Abstract
CD4+ T-follicular helper cells are essential for the survival, proliferation, and differentiation of germinal center B cells and have been implicated in the pathogenesis of follicular lymphoma (FL). To further define the role of these cells in FL, we used multiparameter confocal microscopy to compare the architecture of normal and neoplastic follicles and next generation sequencing to analyze the T-cell receptor repertoire in FL lymph nodes (LN). Multiparameter analysis of LN showed that the proportion of T-follic-ular helper cells (TFH) in normal and neoplastic follicles is the same and that the previously reported increase in TFH numbers in FL is thus due to an increase in the number and not content of follicles. As in normal germinal centers, TFH were shown to have a close spatial correlation with proliferating B cells in neoplastic follicles, where features of immunological synapse formation were observed. The number of TFH in FL correlate with the rate of B-cell proliferation and TFH co-localized to activation induced cytidine deaminase expressing proliferating B cells. T-cell receptor repertoire analysis of FL LN revealed that follicular areas are significantly more clonal when compared to the rest of the LN. These novel findings show that neoplastic follicles and germinal centers share important structural features and provide further evidence that TFH may play a role in driving B-cell proliferation and genomic evolution in TFH. Our results also suggest that targeting this interaction would be an attractive therapeutic option.
Introduction
Follicular lymphoma (FL) is a neoplasm of germinal center B cells that is usually characterized by the t(14;18) translocation and over-expression of BCL2.1,2 The clinical course is variable, prognosis is difficult to predict, and it is typically incurable.3,4 The tumor is infiltrated by numerous subsets of non-malignant T cells.5–8 Gene expression profiling (GEP) studies have shown that prognosis in FL can be correlated with the signature of non-malignant T cells of the microenvironment rather than the tumor itself, indicating that the microenvironment is important in the pathogenesis of this disease.9,10 The relationship between FL B cells and their microenvironment is complex; non-malignant T cells may either promote or inhibit tumor growth whilst the tumor itself can influence the composition of the microenvironment.11,12 Many groups have investigated the impact of microenvironment-related factors on outcome.10,13–16 These studies have, however, yielded contradictory results, most likely because of differences in patient populations studied, therapy administered and technical limitations of single parameter immunohistochemistry (IHC) that preclude accurate identification of cell subsets.
In normal germinal centers (GC), B cells are critically dependent on interactions with CD4pos follicular helper T cells (TFH),17–20 which are characterized by expression of PD-1, ICOS, CXCR5, CXCL13, IL-21 and IL-4 and the transcription factor BCL6.19,21,22 TFH provide signals necessary for the survival and proliferation of GC B cells and induce expression of activation induced cytidine deaminase (AID), a DNA modifying enzyme that initiates somatic hypermutation (SHM) and class switch recombination (CSR) leading to a class-switched, high-affinity antibody response.17,19,20,23
FL follicles and normal GC share a number of features; FL B cells have a similar phenotype and GEP as their normal counterparts and neoplastic follicles contain both follicular dendritic cells (FDC) and T cells. Studies performed on disaggregated FL lymph nodes (LN) have previously demonstrated an enrichment of IL-4-producing TFH in FL with a distinct gene expression profile and the ability to support FL B-cell growth and modify stromal cell function in vitro.24–28 The anatomic relationship between TFH and FL B cells and how closely this mimics the situation in normal GC has, however, not previously been studied.
In this study we compared the architecture of normal GC and neoplastic follicles, specifically focusing on the spatial relationship between B cells and TFH using multiparameter confocal immunofluorescence microscopy and semi-automated image analysis. We found that TFH - as identified by surface expression of CD4, PD1, and ICOS -constitute a similar proportion of CD4pos T cells in FL as they do in normal GC. They colocalize and form synapses with proliferating neoplastic B cells, which also express the DNA modifying enzyme AID. Finally, T-cell receptor (TCR) repertoire analysis revealed that T cells in neoplastic follicles are significantly more clonal than those in interfollicular areas, suggesting a role for antigen stimulation in this process. Overall, our findings further highlight the role of the microenvironment in FL and are relevant to the mode of action of new therapies such as those targeting antigen receptor signaling and the PD1/PDL1 axis.29–32
Methods
Patient samples
Formalin fixed paraffin embedded (FFPE) LN biopsies were obtained from 25 patients with histologically confirmed untreated or relapsed FL including three cases of grade IIIb FL, and eight patients with reactive lymphadenopathy. Patients with relapsed FL had not received any treatment for at least 12 months. Clinical details are presented in the Online Supplementary Tables S1-2. Ethical approval was obtained from the UK national research ethics committee, reference 13/NW/0040.
Immunofluorescent confocal microscopy
FFPE LN sample preparation steps including deparaffinization, antigen retrieval, and staining are described in the Online Supplementary Materials and Methods. All images were acquired on a Nikon Eclipse Ti-E microscope and analyzed using Nikon elements NIS Advanced Research software. Full descriptions of imaging and analysis techniques including the use of binary layers for image analysis are presented in the Online Supplementary Materials and Methods and further explained in the Online Supplementary Figure S1.
Laser micro-dissection, DNA extraction, and TCR sequencing
Follicles were highlighted by conventional IHC staining for BCL6. Follicular and interfollicular areas were dissected from sequential 10 μm FL sections using a laser capture microscope (PALM, Carl Zeiss MicroImaging, Jena, Germany). After DNA extraction, TCR sequences were subject to multiplex PCR amplification prior to next generation sequencing (Adaptive Biotechnologies, Seattle, WA, USA).33 TCRV CDR3 regions and their component V, D and J segments were identified using the IMGT definitions.34 Sequences not corresponding to a CDR3 were discarded and unique clones defined by the presence of more than one identical productive CDR3 DNA sequence. The number and size of each clone was determined and the richness, clonality and overlap of the follicular and interfollicular TCR repertoires determined (see the Online Supplementary Materials and Methods).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software v5 (GraphPad Software Inc, La Jolla, CA, USA). Normally distributed values are presented as the mean (+/- standard deviation), non-normally distributed values are presented as median (+/- interquartile range). Further details of the statistical analysis are presented in the Online Supplementary Materials and Methods.
