Abstract
Although a previous study suggested that erythropoietin-producing hepatoma (EPH) receptors play important roles in tumor progression and the overexpression of EPHs in cancer patients is related to poor prognoses, high-throughput gene expression profiling of EPH family members in different types and subtypes of cancers has so far not been conducted. We herein carried out a series of bioinformatic analyses on expressive profiles of every EPH member across 21 different types of clinical cancers versus matched normal tissues gathered from the Oncomine platform. We validated these results by protein expression study of all EPHs family members by The Human Protein Atlas repository. Our results uncovered the overexpression of most EPH subunits in numerous cancer types, especially the dramatic overexpression of six EPHs members, namely EPHA1, EPHA2, EPHA3, EPHA4 and EPHB1, EPHB2, EPHB3, EPHB4 in bladder, colorectal, esophageal, gastric, and prostate cancers. Furthermore, EPHB2 was specifically highly expressed in cervical cancer, EPHA3 in liver cancer, and EPHB1 in uterine cancer. Collectively, expressive profiles of these EPHs were confirmed and correlated with different cancer subtypes as potential biomarkers. This study provides useful information for further studies on cancer development and clinical treatments.
Keywords: Erythropoietin-producing hepatoma (EPH) receptors, ephrins, erythropoietin-producing hepatocellular type-A (EPHA) receptor, erythropoietin-producing hepatocellular type-B (EPHB) receptor, medical oncology, bioinformatics
Introduction
Erythropoietin-producing hepatoma (EPH) and ephrins have recently become a focal point of research. Mammalian EPH receptors were documented to be the dominant group of tyrosine kinase receptors that are composed of nine A-type EPHs (EPHA1~8, 10), five A-type ephrins (ephrins-A1~5), five B-type EPHs (EPHB1~4, 6), and three B-type ephrins (ephrin-B1~3). The binding complexes of EPHs and ephrins are also known to play important roles in cell-cell communication, as they regulate the actin cytoskeleton, cell structure, and cell motility. Furthermore, other cellular processes, such as cell growth, differentiation, apoptosis, and secretion, are also influenced by these proteins [1,2].
EPHs are involved in many important human physiologic activities such as angiogenesis, pl-asticity and regenerative capacity of the nervous system, glucose and intestinal homeostasis, immune responses, bone formation process, and stem cell flexibility. Besides the physiological activities and effects, the activation and inactivation of the EPH/ephrin system are also involved in many pathophysiological processes such as cancer, diabetes, and Alzheimer’s disease [3,4]. Recently, EPHs garnered attention as potential therapeutic targets in cancer treatment. Numerous studies have revealed correlations between EPH/ephrin levels and tumor angiogenesis. In cancer progression, angiogenesis plays a crucial role in metastasis and invasion. These processes are actualized by the signaling communication between cancer cells and tumor-associated endothelial cells [5,6]. Toma et al. showed that cancer progression and angiogenesis are correlated with EPHB4 expression levels [7]. Another study reported that EPHA1 was significantly overexpressed in metastatic renal cell carcinoma [8]. Despite these meaningful findings, no comprehensive screening method has been exploited to examine EPH member expressions in various types of cancer. The advantage of a high-throughput screening strategy is the ability to explore and collect data from numerous studies in an unbiased way. It can help predict information about cancer progression.
In this study, we addressed the expression profiles of EPH family members in 21 types of cancer from the Oncomine database. To our knowledge, this is the first comprehensive study of gene expression profiling in tumor samples versus corresponding cancer cell lines for all EPH family members. These data may shed new light on novel biomarkers for EPHA/B gene family for use in cancer research.
Material and methods
Analysis of public clinical datasets and gene set enrichment analysis (GSEA)
A meta-analysis of mRNA expression profiles of EPH family members in clinical cancer and matched normal tissues was conducted obeying the PRISMA guidelines (Figure 1) [9]. The Oncomine database (www.oncomine.org) was used to obtain a systematic analysis of different types cancer microarray data [10]. Oncomine has over 700 independent datasets, equivalent to 90,000 microarray experiments. This database covers every major cancer type and many pathological subtypes. Differential expressions of EPHs in cancer versus matched normal tissues were determined by the multiple of change-based standard with linear model correlation. Screening criteria in this study were as follows: a fold change of >2.0, a p value of <0.001, and the percentile ranking of genes of <10%. Oncomine default algorithms (two-tailed Student’s t-test and multiple testing corrections) were used to calculate p values and significant differences in EPH expressions between cancerous and control samples. The false discovery rate (FDR) method was used to perform multiple testing corrections. Corrected p values (Q-values) were calculated as Q = N·P/R, where P = p value, N = total number of genes, and R is the sorted rank of p values. By comparing mRNA expressions in 21 cancer types with the corresponding normal tissues, genes of the EPH receptor family (EPHA1~8 and EPHA10, and EPHB1~4 and EPHB6) were studied across the range of various cancer types and sorted by their sets of origin as we previously described [11,12]. Our data encompassed 68 studies and 10,245 samples in total. In Oncomine, the gene summary view mode was displayed during this analysis, and it also presented expression rankings, which were illustrated by color shading. In particular, a gene’s expression color in cancer was related to the gene rank percentile, from the above-described threshold analysis.
Figure 1.
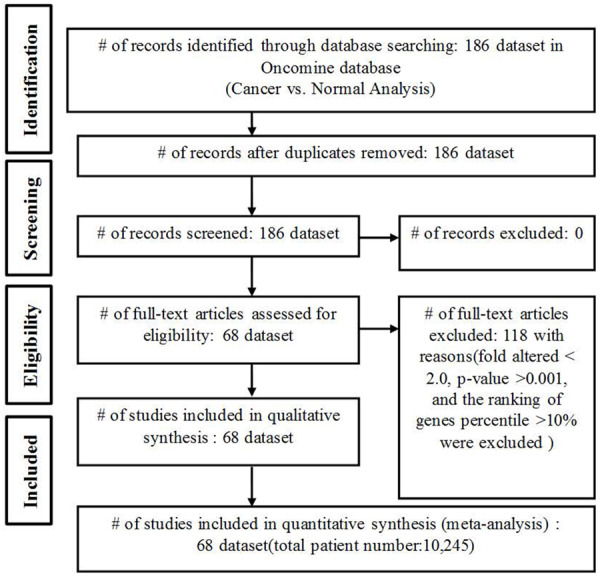
Flow Diagram. Flow chart presenting the identification and collection of studies for the statistical meta-analysis.
Analysis of the human protein atlas database
EPH protein expressions were further evaluated using the publicly available Human Protein Atlas database which contains images of tissue microarrays labeled with antibodies against 11,250 human proteins. These tissue microarrays comprise sections from 46 normal human tissues and more than 20 types of human cancers [13].
Construction of protein-protein interaction (PPI) networks and screening of modules
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org) was used to conduct the protein-protein interaction network. Briefly, the EPH protein symbols were keyed into the search box with multiple proteins/identifiers option. All default parameters from STRING database were selected for this analysis [14]. Subsequently, Cytoscape was used to visualize the network with ClueGO and CluePedia [15-17].
Results
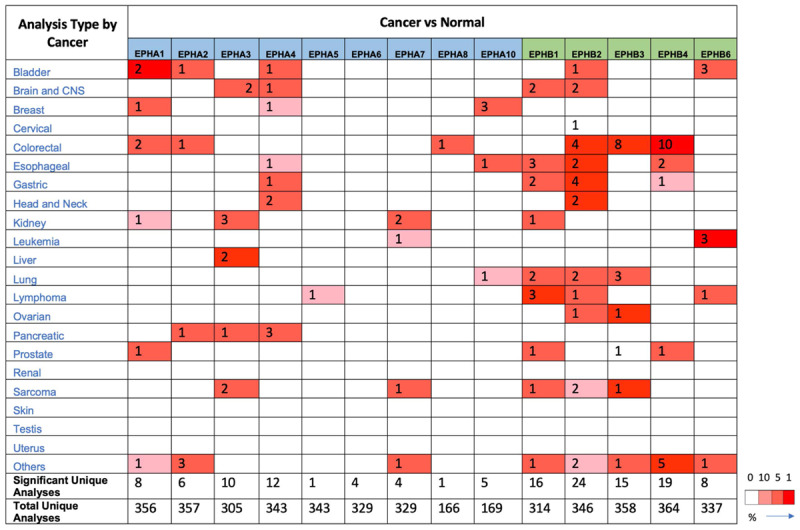
EPH/ephrin receptor expressions in cancer
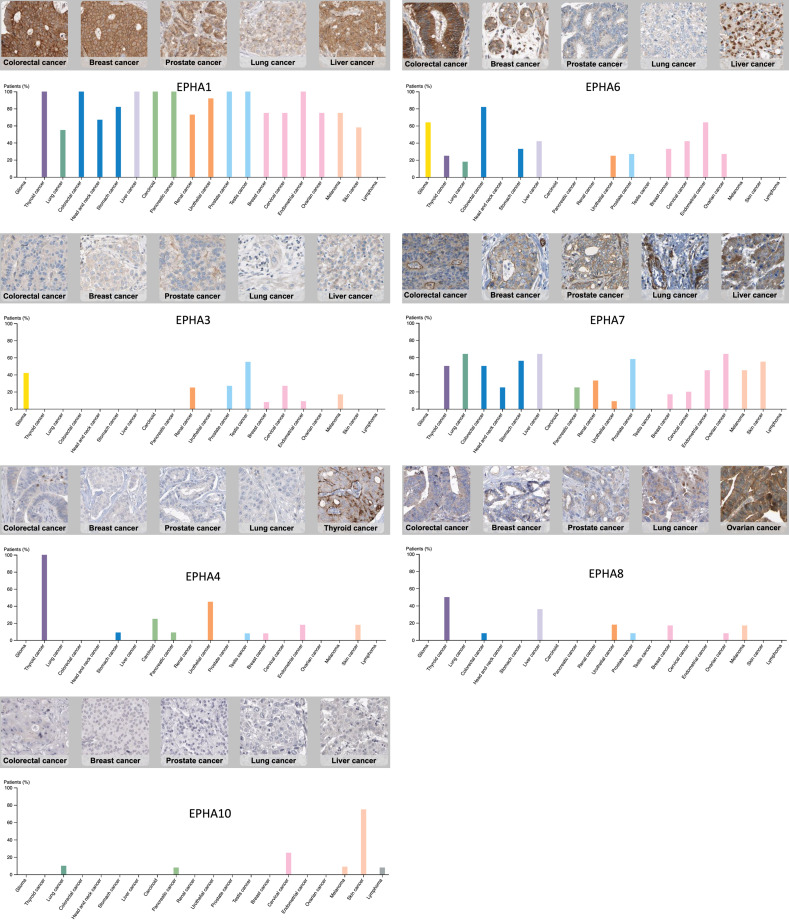
In order to identify expressions of EPH receptors in different cancer subtypes, the web-based high-throughput Oncomine database was utilized [10]. Expression ratios of cancer versus normal tissues are presented in Figure 2, and the stronger intensity of red shows higher overexpression of target genes. The number in each cell reveals the number of analyses that conformed to the selection criteria (a multiple of change of >2.0, a p value of <0.001, and a percentile ranking of genes of <10%). The analyses were classified by the original organ, and all cancerous subtypes were included (e.g., gastric mixed adenocarcinoma or gastric intestinal-type adenocarcinoma). Our bioinformatics data demonstrated that mRNAs of most EPH receptors genes increased in diverse types of cancers. EPHA1 had high expression in prostate carcinoma and infiltrating bladder urothelial tumor tissues (Figure 2). EPHA2 was overexpressed in bladder, colorectal, pancreatic, and vulvar cancers, and seminomas. Expression of EPHA3 increased in brain tumors, kidney, liver, and pancreatic cancers, and sarcomas (Figure 2). Expression of EPHA4 was elevated in bladder, brain, gastric, head and neck, and pancreatic cancers. For EPHB family membranes, 16 of 314 analyses conformed to the selection threshold for EPHB1, 24 of 346 for EPHB2, 15 of 358 for EPHB3, 19 of 364 for EPHB4, and eight of 337 for EPHB6.
