Abstract
Extracellular RNAs (exRNAs) are released by extracellular vesicles, small membranous nanoparticles secreted by all cell types. When transported into cells, exRNAs can modulate gene expression or cellular responses in the target cells since many small RNAs have regulatory functions. Indeed, it is widely acknowledged that endogenous exRNAs in the human body are related to various diseases. However, microbial exRNAs have been less studied, and their connection to host diseases has just begun to be explored. In this review, I will discuss analytical methods for exRNAs and the potential use of exRNAs as disease biomarkers. I also consider current progress in understanding the regulation of host mechanisms by microbial exRNAs as inter-kingdom communication, efforts to utilize extracellular vesicles as therapeutic vehicles loaded with engineered RNA cargos, and a putative connection between microbial exRNA-based regulation of host responses and human diseases such as Alzheimer’s. This overview aims to present novel insights into pathogenesis with regard to the function of microbial exRNAs as “disease-relevant travelers.”
Impact statement
The number of commensal bacteria in the body surpasses the number of actual human cells. Thus, various interactions between microbes and human cells constitute an inevitable phenomenon. Recent evidence has led to bacterial extracellular RNAs (exRNAs) being proposed as good candidates for microbe–host inter-kingdom communication tools as they can modulate the expression of host genes. However, research findings on the relevance of interactions between extracellular RNA and human diseases are still in their infancy. Nevertheless, substantial data suggest that microbial exRNAs are implicated in various human diseases both at local and distant sites. By exploring various scenarios for the involvement of microbial exRNAs in human diseases, we may better understand the role of exRNAs as “communication signals” for diseases and thereby develop novel therapeutic strategies by using them and their carrier extracellular vesicles.
Keywords: Extracellular vesicle, exosome, extracellular RNA, microRNA, small RNA, systemic disease, inter-kingdom communication
Introduction
Extracellular vesicles (EVs) are a group of nano-sized membranous vesicles (typically 40–1000 nm) that are released from both prokaryotic and eukaryotic cells into the extracellular environment.1,2 EV, the current generic term of choice, refers collectively to numerous types of vesicles such as exosomes, ectosomes, apoptotic bodies, bacterial outer membrane vesicles (OMVs); EVs have various cells of origin and show a range of functions.3,4 EVs can transfer numerous types of cargo, including proteins, lipids, DNA, and RNA to distant cells. EVs can internalize in proximal cells via various routes, such as endocytosis, lipid rafts, and/or membrane fusion.5,6 Among various EV cargos, RNAs have gained special attention because RNAs in EVs (extracellular RNAs; exRNAs) are mostly composed of mRNAs, microRNAs (miRNAs), or other small RNAs (sRNAs) that are usually transported by EVs to other cells without losing their biological activities.7 In particular, the biogenesis of sRNAs, which possess regulatory functions, is differentially induced under physiological or environmental stressful conditions such as changes of temperature, osmotic, and redox contions.8,9 Interestingly, such conditions have also been observed in the context of the production of EVs.10 It is thus probable that the release of miRNAs, sRNAs, and misfolded proteins produced under stressful conditions via EVs may alleviate the effects of harmful stress in microbial cells.11
Recently, considerable effort has been devoted to understanding how certain exRNAs are sorted into EVs and how they function in target cells. It seems that selective loading of exRNAs into eukaryotic EVs (i.e. exosomes) is accomplished through special mechanisms. Sequencing motifs present in sRNAs are thought to control sRNA sorting into exosomes in a process mediated by specific enzymes including SYNCRIP (synaptotagmin-binding cytoplasmic RNA-interacting protein; also known as hnRNP-Q or NSAP1) and hnRNPA2B1 (heterogeneous nuclear ribonucleoprotein A2B1); other sorting-involved enzymes continue to be identified.2,12,13 These findings support the idea that specific motifs of sRNAs (or miRNAs) and their interacting proteins ensure that the exosomes carry only a specific subset of RNAs as their cargo, suggesting that EV sRNAs may function as specialized intercellular messenger molecules. Although sorting mechanisms of bacterial RNAs into EVs have not been well studied, exRNAs originating from microbes can bind to host RNA-induced silencing complex (RISC), implying that microbial exRNAs may behave as host gene regulators.2,7
It is widely known that endogenous exRNAs can be used as prognostic biomarkers for disease states and progression.14 However, host gene regulation via microbial EVs and exRNAs has just started to attract wide scrutiny.2 It is well known that endotoxins, exotoxins, and bacterial metabolites released by human commensal microbes participate in systemic diseases by inducing a variety of host immune responses.15,16 Indeed, the number of commensal bacteria is estimated to be higher than the number of human cells,17 and the residential microbes and their byproducts inevitably influence the human body.