Results
Normal and neoplastic follicles contain similar numbers of TFH
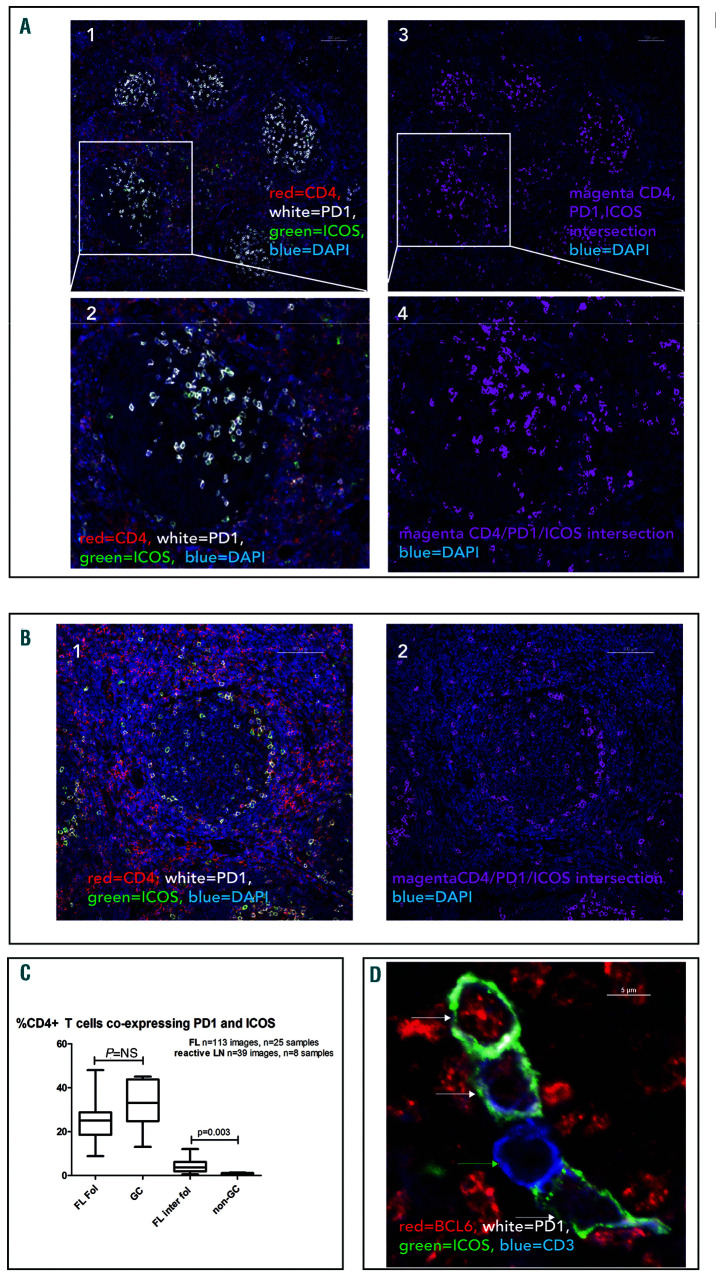
CD4pos T cells were predominantly located in the interfollicular areas of reactive and FL LN but discrete populations were also present within the GC and malignant follicles. We investigated the phenotype of these by staining for CD4, PD-1 and ICOS simultaneously. Within GC of reactive LN 33.05% (24.7-43.7) of CD4pos T cells co-expressed PD-1 and ICOS (TFH phenotype) and these were distributed predominantly in the light zones (Figure 1A). In FL, 25.0% (18.5-28.7) of follicular CD4pos T cells expressed both PD-1 and ICOS and were located at the follicular border or diffusely distributed within the follicles (Figure 1B). The proportion of CD4pos cells co-expressing PD-1 and ICOS was not significantly different between FL follicles and GC (Figure 1C). CD4posPD-1posICOSpos cells were tightly restricted to the GC of reactive LN and FL follicles with only 0.34% (0.26-1.13) and 3.63% (1.89-6.15) of non-GC or interfollicular FL CD4pos cells co-expressing PD-1 and ICOS respectively.
Figure 1.
Distribution of CD4pos PD-1pos ICOSpos cells in normal germinal centers and follicular lymphoma. (A1) Low power view of germinal centers (GC) in a reactive lymph node (LN) showing CD4pos (red) T-cells mainly outside the GC. A population of cells within the GC co-express PD-1 (white) and ICOS (green). (A2) The area highlighted by the white rectangle has been enlarged showing the distribution of CD4/PD1/ICOSpos cells in a normal GC where they are mainly polarized to the light zone (A3) Intersecting binary layer of image (A1) showing CD4/PD-1/ICOSpos cells (magenta) in GC. DAPI staining (blue) highlights cell nuclei. (A4) High power view of the intersecting CD4/PD-1/ICOSpos binary layer. (B1) Representative image of follicular lymphoma (FL) lymph node (LN) showing CD4pos (red) T cells mainly outside the follicles but a population within the follicles co-express PD-1 (white) and ICOS (green). (B2) Same image as (B1) showing only DAPI (blue) and the intersecting binary layer of CD4/PD-1/ICOSpos cells (magenta) which are restricted to the follicles where they are located predominantly in a perifollicular pattern. Scale bars represent 100 μm. (C) There was no significant difference in the proportion of CD4pos cells co-expressing PD-1 and ICOS in normal and neoplastic follicles. There was a small but significant increase in CD4/PD-1/ICOSpos cells in the inter-follicular compartment of FL com-pared to the same area zone of reactive LN. Horizontal lines represent median, boxes represent interquartile range, ‘whiskers’ represent range. (D) Representative, magnified image showing BLC6 expression in T cells in neoplastic follicles. Four CD3pos (blue) cells are shown, three are positive for the transcription factor BCL6 (red) and these are also PD-1pos (white) and ICOSpos (green), indicated with white arrows. One CD3pos cell is negative for BCL6 (green arrow), and this cell does not express PD-1 or ICOS. Overall 89.6% (88.3-91.8) of CD3posPD1posICOSpos cells express BCL6.
In FL, although 46.9% (34.7-51.9) of follicular CD4pos cells expressed PD-1, only about 50% of these co-expressed ICOS indicating that there are at least two distinct populations of CD4posPD-1pos cells within FL follicles, highlighting the importance of using all three parameters for identification of TFH. There was no difference in the proportion of CD4pos cells that co-expressed PD-1 and ICOS by histological grade in FL (Online Supplementary Figure S8), however, as the number and size of neoplastic follicles increase with histological grade, so must the absolute number of TFH.
To investigate differences in T cells located in the follicles and interfollicular areas of FL, the intensity of CD4 and PD-1 expression were measured. CD4 expression was 30.7% lower in follicular CD4pos T cells than in their inter-follicular counterparts suggesting that these represent a distinct population of T cells (Online Supplementary Figure S2). Whilst CD4pos PD-1posICOSpos cells were restricted to the follicles, CD4pos PD-1pos ICOSneg cells were present in the interfollicular area where 9.3% (5.1-26.4) of CD4pos cells expressed PD-1. The intensity of PD-1 expression was significantly higher in follicular PD-1pos T cells than interfollicular PD-1pos T cells (Online Supplementary Figure S2) consistent with them being TFH .35 Additional co-staining experiments demonstrated that these cells had a composite CD3pos, CD8neg, PD-1pos, ICOSpos, BCL6pos, CXCR5pos, TBETneg phenotype further confirming their identity as TFH (Figure 1D and Online Supplementary Figure S2C). Although CXCR5 has frequently been used to identify TFH by flow cytometry, we found that it was unhelpful in identifying this cellular subset by microscopy since most T cells present within these structures were CXCR5pos and it therefore did not help to distinguish them from other GC/follicularly located cells (Online Supplementary Figure S2C). It was not possible to use CD4 in these experiments as the anti-body is the same species as the BCL6. No BCL6pos cells were found to be CD8pos therefore the substitution for CD3 was acceptable (Online Supplementary Figure S2D). The intensity of BCL6 staining in TFH was lower than that observed in FL B-cells but higher than in other T cells (Online Supplementary Figure S2B). Although 25.0% (6.028.0) of ICOSpos T cells and 4.0% (1.0-8.0) of PD-1pos T cells within FL follicles were FOXP3pos, only a minority of dual PD-1posICOSpos T cells expressed FOXP3, (Online Supplementary Figure S3). In comparison to FL, very few FOXP3pos T cells were identified within the GC of reactive LN where they were exclusively located outside the GC (Online Supplementary Figure S3B).
