In vitro fertilization-embryo transfer (IVF-ET) is now widely applied for treating infertility, and unexplained recurrent implantation failure (RIF) has become a substantial challenge. We hypothesize that vaginal microbial dysbiosis is associated with RIF, as it is linked to many female reproductive diseases. In this study, we characterized the vaginal microbiota and metabolomes of patients with unexplained RIF, while patients who achieved clinical pregnancy in the first IVF cycle were set as controls. In general, significant differences were discovered in the vaginal microbiota and metabolomes between the two groups. This study is the first detailed elaboration of the vaginal microbiota and metabolites associated with RIF. We believe that our findings will inspire researchers to consider the dynamics of microbiomes related to the microenvironment as a critical feature for future studies of nosogenesis not only for RIF but also for other reproductive diseases.
KEYWORDS: recurrent implantation failure, RIF, vaginal microbiota, metabolome, in vitro fertilization, IVF, infertility
ABSTRACT
Recurrent implantation failure (RIF) refers to repeated failure to become pregnant after transferring embryos with normal morphology. However, the pathogenesis of RIF remains unrevealed, especially for those without any pathological features. In this study, we characterized the vaginal microbiota and metabolomes of patients with unexplained RIF, while patients who achieved clinical pregnancy in the first frozen embryo transfer (FET) cycle were used as controls. Based on 16S rRNA gene sequencing of the vaginal microbiota, the vaginal Lactobacillus showed a significant positive correlation with the pregnancy rate, and the RIF group presented higher microbial α-diversity than the control group (P value = 0.016). The metabolomic profile identified 2,507 metabolites, of which 37 were significantly different between the two groups (P value < 0.05, variable importance for the projection [VIP] > 1). Among them, 2′,3-cyclic UMP and inositol phosphate were the top two metabolites that were higher in the RIF group, while glycerophospholipids and benzopyran were important metabolites that were lower in the RIF group. A lack of lysobisphosphatidic acid and prostaglandin metabolized from glycerophospholipids will lead to deferred implantation and embryo crowding. Benzopyran, as a selective estrogen receptor modulator, may affect the outcome of pregnancy. All of the changes in metabolite profiles may result in or from the differential microbiota compositions in RIF patients. In conclusion, significant differences were presented in the vaginal microbiota and metabolomes between patients with unexplained RIF and women who became pregnant in the first FET cycle. For the first time, this study elaborates the possible pathogenesis of RIF by investigating the vaginal microbiota and metabolites in RIF patients.
INTRODUCTION
Infertility is defined as the inability to become pregnant by a couple who have normal intercourse without contraception for 1 year (1, 2). The incidence of infertility ranges from 8% to 12% in reproductive-aged couples, which has become a definite global public health issue (3). In vitro fertilization-embryo transfer (IVF-ET) is currently widely used in the treatment of infertility. In 1978, the first IVF baby was born in the United Kingdom. Over the intervening years, the success rate has steadily increased, with a cumulative live birth rate of >52% after 3 cycles of treatment (4). Nevertheless, IVF treatment failure still occurs frequently. Among all repeated IVF failure types, recurrent implantation failure (RIF) is a special subgroup. RIF refers to the women under the age of 40 years who received at least four good-quality embryos in a minimum of three fresh or frozen cycles that still failed to achieve a clinical pregnancy (5). Failed IVF usually occurs in elderly patients and those with a low ovarian reserve or low ovarian response. For them, high-quality embryos will be selected and transferred (5). However, many patients still fail to become pregnant after repeated IVF cycles. RIF has become a new and substantial challenge, as its occurrence rate among infertility patients has continued to increase in recent years (6). RIF also carries a heavy financial burden and deeply impacts the patient’s body and mind.
The investigation and management of RIF usually focuses on the quality of the embryo and endometrial receptivity. Recently, the importance of maternal systemic diseases, such as thyroid, thrombophilia, and immunological disorders, has also been recognized (7–9). Although many RIF patients have undergone plenty of clinical examinations and tried various possible treatments, they are still unable to become pregnant. Moreover, a recent paper reported that nonsenile patients still have a low live-birth rate (36.6%), even when euploid blastocyst transfer cycles are selected by comprehensive chromosome screening (CCS) and after confirming the window of implantation (WOI) by endometrial receptive array (ERA) (10). Even worse, another group of patients with unexplained RIF have no pathological features, for whom the treatment method is more intractable. Hence, the nosogenesis of RIF is complex and requires a multidimensional explanation.
The human commensal microbiome, referred to as “the other human genome,” coexisted and evolved with the human genome to help maintain human health. Microbial dysbiosis and invasion of pathogens can lead to disease and even threaten human life. Metabolic syndrome, diabetes (11, 12), obesity (13, 14), alcoholic liver disease (15), cirrhosis (16), coronary heart disease (17), and some mental diseases (18, 19) have been found to be related to intestinal microbial metabolism. With the development of high-throughput sequencing, the microbiome has also been characterized in the vaginal environment, even in the female upper reproductive tract, which has traditionally been considered sterile (20). Ravel et al. divided the vaginal flora into five community state types (CSTs) by 16S rRNA gene sequencing in 2011, four of which were classified as Lactobacillus-dominated (LD) types (21). Lactobacillus is one of the dominant genera in the vaginal microbiota of healthy females, and its key metabolites, which include lactate acid, can maintain the acidic and anaerobic vaginal environment and protect it from pathogen infection. A series of pregnancy-related diseases, such as premature rupture of membranes, premature delivery, and chorioamnion infection (22–26), diseases associated with infertility, such as diabetes, obesity, pelvic inflammatory disease (27), and sexually transmitted diseases (28), and even cervical cancer (29) have been discovered to be associated with vagina dysbiosis.
A previous systematic review reported that an abnormal vaginal microbiota is associated with tubal factor infertility and early spontaneous abortion in IVF patients. However, that article also pointed out that the quality of evidence was low and needs further research (30). In addition, because microbial compositions change along the female reproductive tract and are associated with pregnancy-related diseases, sexually transmitted diseases, infertility-related metabolic diseases, gynecological tumors, and so on (28, 31–33), we assume that the microbiota and its metabolites might be associated with every step of IVF, including gamete formation, implantation, and delivery (34, 35). In this study, we explored the association of microbial composition and function with IVF/RIF, and microbiomic and metabolomic analyses were both used as advanced tools. Metabolomics is the untargeted identification and quantification of all low-molecular-weight end products of cellular biological processes (36). The levels of metabolites ultimately reflect the integrated response of a biological system and directly influence the host. Additionally, individuals who achieved a successful pregnancy in the first frozen embryo transfer (FET) cycle were used as controls to explore the relationship of the microbial community and metabolites with RIF.
RESULTS
Diversity and composition of the vaginal microbiota.
A total of 67 samples were analyzed by 16S rRNA gene sequencing to investigate the vaginal microbiota, including 27 samples from the RIF group and 40 from the control group. The clinical information of the subjects is shown in Table 1. In total, 2,824,185 reads were obtained from these 67 samples, and on average, 42,152 ± 10,415 reads per sample and 424 ± 7 bp per read were achieved. After clustering, the rarefaction curve of the operational taxonomic unit (OTU) number was almost a straight horizontal line, which demonstrated that the samples were sequenced with enough depth in this study (see Fig. S1 in the supplemental material).
TABLE 1.
