Abstract
Background
Most ALK-positive lung cancers will develop ALK-independent resistance after treatment with next-generation ALK inhibitors. MET amplification has been described in patients progressing on ALK inhibitors, but frequency of this event has not been comprehensively assessed.
Methods
We performed fluorescence in-situ hybridization and/or next-generation sequencing on 207 post-treatment tissue (n=101) or plasma (n=106) specimens from patients with ALK-positive lung cancer to detect MET genetic alterations. We evaluated ALK inhibitor sensitivity in cell lines with MET alterations and assessed antitumor activity of ALK/MET blockade in ALK-positive cell lines and two patients with MET-driven resistance.
Results
MET amplification was detected in 15% of tumor biopsies from patients relapsing on next-generation ALK inhibitors, including 12% and 22% of biopsies from patients progressing on second-generation inhibitors or lorlatinib, respectively. Patients treated with a second-generation ALK inhibitor in the first-line setting were more likely to develop MET amplification than those who had received next-generation ALK inhibitors after crizotinib (p=0.019). Two tumor specimens harbored an identical ST7-MET rearrangement, one of which had concurrent MET amplification. Expressing ST7-MET in the sensitive H3122 ALK-positive cell line induced resistance to ALK inhibitors that was reversed with dual ALK/MET inhibition. MET inhibition re-sensitized a patient-derived cell line harboring both ST7-MET and MET amplification to ALK inhibitors. Two patients with ALK-positive lung cancer and acquired MET alterations achieved rapid responses to ALK/MET combination therapy.
Conclusions
Treatment with next-generation ALK inhibitors, particularly in the first-line setting, may select for MET-driven resistance. Patients with acquired MET alterations may derive clinical benefit from therapies that target both ALK and MET.
Keywords: ALK, Lung Cancer, Resistance, MET Amplification
INTRODUCTION
ALK-rearranged (i.e., ALK-positive) non-small cell lung cancers (NSCLC) depend on constitutive ALK signaling for growth and survival.1 This oncogene dependency underlies the marked sensitivity of these tumors to tyrosine kinase inhibitors (TKIs) targeting ALK.2,3 Historically, the ALK/ROS1/MET TKI crizotinib was the standard first-line therapy for advanced ALK-positive NSCLC.4 However, crizotinib has recently been supplanted by more potent and selective second-generation ALK TKIs (e.g., ceritinib, alectinib, brigatinib).2,3,5 In addition, the third-generation ALK TKI lorlatinib was recently approved for patients previously treated with a second-generation TKI.6
Despite initial sensitivity to ALK TKIs, ALK-positive tumors invariably develop resistance. In approximately one-half of cases, resistance to second-generation ALK TKIs is due to secondary mutations in the ALK kinase domain.7 These tumors remain ALK-dependent and responsive to treatment with lorlatinib.8 The remaining cases have presumably developed ALK-independent resistance mechanisms, most often due to activation of alternative or bypass signaling pathways.7,9 Recent studies of lorlatinib resistance suggest an even more complicated array of resistance mechanisms, including diverse compound ALK mutations in one-third of patients and ALK-independent resistance mechanisms in the remaining two-thirds of patients.9,10
To date, ALK-independent resistance mechanisms remain poorly characterized. In preclinical studies, multiple bypass mechanisms have been described, including activation of MET, EGFR, SRC, and IGF-1R.11–13. Of these, MET is a particularly attractive target given the availability of potent MET TKIs. In addition, MET activation has been previously studied as a bypass pathway in EGFR-mutant NSCLC where MET amplification has been reported in 5–20% of resistant cases.14,15 In phase I/II studies, co-targeting EGFR and MET can re-induce responses in resistant EGFR-mutant tumors with MET amplification.16 In contrast to EGFR-mutant NSCLC, relatively little is known about the contribution of aberrant MET activation to resistance in ALK-positive NSCLC. Several case reports suggest that MET amplification can mediate resistance to ALK TKIs and that the ALK/ROS1/MET TKI crizotinib may be able to overcome MET-driven resistance.17,18 However, larger studies are needed to validate MET as a clinically relevant driver of resistance in ALK-positive NSCLC.
Here we analyze tumor and/or blood specimens from 136 patients with ALK-positive NSCLC relapsing on ALK TKIs. We identify genetic alterations of MET in approximately 15% of resistant cases. In cell lines and in patients, MET activation drives resistance to ALK TKIs but confers sensitivity to dual ALK/MET blockade. These studies support the development of novel combinations targeting ALK and MET in ALK-positive NSCLC.
METHODS
Data Collection
Resistant specimens were obtained from 136 patients with metastatic ALK-positive NSCLC who underwent molecular profiling between 2014 and 2019 (Figure 1A). Medical records from these patients were retrospectively reviewed to extract data on demographics, treatment histories and molecular profiling results. Data were updated as of August 31, 2019. This study was approved by the Massachusetts General Hospital Institutional Review Board. Patient studies were conducted according to the Declaration of Helsinki, the Belmont Report, and the U.S. Common Rule.
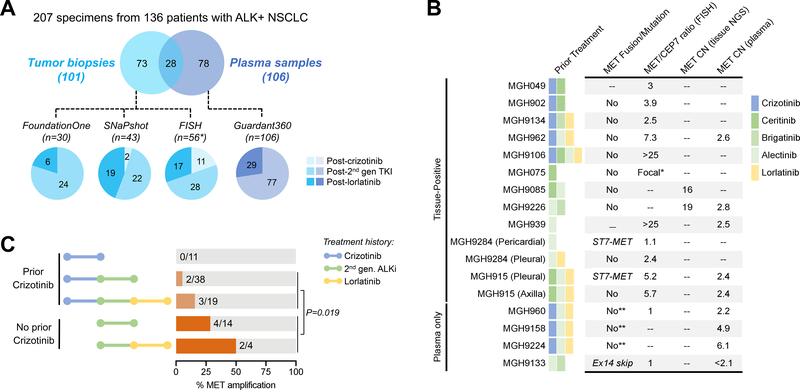
Figure 1. Summary of MET Alterations in Resistant ALK-Positive NSCLC.
(A) Schematic depicts number of specimens analyzed using four different assays. Pie charts are color-coded to reflect whether specimens were tissue (blue) or plasma (purple). The ALK inhibitor received immediately prior to biopsy is shown. The SNaPshot cohort includes specimens genotyped using both SNaPshot and Solid Fusion Assays. Asterisk (*) includes 28 cases analyzed by both FISH and SNaPshot/Solid Fusion Assay. One patient (MGH915) had molecular profiling of two disease sites at relapse on lorlatinib but is only counted once in the FISH cohort given identical findings at both sites. TKI: tyrosine kinase inhibitor; 2nd gen: second-generation (B) Table lists MET alterations identified in resistant specimens. *MGH075 had focal MET amplification that was too variable to estimate copy number or MET/CEP7. **Plasma testing evaluated MET mutations but did not assess for MET rearrangement. MET/CEP7: ratio of MET to centromere 7 probe; CN: copy number; FISH: fluorescence in-situ hybridization; NGS: next-generation sequencing, ex14 skip: exon 14 skipping. (C) Bar graphs illustrate frequency of MET amplification according to prior ALK inhibitors received. The p-value corresponds to the comparison of MET amplification frequency in crizotinib-naïve vs crizotinib-pretreated patients who received next-generation ALK inhibitors. 2nd-gen. ALKi: second-generation ALK inhibitor.
