Mycoplasma bovis causes pneumonia, pharyngitis, otitis, arthritis, mastitis, and reproductive disorders in cattle and bison. Two multilocus sequence typing (MLST) schemes have been developed for M. bovis, with one serving as the PubMLST reference method, but no comparison of the schemes has been undertaken. Although the PubMLST scheme has proven to be highly discriminatory and informative, the recent discovery of isolates missing one of the typing loci, adh-1, raises concern about its suitability for continued use.
KEYWORDS: MLST, Mycoplasma bovis
ABSTRACT
Mycoplasma bovis causes pneumonia, pharyngitis, otitis, arthritis, mastitis, and reproductive disorders in cattle and bison. Two multilocus sequence typing (MLST) schemes have been developed for M. bovis, with one serving as the PubMLST reference method, but no comparison of the schemes has been undertaken. Although the PubMLST scheme has proven to be highly discriminatory and informative, the recent discovery of isolates missing one of the typing loci, adh-1, raises concern about its suitability for continued use. The goal of our study was to compare the performance of the two MLST schemes and identify a new reference scheme capable of fully typing all isolates. We evaluated 448 isolates from diverse geographic and anatomic sites that collectively represent cattle, bison, deer, and a goat. The discrimination indexes (DIs) for the PubMLST and the alternative scheme are 0.909 (91 sequence types [STs]) and 0.842 (77 STs), respectively. Although the PubMLST scheme outperformed the alternative scheme, the adh-1 locus must be retired from the PubMLST scheme if it is to be retained as a reference method. The DI obtained using the six remaining PubMLST loci (0.897, 79 STs) fails to reach the benchmark recommended for a reference method (0.900), mandating the addition of a seventh locus. Comparative analysis of genome sequences from the isolates used here identified the dnaA locus from the alternative scheme as the optimal replacement for adh-1. This revised scheme, which will be implemented as the new PubMLST reference method, has a DI of 0.914 and distinguishes 88 STs from the 448 isolates evaluated.
INTRODUCTION
Mycoplasma bovis is a causative agent of pneumonia, otitis media, arthritis, mastitis and reproductive disorders in cattle and is frequently identified from cases of the polymicrobial syndrome known as bovine respiratory disease complex (1–3). First isolated in the 1960s from a severe outbreak of mastitis in an American dairy herd (4), it has since spread to nearly all countries of the world and has a significant negative impact on cattle health and production (5, 6). Beginning in the early 2000s, disease associated with M. bovis also began to appear in both ranched and free-ranging North American bison. M. bovis is now recognized to be a primary cause of pneumonia, arthritis, necrotic pharyngitis, pleuritis, dystocia and, abortion in these animals, often with extremely high morbidity and mortality (7, 8). The bacterium has additionally been associated with otitis and/or respiratory disease in white-tailed deer and mule deer (9, 10).
The development of measures to more effectively control and manage M. bovis-related disease is hampered by a lack of understanding of the population structure and molecular epidemiology of the bacterium (11). Of the various typing techniques used to characterize bacterial pathogens, multilocus sequence typing (MLST) is often the method of choice. MLST is highly standardized, accurate, robust and scalable, and data are easily shared and compared within and between laboratories (12). Two different MLST schemes have been reported for M. bovis (13, 14), developed in different laboratories at nearly the same time, one of which serves as the current PubMLST reference method (13; https://pubmlst.org/mbovis/). Although the discrimination indices (DI) reported for each method are nearly identical, 0.922 (15) and 0.910 (14), a direct comparison of those values is not informative since they were obtained using different sets of isolates sourced from largely nonoverlapping geographic regions. The PubMLST scheme has proven to be highly informative and discriminatory, but modification of the scheme is necessary due to the recent identification of isolates missing one of the typing loci, adh-1 (10, 16). The goal of this study was to compare the performance of the two MLST schemes using a common set of diverse isolates and to identify a revised reference scheme capable of fully typing all isolates.
MATERIALS AND METHODS
Isolates.
A total of 448 M. bovis isolates obtained from 405 animals were included in this study, with 410 from cattle, 35 from bison, and 1 each from a white-tailed deer, a mule deer, and a goat. Table 1 summarizes the geographic and anatomic origins of the isolates and the clinical status of the source animals at the time samples were collected for culture. These isolates collectively represent North America (n = 241), Europe (n = 36), Asia (n = 48), the Middle East (n = 110), Australia (n = 12), and Africa (n = 1). Most were obtained from animals displaying clinical signs consistent with mycoplasmosis, while 34 were cultured from the nasal cavity, lung, or semen of healthy animals. Dates of isolation range from 1961 to 2017, with 29% of isolates (130/448) acquired over the years 2000 to 2009 and roughly 69% (310/448) obtained in 2010 or more recently. Multiple isolates from the same animal were included only if derived from different anatomic locations or when at least one of the two MLST methods used revealed more than a single sequence type (ST), so as to fully capture the genetic diversity present among available isolates. Additional information pertaining to individual isolates is available in Table S1 in the supplemental material.
TABLE 1.
Host, geographic, and anatomic origins and clinical status of animals from which M. bovis isolates were obtained
| Host (no.) | Geographic origin (no.) | Anatomic origin | Clinical status of source animal (no.) |
|---|---|---|---|
| Bison (35) | United States (19) | Nasal cavity | Healthy (8) |
| Lung | Pneumonia (7), lameness (1), unknown (1) | ||
| Pharynx | Pharyngitis (1) | ||
| Uterus | Unknown (1) | ||
| Canada (16) | Nasal cavity | Healthy (2) | |
| Pharynx | Pharyngitis (1) | ||
| Lunga | Pneumonia (8), abortion (1), septicemia unrelated to M. bovis (1) | ||
| Joint | Pneumonia (1), pneumonia and arthritis (1) | ||
| Placenta | Pneumonia and abortion (1) | ||
| Bovine (410) | United States (51) | Nasal cavity | Healthy (1), pneumonia (1), unknown (20) |
| Lung | Pneumonia (7), pneumonia and arthritis (1) | ||
| Milk | Mastitis (21) | ||
| Canada (153) | Nasal cavity | Healthy (16), pneumonia (6), pneumonia and arthritis (1) | |
| Ear | Otitis (1) | ||
| Lung | Healthy (2), pneumonia (34), pneumonia and arthritis (47) | ||
| Joint | Arthritis (4), pneumonia and arthritis (40) | ||
| Lung or jointb | Pneumonia and arthritis (2) | ||
| France (1) | Unknown | Unknown (1) | |
| Switzerland (3) | Lung | Pneumonia (1), unknown (1) | |
| Milk | Mastitis and pneumonia (1) | ||
| Hungary (15) | Lung | Pneumonia (15) | |
| Lithuania (9) | Lung | Pneumonia (9) | |
| Romania (5) | Lung | Pneumonia (4) | |
| Milk | Mastitis (1) | ||
| Russia (3) | Milk | Mastitis (3) | |
| China (47) | Larynx | Unknown (1) | |
| Lung | Pneumonia (4), pneumonia and arthritis (1), unknown (35) | ||
| Joint | Pneumonia and arthritis (1), unknown (1) | ||
| Milk | Unknown (1) | ||
| Unknown | Unknown (3) | ||
| Japan (1) | Nasal cavity | Unknown (1) | |
| Israel (110) | Nasal cavity | Healthy (1) | |
| Eye | Conjunctivitis (1) | ||
| Larynx | Pneumonia (1) | ||
| Lung | Pneumonia (20) | ||
| Milk | Mastitis (80) | ||
| Fetal tissue | Abortion (1) | ||
| Vulva | Vulvitis (2) | ||
| Semen | Healthy (4) | ||
| Australia (12) | Larynx | Pneumonia (1) | |
| Pharynx | Pneumonia (2) | ||
| Lung | Pneumonia (6) | ||
| Joint | Arthritis (3) | ||
| White-tailed deer (1) | United States (1) | Lung | Pneumonia (1) |
| Mule deer (1) | United States (1) | Lung | Pneumonia and otitis (1) |
| Goat (1) | Ethiopia (1) | Unknown | Unknown (1) |
Includes one lung from a fetus aborted due to M. bovis infection.
