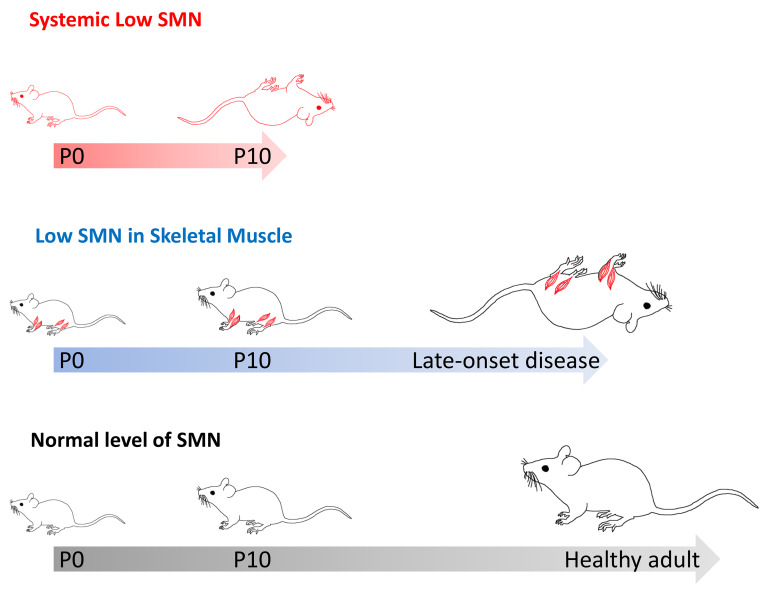
Abstract
Paucity of the survival motor neuron (SMN) protein triggers the oft-fatal infantile-onset motor neuron disorder, spinal muscular atrophy (SMA). Augmenting the protein is one means of treating SMA and recently led to FDA approval of an intrathecally delivered SMN-enhancing oligonucleotide currently in use. Notwithstanding the advent of this and other therapies for SMA, it is unclear whether the paralysis associated with the disease derives solely from dysfunctional motor neurons that may be efficiently targeted by restricted delivery of SMN-enhancing agents to the nervous system, or stems from broader defects of the motor unit, arguing for systemic SMN repletion. We investigated the disease-contributing effects of low SMN in one relevant peripheral organ — skeletal muscle — by selectively depleting the protein in only this tissue. We found that muscle deprived of SMN was profoundly damaged. Although a disease phenotype was not immediately obvious, persistent low levels of the protein eventually resulted in muscle fiber defects, neuromuscular junction abnormalities, compromised motor performance, and premature death. Importantly, restoring SMN after the onset of muscle pathology reversed disease. Our results provide the most compelling evidence yet for a direct contributing role of muscle in SMA and argue that an optimal therapy for the disease must be designed to treat this aspect of the dysfunctional motor unit.
Keywords: Neuroscience
Keywords: Neuromuscular disease
Introduction
Homozygous mutations in the survival of motor neuron 1 (SMN1) gene result in reduced levels of the SMN protein and trigger the infantile-onset neuromuscular disease, spinal muscular atrophy (SMA) (1–3). Complete loss of SMN is embryonic lethal (4). Indeed, patients with SMA invariably express residual protein from an SMN1 paralogue called SMN2. The inability of SMN2 to compensate for SMN1 stems from a silent C→T transition in exon 7 of the paralogue. The single base change alters the efficiency with which exon 7 is spliced into SMN transcripts such that SMN2 expresses mostly an SMNΔ7 isoform (5, 6). The SMNΔ7 mRNA is abundantly expressed. However, the corresponding truncated protein is unstable and degraded (2, 3, 7). Still, the expression of low levels of the FL-SMN protein from SMN2 is sufficient to ensure embryonic development and live births. In contrast, early postnatal development, particularly that of the neuromuscular system, is profoundly affected in the absence of SMN1. Accordingly, most patients with SMA rapidly develop a paralytic phenotype whose signature features include spinal motor neuron loss and proximal muscle atrophy (8). Considering the housekeeping function of the SMN protein in orchestrating the splicing cascade (9), the selective loss of motor neurons in SMA remains largely unexplained.
Although most patients with SMA are severely affected, there is a spectrum of phenotypes that extends to truly mild disease. Mild SMA is generally associated with a greater number of SMN2 copies (10, 11). Indeed, in one instance, a patient with 8 SMN2 copies was reportedly asymptomatic (12). The disease-modifying effect of elevated SMN2 copies, and thus SMN protein, was subsequently demonstrated directly in model mice null for murine Smn but harboring 8 copies of the SMN2 gene (13). The WT phenotype of such mice provided the first real rationale for targeting SMN2 to augment the SMN protein as a means to a treatment. Efforts to implement this strategy were further bolstered when it was shown that a regulatory element, ISS-N1, downstream of SMN2 exon 7 could be effectively targeted to boost FL-SMN levels (14, 15). The outcome of treating severely affected SMA model mice with splice-switching oligonucleotides (SSOs) that targeted the regulatory element was remarkable. Timely administration of the therapeutic oligo–enhanced SMN levels restored neuromuscular function and greatly extended survival (16, 17). The SSO used to target ISS-N1 and modulate SMN2 gene expression was eventually developed into the SMA therapeutic drug, Spinraza (18).
Despite the clinical promise of Spinraza, questions about its long-term efficacy abound (19, 20). A chief concern stems from the spatial requirements for the SMN protein and the manner in which the drug is currently administered — intrathecally. Such manner of treatment is expected to result in robust restoration of the SMN protein to CNS tissue. However, peripheral organs, including muscle, would remain deprived of the potential salutary effects of normal protein concentrations. That low SMN levels have disruptive effects beyond the motor neurons is increasingly evident (21, 22). Of particular interest, owing to the decidedly neuromuscular SMA phenotype, is the potential for such effects in muscle and their contribution to the overall disease. Studies to gauge the contributing role of muscle in SMA have arrived at disparate conclusions (23–30). Here, we have undertaken to resolve the uncertainty in the most direct manner yet by characterizing the disease consequences of selectively depriving muscle of SMN. Using model mice that were selectively depleted of the protein in skeletal muscle, but nevertheless engineered to express 1 or 2 copies of the SMN2 gene — a typical, severe SMA genotype — we showed that low SMN in this tissue was profoundly damaging. In animals with a single SMN2 copy, disease onset was rapid, with a median survival of approximately P21. In contrast, disease in animals bearing 2 copies of the SMN2 gene appeared late, but nevertheless resulted in a broad range of muscle phenotypes, including morphological damage to myofibers and neuromuscular junctions (NMJs), poor motor performance, and an inability to optimally generate muscular force. Akin to the disease in mutants with 1 copy of SMN2, these defects eventually shortened lifespan to approximately 13 months. Notwithstanding these defects, restoring SMN to the skeletal muscle of symptomatic mice proved beneficial. We conclude that muscle is a critical cellular site of action of the SMN protein and that expressing protein from 1 or 2 copies of the SMN2 gene is insufficient to prevent the onset of a severe, cell-autonomous muscle phenotype. Our results imply that the most optimal SMN-enhancing treatment regimens for SMA will be those that restore the protein to skeletal muscle.
Results
MyoD-iCre effects robust, muscle-specific inactivation of the SmnF7 allele.
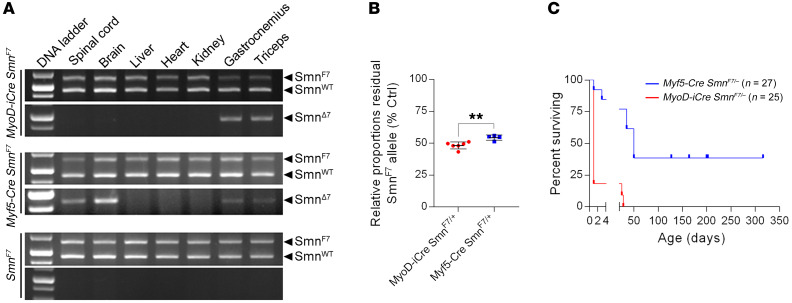
To investigate the cell-autonomous effects of depleting SMN in skeletal muscle, we began by testing which of 2 muscle Cre drivers, MyoD-iCre and Myf5-Cre (31, 32), was better suited for our studies. Each drives expression in muscle progenitor cells as early as E8 (33–35), and has been used to selectively deplete or restore disease-related proteins in skeletal muscle (27, 36). To determine the tissue specificity of the 2 Cre lines, we separately bred the transgenic mice to animals harboring an inducible Smn deletion allele (SmnF7) bearing loxP sites on either side of exon 7 (37, 38). We then used PCR to examine which of the tissues of the double transgenics exhibited evidence of Cre-mediated recombination. As expected, we detected the presence of the deleted (Δ7) allele in the proximal and distal muscles of each of the double transgenic MyoD-iCre SmnF7 and Myf5-Cre SmnF7 animals (Figure 1A). However, and consonant with other reports (27, 39), whereas recombination of the SmnF7 allele was restricted to the skeletal muscle of the MyoD-iCre SmnF7 mice, it was also detected in the CNS tissue of the Myf5-Cre SmnF7 animals; the latter finding precluded the Myf5-Cre line for this study. We nevertheless proceeded to quantify the relative efficiency with which each of the Cre drivers effected SmnF7 inactivation in skeletal muscle. To directly measure how robustly each of the drivers converted the SmnF7 allele to the deleted SmnΔ7 form, we used Q-PCR on genomic DNA from the skeletal muscle of P7 MyoD-iCre SmnF7/+ and Myf5-Cre SmnF7/+ animals, and estimated relative levels of residual SmnF7 allele in each set of mutants. DNA from the muscle tissue of the MyoD-iCre SmnF7/+ mice contained lower levels of the SmnF7 allele than DNA from the muscle of Myf5-Cre SmnF7/+ mice (Figure 1B), suggesting that MyoD-iCre is more robust in inactivating the floxed allele.
Figure 1. Robust, muscle-specific inactivation of the SmnF7 allele with MyoD-iCre.
(A) PCR analysis of genomic DNA from tissue of mice harboring the SmnF7 allele and either MyoD-iCre or Myf5-Cre. The inactivated SmnΔ7 allele is only detected in muscle of the MyoD-iCre mice, whereas it is seen in ectopic locations of Myf5-Cre transgenics. (B) Q-PCR data comparing the efficiency of SmnF7 inactivation in muscle of mice with either MyoD-iCre or Myf5-Cre driver. **P < 0.01, t test, n ≥ 4 mice of each cohort. (C) Kaplan-Meier survival curves of Myf5-Cre SmnF7/– and MyoD-iCre SmnF7/– mice. P < 0.01 between groups, log-rank test.
