Abstract
Introduction
Acinetobacter baumannii is an opportunistic pathogen responsible for nosocomial infections. The emergence of colistin-resistant A. baumannii is a significant threat to public health. The aim of this study was to investigate the molecular characterization and genotyping of clinical A. baumannii isolates in Southwestern Iran.
Methods
A total of 70 A. baumannii isolates were collected from patients admitted to Imam Khomeini Hospital in Ahvaz, Southwestern Iran. Minimum inhibitory concentration test was conducted by using Vitek 2 system. The presence of biofilm-forming genes and colistin resistance-related genes were evaluated by PCR. The isolates were also examined for their biofilm formation ability and the expression of pmrA and pmrB genes. Finally, multilocus sequence typing (MLST) and PCR-based sequence group were used to determine the genetic relationships of the isolates.
Results
Overall, 61 (87.1%) and 9 (12.8%) isolates were multidrug-resistant (MDR) and extensively drug-resistant (XDR), respectively. Colistin and tigecycline with 2 (2.8%) and 32 (45.7%) resistance rates had the highest effect. Among all the isolates, 55 (78.5%), 7 (10%), and 3 (4.3%) were strong, moderate, and weak biofilm producers, respectively. The frequency rates of biofilm-related genes were 64 (91.4%), 70 (100%), 56 (80%), and 22 (31.42%) for bap, ompA, csuE, and blaPER1, respectively. Overexpression of pmrA and pmrB genes was observed in two colistin-resistance isolates, but the expression of these genes did not change in colistin-sensitive isolates. Additionally, 37 (52.8%) and 8 (11.4%) isolates belonged to groups 1 (ICII) and 2 (IC I), respectively. MLST analysis revealed a total of nine different sequence types that six isolates belonged to clonal complex 92 (corresponding to ST801, ST118, ST138, ST 421, and ST735). Other isolates were belonging to ST133 and ST216, and two colistin-resistant (Ab4 and Ab41) isolates were belonging to ST387 and ST1812.
Conclusion
The present study revealed the presence of MDR and XDR A. baumannii isolates harboring biofilm genes and emergence of colistin-resistant isolates in Southwestern Iran. These isolates had high diversity, which was affirmed by typing techniques. The control measures and regular surveillance are urgently needed to preclude the spread of these isolates.
Keywords: Acinetobacter baumannii, drug resistance, colistin, MLST, clonal complex
Introduction
Acinetobacter baumannii is known as an opportunistic nosocomial pathogen in hospitals and is associated with a wide range of infections in healthcare facilities.1 Antibacterial resistance in multidrug-resistant (MDR) A. baumannii isolates is an important issue, and molecular studies are considered serious strategies for controlling the outbreak of these isolates.2 A. baumannii resistance to several antimicrobial drugs has increasingly been elevated in recent decades. This pattern of antibiotic resistance often varies with time and from one area to another or even within the same area.3,4 Nowadays, in developing countries, including Iran, physicians face serious challenges to the treatment of patients infected with MDR A. baumannii, presenting severe healthcare problems owing to treatment failure.5,6
Carbapenem antibiotics, such as imipenem and meropenem, are the most effective therapeutic options for A. baumannii infections. Overuse of broad-spectrum carbapenems and/or cephalosporins is an important risk factor for the development of colonization or infection with carbapenemase-producing A. baumannii strains. The rapid spread of carbapenem-resistant MDR A. baumannii has led to the use of polymyxins, particularly polymyxin E (colistin), which its excessive use has recently given rise to the emergent resistance to this antibiotic.7 Colistin, a cationic polypeptide antibiotic, targets the bacterial outer membrane via an initial charge-based interaction between the positively charged colistin and the negatively charged on the lipid A component of lipopolysaccharide (LPS). Various mechanisms, including mutations in the pmrA and pmrB genes of the PmrAB two-component regulatory system, plasmid-mediated mcr-1 gene, mutations in any of lipid A biosynthesis genes (lpxA, lpxC, and lpxD), and the presence of insertion sequence ISAba11 in either lpxC or lpxA are involved in the emergence of resistance to this antibiotic.8,9 Another leading factor involved in bacterial resistance to varied antibiotics, survival in hospital environments, and chronic infections is the biofilm formation capability of bacteria. Previous studies have shown a positive relationship between the antibiotic resistance and biofilm formation in A. baumannii strains. There are also a variety of virulence factors, including the biofilm-associated protein (BAP), outer membrane protein A (OmpA), chaperon-usher pilus (Csu), and quorum-sensing system, that are engaged in the biofilm formation of A. baumannii. The BAP encoded by the bap gene has a critical function in the biofilm establishment and intercellular adhesion.10,11 The OmpA has negligible contribution to the development of the robust biofilm on the plastic surface. The biofilm-forming ability of A. baumannii is also largely dependent on pili. Therefore, the csuE gene plays a major role in A. baumannii biofilm formation.12 The prevalence of A. baumannii in hospitals has increased dramatically worldwide; therefore, using molecular typing methods is essential for infection control and epidemiological studies. Based on the epidemiological studies and population genetic investigations of A. baumannii, there are several developed typing methods, including multiple-locus variable number tandem repeats (VNTRs) analysis (MLVA), pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and PCR-based sequence group (SG) profiling.13,14
MLVA and MLST schemes provide a genotype in the form of a code that can easily be shared. Both approaches allow for the investigation of the population structure and identification of clonal lineages.15 The aim of this study was to investigate the presence of biofilm-forming genes and colistin resistance-related genes in A. baumannii isolates, which were examined for their ability to form biofilms and to express pmrA and pmrB genes. PCR-based sequence group and MLST were also applied to determine the genetic relationships of the isolates.
Materials and Methods
Bacterial Isolates and Identification
This cross-sectional study was conducted between October 2018 and July 2019 and confirmed by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (No. IR.AJUMS.REC.1397.793). Before initiating this research work, informed consent forms were obtained from all the patients. The study was conducted in accordance with the Declaration of Helsinki. In total, 70 non-repetitive A. baumannii isolates were collected from different clinical samples from patients admitted to Imam Khomeini Hospital in Ahvaz, Southwestern Iran. Patients with concomitant infections not properly treated, human immunodeficiency virus (HIV) infection, and antibiotic treatment less than two days were excluded. All the included isolates were identified based on conventional microbiological tests,16 and final identification was conducted by the PCR of blaOXA-51-like gene and multiplex of gyrB.17 The A. baumannii ATCC 19606 was used as a positive control.
Susceptibility Testing
The minimum inhibitory concentration (MICs) of each antibiotic was determined using the described Vitek 2 system (bioMérieux, Marcy l’Etoile, France). The MICs of all the selected antibiotics were interpreted by the aforesaid system as per CLSI guidelines.18 For tigecycline, no MIC interpretive breakpoint was recommended by the EUCAST and CLSI. A MIC of ≥8 μg/mL was regarded as the resistant breakpoint on the basis of the criteria suggested by Jones et al.19 In addition, a MIC ≥ 4 µg/mL was proposed as the breakpoint of resistance for colistin and a MIC ≥ 8 µg/mL for both imipenem and meropenem.18 MDR was defined as resistance to at least one agent in three or more categories of antibiotics. Isolates of A. baumannii with resistance to at least one agent in all but two or fewer antimicrobial categories were considered as XDR.20 Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as the quality control.
