Abstract
We present an on-chip method for the extraction of RNA within a specific size range from low-abundance samples. We use isotachophoresis (ITP) with an ionic spacer and a sieving matrix to enable size-selection with a high yield of RNA in the target size range. The spacer zone separates two concentrated ITP peaks, the first containing unwanted single nucleotides and the second focusing RNA of the target size range (2 – 35 nt). Our ITP method excludes >90% of single nucleotides and >65% of longer RNAs (>35 nt). Compared to size selection using gel electrophoresis, ITP-based size-selection yields a 2.2-fold increase in the amount of extracted RNAs within the target size range. We also demonstrate compatibility of the ITP-based size-selection with downstream next generation sequencing. On-chip ITP-prepared samples reveal higher reproducibility of transcript-specific measurements compared to samples size selected by gel electrophoresis. Our method offers an attractive alternative to conventional sample preparation for sequencing with shorter assay time, higher extraction efficiency and reproducibility. Potential applications of ITP-based size-selection include sequencing-based analyses of small RNAs from low-abundance samples such as rare cell types, samples from fluorescence activated cell sorting (FACS), or limited clinical samples.
Graphcial Abstract
We present an on-chip method that achieves simultaneous RNA extraction and size selection, and demonstrate its compatibility with high-throughput sequencing.
Introduction
Isotachophoresis (ITP) is an electric field-based focusing and separation technique utilizing two buffers in which one has a high-mobility leading electrolyte (LE) and the second has a relatively low-mobility trailing electrolyte (TE). The self-sharpening and moving interface between the TE and LE is associated with a sharp electric field gradient. Sample molecules with intermediate mobilities between those of TE and LE accumulate at the TE-to-LE interface as peak-mode ITP.1 Anionic peak-mode ITP can selectively preconcentrate nucleic acids while excluding contaminants such as cell debris or proteins.2 With its high extraction efficiency, selectivity, and compatibility with several lysis methods, ITP-based nucleic acid purification has been applied to a broad range of samples including blood, urine, saliva, and cell lysate.3–5
Coupling ITP with sieving matrices enables size-selective purification of nucleic acids.5–9 Sieving matrices decrease nucleic acid mobilities in a size-dependent manner, yet have minimal effect on the mobility of small ions.10 Previous approaches using a sieving matrix for size-selection achieved exclusion of RNAs >40 nt with 5.5% PVP,6,7 miRNA detection with 4% polyacrylamide,9 and exclusion of 66 nt synthetic RNA with 30% Pluronic F-127.5 ITP sample focusing can also be used with ‘spacer ions’ to create separation of mixed samples.11 In the latter mode, multiple peak-mode ITP zones are separated from each other by the spacer ion zones in plateau mode. Previous studies have shown separation of single- and double-stranded DNA by a single spacer zone8 and separation of multiple serum lipoproteins by several spacer zones formed by a carrier ampholyte.12
ITP-extracted nucleic acids are compatible with downstream analyses such as RT-qPCR,13,14 microarray hybridization15 and sequencing.16,17 Among these methods, sequencing offers unmatched advantages including digital quantification, the ability to identify novel transcripts, high throughput, and single-base resolution.18 Many sequencing-based methods have been developed to address a wide range of questions in RNA biology.19 For example, UV Cross-Linking and Immunoprecipitation (CLIP-Seq)20 and RNA immunoprecipitation followed by deep sequencing (RIP-Seq)21 measure protein-RNA interactions. Similarly, sequencing based methods can probe RNA secondary structure22 or unveil mRNA fragment sequences protected by ribosomes during translation.23 All of these approaches require extraction of specific RNA fragment sizes with as high yield as possible. Compared to conventional sample preparation methods such as column-based RNA purification followed by gel electrophoresis, ITP purification has the potential to offer higher yield especially with lower input amounts and shorter nucleic acids.5,17 Furthermore, ITP may provide better consistency, fewer hands-on steps, and faster processing time.
To our knowledge, all previously reported size-selective ITP methods demonstrated exclusion of RNA longer than a certain cutoff size using combinations of TE ions and sieving matrices.5–9 We know of no reported ITP method that is size-selective between both low and high limits of RNA length. Having a lower limit in the size selection is especially critical for sequencing applications since the presence of very small nucleic acid fragments leads to significant contamination in sequencing reads. Importantly, presence of mononucleotides in the sample inhibits the library preparation required in several aforementioned sequencing methods. In these methods, ribonucleases are used to digest mRNAs that are not protected by ribosomes or RNA binding proteins, which results in 3’ phosphate group on the protected RNA fragments. Sequencing library preparation for RNA fragments generated by ribonucleases starts with an initial dephosphorylation. The most commonly used enzyme for the dephosphorylation reaction is T4 PNK.24–28 However, in the presence of ATP, T4 PNK functions as a kinase and instead adds a 5’ phosphate to the substrate.29 Given that intracellular ATP concentration can be as high as 10 mM,30 the removal of this from the lysate is essential for the dephosphorylation step. In conventional methods, size range selection is typically performed by denaturing polyacrylamide gel electrophoresis.24,25 However, despite its ubiquitous adoption, this method is severely limited because it offers low yield, is time-consuming, and cannot be easily parallelized;26 hence alternative methods are needed.
In the current study, we present the first on-chip ITP method for selecting a range of RNA sizes using an ionic spacer, sieving matrix, and a two-step collection method. We demonstrate >90% removal of single nucleotides and >65% removal of RNA longer than 35 nt in the extracted sample. Our method performs RNA extraction and size selection simultaneously in a single on-chip process within 10 min. We also compare our method to size-selection by denaturing gel electrophoresis and demonstrate a 2.2 fold increase in yield. Lastly, we demonstrate the compatibility of ITP-extracted RNA with high-throughput sequencing.