Clinical characteristics of the participants in the two groups whose samples were submitted for 16S rRNA gene sequencing of the vaginal microbiota
| Clinical characteristics | Value |
P value | |
|---|---|---|---|
| RIF group (n = 27) | Control group (n = 40) | ||
| Age (yrs) | 33.4 ± 3.7 | 32.0 ± 4.0 | 0.213 |
| BMI (kg/m2) | 20.9 ± 3.4 | 22.3 ± 7.6 | 0.102 |
| AMH (ng/ml) | 3.0 ± 1.9 | 4.4 ± 3.8 | 0.101 |
| Duration of infertility (yrs) | 4.5 ± 3.0 | 3.7 ± 2. 7 | 0.068 |
| Endometrial thickness (mm) | 11.5 ± 1.8 | 12.1 ± 2.2 | 0.162 |
| No. of oocytes | 12.7 ± 5.9 | 12.2 ± 5.5 | 0.734 |
| No. of embryos | 9.7 ± 5.0 | 9.7 ± 4.4 | 0.847 |
| No. of high-quality embryos | 2.9 ± 2.1 | 2.8 ± 2.5 | 0.568 |
| No. of embryos transferred | 1. 9 ± 0.4 | 2.1 ± 0.4 | 0.044 |
| No. of high-quality embryos transferred | 1.3 ± 0.5 | 1.3 ± 0.9 | 0.576 |
Rarefaction curves of the sequenced samples. Download FIG S1, PDF file, 1.7 MB (1.8MB, pdf) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A total of 804 OTUs were obtained in the two groups. The RIF group contained 730, and the control group contained 429, among which 355 were shared between the two groups. The numbers of OTUs found in each sample of the RIF group and control group were 71.48 ± 37.02 and 42.53 ± 14.51, respectively. The number of OTUs in the RIF group was much larger than that in the control group, indicating that the microbial composition of the RIF group was richer. The α-diversity of the microbiota was calculated by the Shannon-Wiener index (0.80 ± 0.50 for the RIF group and 0.50 ± 0.39 for the control group, P value < 0.01). Although some individuals in both groups showed similar levels of diversity due to the overlap of the Shannon-Wiener index values, the statistical analysis results still demonstrated that the microbial diversity in the vaginal environment was significantly higher in the RIF patients than in the control individuals (Fig. 1).
FIG 1.
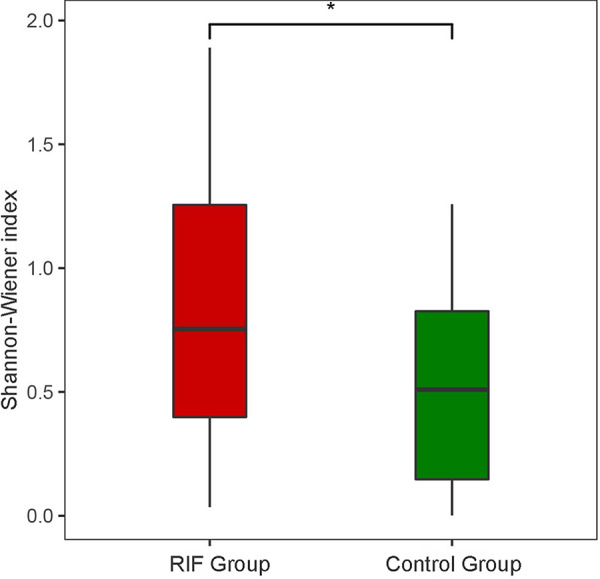
The α-diversity of the vaginal microbiota in the two groups was calculated and is shown by the Shannon-Wiener index.
The taxonomic classification at the phylum level showed similar patterns in the two groups, which were both dominated by Firmicutes, Actinobacteria, Bacteroidetes, Proteobacteria, and Tenericutes (Mollicutes); however, the relative abundances of Firmicutes and Bacteroidetes were significantly different (P value < 0.05) (Fig. 2a). At the genus level, genera with the top 15 abundances are shown in Fig. 2b. Among them, the abundances of Lactobacillus, Gardnerella, Atopobium, Streptococcus, and Prevotella were higher than 1% in both the control and RIF groups, while those of Bifidobacterium, Scardovia, Mycoplasma, and Escherichia were >1% only in the RIF group. There were 26 genera that were significantly different between the two groups, of which 5 were aerobes, 12 were anaerobes, and 9 were unclassified (see Table S1). Lactobacillus, as a main dominant genus, was also dissected in this study, and 12 species were discovered in the samples. The results showed that Lactobacillus crispatus and Lactobacillus iners were the most abundant Lactobacillus species in the RIF and control groups, respectively. The abundance of L. iners was significantly lower in the RIF group than in the control group (P value < 0.05) (see Table S2). However, the other 11 Lactobacillus species showed no significant differences.
FIG 2.
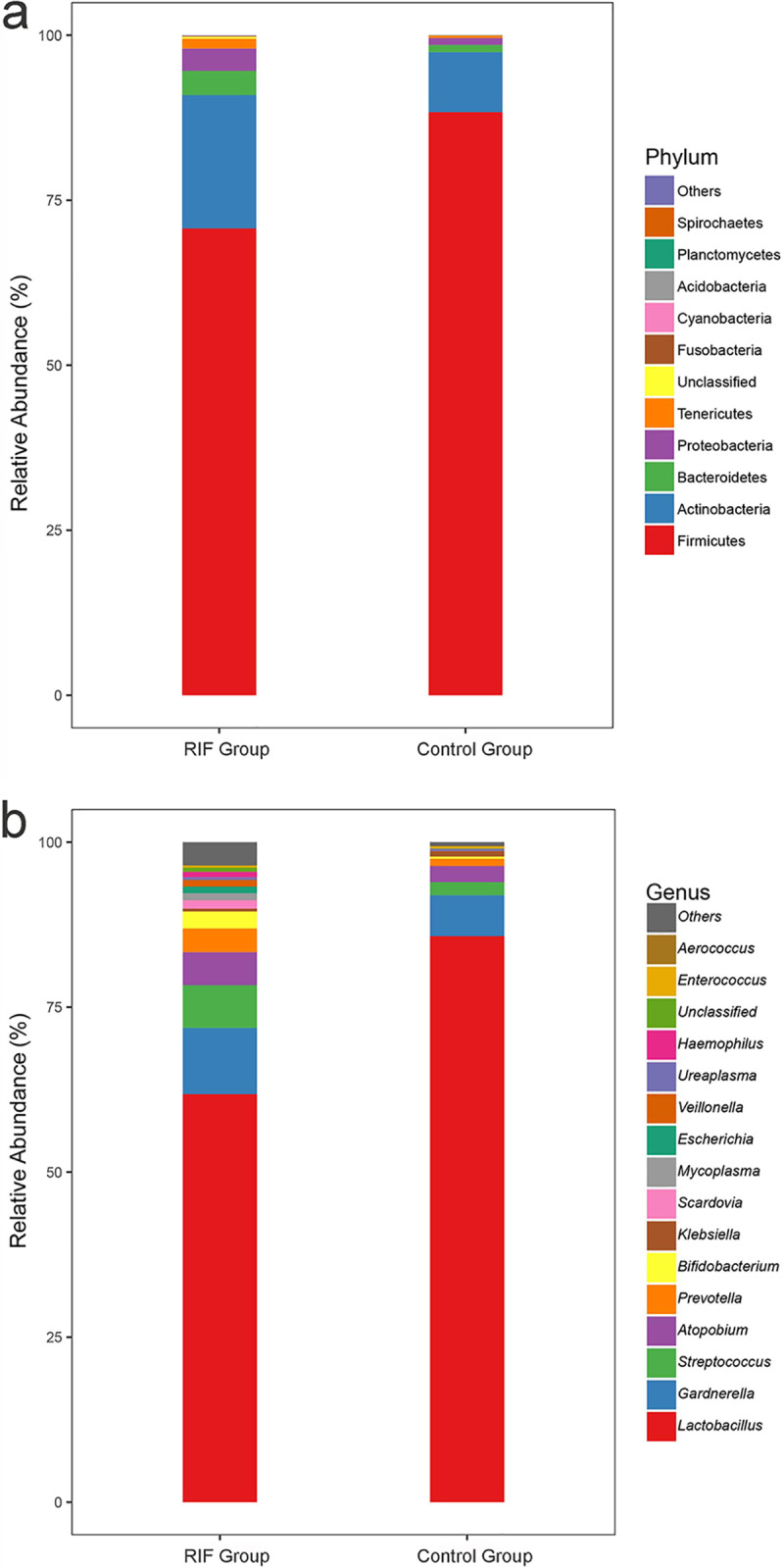
Taxonomic classification of the vaginal microbiota at the phylum level (a) and genus level (b) from the RIF and control groups.
