Abstract
Cell migration is a key process in health and disease. In the last decade an increasing attention is given to chromatin organization in migrating cells. In various types of cells induction of migration leads to a global increase in heterochromatin levels. Heterochromatin is required for optimal cell migration capabilities, since various interventions with heterochromatin formation impeded the migration rate of numerous cell types. Heterochromatin supports the migration process by affecting both the mechanical properties of the nucleus as well as the genetic processes taking place within it. Increased heterochromatin levels elevate nuclear rigidity in a manner that allows faster cell migration in 3D environments. Condensed chromatin and a more rigid nucleus may increase nuclear durability to shear stress and prevent DNA damage during the migration process. In addition, heterochromatin reorganization in migrating cells is important for induction of migration-specific transcriptional plan together with inhibition of many other unnecessary transcriptional changes. Thus, chromatin organization appears to have a key role in the cellular migration process.
Keywords: cell nucleus, chromatin, histones, genome organization, cancer metastasis
Introduction
Chromatin is classically divided to euchromatin and heterochromatin. Euchromatin contains relatively open and active chromatin regions, while heterochromatin includes more condensed, gene-poor and less active chromatin regions (Carlberg and Molnár, 2019). Heterochromatin is subdivided to facultative and constitutive heterochromatin. The former contains repressed genes in a cell type-specific manner, while the latter is formed mainly over repetitive sequences and transposons localized at constant positions in various cell types such as pericentromeric regions, which are transcribed, although, at a very low level (Saksouk et al., 2015; Allshire and Madhani, 2018; Talbert and Henikoff, 2018; Marsano et al., 2019). Heterochromatin formation and maintenance is achieved by a battery of factors including histone variants, non-coding RNAs, DNA and histone modifications, factors that read these modifications, chromatin architectural proteins and chromatin remodeling factors (Allshire and Madhani, 2018). In general, DNA methylation is found in both types of heterochromatin, while facultative heterochromatin is enriched with the histone variant macroH2A, and the histone methylation marks H3K27me3, H2AK119Ub, and to less extent H4K20me1. Constitutive heterochromatin is enriched with H3K9me2/3 and H4K20me2/3 (Fodor et al., 2010; Fadloun et al., 2013; Mozzetta et al., 2015; Saksouk et al., 2015). These modifications promote chromatin condensation through the factors that bind them, which are termed readers (Soshnev et al., 2016). These readers include MeCP2, HP1 proteins, BAHD1 and L3MBTL1 (Canzio et al., 2014; Gozani and Shi, 2014; Mozzetta et al., 2015; Zhao et al., 2016; Tillotson and Bird, 2019). Increased nucleosome compaction in heterochromatin is achieved also by the chromatin architectural protein histone H1 that can be inhibited by phosphorylation (Hergeth and Schneider, 2015; Fyodorov et al., 2018) and chromatin remodeling factors such as ATRX (Clynes et al., 2013; Dyer et al., 2017). Not less important is the eviction of euchromatin markers out of heterochromatin regions (Allshire and Madhani, 2018). Historically, heterochromatin has been studied mainly in relation to regulation of gene expression during differentiation and development and to its supportive roles in cell cycle progression such as the importance of pericentromeric heterochromatin in cell division (Mozzetta et al., 2015; Saksouk et al., 2015; Allshire and Madhani, 2018; Talbert and Henikoff, 2018). However, in the last decade it has become apparent that heterochromatin levels are increased in response to cell migration signals and support better cell migration capabilities (Gerlitz and Bustin, 2011).
In animals, cell migration is a fundamental process in embryogenesis as well as in normal function of various tissues and systems such as regeneration of colon epithelium and the activity of the immune response. Mutations and deregulation of cellular migration processes are linked to various human diseases varied from intellectual disability to cancer metastasis (Nourshargh and Alon, 2014; Jiang and Nardelli, 2016; Reiner et al., 2016; Scarpa and Mayor, 2016; Stouffer et al., 2016; Lambert et al., 2017; Worbs et al., 2017; Chitty et al., 2018; Schumacher, 2019). In recent years it has been appreciated that the cell nucleus, which is the largest and most rigid cellular organelle has to undergo major changes in its position, structure and morphology during cell migration (Wolf et al., 2007, 2013; Friedl and Wolf, 2009; McGregor et al., 2016; Lele et al., 2018; Yamada and Sixt, 2019). Recent reviews covered thoroughly the emerging research field of the cell nucleus during migration while concentrating on the nuclear envelope and its interactions with the cytoskeleton (Krause and Wolf, 2015; Bone and Starr, 2016; Liu et al., 2016; McGregor et al., 2016; Madrazo et al., 2017; Calero-Cuenca et al., 2018; Kengaku, 2018; Lele et al., 2018; Manley et al., 2018; Salvermoser et al., 2018). Here I focused on the major inner nuclear component, chromatin and more specifically on heterochromatin changes and their roles in cell migration.
Heterochromatin Alterations in Migrating Cells
Induction of cell migration was found to trigger global chromatin reorganization in several cell types. Initial comprehensive analysis of global chromatin organization in migrating cells was carried out in mouse melanoma cells. In these cells, induction of migration in the wound healing assay led to a rapid increase in various heterochromatin markers that could be detected already 15–60 min after introducing the migration signals. These markers included the histone modifications H3K9me3, H3K27me3, and H4K20me1, a non-phosphorylated form of histone H1 and DNA methylation (Gerlitz et al., 2007; Gerlitz and Bustin, 2010; Maizels et al., 2017). In addition, chromatin residence time of the chromatin architectural proteins HMGA1 and HMGN2 that are involved in chromatin de-compaction turned shorter, while the chromatin residence time of histone H1 that increases chromatin compaction was prolonged (Gerlitz et al., 2007; Gerlitz and Bustin, 2010). In parallel, migrating cells were found to be more resistant to DNase I treatment compared to non-migrating cells, indicating an elevation in chromatin condensation levels in migrating cells (Gerlitz and Bustin, 2010).
Increased global chromatin condensation in response to migration signals was found in additional cell types: In human breast cancer cells H3K9me3 levels were increased in response to expression of the activated form of Amphiregulin (AR). AR is an EGF family member that upon activation undergoes cleavage and translocation from the plasma membrane to the inner nuclear membrane while activating cell migration (Isokane et al., 2008; Tanaka et al., 2012). In human and mouse CD4+ T-cells induction of migration by Vascular Cell Adhesion Molecule 1 (VCAM1) led to an increase in H3K9me2/3 levels and to a higher resistance of the genome to cleavage by DNase I and MNase (Zhang X. et al., 2016). In rat tenocytes increased DNA methylation levels and genome resistance to DNase I cleavage were identified upon induction of migration by mechano-growth factor E peptide (MGF-C25E) (Zhang B. et al., 2016). In bone marrow-derived mesenchymal stem cells higher H3K27me3 levels and increased resistance to DNase I cleavage were found after induction of migration by the chemokine-like extracellular matrix (ECM)-associated protein Osteopontin (Liu et al., 2018). In vivo, DNA methyltransferases (DNMTs) levels and DNA methylation levels were shown to increase during wound healing of mouse corneal epithelium (Luo et al., 2019) and in colorectal cancer, H3K9me3 levels were found to be higher in the tumor invasive front than in non-invasive parts of it (Yokoyama et al., 2013). Interestingly, heterochromatin reorganization in response to migration signals was found not only in mammalian cells, but also in the filamentous fungus Neurospora crassa, in which accumulation of histone H1 was detected in the leading edge of migrating nuclei (Freitag et al., 2004; Gerlitz et al., 2007), thus heterochromatin reorganization in migrating cells may be an evolutionary conserved feature.
Recently, chromatin in migrating cells has been analyzed using higher resolution next generation sequencing tools. Chromosome conformations were captured by the Hi-C technique in human neutrophil-like cells that migrated through large pores (14 μm in diameter) and through confined pores (5 μm in diameter). As anticipated, migration through narrower pores associated with a higher degree of changes in chromosome conformations. Interestingly, disruptions of short-range interactions and of topologically associating domains (TADs) occurred to a higher extent in heterochromatin regions (compartment B in Hi-C analysis) than in euchromatin (compartment A) (Jacobson et al., 2018). Detailed analysis of heterochromatin was carried out in migrating mouse melanoma cells by a ChIP-seq analysis of the heterochromatin markers H3K9me, H3K27me3, and H4K20me1. Interestingly, upon induction of migration these markers were found to spread over larger genomic regions, while accumulating to a lesser extent, in specific genomic loci to form peaks. Though smaller in number, the migration-specific peaks of H3K9me3 and H4K20me1 accumulated over repetitive regions, while the ones of H3K27me3 accumulated over genes (Segal et al., 2018). Thus, signatures of both facultative and constitutive heterochromatin have been found to be highly dynamic in migrating cells.
Effects of Heterochromatin Levels on Cell Migration Rate
Indications that global chromatin condensation is important for cell migration emerged from numerous experiments in which interference with heterochromatin formation attenuated the migration rate of a vast variety of cells. Knockdown or chemical inhibition of EZH2, which is the catalytic subunit of the H3K27 methyltransferase complex PRC2, inhibited the migration rate of various cell types (Table 1). Interfering with H3K9me2/3 levels by knocking down methyltransferases that generate these modifications such as G9a, SUV39H1, SUV39H2, SETDB1, and SETDB2 or by using chemical inhibitors of G9a and SUV39H1/2 also inhibited the migration rate of many cell types (Table 1). On the other hand, over-expression of H3K9me2/3 methyltransferases was shown to enhance the rate of cell migration (Table 1).