Molecular Testing
MET copy number was quantified in tissue using FISH or the FoundationOne assay as previously described.19 For cases assessed by FISH, amplification was defined as a ratio of MET to centromere 7 (MET/CEP7) of ≥ 2.2.20 Cases with MET/CEP7 ≥ 4 were classified as high-level amplification.20 For tumors genotyped with the FoundationOne assay, MET amplification was defined as ≥ 6 gene copies.19 With the Guardant360 assay, MET amplification was defined as absolute plasma copy number ≥ 2.1.21 As MET copy number analysis based on MGH SNaPshot NGS has not been formally validated, this assay was not used to detect MET amplification. MET rearrangements and mutations and ALK mutations were detected in tissue using either FoundationOne or the MGH SNaPshot/Solid Fusion Assays.19,22 Guardant360 was used to identify MET and ALK mutations in plasma, but is not designed to detect MET rearrangements.21 All patients included in this study provided written informed consent for molecular analysis.
Cell Lines
Using methods previously described,13 we generated cell lines from pleural fluid collected from MGH915 prior to lorlatinib (MGH915–3) and from a xenograft of pleural fluid at relapse on lorlatinib (MGH915–4). The cell lines were sequenced to confirm the presence of ALK rearrangement, MET rearrangement, and MET amplification. The ST7-MET fusion transcript was confirmed by sequencing a 700 bp region spanning the fusion breakpoint in cDNA generated from RNA extracted from MGH915–4. The corresponding ST7-MET fusion sequence was constructed by fusing ST7 exon 1 (NM_021908.3) to exons 2–21 of MET (NM_000245.4) which includes the full-length MET coding sequence. This resulting fusion sequence incorporates the ST7 ATG start codon corresponding to position 41 exon 1 followed by 151 bp of ST7 exon 1 and 14 bp of MET exon 2 5’ to the wild-type MET ATG start codon. Wild-type MET sequence was obtained from an ORF cDNA clone (GenScript OHu28578, NM_001127500.2). The sequence was cloned into pEN TTmcs (Addgene #25755), recombined with pSLIK-Neo (Addgene #25735) by using Gateway LR Clonase Enzyme (Invitrogen), and lentiviral particles were generated in HEK293FT cells by transfection with ViraPower packaging Mix (Invitrogen) following the manufacturers’ instructions. H3122 cells were transduced by lentiviral infection and selected with G418 (Gibco) for 5 days. The expression of ST7-MET was induced by doxycycline followed by addition of lorlatinib to select the ST7-MET-expressing resistant population. After selection, cells were maintained with continuous culture in 1 μg/mL of doxycycline and 300 nM of lorlatinib. For drug sensitivity assays using DOX- controls, doxycycline was removed 3 days prior to the experiment to allow for ST7-MET expression levels to return to baseline.
Drug Sensitivity Assays, Cell Proliferation Assays, and Western Blot Analysis
2,000 cells were plated in triplicate into 96-well plates. Viability was determined by CellTiter-Glo (Promega) three days after drug treatment in drug sensitivity assays. Luminescence was measured with a SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices). GraphPad Prism (GraphPad Software) was used to analyze data. For Western blotting, a total of 0.25–2×106 cells was treated in 6-well plates for 6 hours. Equal amounts of total cell lysate were processed for immunoblotting.
Whole Genome Sequencing
Whole genome sequencing was performed as described in the Supplementary Methods.
Statistical Analysis
Fisher’s exact test was used to compare MET amplification frequency between treatment groups. All p-values were based on a two-sided hypothesis and computed using Stata 12.1.
RESULTS
Study Population
To survey the landscape of molecular alterations associated with resistance to ALK TKIs, we analyzed 207 tissue (n=101) and plasma (n=106) specimens from 136 patients with ALK-positive NSCLC relapsing on ALK TKIs (Figure 1A). Four assays were used for the analysis (see Methods), including two tissue-based next-generation sequencing (NGS) assays (FoundationOne and MGH SNaPshot/Solid Fusion Assay),19,22 a plasma NGS assay (Guardant360),21 and fluorescence in-situ hybridization (FISH) for MET amplification (Supplementary Table 1).
The clinicopathologic characteristics of the study patients were consistent with an ALK-positive NSCLC population (Supplementary Table 2). Of the 207 specimens, twelve (6%) were from patients relapsing on crizotinib who had not been exposed to other ALK TKIs. The majority of specimens (n=136/207, 66%) were obtained at progression on a second-generation ALK TKI. The remaining fifty-nine (29%) specimens were collected from patients relapsing on lorlatinib. Overall, ALK kinase domain mutations were detected in 34 (47%) of 73 tissue biopsies genotyped by NGS, including 25/46 (54%) specimens obtained after second-generation TKIs and 9/25 (36%) post-lorlatinib biopsies (Supplementary Figure 1). These results suggest that a significant fraction of resistant cases lack secondary ALK mutations and likely harbor ALK-independent mechanisms of resistance.
Genetic Alterations of MET
MET Amplification in Tissue Biopsies
To evaluate the role of MET as a resistance mechanism, we first assessed MET copy number in 86 tumor biopsies using FISH or the FoundationOne assay (Figure 1A, Supplementary Table 1). Eleven (13%) biopsies harbored MET amplification (Figure 1B), including four with low-level MET amplification (MET/CEP7 2.4–3.9) and six with high-level MET amplification based on FISH (MET/CEP7 5.2 to >25) or NGS (16–19 MET copies). One sample had focal MET amplification by NGS, and FISH was too variable to estimate copy number. In 3 of 11 cases, there was available tissue to assess MET copy number in a biopsy from an earlier timepoint, none of which had MET amplification, supporting the notion that MET amplification was newly acquired. No co-alterations in other genes potentially associated with resistance or bypass signaling were identified in the 11 biopsies. In addition, MET amplification was mutually exclusive with ALK resistance mutations, with the exception of one case, MGH9226 (Figure 1B, Supplementary Figure 1). This patient developed ALK I1171N (without MET amplification) in plasma after relapsing on first-line alectinib, and then responded to second-line brigatinib for 5.5 months. After relapsing on brigatinib, tumor biopsy demonstrated persistence of ALK I1171N and acquisition of MET amplification, suggesting that MET was driving resistance.