Molecular Detection of Resistance Genes
Biofilm-related genes (bap, ompA, csuE, and blaPER1) and colistin resistance-related genes (mcr-1, mcr-2, pmrA, and pmrB) were detected using PCR.21–23 Bacterial DNA extraction was performed in accordance with the boiling method.24 The PCR assay was carried out in a final volume of 25 μL containing Taq DNA polymerase (1 U; CinnaGen, Iran), dNTPs (100 μM), Taq buffer (5×), DNA template (50 ng), and forward and reverse primers (25 pM). PCR mixtures were subjected to the following thermal cycling: 5 min at 94°C, followed by 35 cycles with denaturation at 94°C for 50 s, annealing at 55–57°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 5 min. Primer sequences used for the detection of the above-mentioned genes are presented in Table 1.
Table 1.
Primers Used for the Amplification of Genes
| Target Genes | Sequence (5/–3/) | Amplicon Size (Base Pairs) | References |
|---|---|---|---|
| bap | F:ATGCCTGAGATACAAATTAT R:GTCAATCGTAAAGGTAACG |
1449 | 21 |
| ompA | F:GTTAAAGGCGACGTAGACG R:CCAGTGTTATCTGTGTGACC |
578 | 21 |
| csuE | F:CATCTTCTATTTCGGTCCC R:CGGTCTGAGCATTGGTAA |
168 | 21 |
| blaPER-1 | F:ATGAATGTCATTATAAAAGC R:AATTTGGGCTTAGGGCAGAA |
925 | 21 |
| mcr-1 | F:AGTCCGTTTGTTCTTGTGGC R:AGATCCTTGGTCTCGGCTTG |
320 | 22 |
| mcr-2 | F:CAAGTGTGTTGGTCGCAGTT R:TCTAGCCCGACAAGCATACC |
715 | 22 |
| pmrA | F:ACTGGACATGTTGCACTCTTGT R:ATGCACTTTTATGAAGTCCCGA |
757 | 23 |
| pmrB | F:TCGGGACTTCATAAAAGTGCAT R:CAGTCACAGGTGTTCGTAATT |
722 | 23 |
| 16S_rRNA | F:CAGCTCGTGTCGTGAGATGT R:CGTAAGGGCCATGATGACTT |
150 | 23 |
| Group1ompAF306 Group1and2ompAR660 |
GATGGCGTAAATCGTGGTA CAACTTTAGCGATTTCTGG |
355 | 27 |
| Group1csuEF Group1csuER |
CTTTAGCAAACATGACCTACC TACACCCGGGTTAATCGT |
702 | 27 |
| Gp1OXA66F89 Gp1OXA66R647 |
GCGCTTCAAAATCTGATGTA GCGTATATTTTGTTTCCATTC |
559 | 27 |
| Group2ompAF378 Group1and2ompAR660 |
GACCTTTCTTATCACAACGA GGCGAACATGACCTATTT |
343 | 27 |
| Group2csuEF Group2csuER |
GGCGAACATGACCTATTT CTTCATGGCTCGTTGGTT |
580 | 27 |
| Gp2OXA69F169 Gp2OXA69R330 |
CATCAAGGTCAAACTCAA TAGCCTTTTTTCCCCATC |
162 | 27 |
Expression of pmrA and pmrB
Quantitative real-time PCR assay was performed for isolates with colistin MICs ≥ 1 µg/mL. RNA extraction was conducted using a High Pure RNA Isolation Kit (Roche, Germany). The integrity and quality of the total RNA assessed with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) were electrophoresed on 1% agarose gel. The RNA was reverse transcribed to cDNA using Prime Script™ 1st strand cDNA Synthesis Kit (Takara Biotechnology Co., LTD., China) according to the instructions provided by the manufacturer. To carry out real-time PCR, we used the specific primers of 16S rRNA (as an internal control), pmrA, and pmrB genes. A. baumannii ATCC 19606 was selected as the reference strain. Real-time PCR amplification reaction was prepared in a 20-μL final volume, with 0.5 μL of each forward and reverse primer (10 nM each), 10 μL of RealQ Plus Master Mix Green (Ampliqon, Denmark), 400 ng of cDNA, and DNase- and RNase-free water up to a 20-μL final volume. This reaction was accomplished in the ABI Thermocycler System (Thermo Fisher Scientific). The reaction conditions were initial denaturation at 95°C for 5 min, 40 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. The relative expression fold changes of mRNAs were calculated using the 2−ΔΔCt method.21
In Vitro Bacterial Biofilm Assay
The biofilm formation capacity of A. baumannii isolates on 96-well polystyrene microtiter plates (MTP) was evaluated using the crystal violet staining (CVS) method, as described elsewhere.25 The results of this assay were interpreted according to the criteria explained before by Zhang et al.26 The isolates were no biofilm producers when A ≤ Ac, weak biofilm producers when Ac < A ≤ 2Ac, moderate biofilm producers when 2Ac < A ≤ 4Ac, and strong biofilm producers when A > 4Ac; A represents optical absorbance and Ac displays OD value. Also, A. baumannii ATCC 19606 and Mueller Hinton Broth were used as positive and negative controls for the evaluation of the biofilm formation, respectively. All experiments were performed in triplicate.
Molecular Typing Methods
International clonal (IC) types were determined based on the absence or presence of the alleles of csuE, ompA, and the intrinsic carbapenemase (blaOXA-51-like)-encoding genes in two multiplex PCRs, as previously described.27 Identification of a strain as a member of Group 1 or Group 2 required the amplification of all fragments in the corresponding multiplex PCR and an absence of any amplification by the other multiplex PCR. Group 3 isolates were defined by the amplification of only the ompA fragment in the Group 2 PCR, and the amplification of only the csuE and blaOXA-51-like fragments in the Group 1 PCR. Isolates pertained to the novel variant of the PCR-based group, according to the new combination of amplified products. The amplification reaction was carried out by using a thermal cycler (Mastercycler Eppendorf, Germany) with an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 1 min. Besides, a final extension step was conducted at 72°C for 5 min. The MLST was also performed the previously described method by Bartual.28 Primer sequences, sequence types (STs), allele sequences, clonal complexes (CCs), and other details are available in the MLST website at http://pubmlst.org. This typing method was carried out for isolates with colistin MICs ≥ 1 µg/mL.
Statistical Analysis
Descriptive data were analyzed using Microsoft Excel and SPSS version 22 statistics software (IBM Corporation, Armonk, NY, USA). Fisher’s exact test was employed to analyze significance. In addition, P-value < 0.05 was considered as significance level. The results are presented as descriptive statistics in terms of relative frequency.
