertler FB. 2008. Ena/VASP: proteins at the tip of the nervous system. Curr Opin Neurobiol 18: 53–59. 10.1016/j.conb.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. 2009. Modulation of gene expression by human cytosolic tRNase ZL through 5′-half-tRNA. PLoS One 4: e5908 10.1371/journal.pone.0005908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5: R1 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson PS, Perfilieva E, Björk-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. 1998. Neurogenesis in the adult human hippocampus. Nat Med 4: 1313–1317. 10.1038/3305 [DOI] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J. 2009. Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682. 10.1016/j.mib.2009.09.009 [DOI] [PubMed] [Google Scholar]
- Garcia-Silva MR, Cabrera-Cabrera F, das Neves RFC, Souto-Padrón T, de Souza W, Cayota A. 2014. Gene expression changes induced by Trypanosoma cruzi shed microvesicles in mammalian host cells: relevance of tRNA-derived halves. Biomed Res Int 2014: 305239 10.1155/2014/305239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert D, Ketting RF, Zischler H, Rosenkranz D. 2015. piRNAs from pig testis provide evidence for a conserved role of the Piwi pathway in post-transcriptional gene regulation in mammals. PLoS One 10: e0124860 10.1371/journal.pone.0124860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert D, Hewel C, Rosenkranz D. 2017. unitas: the universal tool for annotation of small RNAs. BMC Genomics 18: 644 10.1186/s12864-017-4031-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N. 2017. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 14: 1364–1373. 10.1080/15476286.2016.1257470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HCB, Zhang S, Fish L, Tavazoie SF. 2015. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802. 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, et al. 2006. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet 38: 411–413. 10.1038/ng1742 [DOI] [PubMed] [Google Scholar]
- Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimková K, Sommarin MNE, Munita R, Lubas M, et al. 2018. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173: 1204–1216.e26. 10.1016/j.cell.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16: 673–695. 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Bai G, Zhou H, Wei N, White NM, Lauer J, Liu H, Shi Y, Dumitru CD, Lettieri K, et al. 2015. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature 526: 710–714. 10.1038/nature15510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, Zhang Q, Zhang L, Xue Z, Li Y, et al. 2017. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res 77: 3194–3206. 10.1158/0008-5472.CAN-16-3146 [DOI] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, O'Day E, Emara MM, Wagner G, Lieberman J, Anderson P. 2014. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci 111: 18201–18206. 10.1073/pnas.1407361111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn J, Rosenkranz D. 2019. tRNA-derived small RNAs: the good, the bad and the ugly. Med One 4: e190015 10.20900/mo.20190015 [DOI] [Google Scholar]
- Jiang H, Wong WH. 2008. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24: 2395–2396. 10.1093/bioinformatics/btn429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam SP, Sobala A, ten Have S, Hutvagner G. 2017. tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J Proteome Res 16: 413–420. 10.1021/acs.jproteome.6b00267 [DOI] [PubMed] [Google Scholar]
- Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, et al. 2017. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552: 57–62. 10.1038/nature25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S, Yim DG, Lakshmanan V, Tirumalai V, Koh JL, Park JE, Cheong JK, Low JL, Lim MJ, Sze SK, et al. 2019. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep 20: e47789 10.15252/embr.201947789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A. 2014. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12: 78 10.1186/PREACCEPT-5867533061403216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa N, Chen GY, Kadomatsu K, Ikematsu S, Sakuma S, Muramatsu T. 2001. Glypican-2 binds to midkine: the role of glypican-2 in neuronal cell adhesion and neurite outgrowth. Glycoconj J 18: 499–507. 10.1023/A:1016042303253 [DOI] [PubMed] [Google Scholar]
- Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. 2018. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24: 1093–1105. 10.1261/rna.066126.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649. 10.1101/gad.1837609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. 1990. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J 9: 1383–1389. 10.1002/j.1460-2075.1990.tb08253.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, He F, Luo J, Dou S, Wang Y, Guo A, Lu J. 2018. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res 46: 5250–5268. 10.1093/nar/gky189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Choudury SG, Slotkin RK. 2017. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 45: 5142–5152. 10.1093/nar/gkx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. 2013. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci 110: 1404–1409. 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineta K, Nakazawa M, Cebria F, Ikeo K, Agata K, Gojobori T. 2003. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc Natl Acad Sci 100: 7666–7671. 10.1073/pnas.1332513100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten BA, Sardi SP, Koirala S, Nakafuku M, Corfas G. 2006. Notch1 signaling regulates radial glia differentiation through multiple transcriptional mechanisms. J Neurosci 26: 3102–3108. 10.1523/JNEUROSCI.4829-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Li Y, Feng C, Duo S, Ji F, Jiao J. 2018. Nap1l1 controls embryonic neural progenitor cell proliferation and differentiation in the developing brain. Cell Rep 22: 2279–2293. 10.1016/j.celrep.2018.02.019 [DOI] [PubMed] [Google Scholar]
- Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, Grassi A, Zanovello P, Bronte V, Ciminale V, et al. 2014. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol 88: 3612–3622. 10.1128/JVI.02823-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao X-H, Guan B-J, Yuan Y, Jankowsky E, Feng Z, et al. 2014. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 34: 2450–2463. 10.1128/MCB.00136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker G, Sun W, Rosenkranz D, Pelczar P, Opitz L, Efthymiou V, Wolfrum C, Peleg-Raibstein D. 2019. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc Natl Acad Sci 116: 10547–10556. 10.1073/pnas.1820810116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. 2017. LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170: 61–71.e11. 10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351: 391–396. 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short AK, Yeshurun S, Powell R, Perreau VM, Fox A, Kim JH, Pang TY, Hannan AJ. 2017. Exercise alters mouse sperm small noncoding RNAs and induces a transgenerational modification of male offspring conditioned fear and anxiety. Transl Psychiatry 7: e1114 10.1038/tp.2017.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Reina O, Stephan-Otto Attolini C, Ribas de Pouplana L. 2019. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci 116: 8451–8456. 10.1073/pnas.1821120116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sánchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. 2015. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161: 228–239. 10.1016/j.cell.2015.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. 2013. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther 21: 368–379. 10.1038/mt.2012.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Bennasser Y, Watashi K, Le S-Y, Houzet L, Jeang K-T. 2009. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 37: 6575–6586. 10.1093/nar/gkp707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, He X, Liu C, Liu J, Hu Q, Pan T, Duan X, Liu B, Zhang Y, Chen J, et al. 2016. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol 196: 1591–1603. 10.4049/jimmunol.1500805 [DOI] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu W-S, Huang W-C, Weng Z, Lee H-C. 2018a. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359: 587–592. 10.1126/science.aao2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J, et al. 2018b. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 20: 535–540. 10.1038/s41556-018-0087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu S, Chen Y, Fu Y, Silver AJ, Hill MS, Lee I, Lee YS, Bao X. 2017. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen Virol 98: 1600–1610. 10.1099/jgv.0.000852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.