Figure 2.
Expression of erythropoietin-producing hepatoma A/B (EPHA/B) family genes across different cancers. Expressions of EPHA/B family genes in different types of cancers compared to normal patients. Each gene was found in its tissue of origin, and the color gradient correlates with a decreasing gene rank percentile. The search criteria threshold was set to p<0.001 with a multiple of change of >2.0 and gene rank percentile of <10% for screening high-throughput datasets of cancer versus normal cases.
EPHA family member expressions in cancer
The current data revealed that EPHA1 was overexpressed in several types of cancer such as bladder, colon, breast, prostate, and renal cancers (Figure 2). EPHA1 had multiples of increase in bladder cancer tissues of 5.16~12.17, p value changes ranged 2.91E-16~7.24E-24, and EPHA1 ranked in the top 1% in either superficial or infiltrating bladder urothelial carcinoma. The multiples of change of EPHA1 significantly increased in all subtypes of colon, breast, prostate, and renal cancers with gene rankings in the top 9% (Table 1).
Table 1.
Expression of EPHA family members in cancer
| Gene | Cancer | Subtype | N (case) | p value (cancer/normal) | t-test (cancer/normal) | Multiple of change (cancer/normal) | % Gene ranking | Database reference |
|---|---|---|---|---|---|---|---|---|
| EPHA1 | Bladder | Superficial bladder cancer | 157 | 7.24E-24 | 14.661 | 12.168 | 30 (in top 1%) | J Clin Oncol [62] |
| Infiltrating bladder urothelial carcinoma | 157 | 2.91E-16 | 9.620 | 5.164 | 97 (in top 1%) | J Clin Oncol [62] | ||
| Colorectal | Colon adenoma | 64 | 5.32E-14 | 9.912 | 2.243 | 437 (in top 3%) | Mol Cancer Res [63] | |
| Rectal adenoma | 64 | 2.07E-5 | 7.220 | 2.464 | 1334 (in top 7%) | Mol Cancer Res [63] | ||
| Breast | Invasive ductal and lobular carcinoma | 593 | 4.01E-9 | 6.765 | 2.040 | 253 (in top 2%) | Nature [64] | |
| Seminoma | Yolk sac tumor, NOS | 107 | 8.73E-6 | 6.916 | 2.980 | 750 (in top 5%) | Cancer Res [65] | |
| Prostate | Prostate carcinoma | 34 | 7.19E-5 | 4.364 | 4.740 | 444 (in top 6%) | Cancer Res [66] | |
| Renal | Clear cell renal cell carcinoma | 67 | 8.53E-4 | 3.932 | 2.602 | 1746 (in top 9%) | BMC Cancer [67] | |
| EPHA2 | Bladder | Infiltrating bladder urothelial carcinoma | 157 | 6.09E-9 | 6.196 | 2.848 | 592 (in top 5%) | J Clin Oncol [62] |
| Pancreas | Pancreatic carcinoma | 52 | 9.50E-8 | 6.523 | 4.139 | 307 (in top 2%) | Cancer Cell [68] | |
| Colorectal | Rectal mucinous adenocarcinoma | 105 | 1.14E-4 | 8.193 | 2.615 | 618 (in top 4%) | Genome Biol [69] | |
| Seminoma | Yolk sac tumor, NOS | 107 | 1.29E-5 | 8.319 | 3.577 | 843 (in top 5%) | Cancer Res [65] | |
| Teratoma, NOS | 107 | 8.81E-7 | 7.929 | 2.915 | 1200 (in top 7%) | Cancer Res [65] | ||
| Vulva | Vulvar intraepithelial neoplasia | 19 | 2.32E-4 | 4.353 | 2.597 | 869 (in top 5%) | Int J Cancer [70] | |
| EPHA3 | Liver | Hepatocellular carcinoma | 115 | 5.73E-14 | 9.844 | 5.891 | 54 (in top 1%) | Mol Med [71] |
| Cirrhosis | 75 | 4.84E-7 | 6.863 | 5.143 | 82 (in top 1%) | Hepatology [72] | ||
| Brain | Classic medulloblastoma | 85 | 2.15E-8 | 6.567 | 9.620 | 138 (in top 3%) | Nature [73] | |
| Desmoplastic medulloblastoma | 85 | 6.28E-4 | 4.011 | 8.838 | 190 (in top 4%) | Nature [73] | ||
| Sarcoma | Dedifferentiated liposarcoma | 158 | 3.78E-9 | 6.954 | 3.850 | 415 (in top 4%) | Nat Genet [74] | |
| Round cell liposarcoma | 54 | 9.79E-4 | 3.809 | 3.791 | 709 (in top 6%) | Cancer Res [75] | ||
| Renal | Hereditary clear cell renal cell carcinoma | 70 | 3.04E-9 | 7.518 | 2.733 | 515 (in top 5%) | Cancer Res [76] | |
| Non-hereditary clear cell renal cell carcinoma | 70 | 3.27E-7 | 6.231 | 2.350 | 642 (in top 6%) | Cancer Res[76] | ||
| Clear cell renal cell carcinoma | 20 | 2.39E-4 | 4.254 | 5.148 | 1192(in top 10%) | Clin Cancer Res [77] | ||
| Pancreas | Pancreatic ductal adenocarcinoma | 78 | 2.01E-10 | 7.208 | 4.834 | 681 (in top 4%) | Hepatogastroenterology [78] | |
| EPHA4 | Breast | Invasive breast carcinoma stroma | 59 | 3.93E-15 | 13.162 | 3.137 | 962 (in top 6%) | Nat Med [79] |
| Bladder | Infiltrating bladder urothelial carcinoma | 157 | 5.46E-15 | 9.099 | 4.959 | 133 (in top 2%) | J Clin Oncol [62] | |
| Pancreas | Pancreatic ductal adenocarcinoma | 78 | 1.89E-10 | 7.196 | 2.884 | 674 (in top 4%) | Hepatogastroenterology [78] | |
| Pancreatic carcinoma | 52 | 6.24E-6 | 4.859 | 2.147 | 833 (in top 5%) | Cancer Cell [68] | ||
| Pancreatic adenocarcinoma | 27 | 4.28E-4 | 6.646 | 121.141 | 334 (in top 7%) | Cancer Res [80] | ||
| Head-Neck | Head and neck squamous cell carcinoma | 54 | 5.79E-10 | 7.635 | 4.288 | 359 (in top 3%) | Cancer Res [81] | |
| Floor of the mouth carcinoma | 84 | 8.39E-4 | 4.223 | 2.557 | 1934 (in top 10%) | Cancer Res [82] | ||
| Brain | Glioblastoma | 101 | 1.30E-6 | 7.380 | 2.781 | 421 (in top 3%) | Cancer Cell [83] | |
| Gastric | Gastric mixed adenocarcinoma | 69 | 1.32E-6 | 7.196 | 3.984 | 264 (in top 2%) | Eur J Cancer [84] | |
| Myeloma | Monoclonal gammopathy of undetermined significance | 78 | 3.90E-5 | 4.263 | 2.034 | 1885 (in top 10%) | Blood [85] | |
| Melanoma | Benign melanocytic skin nevus | 70 | 8.41E-5 | 5.143 | 6.083 | 493 (in top 4%) | Clin Cancer Res [86] | |
| Esophagus | Esophageal adenocarcinoma | 48 | 4.63E-4 | 3.752 | 2.481 | 1137 (in top 8%) | Gastroenterology [87] | |
| EPHA5 | Lymphoma | Diffuse large B-cell lymphoma | 102 | 9.70E-4 | 3.714 | 2.064 | 317 (in top 8%) | J Exp Med [88] |
| EPHA7 | Leukemia | B-Cell acute lymphoblastic leukemia | 2096 | 1.07E-24 | 12.237 | 2.468 | 1058 (in top 6%) | J Clin Oncol [89] |
| Renal | Papillary renal cell carcinoma | 92 | 3.79E-9 | 14.352 | 5.238 | 571 (in top 5%) | Clin Cancer Res [90] | |
| Clear cell sarcoma of the kidney | 35 | 5.92E-4 | 6.479 | 2.489 | 498 (in top 4%) | Clin Cancer Res [91] | ||
| Parathyroid | Parathyroid gland adenoma | 61 | 2.09E-5 | 4.855 | 3.173 | 251 (in top 2%) | Am J Pathol [92] | |
| EPHA8 | Colorectal | Rectosigmoid adenocarcinoma | 237 | 2.60E-5 | 5.202 | 2.445 | 706 (in top 4%) | Nature [93] |
| EPHA10 | Breast | Invasive breast carcinoma stroma | 59 | 1.95E-15 | 13.532 | 2.230 | 909 (in top 5%) | Nat Med [79] |
| Male breast carcinoma | 593 | 5.94E-8 | 9.165 | 3.480 | 319 (in top 2%) | Nature [64] | ||
| Mixed lobular and ductal breast carcinoma | 593 | 3.07E-4 | 5.030 | 3.417 | 1229 (in top 7%) | Nature [64] | ||
| Lung | Lung adenocarcinoma | 246 | 5.65E-10 | 9.846 | 4.732 | 1378 (in top 8%) | Cancer Res [94] | |
| Esophagus | Barrett’s esophagus | 48 | 2.93E-5 | 4.684 | 3.784 | 400 (in top 3%) | Gastroenterology [87] | |
| Prostate | Prostate carcinoma | 122 | 3.33E-5 | 4.527 | 1.970 | 1068 (in top 6%) | Nature [95] | |
| Prostate carcinoma | 21 | 1.26E-4 | 4.694 | 1.980 | 158 (in top 1%) | Clin Cancer Res [96] |
NOS: not otherwise specified.
