OptrA is an ATP-binding cassette (ABC)-F protein that confers resistance to oxazolidinones and phenicols and can be either plasmid-encoded or chromosomally encoded. Here, we isolated 13 Enterococcus faecalis strains possessing a linezolid MIC of ≥4 mg/liter from nursery pigs in swine herds located across Brazil. Genome sequence comparison showed that these strains possess optrA in different genetic contexts occurring in 5 different E. faecalis sequence type backgrounds.
KEYWORDS: Enterococcus faecalis, oxazolidinones, optrA, Tn554
ABSTRACT
OptrA is an ATP-binding cassette (ABC)-F protein that confers resistance to oxazolidinones and phenicols and can be either plasmid-encoded or chromosomally encoded. Here, we isolated 13 Enterococcus faecalis strains possessing a linezolid MIC of ≥4 mg/liter from nursery pigs in swine herds located across Brazil. Genome sequence comparison showed that these strains possess optrA in different genetic contexts occurring in 5 different E. faecalis sequence type backgrounds. The optrA gene invariably occurred in association with an araC regulator and a gene encoding a hypothetical protein. In some contexts, this genetic island was able to excise and form a covalently closed circle within the cell; this circle appeared to occur in high abundance and to be transmissible by coresident plasmids.
INTRODUCTION
The oxazolidinone phenicol resistance gene optrA codes for an ATP-binding cassette (ABC)-F protein that acts through ribosomal protection (1). The gene optrA was first described in Enterococcus faecalis and Enterococcus faecium from humans and agricultural animals in China (2) and has since been found worldwide in Firmicutes isolates from both settings (3–28). Prior to its discovery, oxazolidinone resistance was known to be mediated by mutations in domain V of 23S rRNA (29–31), mutational changes in ribosomal proteins L3 and L4 (32, 33), and/or by Cfr-type adenosine modification on nucleotide A2503 in the peptidyl transferase center (PTC) of 23S rRNA (34–36). More recently, another ATP-binding cassette (ABC)-F protein, PoxtA, has also been associated with decreased susceptibility to oxazolidinones (28, 37).
OptrA elevates the MIC of oxazolidinones, including tedizolid, which is superior to linezolid in treating infections caused by cfr-positive L3 or 23S rRNA mutant strains of staphylococci and enterococci (38). E. faecalis is a widely found commensal of the gastrointestinal (GI) tracts of humans and most animals, but also a leading cause of hospital-acquired infections (39–41). Enterococci readily acquire and disseminate resistance genes and appear to be the main source for horizontal optrA spread (2–9, 13–15, 17, 18, 20–28). Spread of optrA is of serious concern because oxazolidinones are important last-line drugs for treating infections caused by vancomycin-resistant enterococci (VRE), methicillin-resistant Staphylococcus aureus (MRSA), and methicillin-resistant coagulase-negative staphylococci (MRCoNS).
Here, we report the occurrence of optrA in distinct genetic lineages of linezolid-resistant (LR) E. faecalis isolates from Brazilian swine herds, in genetic contexts that can be mobilized and transferred to new strains. We quantified the evolutionary distances between the Brazilian optrA variants and all other optrA gene sequences in the NCBI database, and we examined the evolutionary trajectory of the gene in the sequence types (STs) of E. faecalis isolated from humans, animals, food products, and environmental sources.
RESULTS
Genetic contexts of optrA in the Brazilian porcine LR E. faecalis isolates.
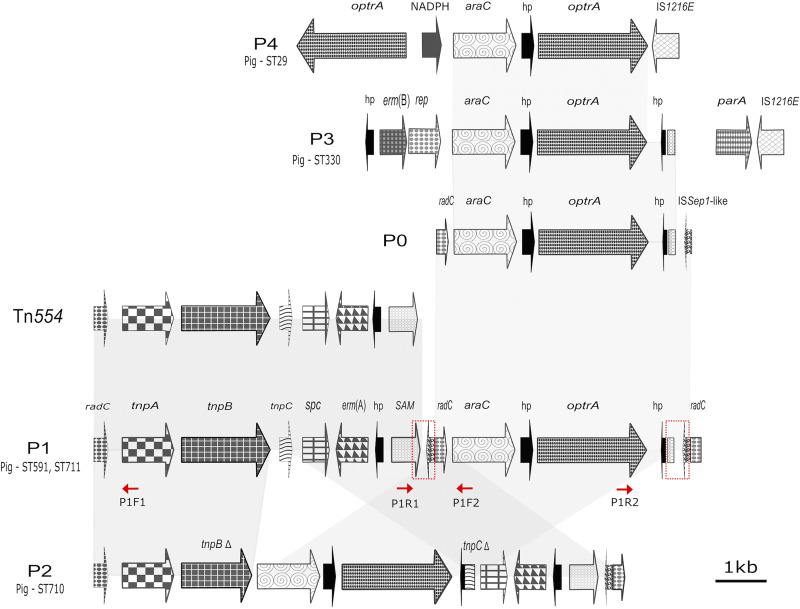
The optrA gene was found in 5 different contexts (patterns P0, P1, P2, P3, and P4) in porcine E. faecalis strains (Table 1; Fig. 1). All carried a core segment of 3,453 bp, composed of the genes encoding an AraC family transcriptional regulator and a small hypothetical protein (hp1) upstream of optrA (core araC-hp1-optrA). In configuration P0 (Fig. 1), a 204-bp fragment of the DNA repair gene radC occurs upstream of araC-hp1-optrA. Downstream of optrA is a gene for a second hypothetical protein (hp2) that has homologues in other E. faecalis and E. faecium strains, as well as in Streptococcus suis isolated from Sus scrofa and in a human-derived Streptococcus pasteurianus strain carrying optrA. Adjacent to hp2 are 72 bp that appear to be derived from the S-adenosylmethionine (SAM)-dependent methyltransferase gene, and, further, 136 bp derived from an ISSep1-like transposase.
TABLE 1.