Results
Study Population, Sampling, and Antibiotic Susceptibility
Seventy non-duplicate isolates of A. baumannii were obtained from the studied hospital and included 42 (60%) from male patients and 28 (40%) from female patients with the mean age of 42.5 ±10 years (ranged 5–80 years). All the 70 isolates originated from the following wards: 31 (44.2%) from the ICU, as well as 5 (7.1%) from pediatric, 8 (11.5%) from urology, 10 (14.3%) from surgery, 6 (8.6%) from infectious diseases, 7 (10%) from general and 3 (4.3%) from neurology units. These isolates were recovered from different clinical specimens, including CSF (5, 7.1%), pleural fluid (7, 10%), urine (8, 11.4%), blood (9, 12.8%), catheter (15, 21.4%), and tracheal aspirates (26, 37.1%). Based on antibiotic susceptibility testing, among 70 A. baumannii isolates screened, 61 (87.1%) isolates were MDR and 9 (12.8%) isolates were extensively drug-resistant (XDR).
The results showed that colistin and tigecycline with the resistance rates of 2.8% and 45.7%, respectively, were the most active agents, while cephalosporins with 100% resistance had no effect on A. baumannii isolates. Also, resistance to other antibiotics was ≥50% (Table 2). Among 70 A. baumannii isolates, 52 isolates (74.2%) were carbapenem-resistant A.baumannii (CRAB). Resistance to colistin was found in 2 isolates (Ab 4 and Ab 41) with MICs 32 and 8 µg/mL, XDR phenotype, and international clones 1 and 2 (Table 2).
Table 2.
Antibiotic Susceptibility Pattern of A. baumannii Isolates
| Antibiotic | LEV | CTZ | CFP | PIP | TGC | AMP/S | COL | CTX | SXT | AMK | CIP | IMI | MER | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptibility pattern |
R N (%) |
65 (92.8) |
70 (100) |
70 (100) |
68 (97.1) |
32 (45.7) |
55 (78.6) |
2 (2.8) |
70 (100) |
67 (95.7) |
59 (84.2) |
65 (92.8) |
50 (71.4) |
52 (74.2) |
|
I N (%) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
2 (2.8) |
0 (0.0) |
3 (4.3) |
0 0.0) |
|
|
S N (%) |
5 (7.1) |
0 (0.0) |
0 (0.0) |
2 (2.8) |
38 (54.3) |
15 (21.4) |
68 (97.1) |
0 (0.0) |
3 (4.3) |
9 (12.8) |
5 (7.1) |
17 (24.2) |
18 (25.7) |
|
| Interpretive categories and MIC breakpoints (µg/mL) | R | ≥8 | ≥32 | ≥32 | ≥128 | ≥8 | ≥32/16 | ≥4 | ≥64 | ≥4/76 | 64 | ≥4 | ≥8 | ≥8 |
| I | 4 | 16 | 16 | 32–64 | – | 16/8 | – | 16–32 | – | 32 | 2 | 4 | 4 | |
| S | ≤2 | ≤8 | ≤8 | ≤16 | <8 | ≤8/4 | ≤2 | ≤8 | ≤ 2/38 | 16 | ≤1 | ≤2 | ≤2 | |
Abbreviations: LEV, levofloxacin; CTZ, ceftazidime; CFP, cefepime; PIP, piperacillin; TGC, tigecycline; Amp/S, ampicillin/sulbactam; COL, colistin; CTX, cefotaxime; SXT, trimethoprim-sulphamethoxazole; AMK, amikacin; CIP, ciprofloxacin; IMI, imipenem; MEM, meropenem; R, resistant; I, intermediate; S, susceptible.
These isolates were strong biofilm producers and had biofilm-related genes and pmrA, pmrB genes. Among the meropenem-resistant and -sensitive isolates, 52 (74.2%) and 18 (25.7%) isolates had the MICs range from ≥8 to 256 µg/mL and from 0.25 to ≤2, respectively. The MICs for imipenem ranged from 0.25 to ≤2 µg/mL in 17 (24.2) isolates, while 50 (71.4%) isolates had MICs ≥ 8–256 µg/mL for this antibiotic.
Distribution of Resistance Genes and Expression of pmrA and pmrB
PCR results showed the presence of the blaOXA-51-like gene in all the isolates. The frequency rates of biofilm-related genes were 64 (91.4%), 70 (100%), 56 (80%), and 22 (31.42%) for bap, ompA, csuE, and blaPER1, respectively (Table 3). Also, 15 (21.42%) isolates had these genes simultaneously, and all the isolates included at least one biofilm-related gene. The relationship between the biofilm formation and related genes in A. baumannii strains with MDR and XDR phenotypes is presented in Table 4. Coexistence genes, bap and ompA, were observed in all the strong and moderate biofilm producers, and more than 55% did not have blaPER1 gene.
Table 3.
Characteristics of A. baumannii Strains
| Strain ID | Biofilm | Phe | MIC | Biofilm Genes | IC | MLST | |||
|---|---|---|---|---|---|---|---|---|---|
| MER | TGC | COL | |||||||
| Ab 1 | S | XDR | 16 | 64 | 0.5 | bap, ompA, csuE | G1 | – | – |
| Ab 2 | S | MDR | 0.25 | 16 | 0.5 | bap, ompA, csuE, blaPER-1 | G1 | – | – |
| Ab 3 | S | XDR | 64 | 2 | 0.5 | bap, ompA, csuE | G2 | – | – |
| Ab 4 | S | XDR | 128 | 8 | 32 | bap, ompA, csuE, blaPER-1 | G2 | 387 | – |
| Ab 5 | W | MDR | 16 | 16 | 0.25 | bap,ompA | G1 | – | – |
| Ab 6 | S | MDR | 0.5 | 4 | 0.25 | bap, ompA, csuE, blaPER-1 | G2 | ||
| Ab 7 | S | MDR | 128 | 0.25 | 1 | bap, ompA, csuE | G15 | 138 | 92 |
| Ab 8 | S | MDR | 16 | 32 | 0.5 | bap, ompA, csuE | G1 | – | – |