Over the past several years, my group has continuously reported miRNA-sized bacterial and EV sRNAs from various human-resident bacteria.18–21 These findings have given us a view of microbial miRNA involvement in host gene regulation or inhibition. Although the field is still in its infancy, this review aims to introduce recent studies on microbial exRNAs and suggest their possible relevance to human systemic diseases.
Purification and analysis of microbial exRNAs
The highest levels of bacterial EVs are produced at the late growth stages of bacteria, when debris such as membrane components and cytosolic proteins from dead cells are most abundant.22 To avoid contamination, it is thus critical to separate intact bacteria and their components during isolation of OMVs, usually by ultracentrifugation or precipitation. Additional purification techniques currently used are density gradient centrifugation, gel filtration, and commercial kits.11,23 The International Society for Extracellular Vesicles (ISEV) provides guidelines for the isolation of EVs, essentially suggested standard methods for researchers.4 After RNase treatment to remove free RNAs that might originate from dead cells, subsequent steps to extract RNA from EVs are necessary for RNA sequencing (RNA-seq) or other experiments requiring complete exclusion of free RNA effects. Among various RNA species, sRNAs are the most common in EVs,24 and small RNA-enrichment purification methods can be used for small regulatory sRNAs.25
Bacterial exRNAs can be sequenced using RNA-seq techniques by first isolating the RNA molecules from EVs.26 To understand exRNA functions in host-pathogen interactions, dual RNA-seq in OMV-infected host cells or tissues has been introduced. This RNA-seq technique allows simultaneous profiling of RNAs of host and bacterial origins from an infected cell.27
This technique allows analysis of intracellular bacteria such as Mycobacterium tuberculosis and Haemophilus influenza after they have infected their host cells in humans.28,29 In addition, some exRNAs have been revealed to have immunological effects after infection of human macrophage-like cells with periodontopathogenic EVs carrying these exRNAs.23 These studies shed light on the microbe–host interactome at a transcriptional level.30 In addition, exRNAs with regulatory functions similar to those of miRNAs in the host cells need to be incorporated into host gene regulatory machinery, such as RISC. Thus, RIP-seq (RNA-immunoprecipitation-sequencing) is a very useful technique for identifying regulatory exRNAs.31
Future microenvironmental microbiome research into the effects of total microbial EVs on host tissues should utilize multi-RNA-seq in conjunction with massive bioinformatics analysis; this will likely shed light on communication mechanisms between microbes and host.32 In addition, the development of new approaches, including nano-flow cytometry, which has been used to analyze EVs from cancer cells,33 is expected to aid in characterizing these highly heterogeneous microbial EVs.
