Abstract

A comprehensive survey on triglycerides (TAGs) of bovine milk was conducted by a combination of exhaustive liquid chromatography (LC) separation, high-resolution mass spectrometry (MS) detection, and automated lipid molecular feature extraction. A total of 220 groups (a series of species having the same chemical formula and mass) and 3454 molecular species of TAGs were identified based on the accurate mass of the parent ion as well as MS2 information. Sixty-five different fatty acids (FAs) were found across these TAG species; C4:0, C6:0, C8:0, C10:0, C12:0, C14:0, C16:0, C18:0, and C18:1 were the most frequent FAs, whereas C11:3, C11:4, C27:0, C27:1, C28:0, and C28:1 were rare FAs in TAG molecules. The number of species identified represents only a small portion of total TAG molecules that can be theoretically synthesized from 65 FAs. Each TAG group contains on average 15–16 isomeric species (species with different FA composition), but positional isomers do not seem to be widespread. As the isomeric species cannot be completely resolved chromatographically, quantification of TAG was conducted at the group level. The most abundant TAG groups in bovine milk include TAG 34:0, TAG 36:0, TAG 38:1, TAG 38:0, and TAG 40:1. This study provides the most comprehensive milk TAG inventory so far that can be used as a reference for studying milk lipids.
Introduction
Lipids are one of the major components of bovine milk alongside proteins and lactose. Milk lipids not only provide energy but also have a profound influence on physicochemical properties of dairy products1−3 as well as on human health.4,5 In addition, some lipid species may be useful as biomarkers of physiological status or even risk of metabolic diseases in dairy cows.6,7
The total fat content and the overall fatty acid (FA) composition are the two most important attributes of milk lipids, which can be determined readily using infrared-based devices8 and GC, respectively.9 However, the biological functions and the nutritional potential of milk lipids are often related to a specific lipid class or even the stereo-structures of lipid molecules.10−12 Hence, a thorough characterization of both major and minor classes of milk lipids at a molecular species level is essential.
The recent advances in separation technology, high-resolution mass spectrometry, and data-processing tools greatly facilitate this task; a number of reviews focusing on analytical platforms applicable to lipidomic analysis of biological systems are available.13−16 Triglycerides (TAGs) are the dominant component of milk lipids, representing >98% of total milk fat.17 It is estimated that around 400 FAs are present in bovine milk,18 from which many thousands of TAG species can theoretically be produced by random combination. However, in the literature, less than 500 species have been described so far with analytical techniques including GC, RPLC-MS, HILIC-LC-MS, and supercritical fluid LC–MS;6,19−26 therefore a large proportion of milk TAG species remains to be characterized. With the aim to build up a complete milk lipid inventory, we have conducted a comprehensive survey of milk lipids by combining exhaustive chromatographic separation with high-resolution mass spectrometry detection and an automated lipid identification tool. A total of 3454 TAG species have been identified in this study; their concentration was also tentatively determined. In this report, we will provide a full description and analysis of the TAG species detected in milk. Indeed, this study surveyed both TAGs and polar lipids. A separate report will summarize the results on polar lipid characterization.
Materials and Methods
Milk Source and Chemicals
Our preliminary tests found that the TAG profile was quite similar between raw milk and commercial milk. As commercial milk samples are bulked milk from many cows and across different herds, they are a better representative of bovine milk. Three brands (A, B, and F) of full cream commercial milk samples obtained from a local supermarket (Victoria, Australia) were used in this study; the total fat content was 3.4% for all the three samples, as shown on the label.
A total of 9 TAG standards (TAG tri-8:0, TAG tri-10:0, TAG tri-12:0, TAG tri-14:0, TAG tri-15:0, TAG tri-16:0, TAG tri-18:0, TAG tri-18:1, and TAG tri-20:0) were purchased from Sigma-Aldrich. Solvents used for milk lipid extraction and mobile phase preparation were of chromatographic grade and were from Fisher Scientific (methanol and isopropanol), Sigma-Aldrich (chloroform), and Merck (acetonitrile and butanol). Ammonium formate (used as a mobile phase additive) was of analytical grade (Sigma-Aldrich).
TAG Standard Preparation and Lipid Extraction
Stock solutions of TAG standards (1 mg/mL) were prepared in chloroform/methanol (2:1, v/v) or in chloroform (for TAG tri-20:0) and stored at −20 °C; working solutions of TAG standards (0.00002, 0.00008, 0.0004, 0.002, and 0.004 mg/mL) were prepared by diluting the stock solution with a solvent mix (butanol/methanol/chloroform, 3:5:4, v/v/v).
For TAG identification, milk lipids were extracted using the Folch method.27 Briefly, 0.5 mL of milk, 0.5 mL of water, and 4 mL of chloroform/methanol (2:1, v/v) were mixed at room temperature in a 15 mL centrifuge tube; lipids were extracted by vortex at full speed for 2 min, followed by centrifugation for 15 min (at 5500 g) to separate the aqueous and organic phases. The organic phase containing the TAG fraction was transferred to a glass vial and dried at 30 °C under N2 to remove the solvents. The remaining lipids were then redissolved in 1 mL of the solvent mix (butanol/methanol/chloroform, 3:5:4, v/v/v), and this crude lipid preparation was used directly for comprehensive TAG identification by LC–MS/MS.
For TAG quantification, milk lipids were extracted using the one-phase extraction method reported by Liu et al.28 Briefly, at room temperature, 100 μL of raw milk was mixed with 1 mL of butanol/methanol/chloroform (3:5:4, v/v/v) in a 2 mL Eppendorf tube, and this mixture was shaken vigorously by vortex for 30 s and then centrifuged for 10 min (at 13,000 g). The supernatant and its 5- and 20-fold dilutions were used for TAG quantification by LC–MS.
TAG Identification by LC–MS/MS
For identification, milk lipids extracted from sample A were separated by non-aqueous reversed phase liquid chromatography (NARP-LC) using an AcclaimTM C30 column (250 × 3 mm, 3 μm, Thermo Fisher Scientific) connected to a Kinetex Core–Shell C18 column (150 × 4.6 mm, 2.6 μm, Phenomenex) on a Vanquish UHPLC system (Thermo Fisher Scientific). The column compartment was maintained at 30 °C and the sample tray at 15 °C. The mobile phase is composed of pure acetonitrile (A) and acetonitrile/isopropanol (10:90, v/v) containing 10 mM ammonium formate (B). The gradient elution was performed by a linear increase of mobile phase B from 0 to 76% over 120 min with a flowrate of 0.45 mL/min. The injection volume was 5 μL.
A Q Exactive Plus mass spectrometer (MS) (Thermo Fisher Scientific) equipped with a heated electrospray ionization source was used for TAG molecule detection. The heated capillary was maintained at 300 °C with a source heater temperature of 325 °C, and the sheath, auxiliary, and sweep gases were respectively at 40, 15, and 4 units. The instrument was operated in positive ion mode (4.2 kV) with a full scan (120–1800 m/z) at a resolution of 70,000 followed by 5 data-dependent MS2 scans at a resolution of 17,500 and collision energy of 25 eV. The precursor isolation width was set to 1.5 Da, and a dynamic exclusion of 8 s was enabled.