| Ab 9 | S | MDR | 8 | 1 | 0.5 | bap, ompA, csuE, blaPER-1 | G1 | – | – |
| Ab 10 | N | XDR | 32 | 0.5 | 0.25 | ompA, csuE, blaPER-1 | G10 | – | – |
| Ab 11 | M | MDR | 0.5 | 64 | 0.125 | bap, ompA, csuE | G1 | – | – |
| Ab 12 | S | MDR | 128 | 0.25 | 0.25 | bap, ompA, csuE, blaPER-1 | G1 | – | – |
| Ab 13 | S | MDR | 0.25 | 2 | 0.5 | bap, ompA, csuE | G1 | – | – |
| Ab 14 | S | MDR | 16 | 32 | 0.5 | bap, ompA | G4 | – | – |
| Ab 15 | S | MDR | 256 | 0.25 | 0.25 | bap, ompA | G10 | – | – |
| Ab 16 | N | MDR | 256 | 1 | 0.25 | ompA, csuE, blaPER-1 | G1 | ||
| Ab 17 | S | MDR | 32 | 0.5 | 0.25 | bap, ompA, csuE | G2 | – | – |
| Ab 18 | S | MDR | 0.5 | 32 | 0.125 | bap, ompA, csuE | G1 | – | – |
| Ab 19 | M | MDR | 0.5 | 0.25 | 0.25 | bap, ompA, csuE | G10 | – | – |
| Ab 20 | S | MDR | 32 | 16 | 0.5 | bap, ompA, csuE | G1 | – | – |
| Ab 21 | S | MDR | 16 | 1 | 0.25 | bap, ompA | G1 | – | – |
| Ab 22 | S | MDR | 128 | 0.5 | 1 | bap, ompA, csuE, blaPER-1 | G1 | 118 | 92 |
| Ab 23 | S | MDR | 256 | 16 | 0.25 | bap, ompA, csuE, blaPER-1 | G15 | – | – |
| Ab 24 | N | MDR | 16 | 2 | 0.25 | ompA, csuE | G 1 | – | – |
| Ab 25 | S | MDR | 2 | 8 | 0.125 | bap, ompA, csuE | G 4 | – | – |
| Ab 26 | S | MDR | 16 | 4 | 2 | bap, ompA | G 1 | 421 | 92 |
| Ab 27 | S | XDR | 16 | 32 | 0.25 | bap, ompA, csuE, blaPER-1 | G 10 | – | – |
| Ab 28 | W | MDR | 64 | 0.5 | 0.25 | ompA, csuE | G 1 | – | – |
| Ab 29 | S | MDR | 0.25 | 8 | 0.25 | bap, ompA, csuE, blaPER-1 | G 1 | – | – |
| Ab 30 | S | MDR | 0.5 | 8 | 0.5 | bap, ompA, csuE | G 2 | – | – |
| Ab 31 | S | MDR | 8 | 16 | 0.25 | bap, ompA | G 1 | – | – |
| Ab 32 | S | MDR | 128 | 1 | 1 | bap, ompA, csuE | G 4 | 216 | 405 |
| Ab 33 | S | MDR | 0.5 | 0.25 | 0.125 | bap, ompA, csuE | G 7 | – | – |
| Ab 34 | M | MDR | 32 | 32 | 0.25 | bap,ompA, csuE, blaPER-1 | G 10 | – | – |
| Ab 35 | S | MDR | 0.5 | 1 | 0.25 | bap, ompA | G 1 | – | – |
| Ab 36 | M | MDR | 16 | 0.5 | 0.5 | bap,ompA, csuE | G 15 | – | – |
| Ab 37 | N | MDR | 256 | 8 | 2 | ompA, csuE, blaPER-1 | G 1 | 735 | 92 |
| Ab 38 | M | MDR | 0.5 | 32 | 0.5 | bap, ompA, csuE, blaPER-1 | G 2 | – | – |
| Ab 39 | S | MDR | 0.25 | 4 | 0.25 | bap, ompA, csuE | G 10 | – | – |
| Ab 40 | S | MDR | 16 | 1 | 0.25 | bap, ompA, csuE | G 1 | – | – |
| Ab 41 | S | XDR | 64 | 16 | 8 | bap, ompA, blaPER-1 | G 1 | 1812 | – |
| Ab 42 | S | MDR | 16 | 0.5 | 0.25 | bap, ompA, csuE | G 1 | – | – |
| Ab 43 | S | MDR | 2 | 4 | 0.25 | bap, ompA, csuE | G10 | – | – |
| Ab 44 | S | MDR | 16 | 4 | 0.5 | bap, ompA, csuE | G 1 | – | – |
| Ab 45 | M | MDR | 128 | 16 | 0.25 | bap,ompA, blaPER-1 | G15 | – | – |
| Ab 46 | S | MDR | 16 | 0.5 | 0.5 | bap, ompA, csuE | G4 | – | – |
| Ab 47 | S | MDR | 2 | 32 | 0.25 | bap, ompA, csuE | G 1 | – | – |
| Ab 48 | S | MDR | 16 | 1 | 0.5 | bap, ompA, csuE, blaPER-1 | G1 | – | – |
| Ab 49 | S | MDR | 16 | 8 | 1 | bap, ompA | G2 | 133 | 225 |
| Ab 50 | S | MDR | 32 | 8 | 0.25 | bap, ompA, csuE | G1 | – | – |
| Ab 51 | S | XDR | 16 | 0.25 | 0.5 | bap, ompA, csuE | G1 | – | – |
| Ab 52 | M | MDR | 16 | 2 | 0.25 | bap,ompA | G7 | – | – |
| Ab 53 | S | XDR | 0.5 | 16 | 0.25 | bap, ompA, csuE | G10 | – | – |
| Ab 54 | S | MDR | 16 | 8 | 0.5 | bap, ompA, csuE, blaPER-1 | G1 | – | – |
| Ab 55 | S | MDR | 64 | 0.5 | 0.5 | bap, ompA, csuE | G 1 | – | – |
| Ab 56 | S | MDR | 16 | 4 | 0.25 | bap, ompA, csuE, blaPER-1 | G 1 | – | – |
| Ab 57 | S | MDR | 32 | 16 | 0.5 | bap, ompA, csuE | G 4 | – | – |
| Ab 58 | S | MDR | 16 | 8 | 0.25 | bap, ompA, csuE | G1 | – | – |
| Ab 59 | S | MDR | 0.25 | 0.5 | 0.5 | bap, ompA, csuE, blaPER-1 | G4 | – | – |
| Ab 60 | S | MDR | 16 | 1 | 0.25 | bap, ompA | G1 | – | – |
| Ab 61 | S | MDR | 128 | 0.5 | 1 | bap, ompA, csuE | G7 | 848 | 92 |
| Ab62 | S | MDR | 16 | 8 | 0.25 | bap, ompA, csuE | G15 | – | – |
| Ab 63 | S | MDR | 32 | 1 | 0.5 | bap, ompA, csuE | G10 | – | – |
| Ab 64 | N | MDR | 16 | 2 | 0.25 | ompA, csuE, blaPER-1 | G2 | – | – |
| Ab 65 | S | MDR | 125 | 32 | 0.25 | bap, ompA, csuE | G1 | – | – |
| Ab 66 | S | XDR | 16 | 32 | 0.5 | bap, ompA | G1 | – | – |
| Ab 67 | S | MDR | 16 | 64 | 0.25 | bap, ompA, csuE | G10 | – | – |
| Ab 68 | S | MDR | 0.25 | 0.5 | 0.25 | bap, ompA, blaPER-1 | G1 | – | – |
| Ab 69 | W | MDR | 16 | 2 | 1 | bap,ompA, csuE | G10 | 118 | 92 |
| Ab 70 | S | MDR | 128 | 16 | 0.25 | bap, ompA, csuE | G1 | – | – |
Abbreviations: Phe, phenotype; MDR, multidrug-resistant; XDR, extensively drug-resistant; M, medium; N, non-biofilm-forming; W, weak; S, strong; MIC, minimum inhibitory concentration; MER, meropenem; TGC, tigecycline; COL, colistin; IC, international clonal lineage.
Table 4.