Microbial exRNAs as disease biomarkers
The finding that endogenous exRNAs function as communication molecules has led to interest in exRNAs as a source of disease biomarkers and therapeutic agents.34 The first phase of the NIH-supported Extracellular RNA Communication Consortium (ERCC1), launched in 2013 to accelerate the study of exRNA biology, focuses on exRNA/EV biogenesis and function as well as the discovery of exRNA disease-biomarkers.35 While most ERCC1 studies have investigated endogenous circulating miRNAs,35,36 microbial exRNAs and their pathogenic roles are poorly understood. However, we now have credible evidence that microbial exRNAs also circulate in the human body, RNA-seq analysis having shown that high levels of bacterial RNA fragments exist in human biofluids,37 although the link between microbial exRNAs and any human disease has not been completely proven. Interestingly, small RNA-seq and bioinformatic analysis of human saliva revealed high abundance of bacterial exRNAs.38,39 Since identification of biomarkers in saliva is relatively easy compared to other invasive approaches, bacterial exRNAs in saliva hold considerable promise for the development of various infectious disease biomarkers. To this end, host salivary exRNA biomarkers were first identified from gingivitis patients; nonetheless, the role of bacterial salivary exRNAs in this disease has not been elucidated.40 Therefore, exRNAs of oral pathogens could be good candidates for disease biomarkers. Streptococcus sanguinis, recognized as a primary agent of early dental caries, contains a number of unique sRNAs in its EVs.18 Similarly, oral periodontal pathogens responsible for peritonitis, including Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, and Treponema denticola, have also been found to contain small miRNA-sized exRNAs in their EVs.19
Other than oral pathogens, a study of fungi that cause paracoccidioidomycosis has revealed the presence of differential levels of sRNA classes in the EVs of fungal isolates, which the authors suspected were responsible for the variable virulence among fungi.41 Moreover, EVs released from human parasitic nematodes harbor miRNAs that are well conserved between the parasite and host. These EVs can be spontaneously internalized by human macrophages, allowing the cargo miRNAs to modulate host gene expression.42
Novel disease biomarkers or therapeutic strategies can be developed by utilizing these exRNA microbial pathogens in the context of their associated diseases.
Microbial exRNAs, host gene regulation, and immune responses
The recent realization that exRNAs enable inter-kingdom or inter-species communication arose when they were found to modulate gene regulation in multiple cell types and diverse species (discussed later in this section; for detailed species and host tissues, see reviews2,7,43). Even though the precise mechanisms have not been elucidated, EVs from both Gram-positive18,44 and Gram-negative bacteria45–48 as well as internal exRNAs have been observed to invade host cells, suggesting their possible function as communication molecules that influence host cell functions. These findings are not limited to bacteria, fungal exRNAs having also been observed to be exported via EVs.49,50
A few years back, indications emerged of host gene regulation mechanisms of sRNAs in intracellular bacteria. Salmonella enterica sRNA was shown to regulate a host gene involved in the signal transduction pathway and Mycobacterium marinum sRNAs were shown to bind the host RISC that can regulate host genes.27,51 Therefore, microbial exRNAs may plausibly be assumed to have similar mechanisms.
Koeppen et al.52 have shown that Pseudomonas aeruginosa EVs have abundant RNA that can downregulate the EV-induced proinflammatory cytokine IL-8 secretion after being transferred into human epithelial cells. This indicates that the small exRNA inhibits innate immune responses by targeting the mitogen-activated protein kinase (MAPK) pathways, which lie upstream of IL-8.52 This exRNA might thus protect pathogens from human defense mechanisms and enhance pathogen penetration into host.
Similarly, my group has reported that three highly expressed assorted exogenous exRNAs of periodontopathogens decrease the secretion of certain cytokines (IL-5, IL-13, and IL-15) in a T cell line in vitro,19 suggesting that periodontal exRNAs may assist in evading host adaptive immune responses.
Microbial exRNAs may thus function in RNA inhibition (RNAi) even in host cells, particularly with respect to immune-related genes. In order to do so as miRNAs in host cells, exRNAs need to be loaded onto host RISC, thus regulating host target transcripts.23 The exRNAs of the oral bacterium A. actinomycetemcomitans can be internalized by human macrophages, exRNAs of A. actinomycetemcomitans EV-origin having been identified by RIP-seq analysis.23 Similarly, small RNAs originating from the intracellular bacterial pathogen M. marinum show characteristics of eukaryotic miRNAs that are able to bind to RISC and repress a target mRNA.51
Although we do not know the exact mechanisms underlying the selective loading of microbial exRNAs into EVs or whether such loading is a random event, it has been reported that EVs of Borrelia burgdorferi, a causative bacterium of Lyme disease, contain more enriched RNA transcripts from plasmids than bacterial cells, suggesting these specially packaged plasmid-encoded exRNA in EVs might be involved in the development and pathogenicity of the disease.53 Moreover, growth stage of bacteria could also affect the composition and selection of cargo into EVs.54 Furthermore, microbial sRNAs seem related to EV generation. Enterobacterial sRNA, MicA showed promoted production of bacterial EVs and MicA-induced EVs activated cytokines (IFN-γ and IL-17) in T cells, although it is not clear whether the effect is driven by exRNAs.55
These studies indicate that host gene regulation by small RNAs originating from microbes is a novel pathogenic mechanism, and that the transfer of microbial exRNAs to host cells represents an additional example of microbe–host interaction.