TAG species identification was achieved using LipidSearch 4.1 Software (Thermo Fisher Scientific), based on accurate-mass information of parent ions as well as MS2 information.29,30 The following parameters were selected to capture the main species of TAG with definite FA composition: precursor ion—M + NH4; precursor tolerance—5 ppm; product tolerance—8 ppm; threshold type—relative; product ion—2.0%; m-score threshold—5.0; recalculate isotope—on; execute quantitation—off; toprank filter—off; main node filter—main isomer peak; FA priority—on; ID quality filter (A).
TAG Quantification by LC–MS
Extracted lipids from the three brands of milk were separated by a Luna Omega C18 column (150 × 2.1 mm, 1.6 μm, Phenomenex) on the same Vanquish UHPLC system. The column compartment was maintained at 55 °C and the sample tray at 15 °C. The mobile phase is composed of water/acetonitrile (40:60, v/v) containing 10 mM ammonium formate (A) and acetonitrile/isopropanol (10:90, v/v) containing 10 mM ammonium formate (B). The gradient elution was performed by a linear increase of mobile phase B from 5 to 100% over 25 min with a flowrate of 0.25 mL/min. The injection volume was 5 μL.
The detection of milk TAG was also by Q Exactive Plus MS operated in full scan (120–1800 m/z) positive ion mode using the same instrument settings as for TAG identification. The raw data acquired were imported into Xcalibur software (Thermo Fisher Scientific) for TAG group identification based on accurate mass of parent ions as well as MS2 information. Quantification of TAG groups was performed with LCquan software (Thermo Fisher Scientific) as well as external calibration curves of TAG standards. Finally, TAG concentrations were converted into μmoles, and the mean values of the three brands of milk samples are presented.
Results and Discussion
Summary of TAG Groups and Species Identified
A TAG group is defined in this study as a series of species having the same chemical formula, same accurate mass, same total acyl carbon number, and same total double bond number, whereas a TAG species is defined as a TAG molecule with definite identity of all the three FAs (irrespective of their stereospecific position). As expected, the number of TAG species identified by LipidSearch varies with the parameter settings. While the top-rank species only display a subset of all species with the highest matching scores, the number of main species identifiable rises when lowering the product ion intensity or m-score threshold or when grade-A (lipid class and FA chains completely identified) and grade-B (lipid class and some FA chains identified) quality ID are combined (Table 1). For all species, the FA position is not defined; so positional isomers are not counted. In addition, the number of TAG groups and species that can be identified depends on the sensitivity of the MS instrument as well as the lipid concentration of the sample.
Table 1. Summary of LipidSearch Results.
| TAG composition | product ion threshold (%) | m-score | gradea | number of groups | number of species |
|---|---|---|---|---|---|
| Top-rank species | 2 | 5 | A | 220 | 1202 |
| main species | 2 | 5 | A | 220 | 3454 |
| main species | 1 | 5 | A | 220 | 3658 |
| main species | 0.5 | 5 | A | 223 | 4021 |
| main species | 2 | 2 | A | 223 | 3707 |
| main species | 2 | 5 | A–B | 233 | 4787 |
A: TAG species with all FAs completely identified; B: TAG species with some FAs identified.
In order to capture the essential lipid species of bovine milk while excluding those of very low/trace level or with incomplete FA identity, a set of parameters of moderate stringency were chosen which allowed us to identify 220 groups and 3454 species (Table 1, bold); the total acyl carbon number (CN) across these TAG groups ranges from 20 to 62, with the total double bond number (DB) varying between 0 and 9 and molar mass between 400 and 1100 Da. Among the 220 TAG groups, 36 are saturated, 37 are mono-unsaturated, 37 are di-unsaturated, and 110 are poly-unsaturated. Because of the very large size of the dataset, only the list of TAG groups is shown in the main text (see the TAG quantification section), whereas the full list of the 3454 TAG species with detailed chemical formula and accurate mass information is provided as the Supporting Information (Table S-1).
Although the 3454 TAG species identified in this study are by no means a complete account of all TAG species present in bovine milk, compared with previous studies, which identified less than 80 TAG groups and between 100 and 300 species,6,19−24,26 this work provides the most comprehensive inventory of bovine milk TAG. Close examination of the large dataset allows us to observe several features that are discussed in the following sections.
FA Composition of TAG Species
FA profiling of milk fat is generally performed by GC, and in most studies around 25 major FAs are quantified and reported.31,32 Examination of the FA composition of the 3454 TAG species reveals that at least 65 FAs are found in these TAG molecules, of which 23 are saturated, 17 are mono-unsaturated, 8 are di-unsaturated, and 17 are poly-unsaturated (Table 2). It is unsurprising to note that the most frequent FAs (present in over 300 out of the 3454 species) are all major FAs of bovine milk, namely, C4:0, C6:0, C8:0, C10:0, C12:0, C14:0, C16:0, C18:0, C18:1, and C18:2, whereas several FAs including C11:2, C11:3, C11:4, C12:4, C27:0, C17:1, C28:0, and C28:1 are only found in less than 10 species. We should point out that the presence of around half of the 65 FAs (Table 2, underlined) has not been reported so far in milk fat. Indeed, it remains a difficult task to identify and quantify such rare FAs by GC, given the limited availability of standards. It is interesting to note that short-chain FAs such as C2:0, C3:0, C5:0, and C7:0 were not found in these 3454 TAG species identified by LipidSearch software.
Table 2. FAs Detected in Milk TAG Molecules and Their Frequency.
| saturated FA (frequency) | mono-unsaturated FA (frequency) | di-unsaturated FA (frequency) | poly-unsaturated FA (frequency) |
|---|---|---|---|
| C4:0 (518) | C10:1 (211) | C10:2(25) | C10:3(23) |
| C6:0 (435) | C11:1(62) | C11:2(6) | C10:4(14) |
| C8:0 (432) | C12:1 (172) | C12:2(66) | C11:3(1) |
| C9:0 (156) | C14:1 (289) | C14:2(90) | C11:4(1) |
| C10:0 (486) | C16:1 (375) | C18:2 (406) | C12:3(17) |
| C11:0 (147) | C17:1 (229) | C20:2 (106) | C12:4(6) |
| C12:0 (390) | C18:1 (630) | C22:2(31) | C14:3(75) |
| C13:0 (162) | C19:1 (172) | C24:2(13) | C14:4(14) |
| C14:0 (497) | C20:1 (107) | C18:3 (281) | |
| C15:0 (324) | C21:1(43) | C18:4(110) | |
| C16:0 (642) | C22:1(40) | C20:3 (143) | |
| C17:0 (276) | C23:1(84) | C20:4 (156) | |
| C18:0 (463) | C24:1(42) | C20:5 (90) | |
| C19:0 (101) | C25:1(28) | C22:3(38) | |
| C20:0 (106) | C26:1(14) | C22:4(74) | |
| C21:0 (49) | C27:1(2) | C22:5 (81) | |
| C22:0 (83) | C28:1(1) | C22:6 (20) | |
| C23:0(50) | |||
| C24:0(69) | |||
| C25:0(15) | |||
| C26:0(30) | |||
| C27:0(2) | |||
| C28:0(2) |
The theoretical number of TAG species that can be synthesized with 65 FAs could reach over 45,000 by random combination even without considering the positional isomers. Although the exact number of TAG groups and species present in bovine milk will never be known, the 3454 species detected in this work are likely to be only the relatively abundant lipids yet a small fraction of the total TAG pool. We believe that a large number of low- or very low-abundance species are still not captured in this inventory, as the number of species identified by LipidSearch rises to 3658 and 4021 when we drop the relative intensity threshold of the product ions from 2 to 1 and 0.5%, respectively (see Table 1). No attempt was made to increase the number of TAG species in this inventory by using a more concentrated sample or by increasing the sensitivity of the LC–MS instrument (e.g., nano-LC). We should also mention here that the current LC–MS/MS method cannot differentiate cis-/trans- and double bond positional isomers of C18:1, nor various isomers of conjugated and non-conjugated C18:2; so the frequency of C18:1 and C18:2 shown in Table 2 is the sum of all isomers.