As a second, indirect means of determining recombination efficiency, we established crosses to generate MyoD-iCre SmnF7/– and Myf5-Cre SmnF7/– mutants. In such mutants, the SMN protein is expected to be completely ablated in muscle cells if SmnF7 is converted to SmnΔ7. Since such ablation is incompatible with the survival of the cell and thus the organism (4), the relative incidence of the respective Cre SmnF7/– mutants and the severity of disease in any resulting animals — as assessed by lifespan — is an indirect but sensitive measure of recombination efficiency at the SmnF7 allele. Given the less than complete conversion of SmnF7 to SmnΔ7, as detected in our PCR experiments, we were not surprised to obtain live and/or stillborn SmnΔ7/– mutants carrying each of the Cre drivers. However, consistent with our Q-PCR results suggesting that MyoD-iCre effects superior recombination, χ2 analysis indicated significantly fewer than expected MyoD-iCre SmnF7/– mutants at birth. In contrast, Myf5-Cre SmnF7/– mutants and their littermates were born at predicted frequencies (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI131989DS1). Moreover, whereas the preponderance (~90%) of MyoD-iCre SmnF7/– mutants were either stillborn or perished at P0, approximately 75% of the Myf5-Cre SmnF7/– mice not only lived to adulthood but also bred vigorously; only 2 MyoD-iCre SmnF7/– mutants lived beyond P1 and neither survived past P30 (Figure 1C). Collectively, the data suggested that the MyoD-iCre transgenic line constitutes a more robust driver than its Myf5-Cre counterpart, and thus is a more prudent choice of reagent with which to evaluate the cell-autonomous effects of low SMN expression in muscle. In a final confirmation of our choice of the MyoD-iCre driver with which to deplete SMN in muscle, we generated ROSA-YFP mice (40) with or without the Cre transgene. As expected, we detected a strong YFP signal in the majority of the muscle fibers of MyoD-iCre ROSA-YFP, but not ROSA-YFP animals (Supplemental Figure 1A). YFP fluorescence was not detected in the spinal cord, brain, or liver tissue of the MyoD-iCre ROSA-YFP mice (Supplemental Figure 1B). Accordingly, the remaining studies were conducted using the MyoD-iCre line.
Severe, rapid-onset disease is observed in single-copy SMN2 mutants depleted of SMN in muscle.
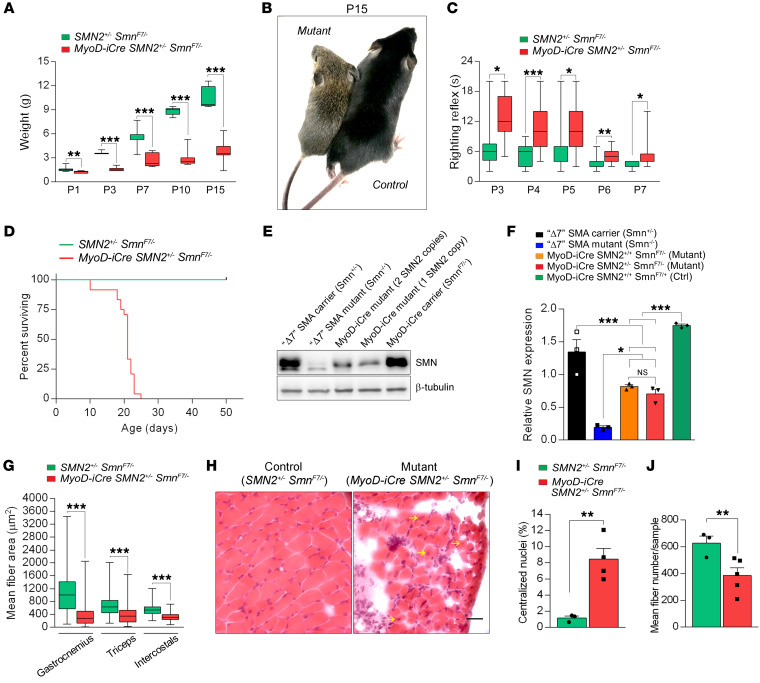
Complete tissue ablation of SMN disrupts snRNP biogenesis and is therefore lethal (4). In addition, a complete ablation of SMN fails to faithfully model human SMA, in which residual levels of the protein derive from 1 or more SMN2 copies. Accordingly, to generate mutants expressing residual SMN only in skeletal muscle, we bred SMA carrier mice homozygous for the SMN2 transgene (13) and heterozygous for the MyoD-iCre driver (MyoD-iCre SMN2+/+ Smn+/–) with SmnF7/F7 animals. MyoD-iCre SMN2+/– SmnF7/– mutants were obtained with the expected frequency of 12.5%, but exhibited an overt phenotype, as assessed by body weight, within 24 hours of birth (Figure 2, A and B). The mutants also exhibited reduced movement, failed a test of motor performance (Figure 2C and Supplemental Video 1), and were moribund by approximately P14. They were frequently found emaciated, disheveled, and exhibiting signs of distressed breathing (Supplemental Video 2). Median survival was shortened to around P21 (Figure 2D). As expected, levels of the SMN protein in the skeletal muscle tissue of P7 mutants were significantly lower and less than half the levels of protein in carrier SMA mice from a commonly employed (41) line of severe (type 1) mutants (Figure 2, E and F). We conclude that the mutants expressed approximately 25% of WT SMN protein and that the overt disease phenotype observed was indeed a consequence of selectively reducing protein in the muscle tissue of the animals.
Figure 2. Severe, early-onset disease in MyoD-iCre SmnF7/– mutants bearing 1 SMN2 copy.
(A) Graph depicts reduced weight of mutants; t test, n ≥ 10 mice of each set. (B) A typical mutant and control littermate reflecting the smaller size of the former. (C) Mutants perform poorly in a righting reflex test; t test, n ≥ 10 mice of each cohort. (D) Kaplan-Meier survival curves depict reduced lifespan of mutants. P < 0.0001 between groups, log-rank test, n = 25 mice from each group. (E) Western blot showed reduced SMN protein in skeletal muscle of a MyoD-iCre SMN2+/– SmnF7/– mutant. (F) Relative SMN levels in skeletal muscle of the mutants and relevant controls; 1-way ANOVA, n ≥ 3 mice of each cohort. (G) Limb and respiratory muscle fibers are markedly reduced in size in P15 mutants; t test, n ≥ 600 fibers from n = 3 mice of each cohort. (H) H&E-stained transverse sections of intercostal muscles from a P15 mutant and control showing tissue loss, and fibers with central nuclei (arrows) in the former. Scale bar: 20 μm. Quantification of (I) central nuclei and (J) fiber numbers in intercostal muscle of P15 mutants and controls. t test, n = 3–5 mice from each group. *P < 0.05; **P < 0.01; ***P < 0.001 for all analyses.
Considering the severe, rapid-onset overt phenotype and the well-known effects of systemic depletion of the SMN protein on the motor unit, we next examined the MyoD-iCre SMN2+/– SmnF7/– mutants for evidence of neuromuscular pathology. We first assessed muscle fiber morphology in representative distal (gastrocnemius), proximal (triceps), and respiratory (intercostal) muscles. Myofiber areas in the mutants were reduced in all 3 muscles at 2 to 3 weeks of age (Figure 2G and Supplemental Figure 2, A–C). A more detailed analysis of the intercostal muscles at P15 demonstrated that this abnormality was accompanied by distinct degenerative pathology characterized by areas devoid of muscle fibers and often harboring infiltrates of monocytes or macrophages, and significant numbers of myofibers containing abnormally localized central nuclei (Figure 2, H and I). Quantification of the myofibers indicated that roughly a third of the cells in this respiratory muscle had been lost (Figure 2J), a likely origin of severe dysfunction of the muscle, labored breathing (also see Supplemental Video 2), and eventual death of the mutant animals. Interestingly, these defects were less significant at P7, but nevertheless suggestive of a postnatal degeneration of muscle in the MyoD-iCre SMN2+/– SmnF7/– mutants (Supplemental Figure 2, D–F). Moreover, the abnormalities were restricted to skeletal muscle because an examination of cardiac muscle, in which MyoD-iCre is not expressed, appeared no different in mutants and controls as determined by myofiber area and the thickness of the wall of the left ventricle (Supplemental Figure 2, G–I).
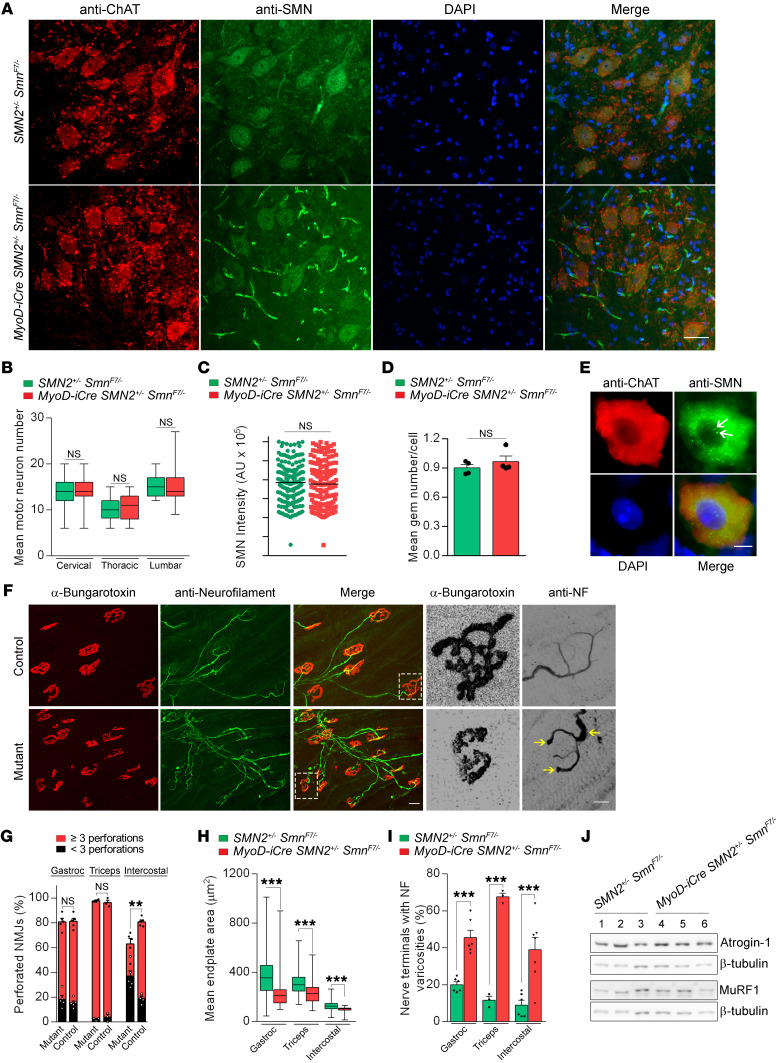
SMA is defined by motor neuron loss. Selectively depleting SMN in these cells is sufficient to trigger their loss (42), but the role of diseased muscle in the neurodegenerative process has not been examined. We therefore quantified and characterized motor neuron somas in our MyoD-iCre SMN2+/– SmnF7/– mutants. Cell body counts indicated that despite low SMN in muscle, there was no loss of the spinal motor neurons (Figure 3, A and B). Moreover, we found neither evidence of reduced SMN staining in the soma of the motor neurons nor a decrease in the number of gems, subnuclear foci in which the protein localizes (Figure 3, C–E, and ref. 43).