Engineering of microbial EVs to deliver therapeutic exRNA cargos
The ability of EVs to transfer cargo also makes it possible to deliver therapeutic molecules, including RNAs, into target cells. Engineering endogenous EVs as carriers of therapeutic RNA cargos to avoid immune rejection or RNase degradation in the host patient would constitute a new modality for RNA therapeutics.1,56 Eukaryotic exosome-mediated transfer of miRNA or miRNA inhibitors has been tested for use in regulating target cells.57,58 However, safety concerns arise since bacterial EVs contain high levels of endotoxins (or lipopolysaccharides; LPS) and other pathogenic materials that can induce various immune responses by the host59; these challenges may limit the therapeutic application of such EVs.
A tumor therapy study used siRNA (small interfering RNA)-loaded Escherichia coli EVs derived from an endotoxin-reduced mutant strain expressing a cancer-specific targeting ligand.60 Injection of the siRNA-containing EVs significantly reduced tumor size in the mouse model by successfully targeting the kinesin spindle protein (KSP) gene, which is related to tumor proliferation.60 My group just recently reported that EVs from bacteria can cross the blood–brain barrier (BBB), implying that bacterial EVs can even be applied to brain diseases, including brain tumors and neuroinflammatory disorders.23 Nonetheless, challenges remain regarding issues such as efficient and specific delivery into target tissues or cells as well as evading degradation in the circulation.56
exRNAs in periodontal pathogens and Alzheimer’s disease
Although systemic invasive activities of EVs across various types of human cells and tissues have been shown, it was until recently unclear whether bacterial EVs could cross the BBB. It remains debatable whether oral microbes can cross the BBB and colonize in the brain. However, P. gingivalis has recently been identified in the brain of Alzheimer’s patients.61 Moreover, Candida albicans (oral commensal yeast) has been detected in a mouse brain that accumulated amyloid beta upon intravenous injection of yeast cells.62 These findings strongly suggest the relevance of oral microbes in Alzheimer’s disease. Since we now have evidence that microbial EVs can cross the BBB along with cargos including RNA,23 many bacterial EVs and their cargo materials may easily reach the brain when there is a wound or bleeding, such as in periodontitis, which is characterized by the degradation of soft connective tissue and alveolar bone, ultimately resulting in bleeding and tooth loss.63 The possible role of periodontal pathogens in Alzheimer’s disease has recently been asserted.64,65 Among the main bacteria implicated in the pathology of periodontal disease, A. actinomycetemcomitans (mentioned earlier in this review) exRNAs of EV (rather than LPS or protein cargos) upregulate the proinflammatory cytokine TNF-α in the mouse brain.23 The function of pathogenic exRNAs is thus not limited to regional effects but can reach any site in the whole body, even the brain, and induce various inflammatory diseases such as Alzheimer’s.
Conclusion and perspectives
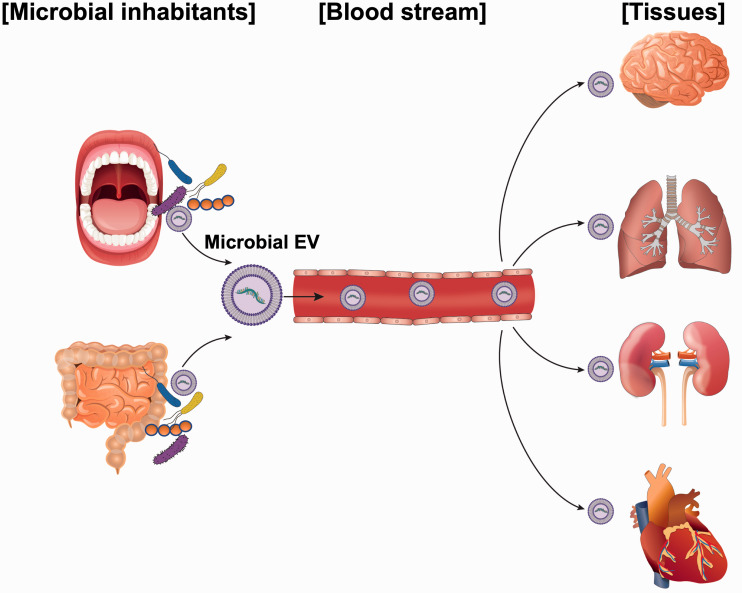
Microbe-to-human communication via functional exRNAs loaded in EVs can be a novel pathogenic mechanism. The ability of EVs to circulate through the bloodstream and reach any place in the human body implies that EVs and their components are relevant to human diseases (Figure 1). Among EV components, small regulatory RNAs or exRNAs have drawn interest in terms of their host effects given the increased awareness of their regulatory functions in the cell. This paper thus aims to introduce relatively new biological findings that might deepen our understanding of microbial exRNAs and extend our knowledge of disease pathogenesis.