Overview of the Method
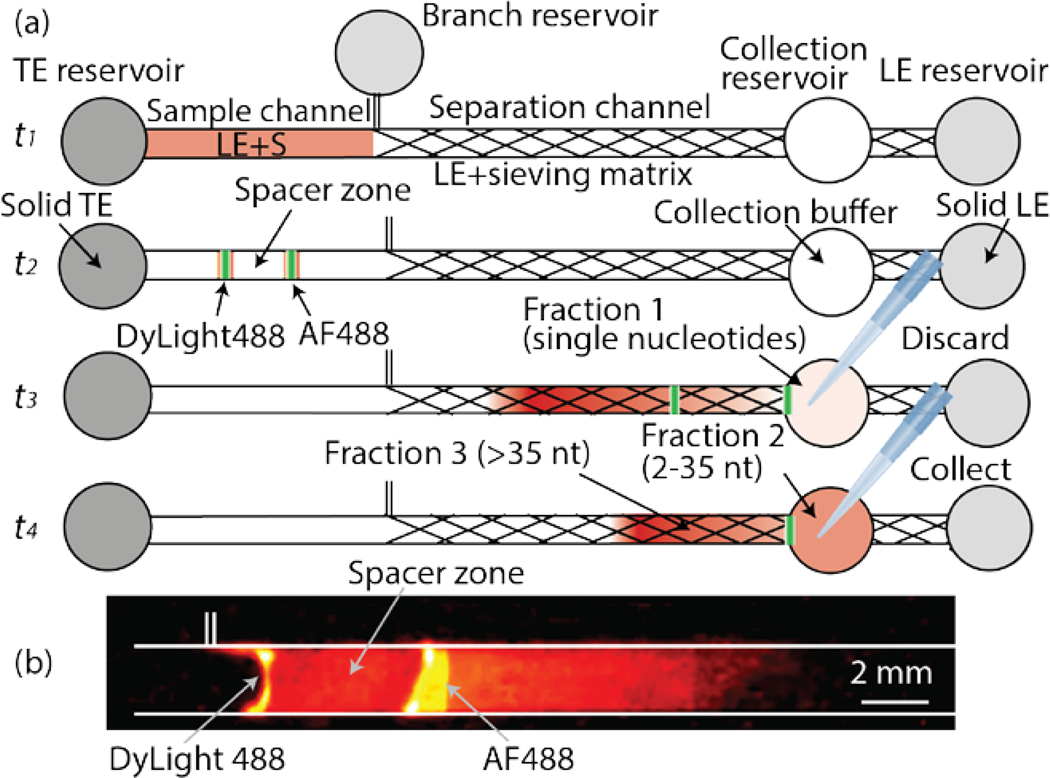
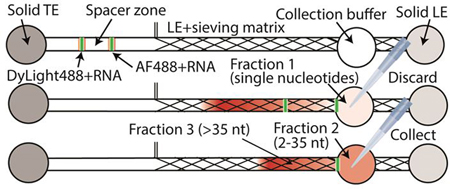
Our chip consists of a 2-mm-wide main and a 300-μm-wide branch channel. These are connected to three electrode reservoirs and one collection reservoir (Figure 1a). All channels have a depth of 300 μm. We refer to the channel section prior to the branch point as the sample channel, and the region beyond the branch point as the separation channel. The detailed dimensions are provided in Electronic Supplementary Information (ESI), Figure S1. Our channel design can process 17 μl of sample volume. As shown in Figure 1a at time t1, the channel is initially filled with sample in the sample channel and LE containing sieving matrix in the separation channel. In the TE, LE, and branch reservoirs, thermal-responsive gel is used to eliminate pressure driven flow in the channel during loading, ITP, and collection steps. The reservoir solutions contain 35% of Pluronic F-127, which behaves as a solid at room temperature, while behaving as a liquid below 4°C.27 At time t2, we apply electrical current to start ITP. A constant current of 300 μA is applied from the LE reservoir to the TE reservoir in the main channel and an additional 30 μA current is applied from the TE reservoir to the branch reservoirs. The minor current in the opposite direction prevents sample loss into the branch channel. Two ITP peaks that contain RNA and fluorescent dyes migrate in the front and the back of the spacer zone. As determined empirically, AF488 and DyLight488 co-migrate with the first and the second ITP peak, respectively. In ESI, we include a video (Video S1) of typical ITP-based size-selection process. Figure S2 presents snapshots from the Video S1 at three sequential times to show the spacer zone formation and size-selective separation process.
Figure 1.
(a) Schematic representation of RNA size-selection. Initially, the chip is loaded with LE including sample (S) in the sample section of the channel and LE including sieving matrix in the separation channel (time t1). At time t2, an electrical current of 300 μA is applied to the main channel and a 30 μA current is applied in the branch channel. The spacer zone forms between ITP peaks of the two fluorescent dyes. At time t3, the first peak arrives at the collection reservoir. Fraction 1 collected from the collection reservoir contains single nucleotides and is discarded. The reservoir is refilled with fresh collection buffer and the same current is applied again. At time t4, the second peak arrives and Fraction 2 containing the target size RNAs is collected. Longer RNA molecules remain in the channel (Fraction 3). (b) Visualization of the two ITP peaks separated by a spacer zone. The first peak is visualized by AF488 and includes single nucleotides. In the second peak, DyLight488 and RNA in the target range of 2 – 35 nt co-focus. The snapshot was captured from Video S1 at 3:20 s.