The current analysis revealed that EPHA2 was overexpressed in pancreas, bladder, colon, and vulvar cancers, and seminomas (Figure 2). EPHA2 had a significant multiple of change of >3.6 and a gene ranking within the top 5% in yolk sac tumors. EPHA2 was also overexpressed with maximum multiples of increase of >2-fold compared to normal tissues, and the gene ranked in the top 5% in infiltrating bladder urothelial carcinoma, rectal mucinous adenocarcinomas, and vulvar intraepithelial neoplasia (Table 1).
Our data also showed that EPHA3 had high expressions in liver, brain, renal, and pancreas cancers and sarcomas, relative to normal matched tissue types (Figure 2). Moreover, it also had significant multiples of increase (>5-fold) in hepatocellular carcinoma and cirrhosis, with the gene ranked in the top 1%. In brain cancer, sarcomas, renal cancer, and pancreas cancer, EPHA3 ranked in top 3%~10% of overexpressed genes with a maximum multiple of change of 9.62 in desmoplastic medulloblastomas (Table 1).
The present data revealed that EPHA4 was significantly overexpressed in seven types of cancers and presented in the top 10% of the majority of commonly altered genes (Figure 2). In invasive breast carcinoma, EPHA4 was found to be significantly overexpressed with a p value of 3.93E-15 and was ranked in the top 6% relative to normal tissues. For infiltrating bladder urothelial carcinoma compared to normal tissues, EPHA4 had a 4.95-fold-increase and was ranked in the top 2%. EPHA4 was overexpressed in pancreas cancer and was ranked within the top 4%~7%. Compared to normal tissues, EPHA4 had gene ranking in the top 3%~10% in head and neck squamous cell carcinomas and floor of the mouth carcinomas. For brain cancer, gastric cancer, myelomas, melanomas, and esophageal cancer, EPHA4 had multiples of change of up to 6-fold with gene ranking in the top 2%~10% (Table 1).
Our results showed that EPHA5 had a 2.06 fold increase in diffuse large B-cell lymphomas relative to normal tissues (Table 1). We found that EPHA7 was overexpressed in kidney cancer, B-cell acute lymphoblastic leukemia, sarcomas, and parathyroid adenomas (Figure 2). EPHA7 had multiples of increase in papillary renal cell carcinoma and clear cell sarcomas of the kidney of 2.489~5.238 (Table 1). For parathyroid adenomas compared to normal tissues, EPHA7 had a 3.173-fold increase with a p value of 2.09E-5, and the gene was ranked in the top 2% (Table 1). We found that EPHA8 had a 2.445-fold increase in rectosigmoid adenocarcinomas with a p value of 2.60E-5, and the gene was ranked in the top 4% (Figure 2, Table 1). We found that EPHA10 not only had high expression in breast cancer but also in lung, esophageal, and prostate cancers (Figure 2, Table 1). EPHA10 was overexpressed in invasive breast carcinoma, male breast carcinoma, and mixed lobular and ductal subtypes (Table 1).
EPHB family members expression in cancer
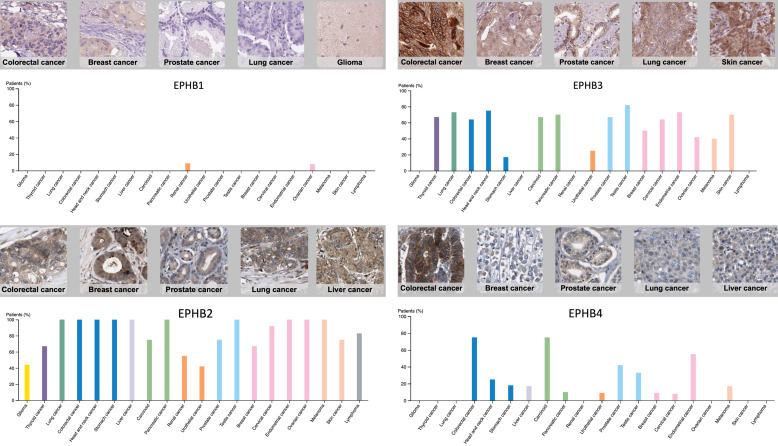
EPHB1 was reported to involve in colorectal cancer [18] and overexpression of EPHB1 was found in patients with gastric cancers [19]. The current data revealed that EPHB1 was overexpressed in brain, esophageal, gastric, kidney, lung, and prostate cancers, lymphomas, sarcomas, and melanomas (Figure 2). EPHB1 was overexpressed in oligodendrogliomas and anaplastic oligodendrogliomas of the brain, in uterus corpus leiomyomas, in diffuse and intestinal subtypes of gastric adenocarcinomas, in Barrett’s esophagitis and esophageal adenocarcinomas, in subtypes of lymphoma (including follicular lymphomas and diffuse large B-cell lymphomas), in benign melanocytic skin nevi, in adenocarcinomas and squamous cell carcinoma of the lungs, in intraepithelial neoplasia of the prostate, and also in clear cell carcinoma of the kidneys (Table 2). Overall, EPHB1 was suggested to be a potential oncogene in cancer development.
Table 2.
Expressions of EPHB family members in cancer
| Gene | Cancer | Subtype | N (case) | p value (cancer/normal) | t-test (cancer/normal) | Multiple of change (cancer/normal) | % Gene ranking | Database reference |
|---|---|---|---|---|---|---|---|---|
| EPHB1 | Brain | Oligodendroglioma | 180 | 7.90E-10 | 6.947 | 4.531 | 501 (in top 3%) | Cancer Cell [97] |
| anaplastic oligodendroglioma | 33 | 3.39E-7 | 6.559 | 2.201 | 379 (in top 2%) | Cancer Res [98] | ||
| Uterus | Uterine corpus leiomyoma | 77 | 4.90E-7 | 5.366 | 2.903 | 318 (in top 2%) | Cancer Res [99] | |
| Gastric | Diffuse gastric adenocarcinoma | 90 | 4.87E-6 | 5.109 | 2.050 | 473 (in top 3%) | Clin Cancer Res [100] | |
| Gastric intestinal type adenocarcinoma | 90 | 7.86E-5 | 4.540 | 2.178 | 467 (in top 3%) | Clin Cancer Res [100] | ||
| Esophagus | Barrett’s esophagus | 118 | 7.23E-6 | 6.398 | 2.369 | 1230 (in top 7%) | PLoS One [101] | |
| Esophageal adenocarcinoma | 48 | 2.68E-5 | 7.013 | 10.052 | 436 (in top 3%) | Gastroenterology [87] | ||
| Barrett’s esophagus | 48 | 4.32E-4 | 3.870 | 2.827 | 783 (in top 6%) | Gastroenterology [87] | ||
| Lymphoma | Follicular lymphoma | 120 | 1.25E-5 | 5.106 | 2.613 | 39 (in top 2%) | Nature [102] | |
| Diffuse large B-cell lymphoma | 120 | 3.04E-4 | 5.286 | 3.006 | 52 (in top 2%) | Nature [102] | ||
| Follicular lymphoma | 102 | 1.10E-4 | 4.555 | 2.088 | 40 (in top 1%) | J Exp Med [88] | ||
| Melanoma | Benign melanocytic skin nevus | 70 | 2.21E-5 | 5.241 | 2.185 | 328 (in top 3%) | Clin Cancer Res [86] | |
| Lung | Lung adenocarcinoma | 73 | 1.01E-4 | 5.192 | 2.608 | 303 (in top 3%) | Proc Natl Acad Sci U S A [103] | |
| Squamous cell lung carcinoma | 73 | 1.35E-4 | 4.645 | 3.360 | 337 (in top 4%) | Proc Natl Acad Sci U S A [103] | ||
| Prostate | Prostatic intraepithelial neoplasia | 101 | 3.44E-4 | 3.899 | 2.616 | 465 (in top 5%) | Nat Genet [104] | |
| Renal | Clear cell sarcoma of the kidney | 35 | 3.79E-4 | 19.043 | 8.371 | 48 (in top 4%) | Clin Cancer Res [91] | |
| EPHB2 | Colorectal | Colon adenoma | 64 | 1.14E-18 | 14.090 | 2.901 | 72 (in top 1%) | Mol Cancer Res [63] |
| Colorectal carcinoma | 82 | 4.91E-15 | 11.319 | 2.266 | 184 (in top 1%) | Clin Exp Metastasis [105] | ||
| Colon adenoma | 40 | 2.96E-8 | 11.028 | 3.221 | 222 (in top 2%) | PLoS One [106] | ||
| Colon carcinoma | 40 | 9.04E-8 | 10.042 | 2.765 | 990 (in top 6%) | PLoS One [106] | ||
| Head-Neck | Tongue squamous cell carcinoma | 58 | 7.10E-13 | 9.363 | 3.708 | 33 (in top 1%) | BMC Cancer [107] | |
| Head and neck squamous cell carcinoma | 38 | 4.79E-5 | 8.795 | 2.652 | 142 (in top 2%) | Oncogene [108] | ||
| Ovarian | Ovarian carcinoma | 195 | 4.75E-12 | 15.823 | 2.250 | 328 (in top 3%) | Cancer Res [109] | |
| Bladder | Infiltrating bladder urothelial carcinoma | 157 | 7.49E-12 | 7.678 | 3.380 | 295 (in top 3%) | J Clin Oncol [62] | |
| Lung | Lung adenocarcinoma | 246 | 1.02E-10 | 9.515 | 2.400 | 1122 (in top 6%) | Cancer Res [94] | |