Records regarding anatomic source are unclear.
MLST schemes.
All isolates were assigned allelic profiles and STs according to the scheme currently used as the PubMLST reference method, based on partial sequences ranging in size from 306 to 570 bp, from the seven housekeeping genes adh-1, gltX, gpsA, gyrB, pta-2, tdk, and tkt (13; https://pubmlst.org/mbovis/). STs for 101 cattle isolates, 31 bison isolates, and the white-tailed deer isolate were reported in prior studies (10, 13, 17; indicated by bold font in Table S1). STs for all remaining isolates were assigned on the basis of output from either the PubMLST automated sequence tag-scanning function (for data derived from genome assemblies) or from sequence queries of the PubMLST Mycoplasma bovis Locus/Sequence Definitions Database (https://pubmlst.org/bigsdb?db=pubmlst_mbovis_seqdef) for data obtained from sequencing PCR amplicons.
Isolates were additionally typed on the basis of target sequences employed in the alternative MLST method of Rosales et al. (14), here designated the Weybridge scheme, based on output from BLAST queries (for genome assemblies) or alignment of target sequences with a locally maintained database of known alleles (for data from PCR amplicon sequencing). Targets include partial sequences ranging in size from 394 to 600 bp from the genes atpA, dnaA, metS, recA, rpoD, tkt, and tufA. Because the report of Rosales et al. (14) does not include a list of representative allele sequences for each target, those sequences must be deduced by cross-referencing the numerical allele assignments found in the supplemental file accompanying their paper (Supplemental File 1) with corresponding nucleotide sequence submissions to GenBank. Our initial attempts to unequivocally discern the sequences corresponding to each allele of the Weybridge scheme were confounded by several inconsistencies and omissions in the originally published data set. Following related discussion with the communicating author, a correction was published (18), but several key pieces of information remained either missing or unclear. A set of individual allele sequences for each MLST target that was subsequently provided failed to resolve the confusion. As a result, it is not possible to know with certainty the nucleotide sequence intended to be represented by each numerical allele identifier. Accordingly, for purposes of the present study, we utilized the following ad hoc system of nomenclature for distinguishing unique alleles of the target sequences used in the Weybridge scheme. Each novel allele encountered for a given MLST target was sequentially assigned a unique upper case alphabetical identifier (i.e., A, B, C, etc.). Each unique combination of alphabetical allelic identifiers was assigned an Arabic numeral to indicate the corresponding ST (Table S2). It is important to emphasize that there is no correspondence between the allelic profiles represented by numerical STs as reported by Rosales et al. (14, 18) and the numerical STs used here for the Weybridge scheme.
MLST data analyses.
For each PubMLST or Weybridge target locus, locus diversity (h) was calculated as h = (1 – ΣPi2)(n/[n − 1]), where Pi is the frequency of the i-th allele and n is the number of STs in the sample population, as reported previously (19). The DI for each MLST scheme was calculated according to the method of Hunter and Gaston (20). Probable genetic relationships among allelic profiles were inferred using PHYLOViZ v.2.0. (21), implementing the goeBURST algorithm (22) with eBURST distance and clonal complexes formed at the single locus variant (SLV) level.
Nucleotide sequence data.
MLST target sequences were acquired from publicly available genome assemblies for 246 of the isolates included in this study (17, 23–30; see Table S1 for GenBank accession numbers). For 190 isolates, target sequences were retrieved from private genome assemblies initially obtained for other purposes, derived from either Illumina sequencing (182 isolates; MiSeq; Illumina, San Diego, CA) or from both PacBio and Illumina reads (8 isolates). Illumina sequencing was carried out using either 2 × 150-bp or 2 × 300-bp paired-end libraries prepared with a Nextera XT DNA library preparation kit (Illumina) as recommended by the manufacturer. PacBio sequencing was carried out by the Yale Center for Genome Analysis (New Haven, CT). Following fragmentation and end repair of genomic DNA, BluePippen size selection was used to enrich for 10- to 20-Kb fragments. Libraries were sequenced using one single-molecule real-time (SMRT) cell per isolate and P6-C4 chemistry on a PacBio RS II instrument. The average depth of coverage across each assembly ranged from 25× to 1,360× (Table S1) with a median of 90×. Genome assemblies were uploaded to the Bacterial Isolate Genome Sequence Database (BIGSdb) on the M. bovis PubMLST site. Analysis of genomes to identify candidate genes for a new typing locus was carried out using the BIGSdb Genome Comparator tool, with PG45 as the reference isolate. The Genome Comparator output includes a list of genes comprising the core genome and the number of alleles found for each gene among the group of isolates analyzed.
Target sequences from the remaining 12 isolates were obtained by sequencing PCR amplicons using the primers and methods previously reported for each MLST scheme (13, 14). Consensus sequences used for allele assignments were derived from a minimum of two high-quality reads with at least one read in each direction. All isolates included in this study have been submitted to the PubMLST Mycoplasma bovis Isolates Database (https://pubmlst.org/bigsdb?db=pubmlst_mbovis_isolates; see Table S1 for database identification [ID] numbers), and related allele sequences can be downloaded from the PubMLST Mycoplasma bovis Locus/Sequence Definitions Database (https://pubmlst.org/bigsdb?db=pubmlst_mbovis_seqdef).
Characterization of the adh-1 region in isolates missing the PubMLST adh-1 locus.
Genome assemblies for the isolates included in this study were uploaded to the BIGSdb on the M. bovis PubMLST website and queried for the presence/absence of adh-1 and neighboring coding DNA sequences (CDSs), as found in the reference isolate PG45 (CP002188.1), using the PubMLST BLAST plugin. Gene content and order in the adh-1 region of isolates missing the PubMLST adh-1 locus were further examined using global assembly alignments constructed with the progressiveMauve algorithm (31) of the Mauve plugin available in Geneious v.11.1.5 (Biomatters Ltd.).
Identification of candidate MLST loci and design of related PCR primers.
Nucleotide sequences from genes evaluated as potential sites of a typing locus to replace the adh-1 locus were aligned using the AlignX program in Vector NTI v.10.3.0 (Invitrogen) and visually examined in continuous increments of up to ∼700 bp to identify the region that discriminates the highest possible number of alleles. The Vector NTI Primer Design program was used to select primers for PCR amplification of each region, with primer sequences limited to gene segments found to be identical in the 436 isolates included in this study for which genome assemblies are available. The melting temperature (Tm) of newly selected primers was restricted to the range of Tms for primers used to amplify and sequence the PubMLST scheme targets, 39.2 to 46.7°C, such that identical cycling conditions can be used to amplify all loci. Locus boundaries were trimmed to be in-frame and ∼50 bp distant from the 3′ ends of each PCR primer.
Data availability.
The sequences representing metS allele H and dnaA allele L of the Weybridge scheme are those found in publicly available genome assemblies for isolates Hubei-1 and SZ-0527, respectively (see Table S1 for GenBank accession numbers). Sequences representing additional alleles reported here for the Weybridge scheme have been submitted to GenBank with accession numbers MN969496 to MN969582.
RESULTS
PubMLST scheme.
Only 80.6% (361/448) of the isolates included in this study can be fully typed according to the PubMLST reference scheme. Automated sequence tag-scanning of genome assemblies for the remaining 87 isolates failed to identify an allele for the MLST target located within the adh-1 gene. Additional details pertaining to the adh-1 region of these nontypeable isolates is provided below.