These findings confirm that the majority of GC or follicular CD4pos cells that strongly express PD-1 and ICOS are TFH and constitute the same proportion of CD4+ cells in normal and neoplastic follicles.
THH co-localize with proliferating B cells
Next, we investigated if there is a spatial relationship between proliferating B cells and TFH in reactive and neo-plastic follicles. An ideal panel of CD20, Ki67, CD4, PD-1 and ICOS was not possible for technical reasons. However, co-staining for CD20, Ki67, and CD3 (Online Supplementary Figure S4) showed that in both normal GC and neoplastic follicles the majority of Ki67pos cells are CD20pos B cells. Also, as most PD-1Hi cells were ICOSpos, it enabled TFH to be identified using just two parameters; CD4 and high PD1 expression.
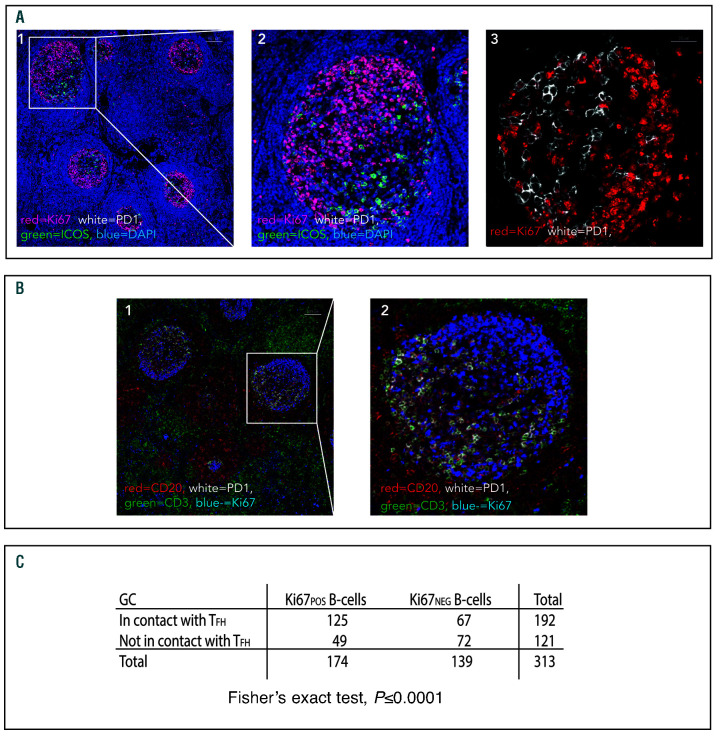
In normal GC a close spatial correlation between Ki67pos cells and CD4posPD-1Hi cells was evident in the light zone of all cases (Figure 2A). Automated image analysis showed that 63.1% ±15.9 of Ki67pos cells in the light zones were in direct contact with CD4posPD-1Hi cells, further-more, the high density of TFH in this compartment meant that the majority of Ki67pos B cells were in close proximity to ≥1 TFH. The majority of these PD-1Hi cells were also ICOSpos with 53.9% ±14.2 of Ki67pos cells in contact with PD-1posICOSpos cells. In GC light zones, Ki67pos B cells were significantly more likely than Ki67neg B cells to be in contact with TFH in all cases studied (P<0.005 in each GC examined) (Figure 2C).
Figure 2.
Close physical association between Ki67pos B cells and follicular helper T cells in normal germinal center light zones. (A1) Representative low power image showing polarization of Ki67pos cells to the dark zones of normal germinal centers. Ki67 (red), PD-1 (white), ICOS (green), DAPI (blue). Scale bar represents 100 μm. The area highlighted by the white rectangle is shown in high power in (A2). (A3) The close association between Ki67pos FL cells (red) and PD-1Hi cells (white) is shown in the light zone of another follicle whereas in the dark zone there is less interaction between Ki67pos cells and PD-1Hi T cells. The scale bar represents 25 μm. (B1) Using a different four-color panel, the Ki67pos cells (blue) were confirmed as CD20pos B cells (red) and the PD-1Hi cells (white) were confirmed as CD3pos T cells (green). The scale bar represents 100 μm and the area highlighted by the white rectangle has been magnified in (B2). Images representative of n=13 images from n=4 reactive lymph node (LN) samples. (C) Contingency table showing that Ki67pos B cells are significantly more likely to be in contact with follicular helper T cells (TFH) than Ki67neg B cells in normal germinal center (GC) light zones, as quantified by manual visual assessment. For all samples analyzed together (n=5, Fisher’s exact test P<0.0001).
In the highly proliferative dark zones, there were few TFH and a very high number of Ki67pos B cells. The closely packed Ki67pos B cells could not be separated by automated image analysis and therefore accurate calculation of the proportion of Ki67pos cells in contact with TFH could not be performed in the dark zones. It is clear from visual inspection, however, that the degree of spatial correlation between these cells is much lower in the dark zones than in the light zones (Figure 2B).