TAG Group Composition
Although most TAG groups contain 30–58 CN and 0–5 DB, those with <30 CN and >5 DB are also not rare, and interestingly, odd CN groups represent as much as 40% of total TAG groups (Table 3). It is known that the presence of short-chain FAs is a distinct feature of bovine milk as compared to human milk.15,21 TAG groups with a total acyl carbon number as low as 20–22 were detected in this study. If the scarcity of TAG groups with total acyl carbon number >60 can be explained by their low melting point, which hinders their secretion from mammary glands, it would be puzzling if TAG 12:0, TAG 14:0, TAG 16:0, and TAG 18:0 were totally absent given the abundance of FA C4:0, C6:0, and C8:0 in bovine milk. Manual inspection of the LC–MS chromatograms and MS2 spectra has revealed the presence of these five TAG groups in the sample, but the level of parent ions and/or product ions are below the threshold for triggering data-dependent MS2 scan or below the threshold for detection by LipidSearch (data not shown). This again indicates that the TAG groups and species listed in Table S-1 is far from exhaustive.
Table 3. TAG Concentration in Bovine Milka.
| TAG | μmol (±SD) | TAG | μmol (±SD) | TAG | μmol (±SD) |
|---|---|---|---|---|---|
| TG 20:0 | 0.50 ± 0.04 | TG 30:0 | 605.23 ± 56.81 | TG 36:5 | 1.00 ± 0.08 |
| TG 22:1 | 0.14 ± 0.02 | TG 31:3 | 0.41 ± 0.04 | TG 36:4 | 6.92 ± 0.74 |
| TG 22:0 | 4.93 ± 0.60 | TG 31:2 | 2.26 ± 0.24 | TG 36:3 | 63.87 ± 6.55 |
| TG 23:0 | 1.40 ± 0.08 | TG 31:1 | 19.87 ± 0.99 | TG 36:2 | 302.12 ± 17.22 |
| TG 24:1 | 2.27 ± 0.29 | TG 31:0 | 103.39 ± 5.03 | TG 36:1 | 1422.68 ± 127.39 |
| TG 24:0 | 37.60 ± 4.11 | TG 32:5 | 0.16 ± 0.01 | TG 36:0 | 2681.86 ± 237.52 |
| TG 25:1 | 0.54 ± 0.03 | TG 32:4 | 1.20 ± 0.16 | TG 37:3 | 11.82 ± 0.88 |
| TG 25:0 | 8.87 ± 0.55 | TG 32:3 | 10.50 ± 1.18 | TG 37:2 | 50.43 ± 2.05 |
| TG 26:3 | 0.17 ± 0.02 | TG 32:2 | 57.44 ± 3.51 | TG 37:1 | 280.79 ± 22.93 |
| TG 26:2 | 1.18 ± 0.07 | TG 32:1 | 332.04 ± 28.67 | TG 37:0 | 411.36 ± 33.09 |
| TG 26:1 | 17.69 ± 1.84 | TG 32:0 | 1006.24 ± 82.32 | TG 38:6 | 0.22 ± 0.03 |
| TG 26:0 | 168.53 ± 17.06 | TG 33:3 | 0.90 ± 0.17 | TG 38:5 | 3.41 ± 0.30 |
| TG 27:2 | 0.19 ± 0.02 | TG 33:2 | 4.86 ± 0.49 | TG 38:4 | 20.33 ± 2.34 |
| TG 27:1 | 2.46 ± 0.20 | TG 33:1 | 43.73 ± 2.40 | TG 38:3 | 152.91 ± 11.47 |
| TG 27:0 | 34.69 ± 1.80 | TG 33:0 | 219.05 ± 10.87 | TG 38:2 | 646.28 ± 34.34 |
| TG 28:3 | 1.36 ± 0.19 | TG 34:5 | 0.52 ± 0.04 | TG 38:1 | 2298.24 ± 220.88 |
| TG 28:2 | 6.82 ± 0.45 | TG 34:4 | 2.50 ± 0.26 | TG 38:0 | 1982.97 ± 185.20 |
| TG 28:1 | 77.12 ± 7.12 | TG 34:3 | 20.23 ± 2.14 | TG 39:5 | 0.41 ± 0.03 |
| TG 28:0 | 364.39 ± 36.18 | TG 34:2 | 102.73 ± 6.84 | TG 39:4 | 4.73 ± 0.24 |
| TG 29:2 | 0.70 ± 0.09 | TG 34:1 | 652.98 ± 53.25 | TG 39:3 | 22.38 ± 1.42 |
| TG 29:1 | 8.78 ± 0.43 | TG 34:0 | 2001.29 ± 170.67 | TG 39:2 | 85.31 ± 0.82 |
| TG 29:0 | 59.53 ± 3.87 | TG 35:4 | 0.56 ± 0.01 | TG 39:1 | 262.15 ± 24.06 |
| TG 30:4 | 0.29 ± 0.03 | TG 35:3 | 2.71 ± 0.13 | TG 39:0 | 255.39 ± 20.41 |
| TG 30:3 | 3.80 ± 0.55 | TG 35:2 | 13.37 ± 0.80 | TG 40:7 | 0.08 ± 0.01 |
| TG 30:2 | 22.61 ± 1.25 | TG 35:1 | 116.64 ± 6.18 | TG 40:6 | 2.16 ± 0.28 |
| TG 30:1 | 172.09 ± 14.90 | TG 35:0 | 546.8 ± 49.20 | TG 40:5 | 20.57 ± 1.32 |
| TG 40:4 | 94.67 ± 8.89 | TG 44:7 | 0.78 ± 0.07 | TG 47:0 | 155.84 ± 11.93 |
| TG 40:3 | 327.11 ± 26.64 | TG 44:6 | 5.88 ± 0.41 | TG 48:6 | 3.61 ± 0.10 |
| TG 40:2 | 954.87 ± 103.60 | TG 44:5 | 18.87 ± 0.82 | TG 48:5 | 15.96 ± 0.71 |
| TG 40:1 | 1565.37 ± 168.62 | TG 44:4 | 29.99 ± 2.17 | TG 48:4 | 45.79 ± 2.17 |