Figure 1.
Graphic representation of the involvement of human commensal microbial exRNAs in systemic diseases. Extracellular RNAs (exRNAs) secreted from regional human commensal bacteria via extracellular vesicles (EVs) can circulate through the bloodstream and reach distant organs, even crossing the blood–brain barrier, then get taken up by target cells. Imported exRNAs, along with EVs, may induce immune response or regulate host gene expression, acting on behalf of microbes to induce host signaling biomolecules. (A color version of this figure is available in the online journal.)
In this review, mostly negative effects of microbial exRNAs on human body are postulated. However, as microbes in the human body always interact with host cells for the body’s physiological homeostasis, positive effects of microbial exRNAs should not be ruled out. Many questions, such as how many EVs and exRNAs are secreted in the human body and how many exRNAs become functional in target cells after integration remain unanswered. Additionally, the potential effects of inter-microbial communication through exRNAs and the way in which exRNAs influence the total microbiome flora should be further explored.
Overall, expanded knowledge of exRNAs in inter-kingdom communication will enable us to elucidate new pathogenic determinants and mechanisms, thus enabling the development of novel therapeutic solutions.
Authors’ contributions
Heon-Jin Lee solely wrote the manuscript and prepared the figure.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Korean Government (2017R1A5A2015391 and 2018R1D1A3B07043539).
ORCID iD
Heon-Jin Lee https://orcid.org/0000-0002-1911-5014
References
- 1.Choi J-W, Um J-H, Cho J-H, Lee H-J. Tiny RNAs and their voyage via extracellular vesicles: secretion of bacterial small RNA and eukaryotic microRNA. Exp Biol Med 2017; 242:1475–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee H-J. Microbe-host communication by small RNAs in extracellular vesicles: vehicles for transkingdom RNA transportation. Int J Mol Sci 2019; 20:1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014; 30:255–89 [DOI] [PubMed] [Google Scholar]
- 4.Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Théry C. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the international society for extracellular vesicles. J Extracell Vesicles 2014; 3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donoghue EJ, Krachler AM. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol 2016; 18:1508–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulcahy LA, Pink RC, Carter D. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014; 3:24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsatsaronis JA, Franch-Arroyo S, Resch U, Charpentier E. Extracellular vesicle RNA: a universal mediator of microbial communication?. Trends Microbiol 2018; 26:401–10 [DOI] [PubMed] [Google Scholar]
- 8.Wagner EGH, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv Genet 2002; 46:361–98 [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol 2004; 58:303–28 [DOI] [PubMed] [Google Scholar]
- 10.Macdonald IA, Kuehn MJ. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J Bacteriol 2013; 195:2971–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klimentová J, Stulík J. Methods of isolation and purification of outer membrane vesicles from Gram-negative bacteria. Microbiol Res 2015; 170:1–9 [DOI] [PubMed] [Google Scholar]
- 12.Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep 2016; 17:799–808 [DOI] [PubMed] [Google Scholar]