All 11 cases with MET amplification were patients relapsing on second- or third-generation ALK TKIs (Figure 1B). MET amplification was identified in six (12%) of 52 biopsies taken after a second-generation ALK TKI and in five (22%) of 23 post-lorlatinib biopsies. MET amplification was not detected in patients relapsing on crizotinib. In addition, six (55%) of the 11 positive cases had never received crizotinib (Figure 1B,C). To determine whether the development of MET amplification may be impacted by prior crizotinib exposure, we evaluated the frequency of MET amplification according to prior TKI therapy. Tumors from patients previously treated with crizotinib followed by next-generation TKI(s) were significantly less likely to harbor MET amplification than those from patients treated only with next-generation TKI(s) (9% vs 33%, p=0.019, Figure 1C). Thus, prior exposure to crizotinib, an ALK inhibitor with documented MET activity, may have suppressed emergence of MET-amplified clones in these patients.
MET Copy Number Gain by Plasma Genotyping
We then analyzed MET copy number in 106 plasma specimens from patients relapsing on next-generation ALK TKIs using the Guardant360 assay (Figure 1A). MET copy number gain was detected in eight (7.5%) plasma specimens (Supplementary Figure 2), one of which was due to polysomy of chromosome 7. The seven cases with focal amplification of MET included two (3%) of 77 plasma specimens from patients relapsing on a second-generation ALK TKI and five (17%) of 29 post-lorlatinib specimens (Figure 1B). Among 23 patients with contemporaneous plasma and tissue specimens, MET amplification was detected in both plasma and tissue in four cases (Supplementary Table 3). Eighteen pairs demonstrated absence of MET amplification. One patient, MGH960, had low-level MET amplification (2.2 copies) in plasma which was not detected in a contemporaneous biopsy. Thus, when tissue was used as the reference, plasma genotyping demonstrated 100% sensitivity, 95% specificity, and 80% positive predictive value for detecting MET amplification. The remaining two patients with high-level MET amplification (4.9–6.1 copies) in plasma did not have a paired tumor biopsy.
Four of the seven specimens with focal MET amplification harbored both an ALK mutation and MET amplification in plasma (Supplementary Figure 2), including MGH9226, as described above. Apart from MGH9226’s ALK I1171N mutation, the remaining three plasma specimens contained the gatekeeper ALK L1196M mutation. ALK L1196M is known to cause resistance to crizotinib but is sensitive to next-generation ALK TKIs.7 Indeed, all three patients had received crizotinib prior to the next-generation ALK TKI(s) and presumably developed this secondary mutation after crizotinib exposure. The known activity of next-generation ALK TKIs against ALK L1196M suggests that MET amplification rather than the ALK mutation was likely mediating drug resistance. In addition to MET amplification (6.1 copies), MGH9224’s plasma demonstrated KRAS amplification (2.6 copies) and a PIK3CA E545K mutation (allelic frequency 0.04%) which may have also contributed to resistance. The remaining plasma specimens did not contain other putative drivers of resistance.
Other Genetic Alterations of MET
We next analyzed resistant specimens for other genetic alterations that could lead to aberrant MET activation. To identify MET mutations, we reviewed molecular profiling results from 179 tissue and plasma specimens genotyped by MGH SNaPshot/Solid Fusion NGS (n= 43), FoundationOne (n=30), or Guardant360 (n=106) (Supplementary Figures 1,2). One plasma specimen from a patient relapsing after sequential alectinib and brigatinib harbored MET exon 14 skipping without MET amplification (Figure 1B). Two additional plasma specimens contained MET mutations (D1101H and G500C, Supplementary Figure 2) of unknown significance. MET mutations were not detected in any of the tissue specimens.
In two (3%) of 73 tissue specimens, we detected a rearrangement fusing exons 2–21 of MET with exon 1 of ST7 (suppressor of tumorigenicity 7), both of which reside in close proximity on chromosome 7q (Figure 2A). Both cases were detected using an RNA-based NGS assay (MGH Solid Fusion Assay).22 In the first case, the ST7-MET fusion was seen in MGH9284’s pericardial fluid at progression on first-line alectinib without concurrent MET amplification. The patient had primary progression on second-line lorlatinib at which time molecular profiling of pleural fluid did not demonstrate the rearrangement but revealed low-level MET amplification (MET/CEP7 2.4, Figure 1B). In the second case, concomitant ST7-MET and high-level MET amplification were detected in MGH915’s pleural fluid after failure of lorlatinib (Figure 1B), neither of which were present prior to lorlatinib. Interestingly, contemporaneous biopsy of an axillary node showed only high-level MET amplification and no detectable ST7-MET.
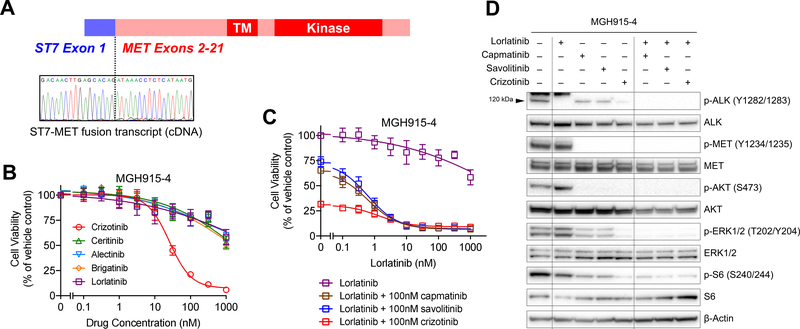
Figure 2. Acquired Resistance Due to ST7-MET and MET Amplification.
(A) Sanger sequencing of the RT-PCR product in MGH915’s lorlatinib-resistant pleural fluid sample shows fusion of ST7 exon 1 to MET exon 2. (B) and (C) Cell viability after 3 days of monotherapy or combination therapy, as indicated. Viability was determined using CellTiter-Glo. Data are mean ± s.e.m. of three biological replicates. (D) Immunoblotting to assess phosphorylation of ALK, MET, and downstream targets in cells treated with lorlatinib and the MET TKIs shown. The arrowhead at 120 kDa indicates the band for phospho-ALK. Cells were treated with each drug at 300 nM for 6 hours.