| TG 40:0 | 1198.55 ± 122.83 | TG 44:3 | 110.98 ± 3.97 | TG 48:3 | 169.17 ± 2.97 |
| TG 41:5 | 1.90 ± 0.13 | TG 44:2 | 382.01 ± 21.05 | TG 48:2 | 535.41 ± 21.90 |
| TG 41:4 | 2.26 ± 0.27 | TG 44:1 | 880.19 ± 65.15 | TG 48:1 | 968.28 ± 88.09 |
| TG 41:3 | 14.17 ± 0.74 | TG 44:0 | 709.80 ± 57.65 | TG 48:0 | 402.76 ± 42.01 |
| TG 41:2 | 70.82 ± 1.34 | TG 45:5 | 1.01 ± 0.02 | TG 49:5 | 1.75 ± 0.05 |
| TG 41:1 | 152.14 ± 9.72 | TG 45:4 | 2.99 ± 0.26 | TG 49:4 | 7.48 ± 0.36 |
| TG 41:0 | 185.17 ± 9.78 | TG 45:3 | 13.33 ± 1.04 | TG 49:3 | 27.75 ± 0.91 |
| TG 42:8 | 0.07 ± 0.01 | TG 45:2 | 50.61 ± 1.54 | TG 49:2 | 113.83 ± 3.88 |
| TG 42:7 | 0.73 ± 0.05 | TG 45:1 | 142.78 ± 6.72 | TG 49:1 | 245.18 ± 20.17 |
| TG 42:6 | 6.02 ± 0.60 | TG 45:0 | 168.30 ± 9.04 | TG 49:0 | 114.71 ± 12.59 |
| TG 42:5 | 32.71 ± 2.78 | TG 46:6 | 2.82 ± 0.11 | TG 50:6 | 5.51 ± 0.32 |
| TG 42:4 | 52.75 ± 4.98 | TG 46:5 | 15.12 ± 0.86 | TG 50:5 | 25.43 ± 0.93 |
| TG 42:3 | 156.66 ± 10.09 | TG 46:4 | 46.87 ± 2.21 | TG 50:4 | 81.26 ± 5.59 |
| TG 42:2 | 493.83 ± 39.70 | TG 46:3 | 153.12 ± 6.81 | TG 50:3 | 292.36 ± 7.47 |
| TG 42:1 | 943.4 ± 92.96 | TG 46:2 | 444.41 ± 25.46 | TG 50:2 | 733.31 ± 53.26 |
| TG 42:0 | 905.92 ± 64.95 | TG 46:1 | 859.45 ± 64.85 | TG 50:1 | 933.12 ± 89.74 |
| TG 43:5 | 2.04 ± 0.10 | TG 46:0 | 575.54 ± 46.77 | TG 50:0 | 287.28 ± 39.31 |
| TG 43:4 | 1.55 ± 0.26 | TG 47:5 | 1.19 ± 0.03 | TG 51:5 | 4.20 ± 0.30 |
| TG 43:3 | 12.50 ± 0.57 | TG 47:4 | 4.92 ± 0.32 | TG 51:4 | 21.32 ± 0.73 |
| TG 43:2 | 45.21 ± 1.55 | TG 47:3 | 18.30 ± 1.77 | TG 51:3 | 59.80 ± 3.00 |
| TG 43:1 | 135.87 ± 5.83 | TG 47:2 | 69.22 ± 2.01 | TG 51:2 | 166.9 ± 14.95 |
| TG 43:0 | 171.47 ± 5.56 | TG 47:1 | 181.66 ± 5.82 | TG 51:1 | 192.19 ± 22.47 |
| TG 51:0 | 51.70 ± 9.74 | TG 55:3 | 12.66 ± 1.21 | TG 58:2 | 4.92 ± 0.69 |
| TG 52:7 | 1.30 ± 0.14 | TG 55:2 | 13.42 ± 2.15 | TG 58:1 | 5.49 ± 0.94 |
| TG 52:6 | 9.95 ± 0.34 | TG 55:1 | 15.02 ± 1.99 | TG 59:5 | 0.53 ± 0.09 |
| TG 52:5 | 41.31 ± 1.13 | TG 55:0 | 3.25 ± 0.42 | TG 59:4 | 0.18 ± 0.03 |
| TG 52:4 | 147.30 ± 3.54 | TG 56:9 | 0.38 ± 0.05 | TG 59:3 | 3.28 ± 0.53 |
| TG 42:3 | 156.66 ± 10.09 | TG 56:8 | 1.98 ± 0.15 | TG 59:2 | 3.27 ± 0.60 |
| TG 52:2 | 831.67 ± 81.24 | TG 56:7 | 7.22 ± 0.37 | TG 60:3 | 1.98 ± 0.25 |
| TG 52:1 | 545.62 ± 82.18 | TG 56:6 | 15.04 ± 1.46 | TG 60:2 | 2.32 ± 0.35 |
| TG 52:0 | 129.01 ± 25.90 | TG 56:5 | 12.86 ± 1.93 | TG 60:1 | 3.42 ± 0.50 |
| TG 53:6 | 1.57 ± 0.07 | TG 56:4 | 8.97 ± 1.04 | TG 61:3 | 0.48 ± 0.07 |
| TG 53:5 | 7.19 ± 0.49 | TG 56:3 | 10.19 ± 1.03 | TG 61:2 | 0.58 ± 0.12 |
| TG 53:4 | 17.02 ± 1.19 | TG 56:2 | 12.05 ± 1.84 | TG 62:2 | 1.00 ± 0.21 |
| TG 53:3 | 48.71 ± 3.42 | TG 56:1 | 10.98 ± 1.94 | ||
| TG 53:2 | 96.49 ± 12.18 | TG 56:0 | 5.76 ± 0.99 | ||
| TG 53:1 | 70.48 ± 11.44 | TG 57:6 | 0.04 ± 0.01 | ||
| TG 53:0 | 15.21 ± 2.54 | TG 57:5 | 0.46 ± 0.07 | ||
| TG 54:7 | 5.54 ± 0.43 | TG 57:4 | 1.26 ± 0.22 | ||
| TG 54:6 | 23.84 ± 4.65 | TG 57:3 | 3.34 ± 0.27 | ||
| TG 54:5 | 89.45 ± 6.39 | TG 57:2 | 1.73 ± 0.14 | ||
| TG 54:4 | 200.54 ± 15.49 | TG 57:1 | 6.66 ± 1.12 | ||
| TG 54:3 | 360.84 ± 44.05 | TG 58:8 | 0.90 ± 0.04 | ||
| TG 54:2 | 307.56 ± 54.80 | TG 58:7 | 2.1 ± 0.17 | ||
| TG 54:1 | 131.94 ± 27.71 | TG 58:6 | 2.12 ± 0.31 | ||
| TG 54:0 | 25.38 ± 6.34 | TG 58:5 | 1.26 ± 0.28 | ||
| TG 55:5 | 2.76 ± 0.28 | TG 58:4 | 1.31 ± 0.10 | ||
| TG 55:4 | 4.59 ± 0.31 | TG 58:3 | 0.55 ± 0.03 |
From TAG 20:0 to TAG 54:0: the average response of TAG 24:0, TAG 30:0, TAG 36:0, TAG 42:0, TAG 45:0, TAG 48:0, and TAG 54:0 was used; from TAG 55:5 to TAG 62:2: the response factor of TAG 60:0 was used.