As the ITP zones enter the separation channel, the sieving matrix reduces the RNA mobility while those of ions are negligibly affected. Consequently, RNA molecules rearrange based on their sizes such that single nucleotides remain focused in the first peak, and ~2–35 nt RNAs are focused in the second peak. RNAs that are longer than 35 nt defocus and travel electrophoretically behind the second peak. When the first peak arrives at the collection reservoir (at time t3), we temporarily suspend the current and collect the contents from the collection reservoir (Fraction 1). Fraction 1 contains single nucleotides and is discarded. We then re-apply current to reinitiate ITP. At time t4 (typically after about a minute of re-applying current) the second peak arrives at the collection reservoir, and the sample containing RNA of the desired size range (Fraction 2) is collected. Longer RNAs remain in the channel and are not collected except when we analyze the contents of Fraction 3, which can be retrieved by applying additional 70 s of electric field.
Materials and Methods
Fabrication of polydimethylsiloxane (PDMS) chips
A clear plastic mold for the single layer PDMS channel was fabricated by 3D printing at Proto Labs, Inc. (Maple Plain, MN) using WaterShed XC 11122 material at a high-resolution specification (with a nominal resolution of 0.002” layers). The mixture of 10:1 (w/w) ratio precursor-to-curing-agent (Sylgard 184, Dow Corning, Menlo Park, CA) was poured on the mold taped on a 10-mm petri dish. After degassing for 20 min in a desiccator chamber connected to a vacuum pump, the petri dish was placed in an oven at 50°C for at least 5 h. We then cut out and peeled off the PDMS slab from the mold, and punched four 6-mm diameter holes to form the TE, LE, branch, and collection reservoirs. The surface of the PDMS slab and a microscope glass slide was cleaned with scotch tape and plasma-treated using a plasma cleaner (PDC-32G, Harrick Plasma, Ithaca, NW) connected to a vacuum pump (PDC-VPE, Harrick Plasma, Ithaca, NW) at high RF power for 90 s. Immediately after the plasma treatment, we bonded the PDMS substrate on the glass slide and waited at least 2 h to ensure a leak-free bond before using the channel.
Preparation of the RNA samples
Human lymphoblastoid cells (LCLs) were grown to a density of ~1 × 106 cells/ml in 15% fetal bovine serum and 1% Pen-Strep. Approximately 10 million cells were pelleted at 4°C (250g) and washed once with ice-cold PBS. Immediately after pelleting, the cells were frozen in liquid nitrogen and stored at −80°C until further processing. Cells were lysed in the presence of 100 μg/ml cycloheximide. Lysis buffer additionally contained 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5mM MgCl2, 1mM DTT, 1% Triton X-100, and 25 U/ml Turbo DNase I. 150 μl of lysis buffer was used for each experiment. Cells were homogenized by repeatedly pipetting the solution using a P1000 pipette. Lysates were incubated on ice for 10 minutes. After centrifugation at 1300g for 10 minutes at 4°C, supernatant was recovered. A combination of 300 Units of RNase T1 (Fermentas) and 500 ng of RNase A (Ambion) was used to digest 7 A260 units of supernatant at room temperature for 30 min. The RNase digestion enriches for ribosome protected fragments of mRNAs in the size range of 17 – 35 nt.28 RNase digestion was stopped with 20 mM Ribonucleoside Vanadyl Complex (NEB: S1402S). Samples were then loaded onto a 34% (w/v) sucrose cushion and centrifuged at 4°C in a TLA120.3 rotor for 4 h at 70,000 rpm. The sucrose layer was discarded and the pellet was resuspended in 700 μl of Qiazol reagent from miRNeasy kit (Qiagen) followed by RNA extraction using manufacturer’s instructions.
ITP buffers and reagents
LE buffer contained 13% polyvinylpyrrolidone (PVP, M.W. 360,000), 8 M urea, 20 mM HCl, 130 mM Bis-Tris (measured pH 7.2). LE buffer was made fresh daily. Reservoir LE buffer consisted of 35% Pluronic F-127, 50 mM HCl, 200 mM Bis-Tris, and reservoir TE buffer was made of 35% Pluronic F-127, 200 mM Bis-Tris, 50 mM MOPS, and 25 mM caproic acid. Solutions containing >25% Pluronic F-127 is liquid at temperature below 4°C and solid otherwise. We stored the solutions containing Pluronic F-127 in a 4°C refrigerator before experiments and on ice during experiments. Pluronic F-127-containing solutions were loaded quickly and carefully while in the cold, liquid state as they solidify quickly at room temperature. Sample buffer included 250 nM Alexa Fluor 488 (AF488), 750 nM DyLight 488, 0.5% PVP, 20 mM HCl, and 130 mM Bis-Tris, and varying contents of RNA. Collection buffer was 20 mM HCl, 130 mM Bis-Tris, 0.1% PVP, and 0.4 U/μl SUPERase In RNase inhibitor.
For the single nucleotide exclusion experiment, we included 500 μM rATP (NEB) and 1 μM synthetic 26 nt RNA in the sample buffer. The sequence of 26 nt synthetic RNA was 5’-AUGUACACGGAGUCGACCCAACGCGA-3’ (IDT). For all other experiments, we used RNA from LCL extracts as detailed above. We purchased PVP, HCl, Bis-Tris, urea, MOPS, caproic acid, Pluronic F-127, NaOH, and Triton X-100 from Sigma-Aldrich. AF488 (A20000), DyLight488 (46402), and SUPERase In (AM2694) were purchased from Thermo Fisher Scientific. All solutions were prepared with UltraPure DNase/RNase free distilled water (10977015, Thermo Fisher Scientific).