Functional Validation of ST7-MET as a Targetable Mechanism of Resistance
To evaluate the functional role of MET in driving resistance to ALK TKIs, we established cell lines from MGH915’s pre-lorlatinib and post-lorlatinib malignant pleural effusion (MGH915–3 and MGH915–4, respectively). Consistent with the pleural fluid analysis, the MGH915–4 cell line harbored both MET amplification (MET/chromosome 7 ratio 4.2) and the ST7-MET rearrangement, but MGH915–3 did not (Supplementary Figure 3A, B). High MET expression was confirmed in MGH915–4 cell line at the mRNA and protein level (Supplementary Figure 3B, C). In cell viability studies, the MGH915–4 cell line was resistant to second- and third-generation ALK TKIs, none of which have MET activity, but was sensitive to crizotinib (Figure 2B). Treatment with MET-specific TKIs capmatinib or savolitinib, none of which have ALK activity, partially suppressed proliferation (Supplementary Figure 3D). However, the combination of the ALK-selective TKI lorlatinib with capmatinib, savolitinib, or crizotinib potently suppressed cell proliferation (Figure 2C, Supplementary Figure 3D). Consistent with the cell growth assays, lorlatinib monotherapy potently inhibited ALK phosphorylation in immunoblotting studies, but failed to inhibit downstream ERK or PI3K/AKT signaling (Figure 2D). Only dual inhibition of ALK and MET by crizotinib or by the combination of lorlatinib plus a MET TKI effectively suppressed both ALK and downstream signaling pathways (Figure 2D, Supplementary Figure 3E).
As MGH915–4 harbors the ST7-MET rearrangement, we next evaluated whether ST7-MET rearrangement could drive resistance to ALK inhibition. The sensitive ALK-positive cell line H3122 was transduced with lentivirus expressing a TET-inducible ST7-MET construct corresponding to the ST7-MET fusion identified in MGH915–4 (Figure 2A, Supplementary Figure 3C). Whereas the H3122 cells without expression of ST7-MET were highly sensitive to lorlatinib and other ALK TKIs, doxycycline-induced expression of ST7-MET in H3122 cells caused resistance to all ALK TKIs except crizotinib (Figure 3A). Treatment with MET TKIs restored sensitivity to lorlatinib (Figure 3B). Similarly, treatment with a selective MET TKI alone did not suppress proliferation of H3122 cells expressing ST7-MET, but the addition of lorlatinib to the MET TKIs resensitized the cells to treatment. (Figure 3B). The combination of lorlatinib plus crizotinib had greater antiproliferative effect than crizotinib alone, likely due to more potent inhibition of ALK (Figure 3B).
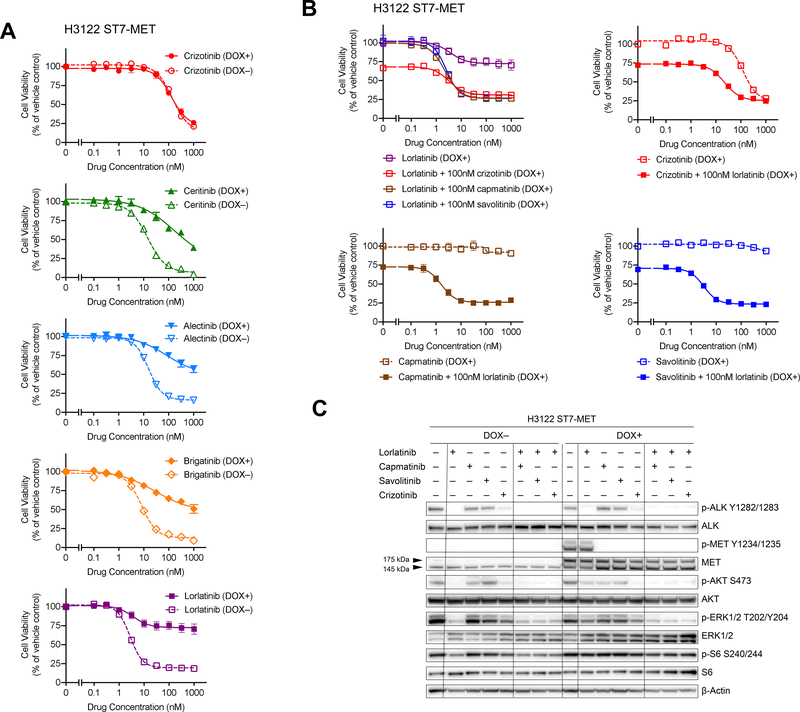
Figure 3. Expression of ST7-MET Confers Resistance to ALK TKIs and Can Be Overcome by Dual ALK and MET Inhibition.
(A) and (B) Cell viability of H3122 cells with or without doxycycline-induced ST7-MET expression (DOX+ and DOX-, respectively) was measured after 3 days of monotherapy or combination therapy, as indicated. Viability was determined using CellTiter-Glo. Data are mean ± s.e.m. of three biological replicates. (C) Immunoblotting to assess phosphorylation of ALK, MET, and downstream targets in cells treated with lorlatinib and the MET TKIs shown. The arrowheads at 175 kDa and 145 kDa indicate the band for pro-MET and MET beta-chain respectively. Cells were treated with each drug at 300 nM for 6 hours. DOX: doxycycline.
We next examined the effect of ST7-MET expression on ALK and MET signaling pathways. In the absence of doxycycline induction of ST7-MET, we observed very little MET phosphorylation, and suppression of ALK phosphorylation with ALK TKIs was sufficient to inhibit downstream MAPK and PI3K signaling (Figure 3C). Upon doxycycline-induced expression of ST7-MET, we observed increased phospho-MET, which could be suppressed with MET TKIs. Consistent with the effects on cell viability, the combination of MET plus ALK inhibitor, but not either agent alone, was sufficient to inhibit downstream signaling (Figure 3C). Of note, we were unable to distinguish between ST7-MET and endogenous MET by western blot due to their similar sizes and lack of suitable antibodies capable of detecting ST7 exon 1. However, upon doxycycline induction of ST7-MET, we detected an increase in two bands corresponding to immature pro-MET (175 kDa) and the mature MET beta-chain (145 kDa), with an increase in phosphorylation of mature MET only after doxycycline induction (Figure 3C). To compare the functional effect of ST7-MET relative to amplification of wild-type MET, we also generated H3122 cells overexpressing wild-type MET. Similar to ST7-MET, doxycycline-induced expression of MET caused resistance to all ALK inhibitors except crizotinib, and the combination of ALK and MET inhibitors suppressed downstream signaling and cell proliferation (Supplementary Figure 4). In our overexpression system, the effect of ST7-MET and wild-type MET were comparable. While the nature of our experimental system precludes the ability to discriminate between increased MET activity resulting from a hyper-functional ST7-MET protein or increased MET expression level, these studies mimic the MGH915–4 clinical scenario and suggest that resistance resulting from increased expression of ST7-MET is potentially targetable.