The Relationship Between Biofilm Formation and Related Genes in A. baumannii Strains with MDR and XDR Phenotypes
| Biofilm Intensity (No.) | bap No (%) | ompA No (%) | csuE No (%) | bla-PER1 No (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |
| Strong (55) | 55 (100) | 0 (0) | 55(100) | 0 (0) | 44 (80) | 11(20) | 15(27.27) | 40(72.73) |
| Moderate (7) | 7 (100) | 0 (0) | 7 (100) | 0 (0) | 5(71.4) | 2(28.57) | 3 (42.85) | 4 (57.14) |
| Weak (3) | 2 (66.6) | 1 (33.3) | 3(100) | 0 (0) | 2 (66.6) | 1 (33.3) | 0 (0) | 3 (100) |
| Non- Biofilm forming (5) | 0 (0) | 5 (100) | 5(100) | 0 (0) | 5 (100) | 0 (0) | 4 (80) | 1(20) |
| Total | 64(91.4) | 6 (8.6) | 70(100) | 0 (0) | 56 (80) | 14 (20) | 22 (31.42) | 48(68.58) |
| P value | <0.001 | – | 0.552 | 0.053 | ||||
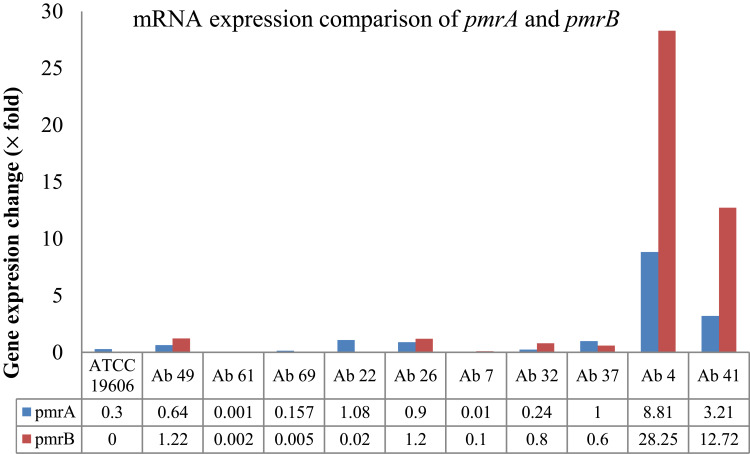
This study found a strong association between the biofilm intensity and presence of bap gene in A. baumannii isolates (P=0.001). Two colistin-resistant isolates contained all three genes (bap, ompA, and blaPER1) involved in the biofilm formation. Besides, all the isolates susceptible to meropenem had bap and ompA genes. PmrA and pmrB genes were detected in all the isolates, but mcr1 and mcr2 were not identified in any of the isolates. In the end, we determined the expression of the pmrA and pmrB genes in two colistin-resistant (Ab 4 and Ab 41) and eight colistin-susceptible isolates (Ab 49, Ab 61, Ab 69, Ab 22, Ab 26, Ab 7, Ab 32, and Ab 37). Overexpression means that the isolates have fourfold increase in the expression level of genes compared with the control strain A. baumannii ATCC 19606. The pmrA and pmrB genes were overexpressed in two colistin-resistant isolates, but their expression did not change in colistin-sensitive isolates (Figure 1). In colistin-resistant isolates, the pmrB gene had higher expression than the pmrA gene.
Figure 1.
Gene expression comparison of pmrA and pmrB in colistin-resistant (Ab 4, Ab41) and susceptible isolates (Ab49, Ab61, Ab69, Ab22, Ab26, Ab7, Ab32, Ab37) and A. baumannii ATCC 19606.
Biofilm Formation Assay
Assessment of biofilm formation was carried out using the MTP method. Among all the isolates, 55 (78.5%), 7 (10%), and 3 (4.3%) were strong, moderate, and weak biofilm producers, respectively, while no biofilm was observed in 5 (7.1%) isolates. The OD570 values for negative and positive controls were 0.044 ± 0.005 and 0.411 ± 0.041 (strong biofilm producer), respectively. Also, of 55 strong biofilm producers, 47 (77%) isolates were MDR, and 8 (88.9%) isolates were XDR. The relationship between the biofilm intensity and antibiotic susceptibility is shown in Table 4. All the weak and moderate biofilm producers were MDR phenotype and meropenem resistant. In addition, the bap and ompA genes were detected in all the strong and moderate biofilm producers.
Molecular Typing
IC type analysis showed six different PCR-based groups (G1, G2, G4, G7, G10, and G15) among A. baumannii isolates. Eight (11.4%) and 37 (52.8%) isolates belonged to group 2 (IC I) and group 1 (IC II), respectively. In addition, 3 (4.2%), 5 (7.1%), 6 (8.5%), and 11 (15.7%) isolates were belonging to four IC variants PCR-based groups, ie, G7, G15, G10, and G4, respectively. All the isolates belonged to IC I were resistant to eight antibiotics, and more than 65% of the isolates belonged to IC II were resistant to all the antibiotics, except for tigecycline and colistin. Among strong biofilm-producing isolates, 6 (10.9%), 31 (56.36%), and 18 (32.72%) were belonging to IC I, IC II, and IC variants, respectively (Table 5).
Table 5.
The Relation Between Biofilm Intensity with Antibiotic Susceptibility, Phenotype, and IC
| Biofilm Intensity (No.) | Antibiotic N (%) | Phenotype N (%) | IC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Colistin | Meropenem | Tigecycline | MDR | XDR | I | II | IV | ||||
| R | S | R | S | R | S | ||||||
| Strong (55) | 2 (3.6) | 53 (96.4) | 40 (72.72) | 15 (27.28) | 26 (47.27) | 29(52.73) | 47 (85.45) | 8(14.54) | 6 | 31 | 18 |
| Moderate(7) | 0 (0) | 7 (100) | 4 (57.15) | 3 (42.85) | 4 (57.14) | 3 (42.86) | 7 (100) | 0(0) | 1 | 1 | 5 |
| Weak(3) | 0 (0) | 3(100) | 3 (100) | 0 (0) | 1(33.33) | 2 (66.67) | 3 (100) | 0(0) | 0 | 2 | 1 |
| Non- Biofilm forming (5) |
0 (0) | 5 (100) | 5(100) | 0 (0) | 1(20) | 4(80) | 4 (80) | 1(20) | 1 | 3 | 1 |
| p-value | 1 | 0.342 | 0.608 | 0.662 | |||||||
| Total | 2 (2.8) | 68 (97.1) | 52 | 18 | 32 | 38 | 61(87.1) | 9 (12.8) | 8 | 37 | 25 |
MLST analysis for isolates with colistin MICs ≥ 1 µg/mL revealed a total of nine different STs. It also showed six isolates belonging to CC 92 (corresponding to ST801, ST118, ST138, ST421, and ST735). These isolates were meropenem resistant (MIC range: 16–256 µg/mL), MDR phenotype, and belonged to IC G1, G7, G10, and G15. Other isolates were, however, belonging to ST133 and ST216 and two colistin-resistant (Ab 4 and Ab 41) isolates to ST387 and ST1812.
Discussion
A. baumannii, due to its multidrug resistance characteristic, has been one of the axes of interest of the medical community in recent years.29,30 Scattered reports of its resistance to the last-line treatment, colistin, have raised further concerns for the medical field.7,8,23,24 A. baumannii has different mechanisms for resistance to colistin. It is also featured by having ability to produce biofilms, which are very effective in maintaining and retaining this bacterium in hospital environments.