TABLE 1.
Studies identifying the dependence of cell migration on heterochromatin levels.
| Heterochromatin alteration and the effect on migration | Cell type (*-primary cells) | Method of manipulation | Migration assay | References |
| H3K27me3 reduction leading to inhibition of migration | Bone marrow-derived mesenchymal stem cells* | EZH2 siRNA | TA | Liu et al., 2018 |
| Mouse embryonic fibroblasts* | EZH2 siRNA | TA and WH | Kottakis et al., 2011 | |
| Endometriotic epithelial cells | EZH2 siRNA and inhibitor (GSK126) | TA and WH | Eskander et al., 2013; Zhang et al., 2017 | |
| Squamous cell carcinoma | EZH2 siRNA and inhibitors (GSK126 and EPZ-6438) | WH | Adhikary et al., 2015 | |
| Immortalized keratinocytes | EZH2 siRNA | WH | Adhikary et al., 2015 | |
| Melanoma | EZH2 siRNA and inhibitors (GSK126 and GSK343) | TA and WH | Luo et al., 2013; Maizels et al., 2017 | |
| Pancreatic cancer cells | EZH2 siRNA | TA and WH | Ma et al., 2018 | |
| Ovarian cancer cells | EZH2 siRNA | TA | Lu et al., 2010; Rao et al., 2010 | |
| Breast carcinoma cells | EZH2 siRNA | TA | Varambally et al., 2008 | |
| Prostate cancer cells | EZH2 siRNA | WH | Varambally et al., 2008 | |
| H3K27me3 enhancement leading to acceleration of migration | Pancreatic cancer cells | EZH2 OE | TA and WH | Ma et al., 2018 |
| H3K9me2/3 reduction leading to inhibition of migration | Vascular smooth muscle cells* | SUV39H1 siRNA | TA | Zhang et al., 2019a |
| Melanoma cells | SUV39H1/2 inhibitor (chaetocin) and SETDB1 siRNA | TA and WH | Gerlitz and Bustin, 2010; Maizels et al., 2017; Orouji et al., 2019 | |
| Liver cancer cells | SETDB1 siRNA | TA | Wong et al., 2016; Zhang et al., 2018 | |
| Gastric cancer cells | SETDB1 siRNA | TA | Nishikawaji et al., 2016 | |
| Lymphocytes | G9a siRNA and inhibitor (BIX01294) | TA and Collagen matrix assay | Zhang X. et al., 2016; Madrazo et al., 2018 | |
| Glioma cells | SETDB1 and SUV39H1 siRNA, SUV39H1/2 inhibitor (chaetocin) | WH | Spyropoulou et al., 2014; Sepsa et al., 2015 | |
| Breast cancer cells | SUV39H1 siRNA and SUV39H1/2 inhibitor (chaetocin) | TA and WH | Yokoyama et al., 2013 | |
| Cervical cancer cells | G9a inhibitor (BIX01294) | TA and WH | Chen et al., 2017 | |
| Lung cancer cells | G9a siRNA, inhibitor (BIX01294) and DN | TA and WH | Chen et al., 2010; Huang et al., 2017 | |
| Colon cancer cells | SETDB1 siRNA | TA | Yu et al., 2019 | |
| Melanoma cells | SUV39H1/2 inhibitor (chaetocin) | TA and WH | Gerlitz and Bustin, 2010; Maizels et al., 2017 | |
| H3K9me2/3 enhancement leading to acceleration of migration | Vascular smooth muscle cells* | SUV39H1 OE | TA | Zhang et al., 2019a |
| Melanoma cells | SETDB1 OE | TA and WH | Orouji et al., 2019 | |
| Liver cancer cells | SETDB1 OE | TA | Zhang et al., 2018 | |
| Gastric cancer cells | SETDB1 OE | TA | Nishikawaji et al., 2016 | |
| Breast cancer cells | SUV39H1 OE | TA | Yokoyama et al., 2013 | |
| Lung cancer cells | G9a OE | TA | Chen et al., 2010 | |
| Colon cancer cells | SETDB1 OE | TA | Yu et al., 2019 | |
| Reduction in DNA methylation leading to inhibition of migration | Cortical interneurons* | DNMT1 KO | Organotypic brain slice culture | Pensold et al., 2017 |
| Corneal epithelial cells* | DNMT1 siRNA | WH | Luo et al., 2019 | |
| Breast cancer cells | DNMT inhibitors (AZA, SGI, C02S) | TA and WH | Shafiei et al., 2008; Su et al., 2018; Yuan et al., 2019 | |
| Prostate cancer cells | DNMT inhibitor (AZA) | WH | Strmiska et al., 2019 | |
| Ovarian cancer cells | DNMT inhibitor (AZA) | TA | Meng et al., 2013 | |
| Lung cancer cells | DNMT1, 3a siRNA, DNMT inhibitor (AZA) | TA and WH | Mateen et al., 2013; Yan et al., 2014; Bu et al., 2018 | |
| Glioma cells | DNMT3a,b siRNA, DNMT inhibitor (AZA) | TA and WH | Wang et al., 2015; Xu et al., 2015; Guo et al., 2017 | |
| Esophageal cancer cells | DNMT inhibitor (AZA) | WH | Ahrens et al., 2015 | |
| Osteosarcoma cells | DNMT inhibitor (AZA) | WH | Gong et al., 2019 | |
| Pancreatic cancer cells | DNMT3b siRNA | TA and WH | Wang et al., 2019 | |
| Colon cancer cells | DNMT inhibitor (AZA) | WH | Oshima et al., 2019 | |
| Trophoblasts | DNMT inhibitor (AZA) | TA | Rahnama et al., 2006 | |
| DNA methylation enhancement leading to acceleration of migration | Lung cancer cells | DNMT3a OE | WH | Yan et al., 2014 |
| Liver cancer cells | DNMT3b OE | WH | Wu et al., 2019 | |
| Histone acetylation elevation leading to inhibition of migration | Bone marrow-derived mesenchymal stem cells* | HDAC inhibitor (TSA) | TA | Liu et al., 2018 |
| Neurons in C. elegans development* | HDAC1 mutations and HDAC inhibitor (TSA) | Whole animal development | Zinovyeva et al., 2006; Nambiar et al., 2007 | |
| Schwann cells* | HDAC inhibitor (TSA) | TA | Wang et al., 2014 | |
| Endothelial cells* | HDAC7 siRNA | WH | Mottet et al., 2007 | |
| Smooth muscle cells* | HDAC4 siRNA and HDAC inhibitor (TSA) | TA | Yang et al., 2012; Usui et al., 2014 | |
| Cardiac fibroblasts* | HDAC1 inhibition (ellagic acid) | TA | Lin et al., 2019 | |
| Dendritic cells* | HDAC inhibitor (TSA) | TA | Kim et al., 2013 | |
| Tenocytes* | HDAC inhibitor (TSA) | WH | Zhang B. et al., 2016 | |
| Melanoma cells | HDAC inhibitor (TSA) | TA and WH | Gerlitz and Bustin, 2010 | |
| Breast cancer cells | HDAC2, 5, 8 siRNA, HDAC inhibitors (MS275, SB939, LBH, Tub, C02S, PCI-34051, VPA) | TA and WH | Jeon and Lee, 2010; Zhang et al., 2012; Hsieh et al., 2016; Li et al., 2016; Su et al., 2018; Yuan et al., 2019 | |
| Ovarian cancer cells | HDAC3, 4 siRNA, HDAC inhibitor (TSA) | TA | Hayashi et al., 2010; Ahn et al., 2012; Meng et al., 2013 | |
| Lung cancer cells | HDAC inhibitor (Silibinin) | TA | Mateen et al., 2013 | |
| Esophageal cancer cells | HDAC inhibitor (MS-275) | WH | Ahrens et al., 2015 | |
| Transformed macrophages | HDAC inhibitor (Butyrate) | TA | Maa et al., 2010 | |
| Oral cancer cells | HDAC2 siRNA | WH | Chang et al., 2011 | |
| Prostate cancer cells | HDAC inhibitor (VPA) | TA | Wedel et al., 2011 | |
| Glioma cells | HDAC3 siRNA | TA and WH | Zhu et al., 2013 | |
| Broad histone methylation inhibition leading to chromatin decondensation and inhibition of migration | Bone marrow-derived mesenchymal stem cells* | DZNep | TA | Liu et al., 2018 |
| Tenocytes* | MTA | WH | Zhang B. et al., 2016 | |
| Chondrosarcoma | DZNep | WH | Girard et al., 2014 | |
| Melanoma cells | MTA | TA and WH | Gerlitz and Bustin, 2010 | |
| Histone H1 alterations leading to inhibition of migration | Melanoma cells | OE of histone H1 DN | TA | Gerlitz et al., 2007 |
| Glioma, osteosarcoma and gastric cancer cells | OE of histone H1 DN | TA | Sang et al., 2019; Zhang et al., 2019b; Xu et al., 2020 |
OE, over expression; DN, over expression of a dominant negative form; TA, transwell assay; WH, wound healing assay; SGI, Guadecitabine/SGI-110; MS275, Entinostat; Tub, Tubastatin A HCL; TSA, Trichostatin A; VPA, Valproic acid; DZNep, 3-Deazaneplanocin-A; MTA, 5′-deoxy-5′-methylthioadenosine.