Clinical Activity of Combined ALK/MET Inhibition in MET-Driven Resistance
Crizotinib Monotherapy
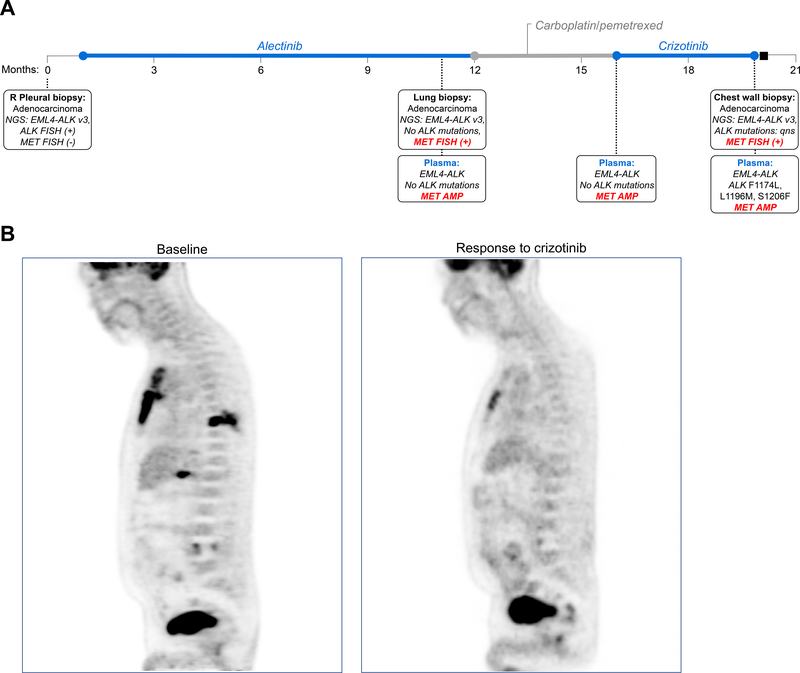
Based on our preclinical findings, we treated two resistant ALK-positive patients with combination ALK/MET TKIs. The first patient, MGH939, received first-line alectinib and relapsed after 9 months (Figure 4A). Biopsy of a progressing lung lesion demonstrated no ALK mutations. However, high-level MET amplification with MET/CEP7 >25 was identified (Figure 1B). Similarly, plasma testing showed no detectable ALK mutations, but evidence of MET amplification (2.5 copies). The patient was treated with carboplatin/pemetrexed for 5 cycles and then transitioned to crizotinib after disease progression. Scans performed five weeks after initiating crizotinib demonstrated significant response to therapy (Figure 4B). However, the patient relapsed 10 weeks later. Biopsy of a chest wall mass confirmed persistent MET amplification. There was insufficient tissue for NGS, but plasma testing revealed multiple ALK resistance mutations that were not detected pre-crizotinib/post-alectinib, in addition to the known MET amplification. These findings suggest that while crizotinib is active against both ALK and MET, its activity may ultimately be limited by secondary ALK mutations.
Figure 4. Clinical Response to Crizotinib in a Patient with ALK-Positive Lung Cancer and Acquired MET amplification:
(A) Timeline illustrates treatments received and molecular profiling results from serial tissue and plasma specimens analyzed during MGH939’s disease course. NGS: next-generation sequencing; v3: EML4-ALK variant 3, or fusion of exon 6 of EML4 to exon 20 of ALK; FISH: fluorescence in-situ hybridization, qns: quantity not sufficient; AMP: amplification. (B) Positron emission computed tomography scans depict metabolic response to treatment in the chest wall and liver during treatment with crizotinib.
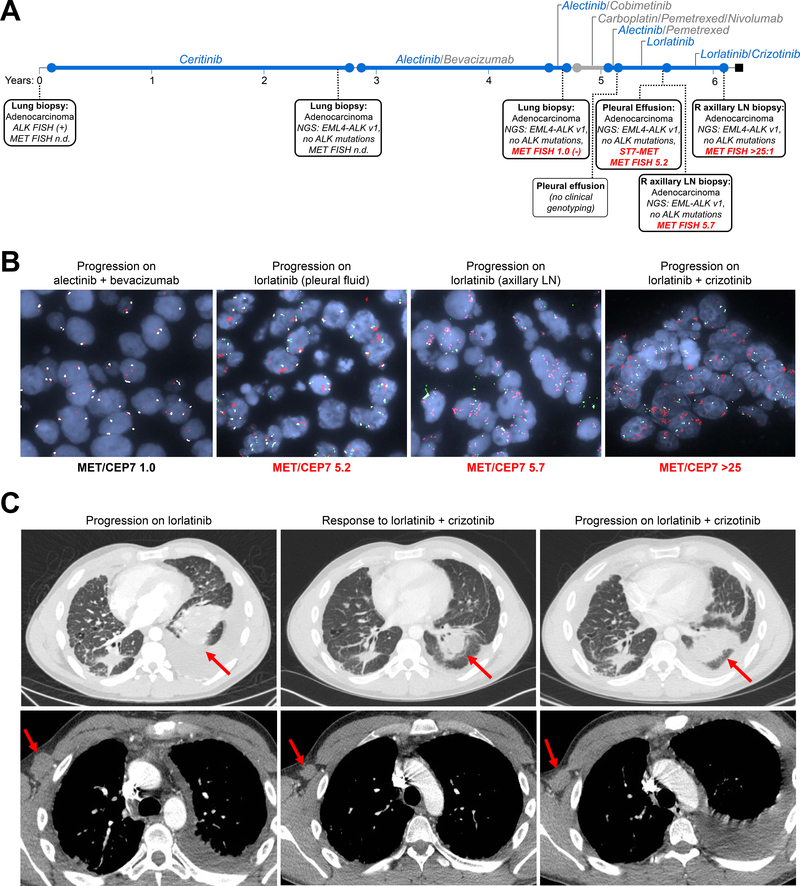
Combination Lorlatinib plus Crizotinib
The second patient, MGH915, was treated with first-line ceritinib with response lasting 31 months. Subsequent therapies included alectinib-based combinations, chemotherapy plus immunotherapy, chemotherapy with alectinib, and lorlatinib (Figure 5A). As noted previously, at the time of relapse on lorlatinib, analysis of pleural fluid demonstrated ST7-MET and high-level MET amplification (MET/CEP7 ratio 5.2, Figure 5B). A concurrent biopsy of a growing axillary node also revealed MET amplification (MET/CEP7 ratio 5.7) but did not demonstrate the MET rearrangement. Based on these findings, the patient received the combination of lorlatinib and crizotinib. Due to potential overlapping toxicities, the lorlatinib dose was reduced to 50 mg when crizotinib 250 mg BID was added. After two weeks on the combination, lorlatinib was escalated to 75 mg daily. The patient tolerated combined therapy well with side effects of grade 1 nausea and grade 1 lipid elevation. He experienced rapid symptomatic improvement, and first restaging scans confirmed improvement in disease (Figure 5C). However, after 3 months, the patient developed progression of lung and axillary metastases (Figure 5C). Biopsy of an axillary node demonstrated persistent high-level MET amplification with MET/CEP7 ratio >25 (Figure 5B).
Figure 5. Clinical Activity of Dual ALK/MET TKIs in MET-Driven Resistance.