The 361 isolates that are fully typeable represent 69 STs, 28 of which have not been previously reported or submitted to the PubMLST Mycoplasma bovis Locus/Sequence Definitions Database (https://pubmlst.org/bigsdb?db=pubmlst_mbovis_seqdef). Partial allelic profiles for the 87 isolates missing the adh-1 locus include 22 different combinations, designated NT1 to NT22 (Table 2). Isolates in this group were found in cattle, bison, and a white-tailed deer originating from geographically diverse regions that include North America, Europe, the Middle East, and Australia. The combination of alleles defining 12 of the partial profiles is novel compared to the 185 STs so far reported here or elsewhere (10, 13, 16, 17, 32, 33) or submitted to the PubMLST Mycoplasma. bovis Locus/Sequence Definitions Database (Table 2). When absence of the adh-1 locus is considered to be a discriminating characteristic for that allele, the PubMLST scheme is able to delineate 91 novel genotypes among the 448 isolates analyzed. Locus-specific statistics are detailed in Table 3. From 10 to 19 alleles were identified per locus (for adh-1 and gyrB, respectively), and the locus diversity ranges from 0.374 (for gltX) to 0.713 (for gpsA) with a mean of 0.510. Again considering the absence of the adh-1 locus to be a legitimate allele assignment, the DI for this method is 0.909, indicating that two randomly selected isolates are expected to represent different STs on 90.9% of the occasions sampled. ST10 is most prevalent among the isolates we evaluated, comprising 125/448 isolates (27.9%), followed by ST2 with 24/448 isolates (5.4%).
TABLE 2.
Partial PubMLST allelic profiles and the host and geographic origin of isolates in which they are found
| Six-locus allelic profile (locus) |
Designationa | No. of isolates | Host(s) of origin | Geographic origin | STs with matching six-locus allelic profiles | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| gltX | gpsA | gyrB | pta-2 | tdk | tkt | |||||
| 3 | 3 | 3 | 2 | 3 | 6 | NT1 | 5 | Cattle | United States, Canada, Switzerland | 5, 38, 39, 62, 76, 155, 163, 166 |
| 3 | 1 | 3 | 5 | 2 | 4 | NT2 | 17 | Cattle | United States | 55 |
| 3 | 3 | 3 | 4 | 3 | 4 | NT3 | 4 | Cattle, bison | Canada | 2, 120 |
| 3 | 1 | 3 | 2 | 2 | 3 | NT4 | 2 | Cattle | United States | 51, 56 |
| 8 | 2 | 1 | 2 | 3 | 4 | NT5 | 1 | Mule deer | United States | 57 |
| 3 | 2 | 3 | 5 | 3 | 4 | NT6 | 7 | Cattle | Israel, Australia | 10, 82, 106, 150 |
| 16 | 6 | 21 | 5 | 10 | 23 | NT7 | 1 | Cattle | Israel | None |
| 3 | 3 | 5 | 2 | 2 | 3 | NT8 | 6 | Cattle | Hungary, Israel | 94 |
| 3 | 3 | 3 | 5 | 3 | 4 | NT9 | 4 | Cattle | Israel | 30, 151 |
| 3 | 2 | 5 | 5 | 3 | 4 | NT10 | 1 | Cattle | Israel | None |
| 3 | 3 | 5 | 20 | 2 | 3 | NT11 | 1 | Cattle | Israel | None |
| 3 | 14 | 11 | 5 | 3 | 4 | NT12 | 2 | Cattle | Israel | None |
| 16 | 6 | 5 | 21 | 2 | 4 | NT13 | 1 | Cattle | Israel | None |
| 3 | 3 | 5 | 22 | 2 | 3 | NT14 | 1 | Cattle | Romania | None |
| 2 | 3 | 3 | 2 | 3 | 2 | NT15 | 1 | Cattle | Hungary | None |
| 3 | 3 | 3 | 5 | 12 | 3 | NT16 | 2 | Cattle | Romania, Israel | None |
| 3 | 4 | 3 | 5 | 3 | 4 | NT17 | 6 | Cattle | United States | None |
| 3 | 3 | 3 | 2 | 3 | 4 | NT18 | 13 | Cattle | Canada | 6, 127 |
| 3 | 4 | 13 | 2 | 3 | 27 | NT19 | 1 | Cattle | Canada | None |
| 3 | 4 | 13 | 2 | 3 | 4 | NT20 | 9 | Cattle | Canada | None |
| 3 | 3 | 3 | 2 | 3 | 1 | NT21 | 1 | Cattle | Canada | 65, 73, 75, 80 |
| 3 | 3 | 3 | 2 | 3 | 28 | NT22 | 1 | Cattle | Canada | None |
NT, nontypeable.
TABLE 3.
Characteristics of the loci used in the two MLST schemes
| Characteristic | Value for PubMLST locus |
Value for Weybridge locus |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| adh-1 | gltX | gpsA | gyrB | pta-2 | tdk | tkt | atpA | dnaA | metS | recA | rpoD | tkt | tufA | |
| Length (bp) | 432 | 423 | 414 | 570 | 378 | 306 | 426 | 589 | 498 | 596 | 463 | 600 | 394 | 499 |
| No. of alleles | 10 | 11 | 12 | 19 | 14 | 13 | 15 | 15 | 13 | 17 | 7 | 11 | 14 | 12 |
| No. of variable sites | 38 | 33 | 55 | 57 | 39 | 33 | 60 | 56 | 58 | 40 | 32 | 28 | 53 | 23 |
| Locus diversity | 0.486 | 0.374 | 0.713 | 0.466 | 0.563 | 0.420 | 0.550 | 0.382 | 0.583 | 0.611 | 0.476 | 0.321 | 0.548 | 0.390 |
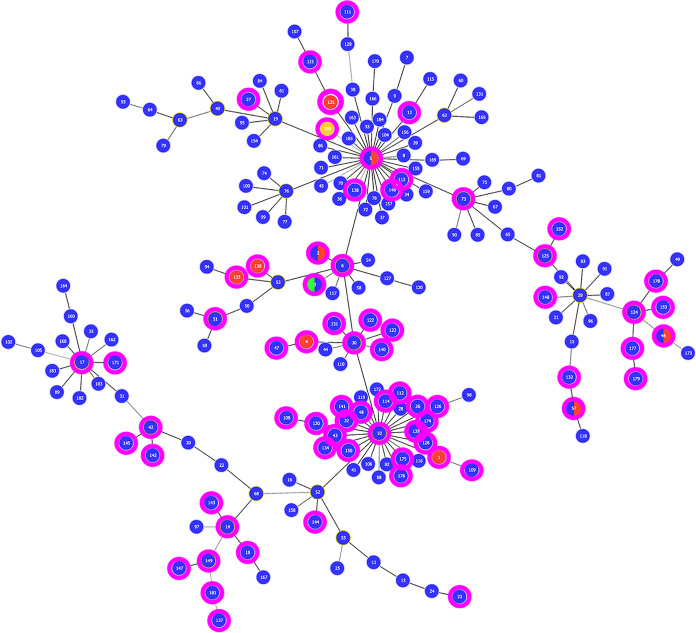
A phylogenetic tree illustrating predicted genetic relationships among the 185 currently defined PubMLST STs is shown in Fig. 1. Although the 361 fully typeable isolates evaluated here represent less than half the STs so far delineated for M. bovis, all major clusters as well as most sublineages having more than a single member are represented. On that basis, we conclude that the isolates included in this study provide a reasonably complete representation of the genetic diversity currently known to exist.
FIG 1.
Minimum spanning tree depicting evolutionary relationships among the 185 PubMLST scheme STs thus far defined for M. bovis. STs are indicated by the numeral in each circle. Circles are colored to indicate the host(s) in which each ST has thus far been found (blue, cattle; red, bison; green, deer; yellow, goat). Magenta borders specify the STs represented by isolates included in this study.
Characterization of the adh-1 region for isolates missing that locus.