| Lung adenocarcinoma | 66 | 7.60E-7 | 5.548 | 2.958 | 264 (in top 3%) | BMC Genomics [110] | ||
| Cervix | Cervical squamous cell carcinoma | 66 | 4.73E-10 | 7.630 | 4.561 | 128 (in top 2%) | Genes Chromosomes Cancer [111] | |
| Lymphoma | Centroblastic lymphoma | 336 | 7.80E-10 | 7.767 | 4.212 | 416 (in top 5%) | Nat Genet [112] | |
| Gastric | Gastric intestinal type adenocarcinoma | 69 | 4.82E-9 | 6.762 | 4.236 | 956 (in top 5%) | Eur J Cancer [84] | |
| Gastric intestinal type adenocarcinoma | 90 | 1.42E-7 | 7.171 | 2.155 | 46 (in top 1%) | Clin Cancer Res [100] | ||
| Gastric mixed adenocarcinoma | 90 | 6.08E-5 | 6.158 | 2.177 | 313 (in top 2%) | Clin Cancer Res [100] | ||
| Gastric cancer | 27 | 1.42E-4 | 4.220 | 3.812 | 360 (in top 2%) | Med Oncol [113] | ||
| Sarcoma | Myxofibrosarcoma | 158 | 2.82E-7 | 6.033 | 2.155 | 1084 (in top 9%) | Nat Genet [74] | |
| Round cell liposarcoma | 54 | 8.21E-4 | 3.742 | 2.288 | 665 (in top 6%) | Cancer Res [75] | ||
| Brain | Glioblastoma | 54 | 6.63E-6 | 5.413 | 2.239 | 743 (in top 6%) | Cancer Res [114] | |
| Meningioma | 18 | 8.07E-4 | 4.871 | 4.265 | 37 (in top 3%) | Am J Pathol [115] | ||
| Esophagus | Barrett’s esophagus | 24 | 3.81E-5 | 5.544 | 6.303 | 27 (in top 1%) | Cancer Res [116] | |
| Esophageal adenocarcinoma | 24 | 2.20E-4 | 4.856 | 7.833 | 164 (in top 2%) | Cancer Res [116] | ||
| Seminoma | Yolk sac tumor, NOS | 107 | 6.91E-5 | 5.585 | 2.350 | 1350 (in top 8%) | Cancer Res [65] | |
| Mesothelioma | Pleural malignant mesothelioma | 54 | 4.38E-4 | 4.421 | 2.444 | 938 (in top 8%) | Am J Pathol [117] | |
| EPHB3 | Colorectal | Colorectal adenocarcinoma | 105 | 3.71E-18 | 12.557 | 3.833 | 65 (in top 1%) | PLoS One [106] |
| Colon adenoma | 64 | 7.26E-16 | 11.608 | 2.321 | 218 (in top 2%) | Mol Cancer Res [63] | ||
| Colon adenocarcinoma | 105 | 8.52E-12 | 9.012 | 2.106 | 206 (in top 2%) | Genome Biol [69] | ||
| Colon mucinous carcinoma | 105 | 8.05E-6 | 6.513 | 2.106 | 947 (in top 5%) | Genome Biol [69] | ||
| Colon adenoma | 40 | 5.62E-11 | 19.664 | 6.410 | 27 (in top 1%) | PLoS One [106] | ||
| Colon adenoma | 40 | 7.05E-11 | 18.096 | 4.881 | 30 (in top 1%) | PLoS One [106] | ||
| Colon adenocarcinoma | 123 | 2.68E-9 | 6.525 | 2.171 | 627 (in top 7%) | Int J Cancer [63] | ||
| Rectal mucinous carcinoma | 237 | 1.61E-4 | 6.917 | 3.393 | 1364 (in top 7%) | Nature [93] | ||
| Ovarian | Ovarian serous Cystadenocarcinoma | 594 | 3.43E-10 | 15.427 | 2.306 | 63 (in top 1%) | Nature [118] | |
| Lung | Squamous cell lung carcinoma | 156 | 5.56E-10 | 8.577 | 2.130 | 964 (in top 5%) | PLoS One [119] | |
| Squamous cell lung carcinoma | 73 | 7.31E-5 | 4.961 | 4.747 | 272 (in top 3%) | Proc Natl Acad Sci U S A [103] | ||
| Squamous cell lung carcinoma | 203 | 6.27E-4 | 3.502 | 5.147 | 389 (in top 5%) | Proc Natl Acad Sci U S A [103] | ||
| Sarcoma | Synovial sarcoma | 54 | 8.17E-7 | 7.323 | 7.234 | 77 (in top 1%) | Cancer Res [75] | |
| Testicular | Testicular seminoma | 74 | 8.38E-7 | 6.034 | 2.418 | 475 (in top 5%) | Proc Natl Acad Sci U S A [120] | |
| Prostate | Prostate adenocarcinoma | 89 | 1.66E-4 | 4.303 | 3.914 | 226 (in top 2%) | Cancer Res [121] | |
| EPHB4 | Colorectal | Colorectal adenocarcinoma | 105 | 1.21E-23 | 15.254 | 2.122 | 20 (in top 1%) | PLoS One [106] |
| Colon adenoma | 64 | 8.31E-22 | 16.394 | 2.589 | 20 (in top 1%) | Mol Cancer Res [63] | ||
| Rectal adenoma | 64 | 4.41E-6 | 10.091 | 2.758 | 831 (in top 5%) | Mol Cancer Res [63] | ||
| Colon adenocarcinoma | 237 | 8.13E-16 | 12.139 | 2.265 | 962 (in top 5%) | Nature [93] | ||
| Cecum adenocarcinoma | 237 | 4.04E-13 | 10.517 | 2.729 | 317 (in top 2%) | Nature [93] | ||
| Rectal adenocarcinoma | 237 | 6.18E-13 | 8.617 | 2.091 | 1483 (in top 8%) | Nature [93] | ||
| Colon mucinous carcinoma | 237 | 1.07E-11 | 9.348 | 2.444 | 408 (in top 2%) | Nature [93] | ||
| Colorectal carcinoma | 82 | 3.99E-12 | 11.872 | 2.525 | 476 (in top 3%) | Clin Exp Metastasis [105] | ||
| Colon carcinoma | 40 | 2.92E-10 | 16.846 | 3.071 | 218 (in top 2%) | PLoS One [106] | ||
| Colon adenoma | 40 | 1.45E-7 | 13.443 | 3.056 | 355 (in top 2%) | PLoS One [106] | ||
| Testicular | Testicular seminoma | 74 | 1.39E-11 | 9.097 | 3.313 | 31 (in top 1%) | Proc Natl Acad Sci U S A [120] | |
| Mixed germ cell tumor, NOS | 107 | 3.43E-9 | 7.921 | 2.246 | 1007 (in top 6%) | Cancer Res [65] | ||
| Seminoma, NOS | 107 | 1.28E-8 | 10.424 | 3.845 | 393 (in top 3%) | Cancer Res [65] | ||
| Yolk sac tumor, NOS | 107 | 9.63E-5 | 5.59;2 | 2.345 | 1476 (in top 9%) | Cancer Res [65] | ||
| Gastric | Gastric intestinal type Adenocarcinoma | 69 | 8.77E-9 | 7.324 | 2.367 | 1057 (in top 6%) | Eur J Cancer [84] | |
| Esophagus | Esophageal squamous cell carcinoma | 34 | 2.90E-6 | 6.257 | 2.054 | 555 (in top 5%) | BMC Genomics [122] | |
| Barrett’s esophagus | 48 | 7.42E-4 | 3.516 | 2.623 | 917 (in top 7%) | Gastroenterology [87] | ||
| Melanoma | Skin basal cell carcinoma | 87 | 8.72E-6 | 8.478 | 2.458 | 360 (in top 2%) | BMC Med Genomics [123] | |
| Prostate | Prostate carcinoma | 101 | 2.58E-5 | 4.437 | 2.363 | 305 (in top 3%) | Nat Genet [104] | |
| EPHB6 | Leukemia | T-cell acute lymphoblastic leukemia | 2096 | 3.83E-55 | 21.261 | 3.924 | 71 (in top 1%) | J Clin Oncol [89] |
| T-cell childhood acute lymphoblastic leukemia | 288 | 1.62E-7 | 10.033 | 4.216 | 173 (in top 2%) | Blood [124] | ||
| T-cell acute lymphoblastic leukemia | 127 | 1.13E-6 | 8.926 | 7.396 | 655 (in top 7%) | Leukemia [125] | ||
| Bladder | Superficial bladder cancer | 157 | 3.90E-13 | 10.463 | 5.740 | 786 (in top 7%) | J Clin Oncol [62] | |
| Superficial bladder cancer | 60 | 2.86E-7 | 6.025 | 2.224 | 477 (in top 4%) | Cancer Res [126] | ||
| Superficial bladder cancer | 54 | 1.16E-6 | 10.625 | 3.650 | 15 (in top 2%) | Clin Cancer Res [127] | ||
| Lymphoma | Mantle cell lymphoma | 336 | 3.65E-6 | 5.395 | 5.250 | 193 (in top 3%) | Nat Genet [112] | |
| Mesothelioma | Pleural malignant mesothelioma | 54 | 1.85E-5 | 5.509 | 2.033 | 354 (in top 3%) | Am J Pathol [117] |
We found that EPHB2 had higher expressions not only in colon and cervix tumors but also in head-neck, ovarian, bladder, lung, gastric, brain, esophagus, brain, and salivary-gland cancers, lymphomas, sarcomas, mesotheliomas, and seminomas (Figure 2). EPHB2 was overexpressed in adenomas and carcinoma of the colon, in squamous cell carcinoma of the tongue, head, and neck, in ovarian carcinoma, in infiltrating uroepithelial carcinoma of the bladder, in adenocarcinoma of the lungs, in squamous carcinoma of the cervix, in centroblastic lymphomas, in intestinal or mixed subtypes of gastric adenocarcinomas, in subtypes of sarcomas (myxofibrosarcomas and round cell liposarcomas), in glioblastomas and meningiomas, in Barrett’s esophagitis and esophageal adenocarcinomas, in yolk sac tumors, and in pleural malignant mesotheliomas (Table 2). All of the increases of cancer/normal multiples of change were significant.