- 13.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 2013; 4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huleihel L, Scarritt ME, Badylak SF. The influence of extracellular RNA on cell behavior in health, disease and regeneration. Curr Pathobiol Rep 2017; 5:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yumoto H, Hirota K, Hirao K, Ninomiya M, Murakami K, Fujii H, Miyake Y. The pathogenic factors from oral streptococci for systemic diseases. Int J Mol Sci 2019; 20:E4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XV, Leonardi I, Iliev ID. Gut mycobiota in immunity and inflammatory disease. Immunity 2019; 50:1365–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016; 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J-W, Kwon T-Y, Hong S-H, Lee H-J. Isolation and characterization of a microRNA-size secretable small RNA in Streptococcus sanguinis. Cell Biochem Biophys 2018; 76:293–301 [DOI] [PubMed] [Google Scholar]
- 19.Choi J-W, Kim SC, Hong S-H, Lee HJ. Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J Dent Res 2017; 96:458–66 [DOI] [PubMed] [Google Scholar]
- 20.Kang S-M, Choi J-W, Lee Y, Hong S-H, Lee H-J. Identification of microRNA-size, small RNAs in Escherichia coli. Curr Microbiol 2013; 67:609–13 [DOI] [PubMed] [Google Scholar]
- 21.Lee H-J, Hong S-H. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol Lett 2012; 326:131–6 [DOI] [PubMed] [Google Scholar]
- 22.Jan AT. Outer membrane vesicles (OMVs) of gram-negative bacteria: a perspective update. Front Microbiol 2017; 8:1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han E-C, Choi S-Y, Lee Y, Park J-W, Hong S-H, Lee H-J. Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice. FASEB J 2019; 33:13412–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosal A, Upadhyaya BB, Fritz JV, Heintz-Buschart A, Desai MS, Yusuf D, Huang D, Baumuratov A, Wang K, Galas D, Wilmes P. The extracellular RNA complement of Escherichia coli. Microbiol Open 2015; 4:252–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi C, Yoon S, Moon H, Bae Y-U, Kim C-B, Diskul-Na-Ayudthaya P, Ngu TV, Munir J, Han J, Park S-B, Moon J-S, Song S, Ryu S. mirRICH, a simple method to enrich the small RNA fraction from over-dried RNA pellets. RNA Biol 2018; 15:763–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habier J, May P, Heintz-Buschart A, Ghosal A, Wienecke-Baldacchino AK, Nolte-’t Hoen ENM, Wilmes P, Fritz JV. Extraction and analysis of RNA isolated from pure bacteria-derived outer membrane vesicles. Methods Mol Biol 2018; 1737:213–30 [DOI] [PubMed] [Google Scholar]
- 27.Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 2016; 529:496–501 [DOI] [PubMed] [Google Scholar]
- 28.Rienksma RA, Suarez-Diez M, Mollenkopf H-J, Dolganov GM, Dorhoi A, Schoolnik GK, Martins Dos Santos VA, Kaufmann SH, Schaap PJ, Gengenbacher M. Comprehensive insights into transcriptional adaptation of intracellular mycobacteria by microbe-enriched dual RNA sequencing. BMC Genom 2015; 16:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baddal B, Muzzi A, Censini S, Calogero RA, Torricelli G, Guidotti S, Taddei AR, Covacci A, Pizza M, Rappuoli R, Soriani M, Pezzicoli A. Dual RNA-seq of nontypeable Haemophilus influenzae and host cell transcriptomes reveals novel insights into host-pathogen cross talk. mBio 2015; 6:e01765–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griesenauer B, Tran TM, Fortney KR, Janowicz DM, Johnson P, Gao H, Barnes S, Wilson LS, Liu Y, Spinola SM. Determination of an interaction network between an extracellular bacterial pathogen and the human host. mBio 2019; 10:e01193–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu Y, Corey DR. RNA sequencing: platform selection, experimental design, and data interpretation. Nucleic Acid Ther 2012; 22:271–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westermann AJ, Barquist L, Vogel J. Resolving host-pathogen interactions by dual RNA-seq. PLoS Pathog 2017; 13:e1006033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi D, Montermini L, Jeong H, Sharma S, Meehan B, Rak J. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano 2019; 13:10499–511 [DOI] [PubMed] [Google Scholar]