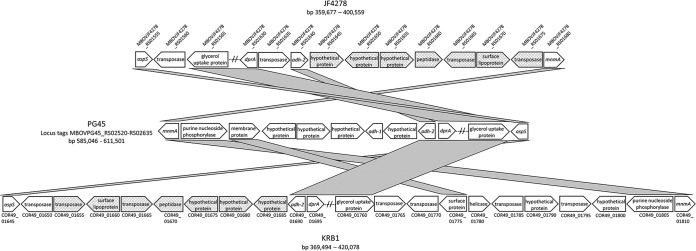
BLAST queries of genome assemblies for the 87 nontypeable isolates using the sequence of the adh-1 open reading frame (ORF) and several 5′ and 3′ neighboring CDSs, as found in the reference isolate PG45 (see the center of Fig. 2 for a schematic representation), indicate that all have a deletion that includes adh-1 and one immediately adjacent 5′ and 3′ CDS (bp 590791 to 592416 of PG45; locus tags MBOVPG45_RS02545, MBOVPG45_RS02550, and MBOVPG45_RS02555). In most isolates, the deletion extends further in the 5′ direction to include one to four additional contiguous CDSs. The adh-2 gene and neighboring downstream CDSs (as found in PG45) are conserved in all nontypeable isolates. Visual examination of genome alignments for assemblies with no contig breaks in this region further reveals a variety of insertions that invariably include one or more transposases characteristic of several different insertion sequence (IS) families. A comparison of local gene content and order in PG45 with that found in two nontypeable isolates for which the region is covered within a single contig is shown in Fig. 2. The deletion in isolate JF4278 comprises seven CDSs upstream of the adh-2 gene. In their place is a roughly 16-Kb insertion with CDSs annotated as transposases, hypothetical proteins, a peptidase, and a surface lipoprotein, most of which are found elsewhere in the genome of some other isolates of M. bovis but not in PG45. Neighboring genes in JF4278 for which there are homologs in PG45 are all found in reverse orientation, since this region of JF4278 is part of a 380-Kb segment of DNA that has undergone an apparent recombination/inversion event relative to PG45. The deletion in KRB1 includes only five genes immediately upstream of adh-2 in PG45. A local recombination/inversion event has positioned the immediately adjacent genes downstream and in the opposite orientation compared to PG45, with the first two separated by an insertion of approximately 7 Kb. A second insertion 5′ of the adh-2 gene is nearly identical to the insertion found 3′ of the adh-2 gene in JF4278, including the 1,029-bp IS30-like transposase flanking one end of each.
FIG 2.
Schematic representation of the gene order and content found in the adh-1 region of PG45 compared to isolates JF4278 and KRB1, both missing the adh-1 PubMLST typing locus. Locus tags for CDSs of JF4428 and KRB1 are given above and below the corresponding block arrows, respectively. Shaded connectors indicate shared CDSs. The orientation of each block arrow denotes the direction of transcription, while hatched arrows represent CDSs shared between JF4278 and KRB1 that are not found here or elsewhere in the PG45 genome.
Weybridge MLST scheme.
All isolates are fully typeable using the seven MLST loci defined for the Weybridge scheme. A total of 77 STs were identified, with each locus having 7 to 17 alleles (recA and metS, respectively; see Table 3 for locus-specific characteristics). Locus diversity varies from 0.321 (for rpoD) to 0.611 (for metS) with a mean of 0.473. The DI obtained with this scheme is 0.842, which falls short of the minimum recommended for a typing scheme, 0.900 (20). The most frequently occurring ST is ST25 (170/448 isolates or 37.9%), followed by ST23 (31/448 isolates or 6.9%).
Comparison of the PubMLST and Weybridge schemes.
The DIs obtained above indicate that the PubMLST scheme out-performed the Weybridge scheme in the analysis undertaken here. Although each scheme was able to further distinguish among isolates having a single ST when typed using the alternative scheme, a breakdown of the distribution of STs between these two methods more specifically reveals the advantages of the PubMLST scheme. There are 19 PubMLST STs for which the Weybridge scheme delineated two or more STs, affecting 261 isolates, compared to 21 Weybridge scheme STs for which the PubMLST scheme delineated two or more STs, affecting 343 isolates (Table 4). Of particular note, the PubMLST scheme was able to further discriminate 35.9% (61/170) of isolates making up the most frequently occurring Weybridge ST (ST25, with 170 isolates). In comparison, the Weybridge scheme further discriminated only 12.8% (16/125) of isolates with the most commonly found PubMLST ST (ST10, with 125 isolates).
TABLE 4.
PubMLST and Weybridge scheme STs for which the alternative method discriminates more than a single ST
| PubMLST STs for which the Weybridge scheme is more discriminatory |
Weybridge STs for which the PubMLST scheme is more discriminatory |
||
|---|---|---|---|
| PubMLST ST (no. of isolates) | Weybridge STs (no. of isolates) | Weybridge ST (no. of isolates) | PubMLST STs (no. of isolates) |
| 2 (24) | 13 (9), 25 (14), 32 (1) | 1 (5) | 17 (4), 171 (1) |
| 3 (5) | 5 (3), 6 (2) | 2 (2) | 73 (1), NT21 (1) |
| 5 (17) | 8 (14), 68 (1), 72 (1), 74 (1) | 6 (6) | 3 (2), 4 (2), 47 (2) |
| 10 (125) | 13 (1), 15 (1), 24 (1), 25 (109), 34 (8), 45 (1), 53 (2), 65 (19), 75 (1) | 7 (3) | 51 (1), NT4 (2) |
| 17 (6) | 1 (4), 40 (1), 73 (1) | 8 (20) | 5 (14), 146 (1), NT1 (5) |
| 27 (2) | 52 (1), 60 (1) | 11 (2) | 6 (1), NT18 (1) |
| 30 (2) | 10 (1), 13 (1) | 13 (12) | 2 (9), 10 (1), 30 (1), NT10 (1) |
| 32 (2) | 25 (1), 31 (1) | 15 (2) | 10 (1), 112 (1) |
| 42 (8) | 50 (6), 58 (2) | 16 (12) | 121 (11), 128 (1) |
| 46 (3) | 30 (1), 56 (2) | 20 (3) | 57 (2), NT5 (1) |
| 121 (12) | 16 (11), 18 (1) | 23 (31) | 126 (2), NT9 (2), NT17 (6), NT18 (12), NT20 (9) |
| 125 (13) | 3 (9), 4 (4) | 25 (170) | 1 (11), 2 (14), 10 (109), 32 (1), 43 (1), 108 (4), 123 (15), 130 (3), 134 (1), 141 (1), 176 (1), NT3 (3), NT6 (6) |
| 128 (3) | 14 (2), 16 (1) | 31 (2) | 26 (1), 32 (1) |
| 137 (4) | 41 (2), 62 (1), 63 (1) | 34 (11) | 10 (8), 114 (1), 174 (1), 175 (1) |
| 140 (7) | 49 (6), 51 (1) | 36 (16) | 124 (3), 153 (1), 177 (11), 179 (1) |
| NT3 (4) | 25 (3), 39 (1) | 43 (22) | 14 (13), 18 (4), 143 (5) |
| NT6 (7) | 25 (6), 53 (1) | 47 (8) | NT8 (6), NT11 (1), NT14 (1) |
| NT9 (4) | 23 (2), 48 (2) | 50 (7) | 42 (6),142 (1) |
| NT18 (13) | 11 (1), 23 (12) | 53 (3) | 10 (2), NT6 (1) |
| 58 (4) | 42 (2), 145 (2) | ||
| 66 (2) | 147 (1), 149 (1) | ||
Despite the apparent advantages of the PubMLST scheme, the absence of the adh-1 locus in some isolates raises concern. If this is a rare occurrence, it might be reasonable to incorporate that information into the nomenclature of the scheme, with the numeral zero used as the adh-1 allele assignment for isolates missing the locus. This approach would restore complete allelic profiles to all isolates and permit the scheme to continue in its original form, thereby preserving the value of the sizeable quantity of data previously collected and submitted to the M. bovis PubMLST database. Until recently, this seemed a reasonable modification, since few isolates missing the adh-1 locus had been encountered. In fact, not until 2018 was this issue documented in the literature (16), when the locus was reported to be missing in a 2008 isolate cultured from the milk of a Swiss cow with mastitis. It now appears that clones missing adh-1 may have first arisen around that time and that their prevalence is growing worldwide. Considering all typing data currently available, reported either here or elsewhere (10, 13, 16, 17, 32, 33) or submitted to the PubMLST Mycoplasma bovis Isolates Database, the adh-1 locus is absent in only 1.4% (5/349) of isolates obtained in 2010 or prior, compared to 14.2% (86/606) of isolates originating after that time. No isolates missing that locus were found among the 137 included in the study describing the development of the PubMLST scheme (13), perhaps because the majority (92/137, or 67.2%) originated in 2010 or earlier. This observation mandates the withdrawal of the adh-1 locus from the PubMLST scheme if it is to be otherwise maintained as a reference typing method.