Inhibition of DNA methylation by 5′-aza-2′-deoxycytidine (AZA) or by knockdown of DNMTs also inhibited cell migration while over-expression of DNMTs was shown to enhance cell migration (Table 1). Interference with histone H1 chromatin binding by over-expression of a dominant form composed of histone H1 C’-terminal part or of phosphor-mimicking forms containing T to E mutations also altered cell migration rate (Table 1). Interference with chromatin condensation can be achieved also by increasing global histone acetylation through inhibition of nuclear histone deacetylases (HDACs) either by chemical inhibitors or by knockdown. As listed in Table 1 and in a recent review (Wawruszak et al., 2019), such manipulations also interfere with cell migration.
In most of the described cases the interventions with heterochromatin formation (e.g., introduction of siRNA or addition of a chemical inhibitor) were introduced ≥24 h before induction of migration. In such cases it is challenging to assess whether migration inhibition was due to failure of the cells to increase heterochromatin levels only upon receiving migration signals or due to alterations in their basal transcriptome. Changes in the basal transcriptome of non-migrating cells can turn it to a less favorable one for migration even before receiving any migration signals. This scenario is supported by the findings that the number of migration-altered genes and the degree of change at their expression levels are limited (Jacobson et al., 2018; Segal et al., 2018) as described below. Moreover, many of these experiments were done in cancer cells, which acquire a migration-supporting transcriptome already during the transformation process (Lamouille et al., 2014; Dhamija and Diederichs, 2016; Huang et al., 2019). Thus, in many cases it is hard to understand if basal heterochromatin levels or migration-induced heterochromatin levels are important for the migration process. Addressing this issue can be achieved by adding chemical inhibitors in parallel to the induction of migration as done only in few cases (Gerlitz and Bustin, 2010; Jeon and Lee, 2010; Wang et al., 2014; Huang et al., 2017; Maizels et al., 2017; Liu et al., 2018). In the future, this issue could be addressed by using degron-based systems (Röth et al., 2019) for rapid depletion of heterochromatin generating enzymes.
Notably, as described above, interference with signatures of both facultative and constitutive heterochromatin can interfere with cell migration rate suggesting that both types of heterochromatin can affect cellular properties important for the migration process.
Heterochromatin Roles in Cell Migration
Heterochromatin Mechanical Roles
Increased heterochromatin levels in migrating cells are spread over large genomic regions as could be detected by immunostaining of heterochromatin markers in various cells such as melanoma cells (Gerlitz et al., 2007; Gerlitz and Bustin, 2010; Maizels et al., 2017) as well as by high resolution mapping of these markers by ChIP-seq analysis in the same melanoma cells (Segal et al., 2018). This pattern supports global changes in the physical properties of the nucleus, since a global increase in heterochromatin levels induced by divalent cations was shown to elevate the stiffness of the nucleus in both isolated nuclei (Dahl et al., 2005) and nuclei in whole cells (Stephens et al., 2019). On the other hand, over-expression of HMGN5 or HMGA1, chromatin architectural proteins that oppose histone H1 chromatin binding and compaction, led to a reduction in nuclear stiffness (Furusawa et al., 2015; Senigagliesi et al., 2019). Chromatin decondensation by chemical inhibitors such as HDAC inhibitors and the methyltransferase inhibitor DZNep also found to reduce nuclear stiffness (Stephens et al., 2017, 2018; Liu et al., 2018; Krause et al., 2019). In agreement with this, atomic force microscopy (AFM) analysis of tenocytes and bone marrow-derived mesenchymal stem cells detected an increase in nuclear stiffness following induction of migration by chemokine-like agents (Zhang B. et al., 2016; Liu et al., 2018). Similar phenomenon was also reported in human and mouse CD4+ T lymphocytes upon activation of migration by VCAM1 (Zhang X. et al., 2016).
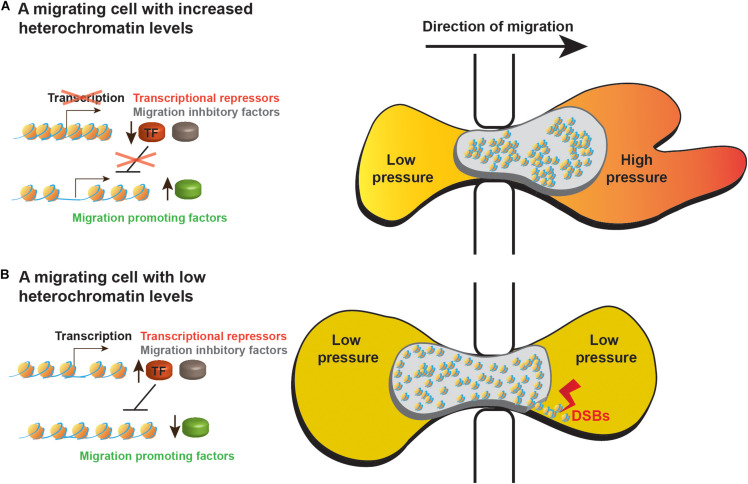
A first indication that indeed global heterochromatinization supports cell migration by altering the nuclear mechanical properties emerged of the finding that a HDAC inhibitor inhibited melanoma cell migration during a short period of time (3 h) in a similar efficiency also when transcription was inhibited (Gerlitz and Bustin, 2010). More recently, a detailed analysis of colon cancer migration through confined spaces revealed that heterochromatin-dependent nuclear stiffness generated a bigger forward jump of the nucleus once it is extracted from a narrow pore (Krause et al., 2019). Thus counter-intuitively, heterochromatin increased nuclear elasticity to generate a better spring-like behavior of the nucleus that can better support movement of the whole cell. An additional 3D migration mode that may benefit from altered nuclear physical properties by global heterochromatin formation is the nuclear piston model, which was identified in primary human cells and can be activated in tumor cells by inhibition of matrix metallopeptidases (MMPs). MMPs cleave the extra cellular matrix to facilitate easier migration of cells. 3D migration by the nuclear piston mechanism involves forward pulling of the nucleus by the actomyosin system in cooperation with the nucleoskeleton linker protein Nesprin 3. Due to the narrow diameter of a cell migrating inside the ECM, nuclear pulling divides the cytoplasm into two compartments. In the anterior compartment, the forward pulling of the nucleus by the actomyosin system increases the intracellular pressure. This pressure was found to promote formation of lobopodial protrusions that support forward movement of the cell (Petrie et al., 2014, 2017). Global heterochromatinization that increases nuclear stiffness may generate a nucleus that will not collapse and will deform only to the right degree that is required to compartmentalize the cytoplasm of a migrating cell (Figure 1). During 2D migration, we hypothesize that increased nuclear stiffness could improve momentum transfer of forces generated by the actomyosin network at the back of the nucleus leading to a more efficient usage of these forces to move the nucleus forward.
FIGURE 1.
A model of heterochromatin roles in migrating cells. Schematic representation of cells migrating through small pores while (A) increasing their heterochromatin levels or (B) leaving their heterochromatin levels low as before receiving the migration signals. Higher heterochromatin levels support better the migration process by the following mechanisms: (i) Heterochromatin-dependent stiffness of the nucleus leads to faster nuclear movement out of the restraining pore. (ii) Increased nuclear stiffness may help the actomyosin network to increase the intracellular pressure in the anterior of the cytoplasm to induce formation of lobopodial protrusions. (iii) Increased nuclear stiffness may protect the nucleus of mechanical insults, preventing nuclear envelope rupture and DNA damage such as double strand breaks (DSBs). (iv) Heterochromatin inhibits transcription of migration inhibitory factors (marked in gray) and of repressors of transcription (TF, marked in red) thus preventing transcriptional inhibition of migration promoting factors (marked in green). (v) Heterochromatin also prevents unnecessary transcriptional alterations.
Higher nuclear stiffness in migrating cells might increase resistance to shear stress that can tear the nucleus. Recent studies on the cell nucleus during 3D migration showed that this process is associated with nuclear blebbing, nuclear envelope rupture and DNA damage that are inversely linked to the diameter of the pores thorough which cells migrate (Denais et al., 2016; Raab et al., 2016; Irianto et al., 2017; Pfeifer et al., 2018; Mistriotis et al., 2019). Notably, chromatin decondensation by chemical inhibition of HDACs or methyltransferases was shown to increase nuclear blebbing, while induction of chromatin condensation by treating cells with a histone demethylase inhibitor was found to reduce nuclear blebbing (Stephens et al., 2018, 2019). Thus, chromatin condensation during cell migration may increase the whole nucleus resistance to shear stress and reduces the susceptibility of DNA to breaks (Figure 1). This hypothesis is supported by the findings that applying mechanical stress on nuclei either by pulling them into small micropipettes or by exposing cells to a biaxial extrinsic cyclic mechanical strain led to global chromatin condensation (Irianto et al., 2016; Le et al., 2016).