- 34.Das S, Extracellular RNA, Communication Consortium Ansel KM, Bitzer M, Breakefield XO, Charest A, Galas DJ, Gerstein MB, Gupta M, Milosavljevic A, McManus MT, Patel T, Raffai RL, Rozowsky J, Roth ME, Saugstad JA, Van Keuren-Jensen K, Weaver AM, Laurent LC. The extracellular RNA communication consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell 2019; 177:231–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ainsztein AM, Brooks PJ, Dugan VG, Ganguly A, Guo M, Howcroft TK, Kelley CA, Kuo LS, Labosky PA, Lenzi R, McKie GA, Mohla S, Procaccini D, Reilly M, Satterlee JS, Srinivas PR, Church ES, Sutherland M, Tagle DA, Tucker JM, Venkatachalam S. The NIH extracellular RNA communication consortium. J Extracell Vesicles 2015; 4:27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinn JF, Patel T, Wong D, Das S, Freedman JE, Laurent LC, Carter BS, Hochberg F, Van Keuren-Jensen K, Huentelman M, Spetzler R, Kalani MY, Arango J, Adelson PD, Weiner HL, Gandhi R, Goilav B, Putterman C, Saugstad JA. Extracellular RNAs: development as biomarkers of human disease. J Extracell Vesicles 2015; 4:27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeri A, Courtright A, Reiman R, Carlson E, Beecroft T, Janss A, Siniard A, Richholt R, Balak C, Rozowsky J, Kitchen R, Hutchins E, Winarta J, McCoy R, Anastasi M, Kim S, Huentelman M, Van Keuren-Jensen K. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci Rep 2017; 7:44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F, Kaczor-Urbanowicz KE, Sun J, Majem B, Lo H-C, Kim Y, Koyano K, Rao SL, Kang SY, Kim SM, Kim KM, Kim S, Chia D, Elashoff D, Grogan TR, Xiao X, Wong D. Characterization of human salivary extracellular RNA by next-generation sequencing. Clin Chem 2018; 64:1085–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaczor-Urbanowicz KE, Kim Y, Li F, Galeev T, Kitchen RR, Gerstein M, Koyano K, Jeong SH, Wang X, Elashoff D, Kang SY, Kim S-M, Kim K, Kim S, Chia D, Xiao X, Rozowsky J, Wong D. Novel approaches for bioinformatic analysis of salivary RNA sequencing data for development. Bioinformatics 2018; 34:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaczor-Urbanowicz KE, Trivedi HM, Lima PO, Camargo PM, Giannobile WV, Grogan TR, Gleber-Netto FO, Whiteman Y, Li F, Lee H-J, Dharia K, Aro K, Martin Carreras-Presas C, Amuthan S, Vartak M, Akin D, Al-Adbullah H, Bembey K, Klokkevold PR, Elashoff D, Barnes VM, Richter R, DeVizio W, Masters JG, Wong D. Salivary exRNA biomarkers to detect gingivitis and monitor disease regression. J Clin Periodontol 2018; 45:806–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peres da Silva R, Longo LGV, Cunha J, Sobreira TJP, Rodrigues ML, Faoro H, Goldenberg S, Alves LR, Puccia R. Comparison of the RNA content of extracellular vesicles derived from Paracoccidioides brasiliensis and Paracoccidioides lutzii. Cells 2019; 8:e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamanian M, Fraser LM, Agbedanu PN, Harischandra H, Moorhead AR, Day TA, Bartholomay LC, Kimber MJ. Release of small RNA-containing exosome-like vesicles from the human filarial parasite Brugia malayi. PLoS Negl Trop Dis 2015; 9:e0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Celluzzi A, Masotti A. How our other genome controls our epi-genome. Trends Microbiol 2016; 24:777–87 [DOI] [PubMed] [Google Scholar]
- 44.Resch U, Tsatsaronis JA, Le Rhun A, Stübiger G, Rohde M, Kasvandik S, Holzmeister S, Tinnefeld P, Wai SN, Charpentier E. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group a streptococcus. mBio 2016; 7:e00267–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blenkiron C, Simonov D, Muthukaruppan A, Tsai P, Dauros P, Green S, Hong J, Print CG, Swift S, Phillips AR. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS One 2016; 11:e0160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho M-H, Chen C-H, Goodwin JS, Wang B-Y, Xie H. Functional advantages of Porphyromonas gingivalis vesicles. PLoS One 2015; 10:e0123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. Bacterial vesicles in marine ecosystems. Science 2014; 343:183–6 [DOI] [PubMed] [Google Scholar]