Figure 3. NMJ defects in MyoD-iCre SmnF7/– mutants with 1 copy of the SMN2 gene.
(A) Transverse sections of the lumbar (L1–L3) spinal cord of a P21 mutant and control. Scale bar: 25 μm. Quantified results of (B) spinal motor neuron counts (L1–L3, T7–T10, C5–C6); (C) SMN signal intensity in the motor neurons; (D) nuclear gem counts in the motor neurons of the mice. NS: P > 0.05, t test, n > 150 cells and n = 4 mice from each group. (E) A lumbar motor neuron from a mutant showing robust SMN signal and the presence of nuclear gems (arrows). Scale bar: 8 μm. (F) NMJs in the intercostal muscles of mutant and control mice. Included are high-magnification images of typical endplates from each animal illustrating reduced complexity and presence of terminal varicosities engorged with NF (arrows) in the mutant. Scale bars: 50 μm and 15 μm, respectively. Graphs of (G) NMJ complexity, (H) endplate size, and (I) NMJ morphology in P21 mutants and controls. **P < 0.01; ***P < 0.001, t test, n ≥ 100 NMJs from n ≥ 3 mice from each group. (J) Atrogene protein expression in mutants and controls showed little change in these markers of denervation. NS: P > 0.05, t test, n = 3 mice.
Although disease originating in the muscle may not affect the proximal motor neuron, chronic myopathy does disrupt the postsynaptic compartment and eventually compromise the morphology and function of the neuromuscular synapse (44). Moreover, it is unclear whether low SMN in muscle contributes to or exacerbates NMJ defects observed in SMA. Accordingly, we carried out a careful morphological analysis of the NMJs in our mutants. We found that acetylcholine receptor (AChR) clusters in all 3 muscles of P17–P21 mutants had acquired the typical pretzel-like structure of mature NMJs (Figure 3F). However, a careful assessment of their relative complexities based on the number of perforations in the clusters indicated that although endplates in the gastrocnemii and triceps were no different between mutants and controls, those of the intercostal were less elaborate in the mutants (Figure 3G). Moreover, consistent with the smaller size of mutant muscle, the clusters in all 3 muscles were found to be relatively diminutive (Figure 3, F and H). An examination of the muscle nerve terminals in the mutant mice revealed an increase in varicosities swollen with neurofilament (NF) protein, a feature reminiscent of defects observed in model mice expressing systemically low SMN protein (refs. 45–48 and Figure 3I). However, evidence of denervation was not detected in either immunohistochemical studies of the NMJ or by quantifying the atrogenes, atrogin-1 and MuRF1, which are routinely upregulated in denervated muscle (Figure 3J). Still, the observed defects in combination with the muscle pathology and overt phenotype of the MyoD-iCre SMN2+/– SmnF7/– mutants lead us to 2 conclusions. First, the results suggest that low SMN in skeletal muscle is sufficient to cause disease. Second, they demonstrate that pathology originating in muscle is capable of triggering, most likely in a retrograde fashion, abnormalities in the nerves. Accordingly, SMN paucity in muscle acts in both a cell-autonomous and non–cell-autonomous fashion to contribute to cellular defects and the overall SMA phenotype.
Myopathy is unmasked after muscle injury in young, phenotypically normal mutants bearing 2 SMN2 copies.
SMA in patients and model mice harboring a single SMN2 copy constitutes the most severe form (type 0) of the disease possible and often results in death in utero (49–51). Thus, the prevalence of this form of the disease is low (52), and the severity of the phenotype in MyoD-iCre SmnF7/– mutants hemizygous for the SMN2 transgene is perhaps not unexpected. To determine the effects of expressing SMN in the mutants’ muscle from 2 copies of the gene, a situation more common among human SMA patients, we generated mice homozygous for SMN2 (MyoD-iCre SMN2+/+ SmnF7/–).
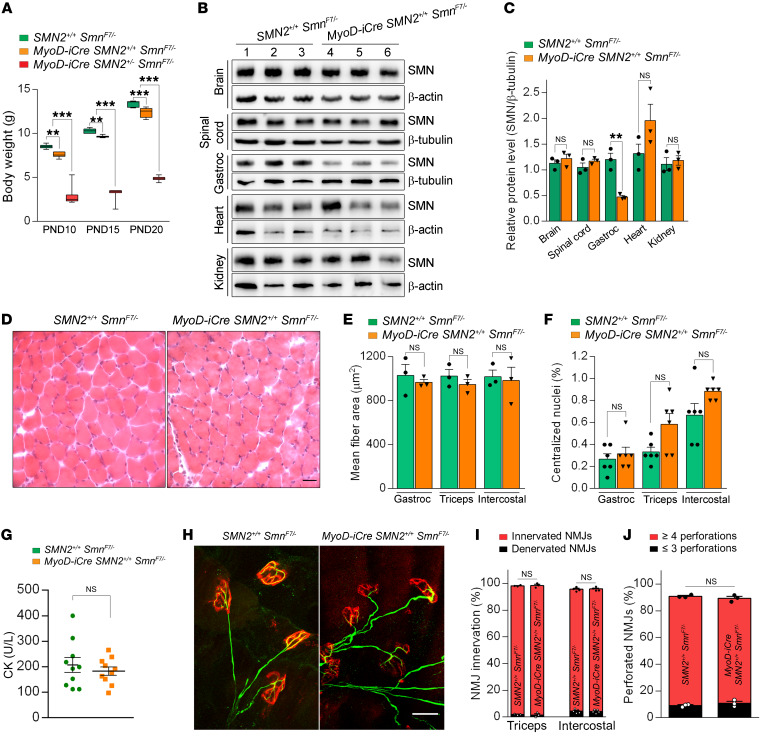
In contrast to the unmistakable, rapid-onset phenotype discerned in MyoD-iCre mutants harboring a single SMN2 copy, we detected only subtle abnormalities in young adult mice expressing 2 copies of the gene. Thus, for instance, although the latter mutants did exhibit a modestly reduced body weight in the second postnatal week of life compared with controls lacking the Cre driver (Figure 4A), aggregate growth curves between P0 and P21 exhibited a clear upward trajectory (Supplemental Figure 3A). Moreover, the mutants displayed no weakness in righting ability as neonates (Supplemental Figure 3B) or on the rotarod at 6 weeks of age (Supplemental Figure 3C), and exhibited no reduction in lifespan between birth and P60 (16 of 16 mutants alive; 18 of 18 controls alive). SMN concentrations in the skeletal muscle of 1-month-old mutants remained low, at approximately 21% of WT levels (Figure 4, B and C); levels of the protein in other tissues were not reduced.
Figure 4. Little cellular pathology in young adult MyoD-iCre SmnF7/– mutants bearing 2 SMN2 copies.
(A) Comparisons of body weights of controls and mutants harboring 1 or 2 SMN2 copies; 1-way ANOVA, n ≥ 10 mice of each cohort. (B) Western blot results showing selective depletion of SMN protein in skeletal muscle of a P30 MyoD-iCre SMN2+/+ SmnF7/– mutant. (C) Quantified levels of SMN protein in P30 mutant and control mice; t test, n ≥ 3 mice from each group. (D) Transverse sections of H&E-stained gastrocnemius muscles from a P30 mutant and control did not reveal major morphological differences between the two. Scale bar: 25 μm. Graphs of (E) myofiber areas, (F) fibers containing central nuclei, and (G) serum creatine kinase (CK) levels in young mutants and controls. NS: P > 0.05, t test, n > 150 fibers from n = 3 mice from each group for results in E and F; NS: P > 0.05, t test, n = 10 mice for CK values. (H) NMJs from the triceps of a P30 mutant and a control showing similar morphology of pre- and postsynaptic compartments in the 2 mice. Scale bar: 30 μm. (I) Graph of the extent of NMJ innervation in P30 mutants and controls. (J) Graph depicting endplate complexity in P30 mutants and control littermates. NS: P > 0.05, Fisher’s exact test, n ≥ 300 NMJs from n = 3 mice from each group of mice for analyses in I and J. **P < 0.01; ***P < 0.001.
Given evidence of neuromuscular pathology in the single-copy SMN2 MyoD-iCre mutants, we proceeded to examine the muscle and distal motor unit of young adult mutants harboring 2 copies of the human transgene. No gross muscle abnormalities were noted. Despite the somewhat smaller weight of the mutants, myofibers in the muscles examined were largely equivalent in size to those of controls (Figure 4, D and E), looked morphologically normal, and did not display pathology in the form of abnormally localized myonuclei in the gastrocnemius and intercostal muscles. The more vulnerable triceps muscle in mutants did harbor greater numbers of central nuclei than controls, but this accounted for less than 1% of total myofibers examined (Figure 4F and Supplemental Figure 4, A–E). Consistent with the absence of major muscle pathology, we found no elevation of muscle creatine kinase (CK) in the serum of the mutant animals (Figure 4G) or damage to the sarcolemma assessed by examining whether circulating serum IgG had permeated the myofibers (Supplemental Figure 4F). Analyses of the expression levels of the myosin heavy chain genes and the myogenic factors Pax7, MyoD, Myf6, and myogenin, known markers of the state of maturity of the myofibers in P7 mutants, revealed no significant change in animals harboring either 1 or 2 SMN2 copies (Supplemental Figure 5, A–L), although MyoD expression trended higher, particularly in the most severe Δ7 SMA mutants, which served as 1 set of controls. Expression changes were also undetectable in P14 mutants (Supplemental Figure 5, M–Q), suggesting that selective depletion of SMN in the skeletal muscle of the intact organism as effected here does not disrupt myogenesis or muscle differentiation.