Revision of the PubMLST reference scheme.
While most PubMLST typing schemes include a minimum of seven loci, a few achieve acceptable discriminatory power using only six targets. Accordingly, we evaluated the discriminatory power of a six-locus scheme that retains all the current PubMLST scheme loci except for adh-1. This approach yielded 79 STs from the 448 isolates included in our study, with a DI of 0.897, just short of the benchmark index of 0.900. The two most frequently encountered STs include 133 isolates with the profile gltX-3, gpsA-2, gyrB-3, pta-2-5, tdk-3, tkt-4, typed as ST10, ST150, or NT6 using the seven-locus scheme, and 28 isolates with the profile gltX-3, gpsA-3, gyrB-3, pta-2-4, tdk-3, tkt-4, typed as ST2 or NT3 using the seven-locus scheme. These data highlight the relatively minimal contribution of the adh-1 locus to the overall discriminatory power of the current PubMLST method. Nonetheless, it would be beneficial to achieve greater resolution by identifying a seventh locus suitable for replacement of the adh-1 locus.
In the search for an additional typing target, we initially focused on the loci defined for the Weybridge scheme, since several exhibit a level of sequence heterogeneity comparable to what is found among the PubMLST targets (see the locus diversity values in Table 3). Should one of the Weybridge scheme loci prove to be beneficial when combined with the six PubMLST scheme targets being retained, an additional benefit is that an allele assignment is already available for many isolates that were previously typed using that method, here and by others (14, 32). It should be noted that the tkt locus used in the Weybridge scheme largely overlaps with the PubMLST tkt target sequence and, therefore, is not suitable as a substitute for the adh-1 locus. A direct comparison of the resolution achieved with each of the tkt loci indicates that the PubMLST target sequence is optimal, based on the number of alleles and the locus diversity obtained for each with the isolates included in this study (Table 3). Moreover, the 55 bp at the 5′ end of the PubMLST tkt target that do not overlap the region of the gene covered by the Weybridge locus are highly informative, as they include 8 polymorphic positions. In contrast, the 22 bp at the 3′ end of the Weybridge scheme tkt target that do not overlap the PubMLST target include only a single polymorphic position thus far found to vary only in a single, unusual isolate already discriminated by other unique PubMLST scheme locus alleles (isolate 8790; see Table S1). Hence, we opt to retain the PubMLST tkt target for use in the revised scheme. Proceeding with the remaining six unique Weybridge loci, the 448 isolates previously evaluated were retyped using schemes in which each Weybridge locus was individually added to the six-locus PubMLST scheme. The greatest number of STs is defined when including either the atpA or dnaA loci, but the effect on discriminatory power is optimal with the dnaA locus whose inclusion raises the DI from 0.897 to 0.914 (Table 5). Furthermore, the two STs found most frequently using the revised scheme that incorporates the dnaA locus both include fewer isolates than are found in the two most common STs for any of the other revised schemes (122 and 22 isolates for dnaA, 130 to 132 and 27 or 28 isolates for the other loci; see Table 5), as well as for the original PubMLST scheme (125 isolates for ST10 and 24 isolates for ST2). Genome sequences are available for 124/125 isolates typed here as ST10 using the original PubMLST scheme. We utilized those data to determine whether supplementing the six-locus PubMLST scheme with a locus from a housekeeping gene other than dnaA might more significantly reduce the number of isolates with the most frequently occurring ST while still maintaining a DI greater than or equal to that obtained when using the Weybridge scheme dnaA locus. From the output of a Genome Comparator analysis of the genome assemblies for those 124 ST10 isolates, we identified every CDS shared among all isolates for which there are two or more alleles. We eliminated from consideration any gene with a function similar to the genes targeted by the six-locus PubMLST scheme and any that are closely linked to them (within ≤20 Kb), those that are incomplete or absent in one or more assemblies, those with insertions or deletions in one or more isolates, and those annotated as either variable or surface lipoproteins, insertion sequences/transposases, or encoding “hypothetical” or “putative” products. The remaining CDSs were sorted on the basis of the number of different alleles found. The four genes for which we identified six or more alleles are listed in Table 6, with the dnaA gene included for comparison. A cutoff of six alleles was used in order to approximate or exceed the number of alleles of the dnaA gene found among this group of isolates (seven alleles). Alignments of the unique alleles for each of these four candidate genes were examined to discern the maximum number of distinct sequences that can be discriminated from any contiguous stretch of up to ∼700 bp, the length we defined as appropriate for use as a typing target. This led to elimination of the 5′-nucleotidase gene (PG45 locus tag MBOVPG45_RS03400) from the list of candidates since the widely scattered spacing of variable positions permits the differentiation of only three alleles from the most informative segment. In comparison, five to six alleles can be defined from the most discriminatory segments of the remaining three genes, while six alleles of the dnaA locus defined by the Weybridge scheme are found among these ST10 isolates. Selection of a gene segment for use as a typing locus further requires that a short stretch of 5′- and 3′-adjacent sequences be identical in all isolates to provide nonvariable sites for the binding of universal PCR primers. Alignments of the three genes still under consideration, as found in all genome assemblies included in our study (436/448 isolates), revealed conserved regions suitable for PCR primer placement bordering each of the gene segments identified above as most discriminatory. The amplicon sizes, locus sizes, and coordinates selected as optimal for each gene are listed in Table 6. However, the alignment for the ptsP gene (PG45 locus tag MBOVPG45_RS03595) revealed the presence of single base pair deletions in four isolates, three from Canada and one from China. The deletion occurs at a different site in each isolate, with two found at different positions within the proposed typing locus, such that it is not possible to select a modified locus that excludes them all. Based on these observations, together with the uncertainty as to whether additional deletions not apparent here might exist in other isolates, the ptsP gene was not considered further.
TABLE 5.
Performance characteristics of the six-locus PubMLST scheme when combined with the additional locus indicated
| Characteristic | Value for locus evaluated using 448 isolates |
Value for locus evaluated using 436 isolates |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| atpAa | dnaAa | metSa | recAa | rpoDa | tufAa | dnaAa | uvrA | MBOVPG45_RS03470 | |
| No. of STs | 88 | 88 | 87 | 86 | 83 | 84 | 87 | 84 | 88 |
| Discrimination index | 0.900 | 0.914 | 0.900 | 0.900 | 0.901 | 0.900 | 0.910 | 0.910 | 0.907 |
| No. of isolates in the two most common STs | 132, 27 | 122, 22 | 131, 28 | 131, 28 | 130, 28 | 131, 28 | 122, 21 | 120, 27 | 123, 27 |
| Locus diversity | 0.382 | 0.583 | 0.611 | 0.476 | 0.321 | 0.390 | 0.576 | 0.167 | 0.447 |
Locus defined for the Weybridge scheme (14).
TABLE 6.
Characteristics of genes and loci considered for replacement of the adh-1 locus in the PubMLST scheme
| CDSs |
Amplicons |
Optimal typing loci |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus tag | Product (abbreviation) | Length (bp) | Genome position | Distance from nearest PubMLST locus | No. of allelesa | Nucleotide positions within the gene | Size (bp) | Nucleotide positions within the gene | Size (bp) | No. of allelesa |
| MBOVPG45_RS02340 | Excinuclease ABC subunit (uvrA) | 2,832 | 542095 | ∼177 Kb from pta-2 | 6 | 1380–2000 | 621 | 1453–1935 | 483 | 5 |
| MBOVPG45_RS03400 | Bifunctional metallophosphatase/5′-nucleotidase | 2,043 | 791046 | ∼49 Kb from tkt | 6 | 700–1324 | 625 | 770–1255 | 485 | 3 |
| MBOVPG45_RS03470 | ABC transporter ATP-binding protein | 1,785 | 806259 | ∼64 Kb from tkt | 7 | 727–1403 | 677 | 796–1326 | 531 | 6 |
| MBOVPG45_RS03595 | Phosphoenolpyruvate-protein phosphotransferase (ptsP) | 1,713 | 851361 | ∼72 Kb from tdk | 11 | 277–946 | 670 | 346–876 | 531 | 6 |
| MBOVPG45_RS00005 | Chromosomal replication initiator protein (dnaA) | 4,377 | 9 | ∼27 Kb from gyrB | 7 | 346–1269b | 924 | 532–1029 | 498 | 6 |
Based on analysis of 124 isolates typed as ST10 using the PubMLST scheme for which genome assemblies are available.