In this study, we investigated the biofilm-producing properties and some colistin resistance mechanisms of clinical A. baumannii isolates collected from Ahvaz City, Southwestern Iran. Based on the results of antibiotic susceptibility test, the tigecycline and colistin with 54.3% and 97.1% sensitivity were the most effective antibiotics against A. baumannii isolates, which confirms previous reports from Iran by Hatami et al2 and Noori et al.31 Besides, 87.1% frequency of MDR A. baumannii isolates in our study was comparable with the results obtained in Iran by Salehi et al32 and Khalilzadegan et al.33 However, studies in Brazil34 and Morocco35 have disclosed the lower rates of MDR A. baumannii than our study. A possible reason for this high level of antibiotic resistance in Iran is non-compliance with antibiotics use standards, overuse and misuse of antibiotics, and easy access to over-the-counter drugs in third world countries.
The current study demonstrated the rate of 2.9% for colistin-resistant A. baumannii isolates, which was lower than the resistance rate (5.6%) reported from north of Iran by Ezadi et al.36 In previous investigations in Pakistan37 and Saudi Arabia,38 no colistin-resistant A. baumannii isolates were found in clinical samples. These diverse resistance rates can be explained by differences in the epidemiology of the regions, the patterns of administration, and use of antibiotics, as well as by dissimilarities in infection treatment regulatory policies.39
According to the information available so far, no evidence of mcr genes have been reported in clinical A. baumannii isolates before 2018.40 In the present study, none of the isolates carried mcr-1 and mcr-2 resistance genes.
Other surveys that have hitherto reported the presence of different mcr genes in the clinical isolates of the A. baumannii include the studies of Hameed et al37 who reported a rate of 1.6% (1/62) for mcr-1 in Pakistan; Al-Kadmy et al41 who detected mcr-1, mcr-2, and mcr-3 genes in 73.5% (89/121), 64.5% (78/121), and 67.8% (82/121) A. baumannii isolates in Iraq; Martins-Sorenson et al42 who found mcr-4.3 in an MDR A. baumannii clinical isolate from a meningitis case in Brazil; Bitar et al43 who pinpointed mcr-4.3 in an A. baumannii isolate from a clinical origin in Czech Republic; and Rahman et al44 who reported 20% frequency rate of mcr-1 in MDR Acinetobacter isolates from India.
In another study, identified mcr genes have mostly been related to Enterobacteriaceae family bacteria or the samples of animal origin.41 In our investigation, the evaluation of pmrA and pmrB gene expression rate in colistin-resistant A. baumannii isolates showed the increased expression rate of both genes compared to colistin-sensitive strains. Park et al45 have also achieved the same result in the PmrAB two-component system in colistin-resistant A. baumannii isolates. However, in an experiment in Iran by Sepahvand et al,46 the expression rate of pmrA gene in colistin-resistant A. baumannii isolates was higher than colistin-susceptible strains, and the expression rate of pmrB gene had no significant change.
Indeed, the increased expression of the pmrA and pmrB genes reduces lipopolysaccharide, resulting in the impaired membrane permeability and colistin ineffectiveness on A. baumannii membrane.46 Additionally, mutations within pmrAB genes contribute to colistin resistance in A. baumannii strains.45,47
One of the major aspects of A. baumannii causing persistent nosocomial infections and antibiotic resistance is the biofilm formation ability of this bacterium in a variety of environments.10 In the current study, the results of MTP assay showed that 78.5% (55/70) of the isolates were strong biofilm producers, which was much higher than previous reports from Thailand with 23.6%10 and China25 with 27.3% frequency rates. The most frequent biofilm-related gene was ompA (100%), followed by bap (91.4%), csuE (80%), and blaPER1 (31.4%), which contradicts Zeighami et al’s48 result that the csuE (100%) was the most prevalent gene.
This investigation suggested a significant association between the biofilm production and presence of bap gene in A. baumannii isolates (P=0.001), which is in line with Monfared et al’s49 and Sung et al’s50 studies. A comparison of biofilm-positive and biofilm-negative A. baumannii isolates revealed that the presence of ompA, csuE, and blaPER-1 was not significantly associated with biofilm production. Existing evidence in this field has shown different results regarding the correlation of bap, ompA, blaPER-1, and csuE with the biofilm formation in A. baumannii isolates.49,51 For instance, insignificant relationship between the biofilm formation and blaPER-1 gene was detected by Bardbari et al,51 while Monfared et al49 observed a positive association between the presence of this gene and the biofilm production.
In this work, we carried out two typing methods, including three-locus dual assay multiplex PCR and MLST. The results of the first method reflected six diverse IC lineages (G1, G2, G4, G7, G10, and G15) among A. baumannii isolates, which G1 (IC II) with the rate of 52.8% was the most frequent group. The predominance of IC II was in good agreement with previous reports from Japan52 and Brazil53 that recognized the IC II as the most common clone in A. baumannii isolates. Meanwhile, the prevalence of IC I with 11.4% frequency rate was lower than the frequency rate (19%) stated by Bahador et al54 in Tehran, Iran. Another difference between this and that study was the strong biofilm production by the majority of our CI I and CI II lineages, while in the Bahador’s study,54 they were moderate biofilm producers.
MLST is a highly informative, reproducible, and portable technique, which puts isolates in a global context and can directly assign them to their CC. Thus, it is regarded as the method of choice for long-term and phylogenetic studies.55 MLST was performed only on isolates with colistin MICs ≥ 1 µg/mL, which suggested a total of nine different STs, including ST801, ST118, ST138, ST421, ST735, ST133, ST216, ST387, and ST812. All ST clones, except for ST387 and ST812, have formerly reported from Iran by Farshadzadeh et al.56 Besides, the ST801, ST118, ST138, ST421, and ST735 clones were categorized as the CC92 lineages, a similar result with the findings of Farshadzadeh et al56 and Hojabri et al57 who identified CC92 as the most prevalent clone in Iran. All the isolates allotted to CC92 were MDR and meropenem resistant. The prevalence of CC92 clone in all around the world may be due to its high adaptation and survival ability in various environments and its resistance to antibiotic pressures, including resistance to carbapenems.58
Conclusion
The present study showed a high frequency of MDR and XDR A. baumannii isolates, with high prevalence of biofilm-forming genes, ompA and bap. Moreover, the emergence of colistin-resistant A. baumannii isolates could be ascribed to the elevated expression of pmrAB gene in our region in Southwestern Iran and also its high diversity, which was verified by typing techniques. Taken together, to better understand these relationships, further studies with a higher number of colistin-resistant isolates are suggested. Moreover, control measures and regular surveillance are urgently needed to preclude the spread of these isolates.
Acknowledgments
This study was a part of the Ph.D. thesis of Saeed Khoshnood. The cooperation of the Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences is acknowledged. Our appreciation also goes to the Vice Chancellor for Research affairs, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, and Tropical and Infectious Diseases Research Center of the University for their financial (Grant No. OG-9739) and executive support.
Disclosure
The authors report no conflict of interest in this study.