RNA size-selection using ITP
We washed PDMS channels first with 1 M NaOH and 0.1% Triton-X100, followed by 1 M HCl and 0.1% Triton-X100. Strong base and acid solutions clean the channel surfaces, and smooth the glass. The 0.1% Triton-X100 prevents bubble formation while filling the channel with ITP buffers. Immediately after washing, we filled the separation channel by capillary force and the pressure driven flow induced by 60 μl of LE buffer loaded in the collection reservoir. Specifically, we monitored the interface of the LE buffer until it arrived near the branch channel. We then removed all liquid from collection reservoir and slowly loaded 17 μl of sample buffer from the branch to fill the sample channel. Once the channels were filled, we added 60 μl of reservoir LE and reservoir TE buffers to the branch reservoir and TE reservoir respectively to block any flow in the channel. Immediately, 60 μl of reservoir LE buffer was loaded in the LE reservoir, which also filled the small section of the channel between the LE and collection reservoirs with the reservoir LE solution. Finally, we loaded 10 μl of collection buffer in the collection reservoir.
After loading, we placed platinum electrodes in the TE and LE reservoirs, and applied 300 μA in the main channel and 30 μA in the branch channel with a high voltage source meter (Keithley 2410, Tektronix, Beaverton, OR). For the current applied in the main channel, any lower current can be used at a cost of increased assay time. We decided to use 300 μA at which we observed no temperature increase due to Joule heating during 10 minutes of our current protocol. We visualized fluorescence from the dyes (AF488 and DyLight 488) with a blue light transilluminator (DR22A, Clare Chemical Research, Dolores, CO). A video of the visualization process was captured by a cell phone camera (iPhone 6S, Apple, Cupertino, CA). The first ITP peak was visualized by AF488 and the second peak was visualized by DyLight488.
Single nucleotide exclusion test
We used a sample containing rATP and 26 nt synthetic RNA to quantify the percent exclusion of single nucleotides by our method. rATP was selected as a control spike-in single nucleotide. Electrophoretic mobilities of mono-, di, and tri-nucleotides are found to be in the range of 6 – 24 cm/(2hr*20V/cm), and the mobility of rATP is within this range (8 cm/(2hr*20V/cm)).34 We performed three replicates of ITP size-selection experiments. After collecting Fraction 1 and Fraction 2, we quantified the RNA concentration in both fractions as well as in the initial sample using A260 absorbance (Nanodrop ND-1000) and fluorescence (Qubit 2.0 Fluorometer) measurements. Because of the mutually exclusive dynamic range of the Qubit (0.02 – 0.5 ng/μl) and Nanodrop (0.5 – 3000 ng/μl), the concentration of 26 nt RNA was only measured by Qubit and that of rATP was only detected by Nanodrop. In this test, we used the same 17 μl volume for the sample input and the collection. After each collection, we validated the output volume was indeed 17 μl by directly measuring the volume using a pipettor.
RNA yield comparison between size-selection using ITP and gel electrophoresis
RNA extracted from LCLs was split equally into six aliquots for three replicates of size-selection each by ITP and by gel electrophoresis. For ITP experiments, we discarded Fraction 1 and collected Fraction 2 after following the ITP protocol described above. Immediately after the collection, we stored ITP-extracted RNA in a −20°C freezer until bioanalyzer analysis. For gel electrophoresis, a 15% (w/v) TBE-Urea gel (Novex, Invitrogen) was used. Custom RNA oligos (26 and 34 nt) were used to demarcate the region for gel extraction. Gel bands were excised on a DarkReader Transilluminator (Clare Chemical Research) and placed into GeBAflex electroelution tubes with 8 kDa molecular weight cutoff (Gerard Biotech). Electroelution was carried out at 140 V for at least 40 minutes. The recovered eluate was filtered through Spin-X 0.22 μm Cellulose Acetate centrifuge filter tubes (Sigma-Aldrich). Lastly, RNA was precipitated overnight at −20°C with isopropanol in the presence of 300 mM sodium acetate (pH 5.5), 10 mM MgCl2, and 22.5 μg of GlycoBlue (Life Technologies). We then immediately placed the extracted RNA in a −20°C freezer. For both methods, we extracted RNA in 10 μl collection buffer. We quantified the RNA concentration from the extracts using Agilent 2100 Bioanalyzer System small RNA Analysis Kit within a week from extraction.
Library preparation and high-throughput sequencing
Both ITP-extracted and gel electrophoresis-prepared samples were treated identically for sequencing library preparation. Specifically, samples were first treated with T4 PNK for dephosphorylation using 1x T4 PNK buffer and 10U of T4 PNK (NEB) in a final volume of 50 μl. Samples were incubated for 1 h at 37°C. RNA was precipitated after dephosphorylation as described above with the exception of the use of 75% ethanol instead of isopropanol. Illumina sequencing libraries were prepared using the Clontech SMARTer smRNA-Seq kit according to the manufacturer’s instructions. Consequently, the sequenced RNA fragments were tailed by a stretch of adenines at the 3’ end. We used an Illumina HiSeq 2500 sequencer with a single-end 50 nt read length to sequence the prepared libraries. We sequenced all six libraries as a pool to avoid batch effects due to run-to-run sequencing variability.