Genera in the vaginal microbiota that were significantly differentially abundant between the RIF and control groups. Download Table S1, DOCX file, 0.1 MB (22.1KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of Lactobacillus species discovered in the samples that were significantly different between the RIF and control groups. Download Table S2, DOCX file, 0.1 MB (21.2KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The potential biomarkers for the predictive model of RIF occurrence were examined by the random forest algorithm among 18 genera with an abundance >0 in both groups out of 26 significantly different genera. Two targeted genera selected by the Gini coefficient, Lactobacillus and Veillonella, were sent for receiver operating characteristic (ROC) analysis (see Table S3). The identified Lactobacillus and Veillonella in patients with a diagnosis of RIF had a sensitivity of 0.627 and 0.560 and specificity of 0.775 and 0.900, respectively (see Fig. S2). Notably, the sensitivity and specificity both improved when the two genera were factored in (sensitivity = 0.633; specificity = 0.925).
ROC curve for Lactobacillus and Veillonella presence in the vagina. Download FIG S2, PDF file, 0.1 MB (125.6KB, pdf) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gini coefficients calculated by a random forest algorithm among 18 genera with an abundance of >0 in two groups out of 26 significantly different genera. Download Table S3, DOCX file, 0.1 MB (15.6KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of the vaginal microbiota in all the samples.
Principal-component analysis (PCA) was applied to illustrate the distribution of the microbial community in the samples (Fig. 3a). The spots belonging to the RIF group were dispersed, and half of them were scattered in the opposite direction of Lactobacillus, which is widely regarded as the probiotic that dominates the healthy female reproductive tract. Additionally, based on linear discriminant analysis (LDA), Lactobacillus was significantly decreased in the RIF group and contributed mostly to group differentiation (Fig. 3b). The dominant genus of both groups was Lactobacillus. The relative abundance of Lactobacillus was 85.766% ± 28.787% in the control group and significantly decreased (P value = 0.013) in the RIF group (61.833% ± 41.849%). Additionally, we defined the subjects for which Lactobacillus accounted for greater than 90% relative abundance as Lactobacillus-dominated (LD) samples and defined those for which Lactobacillus accounted for ≤90% as non-Lactobacillus-dominated (NLD) samples. The pregnancy rates of LD and NLD individuals were 72.723% and 34.723%, respectively, which presented a significant difference (P value = 0.006). This not only showed the link between vaginal Lactobacillus and pregnancy outcomes of the FET but also indicated that the vaginal microbial composition, especially the decrease in Lactobacillus, plays an important role in RIF pathogenesis.
FIG 3.
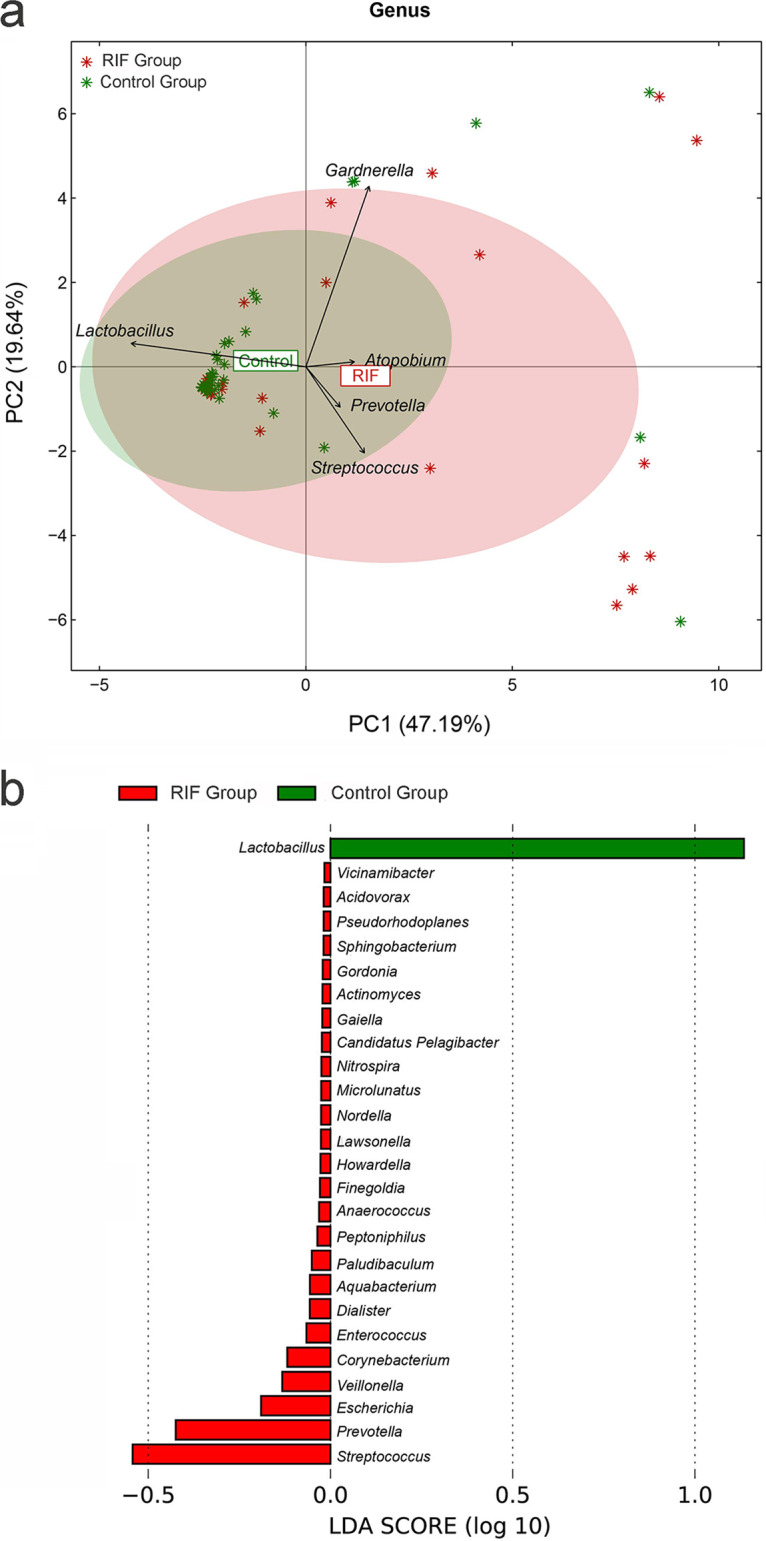
(a) A principal-component analysis was applied to demonstrate the distribution of the vaginal microbial communities in the samples. The arrows indicate the different genera, and their contributions to the explanation of the sample difference are shown by the arrow length. The angle between the arrows represents the positive correlation (<90°) or negative correlation (>90°) among the genera. (b) Linear discriminant analysis of the differentially abundant genera, which indicated their contribution to group differentiation. The green bar indicates that the genus (Lactobacillus) was more abundant in the control group, while red bars indicate that those genera were more abundant in the RIF group.
Comparative metabolomics in the vaginal environment.
Twenty-five samples were subjected to metabolic analysis, including 10 RIF group samples and 15 control samples (Table 2). Both the PCA results of the test and quality control (QC) samples and the correlation analyses of metabolites between the QC samples indicated the stability of the instrument and the good quality of the data (see Fig. S3). In this study, a total of 11,380 peaks of cations were obtained after noise removal; 449 nonredundant metabolites were annotated according to the mass-to-charge ratio (MS1), while 1,541 nonredundant metabolites were detected based on a molecular weight database (MS2). For the anions, 6,658 peaks were obtained, including 146 and 786 nonredundant metabolites according to MS1 and MS2, respectively. After removing the duplicated metabolites, we annotated 573 and 1,934 metabolites based on MS1 and MS2, respectively. As a result, we obtained 2,507 metabolites.
TABLE 2.