We also found that EPHB3 not only had high expression in lung and prostate cancers but also in a variety of cancer subtypes, such as ovarian cancer, sarcomas, and testicular cancer. EPHB3 was present in both colorectal and testicular cancers with gene ranks within the top 1% of upregulated genes (Figure 2). EPHB3 was overexpressed in adenomas, adenocarcinomas, and mucinous carcinoma of the colon and rectum, in serous cystadenocarcinomas of the ovaries, in squamous cell carcinoma of the lungs, in synovial sarcomas, in testicular seminomas, and in prostate adenocarcinomas (Table 2). EPHB3 exhibited the top ranking of expression in all these cancers.
We found that EPHB4 had high expressions in prostate, colorectal, testicular, gastric, and esophageal cancers seminomas, and melanomas (Figure 2). EPHB4 was overexpressed in adenomas, adenocarcinomas, mucinous carcinoma of the colon and rectum, in seminomas, mixed germ cell tumors, and yolk sac tumors of the testes, in the intestinal subtype of gastric adenocarcinomas, in squamous cell carcinoma of the esophagus, in basal cell carcinoma of the skin, and in prostate carcinoma (Table 2). Increased expression of EPHB6 was detected in bladder cancer, leukemia, lymphomas, and pleural malignant mesotheliomas (Figure 2). EPHB6 was mostly overexpressed in T-cell leukemia and superficial bladder cancer (Table 2).
Validation of EPH family member expressions with protein expressions
To further confirm our bioinformatics results analyzed on the Oncomine platform, we used the Human Protein Atlas database to verify EPH receptor members’ protein expressions in a variety of cancer cell lines. Pathology data of clinical human cancer tissues in the Human Protein Atlas collection were analyzed. These data revealed similar protein expression patterns of target genes in different cancer patients. Expressions of EPHA and EPHB family members in various types of cancer, namely colorectal cancer, breast cancer, lung cancer, gliomas, and prostate cancers, were examined by immunohistochemistry (Figures 3, 4). In particular, EPHA1, EPHA6, EPH7, EPHB2, and EPHB3 had strong high expressions throughout many cancer cell lines. These data from the Human Protein Atlas were used to confirm the expressions of EPHA/B proteins from clinical patient tissues. Results of the Human Protein Atlas analysis were consistent with findings from mRNA expressions in the Oncomine analysis.
Figure 3.
Protein expressions of erythropoietin-producing hepatoma A (EPHA) family members in human tumor samples. Protein expression data of EPHA family members were acquired from the Human Protein Atlas. Representative pathology images of immunohistochemical staining for the top four cancers are indicated in the left panel, and the overall protein expression is indicated in the right panel.
Figure 4.
Protein expressions of erythropoietin-producing hepatoma B (EPHB) family members in human tumor samples. Protein expression data of EPHB family members were acquired from the Human Protein Atlas. Representative pathology images of immunohistochemical staining for the top four cancers are indicated in the left panel, and the overall protein expression is indicated in the right panel.
Scoring of genetic associations based on ClueGo and CluePedia
We used the ClueGO and CluePedia databases to query genetic interaction networks associated with EPHA and EPHB family genes. The ClueGO and CluePedia databases incorporate gene-gene interactions from various databases, including Gene Ontology, KEGG, CORUM, and WikiPathways. The various sources of associated data are standardized in the ClueGo and CluePedia databases. A combined score was obtained by computing both known and predicted associations. A higher combined score represents a more-reliable association from more than one type of information. Based on these combined scores, a graphical network of gene-gene interactions was generated for some of the EPHA and EPHB family genes (Figure 5). Strong evidence for interactions among these EPHA and EPHB family genes was supported by STRING, and other networks were validated in previous reports (Figures 6, 7). Hence, our interacting network presents a novel tool for screening potential biomarkers in the EPHA/B gene family.
Figure 5.
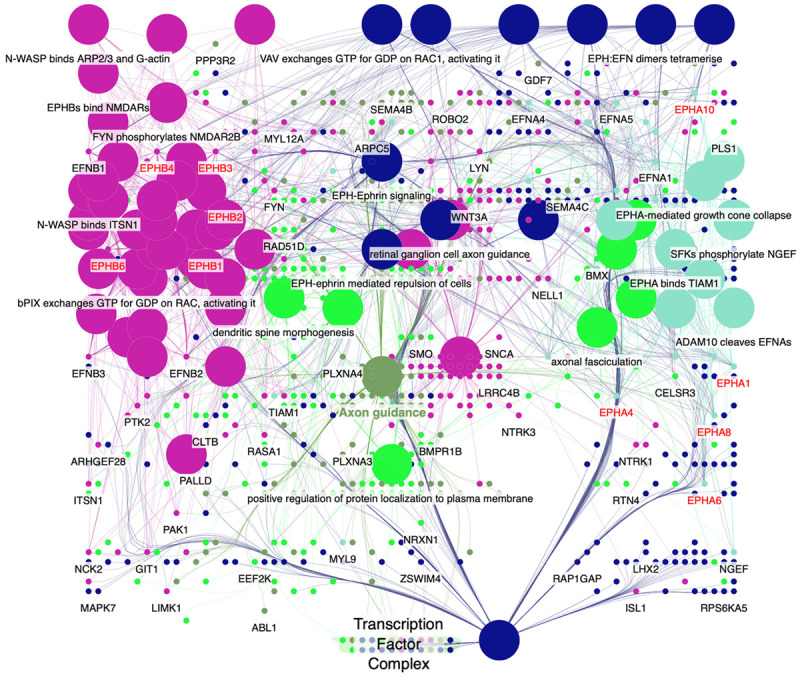
Erythropoietin-producing hepatoma (EPH) member’s interaction network via ClueGo and CluePedia. The interaction network among EPHA and EPHB family members were analyzed with ClueGo and CluePedia with gene ontology. Nodes represent genes and lines represent gene-gene interactions. The network modules were established based on the network structure and biological functions of uploaded EPHA and EPHB member genes.
Figure 6.
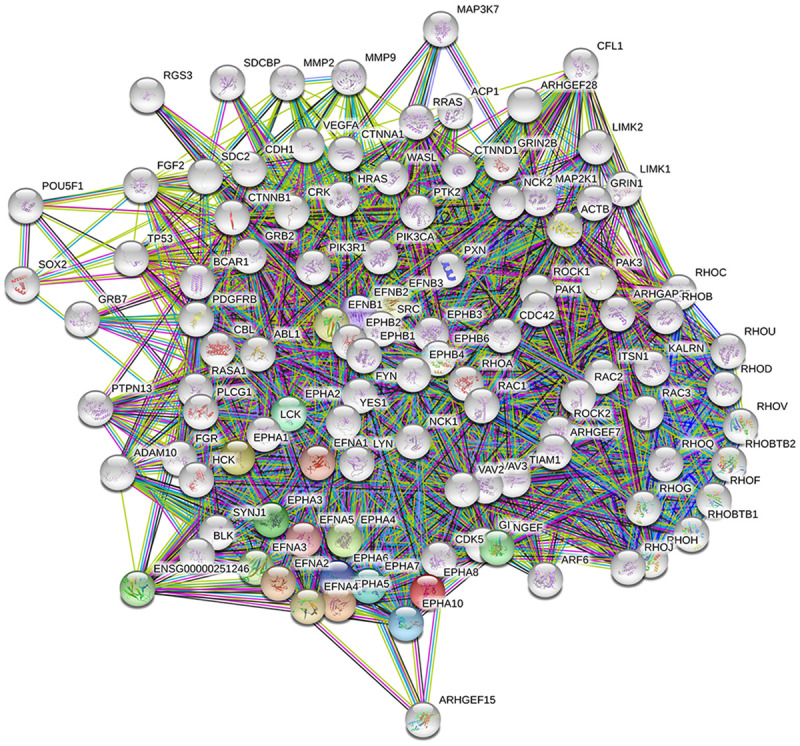
Erythropoietin-producing hepatoma A (EPHA) member’s interaction network via the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database. Protein-protein interactions were constructed with the STRING database. The thickness of the line indicates the strength of data support for protein-protein interactions. Colored nodes represent EPHA member proteins and the first shell of interactors; white nodes represent the second shell of interactors.
Figure 7.
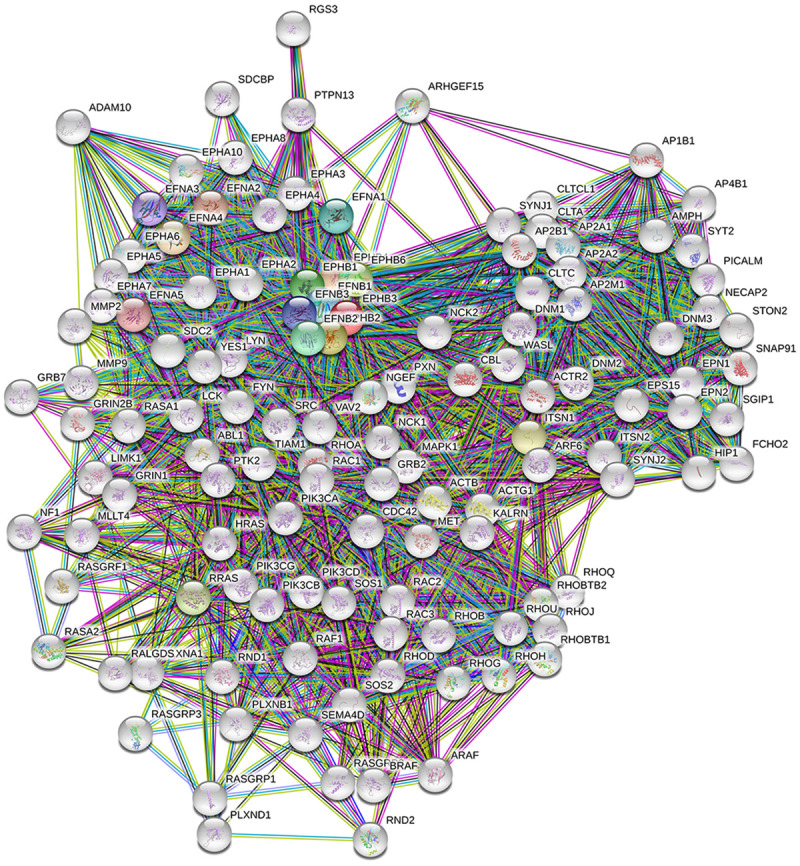
Erythropoietin-producing hepatoma B (EPHB) member’s interaction network via the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database. Protein-protein interactions were constructed with the STRING database. The thickness of the line indicates the strength of data support for protein-protein interactions. Colored nodes represent EPHB member proteins and the first shell of interactors; white nodes represent the second shell of interactors.