As defined for the Weybridge scheme (14).
The 436 isolates for which genome assemblies are available were retyped using individual schemes in which either the uvrA locus (PG45 locus tag MBOVPG45_RS02340) or the ABC transporter ATP-binding protein locus (PG45 locus tag MBOVPG45_RS03470) was individually added to the six loci being retained from the PubMLST scheme. The results were compared to those obtained when the Weybridge scheme dnaA locus was used. Although the ABC transporter ATP-binding protein locus distinguished the greatest number of STs, the DI for this scheme was slightly lower than that for the other two schemes (Table 5). DIs were identical for the schemes including uvrA and dnaA, but the locus diversity value for uvrA is unacceptably low due to the high prevalence of the most common allele, which is found in 398/436 isolates. In comparison, the most common alleles for dnaA and the ABC transporter ATP-binding protein locus are found in 246 and 319 isolates, respectively. These data led us to adopt the dnaA locus as a replacement for the adh-1 locus in the scheme to be used henceforth as the PubMLST reference method, which we refer to here as the revised PubMLST scheme. Related locus definitions, allelic profiles, and STs for isolates included in this study are available on the M. bovis PubMLST website (https://pubmlst.org/mbovis/).
MLST-based phylogenies.
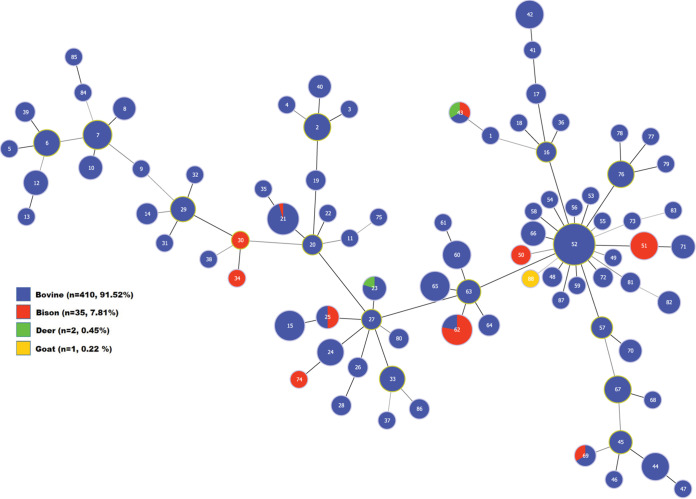
Figure 3 depicts predicted genetic relationships among the 88 STs defined by the revised PubMLST scheme for the 448 isolates included in this study. Relationships among isolates clustered in major lineages are generally similar to those predicted by a phylogenetic tree constructed using the original PubMLST scheme for this same group of isolates (Fig. S1). In both instances, STs that include isolates from bison are scattered among several divergent lineages in which cattle isolates predominate. Neither typing scheme suggests any compelling association between one or a few major lineages and either year of origin (Fig. S2 versus Fig. S6), anatomic site of origin (Fig. S3 versus Fig. S7), or the clinical status of the source animal (Fig. S4 versus Fig. S8). However, both schemes define two clusters each in which joint isolates are noticeably underrepresented, with only 1/51 joint isolates (2.0%) collectively found among the four clusters (Fig. S3 and S7). These same clusters collectively include only 3/99 isolates (3.0%) cultured from animals suffering from arthritis (Fig. S4 and S8). Regarding geographic origin, both schemes segregate all isolates from Asia to a single, major cluster or a related sublineage (for one isolate) that additionally includes isolates from nearby as well as far-removed regions (Fig. S5 and S9). While Middle Eastern and European isolates are found in nearly all lineages, phylogenies for both schemes include a single cluster comprising an unusually high proportion of isolates from those regions; 40/43 isolates (93.0%) and 50/57 isolates (87.7%) for the related clusters arising from the original and revised schemes, respectively. Both schemes consign isolates from Australia to a single, large cluster, but the significance of this observation is uncertain given the small number of Australian isolates evaluated (n = 12).
FIG 3.
Minimum spanning tree depicting evolutionary relationships among the 88 STs defined by the revised PubMLST scheme for M. bovis found among the 448 isolates included in this study. STs are indicated by the numeral in each circle. Circles, or portions thereof, are sized in proportion to the number of isolates with the indicated ST (log scale) and are colored to indicate the host(s) in which each ST was found, as specified in the color key. Circles with green outlines indicate STs predicted to be the founder for the related group.
The phylogenies above arising from the original and revised PubMLST schemes represent considerably fewer isolates and STs than the number currently found in the PubMLST database. To ascertain whether the geographic associations they suggest are supported by a larger and more diverse data set, we compared them with a tree constructed from the 185 STs so far defined for the original scheme and the 920 isolates from which those profiles are derived. At apparent variance with the phylogenies represented in Fig. S5 and S9, Asian isolates are dispersed among several divergent clusters and, in many instances, their predicted closest relatives originate from other world regions (Fig. S10). However, examination of the Asian countries of origin for isolates included in each phylogeny reveals an explanation. The phylogenies depicted in Fig. S5 and S9 include 48 Asian isolates, with 47 from China and one from Japan, while the tree in Fig. S10 includes 138 Asian isolates, with 93 from China and 45 from Japan. All but six Chinese isolates represented in Fig. S10 are members of the ST10 cluster; thus, nearly the entirety of the additional diversity of Asian isolates suggested by this phylogeny is due to Japanese isolates. Considering isolates from Europe and the Middle East as found in the more comprehensive phylogeny (Fig. S10), nearly all are confined to two major lineages, thereby supporting the close genetic relationship between those groups inferred by trees depicted in Fig. S5 and S9.
DISCUSSION
The MLST scheme originally employed on the M. bovis PubMLST site was the first to be proposed as a universal reference method for the bacterium (13), although the Weybridge scheme was published very shortly thereafter (14). The DIs initially reported for each method are similar (0.922 and 0.910, respectively; 14, 15), and both exceed the minimum benchmark for a typing scheme (0.900). Regardless, until now, it was unclear whether one scheme might offer higher discriminatory power than the other since each study evaluated a different set of isolates. On the basis of the data reported here, obtained using a single, diverse set of 448 isolates, we conclude that the originally used PubMLST scheme offers discriminatory power superior to that provided by the Weybridge scheme. Nevertheless, the need to replace the adh-1 locus afforded an opportunity to improve the performance of the PubMLST reference method. To this end, we utilized all available M. bovis genome sequences to assist in identifying a replacement locus able to distinguish among isolates previously typed as the most commonly found ST. It may be possible to further improve discriminatory power by replacing one or more additional loci, but several considerations led us to reject that option. The DI of the revised PubMLST scheme, 0.914, already exceeds the minimum recommended for a reference typing method, 0.900 (20). As more investigators utilize the revised scheme and the number and diversity of fully typed isolates grows, a further increase is to be expected, perhaps even approaching the benchmark for an ideal system, 0.95 (34). Additionally, we wish to preserve the usefulness of as much of the previously gathered data as is reasonably possible. As of February 2020, more than 1,100 isolates and their allelic profiles have been submitted to the M. bovis PubMLST database. Replacing a locus immediately renders all related allele assignments obsolete and abolishes complete allelic profiles and ST assignments for all previously typed isolates. ST assignments can be restored only if investigators choose to obtain and submit data from their isolates for any newly added loci. While the addition of even one new locus imposes a burden on investigators to collect and submit additional data, we judge it to be an acceptable trade-off given the degree of improvement it provides. However, it seems reasonable to avoid further changes that are unlikely to yield any substantial improvement.