Verification of LipidSearch Results
LipidSearch is a powerful tool in automated lipid species identification of milk fat, a daunting task to be completed manually. Table S-1 shows that the most striking feature of milk TAG is the huge number of isomer species within each group (on average 15–16 species per group and as many as >30 for some groups). While identification of the isomer species can be achieved by LipidSearch or other data analysis software with minimal or no chromatographic separation,33,34 the TAG species identified can only be considered as putative in the case of multiple co-eluting isomers. In this study, automated identification by LipidSearch was based on MS/MS2 spectra acquired after an exhaustive NARP-LC separation (Figure S-1, Supporting Information); so confidence in the species identified can be considered as high. Nevertheless, the reliability of the data generated needs to be verified manually given the exact mechanism for biosynthesis of TAG molecules in mammary glands is still obscure.
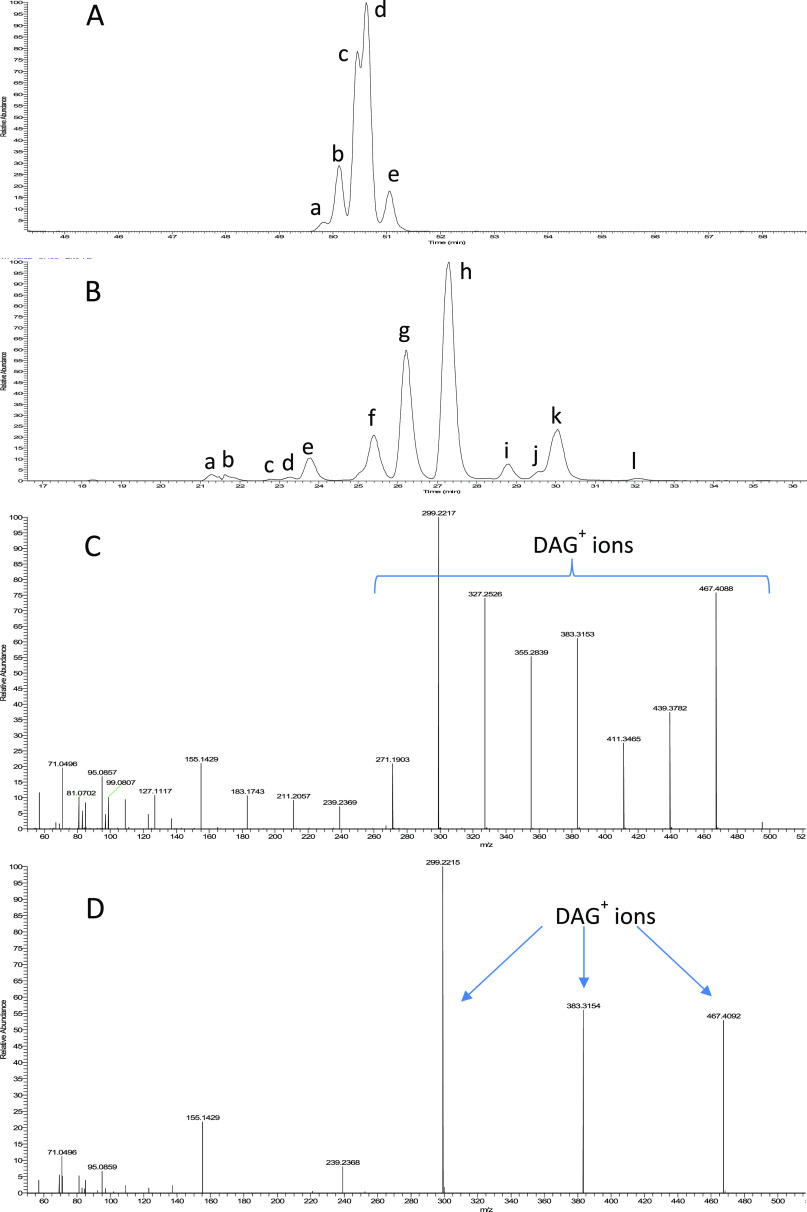
MS2 spectra of highly resolved isomers facilitate considerably manual identification of TAG species. For example, in the case of TAG 30:0, a classic reversed-phase LC (RP-LC) combining a sub-2 μm column with a long gradient elution detected five peaks with peaks a–b and c–d poorly resolved (Figure 1A). The MS2 spectrum of the overlapping peaks c–d is quite complex containing 9 DAG+ ions (Figure 1C), from which it is very difficult to determine with certainty the exact number of isomer species co-eluted and their FA composition, let alone their relative abundance. NARP-LC with 2 columns connected in series is known to provide improved separation of TAG isomers, and such a strategy has been used in several studies.19,21,35 We compared various column combinations, and the best overall performance was obtained by combining a C30 column with a core–shell C18 column. A total of 12 peaks were detected for TAG 30:0 by NARP-LC with several of them baseline resolved (Figure 1B). Only 3 DAG+ ions (m/z 299, 383 467) were found in the MS2 spectrum of the largest peak (h) (Figure 1D), corresponding to the neutral loss of three different FA chains; the mass difference between the parent ion (m/z 572) and the DAG+ ions reflects the identity of the three FAs (C4:0, C10:0, and C16:0). Consequently, this peak contains a single isomer species with a definite FA composition 4:0/10:0/16:0. Similarly, the FA identity of all the other peaks can be readily assigned manually based on DAG+ ion information, including peak k, which contains 2 co-eluting isomers TAG 4:0/8:0/18:0 and TAG 2:0/12:0/16:0. The number of isomer species of this TAG group identified by manual inspection totalled 13, with 3 of them TAG 2:0/10:0/18:0 (peak l), TAG 2:0/12:0/16:0 (peak k), and TAG 2:0/14:0/14:0 (peak j) not detected by LipidSearch due to the relative abundance of one product ion being below the threshold. The presence of FA C2:0 in the TAG structure is an interesting finding, which was rarely reported so far. Indeed, manual inspection of MS2 spectra also revealed the presence of other short-chain FA including C3:0, C5:0, and C7:0 in species belonging to several TAG groups, for example, TAG 27:2, TAG 27:1, TAG 29:3, TAG 29:2, TAG 31:2, and TAG 31:1 (data not shown) but again not captured by LipidSearch.
Figure 1.
Extracted ion chromatograms of TAG 30:0 and MS2 spectra of selected peaks following different chromatographic separation regimes. A: LC separation of milk fat by a Luna Omega C18 column (150 × 2.1 mm, 1.6 μm) in classic RP mode for 80 min; B: LC separation of milk fat by a combination of an AcclaimTM C30 column (250 × 3 mm, 3 μm) and a Kinetex Core–Shell C18 column (150 × 4.6 mm, 2.6 μm) in NARP mode for 120 min; C: MS2 spectrum of peaks c-d from chromatogram A; D: MS2 spectrum of peak h from chromatogram B. The isomer species of all peaks shown in chromatogram B are assigned as 10:0/10:0/10:0 (a), 8:0/10:0/12:0 (b), 8:0/8:0/14:0 (c), 6:0/12:0/12:0 (d), 6:0/10:0/14:0 (e), 6:0/8:0/16:0 (f), 4:0/12:0/14:0 (g), 4:0/10:0/16:0 (h), 6:0/6:0/18:0 (i), 2:0/14:0/14:0 (j), 4:0/8:0/18:0 and 2:0/12:0/16:0 (k), and 2:0/10:0/18:0 (l).