(A) Timeline of treatments and biopsies for MGH915. NGS: next-generation sequencing; n.d.: not performed; v1: EML4-ALK variant 1, or fusion of exon 13 of EML4 to exon 20 of ALK; R: right; LN: lymph node; FISH: fluorescence in-situ hybridization (B) FISH images from serial biopsies demonstrate progressive increase in MET copy number. LN: lymph node; MET/CEP7: ratio of MET to centromere 7 probe. (C) CT scans depicting radiologic response to combined lorlatinib and crizotinib, with decrease in pleural fluid and lung mass (red arrow, top panels) and decrease in right axillary node (bottom panels, red arrow). The patient later relapsed at these same sites due to resistance.
Convergent Evolution of MET Alterations During Treatment with ALK Inhibitors
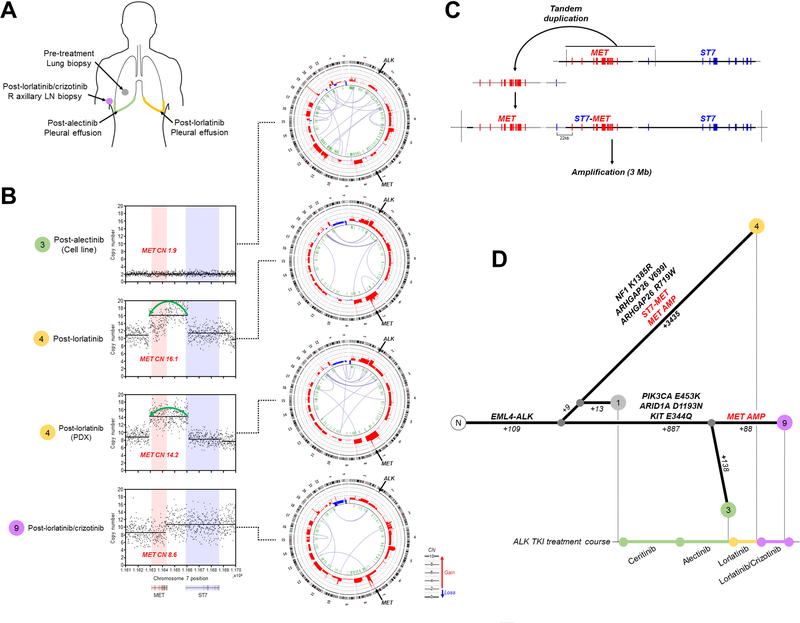
To investigate the evolutionary origin of MET alterations, we performed whole genome sequencing (WGS) on MGH915’s serial tumor specimens and patient-derived models (Figure 6A, Supplementary Figure 5A,B). We first focused on the MET locus and observed that the post-lorlatinib specimens (MGH915–4) harbored a focal amplification (copy number 14–16) extending from the intergenic region preceding the MET gene to intron 1 of ST7 (Figure 6B). This region was contained within a larger ~3 Mb amplification (Supplementary Figure 5C), consistent with an initial tandem duplication of the MET-ST7 locus, followed by amplification of the resulting ST7-MET and wild-type MET loci (~7 copies each) (Figure 6C). The duplication placed exon 1 of ST7 22kb upstream of the MET transcriptional start site, likely generating a chimeric read-through fusion whereby mRNA processing causes the first exon of ST7 to be spliced to MET exon 2. Neither the tandem duplication nor MET amplification were present in the pre-lorlatinib (post-alectinib) pleural effusion (MGH915–3). In the post-lorlatinib/crizotinib combination sample (MGH915–9), we observed an independent MET amplification event that did not involve the amplified 3 Mb region housing the ST7-MET tandem duplication (Figure 6B, Supplementary Figure 5C). When all non-MET mutation clusters and MET alterations were assessed, there was significant overlap between mutations in the post-alectinib pleural effusion (MGH915–3) and the post-lorlatinib/crizotinib axillary node (MGH915–9), but MET amplification only occurred in MGH915–9 (Figure 6D). The post-lorlatinib pleural effusion (MGH915–4), however, represented a distinct branch of the phylogenetic tree which included >3400 private mutations in addition to MET amplification and ST7-MET. Thus, reconstruction of phylogenetic relationships supports convergent evolution of independent genomic events leading to MET-driven acquired resistance to ALK TKIs.
Figure 6. Convergent Evolution Upon MET Activation Drives Resistance to Lorlatinib.
(A) Metastatic disease sites analyzed by whole genome sequencing. (B) Copy number profiles of MET locus demonstrate amplification encompassing MET and first exon of ST7 in post-lorlatinib samples. Circos plots depicting chromosomal structure are shown on right. (C) Proposed sequence of genomic alterations leading to generation of ST7-MET fusion and amplification. The EML4-ALK rearrangement (*) and MET locus (**) are indicated. (D) Phylogenetic relationship of serial tumors samples based on shared and private mutations and MET alterations: MGH915–1 (pre-treatment tumor biopsy), MGH915–3 (cell line derived from post-alectinib pleural effusion), MGH914–4 (post-lorlatinib pleural effusion and patient-derived xenograft), MGH915–9 (post-lorlatinib/crizotinib axillary lymph node biopsy). Number of somatic mutations and selected mutations private to each branch point are indicated.
DISCUSSION
We analyzed >200 tissue and plasma specimens to identify genetic alterations leading to MET bypass signaling in resistant ALK-positive NSCLC. We detected MET amplification and/or a MET fusion in 15% of tumors after failure of a next-generation ALK TKI. Importantly, resistant cell line models and patients harboring MET amplification or rearrangement could be re-sensitized to treatment with dual ALK/MET inhibition.
MET gene amplification was initially discovered as a bypass mechanism in EGFR-mutant NSCLC in 2007 and has subsequently been identified in up to 20% of resistant specimens.14,23 In this study, we demonstrate that the prevalence of MET alterations in patients relapsing on ALK TKIs may be comparable to that seen in EGFR-mutant NSCLC. Interestingly, the incidence of MET amplification in EGFR-mutant NSCLC is higher after exposure to the third-generation EGFR TKI osimertinib compared with less potent EGFR TKIs.14,15 Similarly, in our series we observed that the frequency of MET amplification was highest in tumors resistant to the third generation, broad-spectrum ALK TKI lorlatinib (Figure 1C). These findings suggest that an association may exist between TKI potency and likelihood of developing ALK-independent resistance mechanisms such as MET amplification. In addition to TKI potency, TKI selectivity may also impact the evolution of resistance. In support of this hypothesis, we observed MET amplification in one-third of cases that received front-line second-generation ALK TKIs (which have no activity against MET) compared to less than 10% of patients initially treated with crizotinib.