Alignment and processing of sequence data
We used cutadapt version 1.8.129 to remove the trailing adenines as well as the first three nucleotides that were added during library preparation. The specific parameters were “-u 3 --overlap=4 --minimum-length=21 --quality-cutoff=33”. This excluded reads shorter than 21 nt from our analysis. We filtered out reads mapping to human rRNA and tRNA sequences obtained from UCSC Genome Browser (hg19) repeatmasker track using Bowtie 2 version 2.2.9 with the following parameter “-L 18”. We used the same aligner with the “-L 18 and -norc” options to align the remaining reads to APPRIS principal transcripts (release 12)30 from the GENCODE mRNA annotation v.15.31 We retained alignments with a mapping quality score greater than two and counted the number of matches to each APPRIS transcript using custom scripts.
Statistical analyses
We calculated the number of sequencing reads that mapped to the 5’UTR, 3’UTR and the coding region of each APPRIS transcript. We then used two approaches to assess reproducibility of the sequencing data. The first approach measures the Spearman correlation coefficient of read counts per APPRIS transcript between all pairs of sequencing libraries. The second approach explicitly compares the standard deviation in log-transformed expression values across replicates per APPRIS transcript. Specifically, we inspected the relationship between standard deviation and mean expression. We then fitted a cubic spline to the data and compared the best-fit lines.
Results and Discussion
Single nucleotide exclusion in ITP-extracted RNA
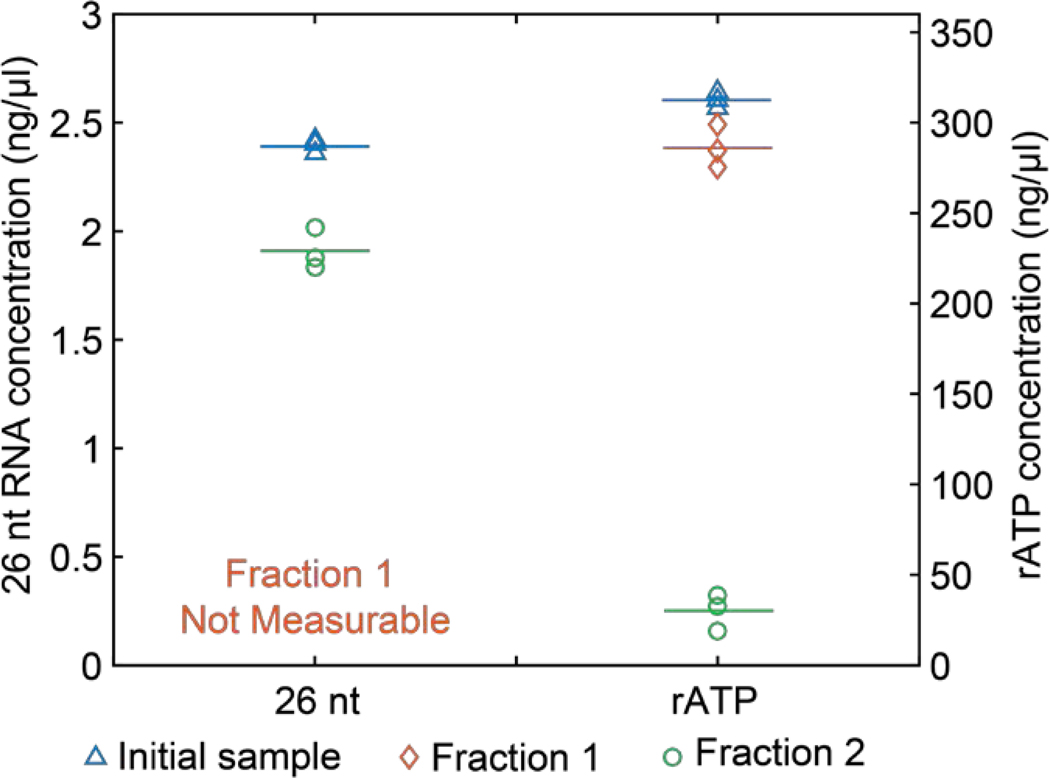
We first tested the fractionation of single nucleotides with the ITP method using a mixture of rATP and 26 nt synthetic RNA. We conducted three replicates where we collected Fraction 1 and Fraction 2 from each experiment. Fraction 1 is supposed to only contain single nucleotides and Fraction 2 is expected to contain the 26 nt synthetic RNA. To verify the fractionation, we measured concentrations of 26 nt RNA and rATP in each fraction. In Figure 2, we present RNA quantification data from Fraction 1 (circles), Fraction 2 (diamonds), and initial sample (triangles) and the average values from each sample with horizontal lines. For rATP (single nucleotides), only 9.6% of the initial concentration was found in the extracted sample (Fraction 2), showing our method effectively excludes the unwanted single nucleotides. On the other hand, 91.6% of rATP was detected from Fraction 1 indicating that a negligible amount of single nucleotides remained in the channel as residues. In sum, we demonstrated efficient removal of unwanted single nucleotides from the sample by focusing them in the first ITP peak and discarding Fraction 1.
Figure 2.
Quantification of 26 nt and rATP in the initial sample and Fraction 1 and 2 collected after ITP size-selection experiments. Each symbol corresponds to an averaged concentration from three technical replicates of nanodrop measurements (for rATP) or three Qubit measurements (for 26 nt). Triangles denote RNA concentration in the initial sample. Diamonds and circles respectively denote measurements from Fraction 1 and 2. Horizontal lines represent the average of three replicates each of which is shown with symbols. Concentration of 26 nt RNA was below the measurable range of Qubit in Fraction 1, and is indicated as ‘not measurable’ in the figure.