Heterochromatin in Transcriptional Control
One of the major roles of heterochromatin is considered to be repression of gene expression and transposons (Allshire and Madhani, 2018), however, a global reduction in transcription levels was found only in breast and ovarian cancer cells that were induced to migrate by an activated form of AR. This reduction was transient and prolonged for only 8 h (Isokane et al., 2008; Tanaka et al., 2012). In other cases such a repression was not identified (Fitsialos et al., 2007; Demuth et al., 2008; Jacobson et al., 2018; Segal et al., 2018). Moreover, active transcription is required for cell migration as the migration process continues for 8 h and more (Gerlitz and Bustin, 2010; Mason et al., 2019). Significantly, induction of migration is associated with specific changes in the cellular transcriptome in the scale of a few hundreds of genes (Fitsialos et al., 2007; Demuth et al., 2008; Jacobson et al., 2018; Segal et al., 2018). Using an EZH2-specific inhibitor to prevent H3K27 methylation upon induction of migration, in melanoma cells, revealed that H3K27 methylation is required for 33% of the 182 transcriptome changes in migrating cells. Surprisingly, H3K27 methylation was also found to prevent changes in 501 other genes that normally do not change upon induction of migration (Segal et al., 2018). Thus, migration-induced heterochromatinization is served not only to induce needed transcriptional changes, but also to prevent or to buffer unnecessary transcriptional changes. These unnecessary transcriptional changes may occur due to activation of transcription factors with multiple target genes of which only a fraction should be altered (Figure 1). A buffering role of heterochromatin in migrating cells could be seen also in migration of neutrophil-like cells, where interference with 3D genome structures occurred to a higher extent in heterochromatin regions than in euchromatin regions (Jacobson et al., 2018).
Overall, recent studies indicate that heterochromatin in migrating cells has physical roles in nuclear biomechanics as well as genetic roles in regulation of transcription. Although it is tempting to speculate that constitutive heterochromatin is important for the former roles, while facultative heterochromatin is important for the later roles, a complete analysis to support such a hypothesis has not been done yet. The findings that heterochromatin is used both to modify transcription and to prevent transcriptional changes suggest that altering the transcriptome of migrating cells should interfere with their migration rate. Especially, if the interference starts hours before induction of migration, thus it can alter the basal transcriptome. Indeed, there are studies in which interference with euchromatin markers also inhibits cell migration (Kim et al., 2018; Wang et al., 2018; Liang et al., 2019).
Concluding Remarks
Cell migration is a key process in metastasis formation in cancer. Indeed, several heterochromatin generating enzymes such as the H3K9 methyltransferases G9a and SETDB1 and the H3K27 methyltransferase EZH2 are considered oncogenes (Tiffen et al., 2015; Kang, 2018; Batham et al., 2019; Cao et al., 2019; Torrano et al., 2019), whereas the H3K27 demethylases UTX and JMJD3 are considered tumor suppressor genes, though exceptions can be found (Arcipowski et al., 2016; Perrigue et al., 2016). Epigenetic drugs that interfere with heterochromatin formation such as DNMT inhibitors and HDAC inhibitors are used in cancer treatment (Castillo-Aguilera et al., 2017; Pechalrieu et al., 2017; Roberti et al., 2019). Unfortunately, a first-order link between heterochromatin and cancer does not always exist. In recent years it has become apparent that cancer cell proliferation and migration may be supported by different transcriptional plans (Nair et al., 2019; Luo et al., 2020) as well as by different global chromatin organization features; in melanoma it seems that euchromatin supports better cell proliferation, whereas increased chromatin condensation (heterochromatin levels) better supports cell migration (Barsotti et al., 2015; Maizels et al., 2017). Thus, targeting cancer cells by a single epigenetic drug might be challenging.
The opposing effects of heterochromatin on cell migration and proliferation suggest that if a heterochromatin marker is kept at the end of the migration process as an epigenetic memory, it may interfere with proliferation. Indeed, in migrating melanoma cells H3K27me3 levels were shown to drop back to basal levels once migration ended (Gerlitz and Bustin, 2010). Still, further studies are required to reveal if epigenetic memory of previous migration episodes can be formed to enhance future migration sessions in non-proliferating cells or in cancer cells in which the proliferation process is not sensitive to high heterochromatin levels as in melanoma cells.
Heterochromatin spatial organization inside the nucleus is not uniform; in most differentiated cells a substantial part of heterochromatin accumulates at the nuclear periphery next to the nuclear envelope (van Steensel and Belmont, 2017). However, in relation to migration it was only found that activation of migration of CD4+ T lymphocytes induced association of the H3K9 methyltransferase G9a with the nuclear envelope protein lamin B1 (Zhang X. et al., 2016). Thus, the spatial organization of heterochromatin in migrating cells is still unknown.
In recent years new links between heterochromatin and the nucleolus have been found. Pericentric heterochromatin is in close association with nucleoli while both structures use similar chromatin architectural proteins for their organization such as cohesion and HDACs (Bersaglieri and Santoro, 2019; Lawrimore and Bloom, 2019). Moreover, knockdown of the nucleolar protein STK35L1 was shown to reduce the migration rate of human endothelial cells (Goyal et al., 2011) and the histone acetyl transferase NAT10 was found to translocate from the nucleolus to the cytoplasm during colorectal transformation (Zhang et al., 2014). Thus, it is worthwhile to look for changes in nucleoli organization in migrating cells and for their roles in the migration process.
An additional important endeavor is to determine if heterochromatin formation upon induction of migration prevents DNA and nuclear damage during the migration process. To better understand the roles of heterochromatin in cell migration it is crucial to enlarge the pool of cell types and histone markers analyzed by next generation sequencing methods upon induction of migration in parallel to transcriptome analysis with and without interference with heterochromatin formation.
These suggested endeavors are important to further establish the emerging notion that chromatin in migrating cells is not a passive passenger, but rather an active player. Heterochromatin formation affects both nuclear mechanical properties and the transcriptome: heterochromatin adjusts the biomechanical properties of the nucleus for more efficient usage of force generated by the cytoskeleton as well as fine-tunes the cellular transcriptome while preventing changes that could impede cell migration rate.
Author Contributions
GG screened the literature and wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to thank Tamar Listovsky, Lukasz Kaczmarczyk, Andrea Fracchia and Rosari Hernandez-Vicens for helpful comments on the manuscript.
Footnotes
Funding. This work was supported by the Israel Cancer Association (20190028) and Ariel University.
References
- Adhikary G., Grun D., Balasubramanian S., Kerr C., Huang J. M., Eckert R. L. (2015). Survival of skin cancer stem cells requires the Ezh2 polycomb group protein. Carcinogenesis 36 800–810. 10.1093/carcin/bgv064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M. Y., Kang D. O., Na Y. J., Yoon S., Choi W. S., Kang K. W., et al. (2012). Histone deacetylase inhibitor, apicidin, inhibits human ovarian cancer cell migration via class II histone deacetylase 4 silencing. Cancer Lett. 325 189–199. 10.1016/j.canlet.2012.06.017 [DOI] [PubMed] [Google Scholar]