Clinical characteristics of the participants in the two groups that were analyzed by metabolic analysis
| Clinical characteristics | Value |
P value | |
|---|---|---|---|
| RIF group (n = 10) | Control group (n = 15) | ||
| Age (yrs) | 33.7 ± 3.8 | 31.8 ± 3.8 | 0.189 |
| BMI (kg/m2) | 21.6 ± 3.5 | 20.9 ± 4.2 | 0.977 |
| AMH (ng/ml) | 3.8 ± 2.3 | 5.8 ± 4.7 | 0.338 |
| Duration of infertility (yrs) | 5.3 ± 3.4 | 3.9 ± 1.4 | 0.480 |
| Endometrial thickness (mm) | 10.2 ± 3.6 | 11.2 ± 2.1 | 0.382 |
| No. of oocytes | 13.5 ± 7.2 | 13.3 ± 6.7 | 0.955 |
| No. of embryos | 10.5 ± 7.2 | 10.5 ± 5.5 | 0.737 |
| No. of high-quality embryos | 2.2 ± 2.9 | 2.6 ± 2.3 | 0.498 |
| No. of embryos transferred | 2.0 ± 0.5 | 2.1 ± 0.5 | 0.658 |
| No. of high-quality embryos transferred | 1.3 ± 0.5 | 1.3 ± 0.9 | 0.628 |
Quality control results of the metabolomics analysis, including the PCA results of test and QC samples (a) and correlation analysis of metabolites between the QC samples (b). Download FIG S3, PDF file, 2.7 MB (2.8MB, pdf) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
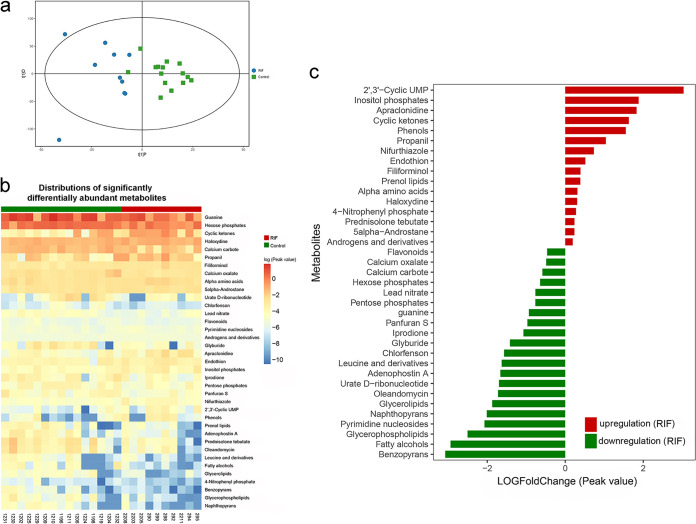
We utilized orthogonal projections to latent structures-discriminant analysis (OPLS-DA) to observe metabolites that were differentially abundant between these two groups and classify them into group-related and group-independent metabolites. The results indicated that the RIF group samples were clustered together and distinct from the grouped control samples (Fig. 4a). Of 2,507 annotated metabolites, 37 metabolites were found to have significant differences between the two groups, with variable importance for the projection (VIP) values of >1 and P values of <0.05 (Fig. 4b; Table S4). To quantify the up/downregulation of differentially abundant metabolites, we calculated the fold change in 37 differentially abundant metabolites. The results revealed that 16 metabolites were significantly upregulated in the RIF group, among which 2′,3-cyclic UMP and inositol phosphate were the top two metabolites, and they were upregulated by 4-fold or more; 21 metabolites were significantly downregulated in the RIF group, and 5 substances were downregulated by 4-fold or more: benzopyran, fatty alcohol, pyrimidine nucleoside, glycerophospholipid, and naphthopyran (Fig. 4c). The network of the metabolites that significantly correlated with each other was also investigated using Spearman’s correlation coefficients (R ≥ 0.6 or R ≤ −0.6) and P values (P<0.05).
FIG 4.
(a) Result of orthogonal projections to latent structures-discriminant analysis of the samples. The x axis shows the predicted principal-component score, which indicates the intergroup difference. The y axis represents the orthogonal principal-component score, which indicates the intragroup difference. (b) Heat map of the differentially abundant metabolites. (c) Quantitative fold change in the differentially abundant metabolites.
Quantities of 37 metabolites that were found to have significant differences between the two groups via the determination of variable importance for the projection (VIP) values and P values. Download Table S4, DOCX file, 0.1 MB (19.6KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Correlation between the vaginal microbiota and metabolome.
To further explore the pathogenic mechanisms of vaginal microbiota in RIF, we examined the correlation between the vaginal microbiota and metabolome. Although a one-to-one relationship between the metabolites and individual bacterial species could not be explained due to the lack of whole-genome information on the bacterial strains and the functional genes of microbial community based on the 16S rRNA sequencing method, the correlations in abundance between differential genera/species and metabolites were analyzed to explore the associated alteration in microbiota and metabolism in RIF patients. The correlations between the differentially abundant genera and metabolites were calculated in the RIF group (Fig. 5). The abundances of four metabolites (benzopyrans, glycerophospholipid, oleandomycin, and prednisolone tebutate) were significantly positively correlated with Lactobacillus, while filiforminol was negatively correlated with Lactobacillus. Among them, benzopyran and glycerophospholipids decreased more than 4-fold in the RIF group (P value < 0.05, R = 0.714), and this was associated with a significant reduction in Lactobacillus relative abundance in the RIF group. This result indicated that benzopyran and glycerophospholipids might function as metabolites during IVF, driven by Lactobacillus or related bacteria in the vaginal microbiota. Interestingly, L. iners was also positively correlated with benzopyran in the RIF group (R = 0.738). For the control group, consistent changes in benzopyrans with the relative abundance of L. iners were identified (R = 0.527).
FIG 5.
Correlation analysis between the differentially abundant metabolites and differential genera in the RIF group. Red on the bar outside the heat map indicates that the metabolite/genus was upregulated in the RIF group (case), while green indicates downregulation.
DISCUSSION
This study is an attempt to explain the influence of the vaginal microbiota and metabolites on RIF patients while using first-cycle successful FET cases as the control. The vaginal microbiota structure of the RIF group was different from that of the control group, containing significantly decreased Lactobacillus and harboring more potential pathogens. The metabolite profile delineated 37 metabolites to be significantly differentially abundant between the two groups, and this may have resulted in or from the changes in the microbiota composition.
Considering that the hormone level, menstrual cycle, cause of infertility, and pregnancy status were closely associated with the female vaginal microbiota and metabolites, these factors were all homogenized in this study by sampling consistently on the same day of embryo transfer in the FET cycle, and only patients with unexplained RIF without any other diseases who were confirmed to not be pregnant in this cycle were recruited. The baseline parameters (age, body mass index [BMI], anti-Müllerian hormone [AMH], duration of infertility, and endometrial thickness) and the number of embryos showed no significant differences in the two groups (Table 1). Notably, although the number of embryos transferred was significantly lower in the RIF group, it should not cause a confounding bias for the results, as it was determined by the clinical treatment characteristics of the RIF patients. To achieve a higher implantation and live-birth rate, doctors usually transfer 1 to 2 blastocysts for RIF patients while transferring 2 to 3 cleavage-stage embryos for the patients in the first FET cycle to avoid cycle cancellation due to blastocyst culture (37–39).
The abundances of 26 genera detected in the present study were significantly different between the two groups (P value < 0.05). In the RIF group, all of the genera were significantly increased, especially the aerobic bacteria (8.593% for the RIF group and 2.349% for the control group, P value < 0.05), whereas only Lactobacillus abundance was significantly reduced. Among the 25 increased genera, many anaerobic bacteria, such as Gardnerella, Prevotella, and Atopobium, are the main pathogens for bacterial vaginosis (BV), while the abundance of many aerobic bacteria, such as Escherichia, Enterococcus, Streptococcus, and Corynebacterium, are closely related to the risk of aerobic vaginitis (AV) and urinary tract infection (UTI). A study of asymptomatic reproductive tract infections in couples undergoing IVF showed that 29.1% (83/285) of the males and 26.3% (75/285) of the females carried at least one potential pathogen for reproductive tract disease, and more positive cases were found in the IVF pregnancy failure group than in the IVF control group. Additionally, Enterococcus faecalis, Escherichia coli, Streptococcus agalactiae, and Gardnerella vaginalis were significantly associated with decreased levels of Lactobacillus (P value < 0.01) (40). Similarly, the abundance of Lactobacillus was negatively related to the abundance of potential BV/AV pathogens in this study, including Gardnerella, Prevotella, Atopobium, and Streptococcus, which indicated the protective function of Lactobacillus. Our results also indicated that Lactobacillus was losing its dominant position in the RIF group, which may be associated with the occurrence of repeated pregnancy failures. Furthermore, the pregnancy rate of NLD subjects was significantly decreased compared with that of LD subjects. Although the relative abundance of Lactobacillus was significantly lower in the RIF group than in the control group, the LDA value was still too low to be identified as a biomarker to predict the occurrence of RIF. However, the ROC curve showed that when Veillonella was factored in, the sensitivity and specificity both increased, demonstrating that the combination of Lactobacillus and Veillonella can be used as a potential biomarker for the prediction of RIF occurrence.