A total of 9 FAs were found in these 13 isomers, namely, C2:0, C4:0, C6:0, C8:0, C10:0, C12:0, C14:0, C16:0, and C18:0. If the 8 major FAs (4:0, 6:0, 8:0, 10:0, 12:0, 14:0, 16:0, and 18:0) were randomly combined for making TAG 30:0 molecules, the isomer species that can be theoretically produced would comprise 4:0/8:0/18:0, 4:0/10:0/16:0, 4:0/12:0/14:0, 6:0/6:0/18:0, 6:/8:0/16:0, 6:0/10:0/14:0, 6:0/12:0/12:0, 8:0/8:0/14:0, 8:0/10:0/12:0, and 10:0/10:0/10:0; all these 10 isomer species were indeed detected by manual inspection as well as LipidSearch. This implies that the type and the total number of isomer species of this TAG group does match the random combination of FAs. If FA C2:0 was also included, the number of theoretical isomers that could be formed for TAG 30:0 would be 13, corresponding exactly to the 13 species detected by manual inspection. This example confirms that LipidSearch is reliable for capturing the main TAG species, but species of low abundance may still be missed out when identification parameters of moderate stringency were applied.
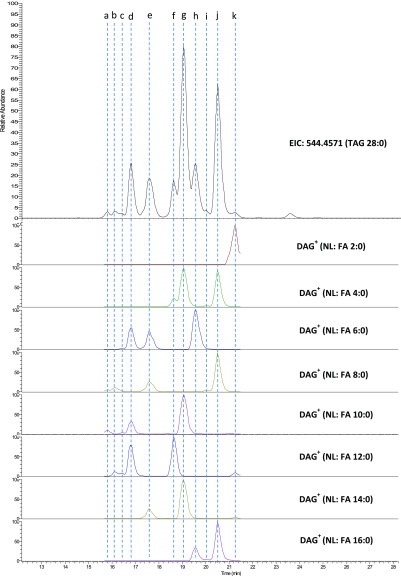
Identification of Positional Isomers
Figure 2 shows the NARP-LC-MS profile of TAG 28:0, from which 11 peaks can be identified (a–k), albeit not fully resolved. Alignment of the peak apex of the extracted parent ion versus the product ion chromatograms allows us to establish with ease the FA composition of the 11 isomers including the over-lapping ones (f–h). A close examination of the FA composition of these 11 peaks indicate that peak c and peak i are, respectively, positional isomers of peak d and peak j, as the MS2 spectra of the positional isomers contain the same DAG+ ions but in different proportions (data not shown). The presence of positional isomers is usually judged based on isomer species having the same FA composition but different retention time. The low abundance of the positional isomers (c and i compared to d and j) does not support random esterification of FAs to the three positions of the glycerol backbone, a fact that is well documented in various types of milk.22,36−38 Consequently, the failure to detect positional isomers for most species is likely due to their low abundance rather than poor chromatographic separation. However, not all positional isomers of TAGs are of low abundance; in the case of the well-known positional isomer pair OPO/OOP, while OPO is the major structure, OOP is of substantial proportion in bovine milk.39,40 Again, using the current parameter settings, LipidSerach is successful in detecting 8 out of the 9 species (except TAG 2:0/12:0/14:0), but is not able to provide detailed information regarding the presence or relative abundance of positional isomers.
Figure 2.
Identification of all isomers of TAG 28:0 by manual alignment of parent and product ion chromatograms. LC separation of TAG 28:0 isomers was carried out by a combination of an AcclaimTM C30 column (250 × 3 mm, 3 μm) and a Kinetex Core–Shell C18 column (150 × 4.6 mm, 2.6 μm) in NARP mode for 120 min. The FA composition of the isomers are assigned as 8:0/10:0/10:0 (a), 8:0/8:0/12:0 (b), 6:0/10:0/12:0 (c), 6:0/10:0/12:0 (d), 6:0/8:0/14:0 (e), 4:0/12:0/12:0 (f), 4:0/10:0/14:0 (g), 6:0/6:0/16:0 (h), 4:0/8:0/16:0 (i), 4:0/8:0/16:0 (j), and 2:0/12:0/14:0 (k).
These two simple examples clearly show that chromatographic separation facilitates considerably the manual identification of TAG species, which is an essential way for verifying and complementing the results generated by LipidSearch. Indeed, manual identification of lipid species is often required when automated data processing tools are not available. In addition, combining exhaustive LC separation with parent ion/product ion chromatogram alignment enables deconvolution of co-eluting isomers and detection of positional isomers. While previous studies strongly advocate the non-random distribution of FA in TAG synthesis,18 our results appear to suggest that what can be theoretically produced is indeed found in milk when it comes to the number and FA composition of isomer species. Based on the present data, we believe that “skewed random” is a more appropriate term to describe FA make-up in milk TAG structures.
It should be mentioned, however, the NARP-LC-MS method showed lower sensitivity (due to larger LC columns and a higher flowrate used). Consequently, for a comprehensive TAG identification, a higher lipid concentration was required, which cannot be achieved easily with the simple one-phase extraction method. Therefore, the Folch lipid extraction method was used for TAG identification, whereas the one-phase method was used for TAG quantification.
Quantification of TAG Groups
As resolution of all isomer species is not possible for most if not all TAG groups, quantification of TAG at the species level is not achievable at this stage. In this study, TAG quantification was performed at the group level using the external calibration method. It is known that MS response of TAG molecules varies with both FA chain length and level of unsaturation,41,42 but currently, it is impossible to procure standards for 220 TAG groups. We have measured the relative response of 9 TAG standards and found a rather similar response from TAG 24:0 to TAG 54:0, or between TAG 54:0 and TAG 54:3, but a slightly lower response with TAG 60:0 (Table S-2, Supporting Information). Consequently, we have used the average response of TAG 24:0, TAG 30:0, TAG 36:0, TAG 42:0, TAG 45:0, TAG 48:0, and TAG 54:0 to estimate the concentration of all TAG groups from TAG 20:0 to TAG 54:0, and the response factor of TAG 60:0 to estimate that from TAG 55:5 to TAG 62:2. Such an approach of using a single or a limited number of standards to quantify or semi-quantify TAGs is widely adopted in lipidomic analysis.43,44 As a result, the TAG quantification results presented in this report can only be considered as an approximate level.