- Ahrens T. D., Timme S., Hoeppner J., Ostendorp J., Hembach S., Follo M., et al. (2015). Selective inhibition of esophageal cancer cells by combination of HDAC inhibitors and Azacytidine. Epigenetics 10 431–445. 10.1080/15592294.2015.1039216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C., Madhani H. D. (2018). Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19 229–244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcipowski K. M., Martinez C. A., Ntziachristos P. (2016). Histone demethylases in physiology and cancer: a tale of two enzymes, JMJD3 and UTX. Curr. Opin. Genet. Dev. 36 59–67. 10.1016/j.gde.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsotti A. M., Ryskin M., Zhong W., Zhang W.-G., Giannakou A., Loreth C., et al. (2015). Epigenetic reprogramming by tumor-derived EZH2 gain-of-function mutations promotes aggressive 3D cell morphologies and enhances melanoma tumor growth. Oncotarget 6 2928–2938. 10.18632/oncotarget.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batham J., Lim P. S., Rao S. (2019). SETDB-1: a potential epigenetic regulator in breast cancer metastasis. Cancers 11:1143. 10.3390/cancers11081143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri C., Santoro R. (2019). Genome organization in and around the nucleolus. Cells 8:579 10.3390/cells8060579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone C. R., Starr D. A. (2016). Nuclear migration events throughout development. J. Cell Sci. 129 1951–1961. 10.1242/jcs.179788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu X., Zhang X., Xu J., Yang H., Zhou X., Wang H., et al. (2018). Inhibition of DNA methyltransferase 1 by RNA interference reverses epithelial-mesenchymal transition in highly metastatic 95D lung cancer cells by inhibiting the Wnt signaling pathway. Oncol. Lett. 15 9242–9250. 10.3892/ol.2018.8449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Cuenca F. J., Janota C. S., Gomes E. R. (2018). Dealing with the nucleus during cell migration. Curr. Opin. Cell Biol. 50 35–41. 10.1016/j.ceb.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Canzio D., Larson A., Narlikar G. J. (2014). Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 24 377–386. 10.1016/j.tcb.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Li L., Yang D., Zeng L., Yewei X., Yu B., et al. (2019). Recent progress in histone methyltransferase (G9a) inhibitors as anticancer agents. Eur. J. Med. Chem. 179 537–546. 10.1016/j.ejmech.2019.06.072 [DOI] [PubMed] [Google Scholar]
- Carlberg C., Molnár F. (eds) (2019). “Chromatin,” in Human Epigenetics: How Science Works, (Cham: Springer International Publishing; ), 15–28. 10.1007/978-3-030-22907-8_2 [DOI] [Google Scholar]
- Castillo-Aguilera O., Depreux P., Halby L., Arimondo P. B., Goossens L. (2017). DNA methylation targeting: the DNMT/HMT crosstalk challenge. Biomolecules 7:3. 10.3390/biom7010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-C., Lin B.-R., Chen S.-T., Hsieh T.-H., Li Y.-J., Kuo M. Y. P. (2011). HDAC2 promotes cell migration/invasion abilities through HIF-1α stabilization in human oral squamous cell carcinoma. J. Oral Pathol. Med. 40 567–575. 10.1111/j.1600-0714.2011.01009.x [DOI] [PubMed] [Google Scholar]
- Chen M. W., Hua K. T., Kao H. J., Chi C. C., Wei L. H., Johansson G., et al. (2010). H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 70 7830–7840. 10.1158/0008-5472.CAN-10-0833 [DOI] [PubMed] [Google Scholar]
- Chen R.-J., Shun C.-T., Yen M.-L., Chou C.-H., Lin M.-C. (2017). Methyltransferase G9a promotes cervical cancer angiogenesis and decreases patient survival. Oncotarget 8 62081–62098. 10.18632/oncotarget.19060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitty J. L., Filipe E. C., Lucas M. C., Herrmann D., Cox T. R., Timpson P. (2018). Recent advances in understanding the complexities of metastasis. F1000Res. 7:F1000 Faculty Rev-1169. 10.12688/f1000research.15064.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes D., Higgs D. R., Gibbons R. J. (2013). The chromatin remodeller ATRX: a repeat offender in human disease. Trends Biochem. Sci. 38 461–466. 10.1016/j.tibs.2013.06.011 [DOI] [PubMed] [Google Scholar]
- Dahl K. N., Engler A. J., Pajerowski J. D., Discher D. E. (2005). Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 89 2855–2864. 10.1529/biophysj.105.062554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth T., Rennert J. L., Hoelzinger D. B., Reavie L. B., Nakada M., Beaudry C., et al. (2008). Glioma cells on the run – the migratory transcriptome of 10 human glioma cell lines. BMC Genomics 9:54. 10.1186/1471-2164-9-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C. M., Gilbert R. M., Isermann P., McGregor A. L., te Lindert M., Weigelin B., et al. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science 352 353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamija S., Diederichs S. (2016). From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer 139 269–280. 10.1002/ijc.30039 [DOI] [PubMed] [Google Scholar]
- Dyer M. A., Qadeer Z. A., Valle-Garcia D., Bernstein E. (2017). ATRX and DAXX: mechanisms and mutations. Cold Spring Harb. Perspect. Med. 7:a026567. 10.1101/cshperspect.a026567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskander R. N., Ji T., Huynh B., Wardeh R., Randall L. M., Hoang B. (2013). Inhibition of enhancer of zeste homolog 2 (EZH2) expression is associated with decreased tumor cell proliferation, migration, and invasion in endometrial cancer cell lines. Int. J. Gynecol. Cancer 23 997–1005. 10.1097/IGC.0b013e318296a265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A., Eid A., Torres-Padilla M.-E. (2013). Mechanisms and dynamics of heterochromatin formation during mammalian development. Curr. Top. Dev. Biol. 104 1–45. 10.1016/B978-0-12-416027-9.00001-2 [DOI] [PubMed] [Google Scholar]
- Fitsialos G., Chassot A.-A., Turchi L., Dayem M. A., LeBrigand K., Moreilhon C., et al. (2007). Transcriptional signature of epidermal keratinocytes subjected to in vitro scratch wounding reveals selective roles for ERK1/2, p38, and phosphatidylinositol 3-kinase signaling pathways. J. Biol. Chem. 282 15090–15102. 10.1074/jbc.M606094200 [DOI] [PubMed] [Google Scholar]
- Fodor B. D., Shukeir N., Reuter G., Jenuwein T. (2010). Mammalian Su(var) genes in chromatin control. Annu. Rev. Cell Dev. Biol. 26 471–501. 10.1146/annurev.cellbio.042308.113225 [DOI] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Raju N. B., Selker E. U., Read N. D. (2004). GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41 897–910. 10.1016/j.fgb.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Friedl P., Wolf K. (2009). Proteolytic interstitial cell migration: a five-step process. Cancer Metastasis Rev. 28 129–135. 10.1007/s10555-008-9174-3 [DOI] [PubMed] [Google Scholar]
- Furusawa T., Rochman M., Taher L., Dimitriadis E. K., Nagashima K., Anderson S., et al. (2015). Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 6:6138. 10.1038/ncomms7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov D. V., Zhou B.-R., Skoultchi A. I., Bai Y. (2018). Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19 192–206. 10.1038/nrm.2017.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G., Bustin M. (2010). Efficient cell migration requires global chromatin condensation. J. Cell Sci. 123 2207–2217. 10.1242/jcs.058271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G., Bustin M. (2011). The role of chromatin structure in cell migration. Trends Cell Biol. 21 6–11. 10.1016/j.tcb.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G., Livnat I., Ziv C., Yarden O., Bustin M., Reiner O. (2007). Migration cues induce chromatin alterations. Traffic 8 1521–1529. 10.1111/j.1600-0854.2007.00638.x [DOI] [PubMed] [Google Scholar]
- Girard N., Bazille C., Lhuissier E., Benateau H., Llombart-Bosch A., Boumediene K., et al. (2014). 3-deazaneplanocin A (DZNep), an inhibitor of the histone methyltransferase EZH2, induces apoptosis and reduces cell migration in chondrosarcoma cells. PLoS One 9:e98176. 10.1371/journal.pone.0098176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H., Tao Y., Mao X., Song D., You D., Ni J. (2019). MicroRNA-29a suppresses the invasion and migration of osteosarcoma cells by regulating the SOCS1/NF-κB signalling pathway through negatively targeting DNMT3B. Int. J. Mol. Med. 44 1219–1232. 10.3892/ijmm.2019.4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal P., Behring A., Kumar A., Siess W. (2011). STK35L1 associates with nuclear actin and regulates cell cycle and migration of endothelial cells. PLoS One 6:e16249. 10.1371/journal.pone.0016249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Shi Y. (2014). “Histone methylation in chromatin signaling,” in Fundamentals of Chromatin, eds Workman J. L., Abmayr S. M. (New York, NY: Springer; ), 213–256. 10.1007/978-1-4614-8624-4_5 [DOI] [Google Scholar]
- Guo Z., Li G., Bian E., Ma C.-C., Wan J., Zhao B. (2017). TGF-β-mediated repression of MST1 by DNMT1 promotes glioma malignancy. Biomed. Pharmacother. 94 774–780. 10.1016/j.biopha.2017.07.081 [DOI] [PubMed] [Google Scholar]
- Hayashi A., Horiuchi A., Kikuchi N., Hayashi T., Fuseya C., Suzuki A., et al. (2010). Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int. J. Cancer 127 1332–1346. 10.1002/ijc.25151 [DOI] [PubMed] [Google Scholar]
- Hergeth S. P., Schneider R. (2015). The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 16 1439–1453. 10.15252/embr.201540749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L., Ma H.-P., Su C.-M., Chang Y.-J., Hung W.-Y., Ho Y.-S., et al. (2016). Alterations in histone deacetylase 8 lead to cell migration and poor prognosis in breast cancer. Life Sci. 151 7–14. 10.1016/j.lfs.2016.02.092 [DOI] [PubMed] [Google Scholar]
- Huang M., Chen Y., Han D., Lei Z., Chu X. (2019). Role of the zinc finger and SCAN domain-containing transcription factors in cancer. Am. J. Cancer Res. 9 816–836. [PMC free article] [PubMed] [Google Scholar]