Demographic data, MLST, and antimicrobial susceptibility profile of optrA-positive E. faecalis isolates from swine in Brazila
| Isolate | State and farm | ST | optrAb | cfr | poxtA | 23S rRNA and L3/L4 mutations | MIC (μg/ml) for: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LZD | TZD | CHL | FFC | TGC | TET | DAP | TYL | STR | SYN | ERY | CIP | PEN | VAN | NIT | GEN | KAN | |||||||
| E. faecalis L10 | SP piggery B | 711 | P1 | − | − | WT | 8 | 1 | 128 | 128 | 0.12 | >32 | 2 | >32 | >2,048 | 16 | >8 | >4 | 2 | 2 | 8 | >1,024 | >1,024 |
| E. faecalis L11 | DF piggery B | 591 | P1 | + | − | WT | 8 | 1 | 128 | 128 | 0.12 | >32 | 2 | 4 | >2,048 | 16 | 0.5 | 2 | 2 | 1 | 32 | <128 | <128 |
| E. faecalis L12 | MG piggery A | 711 | P1 | − | − | WT | 8 | 1 | 128 | 128 | 0.25 | <1 | 2 | >32 | >2,048 | >32 | >8 | 1 | 4 | 2 | 16 | >1,024 | >1,024 |
| E. faecalis L13 | SC piggery A | 591 | P1 | − | − | WT | 8 | 1 | 64 | 128 | 0.25 | >32 | 1 | >32 | >2,048 | >32 | >8 | >4 | 16 | 2 | 16 | >1,024 | >1,024 |
| E. faecalis L15 | DF piggery A | 591 | P1 | + | − | WT | 8 | 1 | 128 | 128 | >0.5 | >32 | 4 | >32 | >2,048 | 8 | 4 | 2 | 4 | 2 | 16 | <128 | 256 |
| E. faecalis L16 | MT piggery A | 591 | P1 | − | − | WT | 16 | 1 | 128 | 128 | 0.12 | >32 | 2 | >32 | >2,048 | 16 | >8 | >4 | 4 | 1 | 16 | >1,024 | >1,024 |
| E. faecalis L18 | SC piggery A | 591 | P1 | − | − | WT | 8 | 1 | 128 | 128 | 0.25 | >32 | 2 | >32 | >2,048 | 16 | >8 | >4 | 2 | 2 | 16 | >1024 | >1,024 |
| E. faecalis L21 | PR piggery A | 591 | P1 | − | − | WT | 8 | 1 | 128 | 128 | 0.12 | >32 | 2 | >32 | >2,048 | 16 | >8 | >4 | 4 | 1 | 16 | >1,024 | >1,024 |
| E. faecalis L8 | SP piggery A | 710 | P2 | − | − | WT | 8 | 1 | 128 | 128 | 0.12 | >32 | 2 | >32 | >2,048 | 8 | 4 | 4 | 1 | 1 | 8 | >1,024 | >1,024 |
| E. faecalis L14 | PR piggery A | 330 | P3 | − | + | WT | 8 | 1 | 128 | 128 | 0.12 | >32 | 2 | >32 | >2,048 | 16 | >8 | >4 | 4 | 1 | 16 | >1,024 | >1,024 |
| E. faecalis L17 | PR piggery A | 330 | P3 | − | + | WT | 8 | 1 | 128 | 128 | 0.12 | >32 | 2 | >32 | >2,048 | 16 | >8 | >4 | 4 | 1 | 16 | >1,024 | >1,024 |
| E. faecalis L19 | PR piggery A | 330 | P3 | − | − | WT | 8 | 1 | 128 | 128 | 0.25 | >32 | 2 | >32 | >2,048 | 16 | 2 | >4 | 4 | 1 | 32 | >1,024 | >1,024 |
| E. faecalis L9 | DF piggery A | 29 | P4 | + | − | WT | 8 | 1 | 128 | 128 | >0.5 | >32 | 4 | >32 | >2,048 | 8 | 4 | 2 | 4 | 2 | 16 | <128 | 256 |
MLST, multilocus sequence typing; WT, wild-type; PR, Paraná; SC, Santa Catarina; DF, Distrito Federal; SP, São Paulo; MG, Minas Gerais; MT, Mato Grosso; LZD, linezolid; TZD, tedizolid; CHL, chloramphenicol; FFC, florfenicol; TGC, tigecycline; TET, tetracycline; DAP, daptomycin; TYL, tylosin; STR, streptomycin; SYN, quinupristin-dalfopristin; ERY, erythromycin; CIP, ciprofloxacin; PEN, penicillin; VAN, vancomycin; NIT, nitrofurantoin; GEN, gentamicin; KAN, kanamycin.
Genetic context of optrA.
FIG 1.
Synteny of optrA loci identified in the Brazilian porcine LR E. faecalis strains. Five different patterns were found (P0, P1, P2, P3, and P4). Areas for recombination identified in the P1 context are marked (red squares). Directions (5′ to 3′) of the primers used to confirm the occurrence of the P0/Tn554/P1-containing circular variants are indicated (red arrows). The primer sets P1F2/P1R2, P1F1/P1R1, and P1F1/P1R2 yielded PCR products corresponding to hp2-ISSep1-like transposase-radC (4,439 bp-P0 circle), SAM-tnpA (6,568 bp-Tn554 circle), and optrA-tnpA (11,007 bp-P1 circle).
In the ST591 and ST711 LR E. faecalis porcine isolates, the basic P0 configuration was inserted downstream of a complete copy of the site-specific transposon, Tn554 (here designated the P1 context), while in ST710 isolate L8, it was inserted inside the transposase genes tnpB and tnpC of Tn554 (termed the P2 context). In the P1 context, the core araC-hp1-optrA is inserted into the extreme 3′ end of Tn554 with an accompanying duplication of the flanking radC and ISSep-1 transposase gene segments from P0. Comparison of the radC fragments flanking P0 within the P1 context suggested it had accumulated sequence from different sources. On the 5′ flank of P0, a 330-bp fragment derived from a radC gene is identical to a homologue from Listeria monocytogenes, while a 227 bp-radC fragment on the 3′ side of P0 is identical to a homologue from Bacterioides and Peptoniphilus species.