We next investigated NMJ morphology. In contrast to the striking defects observed in mutants hemizygous for the SMN2 transgene, the NMJs in the triceps muscles of the 1-month-old mutants homozygous for SMN2 appeared relatively normal (Figure 4H). Specifically, no differences in endplate size (area in μm2 — controls: 218.1 ± 3.9, mutants: 225.3 ± 3.7, n ≥ 150 NMJs from n = 3 mice of each cohort, P = 0.183, t test), innervation (Figure 4I), endplate complexity (Figure 4J), or incidence of NF varicosities were observed (number of endplates with axonal swellings — controls: 7.87 ± 0.69, mutants: 5.25 ± 1.56, n ≥ 150 NMJs from n = 3 mice of each cohort, P = 0.2, t test). As a more sensitive means of detecting potential synaptic defects, we also examined the functional status of the NMJs. Miniature endplate potential (MEPP) frequency, MEPP amplitude, evoked endplate potentials (EPPs), and quantal content were all normal in 4- to 5-week-old MyoD-iCre SMN2+/+ SmnF7/– mutants (Supplemental Figure 6, A–D). Consistent with these results, the cross-sectional area and the mean number of fibers quantified in mutant extensor digitorum longus (EDL) muscles were equivalent to those of controls (Supplemental Figure 6, E and F). Collectively, these results demonstrated that the young adult MyoD-iCre SMN2+/+ SmnF7/– mutants were phenotypically normal and did not exhibit major evidence of muscle or NMJ pathology.
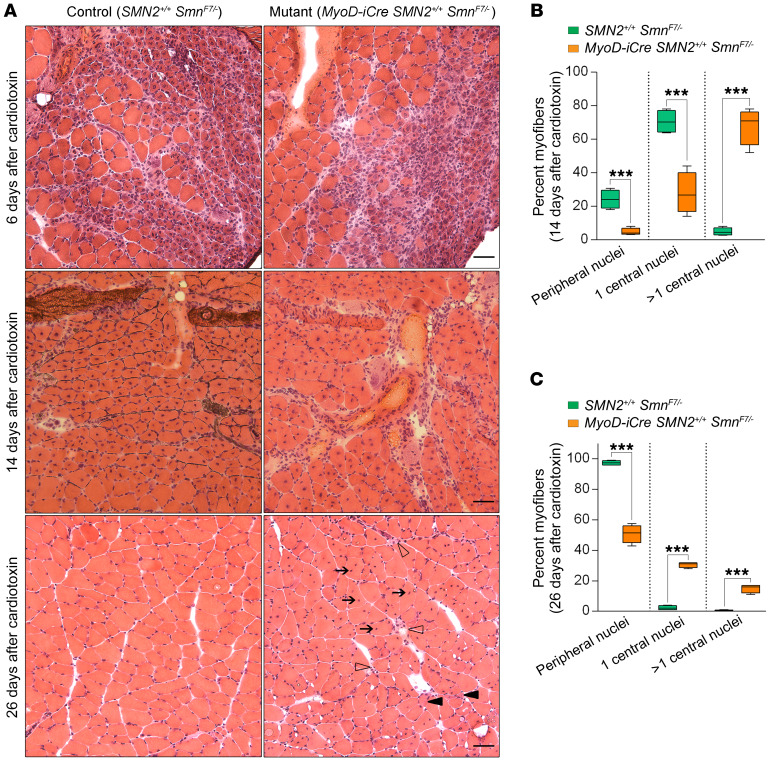
Considering the absence of major defects in young MyoD-iCre SMN2+/+ SmnF7/– mutants, we inquired whether acute injury to the muscle might unmask an underlying pathology. Accordingly, we injected the gastrocnemius of P21 mutants and controls with a myonecrotic agent, cardiotoxin (CTX), and examined the ability of the muscle to regenerate. Six days after exposure to CTX, myofibers from mutants and controls displayed uniform degeneration characterized by necrotic muscle and a great many minute regenerating myofibers with central nuclei (Figure 5A). Two weeks after injury, muscle regeneration was even more obvious. However, quantification of central versus peripherally located nuclei in the new myofibers revealed that mutant muscle was not only characterized by greater numbers of fibers that still harbored centrally located nuclei, but also cells that contained multiple such nuclei (Figure 5, A and B). Examination of the animals 26 days after injection showed that muscle expressing normal levels of SMN had completely recovered, with fewer than 2% of the fibers still harboring central nuclei. In contrast, muscle depleted of the protein continued to exhibit evidence of regeneration; roughly half the fibers in such muscle were still immature, containing 1 or more centrally located nuclei, displaying considerable variation in size, and in a few instances still degenerating (Figure 5, A and C). These results suggest that despite the absence of an obvious myopathy in young MyoD-iCre SMN2+/+ SmnF7/– mutants, an underlying pathology surfaced under conditions of muscle injury, severely compromising the ability of the animals to repair damaged muscle.
Figure 5. Acute injury unmasks muscle pathology in young adult MyoD-iCre SmnF7/– mutants bearing 2 copies of the SMN2 gene.
(A) H&E-stained transverse sections of muscle from mutant and control mice examined 6, 14, and 26 days after injury with CTX. Note persistence of muscle pathology typified by hypotrophic fibers (solid arrowheads), numerous central nuclei (arrows), and degenerating fibers (open arrowheads) at day 26 in the mutant. Scale bars: 25 μm (upper panels), 15 μm (middle panels), and 30 μm (lower panels). Graphs depict the proportion of fibers with central or peripherally located nuclei in mutants and controls (B) 14 days and (C) 26 days after injury. ***P < 0.001, t test, n ≥ 400 fibers from n ≥ 4 mice of each genotype.
Late-onset motor dysfunction and early mortality is observed in 2-copy SMN2 mutants selectively depleted of SMN in skeletal muscle tissue.
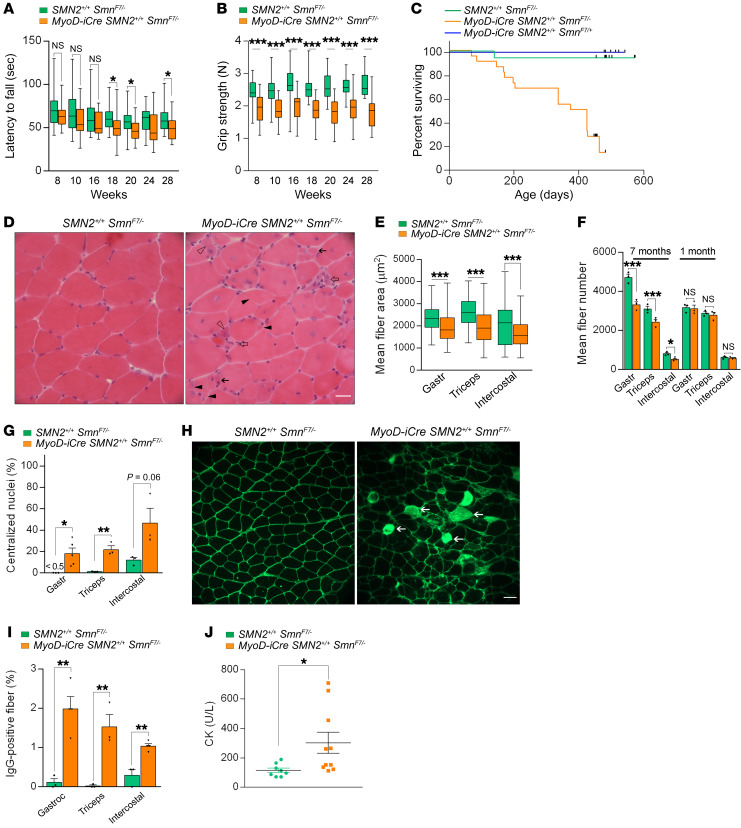
Although we failed to detect major muscle abnormalities or overt disease in young MyoD-iCre SMN2+/+ SmnF7/– mutants, the outcome of our experiments involving CTX and the revelation that acute damage unmasks an underlying pathology predicted that such dysfunction would eventually surface in the form of late-onset disease. Accordingly, we continued to surveil the 2-copy SMN2 mutants, performing a second, broad set of analyses between 6 and 7 months of age. In rotarod assays, we failed to detect marked changes in the mutants at 8, 10, and 16 weeks of age, even though there was a tendency for mutants to underperform controls. However, by 18 weeks of age, the discrepancy became significant and remained so when the animals were tested at 20 and 28 weeks, respectively (Figure 6A). These differences were even more prominent when the results were stratified according to gender and suggested that although mutants generally perform less well than controls, female animals are affected to a greater extent than males on the rotarod (Supplemental Figure 7, A and B).
Figure 6. Late-onset disease in MyoD-iCre SmnF7/– mutants bearing 2 SMN2 copies.
Results of (A) rotarod and (B) grip strength tests demonstrated the late-onset, disease-causing effects of selectively depleting SMN in skeletal muscle; t test, n ≥ 25 mice of each genotype. (C) Kaplan-Meier survival curves depict reduced lifespan of mutants. P < 0.0001 between mutants and the 2 sets of controls; P > 0.05 between the 2 sets of controls, log-rank test. (D) Transverse sections from 6- to 7-month-old mutant and control gastrocnemius muscles illustrate the presence of pathology in mutants. Hypotrophic fibers (solid arrows), regenerating fibers with cytoplasmic basophilia (open arrowhead), split fibers (solid arrowheads), or degenerating fibers (open arrows) are shown. Scale bar: 25 μm. Graphs represent (E) fiber sizes and (F) fiber numbers in muscles from 7-month-old mutants and controls; t test, n ≥ 500 fibers from n ≥ 3 mice of each genotype. (G) Estimates of central nuclei in muscles of 7-month-old mutants or controls; t test, n ≥ 1000 fibers from n ≥ 3 mice of each genotype. (H) Transverse sections of gastrocnemius muscles from 7-month-old mutant and control mice depicting damaged IgG-positive myofibers (arrows). Scale bar: 50 μm. (I) Quantification of damaged fibers from the previous experiment; t test, n ≥ 300 fibers from n ≥ 3 mice of each genotype. (J) Quantified results of serum CK values from 7-month-old mutant and control mice; t test, n ≥ 8 mice of each cohort. *P < 0.05; **P < 0.01; ***P < 0.001.