MET signaling can be activated through a variety of mechanisms, including copy number gain, mutation, fusions, and ligand upregulation.24 In this study, we found that copy number gain was most frequently used to activate MET bypass signaling. The range of MET copies in tissue was broad, spanning MET/CEP7 ratios of 2.4 to greater than 25 (Figure 1B). In cases where MET amplification was detected in paired tissue and plasma specimens, the number of MET copies in plasma was consistently lower than in tissue. Tumor DNA represents a small fraction of total cell-free DNA and some disease sites are more likely to shed tumor DNA than others. Thus, calculation of MET copy number in plasma is challenging. The relationship between MET copy number and dependency on MET bypass signaling in resistant ALK-positive NSCLC remains to be determined. While NSCLCs with de novo high-level MET amplification appear to be more sensitive to crizotinib than NSCLCs with low-level MET amplification, MET/CEP7 ratio of 4–5 is an imperfect threshold for predicting sensitivity to crizotinib and some tumors with low-level amplification respond to crizotinib.20 In the context of acquired resistance, the relationship between MET copies and sensitivity to MET TKIs may be more complex. Specifically, in resistant ALK-positive NSCLCs incompletely inhibited by ALK TKIs, it is conceivable that even low-level MET amplification may be adequate to restore proliferative signals. Furthermore, intratumoral molecular heterogeneity resulting from exposure to multiple lines of therapy may impact response to MET TKIs even among ALK-positive tumors with high-level MET amplification.
In addition to MET amplification, we detected identical ST7-MET rearrangements in two patients relapsing on next-generation ALK TKIs. Rare ST7-MET rearrangements with different breakpoints have been described in gliomas and ovarian cancer, but until now the pathogenicity of these fusions was unknown.25,26 MGH915’s patient-derived cell line harbored both ST7-MET and MET amplification, preventing us from assessing the independent contribution of each MET alteration to ALK TKI resistance. We demonstrate that introducing ST7-MET into a sensitive ALK cell line is sufficient to confer resistance to ALK targeted therapy. However, as engineering the cells to express the ST7-MET rearrangement also increases the total expression ST7-MET, we are unable to conclusively determine whether the fusion alone (in the absence of overexpression) is sufficient to drive resistance. Interestingly, ST7-MET was only identified in one of MGH915’s two contemporaneous biopsies, while high-level MET amplification was present in both of the resistant disease sites. In MGH9284’s case, ST7-MET was identified in pericardial fluid while low-level MET amplification without the rearrangement was detected in pleural fluid two months later. These findings suggest that a single tumor can acquire distinct resistance alterations that converge on MET activation and also highlight the potential for intratumoral heterogeneity of MET genetic alterations.
In two patients with MET-driven resistance, inhibiting both ALK and MET led to clinical responses. However, despite initial responses, both patients relapsed after approximately 3 months. At relapse on crizotinib monotherapy, MGH939’s plasma analysis demonstrated multiple crizotinib-resistant (but next-generation ALK TKI-sensitive) ALK mutations, providing rationale for exploring combinations pairing more potent next generation ALK TKIs with a MET TKI. Indeed, MGH915 did receive combination therapy with lorlatinib and crizotinib, but still had a short-lived response. Repeat biopsy after relapse on the combination revealed persistent MET amplification without additional genetic alterations associated with resistance to ALK and/or MET TKIs (Figure 3B, 4A). Interestingly, the MET/CEP7 ratio increased from 5.7 to >25 at relapse on the lorlatinib/crizotinib combination. As there can be cell-to-cell variability in MET copy number which may be difficult to capture by FISH, it is possible that there was expansion of a more highly-MET amplified subpopulation under the selective pressure of crizotinib. In this scenario, it is conceivable that the MET copy number increase was not the primary driver of relapse in MGH915’s case. Resistant cells may have developed other alterations (including non-genetic aberrations) that decreased sensitivity to dual ALK/MET combination treatment. To inform the clinical development of ALK/MET TKI combinations, additional studies will be required to characterize molecular determinants of resistance to ALK/MET combination therapies, to determine whether sensitivity to combination therapy is impacted by line of therapy, and to assess anti-tumor activity of other more potent MET TKIs like capmatinib and savolitinib (Supplementary Figure 3D). Indeed, there is clinical rationale for exploring ALK/MET combinations besides lorlatinib/crizotinib. For example, while cross-trial comparisons suggest that most adverse events, including nausea/vomiting and edema, occur at comparable rates with the three MET TKIs evaluated in cell lines (crizotinib, capmatinib, savolitinib),4,27,28 the blood-brain barrier penetration of capmatinib is superior to crizotinib. Capmatinib may therefore be a more optimal partner for lorlatinib in patients with brain metastases.
This study has several important limitations. First, due to limited tissue availability, we could not perform analyses for all MET alterations in all specimens. In addition, we used several assays to detect MET alterations, each with its own limitations. Second, patients in our study did not consistently undergo repeat biopsy each time they relapsed on an ALK TKI, precluding definitive timing of the acquisition of the MET alteration in the majority of cases. Third, we limited our analysis to genetic alterations of MET in resistant tumors and did not examine non-genetic mechanisms of MET activation, including upregulation of MET’s ligand hepatocyte growth factor. Fourth, in the WGS analysis, clonal relationships were inferred from patient-derived cell lines and xenografts and biopsies obtained from a variety of sites. It is possible that patient-derived models may not fully recapitulate the spectrum of subclones in the tumor and that spatial heterogeneity contributed to differences in sequential biopsies. Fifth, most of the patients in this study had previously received crizotinib which could have prevented expansion of resistant subclones harboring MET alterations, thereby decreasing the chance of MET activation emerging as a bypass pathway with subsequent next-generation ALK TKIs. As more selective second-generation ALK TKIs have recently replaced crizotinib in the front-line setting, the frequency of MET bypass signaling driving resistance may be higher than estimated in this study. Finally, although we identified rare ST7-MET rearrangements in a subset of cases, we could not fully validate the functional role of the rearrangement or confirm that the rearrangement was as an independent mediator of resistance due to limitations of our experimental system. Additional studies—ideally using patient-derived models that do not harbor concurrent MET amplification—are needed to robustly characterize these rearrangements.
In summary, by performing the most comprehensive analysis of MET alterations in ALK-positive NSCLC to date, we demonstrate that MET is an important bypass mechanism in tumors exposed to next-generation ALK TKIs. The detection of MET amplification in one-third of patients who received next-generation ALK TKIs in the front-line setting provides justification for exploring ALK/MET combinations as an initial treatment strategy for advanced ALK-positive NSCLC.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
With the development of highly potent and selective next-generation ALK inhibitors, resistance is increasingly mediated by ALK-independent mechanisms. Here through molecular profiling of >200 resistant tissue and plasma specimens, including the largest dataset of post-lorlatinib specimens to date, we identified MET amplification in 15% of tumor biopsies from patients relapsing on next-generation ALK inhibitors and detected rare, novel ST7-MET rearrangements in two cases. Upregulation of MET signaling through MET amplification and/or fusion of MET with ST7 decreased sensitivity of ALK-positive cells to ALK inhibitors. In cell lines, combined blockade of ALK and MET overcame resistance driven by MET bypass signaling. Consistent with these findings, two patients with ALK-positive lung cancer and acquired MET alterations achieved rapid clinical and radiographic responses to ALK/MET combination therapy. Our study demonstrates that MET bypass signaling is a recurring and actionable mechanism of resistance to ALK inhibitors, providing rationale for pursuing ALK/MET combination therapy in the clinic.