The P2 context has the core araC-hp1-optrA inserted between tnpB and tnpC of Tn554, truncating both coding sequences but does not have the duplication of radC gene segments seen in P1. Here, the radC fragments at the 5′ and 3′ Tn554 are identical to homologues in E. faecium, suggesting different origins and routes of transmission for P1 and P2 mobile elements.
The P3 and P4 contexts differ from P1 and P2. Rather than being associated with a Tn554 element, araC-hp1-optrA is inserted upstream of IS1216E, with variable gene content flanking the core at either end. The P4 context had a duplication of optrA, which was confirmed by PCR and Sanger sequencing.
Recombination and transposition events involved in the mobilization of optrA.
Recombination and transposition appear to be driving mobilization of P1 and P2. Interestingly, sequencing read counts for the core araC-hp1-optrA occurred at from 1- to 27-fold higher levels than for the host chromosome (see Fig. S1 in the supplemental material), suggesting some mechanism for amplification within the cell. To test for the ability of the optrA element to excise and form a hypothetical circular intermediate as implied, we constructed primers (see Table S1 in the supplemental material) to read outward off the ends of the element. When using genomic DNA from P1 and P2 containing E. faecalis isolates as the template, this primer set (P1F1+P1R1) produced an amplicon proving the existence of the predicted covalently closed circular intermediate (tnpA → SAM). P1 elements also contain a third radC fragment that could enable formation of additional circular intermediates. Primers sets P1F1+P1R2 and P1F2+P1R2 generated amplicons from P1-containing isolates (tnpA → optrA and araC → optrA), confirming that recombination between homologous radC segments can also lead to excision and mobilization of P1 and P0 elements, respectively. Neither P3- or P4-carrying isolates yielded detectable amplicons using any of the above primer combinations, highlighting the importance of the radC segments, the mechanism of Tn554 transposition, or both. The IS1216E transposase at the 3′ end of P3 and P4 elements suggests that their movement may be enabled through transposition rather than recombination.
Mobilization of the optrA-hp1-araC gene cluster in the Brazilian porcine LR E. faecalis strains.
To determine whether optrA in the various contexts can be transmitted to other strains, and, if so, the level of resistance conferred, filter matings were performed using graded levels of chloramphenicol (10, 20, or 25 μg/ml) for selection. Depending on the donor, E. faecalis OG1RF transconjugants were obtained at frequencies from 10−8 to 10−6 per donor cell (Table 2). Transconjugants were verified to contain oxazolidinone and phenicol resistance genes by PCR using primers listed in Table S1.
TABLE 2.
Antimicrobial resistance profiles and transferability of the oxazolidinone resistance genes
| Donor | Resistance genes | Transconjuganta | Transfer rate for donorb: |
Phenicol/oxazolidinone resistance gene(s) present |
MIC (μg/ml) for: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | optrA | poxtA | cfr | fexA | cat | CHL | FFC | LZD | TZD | |||
| L10 | aph(3′)-III, aac(6′)-aph(2″), spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | OG1RF-L10 | 3 × 10−8 | NT | NT | + | + | + | 128 | 64 | 8 | 1.0 | ||
| L11 | aph(3′)-III, aac(6′)-aph(2″), spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | NT | NT | NT | ||||||||||
| L12 | ant(6)-Ia, aph(3′)-III, aac(6′)-aph(2″), spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | NT | NT | NT | ||||||||||
| L13 | ant(6)-Ia, aph(3′)-III, aac(6′)-aph(2″), spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | OG1RF-L13 | 10−9 | NT | NT | + | + | + | 128 | 64 | 8 | 1.0 | ||
| L15 | ant(6)-Ia, aph(3′)-III, aac(6′)-aph(2″), spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | OG1RF-L15 | 5 × 10−8 | 5.2 × 10−9 | 1.6 × 10−9 | +/+/+ | −/−/− | −/−/− | +/+/+ | 128 | 64 | 8 | 1.0 | |
| L16 | ant(6)-Ia, aph(3′)-III, aac(6′)-aph(2″), spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | OG1RF-L16 | 1.1 × 10−8 | NT | NT | + | + | + | ≥256 | 128 | 32 | 1.0 | ||
| L18 | ant(6)-Ia, aph(3′)-III, aac(6′)-aph(2″), spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | NT | NT | NT | ||||||||||
| L21 | ant(6)-Ia, aph(3′)-III, aac(6′)-aph(2″), aadD, spc, lnu(B), lsa(A), erm(A), erm(B), optrA, cat, fex(A), tet(L), tet(M), dfrG | NT | NT | NT | ||||||||||
| L8 | ant(6)-Ia, aph(3′)-III, aac(6′)-aph(2″), spc, lnu(C), lnu(B), lnu(G), lsa(A), erm(A), erm(B), optrA, tet(L), tet(M), dfrG | OG1RF-L8 | NT | NT | SC | + | − | − | 64 | 32 | 8 | 0.5 | ||
| L14 | str, lnu(B), lsa(A), erm(B), optrA, cat, fex(A), tet(L), tet(M) | OG1RF-L14 | 6.4 × 10−7 | 8.2 × 10−8 | 5.8 × 10−8 | +/+/+ | −/−/− | +/+/+ | +/+/+ | 128 | 64 | 8 | 0.5 | |
| L17 | str, lnu(B), lsa(A), erm(B), optrA, cat, fex(A), tet(L), tet(M) | OG1RF-L17 | 3.7 × 10−6 | 1.8 × 10−8 | 1.7 × 10−8 | +/+/+ | −/−/− | +/+/+ | +/+/+ | 128 | 64 | 8 | 0.5 | |
| L19 | str, lnu(B), lsa(A), erm(B), optrA, cat, fex(A), tet(L), tet(M) | OG1RF-L19 | 3.4 × 10−6 | 2.8 × 10−8 | 2.7 × 10−8 | +/+/+ | +/+/+ | +/+/+ | 128 | 64 | 8 | 0.5 | ||
| L9 | str, lsa(A), optrA, cfr, fex(A), tet(L), tet(M), tet(S) | OG1RF-L9 | 4 × 10−7 | NT | SC | +/+ | −/+ | +/+ | −/− | 128 | 64 | 8 | 0.5 | |
Transconjugants were selected on BHI medium containing (A) 25 μg/ml chloramphenicol, 25 μg/ml fusidic acid, and 25 μg/ml rifampin; (B) 20 μg/ml chloramphenicol, 25 μg/ml fusidic acid, and 25 μg/ml rifampin; or (C) 10 μg/ml chloramphenicol, 25 μg/ml fusidic acid, and 25 μg/ml rifampin. PCR detection of the optrA, poxtA, cfr, fexA, and cat genes, in addition to chloramphenicol/florfenicol/linezolid/tedizolid MIC determination, was performed to confirm the transconjugants.