References
- 1.Wareth G, Neubauer H, Sprague LD. Acinetobacter baumannii–a neglected pathogen in veterinary and environmental health in Germany. Vet Res Commun. 2019;43(1):1–6. doi: 10.1007/s11259-018-9742-0 [DOI] [PubMed] [Google Scholar]
- 2.Hatami R. The frequency of multidrug-resistance and extensively drug-resistant Acinetobacter baumannii in west of Iran. J Clin Microbiol Infect Dis. 2018;2:1. [Google Scholar]
- 3.Hassannejad N, Bahador A, Hayati Rudbari N, Modarressi MH, Parivar K. Comparison of OmpA gene-targeted real-time PCR with the conventional culture method for detection of Acinetobacter baumanii in pneumonic BALB/c mice. Iran Biomed J. 2019;23(2):159–164. doi: 10.29252/ibj.23.2.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batarseh A, Al-Sarhan A, Maayteh M, Al-Khatirei S, Alarmouti M. Antibiogram of multidrug resistant Acinetobacter baumannii isolated from clinical specimens at King Hussein Medical Centre, Jordan: a retrospective analysis. East Mediterr Health J. 2015;21(11):11. doi: 10.26719/2015.21.11.828 [DOI] [PubMed] [Google Scholar]
- 5.Rezaei E, Safari H, Naderinasab M, Aliakbarian H. Common pathogens in burn wound and changes in their drug sensitivity. Burns. 2011;37(5):805–807. doi: 10.1016/j.burns.2011.01.019 [DOI] [PubMed] [Google Scholar]
- 6.Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65(2):233–238. doi: 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- 7.Cai Y, Chai D, Wang R, Liang B, Bai N. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother. 2012;67(7):1607–1615. doi: 10.1093/jac/dks084 [DOI] [PubMed] [Google Scholar]
- 8.Ahmed SS, Alp E, Hopman J, Voss A. Global epidemiology on colistin resistant Acinetobacter baumannii. J Infect Dis. 2016. [Google Scholar]
- 9.Moffatt JH, Harper M, Adler B, et al. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55(6):3022–3024. doi: 10.1128/AAC.01732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thummeepak R, Kongthai P, Leungtongkam U, Sitthisak S. Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int Microbiol. 2016;19(2):121–129. doi: 10.2436/20.1501.01.270 [DOI] [PubMed] [Google Scholar]
- 11.Fattahian Y, Rasooli I, Gargari SL, Rahbar MR, Astaneh SD, Amani J. Protection against Acinetobacter baumannii infection via its functional deprivation of biofilm associated protein (Bap). Microb Pathog. 2011;51(6):402–406. doi: 10.1016/j.micpath.2011.09.004 [DOI] [PubMed] [Google Scholar]
- 12.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiol. 2008;154(11):3398–3409. doi: 10.1099/mic.0.2008/019471-0 [DOI] [PubMed] [Google Scholar]
- 13.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One4. 2010;e10034. doi: 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pourcel C, Minandri F, Hauck Y, et al. Identification of variable-number tandem-repeat (VNTR) sequences in Acinetobacter baumannii and interlaboratory validation of an optimized multiple-locus VNTR analysis typing scheme. J Clin Microbiol. 2011;49(2):539–548. doi: 10.1128/JCM.02003-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauck Y, Soler C, Jault P, et al. Diversity of Acinetobacter baumannii in four French military hospitals, as assessed by multiple locus variable number of tandem repeats analysis. PLoS One. 2012;7(9):e44597. doi: 10.1371/journal.pone.0044597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA. Manual of Clinical Microbiology. Washington DC: American Society for Microbiology; 2006. [Google Scholar]
- 17.Higgins PG, Wisplinghoff H, Krut O, Seifert H. A PCR‐based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin Microbiol Infect. 2007;13(12):1199–1201. doi: 10.1111/j.1469-0691.2007.01819.x [DOI] [PubMed] [Google Scholar]
- 18.CLSI. M100-S28. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Eight Informational Supplement. 2018. [Google Scholar]
- 19.Jones RN, Ferraro MJ, Reller LB, Schreckenberger PC, Swenson JM, Sader HS. Multicenter studies of tigecycline disk diffusion susceptibility results for Acinetobacter spp. J Clin Microbiol. 2007;45(1):227–230. doi: 10.1128/JCM.01588-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug resistant, extensively drug resistant and pan drug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiology Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 21.Azizi O, Shahcheraghi F, Salimizand H, et al. Molecular analysis and expression of bap gene in biofilm-forming multi-drug-resistant Acinetobacter baumannii. Rep Biochem Mol Biol. 2016;5(1):62. [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, Duan X, Peng Y, Rui Y. Molecular characteristics of carbapenem-resistant Acinetobacter spp. from clinical infection samples and fecal survey samples in Southern China. BMC Infect Dis. 2019;19(1):900. doi: 10.1186/s12879-019-4423-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haeili M, Kafshdouz M, Feizabadi MM. Molecular mechanisms of colistin resistance among pandrug-resistant isolates of Acinetobacter baumannii with high case-fatality rate in intensive care unit patients. Microb Drug Resist. 2018;24(9):1271–1276. doi: 10.1089/mdr.2017.0397 [DOI] [PubMed] [Google Scholar]
- 24.Gomaa F, Helal Z, Khan M. High prevalence of blaNDM-1, blaVIM, qacE, and qacEΔ1 genes and their association with decreased susceptibility to antibiotics and common hospital biocides in clinical isolates of Acinetobacter baumannii. Microorganisms. 2017;5(2):18. doi: 10.3390/microorganisms5020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Toole GA. Microtiter dish biofilm formation assay. JoVE (J Vis Exp). 2011;(47):e2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Xia J, Xu Y, et al. Biological features of biofilm-forming ability of Acinetobacter baumannii strains derived from 121 elderly patients with hospital-acquired pneumonia. Clin Exp Med. 2016;16(1):73–80. doi: 10.1007/s10238-014-0333-2 [DOI] [PubMed] [Google Scholar]
- 27.Turton JF, Gabriel SN, Valderrey C, et al. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13(8):807–815. doi: 10.1111/j.1469-0691.2007.01759.x [DOI] [PubMed] [Google Scholar]
- 28.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato Y, Unno Y, Miyazaki C, Ubagai T, Ono Y. Multidrug-resistant Acinetobacter baumannii resists reactive oxygen species and survives in macrophages. Sci Rep. 2019;9(1):1–2. doi: 10.1038/s41598-019-53846-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jal’oot AS, Al-Ouqaili MT, Badawy AS. Molecular and bacteriological detection of multi-drug resistant and metallo-β–lactamase producer Acinetobacter baumannii in Ramadi City, West of Iraq. West Iraq Anb Med J. 2016;13:1–3. [Google Scholar]
- 31.Noori M, Mohsenzadeh B, Bahramian A, Shahi F, Mirzaei H, Khoshnood S. Characterization and frequency of antibiotic resistance related to membrane porin and efflux pump genes among Acinetobacter baumannii strains obtained from burn patients in Tehran, Iran. J Acute Dis. 2019;8(2):63–66. [Google Scholar]