Discussion
It is obvious that the EPH and ephrin binding complex functions in the development and progression of different types of tumors. These genes control the proliferation of stem cells and progenitor cells, invasion, and angiogenesis. Functions of EPH/ephrin receptors are distinct; however, all these genes play vital roles in cancer metastasis. Therefore, EPHs and ephrins are proposed to be potential therapeutic targets for cancer treatment [20]. The present study analyzed expression levels of EPHA/B genes in diverse clinical samples and cell lines of various cancers. By determining novel targets of EPHA/B in various types of cancer using high-throughput technology, the present data selected potential targets for future cancer treatment. According to our bioinformatics data, many EPHA/B family genes participate in diverse types of cancer. For instance, colorectal cancer exhibited significant upregulation in EPHA1, EPHA2, EPHA8, EPHB2, EPHB3, and EPHB4. Likewise, EPHA1, EPHA2, EPHA4, EPHB2, and EPHB6 were shown to be highly expressed in bladder tumors. Also, esophageal cancer showed dramatic upregulation of EPHA4, EPHA10, EPHB1, EPHB2, and EPHB4. Gastric cancer showed dramatic upregulation of EPHA4, EPHB1, EPHB2, EPHB3, and EPHB4. Prostate cancer showed dramatic upregulation of EPHA1, EPHA10, EPHB1, EPHB1, EPHB3, and EPHB4. Our study suggested that the high expression of EPHB2 was associated with cervical cancer. EPHA3 and EPHB1 were only respectively upregulated in liver cancer and uterine cancer. Many microarray and RNA-Seq da-tasets were analyzed for expression patterns of EPHA/B through multiple types of cancer. The present study targeted candidates for carcinogenesis of specific cancers, and further studies should be conducted according to these findings.
EPHA/B and their ephrin ligands are known to be involved in tissue boundary formation, vascular development, and axon control [21,22]. EPHs and ephrins are membrane proteins which allow bidirectional signaling between adjacent cells. EPH-ephrin binding can regulate the actin cytoskeleton by affecting G-protein and Rho GTPase signaling to regulate cell morphology, adhesion, and migration [23]. In various cell types, cell motility is controlled by crucial processes, such as microtubular dynamics, polymerization dynamics, and polarization of the cytoskeleton. Expressions of EPH receptors are upregulated in the course of tumor deve-lopment. Overexpression of EPHA receptors is associated with a poor prognosis of cancer patients [24]. EPHB receptors interact with surrounding stromal cells to promote migration and invasion of cancer cells [25]. However, there are no systematic approaches to examine the functions of EPHA/B receptor family genes in diverse types of cancer.
Previous research showed that positive EPHA1 protein staining was significantly linked to more-aggressive renal cell carcinoma [8]. Increased expression in EPHA1 was also detected in prostate cancers [26]. EPHA2 is expressed by most epithelial cells [27]. The independence of EPHA2 with its ephrin ligand suggests its potency with that type of cancer cell development [28]. The EPHA2 staining intensity was dramatically elevated in advanced stages of urothelial carcinoma relative to the normal urothelium [29]. EPHA2 was suggested to play essential roles in stages I and II of colon carcinogenesis [30]. EPHA3 is well known to play oncogenic roles in carcinogenesis, migration, invasion, angiogenesis, and cancer progression [31]. Expression of EPHA3 was correlated with poor survival of liver cancer patients [32]. EPHA4 is known to be dominantly expressed in the nervous system and inhibit axon regeneration [33,34]. In certain types of cancer, inhibition of EPHA4 impedes the progression and invasion of cancer cells [35]. Higher expression of EPHA4 was associated with cancer metastasis [36,37]. EPHA5 is mostly recognized for its critical role in axonal guidance during embryonic development [38]; however, its involvement in cancer is still largely unknown. The expression of EPHA6 was reported to be controlled by HOXA13 in the genital tubercle and its vasculature [39]. Although the biological function of EPHA6 is still largely unknown, EPHA6 was not selected for further examination because its expression data did not satisfy the selection criteria of the present study. Throughout vertebrates and humans, EPHA7 is highly conserved. EPHA7 is also highly present in embryonic tissues, particularly in the central nervous system in the developing stage [40]. But little is known about the role of EPHA7 in cancer development. Recent genetic studies suggested that EPHA8 is involved in regulating cell adhesion and apoptosis [41]. Some findings suggested that the EPHA8 receptor induces axonal projections through regulation of the mitogen-activated protein kinase (MAPK) signaling pathway [42]. A previous study showed that EPHA10 is only expressed in breast cancer but not in normal tissues [43]. Moreover, EPHA10 was also examined for its potential as a therapeutic target [44]. Our data in Figures 2 and 3 and Table 1 further confirmed the significance of EPHA family receptors in various types of cancer.
Overexpression of EPHB1 was found in patients with gastric cancer [19]. EPHB2 overexpression is well documented in various types of human cancers. EPHB2 is known to be involved in the onset of colon cancer [45], cervical cancer and cholangiocarcinoma metastasis [46,47]. EPHB3 was found to engage with the loss of metameric migratory patterns and disorganization of mobility of neural crest cells [48]. Overexpression of EPHB3 improved survival and migration of non-small cell lung cancer cells [49]. EPHB4 is known for playing a vital role in cell signaling and modulates integrin activity to modify the actin skeleton [50]. Upregulation of EPHB4 is associated with the onset and progression of prostate cancer [51,52]. Overexpression of EPHB3 and EPHB4 was detected in prostate cancer and was associated with regional invasion and metastasis [25]. The present study proved the function of EPHB receptors in those cancers (Figure 2, Table 2).
Meanwhile, ClueGo and CluePedia used different types of data and text mining tools to determine relationships between genes. In the literature, the network of SEMA3C, WNT3A, SEMA4B, and ADAM10 was in an intermediate position between EPHB and EPHA family members [53-61]. Our results showed that EPHA and EPHB family genes interacted with SEMA3C, WNT3A, SEMA4B, and ADAM10. The STRING software contains thousands of organisms and genes with millions of gene-gene interactions. Our present data revealed that these relationships might play a crucial role as the genetic backbone of cancer development. In conclusion, our study proved associations between upregulation of EPH receptor family genes in public databases from clinical samples and cancer cell lines. The overexpression of many subunits of the EPHA/B confirmed their function in cancer. The overexpression of EPHA1, EPHA4, EPHB1, EPHB2, EPHB3, and EPHB4 in cancers is a novel feature of this study. Partial inhibition of EPHA1, EPHA4, EPHB1, EPHB2, EPHB3, or EPHB4 may suppress cancer development. Therefore, these EPH receptors may serve as potential therapeutic targets for treating and regulating cancer development.
Acknowledgements
This study was supported in part by the Department of Radiology and Biomedical Imaging, University of California, San Francisco (seed grant 14-29). The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript. The authors also give special thanks for Dan Chamberlin’s English editing from the office of research and development at Taipei Medical University.
Disclosure of conflict of interest
None.