An increase in vaginal microbial diversity in RIF patients was observed in this study, which is consistent with the study by Hyman et al. in 2012 (35). That report was the first study to suggest that the species diversity index of the vaginal microbiota distinguished women who had live births from those who did not (35). Another study also showed that the clinical pregnancy rate and live-birth rate were significantly reduced (P value = 0.01) in a cohort with a vaginal Shannon-Wiener index higher than 0.93 (41). To date, few vaginal microbiota studies have investigated RIF patients, but there are association studies of an abnormal vaginal microbiota with IVF. A review concluded that culture-dependent studies indicated that an abnormal vaginal microbiota was not linked to the outcome of IVF. However, this conclusion was overturned by high-throughput sequencing results, which elaborated that microbiota disorders negatively influenced the IVF outcome (42). Another review demonstrated that the incidence of bacterial vaginosis (BV) was associated with early abortion of IVF pregnancies and infertility due to tubal factors and that it was not associated with the pregnancy rate and live-birth rate (30).
To evaluate the difference in the vaginal metabolites between the RIF and control groups, we identified the fold changes of 37 significantly different metabolites in two groups. In the RIF group, 2′,3-cyclic UMP and inositol phosphate increased by 4-fold or more, while benzopyran, fatty alcohol, pyrimidine nucleoside, glycerophospholipid, and naphthopyran decreased by 4-fold or more. As inositol phosphate is one of the metabolites of glycerophospholipid (43), the significant increase in inositol phosphate should be linked to the decrease in glycerophospholipid, which may result in the reduction of other important metabolites from glycerophospholipid involved in the embryo implantation process due to the lack of substances, such as lysobisphosphatidic acid and prostaglandin (44–49). In addition, the increase in inositol phosphate could promote the internal influx of Ca2+ and lead to contraction of the uterine, which is detrimental to embryo implantation (50). Benzopyran and naphthopyran are selective estrogen receptor modulators that can both influence the outcome of IVF. Interestingly, after analyzing the association between significantly different metabolites and genera, benzopyran and glycerophospholipid were listed in the 4 metabolites that were significantly positively correlated with the abundance of the key vaginal genus Lactobacillus. Thus, these two metabolites are further discussed.
Glycerophospholipids are important structural and regulatory components of biofilms and serve as precursors for many active biomolecules, such as arachidonic acid (AA) and lysobisphosphatidic acid (LPA), which are catalyzed by phospholipase A2 (PLA2) (45, 49). AA then produces prostaglandin (PG) under the action of cyclooxygenase 2 (COX-2). PG and LPA are the terminal products of glycerophospholipids that play key roles in embryo implantation. According to a previous study, LPA plays an essential role in maintaining the normal size and spacing of the embryo, which is positively correlated with embryo implantation in mice (44). LPA3, a G protein-coupled receptor that functions in the uterine epithelium, also regulates the activity of COX-2 and the level of PGs, which could directly affect implantation and decidualization (46, 48). Mice lacking LPA3 showed defects such as deferred on-time implantation, delayed fetal development, embryo crowding, and sharing of one placenta by several embryos. Additionally, deviation in the PG-producing pathway has a significant impact on the implantation process, resulting in a reduction in the likelihood of achieving pregnancy (49). Furthermore, it has been reported that levels of endometrial LPA3 were reduced in RIF patients (49) and that its activation induces decidualization (48). More importantly, phospholipase could be stimulated by lipopolysaccharide, which is the key component of the outer membrane of Gram-negative bacteria, such as Lactobacillus. In addition, glycerophospholipids were positively correlated with the abundance of Lactobacillus. Hence, a balanced vaginal microbiota would result in the normal regulation of glycerophospholipids, which might lead to successful embryo implantation.
Sex hormones are potential metabolites functioning during IVF. In this study, androgens, 5α-androstane, and other androgen derivatives were all upregulated in the RIF group, which may antagonize estrogen secretion, inhibit endometrial hyperplasia, depress ovarian and pituitary function, and, in turn, cause a negative effect on embryo implantation. Benzopyran, flavonoid, and naphthopyran, as selective estrogen receptor modulators (SERMs) (51), were all downregulated in the RIF group. Among them, benzopyran was downregulated more than 4-fold. SERMs such as clomiphene and letrozole, which are commonly used to promote follicular development, bind with high affinity to the estrogen receptor to induce agonistic or antagonistic activities in special tissues (52). Moreover, benzopyran has been identified as a secondary metabolite not only in green plants but also in bacteria and fungi, and it has been routinely employed as an herbal remedy for decades (53). Benzopyran is currently used as an anticoagulant because it is a COX-2-inhibitor (54), but its effect on the reproductive system has not been studied. However, aspirin has been known as an irreversible inhibitor of COX, and its anticoagulant effect plays a significant role in the treatment of recurrent spontaneous abortion (55). Hence, benzopyran may also play a similar role in embryo implantation as aspirin. Furthermore, although the quantity of benzopyran was positively correlated with the abundance of L. iners in our study, the relationship between benzopyran and vaginal microbiota and its specific functions in embryo implantation still need further investigation.
Our discovery of a significant difference in the vaginal microbiota and metabolomes of RIF patients establishes an important theoretical grounding to the potential development of the microbiota and metabolome as biomarkers for predicting RIF occurrence. Although previous studies have demonstrated that the endometrial microbiota is associated with infertility and IVF (56–58), vaginal rather than endometrial samples are more feasible for clinical application due to the lower contamination risk and noninvasive sample process. First, unlike vaginal specimen sampling, endometrial samples are easily contaminated by the cervicovaginal microbiome due to the transvaginal collection and the 4-fold fewer bacteria inhabiting the endometrium than the vagina (37). Second, endometrial biopsy specimens are usually not taken in the WOI of the same cycle because of endometrial damage caused by the invasive procedure; hence, the result from the endometrial biopsy specimen cannot show the condition of the patient at the time of embryo transfer. Moreover, other than endometrial biopsy specimens, endometrial fluid and uterine flushing fluid are the other endometrial samples commonly used in the clinic. As the microecology shows inconsistent characteristics in these three environments, it is hard to distinguish which is more persuasive (59). Nevertheless, despite the significantly different genera and metabolites in RIF patients’ vaginal environments in this study, further verification is still needed. Furthermore, there are limitations of the 16S rRNA sequencing approach, such as the short-read lengths obtained, sequencing errors (60, 61), differences arising from the different regions chosen (62), and difficulties in assessing operational taxonomic units (OTUs) (63). Also, limited microbial taxonomic messages beyond the bacteria will be obtained from 16S rRNA sequences because of the low coverage of chlamydia, archaeal, fungal, chloroplastic, mitochondrial, and eukaryotic rRNA genes (64). Hence, additional biomarkers should be detected but should not be based solely on the taxonomic information from the 16S rRNA sequencing approach. Functional genes should also be complemented and associated with metabolites for microbial metabolic pathway network construction to explore RIF pathogenesis.
Conclusion.
The vaginal microbiota of RIF patients presented higher microbial diversity and lower abundance of Lactobacillus, which significantly associated with the pregnancy rate. LPA and PG metabolized from glycerophospholipid are key factors affecting implantation and decidualization, and benzopyran may contribute to the outcome of pregnancy as a SERM.
MATERIALS AND METHODS
Criteria for recruitment of the research subjects and ethical approval.
Patients who underwent frozen embryo transfer (FET) in the Department of Reproductive Medicine, Peking University Shenzhen Hospital, from May 2018 to December 2018 were selected and divided into the unexplained RIF group (RIF group) and successful pregnancy in the first FET cycle group (control group). All of the women were less than 40 years of age, and their functional ovarian reserve was assessed by levels of follicle stimulation hormone (FSH) of <12 mIU/ml and AMH of >1.1 ng/ml. The inclusion criteria for the RIF group were patients who (i) received at least four good-quality embryos in a minimum of three fresh or frozen cycles but had still not become pregnant and (ii) were not pregnant in this FET cycle. The patients who were counted in the control group were those undergoing an embryo transfer for the first time, and this cycle was FET. In addition, these patients were confirmed to be pregnant in this FET cycle.