- Huang T., Zhang P., Li W., Zhao T., Zhang Z., Chen S., et al. (2017). G9A promotes tumor cell growth and invasion by silencing CASP1 in non-small-cell lung cancer cells. Cell Death Dis. 8:e2726. 10.1038/cddis.2017.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J., Pfeifer C. R., Bennett R. R., Xia Y., Ivanovska I. L., Liu A. J., et al. (2016). Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol. Biol. Cell 27 4011–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J., Xia Y., Pfeifer C. R., Athirasala A., Ji J., Alvey C., et al. (2017). DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr. Biol. 27 210–223. 10.1016/j.cub.2016.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isokane M., Hieda M., Hirakawa S., Shudou M., Nakashiro K., Hashimoto K., et al. (2008). Plasma-membrane-anchored growth factor pro-amphiregulin binds A-type lamin and regulates global transcription. J. Cell Sci. 121 3608–3618. 10.1242/jcs.031443 [DOI] [PubMed] [Google Scholar]
- Jacobson E. C., Perry J. K., Long D. S., Olins A. L., Olins D. E., Wright B. E., et al. (2018). Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells. BMC Biol. 16:142. 10.1186/s12915-018-0608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon H. W., Lee Y. M. (2010). Inhibition of histone deacetylase attenuates hypoxia-induced migration and invasion of cancer cells via the restoration of RECK expression. Mol. Cancer Ther. 9 1361–1370. 10.1158/1535-7163.MCT-09-0717 [DOI] [PubMed] [Google Scholar]
- Jiang X., Nardelli J. (2016). Cellular and molecular introduction to brain development. Neurobiol. Dis. 92 3–17. 10.1016/j.nbd.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.-K. (2018). Surveillance of retroelement expression and nucleic-acid immunity by histone methyltransferase SETDB1. Bioessays 40:1800058. 10.1002/bies.201800058 [DOI] [PubMed] [Google Scholar]
- Kengaku M. (2018). Cytoskeletal control of nuclear migration in neurons and non-neuronal cells. Proc. Jpn. Acad. Ser. B 94 337–349. 10.2183/pjab.94.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jeong Y., Kwon K., Ismail T., Lee H.-K., Kim C., et al. (2018). Physiological effects of KDM5C on neural crest migration and eye formation during vertebrate development. Epigenetics Chromatin 11:72. 10.1186/s13072-018-0241-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. H., Han S.-B., Lee J. K. (2013). Histone deacetylase inhibitors suppress CXCR4-mediated dendritic cell migration by regulation of maturation process. Cell. Immunol. 284 139–145. 10.1016/j.cellimm.2013.07.014 [DOI] [PubMed] [Google Scholar]
- Kottakis F., Polytarchou C., Foltopoulou P., Sanidas I., Kampranis S. C., Tsichlis P. N. (2011). FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 Pathway. Mol. Cell 43 285–298. 10.1016/j.molcel.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Wolf K. (2015). Cancer cell migration in 3D tissue: negotiating space by proteolysis and nuclear deformability. Cell Adh. Migr. 9 357–366. 10.1080/19336918.2015.1061173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Yang F. W., te Lindert M., Isermann P., Schepens J., Maas R. J. A., et al. (2019). Cell migration through three-dimensional confining pores: speed accelerations by deformation and recoil of the nucleus. Philos. Trans. R. Soc. B Biol. Sci. 374:20180225 10.1098/rstb.2018.0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A. W., Pattabiraman D. R., Weinberg R. A. (2017). Emerging biological principles of metastasis. Cell 168 670–691. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15 178–196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore C. J., Bloom K. (2019). Common features of the pericentromere and nucleolus. Genes 10:1029. 10.3390/genes10121029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H. Q., Ghatak S., Yeung C.-Y. C., Tellkamp F., Günschmann C., Dieterich C., et al. (2016). Mechanical regulation of transcription controls polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18 864–875. 10.1038/ncb3387 [DOI] [PubMed] [Google Scholar]
- Lele T. P., Dickinson R. B., Gundersen G. G. (2018). Mechanical principles of nuclear shaping and positioning. J. Cell Biol. 217 3330–3342. 10.1083/jcb.201804052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Liu Z., Li M., Zhou S., Xu Y., Xiao Y., et al. (2016). HDAC5, a potential therapeutic target and prognostic biomarker, promotes proliferation, invasion and migration in human breast cancer. Oncotarget 7 37966–37978. 10.18632/oncotarget.9274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Yao Q., Wei D., Liu M., Geng F., Wang Q., et al. (2019). KDM6B promotes ovarian cancer cell migration and invasion by induced transforming growth factor-β1 expression. J. Cell. Biochem. 120 493–506. 10.1002/jcb.27405 [DOI] [PubMed] [Google Scholar]
- Lin C., Wei D., Xin D., Pan J., Huang M. (2019). Ellagic acid inhibits proliferation and migration of cardiac fibroblasts by down-regulating expression of HDAC1. J. Toxicol. Sci. 44 425–433. 10.2131/jts.44.425 [DOI] [PubMed] [Google Scholar]
- Liu L., Luo Q., Sun J., Ju Y., Morita Y., Song G. (2018). Chromatin organization regulated by EZH2-mediated H3K27me3 is required for OPN-induced migration of bone marrow-derived mesenchymal stem cells. Int. J. Biochem. Cell Biol. 96 29–39. 10.1016/j.biocel.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Liu L., Luo Q., Sun J., Song G. (2016). Nucleus and nucleus-cytoskeleton connections in 3D cell migration. Exp. Cell Res. 348 56–65. 10.1016/j.yexcr.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Lu C., Han H. D., Mangala L. S., Ali-Fehmi R., Newton C. S., Ozbun L., et al. (2010). Regulation of tumor angiogenesis by EZH2. Cancer Cell 18 185–197. 10.1016/j.ccr.2010.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Balsa E., Perry E. A., Liang J., Tavares C. D., Vazquez F., et al. (2020). H3K27me3-mediated PGC1α gene silencing promotes melanoma invasion through WNT5A and YAP. J. Clin. Invest. 130 853–862. 10.1172/JCI130038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Merz P. R., Chen Y., Dickes E., Pscherer A., Schadendorf D., et al. (2013). MiR-101 inhibits melanoma cell invasion and proliferation by targeting MITF and EZH2. Cancer Lett. 341 240–247. 10.1016/j.canlet.2013.08.021 [DOI] [PubMed] [Google Scholar]
- Luo G., Jing X., Yang S., Peng D., Dong J., Li L., et al. (2019). DNA methylation regulates corneal epithelial wound healing by targeting miR-200a and CDKN2B. Invest. Ophthalmol. Vis. Sci. 60 650–660. 10.1167/iovs.18-25443 [DOI] [PubMed] [Google Scholar]
- Ma J., Zhang J., Weng Y.-C., Wang J.-C. (2018). EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal transition and lymph node metastasis of pancreatic cancer. Mol. Cells 41 868–880. 10.14348/MOLCELLS.2018.0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maa M.-C., Chang M. Y., Hsieh M.-Y., Chen Y.-J., Yang C.-J., Chen Z.-C., et al. (2010). Butyrate reduced lipopolysaccharide-mediated macrophage migration by suppression of Src enhancement and focal adhesion kinase activity. J. Nutr. Biochem. 21 1186–1192. 10.1016/j.jnutbio.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Madrazo E., Conde A. C., Redondo-Muñoz J. (2017). Inside the cell: integrins as new governors of nuclear alterations? Cancers 9:82. 10.3390/cancers9070082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrazo E., Ruano D., Abad L., Alonso-Gómez E., Sánchez-Valdepeñas C., González-Murillo Á., et al. (2018). G9a correlates with VLA-4 integrin and influences the migration of childhood acute lymphoblastic leukemia cells. Cancers 10:325. 10.3390/cancers10090325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels Y., Elbaz A., Hernandez-Vicens R., Sandrusy O., Rosenberg A., Gerlitz G. (2017). Increased chromatin plasticity supports enhanced metastatic potential of mouse melanoma cells. Exp. Cell Res. 357 282–290. 10.1016/j.yexcr.2017.05.025 [DOI] [PubMed] [Google Scholar]
- Manley H. R., Keightley M. C., Lieschke G. J. (2018). The neutrophil nucleus: an important influence on neutrophil migration and function. Front. Immunol. 9:2867. 10.3389/fimmu.2018.02867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsano R. M., Giordano E., Messina G., Dimitri P. (2019). A new portrait of constitutive heterochromatin: lessons from Drosophila melanogaster. Trends Genet. 35 615–631. 10.1016/j.tig.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Mason D. E., Collins J. M., Dawahare J. H., Nguyen T. D., Lin Y., Voytik-Harbin S. L., et al. (2019). YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J. Cell Biol. 218 1369–1389. 10.1083/jcb.201806065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateen S., Raina K., Agarwal C., Chan D., Agarwal R. (2013). Silibinin synergizes with histone deacetylase and DNA methyltransferase inhibitors in upregulating e-cadherin expression together with inhibition of migration and invasion of human non-small cell lung cancer cells. J. Pharmacol. Exp. Ther. 345 206–214. 10.1124/jpet.113.203471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A. L., Hsia C.-R., Lammerding J. (2016). Squish and squeeze—the nucleus as a physical barrier during migration in confined environments. Curr. Opin. Cell Biol. 40 32–40. 10.1016/j.ceb.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Sun G., Zhong M., Yu Y., Brewer M. A. (2013). Anticancer efficacy of cisplatin and trichostatin A or 5-aza-2’-deoxycytidine on ovarian cancer. Br. J. Cancer 108 579–586. 10.1038/bjc.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistriotis P., Wisniewski E. O., Bera K., Keys J., Li Y., Tuntithavornwat S., et al. (2019). Confinement hinders motility by inducing RhoA-mediated nuclear influx, volume expansion, and blebbing. J. Cell Biol. 218 4093–4111. 10.1083/jcb.201902057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet D., Bellahcène A., Pirotte S., Waltregny D., Deroanne C., Lamour V., et al. (2007). Histone deacetylase 7 silencing alters endothelial cell migration, a key step in angiogenesis. Circ. Res. 101 1237–1246. 10.1161/CIRCRESAHA.107.149377 [DOI] [PubMed] [Google Scholar]