We next estimated the 26 nt RNA extraction efficiency. We found that the 26 nt RNA concentration in Fraction 2 compared to that of the initial sample was 79.7%. Our results are consistent with previously reported recovery efficiencies of ~80% for DNA input concentrations ranging from 0.25 to 250 ng using ITP.32 High-efficiency of ITP extraction compares favorably to gel extraction which suffers from low yields especially with low input samples.
Size distribution of extracted RNAs
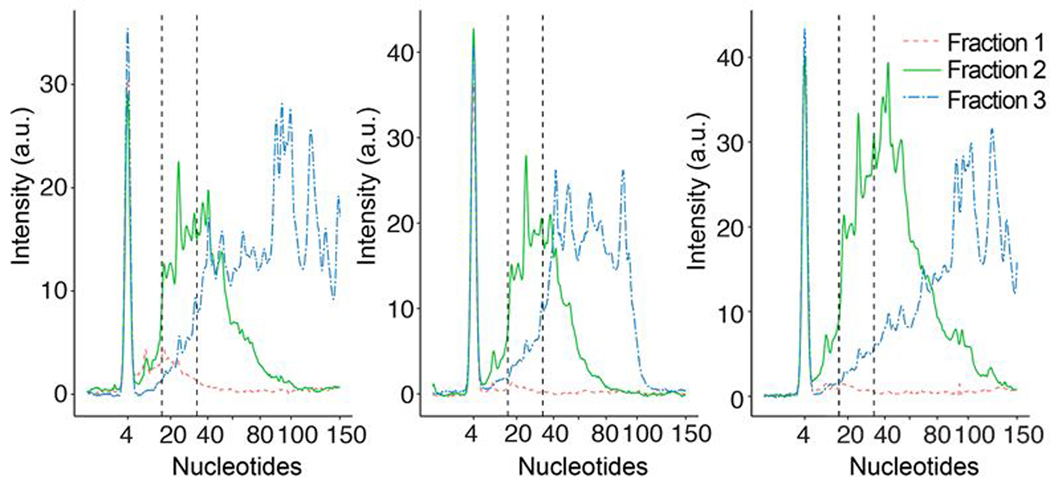
We performed three replicates of ITP extraction to estimate the size distribution of the ITP-extracted RNA. For these experiments, we used RNA from lymphoblastoid cells treated with RNase A and RNase T1 (see Methods). This RNA source is a complex mixture and is enriched for the 17 – 35 nt size range of RNAs.28 We collected three fractions (Fraction 1, 2, and Fraction 3 as defined above) after each ITP size-selection experiment. Each fraction was quantified with Agilent 2100 Bioanalyzer small RNA kit to determine its size distribution. In Figure 3, we show electropherograms of three fractions from each ITP experiment. Several characteristic patterns are shared between the replicates. In Fraction 1, low amounts of small RNAs within the Bioanalyzer’s measurement range of 4 – 150 nt were found, which confirms that our method enriches only single nucleotides in Fraction 1. In addition, RNAs with size between 100 and 150 nt comprised at most 4% of the total signal in Fraction 2. Size distribution of the initial lymphoblastoid RNA sample extracted by miRNeasy kit is provided in Figure S4 as a bioanalyzer electropherogram.
Figure 3.
Bioanalyzer electropherograms from three replicates of ITP size-selection experiments. Dotted, solid, and dash-dotted lines respectively denote the results from Fraction 1, 2, and 3 as defined previously. The peaks at 4 nt from all fractions denote the signal from a marker RNA used in the bioanalyzer kit.
We observed replicate to replicate variability in the size selection. For example, the Fraction 3 of the center panel included negligible amount of RNA >100 nt while that of other replicates detected RNA >100 nt. We hypothesize that in the experiment associated with the center panel, RNAs electro-migrated slower than other ITP experiments. This hypothesis is supported by the narrower RNA size range observed in Fraction 2 of the center panel compared to others. The slower RNA migration can happen if LE containing the sieving matrix smears into the sample channel during the loading step. For such cases, 70 s for Fraction 3 recovery is insufficient for RNAs larger than 100 nt to arrive the collection reservoir. In addition, indirect monitoring of the RNA locations using fluorescence dyes may contribute to run-to-run variability. The repeatability of our method may be improved with the use of labeled RNA markers with specific sizes.
We further analyzed the data by quantifying the fluorescence signal from two size groups: 17–35 nt and 36 – 150 nt. We chose the size range of 17 – 35 nt since it coincides with the size range that is relevant for many sequencing analyses including ribosome profiling and RNA-binding protein footprinting techniques. RNAs longer than 35 nt represent undesired longer fragments that we aim to remove. For each size range, we calculated the mean percentage of RNAs contained in each fraction such that the sum of the values from all three fractions constitutes 100%. As shown in Figure S3, 75.3% of the total signal in the size range of 17 – 35 nt was from the ITP-extracted sample (Fraction 2). The observed percentage is consistent with the recovery efficiency estimated using the synthetic 26 nt RNA as described above. In the size range of 36 – 150 nt, we found 68.5% of the signal was from Fraction 3 and the rest of the signal was predominantly from Fraction 2. We attribute the presence of longer RNAs in Fraction 2 to a varying range of mobilities due to both RNA secondary structures33 and long RNAs outpacing the ITP interface due to their starting location far ahead (near branch channel) of the initial ITP interface (TE reservoir). This observation suggests that the percentage of long RNAs in the collected sample may be reduced further by having a longer separation channel compared to sample channel, at the cost of longer assay time and potential joule heating problems.