In a grip strength test, dysfunction became apparent even earlier. Mutants underperformed at all time points examined (Figure 6B). Moreover, both genders appeared equally affected (Supplemental Figure 7, C and D). Finally, an assessment of lifespan demonstrated that low SMN in skeletal muscle also caused early mortality in 2-copy SMN2 mutants. Whereas approximately 95% of control (SMN2+/+ SmnF7/–) animals remained alive at roughly 18 months of age, only 14% of the mutants were still viable at this age (Figure 6C). As expected, we found no difference between the SMN2+/+ SmnF7/– controls and a second set of controls that harbored the MyoD-iCre driver, but also carried 1 WT SMN allele (MyoD-iCre SMN2+/+ SmnF7/+), thus ensuring heterozygous levels of the protein in muscle (Figure 6C). Combined with the behavioral analysis of young adult mutants, these results suggest that persistent low SMN expression in skeletal muscle from 2 SMN2 copies was sufficient to trigger an overt disease phenotype, albeit in the form of late-onset dysfunction.
Myopathy is a late-onset, cell-autonomous consequence of low SMN in skeletal muscle.
Considering evidence of overt disease in the older MyoD-iCre SMN2+/+ SmnF7/– mutants, we viewed a reassessment of potential neuromuscular pathology in the animals as especially important. Accordingly, we once again began our analysis by examining muscle morphology. In contrast to observations in young adult mutants, the results of the analyses of older (6- to 7-month-old) mice were striking. In all 3 muscles examined, we found obvious evidence of pathology characterized by necrotic fibers infiltrated by inflammatory cells, regenerating fibers exhibiting cytoplasmic basophilia, angular fibers that appeared to have arisen from the splitting of a larger fiber, and the occasional presence of groups of hypotrophic fibers within what appeared to have once been an individual endomysium (Figure 6D and Supplemental Figure 7, E and F). Evidence of pathology in mutant muscle was also demonstrated in the form of increased puncta that stained positive for the microglial/macrophage marker, Iba-1 (Supplemental Figure 7G). Secondly, we found that mutant myofibers were not only generally smaller (Figure 6E and Supplemental Figure 8, A–C), but also inconsistently sized. Third, we found that mutant muscles had significantly fewer fibers (Figure 6F), many with 1 or more centrally located nuclei (Figure 6G). Loss of myofibers is clearly a late event, as we found equivalent numbers of fibers in mutant and control muscles at 1 month of age (Figure 6F). Finally, an assessment of the gross size of the gastrocnemii showed that mutant muscle weighed significantly less than control muscle (weight in mg: controls — 126 ± 7.3, mutants — 93.08 ± 4.5, P < 0.01, n ≥ 7 mice, t test). Pathology in the more vulnerable flexor digitorum brevis (FDB) muscle was even more severe. In mutants, only remnants of the muscle were found, suggesting almost complete loss or degeneration (Supplemental Figure 8D).
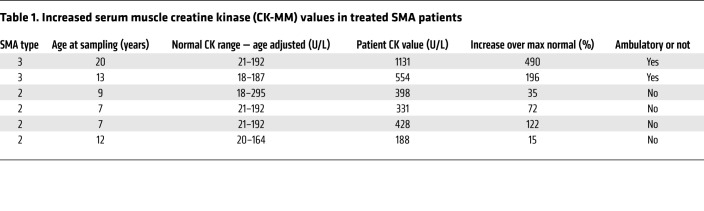
Given the ongoing muscle degenerative process in the older MyoD-iCre SMN2+/+ SmnF7/– mutants and as a second means of confirming the pathology, we assessed individual myofiber integrity by quantifying penetration by serum IgG and serum CK levels. Consistent with the observed pathology, and in contrast to that observed in young animals, a significantly greater number of mutant fibers contained IgG (Figure 6, H and I), akin to what is observed in muscular dystrophy, albeit to a lesser extent (Supplemental Figure 4F). Mutant CK levels were also elevated (Figure 6J). Not surprisingly, muscle pathology characterized by elevated serum CK and smaller myofibers with increased central nuclei was also observed in older mutants selectively depleted of SMN in fused myotubes — by means of an HSA-Cre driver (24) — instead of muscle progenitors (Supplemental Figure 9, A–D). To ascertain the relevance of these findings in our model mice to potential muscle pathology in human SMA, we examined serum CK values in a sample of convenience of 6 patients, all of whom had been treated for at least a year with Spinraza. In each instance, CK values were elevated and exceeded maximum normal values by 15% to 500% (Table 1). Collectively, these results strengthen observations of overt disease in older MyoD-iCre SMN2+/+ SmnF7/– mutants, provide a cellular basis for motor dysfunction and early mortality, and bolster the notion that skeletal muscle is a critical cellular site of action of the SMN protein. Deficiency of the protein, even in the presence of 2 SMN2 copies, is detrimental to muscle as the organism ages.
Table 1. Increased serum muscle creatine kinase (CK-MM) values in treated SMA patients.
Functional and structural defects of the NMJs are cell-autonomous consequences of low SMN in skeletal muscle.
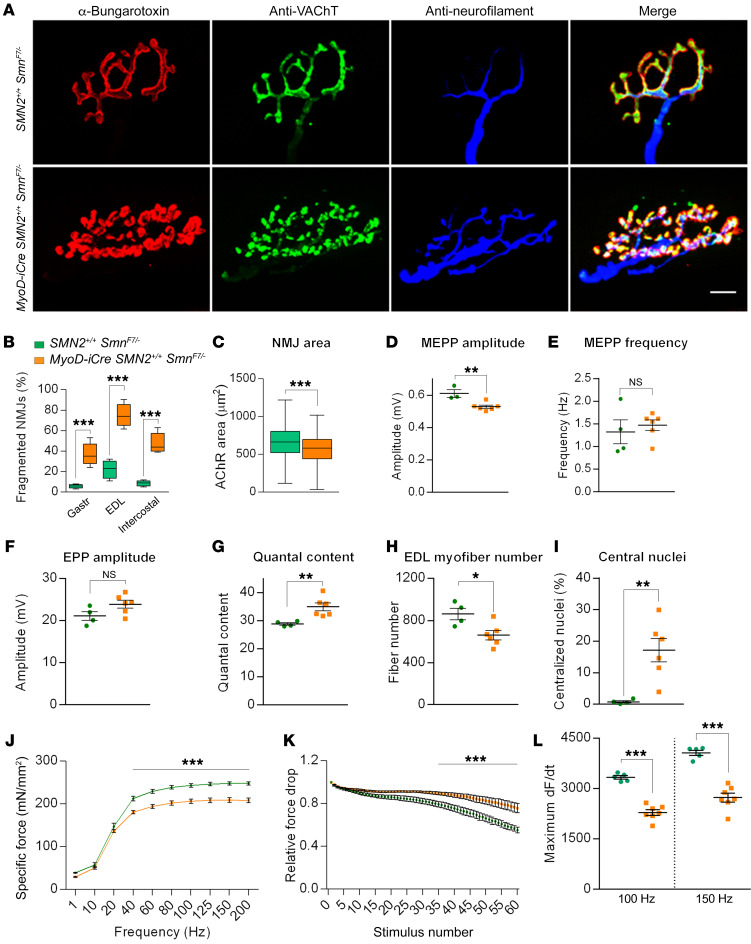
We next examined the NMJs in the 7-month-old MyoD-iCre SMN2+/+ SmnF7/– mice. The most striking outcome of our morphological analyses was the extent to which mutant AChR clusters were disassembled and fragmented (Figure 7, A and B). Similar defects were observed in 6-month-old HSA-Cre SMN2+/+ SmnF7/– mutants (Supplemental Figure 9, E and F). Moreover, in contrast to young MyoD-iCre SMN2+/+ SmnF7/– mutants in which the endplates were no different in size and structure to those of controls, endplates in the older mice were not only significantly smaller (Figure 7C), but also less elaborate — as assessed by quantifying the proportion of AChR clusters in the gastrocnemius possessing the typical pretzel-like structure with 4 or more perforations (controls — 86.33% ± 3.1%, mutants — 3.33% ± 1.85%, P < 0.001, n ≥ 100 NMJs in n = 3 mice of each cohort, t test).
Figure 7. NMJ pathology and neuromuscular dysfunction in MyoD-iCre SmnF7/– mutants bearing 2 SMN2 copies.
(A) NMJs from the EDL muscles of 7-month-old mice show profound fragmentation of endplates in mutants. Scale bar: 10 μm. (B) Quantified representation of fragmented NMJs in 7-month-old mutant and control mice; t test, n ≥ 500 endplates from n ≥ 3 mice of each genotype. (C) Graph depicts NMJ size in gastrocnemius muscles of 7-month-old mutants and controls; t test, n ≥ 420 endplates from n = 3 mice of each genotype. Quantified depictions of (D) MEPP amplitude, (E) MEPP frequency, (F) EPP amplitude, and (G) quantal content in EDL muscles of 7-month-old mutant and control mice. NS: P > 0.05, t test, n ≥ 10 endplates per animal from n ≥ 4 mice of each genotype. Graphs represent (H) myofiber numbers and (I) the proportion of fibers with central nuclei in EDL muscles of analyzed mice; t test, n ≥ 4 mice of each genotype. Quantified depictions of (J) peak specific force, (K) relative force drop, and (L) rate of maximal force production in soleus muscles of 5- to 7-month-old mice; 2-way ANOVA for results in J and t tests for results in K and L, n ≥ 5 mice of each cohort. *P < 0.05; **P < 0.01; ***P < 0.001.
To determine whether the morphological abnormalities correlated with impaired neurotransmission, we also assessed NMJ function by electrophysiological means. Although there were no differences in EPP amplitudes and MEPP frequencies in the EDL muscles of mutants and controls, MEPP amplitudes were significantly lower in the MyoD-iCre SMN2+/+ SmnF7/– mice, consistent with a paucity of postsynaptic AChRs (Figure 7, D–F). This led to a marked increase in mean quantal content at mutant endplates (Figure 7G), suggesting a compensatory increase in presynaptic neurotransmitter release in response to lower postsynaptic AChR concentrations. Mutant EDL muscle was, furthermore, reduced in size (muscle cross-sectional area in μm2: controls — 584,300 ± 33,470, mutants — 406,900 ± 36,780, P < 0.05, t test of n ≥ 4 animals of each cohort) and consisted of fewer fibers, many with central rather than peripherally located nuclei (Figure 7, H and I). The muscle defects noted above did not affect spinal motor neuron numbers in the mutants (mean number per section: mutants — 14.5 ± 0.25, controls — 12.8 ± 1.08, P > 0.05, n = 3 mice, t test). These results strengthen the observation that morphological and motor dysfunction result from low SMN in skeletal muscle, and further suggest that defects originating in the postsynaptic compartment can have a retrograde effect on motor neuron function.