Patients with simple male factor infertility, ovulation and menstrual disorders, genital tract organic lesions, and systemic diseases were excluded. All patients were confirmed to have a normal uterine cavity by hysteroscopy and a normal chromosome karyotype by chromosome G-banding and karyotype analysis. The patients all underwent examinations of the cervical mucus and vaginal secretions at 1 month and 7 days before embryo transfer. Patients showing bacterial vaginosis, vulvovaginal candidiasis, trichomonas vaginitis, Chlamydia trachomatis, Ureaplasma urealyticum, or Neisseria gonorrhoeae infection, or other vaginal subjective symptoms, such as vaginal itching and abnormal discharge, at the examination at either 1 month or 7 days before transfer were excluded. Patients receiving any antibiotics (oral or topical) or vaginal douching or engaging in sexual behavior within 2 weeks before sample collection were also excluded.
The study was approved by the Ethics Committee of Peking University Shenzhen Hospital on 10 May 2018. The study is sponsored by the Peking University Shenzhen Hospital (2018 no. 017) and was conducted as a single-center study without any investigational product. All enrolled subjects provided written consent and gave permission for access to medical records to obtain their related clinical information and vaginal specimens.
Sample processing.
Vaginal specimens were collected on the day of embryo transfer before the operation. A sterile sample collection kit (CY-90003T, iCleanhcy; Huachenyang [Shenzhen] Technology Co., Ltd., China) with a separated package was used in the sample collection, and it comprised a long-handle swab connected to a sealed cap and inserted in a collection tube. Each sample kit was weighed before sample collection as the dry weight. During the sampling, the swab was carefully taken out of the tube and immersed into the upper third of the vagina and maneuvered in a circle by holding the cap constantly to avoid contamination of the swab. After sampling, the swab with vaginal discharge was placed back into the tube and weighed as the wet weight. The net weight of the vaginal discharge was recorded as the wet weight minus the dry weight. Two parallel samples were taken from each patient, immediately placed in an ice box, and then transferred to a −80°C freezer within 20 min for subsequent 16S rRNA gene sequencing and metabolic analysis (Shenzhen We-Health Gene Company).
Total DNA extraction and 16S rRNA sequencing.
DNA extraction was conducted with a DNeasy PowerSoil kit (Qiagen, Hilden, Germany) followed by DNA purification with VAHTS DNA Clean Beads (Vazyme, Nanjing, China) according to the manufacturers’ instructions. The V3-V4 region of the 16S rRNA gene was amplified by PCR with the universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) using a TransStart FastPfu DNA polymerase kit on an ABI GeneAmp 9700 PCR instrument. The PCR system was 50 μl in total, and it contained 4 μl of 5× FastPfu buffer, 2 μl of 2.5 mM deoxynucleoside triphosphates (dNTPs), 0.8 μl of forward primer, 0.8 μl of reverse primer, 0.4 μl of FastPfu Polymerase, 0.2 μl of bovine serum albumin (BSA), 10 ng of template DNA, and double-distilled water (ddH2O) to make up the total volume. PCR was performed using the following conditions: 3 min of denaturation at 95°C, 29 cycles of denaturation at 95°C for 30 s, annealing at 45°C for 30 s, and elongation at 72°C for 45 s, and a final extension at 72°C for 10 min. The 16S rRNA gene amplicons were purified and used for MiSeq sequencing library construction according to the manufacturer’s instructions for the MiSeq reagent kit v2.
Microbiota analysis.
Clean paired-end sequences were filtered by eliminating low-quality base pairs, contaminated reads, N-containing reads, and low-complexity sequences. The operational taxonomic units (OTUs) were clustered under the 97% cutoff and annotated by the Ribosomal Database Project after merging reads via overlaps. The α-diversity inferred by the Shannon-Wiener index was calculated by the R package vegan; meanwhile, a principal-component analysis (PCA) was performed by the ade4 package. Linear discriminant analysis (LDA) was performed using Galaxy software. A random forest model was constructed between the RIF group and control group with 5-fold cross-validation using the R package randomForest (65). The genera selected by Gini coefficient were used for a receiver operating characteristic (ROC) analysis, and the area under the curve (AUC) was calculated.
Metabolome analysis.
Metabolites were extracted from swabs by adding 20 μl isolation liquid (methanol/acetonitrile/water [2:2:1]) to each 1 mg vaginal discharge. Then, 10 μl of isolates from all samples were mixed as quality control (QC) samples for stability evaluation during the experiment (66, 67), 2 μl of isolates of each sample was analyzed by liquid chromatography (LC) and mass spectrometry (MS) (Q-Exactive Orbitrap; Thermo-Fisher Scientific, USA), and the signals of metabolites in all samples were examined. Using ProteoWizard (v3.0.9134) and the XCMS package in R (v3.2), the MS raw data were converted to a data matrix that contained the retention time (RT), mass-to-charge ratio (m/z) value, and peak intensity. All output data were normalized by the internal standard normalization method and are presented as the peak value (peak area of the test sample/peak area of internal standard sample). According to the peak values of metabolites, their variable importance in the projection (VIP) was calculated by orthogonal projections to latent structures-discriminant analysis (OPLS-DA). Student's t test was applied to detect differentially enriched metabolites between the control and RIF groups (VIP > 1, P value < 0.05). Correlations among the metabolites were evaluated by Spearman’s correlation analysis, and the network was visualized by Cytoscape software (v3.4.0). In addition, a Spearman correlation analysis was also applied to investigate the relationship between the vaginal microbiota and metabolites.
Data availability.
The 16S rRNA gene sequencing data for the 67 vaginal microbiota samples analyzed in this study have been deposited with the National Center for Biotechnology Information (NCBI) under reference number PRJNA590580. The metabolome profiles of the final identified 2,507 metabolites are shown in Table S5 in the supplemental matieral.
Metabolome profile of the final identified 2,507 metabolites. Download Table S5, XLSX file, 0.1 MB (129.9KB, xlsx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank the research assistant Liting Huang and nurses Dongxia Lin, Dan Liu, HuiChang Peng, and Liyan Xu, etc., for their contributions to the collection and transportation of specimens. We also thank the people at Shenzhen We-Health Gene Company for their assistance with specimen detection and analysis. More importantly, we thank all the patients who participated in this project.
The study was funded by the Shenzhen Municipal Government (JCYJ20180228162311024, SZSM201612065, and SZXJ2017003), Shenzhen, China.
S.F. and M.F. contributed to the conception and design of the study. M.F. and W.Q. contributed to obtaining ethical approval. S.F. was responsible for the funding support. M.F. directed the experiments and data analyses. M.F., W.Q., and Y.L. conducted the collection of samples. M.F., X.Z., and S.F. analyzed the data and generated the figures and tables. X.Z. drafted the manuscript, and X.Z. and M.F. revised it. All authors have read and approved the final manuscript.
Footnotes
Citation Fu M, Zhang X, Liang Y, Lin S, Qian W, Fan S. 2020. Alterations in vaginal microbiota and associated metabolome in women with recurrent implantation failure. mBio 11:e03242-19. https://doi.org/10.1128/mBio.03242-19.
Contributor Information
Paul L. Fidel, LA State University Health Sciences Center.
Xiaorong Lin, University of Georgia.