NT, no transconjugant; SC, small colonies (not countable).
Transfer of P1 from L15 donor to transconjugant E. faecalis OG1RF-L15 was mediated by an 82,898-bp plasmid, pL15 (GenBank accession number CP042214) (Fig. S2). An intact P1 element was identified in the transconjugant on pL15 and included the disrupted gene fragments of radC at its ends. pL15 carries a complete copy of a Tn916-like transposon, Tn6248, harboring the chloramphenicol acetyltransferase gene, cat, in addition to tetracycline resistance determinants tet(L) and tet(M), which encode an efflux protein from the major facilitator superfamily, and a ribosome protective protein, respectively. Tn6248 from pL15 exhibits 99% DNA identity to a Tn6248 described in an E. faecium isolate from swine in China (GenBank accession number KP834592). Plasmid pL15 also includes erm(A) and erm(B), which confer resistance to macrolides, lincosamides, and streptogramin B, as well as the spectinomycin resistance gene spc. The pL15 rep gene shares 97% DNA identity to repA (Rep9) encoded by the E. faecalis pheromone-responsive plasmid pAD1 (42). pL15 was carried by all naturally occurring ST591 and ST711 LR E. faecalis strains investigated in this study.
Transfer of P2 from LR E. faecalis L8 into OG1RF-L8 was mediated by a 91,525-bp plasmid, pL8 (GenBank accession number CP042217). pL8 possesses a rep9 gene that is 96% identical to that of E. faecalis pheromone-responsive plasmid pPD1 (43). Unlike all other isolates, cat and fexA genes do not occur in the donor LR E. faecalis L8, leaving optrA as the only known mechanism of chloramphenicol resistance in the donor and transconjugant. P2-containing transconjugants could only be selected using 10 μg/ml chloramphenicol, indicating that optrA alone provides lower-level resistance than the combination of optrA and cat, which occur together in isolates carrying pL15.
Intact P3 (GenBank accession number CP043725) was not mobilized from L14, L17, or L19 donors selected with 10, 20, or 25 μg/ml chloramphenicol. However, optrA was confirmed in transconjugants by PCR and Sanger sequencing, indicating that optrA can recombine and transfer independently of the P3 element. Lastly, P4 (GenBank accession number CP041776) was transferred intact from LR E. faecalis L9 and appeared to be inserted into a plasmid of >60 kb, which could not be closed by de novo assembly.
optrA from Brazilian porcine E. faecalis isolates shows signatures of both horizontal and vertical inheritance.
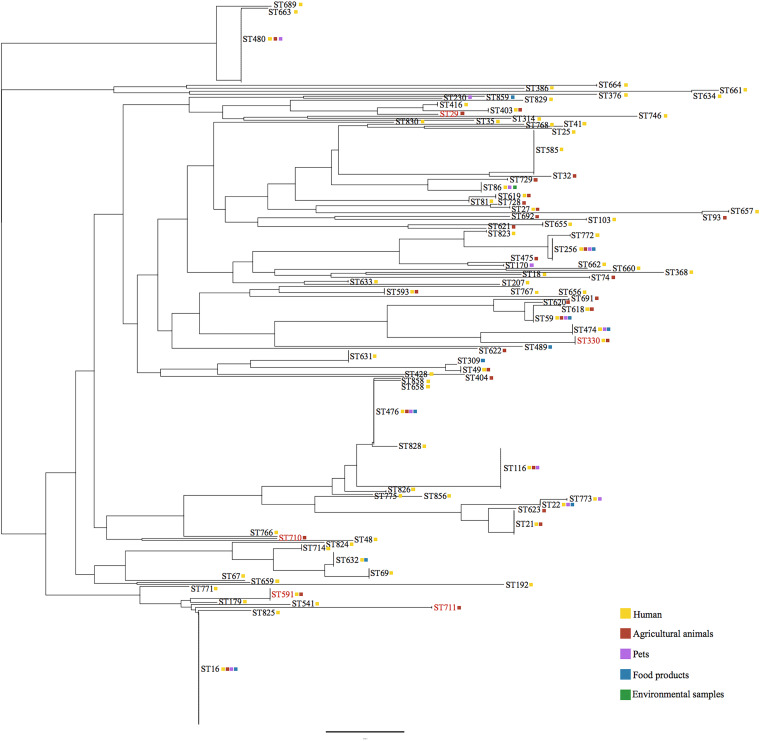
Globally, optrA occurs in multiple clonal clusters (CC) of E. faecalis that have been isolated from humans, animals, food products, and environmental sources (Fig. 2). In swine herds at various locations in Brazil, optrA was identified in genetically distant E. faecalis sequence types (ST29, ST330, ST591, ST710, and ST711). ST591, most common in this study, occurs in a CC that includes ST16 as a potential ancestral type (AT). Worldwide, ST16 accounts for the largest number of optrA-carrying E. faecalis isolates, most of them of human origin in China. ST591, a single-locus variant (SLV) of ST16, is the greatest source of optrA in animal hosts in this CC. Of the 31 STs that currently constitute this group, optrA has been identified in ST179 and ST541, in addition to ST16 and ST591.