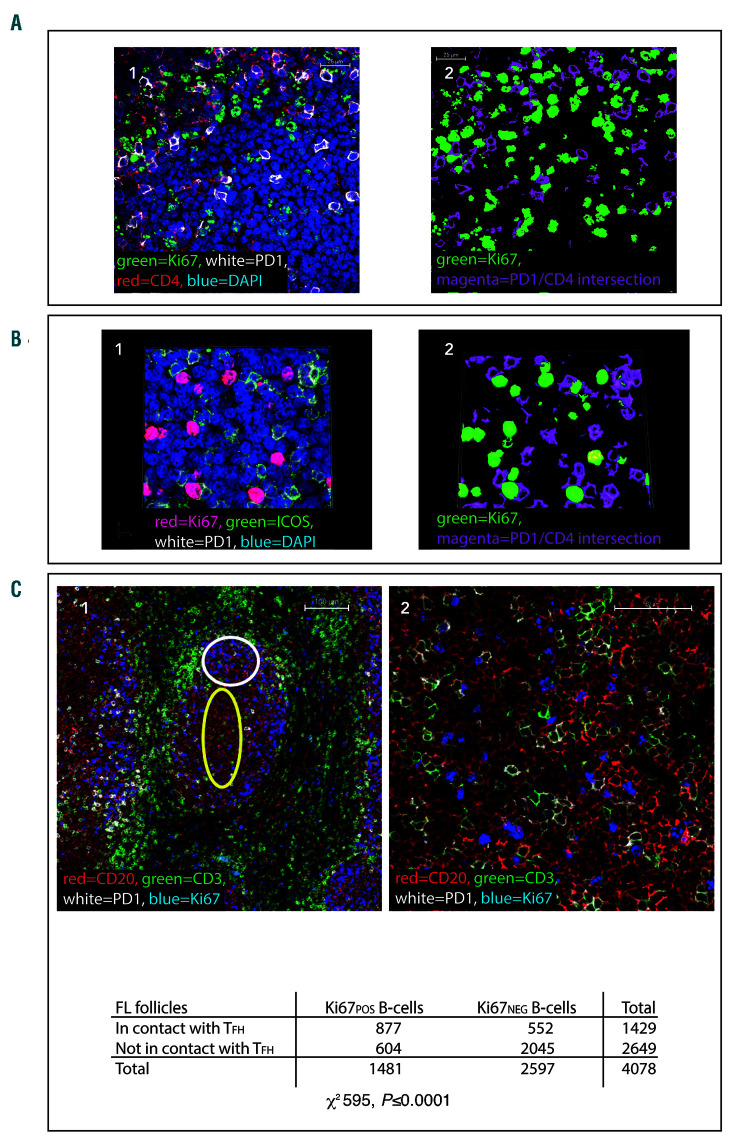
A close spatial relationship between Ki67pos B cells and CD4posPD-1Hi T cells was also found in FL and, in contrast to normal GC, all areas with high Ki67 also had increased numbers of TFH cells. In FL 41.0% ± 13.6 of CD20posKi67pos cells were found to be in direct contact with CD4posPD-1Hi cells, although the level of co-localization was significantly lower than in GC (P=0.003), there was a high level of co-localization in both settings (Figure 3A). High power images and 3D Z-stack reconstructions revealed that there was very close contact between these cells, and Ki67pos cells were frequently observed to be in contact with more than one CD4posPD-1Hi cell simultaneously. Staining for Ki67, PD-1, and ICOS revealed that 84.7% ± 11.1 of the PD-1HiCD4pos cells in contact with Ki67pos cells were also ICOSpos and therefore likely to be TFH (Figure 3B).
Figure 3.
Ki67pos cells are in close proximity to follicular helper T cells (TFH) in follicular lymphoma lymph nodes. (A1) Representative image of a neoplastic follicle showing Ki67pos cells (green) in close proximity to CD4pos (red), PD-1Hi (white) T cells. The scale bar represents 25 μm. (A2) Binary image of (A1), the binary layers of Ki67 (green) and the CD4-PD-1Hi intersection (magenta) are shown highlighting the close association of Ki67pos cells to PD-1Hi T cells. (B1) Representative image demonstrating that the majority of the PD-1Hi cells in contact with Ki67pos cells (red) are also positive for ICOS (green). (B2) This is highlighted in the binary layer 3D reconstruction of the same image, PD-1/ICOSpos (magenta) and Ki67 (green). Images representative of n=100 images from n=23 follicular lymphoma (FL) samples (4A), n=43 images from n=13 samples (4B). (C1) Ki67=blue, CD20=red, PD-1=white, CD3=green. Low power image (x10) showing Ki67pos and Ki67neg CD20pos B-cell co-localisation with PD1Hi CD3pos T cells in FL. Within the follicles there are areas of low proliferation (low Ki67=blue) where there are few PD1Hi (white) CD3pos T cells (green) - area highlighted by yellow oval, whereas in areas where there is high Ki67, there are more PD1Hi, CD3pos T cells (area highlighted by white circle) and they are frequently in contact with Ki67pos CD20pos FL B cells. Scale bar represents 100 μm. (C2) High power image (x60) in which the close correlation of Ki67pos (blue) B cells with PD1Hi (white) CD3pos (green) cells can be seen, whilst the CD20pos (red), Ki67neg cells are less frequently in contact with follicular helper T cells (TFH). Scale bar represents 50 μm. (C3) contingency tables showing that Ki67pos B cells are significantly more likely to be in contact with TFH than Ki67neg B-cells in FL (for all samples analyzed together [n=25 images from n=7 follicular lymphoma specimens] χ2 595, P<0.0001).
Ki67posCD20pos FL B cells were significantly more likely than Ki67negCD20pos FL B cells to be in direct contact with TFH in each case examined, (P<0.0001 for each specimen) (Figure 3C).
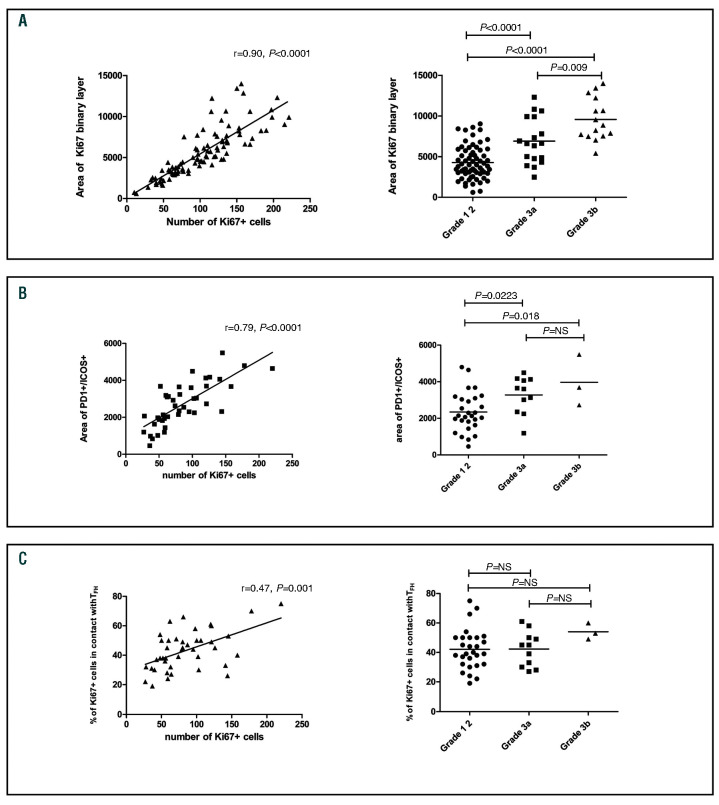
Relationship between Ki67, TFH cells and histological grade
The area of the Ki67 binary layer and the corresponding number of Ki67pos cells counted by automated analysis increased with histological grade (Figure 4A). The area of the PD-1posICOSpos intersection was closely correlated with the number of Ki67pos cells and histological grade demonstrating that, in higher grade cases with a higher proliferation rate, the absolute number of TFH is increased (Figure 4B). The degree of colocalization between Ki67+ B-cells and TFH was, however, similar across all histological grades. Thus, in cases with low Ki67 there were few TFH and in cases with high Ki67 there were more TFH, but the extent of co-localization remained relatively constant (Figure 4C). There was also a correlation between the number of Ki67pos cells and the number of TFH in normal GC (r=0.55, P=0.019, n=17 GC from n=4 samples; Online Supplementary Figure S7).
Figure 4.