In addition, we carried out experiments which demonstrate the efficacy of our methods in size selection of RNA from cell lysate (data shown in ESI). We applied ITP extraction to micrococcal nuclease (MNase)-digested Chronic myelogenous leukemia cell lysate (K562). Since we used an endonuclease, the digested lysates included significant amounts of mono- and oligo-nucleotides that were initially part of the mRNAs that were not protected by ribosomes or RNA binding proteins. The sample preparation protocol and the experimental data are presented in Section S1 of ESI. Figure S5 includes the bioanalyzer electropherograms of Fraction 2 collected from three replicates of ITP size selection experiment using the digested cell lysate. This data provides evidence that our method is directly applicable to complex RNase-digested cell lysates for simultaneous purification and size selection.
Yield and size-selection comparison between ITP and gel electrophoresis methods
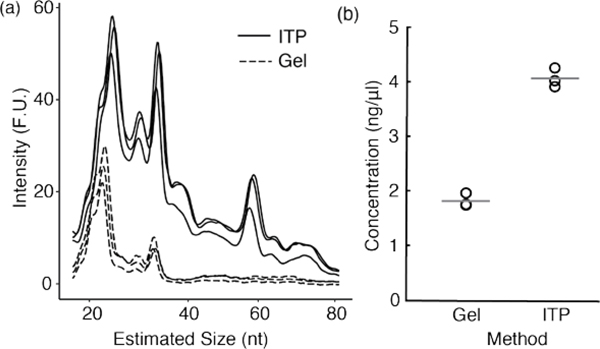
In Figure 4, we show size distribution and yield of RNAs purified by ITP or by gel electrophoresis methods. We quantified the size-selected RNA from three replicates of each method with the Agilent 2100 Bioanalyzer. Each ITP experiment took 10 min to simultaneously perform both size-selection and extraction whereas gel electrophoresis method required a series of separation, elution, and precipitation experiments, which took a minimum of 6 h in total.
Figure 4.
Yield comparison and size distribution for gel electrophoresis and ITP-based methods. (a) RNA size distribution was determined using Agilent Bioanalyzer 2100. The solid lines represent three replicates from ITP size selection method and the dotted lines denote the three replicates from gel electrophoresis method. (b) Concentration of extracted RNA in the desired size range of 17 – 35 nt was quantified. Gray lines indicate the mean values of the three replicates from each method.
ITP size selection excluded the majority of RNAs >35nt as they remained in the channel as discussed in Figure 3. Yet, a broader size range of RNA was observed in the ITP sample (Fraction 2) compared to the gel electrophoresis method (Figure 4a). To quantify the yield of RNA extraction, we calculated the RNA concentration in the size range of 17 – 35 nt using bioanalyzer software (2100 Expert). Compared to gel electrophoresis method, ITP method yielded 2.2 fold higher amount of RNA in the desired size range (Figure 4b). The loss in the gel electrophoresis method is mainly attributed to the gel extraction step which is highly dependent on RNA size and sample amount.
High-throughput sequencing of RNAs from ITP and gel electrophoresis methods
Given the advantages of sequencing over methods such as qPCR and microarrays, we sought to establish the compatibility of our ITP size-selection method with downstream high-throughput sequencing. Hence, we compared high-throughput sequencing data generated from RNAs extracted by gel electrophoresis and those prepared by ITP size-selection method. Specifically, we prepared three replicates of sequencing libraries for each method simultaneously to minimize any differences due to processing steps. Furthermore, In total, we obtained 276M reads ranging between 27.2M-70.5M per library.
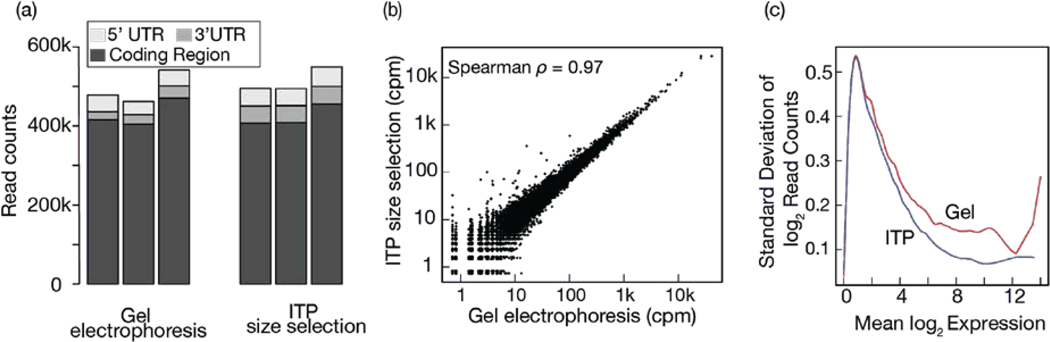
For these experiments, we used RNA extracts from cells that were treated with ribonucleases (Methods). This sample preparation enriches for RNA fragments in the 17–35 nt range as ribosomes protect these mRNA fragments from nuclease digestion. Consequently, we expected a large fraction of the reads to map to the coding regions of transcripts as opposed to 5’ or 3’ untranslated regions. As expected, more than 82% of the transcriptome mapping reads mapped to the coding regions (Figure 5a). We note that the mappability of reads to transcriptome was low due to significant portion of reads mapped to rRNA in both methods (range 89.8 – 92.9%). This is consistent with previous studies that do not employ rRNA depletion where approximately 80–95% of all reads are typically assigned to rRNA fragments.25,40 Although rRNA depletion method can be easily incorporated, we intentionally decided to not include this step in order to reflect the entire pool of captured RNAs and to avoid potential sequence-specific biases due to additional selection.41
Figure 5.