In a concluding set of functional experiments, we performed ex vivo contractility measurements in soleus muscles from 5- to 7-month-old MyoD-iCre SMN2+/+ SmnF7/– mutants. We found that maximal specific force generated during single-twitch and low-frequency (10–20 Hz) stimulations did not differ between mutant and control samples. However, a significant reduction in peak specific force was detected when mutant muscles were stimulated with frequencies that resulted in a summating contraction (40–200 Hz) (Figure 7J). No shift in force-frequency relationship was observed between control and mutant muscles. We also subjected the muscles to repetitive, moderate-frequency stimulation to assess susceptibility to fatigue. Interestingly, mutant muscle exhibited a modest, but significant, decrease in fatigability during the final approximately 25 stimulus trains (Figure 7K). To investigate the cellular basis of this observation, myofibrils extracted from whole soleus muscle were analyzed for relative levels of the myosin heavy chain isoforms. Consistent with an increased resistance to fatigue, we found that soleus muscles from MyoD-iCre SMN2+/+ SmnF7/– mice expressed considerably more type I (MyHc7) myosin characteristically found in slow-twitch, fatigue-resistant myofibers (Supplemental Figure 10, A and B). Finally, considering these observed changes in peak specific tetanic force and slow type I myosin content, we analyzed the kinetics of force development (dF/dt) in soleus muscles from control and mutant mice. Congruent with the observed increase in MyHc7 content, and despite being normally sized (weight in mg: controls — 6.15 ± 0.22, mutants — 6.15 ± 0.38; P = 1, n ≥ 4 mice, t test), maximal dF/dt was reduced in soleus muscle expressing low SMN (Figure 7L). Collectively, these results provide strong evidence of significant morphological and functional deficits of muscle arising from a cell-autonomous effect of SMN paucity in this tissue.
Mitigation of neuromuscular pathology is observed after postsymptomatic repletion of the SMN protein.
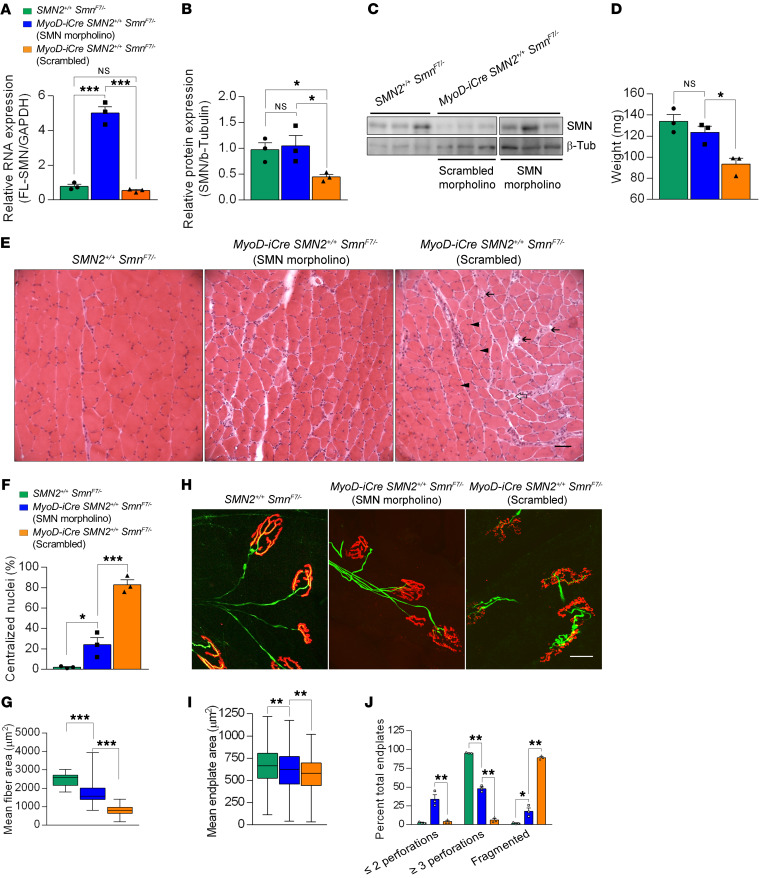
Early postnatal repletion of SMN effectively prevents SMA onset (53, 54). To determine whether restoring SMN also benefits postsymptomatic mice selectively depleted of the protein in muscle, we systemically administered either an SMN2 splice-switching morpholino (MO) or one with a scrambled sequence to 7-month-old MyoD-iCre SMN2+/+ SmnF7/– mutants. Nine weeks later, muscle SMN levels were assessed. Consistent with prior reports, we found that FL-SMN transcripts and total levels of SMN protein were significantly higher in the SMN MO–treated mutants than in the cohort treated with the scrambled molecule (Figure 8, A–C). To determine whether the increase in muscle SMN had mitigated the muscle pathology typically observed in the mutants, we dissected out the gastrocnemii from the various cohorts, at the same time as SMN levels were examined, to assess the muscles. Measurements of muscle wet weight demonstrated that whereas muscles from scrambled MO–treated mutants remained significantly smaller than those of controls, muscles from mutants restored for SMN were larger and no different in weight than control muscle (Figure 8D). This was accompanied by fewer cellular defects in the SMN MO–treated mutants; myofibers in these animals exhibited larger fiber areas compared with those in mutants administered the scrambled MO. Additionally, fewer SMN MO–treated myofibers contained centralized nuclei compared with those in scrambled MO–treated mutants (Figure 8, E–G). Finally, measurements of endplate size and complexity in the 3 cohorts of mice demonstrated that both of these parameters were partially normalized in SMN MO–treated mutants (Figure 8, H–J). Specifically, NMJs in mutants restored for SMN were not only larger (Figure 8I), but also less fragmented (Figure 8, H and J). Moreover, the intact endplates in mutants restored for SMN were more elaborate, as assessed by a quantification of pretzel-like structures with 3 or more distinct perforations (Figure 8J). We conclude that restoring SMN even after muscle pathology becomes evident arrests and/or reverses the myopathy attributed to low protein in the tissue. Accordingly, repletion of SMN to the muscle of SMA subjects is expected to have an important mitigating effect on the overall disease phenotype.
Figure 8. SMN repletion mitigates muscle defects in symptomatic mutants.
Levels of the (A) SMN2-derived FL-SMN transcripts and (B) SMN protein in gastrocnemius muscles of controls and mutants either administered an SMN2 splice-switching MO or a nonspecific MO at 7 months of age; 1-way ANOVA, n = 3 mice of each genotype. (C) Western blot probed for SMN protein in skeletal muscle after treatment with SMN-MO or scrambled MO. (D) Muscle weights in mice treated as described in the previous panels; 1-way ANOVA, n ≥ 3 mice in each group. (E) Transverse sections of gastrocnemius muscles from treated mutants and controls. Less pathology in the form of central nuclei (arrowheads), hypotrophic fibers (open arrows), and degenerating fibers (solid arrows) observed in SMN-MO treated muscle. Scale bar: 100 μm. Quantified results of (F) central nuclei and (G) myofiber areas in muscles from the mice; 1-way ANOVA, n ≥ 300 fibers from n ≥ 3 mice of each cohort. (H) Endplates from the 3 sets of mice; fewer fragmented NMJs are seen in the SMN-MO treated mutant than in its counterpart administered scrambled MO. Scale bar: 25 μm. (I) NMJ area measurements and (J) NMJ complexity analysis in gastrocnemius muscles of treated mutants or littermate controls; 1-way ANOVA, n ≥ 400 endplates from n ≥ 3 mice of each genotype for results in I and J. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
SMA is a paralytic infantile-onset disorder caused by low SMN protein. Untreated, most patients with SMA perish before 2 years of age. Restoring SMN in model mice prevents and/or reverses SMA onset (54, 55) and was therefore rapidly adopted as a means to a viable treatment. However, the repletion studies also made it clear that the success of the strategy critically depends on when and in which tissues SMN is restored. That CNS cells and, in particular, spinal motor neurons are especially vulnerable to low SMN, and therefore must express adequate SMN levels to thwart disease onset is widely accepted. These cells are efficiently targeted by intrathecal administration of SMN-restoring agents such as Spinraza. Indeed, the initial outcome of treating patients in this manner has left no doubt about the clinical efficacy of targeting the CNS. Yet, lingering questions about restricted intrathecal delivery of the drug and the long-term effects of depriving the periphery of adequate SMN remain. Here we used what we believe is a novel a line of model mice to demonstrate the outcome of depriving one important peripheral organ, skeletal muscle, of adequate SMN. This tissue is especially relevant given the vulnerability of the motor unit in SMA. Four main findings emerged from our studies. Of greatest import is revealing how detrimental SMN deficiency is to skeletal muscle. We found defects in individual myofibers and NMJs, abnormalities and dysfunction of entire muscles, and eventually an overt phenotype that compromised survival. This pathology emerged despite normal levels of SMN in other tissues. Second, we demonstrated that the extent of disease correlates with the number of SMN2 copies and thus absolute levels of functional SMN protein in the tissue. Whereas mutants expressing only 1 copy of the SMN2 gene rapidly developed disease, those with 2 copies exhibited a much later-onset phenotype. Nevertheless, the cellular pathology underlying even late-onset disease could be unmasked at early stages under conditions of acute muscle injury. Third, our results unequivocally demonstrated that at least some aspect of the overall SMA phenotype stems from a muscle-autonomous effect. Depleting SMN in this tissue is sufficient to trigger pathology. Moreover, although the resulting abnormalities were most apparent in the muscle itself, defects were also eventually detected in the presynapse in the form of NF-containing axonal varicosities and an attempt at compensatory neurotransmission. Finally, we showed that restoring SMN to muscle even after onset of pathology can have a mitigating effect. This is particularly reassuring from a clinical standpoint. We conclude that the SMN protein is intrinsically important to muscle function. SMN restorative therapies that deprive this tissue of adequate protein are unlikely to achieve maximum benefit. Rather, our results have raised the prospect of intrathecally treated subjects possibly developing severe and potentially life-threatening myopathies over time. Zolgensma, an AAV9-mediated SMN-enhancing agent, which was recently approved for the treatment of SMA, addresses this concern because it is delivered systemically and thus likely targets muscle. However, such treatment is currently only approved for patients under the age of 2 years; older patients are limited to intrathecal therapy.