The clustering we observed between isolates from Europe and the Middle East (the latter all originating from Israel; see Table S1) was also noted by others (30) in a recent study using genome-wide single nucleotide polymorphism (SNP) analysis to compare Israeli isolates with those from Europe and Australia, the source of most cattle imported by Israel. That investigation identified isolates from Europe as being the closest relatives of isolates classified as belonging to one of two cohorts of Israeli isolates. Our data are also in agreement with their conclusion that a close genetic relationship exists between a second group of Israeli isolates and those from Australia. The STs we identified among Australian isolates (ST10 and NT6 for the original PubMLST scheme; ST52 for the revised PubMLST scheme) also predominate among Israeli isolates, accounting for 52/110 (47.3%) as typed by the original PubMLST scheme and 51/110 (46.4%) using the revised PubMLST scheme (Fig. S5 and S9). These observations are consistent with an epidemiologic link between M. bovis clones circulating in Israel and those found in countries from which much of Israel’s cattle population is imported. A further area of agreement between our study and theirs is the finding that little genetic diversity exists among Australian isolates. With respect to our study, this might be explained by the small number of isolates we evaluated (n = 12). However, Yair et al. (30) reached a similar conclusion based on genome-wide SNP analysis of 27 isolates. Furthermore, a comprehensive comparison of genome sequence data from 75 Australian isolates, collected between 2006 and 2015, from a variety of anatomic sites and geographic locales and from both healthy and diseased cattle, detected minimal genetic variation and suggests that the M. bovis population circulating within the country is composed of essentially a single strain (35). Finally, Yair et al. (30) observed that the six Chinese isolates they evaluated formed a tight cluster with a high degree of genetic relatedness. The phylogenies we generated here for both the original (Fig. S5 and S10) and revised PubMLST schemes (Fig. S9) similarly indicate a close relationship among the 93 Chinese isolates they collectively represent, which include the six mentioned above. These areas of concordance between MLST and genome-based approaches highlight the utility of MLST as a reliable tool to survey population structure and diversity for M. bovis. Comparative analysis of genome sequences provides a high degree of resolution but requires significant expertise and expenditure of both time and money to sequence and assemble genomes and to analyze the data. In contrast, MLST is a rapid, simple, low cost, and highly discriminatory methodology that is within reach of most laboratories.
The information uncovered here with respect to the adh-1 gene in M. bovis raises some interesting questions for further investigation. Many bacteria produce alcohol dehydrogenases that participate in glycolysis and gluconeogenesis. Some pathogens utilize the enzyme to convert ethanol to acetaldehyde, which can exert toxic effects on host tissues (36). No specific role for alcohol dehydrogenase during in vivo infection has yet been defined for M. bovis, though proteomics data suggest it may contribute to virulence (37), and an adh-1 knockout mutant displays reduced adherence to an in vitro cell line compared to the wild-type parent (38). Deletion of the adh-1 locus was reported previously for two of the isolates in our study (10, 16), but the number of additional isolates encountered here with similar deletions was initially somewhat surprising given the selective pressure to maintain housekeeping genes that encode proteins supporting basic cellular functions. However, we noted that a 1,041-bp CDS also annotated as an alcohol dehydrogenase found nearby in PG45 (adh-2 gene, locus tag MBOVPG45_RS02560) is also present in this region of the genome of all nontypeable isolates. A search of the Pfam database identifies two functional domains in both the Adh-1 and Adh-2 proteins of PG45—an alcohol dehydrogenase GroES-like domain (PF08240.12) for amino acids 27 to 136 (Adh-1) and 24 to 133 (Adh-2) and a zinc-binding dehydrogenase (PF00107.26) for amino acids 177 to 310 (Adh-1) and 174 to 308 (Adh-2). Thus, the functions of Adh-1 and Adh-2 are perhaps redundant or overlapping such that one protein may be capable of compensating for the absence of the other. Three different alleles of adh-2 exist among the nontypeable isolates evaluated here, each predicted to encode a functional protein either identical to PG45 Adh-2 (2 isolates) or with one or two amino acid substitutions (83 and 2 isolates, respectively), only one of which lies within a functional domain. Specifically, for two isolates, an aspartate residue in the zinc-binding dehydrogenase region is predicted to be replaced with a similarly acidic glutamate. Assessment of the relative levels of expression of adh-1 and adh-2 in isolates possessing functional copies of both genes compared to the level of adh-2 expression in isolates with naturally occurring adh-1 deletions, in conjunction with in vitro cell binding studies, may shed further light on the biological functions of the products of these two genes.
Concurrent with the publication of this study, the revised PubMLST scheme described here, encompassing 3,015 bp from loci within the dnaA, gltX, gpsA, gyrB, pta-2, tdk, and tkt genes, will be implemented as the default scheme for the M. bovis PubMLST database. The originally used scheme and all related data will continue to be available in the database (as the “legacy” scheme) for a period of 6 months. During this interim period, users of the database can elect to view information pertinent to only the legacy scheme, only the newly installed reference scheme, or both (see https://bigsdb.readthedocs.io/en/latest/data_query.html#modifying-locus-and-scheme-display-options for related information). We recognize the inconvenience and disruption of modifying the reference method, particularly considering the large amount of data thus far submitted to the database. Obtaining dnaA locus sequences will be a relatively trivial task in the case of isolates for which genome assemblies are available, but there are likely many isolates currently in the database for which a dnaA allele assignment will require the sequencing of newly generated PCR amplicons. We encourage all those who previously submitted isolates to the database to obtain and submit sequences for the newly added dnaA locus so as to maintain the value and integrity of the database moving forward.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karen Gesy for her efforts in growing and extracting DNA from a portion of the M. bovis isolates used in this study.
Financial support for this work was provided by the USDA Agricultural Research Service, the Israeli Office of the Chief Scientist Ministry of Agriculture and Rural Development (grant 33-08-0006), the Saskatchewan Ministry of Agriculture, and the Canada-Saskatchewan Growing Forward 2 bilateral agreement 20160253.