References
- 1.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–80. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer S, Hafner C, Guba M, Flegel S, Geissler EK, Becker B, Koehl GE, Orsó E, Landthaler M, Vogt T. Ephrin-B2 overexpression enhances integrin-mediated ECM-attachment and migration of B16 melanoma cells. Int J Oncol. 2005;27:1197–206. [PubMed] [Google Scholar]
- 3.Darling TK, Lamb TJ. Lamb, emerging roles for Eph receptors and ephrin ligands in immunity. Front Immunol. 2019;10:1473. doi: 10.3389/fimmu.2019.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang JS, Wei HX, Chen PP, Wu G. Roles of Eph/ephrin bidirectional signaling in central nervous system injury and recovery. Exp Ther Med. 2018;15:2219–2227. doi: 10.3892/etm.2018.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–75. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 6.Liang LY, Patel O, Janes PW, Murphy JM, Lucet IS. Eph receptor signalling: from catalytic to non-catalytic functions. Oncogene. 2019;38:6567–6584. doi: 10.1038/s41388-019-0931-2. [DOI] [PubMed] [Google Scholar]
- 7.Lisle JE, Mertens-Walker I, Stephens CR, Stansfield SH, Clements JA, Herington AC, Stephenson SA. Murine, but not human, ephrin-B2 can be efficiently cleaved by the serine protease kallikrein-4: implications for xenograft models of human prostate cancer. Exp Cell Res. 2015;333:136–46. doi: 10.1016/j.yexcr.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Toma MI, Erdmann K, Diezel M, Meinhardt M, Zastrow S, Fuessel S, Wirth MP, Baretton GB. Lack of ephrin receptor A1 is a favorable independent prognostic factor in clear cell renal cell carcinoma. PLoS One. 2014;9:e102262. doi: 10.1371/journal.pone.0102262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, Moher D, Peters MDJ, Horsley T, Weeks L, Hempel S, Akl EA, Chang C, McGowan J, Stewart L, Hartling L, Aldcroft A, Wilson MG, Garritty C, Lewin S, Godfrey CM, Macdonald MT, Langlois EV, Soares-Weiser K, Moriarty J, Clifford T, Tunçalp Ö, Straus SE. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018;169:467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 10.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phan NN, Wang CY, Chen CF, Sun Z, Lai MD, Lin YC. Voltage-gated calcium channels: Novel targets for cancer therapy. Oncol Lett. 2017;14:2059–2074. doi: 10.3892/ol.2017.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang CY, Li CY, Hsu HP, Cho CY, Yen MC, Weng TY, Chen WC, Hung YH, Lee KT, Hung JH, Chen YL, Lai MD. PSMB5 plays a dual role in cancer development and immunosuppression. Am J Cancer Res. 2017;7:2103–2120. [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheng Z, Wang J, Dong Y, Ma H, Zhou H, Sugimura H, Lu G, Zhou X. EphB1 is underexpressed in poorly differentiated colorectal cancers. Pathobiology. 2008;75:274–80. doi: 10.1159/000151707. [DOI] [PubMed] [Google Scholar]
- 19.Aquea G, Bresky G, Lancellotti D, Madariaga JA, Zaffiri V, Urzua U, Haberle S, Bernal G. Increased expression of P2RY2, CD248 and EphB1 in gastric cancers from Chilean patients. Asian Pac J Cancer Prev. 2014;15:1931–1936. doi: 10.7314/apjcp.2014.15.5.1931. [DOI] [PubMed] [Google Scholar]
- 20.Boyd AW, Bartlett PF, Lackmann M. Therapeutic targeting of EPH receptors and their ligands. Nat Rev Drug Discov. 2014;13:39–62. doi: 10.1038/nrd4175. [DOI] [PubMed] [Google Scholar]
- 21.Kullander K, Klein R. Mechanisms and functions of EPH and ephrin signalling. Nat Rev Mol Cell Biol. 2002;3:475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 22.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Noren NK, Pasquale EB. Eph receptor-ephrin bidirectional signals that target Ras and Rho proteins. Cell Signal. 2004;16:655–666. doi: 10.1016/j.cellsig.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Zeng G, Hu Z, Kinch MS, Pan CX, Flockhart DA, Kao C, Gardner TA, Zhang S, Li L, Baldridge LA, Koch MO, Ulbright TM, Eble JN, Cheng L. High-level expression of EphA2 receptor tyrosine kinase in prostatic intraepithelial neoplasia. Am J Pathol. 2003;163:2271–2276. doi: 10.1016/S0002-9440(10)63584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Astin JW, Batson J, Kadir S, Charlet J, Persad RA, Gillatt D, Oxley JD, Nobes CD. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat Cell Biol. 2010;12:1194–U175. doi: 10.1038/ncb2122. [DOI] [PubMed] [Google Scholar]
- 26.Peng L, Wang H, Dong Y, Ma J, Wen J, Wu J, Wang X, Zhou X, Wang J. Increased expression of EphA1 protein in prostate cancers correlates with high Gleason score. Int J Clin Exp Pathol. 2013;6:1854–60. [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz JC, Robertson EJ. The expression of the receptor-protein tyrosine kinase gene, eck, is highly restricted during early mouse development. Mech Dev. 1994;46:87–100. doi: 10.1016/0925-4773(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 28.Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, Dhe-Paganon S. Architecture of Eph receptor clusters. Proc Natl Acad Sci U S A. 2010;107:10860–5. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham S, Knapp DW, Cheng L, Snyder PW, Mittal SK, Bangari DS, Kinch M, Wu L, Dhariwal J, Mohammed SI. Expression of EphA2 and Ephrin A-1 in carcinoma of the urinary bladder. Clin Cancer Res. 2006;12:353–60. doi: 10.1158/1078-0432.CCR-05-1505. [DOI] [PubMed] [Google Scholar]
- 30.Kataoka H, Igarashi H, Kanamori M, Ihara M, Wang JD, Wang YJ, Li ZY, Shimamura T, Kobayashi T, Maruyama K, Nakamura T, Arai H, Kajimura M, Hanai H, Tanaka M, Sugimura H. Correlation of EPHA2 overexpression with high microvessel count in human primary colorectal cancer. Cancer Sci. 2004;95:136–41. doi: 10.1111/j.1349-7006.2004.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Li H, Wu R, Li S, Wang P, Wang H, Wang J, Zhou J. Antitumor effects of oncolytic adenovirus-carrying siRNA targeting potential oncogene EphA3. PLoS One. 2015;10:e0126726. doi: 10.1371/journal.pone.0126726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasri B, Inokuchi M, Ishikawa T, Uetake H, Takagi Y, Otsuki S, Kojima K, Kawano T. High expression of EphA3 (erythropoietin-producing hepatocellular A3) in gastric cancer is associated with metastasis and poor survival. BMC Clin Pathol. 2017;17:8. doi: 10.1186/s12907-017-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabes J, Anderson P, Brennan C, Bolsover S. Regeneration-enhancing effects of EphA4 blocking peptide following corticospinal tract injury in adult rat spinal cord. Eur J Neurosci. 2007;26:2496–2505. doi: 10.1111/j.1460-9568.2007.05859.x. [DOI] [PubMed] [Google Scholar]
- 34.Goldshmit Y, Galea MP, Wise G, Bartlett PF, Turnley AM. Axonal regeneration and lack of astrocytic gliosis in EphA4-deficient mice. J Neurosci. 2004;24:10064–10073. doi: 10.1523/JNEUROSCI.2981-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamberto I, Qin H, Noberini R, Premkumar L, Bourgin C, Riedl SJ, Song J, Pasquale EB. Distinctive binding of three antagonistic peptides to the ephrin-binding pocket of the EphA4 receptor. Biochem J. 2012;445:47–56. doi: 10.1042/BJ20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oki M, Yamamoto H, Taniguchi H, Adachi Y, Imai K, Shinomura Y. Overexpression of the receptor tyrosine kinase EphA4 in human gastric cancers. World J Gastroenterol. 2008;14:5650–5656. doi: 10.3748/wjg.14.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyazaki K, Inokuchi M, Takagi Y, Kato K, Kojima K, Sugihara K. EphA4 is a prognostic factor in gastric cancer. BMC Clin Pathol. 2013;13:19. doi: 10.1186/1472-6890-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou R. Regulation of topographic projection by the Eph family receptor Bsk (EphA5) and its ligands. Cell Tissue Res. 1997;290:251–259. doi: 10.1007/s004410050929. [DOI] [PubMed] [Google Scholar]
- 39.Shaut CA, Saneyoshi C, Morgan EA, Knosp WM, Sexton DR, Stadler HS. HOXA13 directly regulates EphA6 and EphA7 expression in the genital tubercle vascular endothelia. Dev Dyn. 2007;236:951–960. doi: 10.1002/dvdy.21077. [DOI] [PubMed] [Google Scholar]
- 40.Stanic K, Vera A, González M, Recabal A, Astuya A, Torrejón M, Montecinos H, Caprile T. Complementary expression of EphA7 and SCO-spondin during posterior commissure development. Front Neuroanat. 2014;8:49. doi: 10.3389/fnana.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim Y, Park E, Noh H, Park S. Expression of EphA8-Fc in transgenic mouse embryos induces apoptosis of neural epithelial cells during brain development. Dev Neurobiol. 2013;73:702–712. doi: 10.1002/dneu.22092. [DOI] [PubMed] [Google Scholar]
- 42.Gu C, Shim S, Shin J, Kim J, Park J, Han K, Park S. The EphA8 receptor induces sustained MAP kinase activation to promote neurite outgrowth in neuronal cells. Oncogene. 2005;24:4243–4256. doi: 10.1038/sj.onc.1208584. [DOI] [PubMed] [Google Scholar]
- 43.Nagano K, Kanasaki S, Yamashita T, Maeda Y, Inoue M, Higashisaka K, Yoshioka Y, Abe Y, Mukai Y, Kamada H, Tsutsumi Y, Tsunoda S. Expression of Eph receptor A10 is correlated with lymph node metastasis and stage progression in breast cancer patients. Cancer Med. 2013;2:972–977. doi: 10.1002/cam4.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagano K, Maeda Y, Kanasaki S, Watanabe T, Yamashita T, Inoue M, Higashisaka K, Yoshioka Y, Abe Y, Mukai Y, Kamada H, Tsutsumi Y, Tsunoda S. Ephrin receptor A10 is a promising drug target potentially useful for breast cancers including triple negative breast cancers. J Control Release. 2014;189:72–79. doi: 10.1016/j.jconrel.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/EphrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 46.Gao Q, Liu W, Cai J, Li M, Gao Y, Lin W, Li Z. EphB2 promotes cervical cancer progression by inducing epithelial-mesenchymal transition. Hum Pathol. 2014;45:372–381. doi: 10.1016/j.humpath.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Khansaard W, Techasen A, Namwat N, Yongvanit P, Khuntikeo N, Puapairoj A, Loilome W. Increased EphB2 expression predicts cholangiocarcinoma metastasis. Tumour Biol. 2014;35:10031–10041. doi: 10.1007/s13277-014-2295-0. [DOI] [PubMed] [Google Scholar]
- 48.Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr Biol. 1997;7:571–580. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- 49.Ji XD, Li G, Feng YX, Zhao JS, Li JJ, Sun ZJ, Shi S, Deng YZ, Xu JF, Zhu YQ, Koeffler HP, Tong XJ, Xie D. EphB3 is overexpressed in non-small-cell lung cancer and promotes tumor metastasis by enhancing cell survival and migration. Cancer Res. 2011;71:1156–1166. doi: 10.1158/0008-5472.CAN-10-0717. [DOI] [PubMed] [Google Scholar]
- 50.Yang NY, Pasquale EB, Owen LB, Ethell IM. The EphB4 receptor-tyrosine kinase promotes the migration of melanoma cells through Rho-mediated actin cytoskeleton reorganization. J Biol Chem. 2006;281:32574–32586. doi: 10.1074/jbc.M604338200. [DOI] [PubMed] [Google Scholar]
- 51.Lee YC, Perren JR, Douglas EL, Raynor MP, Bartley MA, Bardy PG, Stephenson SA. Investigation of the expression of the EphB4 receptor tyrosine kinase in prostate carcinoma. BMC Cancer. 2005;5:119. doi: 10.1186/1471-2407-5-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mertens-Walker I, Fernandini BC, Maharaj MS, Rockstroh A, Nelson CC, Herington AC, Stephenson SA. The tumour-promoting receptor tyrosine kinase, EphB4, regulates expression of Integrin-beta8 in prostate cancer cells. BMC Cancer. 2015;15:164. doi: 10.1186/s12885-015-1164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T, Barany F, Paty P, Notterman D, Domany E, Ben-Ze’ev A. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 2007;67:7703–7712. doi: 10.1158/0008-5472.CAN-07-0991. [DOI] [PubMed] [Google Scholar]