REFERENCES
- 1.Zegers-Hochschild F, Adamson GD, de Mouzon J, Ishihara O, Mansour R, Nygren K, Sullivan E, Van der Poel S, International Committee for Monitoring Assisted Reproductive Technology, World Health Organization. 2009. The international committee for monitoring assisted reproductive technology (ICMART) and the world health organization (WHO) revised glossary on ART terminology. Hum Reprod 24:2683–2687. doi: 10.1093/humrep/dep343. [DOI] [PubMed] [Google Scholar]
- 2.Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, Simpson JL, van der Poel S. 2017. The international glossary on infertility and fertility care. Hum Reprod 32:1786–1801. doi: 10.1093/humrep/dex234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inhorn MC, Patrizio P. 2015. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update 21:411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 4.Gnoth C, Maxrath B, Skonieczny T, Friol K, Godehardt E, Tigges J. 2011. Final ART success rates: a 10 years survey. Hum Reprod 26:2239–2246. doi: 10.1093/humrep/der178. [DOI] [PubMed] [Google Scholar]
- 5.Coughlan C, Ledger W, Wang Q, Liu F, Demirol A, Gurgan T, Cutting R, Ong K, Sallam H, Li T. 2014. Recurrent implantation failure: definition and management. Reprod Biomed Online 28:14–38. doi: 10.1016/j.rbmo.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Bashiri A, Halper KI, Orvieto R. 2018. Recurrent implantation failure-update overview on etiology, diagnosis, treatment and future directions. Reprod Biol Endocrinol 16:121. doi: 10.1186/s12958-018-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellver J, Soares SR, Alvarez C, Munoz E, Ramírez A, Rubio C, Serra V, Remohí J, Pellicer A. 2008. The role of thrombophilia and thyroid autoimmunity in unexplained infertility, implantation failure and recurrent spontaneous abortion. Hum Reprod 23:278–284. doi: 10.1093/humrep/dem383. [DOI] [PubMed] [Google Scholar]
- 8.Tan BK, Vandekerckhove P, Kennedy R, Keay SD. 2005. Investigation and current management of recurrent IVF treatment failure in the UK. BJOG 112:773–780. doi: 10.1111/j.1471-0528.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- 9.Christiansen OB, Nielsen HS, Kolte AM. 2006. Future directions of failed implantation and recurrent miscarriage research. Reprod Biomed Online 13:71–83. doi: 10.1016/s1472-6483(10)62018-4. [DOI] [PubMed] [Google Scholar]
- 10.Tan J, Kan A, Hitkari J, Taylor B, Tallon N, Warraich G, Yuzpe A, Nakhuda G. 2018. The role of the endometrial receptivity array (ERA) in patients who have failed euploid embryo transfers. J Assist Reprod Genet 35:683–692. doi: 10.1007/s10815-017-1112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, et al. . 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 12.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan SH, Lobley G, Holtrop G, Ince J, Johnstone A, Louis P, Flint HJ. 2008. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. nature 444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 15.Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, Puchois V, Martin JC, Lepage P, Le Roy T, Lefèvre L, Langelier B, Cailleux F, González-Castro AM, Rabot S, Gaudin F, Agostini H, Prévot S, Berrebi D, Ciocan D, Jousse C, Naveau S, Gérard P, Perlemuter G. 2016. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65:830–839. doi: 10.1136/gutjnl-2015-310585. [DOI] [PubMed] [Google Scholar]
- 16.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. 2014. Alterations of the human gut microbiome in liver cirrhosis. Nature 513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 17.Tang WHW, Li DY, Hazen SL. 2019. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol 16:137–154. doi: 10.1038/s41569-018-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster JA, Neufeld K-A. 2013. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci 36:305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, Li F, Yu X, Feng Q, Wang Z, Xie H, Chen X, Zeng C, Wen B, Zeng L, Du H, Tang H, Xu C, Xia Y, Xia H, Yang H, Wang J, Wang J, Madsen L, Brix S, Kristiansen K, Xu X, Li J, Wu R, Jia H. 2017. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun 8:875. doi: 10.1038/s41467-017-00901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donders G, Van Calsteren K, Bellen G, Reybrouck R, Van den Bosch T, Riphagen I, Van Lierde S. 2009. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG 116:1315–1324. doi: 10.1111/j.1471-0528.2009.02237.x. [DOI] [PubMed] [Google Scholar]
- 23.Amabebe E, Reynolds S, Stern VL, Parker JL, Stafford GP, Paley MN, Anumba DO. 2016. Identifying metabolite markers for preterm birth in cervicovaginal fluid by magnetic resonance spectroscopy. Metabolomics 12:67. doi: 10.1007/s11306-016-0985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onderdonk AB, Delaney ML, Fichorova RN. 2016. The human microbiome during bacterial vaginosis. Clin Microbiol Rev 29:223–238. doi: 10.1128/CMR.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiller S. 1995. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. N Engl J Med 333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 26.Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, Vo KC, Caughey AB, Hilton JF, Davis RW, Giudice LC. 2014. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci 21:32–40. doi: 10.1177/1933719113488838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma H, Tal R, Clark NA, Segars JH. 2014. Microbiota and pelvic inflammatory disease. Semin Reprod Med 32:43–49. doi: 10.1055/s-0033-1361822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brotman RM. 2011. Vaginal microbiome and sexually transmitted infections: an epidemiologic perspective. J Clin Invest 121:4610–4617. doi: 10.1172/JCI57172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JE, Lee S, Lee H, Song Y-M, Lee K, Han MJ, Sung J, Ko G. 2013. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 8:e63514. doi: 10.1371/journal.pone.0063514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haahr T, Zacho J, Bräuner M, Shathmigha K, Skov Jensen J, Humaidan P. 2019. Reproductive outcome of patients undergoing in vitro fertilisation treatment and diagnosed with bacterial vaginosis or abnormal vaginal microbiota: a systematic PRISMA review and meta‐analysis. BJOG 126:200–207. doi: 10.1111/1471-0528.15178. [DOI] [PubMed] [Google Scholar]
- 31.Dunlop AL, Mulle JG, Ferranti EP, Edwards S, Dunn AB, Corwin EJ. 2015. The maternal microbiome and pregnancy outcomes that impact infant health: a review. Adv Neonatal Care 15:377–385. doi: 10.1097/ANC.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benner M, Ferwerda G, Joosten I, Van der Molen RG. 2018. How uterine microbiota might be responsible for a receptive, fertile endometrium. Hum Reprod Update 24:393–415. doi: 10.1093/humupd/dmy012. [DOI] [PubMed] [Google Scholar]
- 33.Chase D, Goulder A, Zenhausern F, Monk B, Herbst-Kralovetz M. 2015. The vaginal and gastrointestinal microbiomes in gynecologic cancers: a review of applications in etiology, symptoms and treatment. Gynecol Oncol 138:190–200. doi: 10.1016/j.ygyno.2015.04.036. [DOI] [PubMed] [Google Scholar]
- 34.Franasiak JM, Scott RT Jr.. 2015. Reproductive tract microbiome in assisted reproductive technologies. Fertil Steril 104:1364–1371. doi: 10.1016/j.fertnstert.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Hyman RW, Herndon CN, Jiang H, Palm C, Fukushima M, Bernstein D, Vo KC, Zelenko Z, Davis RW, Giudice LC. 2012. The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J Assist Reprod Genet 29:105–115. doi: 10.1007/s10815-011-9694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholson JK, Lindon JC, Holmes E. 1999. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 37.Moreno I, Simon C. 2018. Relevance of assessing the uterine microbiota in infertility. Fertil Steril 110:337–343. doi: 10.1016/j.fertnstert.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 38.Gardner DK, Schoolcraft WB, Wagley L, Schlenker T, Stevens J, Hesla J. 1998. A prospective randomized trial of blastocyst culture and transfer in in-vitro fertilization. Hum Reprod 13:3434–3440. doi: 10.1093/humrep/13.12.3434. [DOI] [PubMed] [Google Scholar]
- 39.Practice Committee of the American Society for Reproductive Medicine, Practice Committee of the Society for Assisted Reproductive Technology. 2006. Guidelines on number of embryos transferred. Fertil Steril 86:S52–S52. doi: 10.1016/j.fertnstert.2006.07.1473. [DOI] [PubMed] [Google Scholar]