SMN repletion to treat SMA must consider the spatial and temporal requirements for the protein. In this context, a potential cell-autonomous, disease-causing effect of low SMN in skeletal muscle has been much debated. The ambivalence has largely stemmed from the nature of SMA, which involves ubiquitous low levels of the SMN protein, the primary vulnerability of spinal motor neurons to SMN paucity, and the manner in which denervation affects muscle. Any effects of SMA on muscle could therefore be construed as emerging secondary to motor neuron loss. Still, a great many studies have inferred an independent role for muscle in SMA. These studies ranged from analyses of cells in culture demonstrating a damaging effect of SMA muscle on nerve–muscle cocultures (23, 56) to assessments of intact muscle and markers of myogenesis in autopsy tissue and model mice (26, 57, 58). However, since the experiments relied on patients’ cells or mice ubiquitously depleted for SMN, it was impossible to truly separate effects in motor neurons from those intrinsic to muscle. The most direct attempts to determine potential cell-autonomous effects of low muscle SMN on the SMA phenotype concluded that low protein in this tissue contributes little to disease (25, 28).
Our results are in sharp contrast to those of Iyer et al. (28). One explanation for the difference in outcomes stems from our choice of Cre drivers. The robustness and specificity of such drivers are key to enabling one to accurately predict outcomes in the context of disease. In this respect, not only did we find the MyoD-iCre line to deplete SMN more efficiently than the Myf5-Cre driver, but also to do so with greater tissue specificity. Particularly telling were our results demonstrating that almost half of all Myf5-Cre SmnF7/– mutants not only lived to adulthood, but also were robust enough to breed. This strongly suggests that the Myf5-Cre line failed to efficiently drive recombination at the SmnF7 allele, and it is therefore no wonder that adding SMN2 to Myf5-Cre SmnF7/– mice would further mask any symptoms of disease — akin to what was observed by Iyer et al. A related explanation for the contrasting findings is the allelic background adopted in the 2 studies. Whereas the model mice used by Iyer et al. harbored and expressed an artificial SMNΔ7 transgene, ours did not. Notwithstanding reliable data demonstrating that the SMNΔ7 protein is on its own nonfunctional, it is nevertheless able, in concert with FL-SMN, to bestow partial function to the SMN complex and thus significantly mitigate disease (41, 59). A final explanation for the inability to detect dysfunction in the mice generated by Iyer et al. likely lies in the fact that the preponderance of analyses were conducted in relatively young (8 weeks), 2-copy SMN2 mutants. Indeed, we too failed to detect evidence of cellular pathology or overt disease in our young adult MyoD-iCre mutants harboring 2 SMN2 copies. A disease phenotype only emerged at 6 to 7 months of age. We suspect this too would have been the case had Iyer and colleagues analyzed older mutants. The difference in our respective findings does not negate the assertion by Iyer et al. that exclusively treating muscle tissue in SMA will fail to produce clinical benefit. This likely remains the case, with optimal clinical benefit accruing only when the periphery and CNS are restored for SMN.
Our results are not entirely unexpected considering the predominantly neuromuscular SMA phenotype and inferences of an independent role for muscle in contributing to disease. Still, to our knowledge, ours are the first results to use a mammalian model expressing SMN2 to directly show that low SMN in skeletal muscle produces disease in a cell-autonomous fashion. This is especially pertinent from a clinical standpoint, given the therapeutic promise of SMN-restoring agents. Two of these, Spinraza and Zolgensma, have now received regulatory approval for the treatment of SMA. However, their use is subject to caveats already alluded to. In light of our results, this combination of factors warrants concern for the many hundreds of patients currently undergoing treatment, particularly if limited to the CNS. If, as our preclinical results predict, muscle pathology is a late-onset consequence of low SMN in the tissue, there is every possibility that the initial benefits of intrathecal treatment will eventually yield to a chronic and insidious late-onset muscle disorder. The generally elevated serum CK values in our Spinraza-treated SMA patients and the particularly high levels of this biomarker in the ambulatory versus nonambulatory patients sampled supported this assertion; activity-dependent use of SMN-deprived muscle in the ambulatory patients likely accelerated damage to the tissue. Dysfunction may be arrested and/or reversed, as demonstrated in our model, by augmenting SMN in muscle, but is once again likely to depend on the timing of intervention. Muscle once lost in its entirety will be hard to replace.
Although the muscle defects in our model mice and the therapeutic implications of such damage are obvious, precisely how low SMN triggers pathology remains to be explored. Some have suggested disruptions in the process of myogenesis (29, 57, 60, 61). Although this may be true in the most severe form of SMA, it appeared to not be the case in our mutants, particularly those expressing 2 SMN2 copies. In such mutants examined at P7, we found few muscle defects. Rather, our data suggested defects in muscle maintenance. Defects of muscle maintenance in SMA have hitherto not been reported or emphasized — a likely consequence of the severity of the phenotype and short lifespans of the model mice examined thus far. Nevertheless, maintenance defects remain a distinct possibility and may originate in damaged myofibers and/or satellite cells. Satellite cells are particularly active during early postnatal muscle development when they serve as sources of new myonuclei (62). After prepubertal life in mice (4–6 weeks of age), a period when SMN levels also decline considerably (63), the numbers of these cells fall dramatically as myofibers reach their adult dimensions. We speculate that it is during subsequent weeks that myofibers expressing low SMN degenerate in substantial numbers. Satellite cells may initially satisfy the demand to replenish degenerating fibers. However, an accelerating process of degeneration accompanied by an intrinsic dysfunction in SMN-depleted satellite cells eventually overwhelms the capacity of these stem cells to repopulate muscle. Satellite cell dysfunction might be inferred from the discrepancy between the far greater numbers of fibers exhibiting central nuclei relative to those actively degenerating — as assessed by serum IgG uptake (Figure 6I). One intriguing explanation for such disparity might lie in the normal ability of SMN-depleted satellite cells to respond to fiber damage, but a subsequent failure of the cells to return to a quiescence. The resulting constitutively active state of these cells could hasten their exhaustion and thus overall muscle fiber loss. Our CTX experiments further suggested that SMN levels severely affected muscle regeneration after injury. If this is true in humans, patients treated with drugs that restore SMN exclusively to the CNS will have to exercise care. One apparent conundrum emerging from our observations is why muscle pathology akin to what we have observed has not been reported widely in classical SMA. We think the answer lies primarily in the masking effects of early motor neuron loss. A second intriguing hypothesis centers on the cross-talk between muscle and nerve. In classical SMA, the health of both the pre- and postsynaptic compartments is likely compromised. Selectively rescuing the motor neuron, as might be expected in intrathecally treated patients, may result in the rescued presynaptic compartment imposing an undue burden on muscle. The molecular determinants underlying such disruptions in signaling at the NMJ remain to be explored in the context of SMA. Our model mice will serve as a useful tool in investigating this and broader questions centering on the required signaling between motor neurons and muscle to ensure the health and viability of both cell types.
Methods
Mice.
ROSA26-flox-STOP-flox-YFP mice (The Jackson Laboratory, stock no. 006148) were genotyped according to protocols at https://www.jax.org/ MyoD-iCre or Myf5-Cre SMA mice without SMN2 (MyoD-Cre SmnF7/– or Myf5-Cre SmnF7/–) and controls (SmnF7/–) were generated by breeding MyoD-iCre (The Jackson Laboratory, stock no. 014140) or Myf5-Cre (The Jackson Laboratory, stock no. 007893) mice heterozygous for murine Smn (Smn+/–; The Jackson Laboratory, stock no. 006214) with SmnF7/F7 animals (SmnF7, The Jackson Laboratory, stock no. 006138). MyoD-Cre SMA mutants with 1 or 2 copies of SMN2 and controls (SMN2+/– SmnF7/–, SMN2+/+ SmnF7/–, or MyoD-Cre SMN2+/+ SmnF7/+) were generated by introducing the SMN2 transgene (The Jackson Laboratory, stock no. 005024) onto animals with the Cre driver and floxed allele. All analyses on 2-copy SMN2 mice were carried out with the investigator blinded to phenotype. A similar protocol was followed for the 1-copy SMN2 mice, except for behavioral studies where an overt phenotype in the mutants precluded blinding. See the Supplemental Methods for details of genotyping primers.
Motor behavior assays.
The rotarod and grip strength tests were performed using the LE8200 rotarod (Harvard Apparatus/Panlab Inc.) and BIOSEB Grip Strength Tester BIO-GS3 (Bioseb Inc.), respectively, as per the manufacturer’s instructions. Details of these methods are described in the Supplemental Methods.
Serum CK assay.
Mice were euthanized with CO2 gas and 500 μL of blood collected from the right ventricle using a 21-gauge needle. Serum CK level was measured by the Institute of Comparative Medicine of Columbia University using an Element DC veterinary chemistry analyzer (Heska).
Motor neuron, NMJ, and muscle histology.
Motor neuron, NMJ, and muscle histology was essentially carried out as previously described (42, 45). Additional details are presented in the Supplemental Methods.
PCR, Western blots, and silver-stained gel analyses.
Tissues were lysed using TRIzol (Invitrogen) according to the manufacturer’s instructions. Following cDNA synthesis, quantitative PCR was performed in triplicate on a MasterCycler Real Plex4 (Eppendorf). Primer sequences used for the qRT-PCR are indicated in the Supplemental Methods. Western blotting was performed as described (13); myosin heavy chain isoform analysis was carried out according to a prior report (64); also see Supplemental Methods.
CTX and MO experiments.
Mice were anesthetized with 1%–2% isoflurane, and myonecrosis induced with CTX. A 30-gauge needle was used to inject 20 μL of a 10 μM solution of CTX, in PBS, into the belly of the gastrocnemius. Cardiotoxin was delivered uniformly into the muscles of the right hindlimb. The left hindlimb was injected with identical volumes of PBS. Mice were assessed at baseline point (no injections) and at various time points after CTX and PBS injections, and muscles of the hindlimb removed, weighed, and immediately used to prepare tissue lysates or cryosections. SMN and scrambled MOs used were identical to those in a prior report (16). Each MO was resuspended in sterile water and aliquoted to a final concentration of 0.5 mM. Mice were administered the MOs by i.p. injection (12.5 mg/kg) at P210.