Association between Ki67 and number of follicular helper T cells in follicular lymphoma. (A) The area of the Ki67 binary layer correlates closely with the number of Ki67pos cells automatically counted (left) and the area of the Ki67 binary layer is significantly higher in grade IIIa or IIIb disease than in grade I-II disease (right), n=99 images from n=23 samples. (B) The number of Ki67pos cells correlates closely with the number of PD1pos ICOSpos cells (left) and there are significantly more follicular helper T cells (TFH) (as represented by increased area of PD1/ICOS intersection) in grade IIIa and IIIb disease than in grade I-II disease (right), n=42 images from n=13 samples. (C) The degree of TFH – Ki67 interaction is weakly associated with the number of Ki67pos cells (left), and the proportion of Ki67pos cells in contact with TFH does not differ significantly according to histological grade disease (right) (n=42 images from n=13 samples).
Proliferating cells in contact with TFH express AID
Since TFH have been implicated in initiating SHM and CSR through induction of AID in GC,20 we investigated if there was a spatial relationship between TFH and AIDpos cells. AID was restricted to Ki67pos cells in FL, 63% ±8.8 of which were AIDpos and 39.8% ±9.7 of AIDposKi67pos cells were in direct contact with PD-1Hi cells. As we had established that most PD-1Hi cells in contact with Ki67pos cells were ICOSpos, we can predict that the majority of PD-1Hi cells in contact with AIDposKi67pos cells were TFH (Online Supplementary Figure S5). AID was similarly restricted to Ki67pos cells of GC where close association with PD-1Hi cells was evident in the light zones (Online Supplementary Figure S5C).
PD-L1
PD-L1 has previously been reported to be absent from the surface of FL B-cells36 and we found no evidence that PD-L1 was strongly expressed on the Ki67pos cells in con-tact with PD-1Hi cells. Instead, PD-L1 was expressed mainly on interfollicular CD23neg cells (Online Supplementary Figure S6). The identity of these cells was not further investigated in this study.
Features of synapse formation
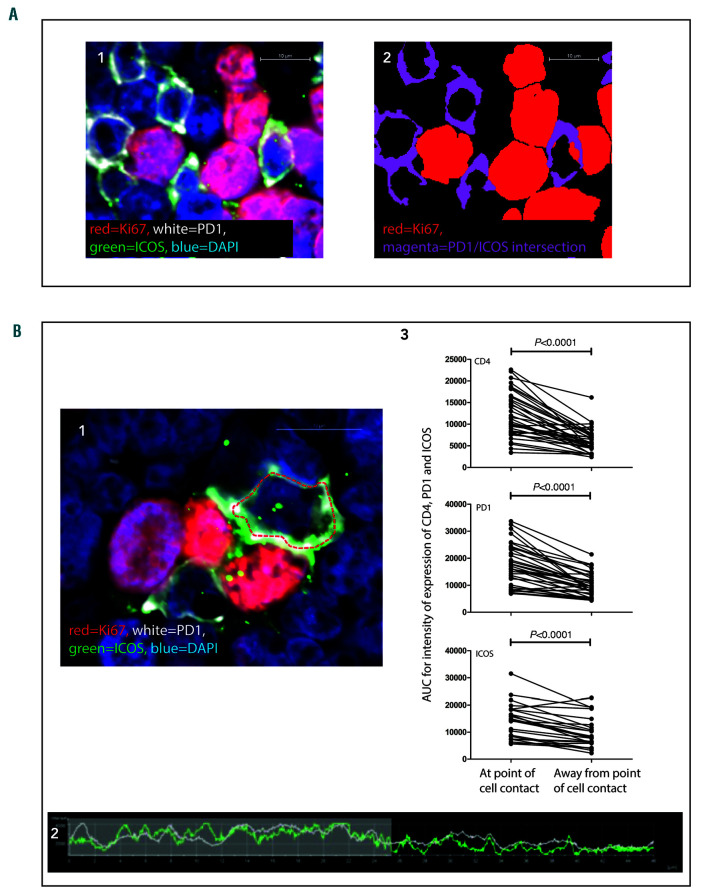
The close spatial relationship between Ki67pos FL B cells and TFH was further investigated in high power images where morphological features indicating the formation of immunological synapses were identified. Features included: TFH cell membrane projections encompassing the Ki67pos cells, overlapping of the B- and T-cell membranes, distortion of T-cell nuclei away from points of cell contact and significantly increased intensity of CD4, PD-1, and ICOS expression at points of cell contact (Figure 5).37 The intensity of expression of CD4, PD-1, and ICOS at points where TFH were in contact with Ki67pos cells was formally quantified by defining the perimeter of the T-cell membrane and measuring the intensity of fluorescence at each pixel around the perimeter. The area under the curve for intensity at the point of cell contact was compared with an equivalent length of cell membrane at the opposite pole (Figure 5B). Similar features indicating synapse formation were identified in GC light zones but the high number of closely-packed Ki67pos cells and TFH in GC precluded the same ana-lytic method being used because TFH in GC were usually in contact with more than one Ki67pos cell simultaneously.
Figure 5.
Close contact between Ki67pos cells and follicular helper T cells in follicular lymphoma: evidence for immune synapse formation. (A1) A Ki67pos cell (red) is seen to be in contact with 4 PD-1pos (white) ICOSpos (green) cells simultaneously. The PD-1posICOSpos cells are closely associated with the Ki67pos cell. (A2) Binary layer image of A1, the binary layers of Ki67 (red) and PD-1/ICOS intersection (magenta) are shown highlighting the close spatial association. Scale bars represent 10 μm. (B1) The follicular helper T cells (TFH) form projections encompassing the Ki67pos cells. Scale bar represents 10 μm. The perimeter of the TFH has been highlighted by the red dotted line and the intensity of CD4, PD-1 and ICOS have been measured around this line. CD4, PD1 and ICOS are more concentrated at the pole in contact with the Ki67pos cell as seen in the representative graph of intensity of expression around the dotted line, the area of cell contact is highlighted in the shaded area (B2). (B3) CD4, PD1, and ICOS all have significantly higher intensity of expression at the sites of cell contact than at the opposite pole, paired t-tests, n=61 cell con-tacts, from highly magnified images in nine follicular lymphoma specimens stained with CD4/PD-1/Ki67, or PD-1/ICOS/Ki67. AUC: area under the curve.