- Mozzetta C., Boyarchuk E., Pontis J., Ait-Si-Ali S. (2015). Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 16 499–513. 10.1038/nrm4029 [DOI] [PubMed] [Google Scholar]
- Nair N. U., Das A., Rogkoti V.-M., Fokkelman M., Marcotte R., de Jong C. G., et al. (2019). Migration rather than proliferation transcriptomic signatures are strongly associated with breast cancer patient survival. Sci. Rep. 9:10989. 10.1038/s41598-019-47440-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R. M., Ignatius M. S., Henion P. D. (2007). Zebrafish colgate/hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mech. Dev. 124 682–698. 10.1016/j.mod.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawaji T., Akiyama Y., Shimada S., Kojima K., Kawano T., Eishi Y., et al. (2016). Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer. Oncotarget 7 67251–67265. 10.18632/oncotarget.11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourshargh S., Alon R. (2014). Leukocyte migration into inflamed tissues. Immunity 41 694–707. 10.1016/j.immuni.2014.10.008 [DOI] [PubMed] [Google Scholar]
- Orouji E., Federico A., Larribère L., Novak D., Lipka D. B., Assenov Y., et al. (2019). Histone methyltransferase SETDB1 contributes to melanoma tumorigenesis and serves as a new potential therapeutic target. Int. J. Cancer 145 3462–3477. 10.1002/ijc.32432 [DOI] [PubMed] [Google Scholar]
- Oshima G., Poli E. C., Bolt M. J., Chlenski A., Forde M., Jutzy J. M. S., et al. (2019). DNA methylation controls metastasis-suppressive 14q32-encoded miRNAs. Cancer Res. 79 650–662. 10.1158/0008-5472.CAN-18-0692 [DOI] [PubMed] [Google Scholar]
- Pechalrieu D., Etievant C., Arimondo P. B. (2017). DNA methyltransferase inhibitors in cancer: from pharmacology to translational studies. Biochem. Pharmacol. 129 1–13. 10.1016/j.bcp.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Pensold D., Symmank J., Hahn A., Lingner T., Salinas-Riester G., Downie B. R., et al. (2017). The DNA methyltransferase 1 (DNMT1) controls the shape and dynamics of migrating POA-derived interneurons fated for the murine cerebral cortex. Cereb. Cortex 27 5696–5714. 10.1093/cercor/bhw341 [DOI] [PubMed] [Google Scholar]
- Perrigue P. M., Najbauer J., Barciszewski J. (2016). Histone demethylase JMJD3 at the intersection of cellular senescence and cancer. Biochim. Biophys. Acta 1865 237–244. 10.1016/j.bbcan.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Petrie R. J., Harlin H. M., Korsak L. I. T., Yamada K. M. (2017). Activating the nuclear piston mechanism of 3D migration in tumor cells. J. Cell Biol. 216 93–100. 10.1083/jcb.201605097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R. J., Koo H., Yamada K. M. (2014). Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science 345 1062–1065. 10.1126/science.1256965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer C. R., Xia Y., Zhu K., Liu D., Irianto J., García V. M. M., et al. (2018). Constricted migration increases DNA damage and independently represses cell cycle. Mol. Biol. Cell 29 1948–1962. 10.1091/mbc.E18-02-0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H. R., Vargas P., Jimenez A. J., et al. (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352 359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Rahnama F., Shafiei F., Gluckman P. D., Mitchell M. D., Lobie P. E. (2006). Epigenetic regulation of human trophoblastic cell migration and invasion. Endocrinology 147 5275–5283. [DOI] [PubMed] [Google Scholar]
- Rao Z.-Y., Cai M.-Y., Yang G.-F., He L.-R., Mai S.-J., Hua W.-F., et al. (2010). EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-β1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis 31 1576–1583. 10.1093/carcin/bgq150 [DOI] [PubMed] [Google Scholar]
- Reiner O., Karzbrun E., Kshirsagar A., Kaibuchi K. (2016). Regulation of neuronal migration, an emerging topic in autism spectrum disorders. J. Neurochem. 136 440–456. 10.1111/jnc.13403 [DOI] [PubMed] [Google Scholar]
- Roberti A., Valdes A. F., Torrecillas R., Fraga M. F., Fernandez A. F. (2019). Epigenetics in cancer therapy and nanomedicine. Clin. Epigenet. 11:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röth S., Fulcher L. J., Sapkota G. P. (2019). Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci. 76 2761–2777. 10.1007/s00018-019-03112-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N., Simboeck E., Déjardin J. (2015). Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8:3. 10.1186/1756-8935-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvermoser M., Begandt D., Alon R., Walzog B. (2018). Nuclear deformation during neutrophil migration at sites of inflammation. Front. Immunol. 9:2680. 10.3389/fimmu.2018.02680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang B., Sun J., Yang D., Xu Z., Wei Y. (2019). Ras-AKT signaling represses the phosphorylation of histone H1.5 at threonine 10 via GSK3 to promote the progression of glioma. Artif. Cells Nanomed. Biotechnol. 47 2882–2890. 10.1080/21691401.2019.1638795 [DOI] [PubMed] [Google Scholar]
- Scarpa E., Mayor R. (2016). Collective cell migration in development. J. Cell Biol. 212 143–155. 10.1083/jcb.201508047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher L. (2019). Collective cell migration in development. Adv. Exp. Med. Biol. 1146 105–116. 10.1007/978-3-030-17593-1_7 [DOI] [PubMed] [Google Scholar]
- Segal T., Salmon-Divon M., Gerlitz G. (2018). The heterochromatin landscape in migrating cells and the importance of H3K27me3 for associated transcriptome alterations. Cells 7:205. 10.3390/cells7110205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senigagliesi B., Penzo C., Severino L. U., Maraspini R., Petrosino S., Morales-Navarrete H., et al. (2019). The high mobility group A1 (HMGA1) chromatin architectural factor modulates nuclear stiffness in breast cancer cells. Int. J. Mol. Sci. 20:2733. 10.3390/ijms20112733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepsa A., Levidou G., Gargalionis A., Adamopoulos C., Spyropoulou A., Dalagiorgou G., et al. (2015). Emerging role of linker histone variant H1x as a biomarker with prognostic value in astrocytic gliomas. A multivariate analysis including trimethylation of H3K9 and H4K20. PLoS One 10:e0115101. 10.1371/journal.pone.0115101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiei F., Rahnama F., Pawella L., Mitchell M. D., Gluckman P. D., Lobie P. E. (2008). DNMT3A and DNMT3B mediate autocrine hGH repression of plakoglobin gene transcription and consequent phenotypic conversion of mammary carcinoma cells. Oncogene 27 2602–2612. 10.1038/sj.onc.1210917 [DOI] [PubMed] [Google Scholar]
- Soshnev A. A., Josefowicz S. Z., Allis C. D. (2016). Greater than the sum of parts: complexity of the dynamic epigenome. Mol. Cell 62 681–694. 10.1016/j.molcel.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulou A., Gargalionis A., Dalagiorgou G., Adamopoulos C., Papavassiliou K. A., Lea R. W., et al. (2014). Role of histone lysine methyltransferases SUV39H1 and SETDB1 in gliomagenesis: modulation of cell proliferation, migration, and colony formation. Neuromolecular Med. 16 70–82. 10.1007/s12017-013-8254-x [DOI] [PubMed] [Google Scholar]
- Stephens A. D., Banigan E. J., Adam S. A., Goldman R. D., Marko J. F. (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 28 1984–1996. 10.1091/mbc.E16-09-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A. D., Liu P. Z., Banigan E. J., Almassalha L. M., Backman V., Adam S. A., et al. (2018). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 29 220–233. 10.1091/mbc.E17-06-0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A. D., Liu P. Z., Kandula V., Chen H., Almassalha L. M., Herman C., et al. (2019). Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30 2320–2330. 10.1091/mbc.E19-05-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer M. A., Golden J. A., Francis F. (2016). Neuronal migration disorders: focus on the cytoskeleton and epilepsy. Neurobiol. Dis. 92 18–45. 10.1016/j.nbd.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strmiska V., Michalek P., Lackova Z., Guran R., Krizkova S., Vanickova L., et al. (2019). Sarcosine is a prostate epigenetic modifier that elicits aberrant methylation patterns through the SAMe-Dnmts axis. Mol. Oncol. 13 1002–1017. 10.1002/1878-0261.12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Hopfinger N. R., Nguyen T. D., Pogash T. J., Santucci-Pereira J., Russo J. (2018). Epigenetic reprogramming of epithelial mesenchymal transition in triple negative breast cancer cells with DNA methyltransferase and histone deacetylase inhibitors. J. Exp. Clin. Cancer Res. 37:314. 10.1186/s13046-018-0988-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S. (2018). Transcribing centromeres: noncoding RNAs and kinetochore assembly. Trends Genet. 34 587–599. 10.1016/j.tig.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Nishioka Y., Yokoyama Y., Higashiyama S., Matsuura N., Matsuura S., et al. (2012). Nuclear envelope-localized EGF family protein amphiregulin activates breast cancer cell migration in an EGF-like domain independent manner. Biochem. Biophys. Res. Commun. 420 721–726. 10.1016/j.bbrc.2012.03.045 [DOI] [PubMed] [Google Scholar]