FIG 2.
Multilocus sequence typing (MLST)-based phylogeny of optrA-carrying E. faecalis. The distribution of the optrA gene in the STs of E. faecalis isolated in Brazil and in other countries is color coded as follows: human (clinical samples) in yellow, agricultural animals in red, pets in pink, food products in blue, and environmental samples in green. STs identified in Brazil are highlighted in red.
ST711, also a carrier of P1, is not closely related to ST591 and is a singleton by eBURST analysis. ST710, which carries the P2 context, is also a singleton. Data for the CC that includes ST330 is rarer in the MLST database, and, so far, optrA is restricted to ST330 from animal hosts (swine and chicken), or to isolates of its double-locus variant (DLV) ST474 from humans. Lastly, ST29 is within a CC that has ST403 as a potential AT. ST403 was first identified in 2013 as a high-level gentamicin-resistant (HLGR) E. faecalis isolate from chicken meat in South Korea, and it now joins 5 SLV (ST29, ST244, ST292, ST416, and ST734), and 2 DLV (ST361 and ST400) identified in poultry, wild birds, rodents, and nonhospitalized humans. ST403-associated lineages, including the ones which carry optrA (ST29, ST403, and ST416), appear to be well adapted to animal hosts.
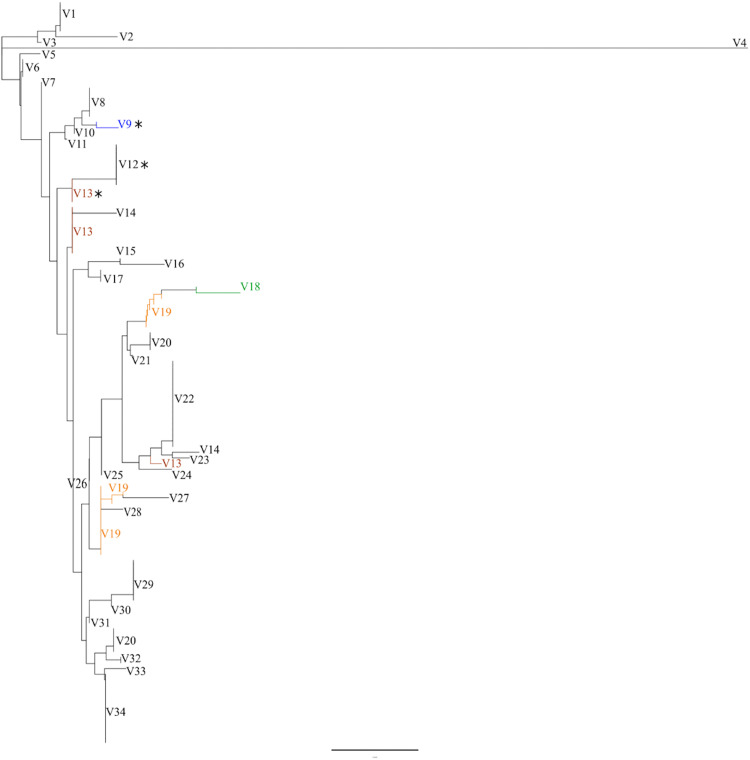
To determine the relatedness of the optrA genes studied here to homologous sequences in GenBank, single-nucleotide polymorphisms (SNPs) were mapped, and a SNP-based phylogeny (Fig. 3) was generated. This analysis shows that E. faecalis is the main reservoir of optrA diversity; 39 of all 49 optrA variants occur in multiple STs of E. faecalis. Seven optrA variants carried by E. faecalis also occur in strains of E. faecium, Enterococcus avium, S. suis, and Staphylococcus sciuri isolated in 6 different countries. Three OptrA protein variants encoded by four optrA variants were identified in the LR E. faecalis isolates from Brazil studied here. While V12 OptrA was found in the ST591 and ST711 E. faecalis isolates from various pig farms (P1 optrA context), V13 OptrA was unique to the ST330 lineage (P3 optrA context) at Paraná (PR) piggery A. V9 OptrA could be identified in distantly related STs of E. faecalis (ST29 and ST710), which came from different states of Brazil (P4 and P2 optrA contexts, respectively). The nature of these variations is summarized in Table S2.
FIG 3.
optrA alleles SNP-based phylogeny. optrA DNA sequences from the porcine LR E. faecalis isolated in Brazil were compared to all optrA variants available so far in the NCBI database. STs, sources, geographical locations, and GenBank accession numbers are indicated if known (see Table S2 in the supplemental material). A total of 34 OptrA variants are indicated by sequential numbering from top to bottom. The Brazilian OptrA variants (V9, V12, and V13) are marked with asterisks. optrA variants in which SNPs did not yield variations in the 655-amino acid sequence are highlighted by colors. Compared to the OptrA variant first described in China (ST116 E. faecalis E349) (2), variations in the 655-amino acid sequence were detected in V12, V13, and V9 OptrA variants as follows: V12 (Tyr176Asp, Ala350Val, and Gly393Asp), V13 (Tyr176Asp, and Gly393Asp), and V9 (Lys3Glu, Tyr176Asp, Gly393Asp, and Ile622Met).
DISCUSSION
Brazil is the fourth largest pork producer in the world, and Brazilian swine production currently represents 10% of global exports. Brazil imports breeding animals exclusively from the United States or Europe. There are no import records of meat from China, nor does Brazil have a history of exporting live pigs to any country. Although neither Brazilian nor Chinese swine production is directly tied to the import or export of animals, China is the largest importer of Brazilian meat. However, veterinary antimicrobial stewardship programs in Brazil and China have been similar over the last 20 years, mainly with respect to the phenicol, pleuromutilin, and macrolide classes, which have been largely used to control diseases, as well as growth promoters in swine production.