TCR repertoire within follicles shows evidence of antigen restriction
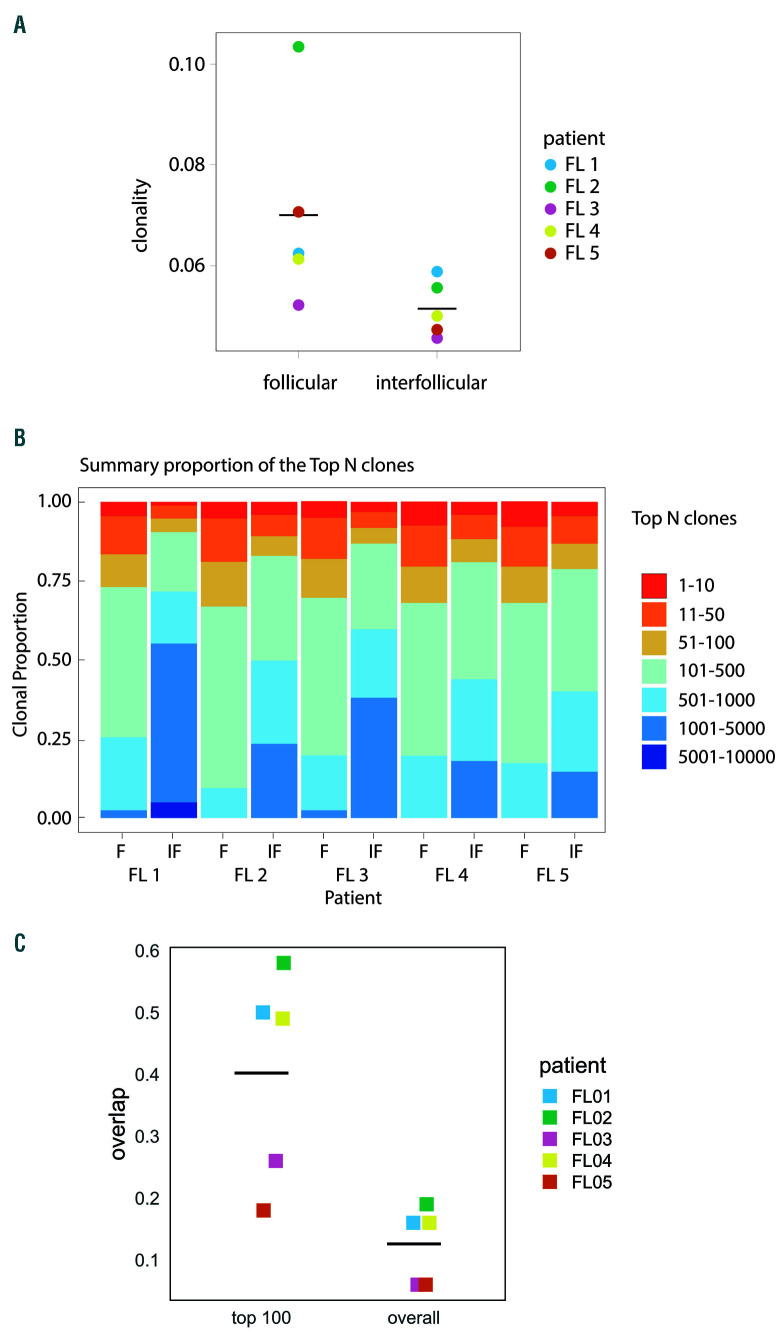
In view of the close spatial relationship observed between T cells and proliferating tumor cells, we investigated whether T cells within the follicles show evidence of antigen restriction by performing TCRV next generation sequencing of genomic DNA from laser dissected follicular and interfollicular areas from five FL samples. The degree of restriction of the TCRV repertoires in FL neo-plastic follicles and interfollicular areas was assessed in several ways. First, we estimated the richness of the repertoire in each compartment by determining the number of different clones present per ng of input DNA which, since we were analysing genomic DNA, was proportionate to the total cell number. The interfollicular areas contained more T-cell clones per ng of input DNA than the intrafollicular regions, however, this did not quite reach statistical significance (P=0.06, Online Supplementary Table S4). We also calculated the clonality index38 (see the Online Supplementary Materials and Methods for further details). In each of the five cases examined, the clonality of the follicular T cells was greater than in the interfollicular areas (P=0.0317, Figure 6A). We also calculated the proportion of the TCR repertoire in each compartment that was accounted for by high frequency clones.39 Compared to the interfollicular areas, the follicular regions were dominated by high frequency clones (Figure 6B). For example, the top 50 most frequent clones made up a mean of 19% of all clones in the follicular areas (95% CI: 17-21) compared to 9.8% in the interfollicular region (95% CI: 6.1-13.4) P=0.0002, n=5.
Figure 6.
Evidence of T-cell receptor repertoire restriction in follicular lymphoma. (A) The clonality of the T-cell receptors (TCR) in intrafollicular areas was higher than in the interfollicular compartment in all cases examined, horizontal bars represent mean of all samples, median clonality 0.062 versus 0.049 respectively, Mann Whitney, P=0.0317. (B) Summary of TCR repertoire data showing the proportion of the total population accounted for by high frequency clones in the follicular and interfollicular regions of follicular lymphoma lymph nodes. In each case, the more frequent clones predominate in the follicular regions compared to the interfollicular areas. (C) The level of overlap of clonotypes between follicular and interfollicular compartments for all clones in paired samples (all clones) and for the 100 most frequent clones (top 100 clones). Horizontal bars represent the mean overlap (0.125 for all clones and 0.22 for top 100 clones)
As expected from their different phenotypes, the clones present in the follicular and interfollicular areas of the same sample were markedly different, indicating that the TCR repertoires of the follicular and interfollicular areas are distinct (Figure 6C).
Discussion
In this study we compared the structure of neoplastic follicles in FL with GC in reactive LN, focusing on TFH, their relationship with proliferating B cells and TCR repertoire. Although TFH have previously been reported to be present in the FL microenvironment,24–27 these studies were performed on disaggregated LN and this is the first time that their spatial organization has been investigated in situ in this way. Using multi-parameter immunofluorescent confocal microscopy, we demonstrated that TFH – as identified by surface expression of CD4, PD1, and ICOS - constitute a similar proportion of CD4pos T cells in FL as in reactive LN and form synapses with proliferating Ki67pos tumor cells which express the DNA modifying enzyme AID. The number of TFH in neoplastic follicles correlates with the level of tumor proliferation and histological grade, and there is evidence for antigen restriction, as supported by the more clonal TCR repertoire found within neoplastic follicles compared to interfollicular areas.
These findings are novel and of significance for a number of reasons. First, in contrast to previous work on disaggregated FL LN, which showed an increase in the total number of TFH,24,25,40 we found that FL follicles contain TFH in similar proportions to normal reactive GC. This discrepancy likely relates to the fact that in FL, LN architecture is usually effaced by many closely packed follicles, whereas in normal tissues the interfollicular areas, which contain many fewer TFH, are more extensive. Thus, although the overall TFH content of FL LN is increased compared to normal,25 this is because of the larger number of follicles in the tumor and when neoplastic and normal follicles are compared directly, the numbers are the same. This finding underlines the need to complement data obtained from disaggregated tissues with anatomic studies.