- 32.Salehi B, Goudarzi H, Nikmanesh B, Houri H, Alavi-Moghaddam M, Ghalavand Z. Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran, Iran. J Infect Chem. 2018;24(7):515–523. doi: 10.1016/j.jiac.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 33.Khalilzadegan S, Sade M, Godarzi H, et al. Beta-lactamase encoded genes blaTEM and blaCTX among Acinetobacter baumannii species isolated from medical devices of intensive care units in Tehran hospitals. Jundishapur J Microbiol. 2016;9(5):e14990. doi: 10.5812/jjm.14990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira AE, Marchetti DP, Cunha GR, et al. Molecular characterization of clinical multiresistant isolates of Acinetobacter sp. from hospitals in Porto Alegre, State of Rio Grande do Sul, Brazil. Rev Soc Bras Med Trop. 2011;44(6):725–730. doi: 10.1590/S0037-86822011000600014 [DOI] [PubMed] [Google Scholar]
- 35.Uwingabiye J, Frikh M, Lemnouer A, et al. Acinetobacter infections prevalence and frequency of the antibiotics resistance: comparative study of intensive care units versus other hospital units. Pan Afr Med J. 2016;23:191. doi: 10.11604/pamj.2016.23.191.7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ezadi F, Jamali A, Heidari A, Javid N, Ardebili A. Heteroresistance to colistin in oxacillinases-producing carbapenem-resistant Acinetobacter baumannii clinical isolates from Gorgan, Northern Iran. J Glob Antimicrob Resist. 2019. doi: 10.1016/j.jgar.2019.11.010 [DOI] [PubMed] [Google Scholar]
- 37.Hameed F, Khan MA, Muhammad H, Sarwar T, Bilal H, Rehman TU. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev Soc Bras Med Trop. 2019;52:e20190237. doi: 10.1590/0037-8682-0237-2019 [DOI] [PubMed] [Google Scholar]
- 38.Abdalhamid B, Hassan H, Itbaileh A, Shorman M. Characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates in a tertiary care hospital in Saudi Arabia. New Microbiol. 2014;37(1):65–73. [PubMed] [Google Scholar]
- 39.Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules. 2018;23(4):795. doi: 10.3390/molecules23040795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dortet L, Potron A, Bonnin RA, et al. Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci Rep. 2018;8(1):1–5. doi: 10.1038/s41598-018-35041-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Kadmy IM, Ibrahim SA, Al-Saryi N, Aziz SN, Besinis A, Hetta HF. Prevalence of genes involved in colistin resistance in Acinetobacter baumannii: first report from Iraq. Microb Drug Resist. 2019. doi: 10.1089/mdr.2019.0243 [DOI] [PubMed] [Google Scholar]
- 42.Martins-Sorenson N, Snesrud E, Xavier DE, et al. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J Antimicrob Chemother. 2020;75(1):60–64. doi: 10.1093/jac/dkz413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bitar I, Medvecky M, Gelbicova T, et al. Complete nucleotide sequences of mcr-4.3-carrying plasmids in Acinetobacter baumannii sequence type 345 of human and food origin from the Czech Republic, the First Case in Europe. Antimicrob Agents Chemother. 2019;63(10):e01166–19. doi: 10.1128/AAC.01166-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahman M, Ahmad S. 549. First report for emergence of chromosomal borne colistin resistance gene mcr-1 in a clinical acinetobacter baumannii isolates from India. Open Forum Infect Dis. 2019;6(Supplement_2):S261–S262. doi: 10.1093/ofid/ofz360.618 [DOI] [Google Scholar]
- 45.Park YK, Choi JY, Shin D, Ko KS. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 2011;37(6):525–530. doi: 10.1016/j.ijantimicag.2011.02.008 [DOI] [PubMed] [Google Scholar]
- 46.Sepahvand S, Davarpanah MA, Roudgari A, Bahador A, Karbasizade V, Jahromi ZK. Molecular evaluation of colistin-resistant gene expression changes in Acinetobacter baumannii with real-time polymerase chain reaction. Infect Drug Resist. 2017;10:455–462. doi: 10.2147/IDR.S141196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lean SS, Suhaili Z, Ismail S, et al. Prevalence and genetic characterization of polymyxin-resistant Acinetobacter baumannii isolated from a tertiary hospital in Terengganu, Malaysia. ISRN Microbiol. 2014;2014:953417. doi: 10.1155/2014/953417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeighami H, Valadkhani F, Shapouri R, Samadi E, Haghi F. Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients. BMC Infect Dis. 2019;19(1):629. doi: 10.1186/s12879-019-4272-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monfared AM, Rezaei A, Poursina F, Faghri J. Detection of genes involved in biofilm formation in MDR and XDR Acinetobacter baumannii isolated from human clinical specimens in Isfahan, Iran. Arch Clin Infect Dis. 2019;14(2):e85766. [Google Scholar]
- 50.Sung JY, Koo SH, Kim S, Kwon GC. Persistence of multidrug resistant Acinetobacter baumannii isolates harboring blaOXA-23 and bap for 5 years. J Microbiol Biotechnol. 2016;26(8):1481–1489. doi: 10.4014/jmb.1604.04049 [DOI] [PubMed] [Google Scholar]
- 51.Bardbari AM, Arabestani MR, Karami M, Keramat F, Alikhani MY, Bagheri KP. Correlation between ability of biofilm formation with their responsible genes and MDR patterns in clinical and environmental Acinetobacter baumannii isolates. Microb Pathog. 2017;108:122–128. doi: 10.1016/j.micpath.2017.04.039 [DOI] [PubMed] [Google Scholar]
- 52.Matsui M, Suzuki M, Suzuki M, et al. Distribution and molecular characterization of Acinetobacter baumannii international clone II lineage in Japan. Antimicrob Agents Chemother. 2018;62(2):e02190–17. doi: 10.1128/AAC.02190-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martins N, Dalla-Costa L, Uehara AA, Riley LW, Moreira BM. Emergence of Acinetobacter baumannii international clone II in Brazil: reflection of a global expansion. Infect Genet Evol. 2013;20:378–380. doi: 10.1016/j.meegid.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bahador A, Farshadzadeh Z, Raoofian R, et al. Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: analysis of genotype, antimicrobial susceptibility, and biofilm formation. Ann Clin Microbiol Antimicrob. 2018;17(1):24. doi: 10.1186/s12941-018-0277-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rafei R, Dabboussi F, Hamze M, et al. Molecular analysis of Acinetobacter baumannii strains isolated in Lebanon using four different typing methods. PLoS One. 2014;9(12):e115969. doi: 10.1371/journal.pone.0115969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farshadzadeh Z, Hashemi FB, Rahimi S, et al. Wide distribution of carbapenem resistant Acinetobacter baumannii in burns patients in Iran. Front Microbiol. 2015;6:1146. doi: 10.3389/fmicb.2015.01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hojabri Z, Pajand O, Bonura C, Aleo A, Giammanco A, Mammina C. Molecular epidemiology of Acinetobacter baumannii in Iran: endemic and epidemic spread of multiresistant isolates. J Antimicrob Chemother. 2014;69(9):2383–2387. doi: 10.1093/jac/dku045 [DOI] [PubMed] [Google Scholar]
- 58.Chang Y, Luan G, Xu Y, et al. Characterization of carbapenem-resistant Acinetobacter baumannii isolates in a Chinese teaching hospital. Front Microbiol. 2015;6:910. doi: 10.3389/fmicb.2015.00910 [DOI] [PMC free article] [PubMed] [Google Scholar]