- 48.Sjöström AE, Sandblad L, Uhlin BE, Wai SN. Membrane vesicle-mediated release of bacterial RNA. Sci Rep 2015; 5:15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peres da Silva R, Puccia R, Rodrigues ML, Oliveira DL, Joffe LS, César GV, Nimrichter L, Goldenberg S, Alves LR. Extracellular vesicle-mediated export of fungal RNA. Sci Rep 2015; 5:7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rayner S, Bruhn S, Vallhov H, Andersson A, Billmyre RB, Scheynius A. Identification of small RNAs in extracellular vesicles from the commensal yeast Malassezia sympodialis. Sci Rep 2017; 7:39742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuse Y, Finethy R, Saka HA, Xet-Mull AM, Sisk DM, Smith KL, Lee S, Coers J, Valdivia RH, Tobin DM, Cullen BR. Search for microRNAs expressed by intracellular bacterial pathogens in infected mammalian cells. PLoS One 2014; 9:e10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, Demers EG, Dolben EL, Hammond JH, Hogan DA, Stanton BA. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog 2016; 12:e1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malge A, Ghai V, Reddy PJ, Baxter D, Kim TK, Moritz RL, Wang K. mRNA transcript distribution bias between Borrelia burgdorferi bacteria and their outer membrane vesicles. FEMS Microbiol Lett 2018; 365:fny135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zavan L, Bitto NJ, Johnston EL, Greening DW, Kaparakis-Liaskos M. Helicobacter pylori growth stage determines the size, protein composition, and preferential cargo packaging of outer membrane vesicles. Proteomics 2019; 19:e1800209. [DOI] [PubMed] [Google Scholar]
- 55.Choi H-I, Kim M, Jeon J, Han JK, Kim K-S. Overexpression of MicA induces production of OmpC-enriched outer membrane vesicles that protect against Salmonella challenge. Biochem Biophys Res Commun 2017; 490:991–6 [DOI] [PubMed] [Google Scholar]
- 56.Melling GE, Carollo E, Conlon R, Simpson JC, Carter D. The challenges and possibilities of extracellular vesicles as therapeutic vehicles. Eur J Pharm Biopharm 2019; 144:50–6 [DOI] [PubMed] [Google Scholar]
- 57.Choi S-Y, Han E-C, Hong S-H, Kwon T-G, Lee Y, Lee H-J. Regulating osteogenic differentiation by suppression of exosomal microRNAs. Tissue Eng Part A 2019; 25:1146–54 [DOI] [PubMed] [Google Scholar]
- 58.Momen-Heravi F, Bala S, Bukong T, Szabo G. Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine 2014; 10:1517–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 2015; 13:605–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gujrati V, Kim S, Kim S-H, Min JJ, Choy HE, Kim SC, Jon S. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano 2014; 8:1525–37 [DOI] [PubMed] [Google Scholar]
- 61.Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, Nguyen M, Haditsch U, Raha D, Griffin C, Holsinger LJ, Arastu-Kapur S, Kaba S, Lee A, Ryder MI, Potempa B, Mydel P, Hellvard A, Adamowicz K, Hasturk H, Walker GD, Reynolds EC, Faull RLM, Curtis MA, Dragunow M, Potempa J. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv 2019; 5:eaau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y, Du S, Johnson JL, Tung H-Y, Landers CT, Liu Y, Seman BG, Wheeler RT, Costa-Mattioli M, Kheradmand F, Zheng H, Corry DB. Microglia and amyloid precursor protein coordinate control of transient Candida cerebritis with memory deficits. Nat Commun 2019; 10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishihara T, Koseki T. Microbial etiology of periodontitis. Periodontol 2000 2004; 36:14–26 [DOI] [PubMed] [Google Scholar]
- 64.Pritchard AB, Crean S, Olsen I, Singhrao SK. Periodontitis, microbiomes and their role in Alzheimer’s disease. Front Aging Neurosci 2017; 9:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaur S, Agnihotri R. Alzheimer’s disease and chronic periodontitis: is there an association? Geriatr Gerontol Int 2015; 15:391–404 [DOI] [PubMed] [Google Scholar]