Our use of multiparameter microscopy permitted the spatial relationship between TFH and B cells to be closely investigated and this also provided new insights. TFH are essential for providing normal GC B cells with signals necessary for their survival, proliferation and maturation.19,41 To our knowledge, this is the first time that the intimate relationship between Ki67pos B cells and TFH has been demonstrated in situ in human LN in this way and our observations are in keeping with the pivotal role they play in the normal GC reaction. Importantly, we also found that the close spatial association of Ki67pos B cells and TFH is recapitulated in FL, as 41% of Ki67pos FL B cells were in direct contact with TFH and were significantly more likely to be in direct contact with TFH than non-proliferating cells. The observed close spatial correlation between the two cell types is thus not due to chance and suggests that TFH are involved in functionally important interactions with the tumor. This corroborates and advances findings from previous in vitro experiments which showed that FL TFH provide signals for B-cell survival.25,27
We also found a correlation between the numbers of TFH and Ki67pos B cells in both normal GC and neoplastic follicles and that, in FL, the number of TFH increase with histological grade. The relationship between number of TFH and rate of B-cell proliferation observed in our study has not been reported previously in FL but is consistent with previous data showing that the regulation of GC size and B-cell number is critically dependent on the number of TFH.17,19,23 This adds to the evidence that TFH are central to the pathogenesis of FL, just as they are essential in the normal GC reaction. Furthermore, the degree of co-localization (the proportion of Ki67pos B cells in contact with one or more TFH) remained constant as histological grade increased, with no significant change in the proportion of Ki67pos cells in con-tact with TFH in grade IIIa or IIIb disease compared to grade I-II disease, suggesting that interaction with TFH remains important regardless of histological grade.
Our studies also underline the crucial importance of using a multi-parameter approach to define and quantify the complex T-cell subsets present in the FL microenvironment. No single antigen or transcription factor specifically identifies TFH and this is the first study reporting the presence of TFH in FL in situ using techniques that overcome the limitations of traditional IHC. By using co-staining for ICOS and BCL6 we were able to show that only half of the PD1 expressing cells neoplastic follicles are TFH. Single parameter analysis of PD1 would therefore lead to significant overestimate of TFH numbers perhaps explaining, at least in part, why previous IHC studies have yielded divergent results with regards to the impact of different T-cell infiltrates on prognosis.10,13–16
Previous in vitro studies have shown that peripheral blood T cells in FL are dysfunctional and form impaired synapses with B cells.40,42 In the present study, however, we found features that suggest normal synapse formation between Ki67pos tumor cells and TFH within the LN.43 This divergence from previous research may be because we examined the interactions between TFH and Ki67pos cells in situ in human tissue rather than in an ex vivo system using peripheral blood derived cells. It also remains possible that there are other subsets of non TFH cells in the FL microenvironment that are dysfunctional and have an impaired ability to form immunological synapses.
In addition to promoting GC B-cell proliferation, interaction with TFH cells also induces AID expression which induces somatic hypermutation and class switch recombination. Off-target action of AID has previously been proposed to lead to the accumulation of mutations required for germinal center-derived lymphomas to develop or progress and has been associated with transformation of FL.44,45 The close spatial association between TFH and AIDposKi67pos FL B cells observed in the present study is compatible with this theory.
Finally, next generation sequencing analysis of the TCR repertoire of follicular and interfollicular areas of FL LN showed that the neoplastic follicles are significantly more clonal and dominated by high frequency clones compared to the interfollicular regions. As expected from their divergent phenotype, very little repertoire overlap between the two compartments was present. PCR-based analyses of TCR repertoire on small samples are known to suffer from a number of potential limitations including sampling effects and errors introduced during the amplification process, which may lead to apparent skewing.46 Whilst we cannot completely exclude these possibilities, we minimized the risk by direct, intra-patient comparison in the same assay run, and, of note, our findings were consistent in all five cases studied. Another possibility is that the demonstrated differences in TCR repertoire relate to the greater number of T cells found in the interfollicular regions compared to the follicles. Whilst T-cell numbers undoubtedly do differ between these two areas, significant difference in the clonality index, which takes into account the number of unique clones present, were observed (Figure 6A). Furthermore, the repertoire of the intrafollicular area was strikingly dominated by high frequency clones; for example, the top 50 clones accounted for a mean of 19% (CI: 17-21%) of all clones present, compared to 9.7%(CI: 6.1-13.4%) in the interfollicular areas (P=0.0002).
Taken together, these findings suggest that the interactions between B cells and activated TFH that induce B-cell proliferation and differentiation and lead to the generation of high affinity antibody in normal GC may be recapitulated within the follicles of FL.19,41 Since TFH may be involved in processes fundamental to disease progression, such as clonal expansion and genomic evolution of the tumor, our results suggest that they would be an attractive target for novel therapies. This is especially relevant in the era of drugs that target antigen receptor signaling such as PI3-kinase inhibitors, which affect both B- and T-cell receptor pathways. Our results are also relevant to understanding the mechanism of action of drugs that target PD-1 expressing cells, which have been shown to be effective in FL and other lymphomas.29–31 It is clear that whilst some of the PD-1 expressing cells in the FL LN are indeed TFH, many are not and these may represent exhausted effector cells. Blockade of PD-1 in the latter case may unmask antitumor immunity and lead to disease regression. The impact of interrupting PD-1 function in TFH is, however, less clear as the role of the PD-1 axis in TFH function is not fully established. These findings add another level of complexity to our understanding of the FL tumor microenvironment and underline the necessity of using multi-dimensional methods in future studies.
Supplementary Material
Acknowledgments
We are grateful for the technical assistance and advice of Adaptive Biotechnologies and of Jon Harris and Jan Soetaert from the Nikon Imaging Centre at Kings’ College London.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/106/6/1593
Funding This research was funded by grants from the British Society of Haematology in partnership with the Roger Counter Foundation, and Bloodwise.