- 40.Ricci S, De Giorgi S, Lazzeri E, Luddi A, Rossi S, Piomboni P, De Leo V, Pozzi G. 2018. Impact of asymptomatic genital tract infections on in vitro fertilization (IVF) outcome. PLoS One 13:e0207684. doi: 10.1371/journal.pone.0207684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haahr T, Al Humaidan PSH, Elbæk H, Alsbjerg B, Rygaard K, Lilje B, Andersen P, Ng K, Jensen J. 2017. Vaginal microbiota and IVF outcomes-Development of a simple diagnostic tool to predict patients at risk of a poor reproductive outcome. ReproYoung Conference 2017, Båstad, Sweden. [Google Scholar]
- 42.Bracewell-Milnes T, Saso S, Nikolaou D, Norman‐Taylor J, Johnson M, Thum MY. 2018. Investigating the effect of an abnormal cervico‐vaginal and endometrial microbiome on assisted reproductive technologies: a systematic review. Am J Reprod Immunol 80:e13037. doi: 10.1111/aji.13037. [DOI] [PubMed] [Google Scholar]
- 43.Freitas AC, Bocking A, Hill JE, Money DM, the VOGUE Research Group. 2018. Increased richness and diversity of the vaginal microbiota and spontaneous preterm birth. Microbiome 6:117. doi: 10.1186/s40168-018-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye X, Hama K, Contos JJA, Anliker B, Inoue A, Skinner MK, Suzuki H, Amano T, Kennedy G, Arai H, Aoki J, Chun J. 2005. LPA 3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature 435:104–108. doi: 10.1038/nature03505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah BH, Catt KJ. 2005. Roles of LPA3 and COX-2 in implantation. Trends Endocrinol Metab 16:397–399. doi: 10.1016/j.tem.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Mizugishi K, Li C, Olivera A, Bielawski J, Bielawska A, Deng C-X, Proia RL. 2007. Maternal disturbance in activated sphingolipid metabolism causes pregnancy loss in mice. J Clin Invest 117:2993–3006. doi: 10.1172/JCI30674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mills GB, Moolenaar WH. 2003. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer 3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 48.Aikawa S, Kano K, Inoue A, Wang J, Saigusa D, Nagamatsu T, Hirota Y, Fujii T, Tsuchiya S, Taketomi Y, Sugimoto Y, Murakami M, Arita M, Kurano M, Ikeda H, Yatomi Y, Chun J, Aoki J. 2017. Autotaxin–lysophosphatidic acid–LPA3 signaling at the embryo‐epithelial boundary controls decidualization pathways. EMBO J 36:2146–2160. doi: 10.15252/embj.201696290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Achache H, Tsafrir A, Prus D, Reich R, Revel A. 2010. Defective endometrial prostaglandin synthesis identified in patients with repeated implantation failure undergoing in vitro fertilization. Fertil Steril 94:1271–1278. doi: 10.1016/j.fertnstert.2009.07.1668. [DOI] [PubMed] [Google Scholar]
- 50.Berridge MJ. 2016. The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol Rev 96:1261–1296. doi: 10.1152/physrev.00006.2016. [DOI] [PubMed] [Google Scholar]
- 51.Goldstein SR, Siddhanti S, Ciaccia AV, Plouffe L Jr.. 2000. A pharmacological review of selective oestrogen receptor modulators. Hum Reprod Update 6:212–224. doi: 10.1093/humupd/6.3.212. [DOI] [PubMed] [Google Scholar]
- 52.Shelly W, Draper MW, Krishnan V, Wong M, Jaffe RB. 2008. Selective estrogen receptor modulators: an update on recent clinical findings. Obstet Gynecol Surv 63:163–181. doi: 10.1097/OGX.0b013e31816400d7. [DOI] [PubMed] [Google Scholar]
- 53.Murray R. 2002. The naturally occurring coumarins. Fortschr Chem Org Naturst 83:1–619. doi: 10.1007/978-3-7091-6172-2_1. [DOI] [PubMed] [Google Scholar]
- 54.Prasanna S, Manivannan E, Chaturvedi SC. 2004. QSAR analysis of 2,3-diaryl benzopyrans/pyrans as selective COX2 inhibitors based on semiempirical AM1 calculations. QSAR Comb Sci 23:621–628. doi: 10.1002/qsar.200430887. [DOI] [Google Scholar]
- 55.Andreoli L, Bertsias GK, Agmon-Levin N, Brown S, Cervera R, Costedoat-Chalumeau N, Doria A, Fischer-Betz R, Forger F, Moraes-Fontes MF, Khamashta M, King J, Lojacono A, Marchiori F, Meroni PL, Mosca M, Motta M, Ostensen M, Pamfil C, Raio L, Schneider M, Svenungsson E, Tektonidou M, Yavuz S, Boumpas D, Tincani A. 2017. EULAR recommendations for women’s health and the management of family planning, assisted reproduction, pregnancy, and menopause in patients with systemic lupus erythematosus and/or the antiphospholipid syndrome. Ann Rheum Dis 76:476–485. doi: 10.1136/annrheumdis-2016-209770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franasiak JM, Werner MD, Juneau CR, Tao X, Landis J, Zhan Y, Treff NR, Scott RT. 2016. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet 33:129–136. doi: 10.1007/s10815-015-0614-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verstraelen H, Vilchez-Vargas R, Desimpel F, Jauregui R, Vankeirsbilck N, Weyers S, Verhelst R, De Sutter P, Pieper DH, Van De Wiele T. 2016. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1–2 region of the 16S rRNA gene. Peerj 4:e1602. doi: 10.7717/peerj.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreno I, Codoñer FM, Vilella F, Valbuena D, Martinez-Blanch JF, Jimenez-Almazán J, Alonso R, Alamá P, Remohí J, Pellicer A, Ramon D, Simon C. 2016. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol 215:684–703. doi: 10.1016/j.ajog.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Wong KK-W, Ko E-L, Chen X, Huang J, Tsui S-W, Li TC, Chim SS-C. 2018. Systematic comparison of bacterial colonization of endometrial tissue and fluid samples in recurrent miscarriage patients: implications for future endometrial microbiome studies. Clin Chem 64:1743–1752. doi: 10.1373/clinchem.2018.289306. [DOI] [PubMed] [Google Scholar]
- 60.Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 61.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS. 2009. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75:5227–5236. doi: 10.1128/AEM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mori H, Maruyama F, Kato H, Toyoda A, Dozono A, Ohtsubo Y, Nagata Y, Fujiyama A, Tsuda M, Kurokawa K. 2014. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res 21:217–227. doi: 10.1093/dnares/dst052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 66.Cai Y, Weng K, Guo Y, Peng J, Zhu Z-J. 2015. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing. Metabolomics 11:1575–1586. doi: 10.1007/s11306-015-0809-4. [DOI] [Google Scholar]
- 67.Wang J, Zhang T, Shen X, Liu J, Zhao D, Sun Y, Wang L, Liu Y, Gong X, Liu Y, Zhu Z-J, Xue F. 2016. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics 12:116. doi: 10.1007/s11306-016-1050-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction curves of the sequenced samples. Download FIG S1, PDF file, 1.7 MB (1.8MB, pdf) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genera in the vaginal microbiota that were significantly differentially abundant between the RIF and control groups. Download Table S1, DOCX file, 0.1 MB (22.1KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of Lactobacillus species discovered in the samples that were significantly different between the RIF and control groups. Download Table S2, DOCX file, 0.1 MB (21.2KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ROC curve for Lactobacillus and Veillonella presence in the vagina. Download FIG S2, PDF file, 0.1 MB (125.6KB, pdf) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gini coefficients calculated by a random forest algorithm among 18 genera with an abundance of >0 in two groups out of 26 significantly different genera. Download Table S3, DOCX file, 0.1 MB (15.6KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quality control results of the metabolomics analysis, including the PCA results of test and QC samples (a) and correlation analysis of metabolites between the QC samples (b). Download FIG S3, PDF file, 2.7 MB (2.8MB, pdf) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantities of 37 metabolites that were found to have significant differences between the two groups via the determination of variable importance for the projection (VIP) values and P values. Download Table S4, DOCX file, 0.1 MB (19.6KB, docx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolome profile of the final identified 2,507 metabolites. Download Table S5, XLSX file, 0.1 MB (129.9KB, xlsx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The 16S rRNA gene sequencing data for the 67 vaginal microbiota samples analyzed in this study have been deposited with the National Center for Biotechnology Information (NCBI) under reference number PRJNA590580. The metabolome profiles of the final identified 2,507 metabolites are shown in Table S5 in the supplemental matieral.
Metabolome profile of the final identified 2,507 metabolites. Download Table S5, XLSX file, 0.1 MB (129.9KB, xlsx) .
Copyright © 2020 Fu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.