Because of the wide concentration range of various TAG groups, 3 dilutions of the lipid extract were measured to ensure that the concentration of each group is within the linear range (between 10 ng and 10 μg/mL). Table 3 shows that the concentrations of the 220 TAG groups in bovine milk spans from 0.07 to over 2000 μmol. The most abundant TAG groups (>800 μmol) include TAG 32:0, TAG 34:0, TAG 36:1, TAG 36:0, TAG 38:1, TAG 38:0, TAG 40:1 and TAG 40:0, TAG 42:1, TAG 42:0, TAG 44:1, TAG 46:1, TAG 48:1, TAG 50:1, and TAG 52:2, consistent with previous reports,20,22,40,45,46 although the abundance order of these major TAG groups differs across different studies probably due to milk samples being of various origins. By contrast, TAG groups with CN > 54 are generally present at low concentration (<20 μmol) and ignored in most studies. Overall odd-CN groups are less abundant compared to even-CN ones and groups with total DB > 4 are mostly at a concentration <50 μmol. By contrast, only a small variation was observed across the three brands of milk; this can be explained by the fact that all the three brands are probably derived from raw milk of the same region. The total amount of TAG species added together is approximately 30 mg/mL of milk for all the three brands, a value very close to the theoretical fat content of the samples (34 mg/mL). This indicates that our method used for TAG quantification is of adequate accuracy.
Conclusions
Bovine milk TAGs are an immense assembly of molecules of varying FA composition. The 3454 species detected in this work represent the most comprehensive coverage of milk TAGs to date. However, these species are likely to be only a relatively abundant and small portion of all TAG molecules present in milk. In addition to the dozen major FAs, many low-abundance FAs including C2:0 and C3:0 were found to participate in TAG synthesis for the first time. A non-complete random rule appears to govern FA esterification in TAG molecules. The molar concentration of 220 TAG groups was also tentatively determined using currently available standards. This dataset is expected to be a valuable reference inventory for researchers involved in dairy research.
Acknowledgments
C.L.’s PhD studentship is funded by DaiyFeedbase program.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01841.
The authors declare no competing financial interest.
Supplementary Material
References
- Dimick P. S.; Reddy S. Y.; Ziegler G. R. Chemical and thermal characteristics of milk-fat fractions isolated by a melt crystallization. J. Am. Oil Chem. Soc. 1996, 73, 1647–1652. 10.1007/bf02517966. [DOI] [Google Scholar]
- Narine S. S.; Marangoni A. G. Relating structure of fat crystal networks to mechanical properties. Food Res. Int. 1999, 32, 227–248. 10.1016/s0963-9969(99)00078-2. [DOI] [PubMed] [Google Scholar]
- Smiddy M. A.; Huppertz T.; van Ruth S. M. Triacylglycerol and melting profiles of milk fat from several species. Int. Dairy J. 2012, 24, 64–69. 10.1016/j.idairyj.2011.07.001. [DOI] [Google Scholar]
- Haug A.; Høstmark A. T.; Harstad O. M. Bovine milk in human nutrition – a review. Lipids Health Dis. 2007, 6, 25. 10.1186/1476-511x-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küllenberg D.; Taylor L. A.; Schneider M.; Massing U. Health effects of dietary phospholipids. Lipids Health Dis. 2012, 11, 3. 10.1186/1476-511x-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Ezernieks V.; Wang J.; Arachchillage N. W.; Garner J. B.; Wales W. J.; Cocks B. G.; Rochfort S. Heat stress in dairy cattle alters lipid composition of milk. Sci. Rep. 2017, 7, 961. 10.1038/s41598-017-01120-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke T. D. W.; Rochfort S.; Wales W. J.; Bonfatti V.; Marett L.; Pryce J. E. Metabolic profiling of early-lactation dairy cows using milk mid-infrared spectra. J. Dairy Sci. 2019, 102, 1747–1760. 10.3168/jds.2018-15103. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Otero J. L.; Hermida M.; Centeno J. Analysis of dairy products by near-infrared spectroscopy: A review. J. Agric. Food Chem. 1997, 45, 2815–2819. 10.1021/jf960744p. [DOI] [Google Scholar]
- Amores G.; Virto M. Total and free fatty acids analysis in milk and dairy fat. Separations 2019, 6, 14. 10.3390/separations6010014. [DOI] [Google Scholar]
- Lien E. L. The role of fatty acid composition and positional distribution in fat absorption in infants. J. Pediatr. 1994, 125, S62–S68. 10.1016/s0022-3476(06)80738-9. [DOI] [PubMed] [Google Scholar]
- Zou X.; Huang J.; Jin Q.; Guo Z.; Liu Y.; Cheong L.; Xu X.; Wang X. Lipid composition analysis of milk fats from different mammalian species: Potential for use as human milk fat substitutes. J. Agric. Food Chem. 2013, 61, 7070–7080. 10.1021/jf401452y. [DOI] [PubMed] [Google Scholar]
- Contarini G.; Povolo M. Phospholipids in milk fat: composition, biological and technological significance, and analytical strategies. Int. J. Mol. Sci. 2013, 14, 2808–2831. 10.3390/ijms14022808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T.; Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Anal. Chem. 2014, 61, 192–206. 10.1016/j.trac.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köfeler H. C.; Fauland A.; Rechberger G. N.; Trötzmüller M. Mass spectrometry based lipidomics: an overview of technological platforms. Metabolites 2012, 2, 19–38. 10.3390/metabo2010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Rochfort S.; Cocks B. Milk lipidomics: What we know and what we don’t. Prog. Lipid Res. 2018, 71, 70–85. 10.1016/j.plipres.2018.06.002. [DOI] [PubMed] [Google Scholar]
- Li M.; Yang L.; Bai Y.; Liu H. Analytical methods in lipidomics and their applications. Anal. Chem. 2014, 86, 161–175. 10.1021/ac403554h. [DOI] [PubMed] [Google Scholar]
- Lindmark Månsson H. Fatty acids in bovine milk fat. Food Nutr. Res. 2008, 52, 1821. 10.3402/fnr.v52i0.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. G. The composition of bovine milk lipids: January 1995 to December 2000. J. Dairy Sci. 2002, 85, 295–350. 10.3168/jds.s0022-0302(02)74079-4. [DOI] [PubMed] [Google Scholar]
- Beccaria M.; Sullini G.; Cacciola F.; Donato P.; Dugo P.; Mondello L. High performance characterization of triacylglycerols in milk and milk-related samples by liquid chromatography and mass spectrometry. J. Chromatogr. A 2014, 1360, 172–187. 10.1016/j.chroma.2014.07.073. [DOI] [PubMed] [Google Scholar]