Electrophysiology.
NMJ electrophysiology: NMJ electrophysiology was essentially performed as previously described (46). The ex vivo muscle contractility experiments were carried out based on methods optimized and previously used (64). Modifications to the respective methods for this study are detailed in the Supplemental Methods.
Statistics.
Kaplan-Meier survival curves were assessed for differences using the log-rank test equivalent to the Mantel-Haenszel test. The unpaired, 2-tailed Student’s t test or 1-way ANOVA followed by Tukey’s post hoc comparison, where indicated, were used to compare means for statistical differences. In instances where categorical binomial variables had to be analyzed, the Fisher’s exact test was employed. Data are represented as mean ± SEM unless otherwise indicated. P values less than 0.05 were considered significant. Statistical analyses were performed with Prism version 6.0 (GraphPad Software).
Study approval.
All animal procedures adhered to protocols described in the Guide for the Care and Use of Lab Animals (National Academies Press, 2011) and were approved by Columbia University’s IACUC. The subjects of this study were randomly selected, mixed background, male and female mice housed in a controlled environment on a 12-hour light/12-hour dark cycle with food and water.
Author contributions
JKK, NNJ, and MRF performed most of the experiments described here. ZF assessed the functional status of the NMJs while CPK supervised these experiments and provided intellectual input. CAC contributed to the patient data described in this study. LWL carried out the ex vivo muscle contractility studies and RTD directed this aspect of the project. URM conceptualized the experiments, directed the overall project, and analyzed and interpreted the data. JKK and URM wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
We thank P. Faust and members of the Columbia MNC for advice and suggestions. We are also grateful to D.C. De Vivo for his support during the course of this study. This project was funded by grants from the AFM-France, Cure SMA, and the NIH (R21 NS099921, R01 NS057482, and R56 NS104218) to URM, NIH R01 AR059646-08 to RTD, and the SMA Foundation to CPK.
Version 1. 02/10/2020
Electronic publication
Version 2. 02/11/2020
Labeling error corrected in Figure 2E
Version 3. 03/02/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(3):1271–1287.https://doi.org/10.1172/JCI131989.
Contributor Information
Jeong-Ki Kim, Email: jk3678@cumc.columbia.edu.
Zhihua Feng, Email: zfeng@usc.edu.
Lan Wei-Lapierre, Email: Lan_wei-Lapierre@urmc.rochester.edu.
Chien-Ping Ko, Email: cko@usc.edu.
References
- 1.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre S, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16(3):265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 3.Coovert DD, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6(8):1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 4.Schrank B, et al. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94(18):9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monani UR, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 6.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24(5):438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3(2):97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 9.Eggert C, Chari A, Laggerbauer B, Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol Med. 2006;12(3):113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10.McAndrew PE, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60(6):1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70(2):358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitali T, et al. Detection of the survival motor neuron (SMN) genes by FISH: further evidence for a role for SMN2 in the modulation of disease severity in SMA patients. Hum Mol Genet. 1999;8(13):2525–2532. doi: 10.1093/hmg/8.13.2525. [DOI] [PubMed] [Google Scholar]
- 13.Monani UR, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(–/–) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9(3):333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 14.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26(4):1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua Y, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24(15):1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porensky PN, et al. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet. 2012;21(7):1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Y, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478(7367):123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel RS, et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377(18):1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 19.Kim JK, Monani UR. Augmenting the SMN protein to treat infantile spinal muscular atrophy. Neuron. 2018;97(5):1001–1003. doi: 10.1016/j.neuron.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Sumner CJ, Crawford TO. Two breakthrough gene-targeted treatments for spinal muscular atrophy: challenges remain. J Clin Invest. 2018;128(8):3219–3227. doi: 10.1172/JCI121658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamilton G, Gillingwater TH. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19(1):40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Nash LA, Burns JK, Chardon JW, Kothary R, Parks RJ. Spinal muscular atrophy: more than a disease of motor neurons? Curr Mol Med. 2016;16(9):779–792. doi: 10.2174/1566524016666161128113338. [DOI] [PubMed] [Google Scholar]
- 23.Guettier-Sigrist S, Hugel B, Coupin G, Freyssinet JM, Poindron P, Warter JM. Possible pathogenic role of muscle cell dysfunction in motor neuron death in spinal muscular atrophy. Muscle Nerve. 2002;25(5):700–708. doi: 10.1002/mus.10081. [DOI] [PubMed] [Google Scholar]
- 24.Cifuentes-Diaz C, et al. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J Cell Biol. 2001;152(5):1107–1114. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavrilina TO, et al. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum Mol Genet. 2008;17(8):1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyer JG, Murray LM, Scott K, De Repentigny Y, Renaud JM, Kothary R. Early onset muscle weakness and disruption of muscle proteins in mouse models of spinal muscular atrophy. Skelet Muscle. 2013;3(1):24. doi: 10.1186/2044-5040-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez TL, et al. Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. J Neurosci. 2012;32(25):8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer CC, et al. Low levels of survival motor neuron protein are sufficient for normal muscle function in the SMNΔ7 mouse model of SMA. Hum Mol Genet. 2015;24(21):6160–6173. doi: 10.1093/hmg/ddv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bricceno KV, et al. Survival motor neuron protein deficiency impairs myotube formation by altering myogenic gene expression and focal adhesion dynamics. Hum Mol Genet. 2014;23(18):4745–4757. doi: 10.1093/hmg/ddu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripolone M, et al. Impaired muscle mitochondrial biogenesis and myogenesis in spinal muscular atrophy. JAMA Neurol. 2015;72(6):666–675. doi: 10.1001/jamaneurol.2015.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto M, et al. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47(2):107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tallquist MD, et al. Retention of PDGFR-β function in mice in the absence of phosphatidylinositol 3′-kinase and phospholipase Cgamma signaling pathways. Genes Dev. 2000;14(24):3179–3190. doi: 10.1101/gad.844700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111(4):1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- 34.Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev Biol. 2009;332(1):131–141. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10(10):1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 36.Beedle AM, et al. Mouse fukutin deletion impairs dystroglycan processing and recapitulates muscular dystrophy. J Clin Invest. 2012;122(9):3330–3342. doi: 10.1172/JCI63004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27(19):e27. doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frugier T, et al. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2000;9(5):849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 39.Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135(9):1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- 40.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 42.Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci. 2010;30(36):12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15(14):3555–3565. doi: 10.1002/j.1460-2075.1996.tb00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grady RM, Zhou H, Cunningham JM, Henry MD, Campbell KP, Sanes JR. Maturation and maintenance of the neuromuscular synapse: genetic evidence for roles of the dystrophin — glycoprotein complex. Neuron. 2000;25(2):279–293. doi: 10.1016/S0896-6273(00)80894-6. [DOI] [PubMed] [Google Scholar]
- 45.Kariya S, et al. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(16):2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling KK, Lin MY, Zingg B, Feng Z, Ko CP. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One. 2010;5(11):e15457. doi: 10.1371/journal.pone.0015457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong L, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29(3):842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(7):949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 49.Jha NN, Kim JK, Monani UR. Motor neuron biology and disease: A current perspective on infantile-onset spinal muscular atrophy. Future Neurol. 2018;13(3):161–172. doi: 10.2217/fnl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacLeod MJ, Taylor JE, Lunt PW, Mathew CG, Robb SA. Prenatal onset spinal muscular atrophy. Eur J Paediatr Neurol. 1999;3(2):65–72. doi: 10.1016/S1090-3798(99)80015-4. [DOI] [PubMed] [Google Scholar]
- 51.Sleigh JN, Gillingwater TH, Talbot K. The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy. Dis Model Mech. 2011;4(4):457–467. doi: 10.1242/dmm.007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oskoui M, Darras BT, De Vivo DC. Spinal muscular atrophy: 125 years later and on the verge of a cure. In: Sumner CJ, Paushkin S, Ko CP, eds. Spinal Muscular Atrophy: Disease Mechanisms and Therapy. New York, New York, USA; 2017:3–19. [Google Scholar]
- 53.Foust KD, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28(3):271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Lutz CM, et al. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J Clin Invest. 2011;121(8):3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le TT, et al. Temporal requirement for high SMN expression in SMA mice. Hum Mol Genet. 2011;20(18):3578–3591. doi: 10.1093/hmg/ddr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braun S, Croizat B, Lagrange MC, Warter JM, Poindron P. Constitutive muscular abnormalities in culture in spinal muscular atrophy. Lancet. 1995;345(8951):694–695. doi: 10.1016/S0140-6736(95)90869-2. [DOI] [PubMed] [Google Scholar]
- 57.Boyer JG, et al. Myogenic program dysregulation is contributory to disease pathogenesis in spinal muscular atrophy. Hum Mol Genet. 2014;23(16):4249–4259. doi: 10.1093/hmg/ddu142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martínez-Hernández R, Bernal S, Alias L, Tizzano EF. Abnormalities in early markers of muscle involvement support a delay in myogenesis in spinal muscular atrophy. J Neuropathol Exp Neurol. 2014;73(6):559–567. doi: 10.1097/NEN.0000000000000078. [DOI] [PubMed] [Google Scholar]
- 59.Lorson CL, et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19(1):63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 60.Hayhurst M, Wagner AK, Cerletti M, Wagers AJ, Rubin LL. A cell-autonomous defect in skeletal muscle satellite cells expressing low levels of survival of motor neuron protein. Dev Biol. 2012;368(2):323–334. doi: 10.1016/j.ydbio.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hellbach N, et al. Impaired myogenic development, differentiation and function in hESC-derived SMA myoblasts and myotubes. PLoS One. 2018;13(10):e0205589. doi: 10.1371/journal.pone.0205589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bachman JF, et al. Prepubertal skeletal muscle growth requires Pax7-expressing satellite cell-derived myonuclear contribution. Development. 2018;145(20):dev167197. doi: 10.1242/dev.167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jablonka S, et al. Co-regulation of survival of motor neuron (SMN) protein and its interactor SIP1 during development and in spinal muscular atrophy. Hum Mol Genet. 2001;10(5):497–505. doi: 10.1093/hmg/10.5.497. [DOI] [PubMed] [Google Scholar]
- 64.Wei-Lapierre L, Carrell EM, Boncompagni S, Protasi F, Dirksen RT. Orai1-dependent calcium entry promotes skeletal muscle growth and limits fatigue. Nat Commun. 2013;4:2805. doi: 10.1038/ncomms3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.