Mention of trade names, proprietary products, or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Maunsell FP, Woolums AR, Francoz D, Rosenbusch RF, Step DL, Wilson DJ, Janzen ED. 2011. Mycoplasma bovis infections in cattle. J Vet Intern Med 25:772–783. doi: 10.1111/j.1939-1676.2011.0750.x. [DOI] [PubMed] [Google Scholar]
- 2.Caswell JL, Bateman KG, Cai HY, Castillo-Alcala F. 2010. Mycoplasma bovis in respiratory disease of feedlot cattle. Vet Clin North Am Food Anim Pract 26:365–379. doi: 10.1016/j.cvfa.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Ridley A, Hately G. 2018. Mycoplasma bovis investigations in cattle. Vet Rec 183:256–258. [DOI] [PubMed] [Google Scholar]
- 4.Hale HH, Helmboldt CF, Plastridge WN, Stula EF. 1962. Bovine mastitis caused by a Mycoplasma species. Cornell Vet 52:582–591. [PubMed] [Google Scholar]
- 5.Nicholas RAJ. 2011. Bovine mycoplasmosis: silent and deadly. Vet Rec 168:459–462. doi: 10.1136/vr.d2468. [DOI] [PubMed] [Google Scholar]
- 6.Government of New Zealand. 2017. Analysis of risk pathways for the introduction of Mycoplasma bovis into New Zealand. Ministry for Primary Industries, Wellington, New Zealand: https://www.biosecurity.govt.nz/dmsdocument/28050/direct. [Google Scholar]
- 7.Epp T, Uehlinger FD, Wojnarowicz C, Malhi PS, Sayi S, Woodbury MR. 2018. Observations of mortality in farmed bison in the Canadian prairies: 2103–2016. Prev Vet Med 157:1–7. doi: 10.1016/j.prevetmed.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 8.United States Department of Agriculture. 2013. Mycoplasma bovis: an emerging pathogen in ranched bison. USDA, APHIS, VS, Center for Epidemiology and Animal Health, Fort Collins, CO: Document 222.0913 https://www.aphis.usda.gov/animal_health/nahms/bison/downloads/bison14/Bison14_Mbovis_1.pdf. [Google Scholar]
- 9.Dyer NW, Krogh DF, Schaan LP. 2004. Pulmonary mycoplasmosis in farmed white-tailed deer (Odocoileus virginianus). J Wildl Dis 40:366–370. doi: 10.7589/0090-3558-40.2.366. [DOI] [PubMed] [Google Scholar]
- 10.Register KB, Jelinski MD, Waldner M, Boatwright WD, Anderson TK, Hunter DL, Hamilton RG, Burrage P, Shury T, Bildfell R, Wolff PL, Miskimins D, Derscheid RJ, Woodbury MR. 2019. Comparison of multilocus sequence types found among North American isolates of Mycoplasma bovis from cattle, bison, and deer, 2007-2017. J Vet Diagn Invest 31:899–904. doi: 10.1177/1040638719874848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calcutt MJ, Lysnyansky I, Sachse K, Fox LK, Nicholas RAJ, Ayling RD. 2018. Gap analysis of Mycoplasma bovis disease, diagnosis and control: an aid to identify future development requirements. Transbound Emerg Dis 65:91–109. doi: 10.1111/tbed.12860. [DOI] [PubMed] [Google Scholar]
- 12.Ibarz Pavón AB, Maiden MC. 2009. Multilocus sequence typing. Methods Mol Biol 551:129–140. doi: 10.1007/978-1-60327-999-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Register KB, Thole L, Rosenbush RF, Minion FC. 2015. Multilocus sequence typing of Mycoplasma bovis reveals host-specific genotypes in cattle versus bison. Vet Microbiol 175:92–98. doi: 10.1016/j.vetmic.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Rosales RS, Churchward CP, Schnee C, Sachse K, Lysnyansky I, Catania S, Iob L, Ayling RD, Nicholas RA. 2015. Global MLST analysis of Mycoplasma bovis isolates reveals two main population clusters. J Clin Microbiol 53:789–794. doi: 10.1128/JCM.01910-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Register KB, Thole L, Rosenbush RF, Minion FC. 2019. Corrigendum to ‘multilocus sequence typing of Mycoplasma bovis reveals host-specific genotypes in cattle versus bison’ Vet Microbiol 175 (1) (2015) 92–98. Vet Microbiol 228:264–265. doi: 10.1016/j.vetmic.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Josi C, Bürki S, Stojiljkovic A, Wellnitz O, Stoffel MH, Pilo P. 2018. Bovine epithelial in vitro infection models for Mycoplasma bovis. Front Cell Infect Microbiol 8:329. doi: 10.3389/fcimb.2018.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morimoto M, Kenri T, Ohmori T, Teshima K, Shibuya K, Sasakawa C, Suzuki M. 2019. Complete genome sequence of Mycoplasma bovis strain KG4397, isolated from cattle in Japan. Microbiol Resour Announc 8. doi: 10.1128/MRA.00838-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosales RS, Churchward CP, Schnee C, Sachse K, Lysnyansky I, Catania S, Iob L, Ayling RD, Nicholas RAJ. 2017. Correction for Rosales et al., “global multilocus sequence typing analysis of Mycoplasma bovis isolates reveals two main population clusters. J Clin Microbiol 55:1596–1597. doi: 10.1128/JCM.00230-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackall PJ, Trott DJ, Rapp-Gabrielson V, Hampson DJ. 1997. Analysis of Haemophilus parasuis by multilocus enzyme electrophoresis. Vet Microbiol 56:125–134. doi: 10.1016/S0378-1135(96)01342-9. [DOI] [PubMed] [Google Scholar]
- 20.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol 26:2465–2466. doi: 10.1128/JCM.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carrico JA, Vaz C. 2017. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 22.Francisco AP, Bugalho M, Ramirez M, Carrico JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Hao H, Zhao P, Gao P, He Y, Ji W, Wang Z, Lu Z, Liu Y, Chu Y. 2017. Complete genome sequence of Mycoplasma bovis strain 08M. Genome Announc 5:e00324-17. doi: 10.1128/genomeA.00324-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Josi C, Bürki S, Vidal S, Dordet-Frisoni E, Citti C, Falquet L, Pilo P. 2019. Large-scale analysis of the Mycoplasma bovis genome identified non-essential, adhesion- and virulence-related genes. Front Microbiol 10:2085. doi: 10.3389/fmicb.2019.02085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zheng H, Liu Y, Jiang Y, Xin J, Chen W, Song Z. 2011. The complete genome sequence of Mycoplasma bovis strain Hubei-1. PLoS One 6:e20999. doi: 10.1371/journal.pone.0020999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen JR, Noyes N, Young AE, Prince DJ, Blanchard PC, Lehenbauer TW, Aly SS, Davis JH, O’Rourke SM, Abdo Z, Belk K, Miller MR, Morley P, Van Eenennaam A. 2017. Whole-genome sequencing and concordance between antimicrobial susceptibility genotypes and phenotypes of bacterial isolates associated with bovine respiratory disease. G3 (Bethesda) 7:3059–3071. doi: 10.1534/g3.117.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi J, Guo A, Cui P, Chen Y, Mustafa R, Ba X, Hu C, Bai Z, Chen X, Shi L, Chen H. 2012. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS One 7:e38239. doi: 10.1371/journal.pone.0038239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun P, Luo H, Zhang X, Xu J, Guo Y, He S. 2018. Whole-genome sequence of Mycoplasma bovis strain Ningxia-1. Genome Announc 6:e01367-17. doi: 10.1128/genomeA.01367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise KS, Calcutt MJ, Foecking MF, Roske K, Madupu R, Methe BA. 2011. Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523). Infect Immun 79:982–983. doi: 10.1128/IAI.00726-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yair Y, Borovok I, Mikula I, Falk R, Fox LK, Gophna U, Lysnyansky I. 2020. Genomics-based epidemiology of bovine Mycoplasma bovis strains in Israel. BMC Genomics 21:70. doi: 10.1186/s12864-020-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menghwar H, He C, Zhang H, Zhao G, Zhu X, Khan FA, Faisal M, Rasheed MA, Zubair M, Memon AM, Ridley A, Robertson ID, Chen Y, Guo A. 2017. Genotype distribution of Chinese Mycoplasma bovis isolates and their evolutionary relationship to strains from other countries. Microb Pathog 111:108–117. doi: 10.1016/j.micpath.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 33.Bürki S, Spergser J, Bodmer M, Pilo P. 2016. A dominant lineage of Mycoplasma bovis is associated with an increased number of severe mastitis cases in cattle. Vet Microbiol 196:63–66. doi: 10.1016/j.vetmic.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 34.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13:1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 35.Parker AM, Shukla A, House JK, Hazelton MS, Bosward KL, Kokotovic B, Sheehy PA. 2016. Genetic characterization of Australian Mycoplasma bovis isolates through whole genome sequencing analysis. Vet Microbiol 196:118–125. doi: 10.1016/j.vetmic.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Salaspuro M. 1997. Microbial metabolism of ethanol and acetaldehyde and clinical consequences. Addict Biol 2:35–46. doi: 10.1080/13556219772840. [DOI] [PubMed] [Google Scholar]
- 37.Khan FA, Rasheed MA, Faisal M, Menghwar H, Zubair M, Sadique U, Chen H, Guo A. 2017. Proteomics analysis and its role in elucidation of functionally significant proteins in Mycoplasma bovis. Microb Pathog 111:50–59. doi: 10.1016/j.micpath.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 38.Zhu X, Dong Y, Baranowski E, Li X, Zhao G, Hao Z, Zhang H, Chen Y, Hu C, Chen H, Citti C, Guo A. 2020. Mbov_0503 encodes a novel cytoadhesin that facilitates Mycoplasma bovis interaction with tight junctions. Microorganisms 8:164. doi: 10.3390/microorganisms8020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences representing metS allele H and dnaA allele L of the Weybridge scheme are those found in publicly available genome assemblies for isolates Hubei-1 and SZ-0527, respectively (see Table S1 for GenBank accession numbers). Sequences representing additional alleles reported here for the Weybridge scheme have been submitted to GenBank with accession numbers MN969496 to MN969582.