The pig farms selected for sample collection in this study utilize similar animal husbandry practices, biosecurity, and antimicrobial use, despite their geographical diversity. Even though chloramphenicol has been prohibited for veterinary use and animal feeding since 2003 in Brazil, florfenicol has been widely used for the treatment of gastrointestinal tract and respiratory infections in food-producing animals, for prevention of respiratory disease in immature pigs, and also for treatment of genitourinary tract infections in adult sows on pig farms throughout the country. The selective pressure due to the use of florfenicol has likely selected for resistance carriage by some ubiquitous constituent of the swine gut microbiota. E. faecalis so far is the main bacterium found to carry transmissible optrA (2–9, 13–15, 17, 18, 20–28). ST591 E. faecalis was identified here as being quite widespread in various swine herds located in different Brazilian states and may play a major role in optrA transmission.
Tn554 appears to be central in acquiring and transferring the P0 context in LR E. faecalis lineages ST591 and ST711. Flanking radC segments have been used for Tn554 recombination, favoring the mobilization of P0 and P1 circular intermediates. The ability of the araC-hp1-optrA gene cluster to excise and form a covalently closed circle appears to enhance its ability to integrate into the chromosome or other mobile genetic elements, as has been characterized elsewhere (44–46). The high numbers of read counts for the core araC-hp1-optrA, compared to sequences from the chromosome or other elements in the cell, suggest it is stable and possesses a mechanism for amplification within the cell. It is currently unclear what role phenicol selection may play in promoting excision, circularization, or potential amplification. In addition to Tn554, IS1216E appears to be involved in the mobilization of optrA-carrying segments in the Brazilian porcine LR E. faecalis lineages. The host range of Tn554 or IS1216E is unknown, making it difficult to estimate the risk for optrA spread beyond enterococci to other genera.
MATERIALS AND METHODS
Sample collection and bacterial strains.
From January 2012 to January 2013, rectal swabs were collected from 10 randomly selected piglets (45 days old) in each of 31 swine herds in 7 Brazilian states. Of those 310 swabs, 171 yielded colonies on bile esculin azide agar. Of 364 colonies screened by PCR using species-specific primers, 245 colonies were confirmed as E. faecalis, including 13 with MICs for linezolid of ≥4 mg/liter by broth microdilution. These 13 LR E. faecalis strains were isolated from healthy pigs belonging to 8 of the 31 swine herds, located in 6 different states of Brazil, and were selected for further study.
Genome sequencing and analysis.
DNA of the 13 LR E. faecalis strains and E. faecalis OG1RF transconjugants was extracted using the DNeasy blood and tissue kit (Qiagen, USA). Genome sequencing was performed on an Illumina MiSeq instrument (OGI core, Massachusetts Eye and Ear Infirmary Ocular Genomics Institute, Boston, MA, USA). Libraries were prepared using the Illumina Nextera XT kit, with modifications for 2 × 250-bp paired-end reads (Illumina Inc., USA). CLC Genomics Workbench 8.0.3 was used to generate assemblies de novo. The Rapid Annotation using Subsystem Technology (RAST) server and Prokaryotic Genome Annotation Pipeline (NCBI PGAP) were accessed for genome annotation. BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Center for Genomic Epidemiology (http://www.genomicepidemiology.org) utilities were used for comparisons and data analysis.
Filter mating and susceptibility testing.
Filter mating was performed using the LR E. faecalis strains as donors and E. faecalis OG1RF as the recipient, essentially as described previously (47). The filters were then washed in 5 ml of phosphate-buffered saline (PBS), and aliquots were spread on brain heart infusion (BHI) agar containing 25 μg/ml fusidic acid and 25 μg/ml rifampin to select for the OG1RF background and 10, 20 or 25 μg/ml of chloramphenicol to select for acquisition of the optrA gene. Donors and transconjugants were quantified by track dilution (48), and conjugation efficiency (CFU transconjugants per CFU donors) was calculated.
To confirm the presence in transconjugants of transferred genes inferred from the resistance phenotypes, PCR assays for optrA, cfr, poxtA, fexA, and cat, in addition to MIC determinations of chloramphenicol, florfenicol, linezolid, and tedizolid, were performed. Antimicrobial susceptibility of donors and transconjugants was performed using broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) guidelines (49, 50).
Data availability.