- Tiffen J., Gallagher S. J., Hersey P. (2015). EZH2: an emerging role in melanoma biology and strategies for targeted therapy. Pigment Cell Melanoma Res. 28 21–30. 10.1111/pcmr.12280 [DOI] [PubMed] [Google Scholar]
- Tillotson R., Bird A. (2019). The molecular basis of MeCP2 function in the brain. J. Mol. Biol. 10.1016/j.jmb.2019.10.004 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Torrano J., Al Emran A., Hammerlindl H., Schaider H. (2019). Emerging roles of H3K9me3, SETDB1 and SETDB2 in therapy-induced cellular reprogramming. Clin. Epigenetics 11:43. 10.1186/s13148-019-0644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Morita T., Okada M., Yamawaki H. (2014). Histone deacetylase 4 controls neointimal hyperplasia via stimulating proliferation and migration of vascular smooth muscle cells. Hypertension 63 397–403. 10.1161/HYPERTENSIONAHA.113.01843 [DOI] [PubMed] [Google Scholar]
- van Steensel B., Belmont A. S. (2017). Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169 780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S., Cao Q., Mani R.-S., Shankar S., Wang X., Ateeq B., et al. (2008). Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322 1695–1699. 10.1126/science.1165395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-C., Wang Z., Fan Y.-X., Si Y.-Q., Wang J.-X. (2015). DNA methyltransferase 3b silencing affects locus-specific DNA methylation and inhibits proliferation, migration and invasion in human hepatocellular carcinoma SMMC-7721 and BEL-7402 cells. Oncol. Lett. 9 2499–2506. 10.3892/ol.2015.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Z., Huang L., Wu C., Zhang B. (2019). MiR-29b suppresses proliferation and mobility by targeting SOX12 and DNMT3b in pancreatic cancer. Anticancer Drugs 30 281–288. 10.1097/CAD.0000000000000719 [DOI] [PubMed] [Google Scholar]
- Wang P., Dreger M., Madrazo E., Williams C. J., Samaniego R., Hodson N. W., et al. (2018). WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proc. Natl. Acad. Sci. U.S.A. 115 8581–8586. 10.1073/pnas.1719405115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu X., Zhong Y., Shen J., Wu X., Ju S., et al. (2014). Effects of histone deacetylase inhibition on the survival, proliferation and migration of Schwann cells, as well as on the expression of neurotrophic factors and genes associated with myelination. Int. J. Mol. Med. 34 599–605. 10.3892/ijmm.2014.1792 [DOI] [PubMed] [Google Scholar]
- Wawruszak A., Kalafut J., Okon E., Czapinski J., Halasa M., Przybyszewska A., et al. (2019). Histone deacetylase inhibitors and phenotypical transformation of cancer cells. Cancers 11:148. 10.3390/cancers11020148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel S., Hudak L., Seibel J.-M., Makarević J., Juengel E., Tsaur I., et al. (2011). Impact of combined HDAC and mTOR inhibition on adhesion, migration and invasion of prostate cancer cells. Clin. Exp. Metastasis 28 479–491. 10.1007/s10585-011-9386-8 [DOI] [PubMed] [Google Scholar]
- Wolf K., te Lindert M., Krause M., Alexander S., te Riet J., Willis A. L., et al. (2013). Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201 1069–1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Wu Y. I., Liu Y., Geiger J., Tam E., Overall C., et al. (2007). Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9 893–904. 10.1038/ncb1616 [DOI] [PubMed] [Google Scholar]
- Wong C.-M., Wei L., Law C.-T., Ho D. W.-H., Tsang F. H.-C., Au S. L.-K., et al. (2016). Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology 63 474–487. 10.1002/hep.28304 [DOI] [PubMed] [Google Scholar]
- Worbs T., Hammerschmidt S. I., Förster R. (2017). Dendritic cell migration in health and disease. Nat. Rev. Immunol. 17 30–48. 10.1038/nri.2016.116 [DOI] [PubMed] [Google Scholar]
- Wu H., Zhang W., Wu Z., Liu Y., Shi Y., Gong J., et al. (2019). miR-29c-3p regulates DNMT3B and LATS1 methylation to inhibit tumor progression in hepatocellular carcinoma. Cell Death Dis. 10:48. 10.1038/s41419-018-1281-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sun J., Shi C., Sun C., Yu L., Wen Y., et al. (2015). miR-29s inhibit the malignant behavior of U87MG glioblastoma cell line by targeting DNMT3A and 3B. Neurosci. Lett. 590 40–46. 10.1016/j.neulet.2015.01.060 [DOI] [PubMed] [Google Scholar]
- Xu J., Tian F., Chen X., Liu Z., Wu C., Zhao Z. (2020). Ras-ERK1/2 signaling participates in the progression of gastric cancer through repressing Aurora B-mediated H1.4 phosphorylation at Ser27. J. Cell. Physiol. 10.1002/jcp.29432 Epub ahead of print]. 31907925 [DOI] [Google Scholar]
- Yamada K. M., Sixt M. (2019). Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 20 738–752. 10.1038/s41580-019-0172-9 [DOI] [PubMed] [Google Scholar]
- Yan F., Shen N., Pang J., Xie D., Deng B., Molina J. R., et al. (2014). Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis. 5:e1413. 10.1038/cddis.2014.380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Lu Z., Ramchandran R., Longo L. D., Raj J. U. (2012). Pulmonary artery smooth muscle cell proliferation and migration in fetal lambs acclimatized to high-altitude long-term hypoxia: role of histone acetylation. Am. J. Physiol. Lung Cell. Mol. Physiol. 303 L1001–L1010. 10.1152/ajplung.00092.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y., Hieda M., Nishioka Y., Matsumoto A., Higashi S., Kimura H., et al. (2013). Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci. 104 889–895. 10.1111/cas.12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Ye F., Li Y.-Y., Zhan Y.-Z., Liu Y., Yan H.-M., et al. (2019). Histone methyltransferase SETDB1 promotes colorectal cancer proliferation through the STAT1-CCND1/CDK6 axis. Carcinogenesis 10.1093/carcin/bgz131 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Yuan Z., Chen S., Gao C., Dai Q., Zhang C., Sun Q., et al. (2019). Development of a versatile DNMT and HDAC inhibitor C02S modulating multiple cancer hallmarks for breast cancer therapy. Bioorg. Chem. 87 200–208. 10.1016/j.bioorg.2019.03.027 [DOI] [PubMed] [Google Scholar]
- Zhang B., Luo Q., Chen Z., Shi Y., Ju Y., Yang L., et al. (2016). Increased nuclear stiffness via FAK-ERK1/2 signaling is necessary for synthetic mechano-growth factor E peptide-induced tenocyte migration. Sci. Rep. 6:18809. 10.1038/srep18809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hou W., Wang H.-L., Liu H.-J., Jia X.-Y., Zheng X.-Z., et al. (2014). GSK-3 -regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin. Cancer Res. 20 4717–4729. 10.1158/1078-0432.CCR-13-3477 [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen J., Yang J., Xu C., Hu Q., Wu H., et al. (2019a). Suv39h1 downregulation inhibits neointimal hyperplasia after vascular injury. Atherosclerosis 288 76–84. 10.1016/j.atherosclerosis.2019.06.909 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu W., Dong H., Wang W. (2019b). K-Ras G12V/Y40C -PI3K/AKT pathway regulates H1.4 S35ph through PKA to promote the occurrence and development of osteosarcoma cancer. Artif. Cells Nanomed. Biotechnol. 47 2048–2057. 10.1080/21691401.2019.1617726 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang G., Wang L., Song C., Leng Y., Wang X., et al. (2012). VPA inhibits breast cancer cell migration by specifically targeting HDAC2 and down-regulating Survivin. Mol. Cell. Biochem. 361 39–45. 10.1007/s11010-011-1085-x [DOI] [PubMed] [Google Scholar]
- Zhang Q., Dong P., Liu X., Sakuragi N., Guo S.-W. (2017). Enhancer of Zeste homolog 2 (EZH2) induces epithelial-mesenchymal transition in endometriosis. Sci. Rep. 7:6804. 10.1038/s41598-017-06920-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Cook P. C., Zindy E., Williams C. J., Jowitt T. A., Streuli C. H., et al. (2016). Integrin α4β1 controls G9a activity that regulates epigenetic changes and nuclear properties required for lymphocyte migration. Nucleic Acids Res. 44 3031–3044. 10.1093/nar/gkv1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang J., Li Q., Chen K., Liang Y., Zhan Z., et al. (2018). Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer 18:539. 10.1186/s12885-018-4464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Zhang X., Guan H., Xiong X., Shi X., Deng H., et al. (2016). The BAH domain of BAHD1 is a histone H3K27me3 reader. Protein Cell 7 222–226. 10.1007/s13238-016-0243-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Wan H., Xue C., Jiang T., Qian C., Zhang Y. (2013). Histone deacetylase 3 implicated in the pathogenesis of children glioma by promoting glioma cell proliferation and migration. Brain Res. 1520 15–22. 10.1016/j.brainres.2013.04.061 [DOI] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Graham S. M., Cloud V. J., Forrester W. C. (2006). The C. elegans histone deacetylase HDA-1 is required for cell migration and axon pathfinding. Dev. Biol. 289 229–242. 10.1016/j.ydbio.2005.10.033 [DOI] [PubMed] [Google Scholar]