- Foroutan A.; Guo A. C.; Vazquez-Fresno R.; Lipfert M.; Zhang L.; Zheng J.; Badran H.; Budinski Z.; Mandal R.; Ametaj B. N.; Wishart D. S. Chemical composition of commercial cow’s milk. J. Agric. Food Chem. 2019, 67, 4897–4914. 10.1021/acs.jafc.9b00204. [DOI] [PubMed] [Google Scholar]
- Gastaldi D.; Medana C.; Giancotti V.; Aigotti R.; Dal Bello F.; Baiocchi C. HPLC-APCI analysis of triacylglycerols in milk fat from different sources. Eur. J. Lipid Sci. Technol. 2011, 113, 197–207. 10.1002/ejlt.201000068. [DOI] [Google Scholar]
- Gresti J.; Bugaut M.; Maniongui C.; Bezard J. Composition of molecular species of triacylglycerols in bovine milk Fat. J. Dairy Sci. 1993, 76, 1850–1869. 10.3168/jds.s0022-0302(93)77518-9. [DOI] [PubMed] [Google Scholar]
- Mottram H. R.; Evershed R. P. Elucidation of the composition of bovine milk fat triacylglycerols using high-performance liquid chromatography–atmospheric pressure chemical ionisation mass spectrometry. J. Chromatogr. A 2001, 926, 239–253. 10.1016/s0021-9673(01)01048-2. [DOI] [PubMed] [Google Scholar]
- Myher J. J.; Kuksis A.; Marai L.; Sandra P. Identification of the more complex triacylglycerols in bovine milk fat by gas chromatography—mass spectrometry using polar capillary columns. J. Chromatogr. A 1988, 452, 93–118. 10.1016/s0021-9673(01)81440-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Sala P.; Hierro M. T. G.; Martínez-Castro I.; Santa-María G. Triglyceride composition of ewe, cow, and goat milk fat. J. Am. Oil Chem. Soc. 1996, 73, 283–293. 10.1007/bf02523421. [DOI] [Google Scholar]
- Zhou Q.; Gao B.; Zhang X.; Xu Y.; Shi H.; Yu L. Chemical profiling of triacylglycerols and diacylglycerols in cow milk fat by ultra-performance convergence chromatography combined with a quadrupole time-of-flight mass spectrometry. Food Chem. 2014, 143, 199–204. 10.1016/j.foodchem.2013.07.114. [DOI] [PubMed] [Google Scholar]
- Folch J.; Lees M.; Stanley G. H. S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [PubMed] [Google Scholar]
- Liu Z.; Rochfort S.; Cocks B. G. Optimization of a single phase method for lipid extraction from milk. J. Chromatogr. A 2016, 1458, 145–149. 10.1016/j.chroma.2016.06.055. [DOI] [PubMed] [Google Scholar]
- Breitkopf S. B.; Ricoult S. J. H.; Yuan M.; Xu Y.; Peake D. A.; Manning B. D.; Asara J. M. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 2017, 13, 30. 10.1007/s11306-016-1157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T.; Uchikata T.; Sakamoto S.; Yokoi Y.; Fukusaki E.; Bamba T. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J. Chromatogr. A 2013, 1292, 211–218. 10.1016/j.chroma.2013.01.078. [DOI] [PubMed] [Google Scholar]
- Moate P. J.; Williams S. R. O.; Torok V. A.; Hannah M. C.; Ribaux B. E.; Tavendale M. H.; Eckard R. J.; Jacobs J. L.; Auldist M. J.; Wales W. J. Grape marc reduces methane emissions when fed to dairy cows. J. Dairy Sci. 2014, 97, 5073–5087. 10.3168/jds.2013-7588. [DOI] [PubMed] [Google Scholar]
- O’Donnell-Megaro A. M.; Barbano D. M.; Bauman D. E. Survey of the fatty acid composition of retail milk in the United States including regional and seasonal variations. J. Dairy Sci. 2011, 94, 59–65. 10.3168/jds.2010-3571. [DOI] [PubMed] [Google Scholar]
- Wang M.; Wang C.; Han R. H.; Han X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 2016, 61, 83–108. 10.1016/j.plipres.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Wang X.; Jiao Y.; Liu X. Assessment of potential false positives via orbitrap-based untargeted lipidomics from rat tissues. Talanta 2018, 178, 287–293. 10.1016/j.talanta.2017.09.046. [DOI] [PubMed] [Google Scholar]
- Lísa M.; Holčapek M.; Boháč M. Statistical evaluation of triacylglycerol composition in plant oils based on high-performance liquid chromatography–atmospheric pressure chemical ionization mass spectrometry data. J. Agric. Food Chem. 2009, 57, 6888–6898. 10.1021/jf901189u. [DOI] [PubMed] [Google Scholar]
- Fontecha J.; Goudjil H.; Ríos J. J.; Fraga M. J.; Juárez M. Identity of the major triacylglycerols in ovine milk fat. Int. Dairy J. 2005, 15, 1217–1224. 10.1016/j.idairyj.2004.11.013. [DOI] [Google Scholar]
- Fontecha J.; Ríos J. J.; Lozada L.; Fraga M. J.; Juárez M. Composition of goat’s milk fat triglycerides analysed by silver ion adsorption-TLC and GC–MS. Int. Dairy J. 2000, 10, 119–128. 10.1016/s0958-6946(00)00026-1. [DOI] [Google Scholar]
- Haddad I.; Mozzon M.; Strabbioli R.; Frega N. G. A comparative study of the composition of triacylglycerol molecular species in equine and human milks. Dairy Sci. Technol. 2012, 92, 37–56. 10.1007/s13594-011-0042-5. [DOI] [Google Scholar]
- Gotoh N.; Matsumoto Y.; Nagai T.; Mizobe H.; Yoshinaga K.; Kojima K.; Kuroda I.; Kitamura Y.; Shimizu T.; Ishida H.; Wada S. Actual ratio of triacylglycerol positional isomers in milk and cheese. J. Oleo Sci. 2012, 61, 173–180. 10.5650/jos.61.173. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Wang J.; Cocks B. G.; Rochfort S. Seasonal variation of triacylglycerol profile of bovine milk. Metabolites 2017, 7, 24. 10.3390/metabo7020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Gross R. W. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 2001, 295, 88–100. 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- Holčapek M.; Lísa M.; Jandera P.; Kabátová N. Quantitation of triacylglycerols in plant oils using HPLC with APCI-MS, evaporative light-scattering, and UV detection. J. Sep. Sci. 2005, 28, 1315–1333. 10.1002/jssc.200500088. [DOI] [PubMed] [Google Scholar]
- Teng F.; Reis M. G.; Yang L.; Ma Y.; Day L. In-depth lipidomic analysis of tri-, di-, and mono-acylglycerol released from milk fat after in vitro digestion. Food Chem. 2019, 297, 124976. 10.1016/j.foodchem.2019.124976. [DOI] [PubMed] [Google Scholar]
- Li Q.; Zhao Y.; Zhu D.; Pang X.; Liu Y.; Frew R.; Chen G. Lipidomics profiling of goat milk, soymilk and bovine milk by UPLC-Q-Exactive Orbitrap Mass Spectrometry. Food Chem. 2017, 224, 302–309. 10.1016/j.foodchem.2016.12.083. [DOI] [PubMed] [Google Scholar]
- Tzompa-Sosa D. A.; van Valenberg H. J. F.; van Aken G. A.; Bovenhuis H. Milk fat triacylglycerols and their relations with milk fatty acid composition, DGAT1 K232A polymorphism, and milk production traits. J. Dairy Sci. 2016, 99, 3624–3631. 10.3168/jds.2015-10592. [DOI] [PubMed] [Google Scholar]
- Sokol E.; Ulven T.; Faergeman N. J.; Ejsing C. S. Comprehensive and quantitative profiling of lipid species in human milk, cow milk and a phospholipid-enriched milk formula by GC and MS/MSALL. Eur. J. Lipid Sci. Technol. 2015, 117, 751–759. 10.1002/ejlt.201400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.