Complete genome sequences of E. faecalis strains L8, L9, L12, L14, and L15 have been deposited in GenBank under the nucleotide sequence accession numbers CP042216, CP018004, CP018102, CP043724, and CP042213, respectively. CP042217, CP041776, CP042214, and CP043725 have been assigned to pL8, optrA-carrying partial sequence pL9, pL15, and optrA-carrying partial sequence pL14, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by the Harvard-wide Program on Antibiotic Resistance, by NIH/NIAID grant AI083214, and by FAPESP scholarship 2014/27267-0.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sharkey LKR, O’Neill AJ. 2018. Antibiotic resistance ABC-F proteins: bringing target protection into the limelight. ACS Infect Dis 4:239–246. doi: 10.1021/acsinfecdis.7b00251. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, Wang D, Wang Z, Shen Y, Li Y, Feßler AT, Wu C, Yu H, Deng X, Xia X, Shen J. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 3.He T, Shen Y, Schwarz S, Cai J, Lv Y, Li J, Feßler AT, Zhang R, Wu C, Shen J, Wang Y. 2016. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J Antimicrob Chemother 71:1466–1473. doi: 10.1093/jac/dkw016. [DOI] [PubMed] [Google Scholar]
- 4.Cui L, Wang Y, Lv Y, Wang S, Song Y, Li Y, Liu J, Xue F, Yang W, Zhang J. 2016. Nationwide surveillance of novel oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Antimicrob Agents Chemother 60:7490–7493. doi: 10.1128/AAC.01256-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D, Li X-Y, Schwarz S, Yang M, Zhang S-M, Hao W, Du X-D. 2019. Tn6674 is a novel enterococcal optrA-carrying multiresistance transposon of the Tn554 family. Antimicrob Agents Chemother 63:e00809-19. doi: 10.1128/AAC.00809-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang Z-Z, Lei C-W, Kong L-H, Wang Y-L, Ye X-L, Ma B-H, Wang X-C, Li C, Zhang Y, Wang H-N. 2019. Detection of transferable oxazolidinone resistance determinants in Enterococcus faecalis and Enterococcus faecium of swine origin in Sichuan Province, China. J Glob Antimicrob Resist 19:333–337. doi: 10.1016/j.jgar.2019.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Hao W, Shan X, Li D, Schwarz S, Zhang S-M, Li X-S, Du X-D. 2019. Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J Antimicrob Chemother 74:1771–1775. doi: 10.1093/jac/dkz109. [DOI] [PubMed] [Google Scholar]
- 8.Zhou W, Gao S, Xu H, Zhang Z, Chen F, Shen H, Zhang C. 2019. Distribution of the optrA gene in Enterococcus isolates at a tertiary care hospital in China. J Glob Antimicrob Resist 17:180–186. doi: 10.1016/j.jgar.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y, Fan R, Wang Y, Lei L, Feßler AT, Wang Z, Wu C, Schwarz S, Wang Y. 2019. Analysis of combined resistance to oxazolidinones and phenicols among bacteria from dogs fed with raw meat/vegetables and the respective food items. Sci Rep 9:15500. doi: 10.1038/s41598-019-51918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan R, Li D, Wang Y, He T, Feßler AT, Schwarz S, Wu C. 2016. Presence of the optrA gene in methicillin-resistant Staphylococcus sciuri of porcine origin. Antimicrob Agents Chemother 60:7200–7205. doi: 10.1128/AAC.01591-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo D, Liu Y, Han C, Chen Z, Ye X. 2018. Phenotypic and molecular characteristics of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolated from pigs: implication for livestock-association markers and vaccine strategies. Infect Drug Resist 11:1299–1307. doi: 10.2147/IDR.S173624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang Y, Li D, Hao W, Schwarz S, Shan X, Liu B, Zhang S-M, Li X-S, Du X-D. 2019. A prophage and two ICESa2603-family integrative and conjugative elements (ICEs) carrying optrA in Streptococcus suis. J Antimicrob Chemother 74:2876–2879. doi: 10.1093/jac/dkz309. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda M, Sekizuka T, Matsui H, Suzuki K, Seki H, Saito M, Hanaki H. 2018. Complete genome sequence and characterization of linezolid-resistant Enterococcus faecalis clinical isolate KUB3006 carrying a cfr(B)-transposon on its chromosome and optrA-plasmid. Front Microbiol 9:2576. doi: 10.3389/fmicb.2018.02576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Said HS, Abdelmegeed ES. 2019. Emergence of multidrug resistance and extensive drug resistance among enterococcal clinical isolates in Egypt. Infect Drug Resist 12:1113–1125. doi: 10.2147/IDR.S189341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elghaieb H, Tedim AP, Abbassi MS, Novais C, Duarte B, Hassen A, Peixe L, Freitas AR. 2020. From farm to fork: identical clones and Tn6674-like elements in linezolid-resistant Enterococcus faecalis from food-producing animals and retail meat. J Antimicrob Chemother 75:30–35. doi: 10.1093/jac/dkz419. [DOI] [PubMed] [Google Scholar]
- 16.Brenciani A, Morroni G, Vincenzi C, Manso E, Mingoia M, Giovanetti E, Varaldo PE. 2016. Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J Antimicrob Chemother 71:1118–1119. doi: 10.1093/jac/dkv438. [DOI] [PubMed] [Google Scholar]
- 17.Cavaco LM, Korsgaard H, Kaas RS, Seyfarth AM, Leekitcharoenphon P, Hendriksen RS. 2017. First detection of linezolid resistance due to the optrA gene in enterococci isolated from food products in Denmark. J Glob Antimicrob Resist 9:128–129. doi: 10.1016/j.jgar.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Deshpande LM, Castanheira M, Flamm RK, Mendes RE. 2018. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: results from the SENTRY Antimicrobial Surveillance Program. J Antimicrob Chemother 73:2314–2322. doi: 10.1093/jac/dky188. [DOI] [PubMed] [Google Scholar]
- 19.Morroni G, Brenciani A, Antonelli A, D’Andrea MM, Di Pilato V, Fioriti S, Mingoia M, Vignaroli C, Cirioni O, Biavasco F, Varaldo PE, Rossolini GM, Giovanetti E. 2018. Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an Enterococcus faecium clinical isolate. Front Microbiol 9:2189. doi: 10.3389/fmicb.2018.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender JK, Fleige C, Lange D, Klare I, Werner G. 2018. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int J Antimicrob Agents 52:819–827. doi: 10.1016/j.ijantimicag.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Càmara J, Camoez M, Tubau F, Pujol M, Ayats J, Ardanuy C, Domínguez MÁ. 2019. Detection of the novel optrA gene among linezolid-resistant enterococci in Barcelona, Spain. Microb Drug Resist 25:87–93. doi: 10.1089/mdr.2018.0028. [DOI] [PubMed] [Google Scholar]