- 54.Moss ML, Stoeck A, Yan W, Dempsey PJ. ADAM10 as a target for anti-cancer therapy. Curr Pharm Biotechnol. 2008;9:2–8. doi: 10.2174/138920108783497613. [DOI] [PubMed] [Google Scholar]
- 55.Nicolini A, Ferrari P, Rossi G. Mucins and cytokeratins as serum tumor markers in breast cancer. Adv Exp Med Biol. 2015;867:197–225. doi: 10.1007/978-94-017-7215-0_13. [DOI] [PubMed] [Google Scholar]
- 56.Song J, Li Y. miR-25-3p reverses epithelial-mesenchymal transition via targeting Sema4C in cisplatin-resistance cervical cancer cells. Cancer Sci. 2017;108:23–31. doi: 10.1111/cas.13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jian H, Zhao Y, Liu B, Lu S. SEMA4b inhibits MMP9 to prevent metastasis of non-small cell lung cancer. Tumour Biol. 2014;35:11051–11056. doi: 10.1007/s13277-014-2409-8. [DOI] [PubMed] [Google Scholar]
- 58.Jian H, Zhao Y, Liu B, Lu S. SEMA4B inhibits growth of non-small cell lung cancer in vitro and in vivo. Cell Signal. 2015;27:1208–1213. doi: 10.1016/j.cellsig.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 59.Verras M, Brown J, Li X, Nusse R, Sun Z. Wnt3a growth factor induces androgen receptor-mediated transcription and enhances cell growth in human prostate cancer cells. Cancer Res. 2004;64:8860–8866. doi: 10.1158/0008-5472.CAN-04-2370. [DOI] [PubMed] [Google Scholar]
- 60.Qi L, Sun B, Liu Z, Cheng R, Li Y, Zhao X. Wnt3a expression is associated with epithelial-mesenchymal transition and promotes colon cancer progression. J Exp Clin Cancer Res. 2014;33:107. doi: 10.1186/s13046-014-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He S, Lu Y, Liu X, Huang X, Keller ET, Qian CN, Zhang J. Wnt3a: functions and implications in cancer. Chin J Cancer. 2015;34:50. doi: 10.1186/s40880-015-0052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 63.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, Luz J, Ranalli TV, Gomes V, Pastorelli A, Faggiani R, Anti M, Jiricny J, Clevers H, Marra G. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, Reuter VE, Bosl GJ, Chaganti RS. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 66.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 67.Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, Kong S, Sakthivel B, Xu H, Reichling T, Azhar M, Boivin GP, Roberts RB, Bissahoyo AC, Gonzales F, Bloom GC, Eschrich S, Carter SL, Aronow JE, Kleimeyer J, Kleimeyer M, Ramaswamy V, Settle SH, Boone B, Levy S, Graff JM, Doetschman T, Groden J, Dove WF, Threadgill DW, Yeatman TJ, Coffey RJ Jr, Aronow BJ. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santegoets LA, Seters Mv, Helmerhorst TJ, Heijmans-Antonissen C, Hanifi-Moghaddam P, Ewing PC, van Ijcken WF, van der Spek PJ, van der Meijden WI, Blok LJ. HPV related VIN: highly proliferative and diminished responsiveness to extracellular signals. Int J Cancer. 2007;121:759–766. doi: 10.1002/ijc.22769. [DOI] [PubMed] [Google Scholar]
- 71.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P, Fisher R. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, Bottinger E, Friedman S, Waxman S, Llovet JM. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 73.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 74.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, Chiang DY, Reva B, Mermel CH, Getz G, Antipin Y, Beroukhim R, Major JE, Hatton C, Nicoletti R, Hanna M, Sharpe T, Fennell TJ, Cibulskis K, Onofrio RC, Saito T, Shukla N, Lau C, Nelander S, Silver SJ, Sougnez C, Viale A, Winckler W, Maki RG, Garraway LA, Lash A, Greulich H, Root DE, Sellers WR, Schwartz GK, Antonescu CR, Lander ES, Varmus HE, Ladanyi M, Sander C, Meyerson M, Singer S. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Detwiller KY, Fernando NT, Segal NH, Ryeom SW, D’Amore PA, Yoon SS. Analysis of hypoxia-related gene expression in sarcomas and effect of hypoxia on RNA interference of vascular endothelial cell growth factor A. Cancer Res. 2005;65:5881–9. doi: 10.1158/0008-5472.CAN-04-4078. [DOI] [PubMed] [Google Scholar]
- 76.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WG Jr, Signoretti S. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, Sun LZ, Ahlquist DA, Wood CG, Copland JA. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 78.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 79.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 80.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:3445–3445. [PubMed] [Google Scholar]
- 81.Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, Gaffney PM. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- 82.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, Ahlquist P. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 84.D’Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, Palombo F, Giuliani A, Dogliotti E. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 85.Zhan FH, et al. A gene expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2006;108:969a. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 87.Hao Y, Triadafilopoulos G, Sahbaie P, Young HS, Omary MB, Lowe AW. Gene expression profiling reveals stromal genes expressed in common between Barrett’s esophagus and adenocarcinoma. Gastroenterology. 2006;131:925–933. doi: 10.1053/j.gastro.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, Yang L, Pickeral OK, Rassenti LZ, Powell J, Botstein D, Byrd JC, Grever MR, Cheson BD, Chiorazzi N, Wilson WH, Kipps TJ, Brown PO, Staudt LM. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK, Mills KI, Gilkes A, Chiaretti S, Shurtleff SA, Kipps TJ, Rassenti LZ, Yeoh AE, Papenhausen PR, Liu WM, Williams PM, Foà R. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the international microarray innovations in leukemia study group. J. Clin. Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, Jonas D, Libermann TA. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 91.Cutcliffe C, Kersey D, Huang CC, Zeng Y, Walterhouse D, Perlman EJ Renal Tumor Committee of the Children’s Oncology Group. Clear cell sarcoma of the kidney: up-regulation of neural markers with activation of the sonic hedgehog and Akt pathways. Clin Cancer Res. 2005;11:7986–7994. doi: 10.1158/1078-0432.CCR-05-1354. [DOI] [PubMed] [Google Scholar]
- 92.Morrison C, Farrar W, Kneile J, Williams N, Liu-Stratton Y, Bakaletz A, Aldred MA, Eng C. Molecular classification of parathyroid neoplasia by gene expression profiling. Am J Pathol. 2004;165:565–576. doi: 10.1016/S0002-9440(10)63321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 95.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arredouani MS, Lu B, Bhasin M, Eljanne M, Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA, Sanda MG. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794–5802. doi: 10.1158/1078-0432.CCR-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 98.French PJ, Swagemakers SM, Nagel JH, Kouwenhoven MC, Brouwer E, van der Spek P, Luider TM, Kros JM, van den Bent MJ, Sillevis Smitt PA. Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Res. 2005;65:11335–11344. doi: 10.1158/0008-5472.CAN-05-1886. [DOI] [PubMed] [Google Scholar]
- 99.Crabtree JS, Jelinsky SA, Harris HA, Choe SE, Cotreau MM, Kimberland ML, Wilson E, Saraf KA, Liu W, McCampbell AS, Dave B, Broaddus RR, Brown EL, Kao W, Skotnicki JS, Abou-Gharbia M, Winneker RC, Walker CL. Comparison of human and rat uterine leiomyomata: identification of a dysregulated mammalian target of rapamycin pathway. Cancer Res. 2009;69:6171–6178. doi: 10.1158/0008-5472.CAN-08-4471. [DOI] [PubMed] [Google Scholar]
- 100.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, Noh SH, Park ES, Chu IS, Hong WK, Ajani JA, Lee JS. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim SM, Park YY, Park ES, Cho JY, Izzo JG, Zhang D, Kim SB, Lee JH, Bhutani MS, Swisher SG, Wu X, Coombes KR, Maru D, Wang KK, Buttar NS, Ajani JA, Lee JS. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS One. 2010;5:e15074. doi: 10.1371/journal.pone.0015074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 103.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 105.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 106.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, Kraus DH, Patel S, Shaha AR, Wong RJ, Huryn JM, Shah JP, Singh B. Oral tongue cancer gene expression profiling: identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, Poch O, Abecassis J, Wasylyk B. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 109.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC, Boyd J, Birrer MJ. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Murty VV. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- 112.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 113.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 114.Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 115.Watson MA, Gutmann DH, Peterson K, Chicoine MR, Kleinschmidt-DeMasters BK, Brown HG, Perry A. Molecular characterization of human meningiomas by gene expression profiling using high-density oligonucleotide microarrays. Am J Pathol. 2002;161:665–672. doi: 10.1016/S0002-9440(10)64222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ, Weichselbaum RR, Khodarev NN. Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–54. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 117.Gordon GJ, Rockwell GN, Jensen RV, Rheinwald JG, Glickman JN, Aronson JP, Pottorf BJ, Nitz MD, Richards WG, Sugarbaker DJ, Bueno R. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol. 2005;166:1827–40. doi: 10.1016/S0002-9440(10)62492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100:13350–5. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA, Ambs S. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 122.Hu N, Clifford RJ, Yang HH, Wang C, Goldstein AM, Ding T, Taylor PR, Lee MP. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, Shevde LA, Li W, Eschrich S, Daud A, Ju J, Matta J. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Coustan-Smith E, Song G, Clark C, Key L, Liu P, Mehrpooya M, Stow P, Su X, Shurtleff S, Pui CH, Downing JR, Campana D. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2011;117:6267–76. doi: 10.1182/blood-2010-12-324004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Andersson A, Ritz C, Lindgren D, Edén P, Lassen C, Heldrup J, Olofsson T, Råde J, Fontes M, Porwit-Macdonald A, Behrendtz M, Höglund M, Johansson B, Fioretos T. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 126.Dyrskjøt L, Kruhøffer M, Thykjaer T, Marcussen N, Jensen JL, Møller K, Ørntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–8. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 127.Modlich O, Prisack HB, Pitschke G, Ramp U, Ackermann R, Bojar H, Vögeli TA, Grimm MO. Identifying superficial, muscle-invasive, and metastasizing transitional cell carcinoma of the bladder: use of cDNA array analysis of gene expression profiles. Clin Cancer Res. 2004;10:3410–21. doi: 10.1158/1078-0432.CCR-03-0134. [DOI] [PubMed] [Google Scholar]