References
- 1.Harris N, Swerdlow S, Jaffe E, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. 4th ed. Lyon: International Agency for Research on Cancer, 2008. [Google Scholar]
- 2.Marafioti T, Copie-Bergman C, Calaminici M, et al. Another look at follicular lymphoma: immunophenotypic and molecular analyses identify distinct follicular lymphoma subgroups. Histopathology. 2013;62(6):860–875. [DOI] [PubMed] [Google Scholar]
- 3.Ardeshna KM, Smith P, Norton A, et al. Long-term effect of a watch and wait policy versus immediate systemic treatment for asymptomatic advanced-stage non-Hodgkin lymphoma: a randomised controlled trial. Lancet. 2003;362(9383):516–522. [DOI] [PubMed] [Google Scholar]
- 4.Montoto S, Davies AJ, Matthews J, et al. Risk and clinical implications of transformation of follicular lymphoma to diffuse large B-cell lymphoma. J Clin Oncol. 2007; 25(17):2426–2433. [DOI] [PubMed] [Google Scholar]
- 5.Ame-Thomas P, Tarte K. The yin and the yang of follicular lymphoma cell niches: role of microenvironment heterogeneity and plasticity. Semin Cancer Biol. 2014; 24: 23–32. [DOI] [PubMed] [Google Scholar]
- 6.de Jong D, Koster A, Hagenbeek A, et al. Impact of the tumor microenvironment on prognosis in follicular lymphoma is dependent on specific treatment protocols. Haematologica. 2009; 94(1): 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farinha P, Al-Tourah A, Gill K, Klasa R, Connors JM, Gascoyne RD. The architectural pattern of FOXP3-positive T-cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood. 2010; 115(2): 289–295. [DOI] [PubMed] [Google Scholar]
- 8.Farinha P, Masoudi H, Skinnider BF, et al. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood. 2005; 106(6): 2169–2174. [DOI] [PubMed] [Google Scholar]
- 9.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21): 2159–2169. [DOI] [PubMed] [Google Scholar]
- 10.Glas AM, Knoops L, Delahaye L, et al. Gene-expression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J Clin Oncol. 2007;25(4):390–398. [DOI] [PubMed] [Google Scholar]
- 11.Yang ZZ, Novak AJ, Stenson MJ, Witzig TE, Ansell SM. Intratumoral CD4+CD25+ regulatory T-cell-mediated suppression of infiltrating CD4+ T-cells in B-cell non-Hodgkin lymphoma. Blood. 2006;107(9):3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiaii S, Clear AJ, Ramsay AG, et al. Follicular lymphoma cells induce changes in T-cell gene expression and function: potential impact on survival and risk of transformation. J Clin Oncol. 2013;31(21):2654–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee AM, Clear AJ, Calaminici M, et al. Number of CD4+ cells and location of fork-head box protein P3-positive cells in diagnostic follicular lymphoma tissue microarrays correlates with outcome. J Clin Oncol. 2006;24(31):5052–5059. [DOI] [PubMed] [Google Scholar]
- 14.Wahlin BE, Aggarwal M, Montes-Moreno S, et al. A unifying microenvironment model in follicular lymphoma: outcome is predicted by programmed death-1--positive, regulatory, cytotoxic, and helper T-cells and macrophages. Clin Cancer Res. 2010;16(2):637–650. [DOI] [PubMed] [Google Scholar]
- 15.Carreras J, Lopez-Guillermo A, Roncador G, et al. High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J Clin Oncol. 2009;27(9):1470–1476. [DOI] [PubMed] [Google Scholar]
- 16.Richendollar BG, Pohlman B, Elson P, Hsi ED. Follicular programmed death 1-positive lymphocytes in the tumor microenvironment are an independent prognostic factor in follicular lymphoma. Hum Pathol. 2011;42(4):552–557. [DOI] [PubMed] [Google Scholar]
- 17.Rolf J, Bell SE, Kovesdi D, et al. Phosphoinositide 3-kinase activity in T-cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185(7):4042–4052. [DOI] [PubMed] [Google Scholar]
- 18.King C. New insights into the differentiation and function of T follicular helper cells. Nat Rev Immunol. 2009; 9(11): 757–766. [DOI] [PubMed] [Google Scholar]
- 19.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014; 41(4): 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linterman MA, Liston A, Vinuesa CG. T-follicular helper cell differentiation and the co-option of this pathway by non-helper cells. Immunol Rev. 2012;247(1):143–159. [DOI] [PubMed] [Google Scholar]
- 21.Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. Follicular helper T-cells: lineage and location. Immunity. 2009;30(3):324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, Rao S, Tsai LM, et al. The transcriptional repressor BCL6 directs T follicular helper cell lineage commitment. Immunity. 2009;31(3):457–468. [DOI] [PubMed] [Google Scholar]
- 23.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27(2):190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pangault C, Ame-Thomas P, Ruminy P, et al. Follicular lymphoma cell niche: identification of a preeminent IL-4-dependent T(FH)-B cell axis. Leukemia. 2010; 24(12): 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ame-Thomas P, Le Priol J, Yssel H, et al. Characterization of intratumoral follicular helper T-cells in follicular lymphoma: role in the survival of malignant B cells. Leukemia. 2012;26(5):1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang ZZ, Grote DM, Ziesmer SC, Xiu B, Novak AJ, Ansell SM. PD-1 expression defines two distinct T-cell sub-populations in follicular lymphoma that differentially impact patient survival. Blood Cancer J. 2015;5:e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ame-Thomas P, Hoeller S, Artchounin C, et al. CD10 delineates a subset of human IL-4 producing follicular helper T-cells involved in the survival of follicular lymphoma B cells. Blood. 2015;125(15):2381–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey S, Mourcin F, Marchand T, et al. IL-4/CXCL12 loop is a key regulator of lymphoid stroma function in follicular lymphoma. Blood. 2017;129(18):2507–2518. [DOI] [PubMed] [Google Scholar]
- 29.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkes EA, Grigg A, Chong G. Programmed cell death-1 inhibition in lymphoma. Lancet Oncol. 2015;16(5):e234–245. [DOI] [PubMed] [Google Scholar]
- 31.Westin JR, Chu F, Zhang M, et al. Safety and activity of PD1 blockade by pidilizumab in combination with rituximab in patients with relapsed follicular lymphoma: a single group, open-label, phase 2 trial. Lancet Oncol. 2014;15(1):69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopal AK, Kahl BS, de Vos S, et al. PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med. 2014;370(11):1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T-cells. Blood. 2009;114(19):4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/Junction Analysis: the first tool for the analysis of the immunoglobulin and T-cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20 Suppl 1:i379–385. [DOI] [PubMed] [Google Scholar]
- 35.Haynes NM, Allen CDC, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1 high germinal center-associated sub-population. J Immunol. 2007;179(8):5099–5108. [DOI] [PubMed] [Google Scholar]
- 36.Andorsky DJ, Yamada RE, Said J, Pinkus GS, Betting DJ, Timmerman JM. Programmed death ligand 1 is expressed by non-hodgkin lymphomas and inhibits the activity of tumor-associated T-cells. Clin Cancer Res. 2011;17(13):4232–4424. [DOI] [PubMed] [Google Scholar]
- 37.Kupfer A, Kupfer H. Imaging immune cell interactions and functions: SMACs and the Immunological Synapse. Semin Immunol. 2003;15(6):295–300. [DOI] [PubMed] [Google Scholar]
- 38.Sherwood AM, Emerson RO, Scherer D, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T-cell receptor sequences that differ from the T-cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62(9):1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nazarov VI, Pogorelyy MV, Komech EA, et al. TcR: an R package for T-cell receptor repertoire advanced data analysis. BMC Bioinformatics. 2015;16:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myklebust JH, Irish JM, Brody J, et al. High PD-1 expression and suppressed cytokine signaling distinguish T-cells infiltrating follicular lymphoma tumors from peripheral T-cells. Blood. 2013;121(8):1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. [DOI] [PubMed] [Google Scholar]
- 42.Ramsay AG, Clear AJ, Kelly G, et al. Follicular lymphoma cells induce T-cell immunologic synapse dysfunction that can be repaired with lenalidomide: implications for the tumor microenvironment and immunotherapy. Blood. 2009; 114(21): 4713–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barcia C, Thomas CE, Curtin JF, et al. In vivo mature immunological synapses forming SMACs mediate clearance of virally infected astrocytes from the brain. J Exp Med. 2006;203(9):2095–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasqualucci L, Bhagat G, Jankovic M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40(1):108–112. [DOI] [PubMed] [Google Scholar]
- 45.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6(1):130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Best K, Oakes T, Heather JM, Shawe-Taylor J, Chain B. Computational analysis of stochastic heterogeneity in PCR amplification efficiency revealed by single molecule barcoding. Sci Rep. 2015;5:14629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.