- 22.Tsilipounidaki K, Gerontopoulos A, Papagiannitsis C, Petinaki E. 2019. First detection of an optrA-positive, linezolid-resistant ST16 Enterococcus faecalis from human in Greece. New Microbes New Infect 29:100515. doi: 10.1016/j.nmni.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sassi M, Guérin F, Zouari A, Beyrouthy R, Auzou M, Fines-Guyon M, Potrel S, Dejoies L, Collet A, Boukthir S, Auger G, Bonnet R, Cattoir V. 2019. Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006–16. J Antimicrob Chemother 74:1469–1472. doi: 10.1093/jac/dkz097. [DOI] [PubMed] [Google Scholar]
- 24.Argudín MA, Youzaga S, Dodémont M, Heinrichs A, Roisin S, Deplano A, Nonhoff C, Hallin M. 2019. Detection of optrA-positive enterococci clinical isolates in Belgium. Eur J Clin Microbiol Infect Dis 38:985–987. doi: 10.1007/s10096-019-03504-3. [DOI] [PubMed] [Google Scholar]
- 25.Cavaco LM, Bernal JF, Zankari E, Léon M, Hendriksen RS, Perez-Gutierrez E, Aarestrup FM, Donado-Godoy P. 2017. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother 72:678–683. doi: 10.1093/jac/dkw490. [DOI] [PubMed] [Google Scholar]
- 26.Tyson GH, Sabo JL, Hoffmann M, Hsu C-H, Mukherjee S, Hernandez J, Tillman G, Wasilenko JL, Haro J, Simmons M, Wilson Egbe W, White PL, Dessai U, Mcdermott PF. 2018. Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J Antimicrob Chemother 73:3254–3258. doi: 10.1093/jac/dky369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freitas AR, Elghaieb H, León-Sampedro R, Abbassi MS, Novais C, Coque TM, Hassen A, Peixe L. 2017. Detection of optrA in the African continent (Tunisia) within a mosaic Enterococcus faecalis plasmid from urban wastewaters. J Antimicrob Chemother 72:3245–3251. doi: 10.1093/jac/dkx321. [DOI] [PubMed] [Google Scholar]
- 28.Elghaieb H, Freitas AR, Abbassi MS, Novais C, Zouari M, Hassen A, Peixe L. 2019. Dispersal of linezolid-resistant enterococci carrying poxtA or optrA in retail meat and food-producing animals from Tunisia. J Antimicrob Chemother 74:2865–2869. doi: 10.1093/jac/dkz263. [DOI] [PubMed] [Google Scholar]
- 29.Xiong L, Kloss P, Douthwaite S, Andersen NM, Swaney S, Shinabarger DL, Mankin AS. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J Bacteriol 182:5325–5331. doi: 10.1128/jb.182.19.5325-5331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillai SK, Sakoulas G, Wennersten C, Eliopoulos GM, Moellering RC Jr, Ferraro MJ, Gold HS. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J Infect Dis 186:1603–1607. doi: 10.1086/345368. [DOI] [PubMed] [Google Scholar]
- 31.Prystowsky J, Siddiqui F, Chosay J, Shinabarger DL, Millichap J, Peterson LR, Noskin GA. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob Agents Chemother 45:2154–2156. doi: 10.1128/AAC.45.7.2154-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke JB, Hilgers M, Shaw KJ. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother 53:5275–5278. doi: 10.1128/AAC.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother 56:603–612. doi: 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, Zhang Q, Wu C, Shen J. 2013. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother 57:42–48. doi: 10.1128/AAC.01605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 36.Deshpande LM, Ashcraft DS, Kahn HP, Pankey G, Jones RN, Farrell DJ, Mendes RE. 2015. Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 59:6256–6261. doi: 10.1128/AAC.01473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antonelli A, D’Andrea MM, Brenciani A, Galeotti CL, Morroni G, Pollini S, Varaldo PE, Rossolini GM. 2018. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J Antimicrob Chemother 73:1763–1769. doi: 10.1093/jac/dky088. [DOI] [PubMed] [Google Scholar]
- 38.Locke JB, Zurenko GE, Shaw KJ, Bartizal K. 2014. Tedizolid for the management of human infections: in vitro characteristics. Clin Infect Dis 58 Suppl 1:S35–42. doi: 10.1093/cid/cit616. [DOI] [PubMed] [Google Scholar]
- 39.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lebreton F, Manson AL, Saavedra JT, Straub TJ, Earl AM, Gilmore MS. 2017. Tracing the enterococci from Paleozoic origins to the hospital. Cell 169:849–861.e13. doi: 10.1016/j.cell.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clewell DB. 2007. Properties of Enterococcus faecalis plasmid pAD1, a member of a widely disseminated family of pheromone-responding, conjugative, virulence elements encoding cytolysin. Plasmid 58:205–227. doi: 10.1016/j.plasmid.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Fujimoto S, Tomita H, Wakamatsu E, Tanimoto K, Ike Y. 1995. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J Bacteriol 177:5574–5581. doi: 10.1128/jb.177.19.5574-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locke JB, Rahawi S, Lamarre J, Mankin AS, Shaw KJ. 2012. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob Agents Chemother 56:332–340. doi: 10.1128/AAC.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazimierczak KA, Flint HJ, Scott KP. 2006. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob Agents Chemother 50:2632–2639. doi: 10.1128/AAC.01587-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmieri C, Mingoia M, Massidda O, Giovanetti E, Varaldo PE. 2012. Streptococcus pneumoniae transposon Tn1545/Tn6003 changes to Tn6002 due to spontaneous excision in circular form of the erm(B) and aphA3-containing macrolide-aminoglycoside-streptothricin (MAS) element. Antimicrob Agents Chemother 56:5994–5997. doi: 10.1128/AAC.01487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaworski DD, Clewell DB. 1994. Evidence that coupling sequences play a frequency-determining role in conjugative transposition of Tn916 in Enterococcus faecalis. J Bacteriol 176:3328–3335. doi: 10.1128/jb.176.11.3328-3335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jett BD, Hatter KL, Huycke MM, Gilmore MS. 1997. Simplified agar plate method for quantifying viable bacteria. Biotechniques 23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 49.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and diffusion susceptibility tests for bacteria isolated from animals: second informational supplement VET01-S2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 50.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete genome sequences of E. faecalis strains L8, L9, L12, L14, and L15 have been deposited in GenBank under the nucleotide sequence accession numbers CP042216, CP018004, CP018102, CP043724, and CP042213, respectively. CP042217, CP041776, CP042214, and CP043725 have been assigned to pL8, optrA-carrying partial sequence pL9, pL15, and optrA-carrying partial sequence pL14, respectively.