Acknowledgments
FUNDING:
This work was support by a Conquer Cancer/Amgen Young Investigator Award (I.D.J), an institutional research grant from American Cancer Society (I.D.J.), a National Cancer Institute Career Development Award (K12CA087723-16 to I.D.J.), a Lung Cancer Research Foundation Annual Grant Program award (S.Y.), a Conquer Cancer/AstraZeneca Young Investigator Award (J.J.L.), grants from the National Cancer Institute (R01CA164273 to A.T.S. and R01CA225655 to J.K.L.), by Be a Piece of the Solution, and by Targeting a Cure for Lung Cancer Research Fund at MGH.
CONFLICTS OF INTEREST/DISCLOSURES:
IDJ has received honoraria from Foundation Medicine, consulting fees from Boehringer Ingelheim, research support from Array, Genentech, Novartis, Pfizer, and Guardant Health, and travel support from Array and Pfizer. JJL has received honoraria or served as a compensated consultant for Chugai, Boehringer Ingelheim, and Pfizer; institutional research support from Loxo Oncology and Novartis; and CME honorarium from OncLive. RJN is an employee of Guardant Health. SRD provides independent image analysis through the institution for clinical research trials sponsored by Merck, Pfizer, Bristol Myers Squibb, Novartis, Roche, Polaris, Cascadian, Abbvie, Gradalis, Bayer, Zai Laboratories; and has received honoraria from Siemens Medical Solutions. AS has served on advisory boards for AstraZeneca and Takeda and as a consultant for Genentech, AstraZeneca, and Medtronic. JFG has served as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Loxo, Pfizer, Incyte, Novartis, Merck, Agios, Amgen, Jounce, Regeneron, Oncorus, Helsinn, Jounce, Array, and Clovis Oncology, has an immediate family member who is an employee of Ironwood Pharmaceuticals, has received research funding from Novartis, Genentech/Roche, and Ariad/Takeda, and institutional research support from Tesaro, Moderna, Blueprint, BMS, Jounce, Array, Adaptimmune, Novartis, Genentech/Roche, Alexo and Merck. ATS has served as a compensated consultant or received honoraria from Achilles, Archer, ARIAD, Bayer, Blueprint Medicines, Chugai, Daiichi Sankyo, EMD Serono, Foundation Medicine, Genentech/Roche, Guardant, Ignyta, KSQ Therapeutics, LOXO, Natera, Novartis, Pfizer, Servier, Syros, Taiho Pharmaceutical, Takeda, and TP Therapeutics, has received research (institutional) funding from Daiichi Sankyo, Ignyta, Novartis, Pfizer, Roche/Genentech, and TP Therapeutics; has received travel support from Pfizer and Genentech/Roche; and is an employee of Novartis. ANH has received research grants/funding from Pfizer, Novartis, Amgen, Eli Lilly, Relay Therapeutics, and Roche/Genentech. The remaining authors have no disclosures to report.
References
- 1.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. [DOI] [PubMed] [Google Scholar]
- 2.Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2018;379(21):2027–2039. [DOI] [PubMed] [Google Scholar]
- 3.Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2017;377(9):829–838. [DOI] [PubMed] [Google Scholar]
- 4.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. [DOI] [PubMed] [Google Scholar]
- 5.Soria JC, Tan DS, Chiari R, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet. 2017;389(10072):917–929. [DOI] [PubMed] [Google Scholar]
- 6.Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol. 2017;18(12):1590–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016;6(10):1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AT, Solomon BJ, Besse B, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol. 2019;37(16):1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoda S, Lin JJ, Lawrence MS, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound. Cancer Discov. 2018;8(6):714–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recondo G, Mezquita L, Facchinetti F, et al. Diverse Resistance Mechanisms to the Third-Generation ALK Inhibitor Lorlatinib in ALK-Rearranged Lung Cancer. Clin Cancer Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrustanovic G, Olivas V, Pazarentzos E, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21(9):1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isozaki H, Ichihara E, Takigawa N, et al. Non-Small Cell Lung Cancer Cells Acquire Resistance to the ALK Inhibitor Alectinib by Activating Alternative Receptor Tyrosine Kinases. Cancer Res. 2016;76(6):1506–1516. [DOI] [PubMed] [Google Scholar]
- 13.Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in. Cancer Discov. 2018;8(12):1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YL, Zhang L, Kim DW, et al. Phase Ib/II Study of Capmatinib (INC280) Plus Gefitinib After Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients With EGFR-Mutated, MET Factor-Dysregulated Non-Small-Cell Lung Cancer. J Clin Oncol. 2018;36(31):3101–3109. [DOI] [PubMed] [Google Scholar]
- 17.Gouji T, Takashi S, Mitsuhiro T, Yukito I. Crizotinib can overcome acquired resistance to CH5424802: is amplification of the MET gene a key factor? J Thorac Oncol. 2014;9(3):e27–28. [DOI] [PubMed] [Google Scholar]
- 18.Sakakibara-Konishi J, Kitai H, Ikezawa Y, et al. Response to Crizotinib Re-administration After Progression on Lorlatinib in a Patient With ALK-rearranged Non-small-cell Lung Cancer. Clin Lung Cancer. 2019;20(5):e555–e559. [DOI] [PubMed] [Google Scholar]
- 19.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camidge D, GA O, Clark J, et al. Crizotinib in patients with MET-amplified non-small cell lung cancer: Updated safety and efficacy findings from a phase 1 trial. In. Vol 36. Journal of Clinical Oncology 2018:9062. [Google Scholar]
- 21.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res. 2018;24(15):3539–3549. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. [DOI] [PubMed] [Google Scholar]
- 23.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. [DOI] [PubMed] [Google Scholar]
- 24.Drilon A, Cappuzzo F, Ou SI, Camidge DR. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol. 2017;12(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Earp MA, Raghavan R, Li Q, et al. Characterization of fusion genes in common and rare epithelial ovarian cancer histologic subtypes. Oncotarget. 2017;8(29):46891–46899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson SD, Zhou S, Huse JT, et al. Targetable Gene Fusions Associate With the IDH Wild-Type Astrocytic Lineage in Adult Gliomas. J Neuropathol Exp Neurol. 2018;77(6):437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan HK, Millward M, Hua Y, et al. First-in-Human Phase I Study of the Selective MET Inhibitor, Savolitinib, in Patients with Advanced Solid Tumors: Safety, Pharmacokinetics, and Antitumor Activity. Clin Cancer Res. 2019;25(16):4924–4932. [DOI] [PubMed] [Google Scholar]
- 28.Wolf J, Seto T, Han J-Y, et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. Journal of Clinical Oncology 37, no. 15_suppl(May 20, 2019) 9004–9004.. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.