Comparison between high-throughput sequencing results of RNA fragments recovered by ITP and gel electrophoresis methods. (a) Reads mapping to the coding region, 3’ and 5’ UTR were counted and plotted for both methods (three replicates each). (b) The mean number of reads per transcript across the three replicates of each method was calculated and compared. See Figure S4 for replicate-level comparisons. (c) For each transcript, the mean and standard deviation of log2 read count were calculated across the replicates. A cubic spline was fitted and plotted for each method. See Figure S11 for the individual data points showing each transcript.
We next compared transcript-level quantification obtained by the two methods using the reads mapping to the coding regions. Specifically, we selected the subset of transcripts with read counts per million (cpm) greater than one in at least two of the six libraries. The mean number of reads per transcript correlated strongly (Spearman rank correlation: 0.97) between libraries prepared from ITP and gel electrophoresis methods (Figure 5b). When we analyzed the pairwise rank correlations between replicates of each method, we similarly observed very high Spearman rank correlations ranging from 0.93 to 0.95 (Figures S4). We include systematic identification of transcripts with the high deviations between the quantifications from the two methods in Section S2 of ESI. In short, we used an MA-plot (Figure S9) to identify 230 transcripts with the largest deviations between two methods. Functional enrichment analysis of these “outliers” suggested higher read counts for transcripts associated with nucleosome (Table S1). In a plot of M-values as a function of transcript length (Figure S10), we observed very week relationship (Spearman correlation ρ= 0.12)
While correlation coefficients are ubiquitously adopted in the literature for assessing reproducibility, they are susceptible to dynamic-range-dependent biases. Hence, we evaluated reproducibility by analyzing the mean to variance relationship of the quantifications for each transcript. We fitted splines to standard deviation of log2 read counts as a function of their mean. In Figure 5c, we plotted these best-fit lines for the gel electrophoresis and ITP method, and individual data points for all transcripts are presented in Figure S5. A method with higher reproducibility should yield lower standard deviation of read counts across the range of mean of log2 read counts. We observed that results from ITP method had higher reproducibility across the three replicates for the entire range of mean transcript expression (Figure 5c). We attribute the higher reproducibility of ITP compared to gel extraction to the higher yield of ITP. With higher yield, we sample more molecules from RNA prepared by ITP extraction, and thus the uncertainty of the mean is expected to be smaller for the same underlying distribution.
Detailed analyses on the sequencing read lengths can be found in ESI Section S3. In Figure S6, we provide the size distribution of all sequencing reads after the removal of the adapter sequence. Both methods exhibit specific peaks predominantly at 23nt and 35–36 nt. These corresponded to rRNA contaminants, which were bioinformatically filtered in ribosome profiling data. We also observed that ITP samples had a higher proportion of reads of length <20 nt, which are removed from analysis as described in Materials and Methods. In Figure S7, we present size distribution of only the reads that aligned to the transcriptome for the two methods. We found that sequencing libraries from both methods contained only a small number of reads longer than 35 nt (<3% for ITP; <2% for gel electrophoresis) as expected.
Conclusion
Polyacrylamide gel electrophoresis-based size-selection remains the most highly used method for preparation of small RNAs for deep sequencing. However, the method is time-consuming and leads to significant loss of input material. There is hence an urgent need for alternative methods.26 Here, we have reported the first on-chip method utilizing ITP for improved yield in specific size-selection of RNAs and demonstrated the compatibility of our method with high-throughput sequencing. Our method is designed to select RNAs in the size range of 2 – 35 nt, and our experiments demonstrate >90% exclusion of single nucleotides and >65% exclusion of RNAs larger than 35 nt. We predict that the cutoff lengths can be further optimized with different sieving matrices and/or different ITP chemistry. In our comparison experiments, gel electrophoresis provided better size separation resolution while ITP method yielded 2.2-fold higher RNA amount. We expect the difference in RNA yield to be even greater for lower input samples given that ITP recovery efficiency is effectively independent of sample amount. Lastly, we demonstrated compatibility of our method with high throughput sequencing revealing on-chip ITP method to be a more reproducible and efficient alternative to gel electrophoresis. Our method offers an attractive alternative for small RNA size selection with higher extraction efficiency, reproducibility, and reduced time requirements (assay time of <10 min). We envision potential applications of our method in next generation sequencing of size-selected RNAs, particularly to low-abundance samples such as rare cell types, samples from FACS, and precious clinical samples.
Supplementary Material
Acknowledgements
This work used the Genome Sequencing Service Center by Stanford Center for Genomics and Personalized Medicine Sequencing Center, supported by NIH S10OD020141 instrumentation grant. This work was supported in part by NIH grant CA204522 (CC). CC is a CPRIT Scholar in Cancer Research supported by CPRIT Grant RR180042. CMH acknowledges support from the National Institute of Standards and Technology (NIST) NRC Postdoctoral Associateship Program. CMH, SAM, further acknowledge support from the NIST Joint Initiative for Metrology in Biology at Stanford.
Certain commercial equipment, instruments or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology (NIST), nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
References
- 1.Khurana TK and Santiago JG, Anal. Chem, 2008, 80, 6300–6307. [DOI] [PubMed] [Google Scholar]
- 2.Rogacs A, Marshall LA and Santiago JG, J. Chromatogr. A, 2014, 1335, 105–120. [DOI] [PubMed] [Google Scholar]
- 3.Bercovici M, Kaigala GV, Mach KE, Han CM, Liao JC and Santiago JG, Anal. Chem, 2011, 83, 4110–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Persat A, Marshall LA and Santiago JG, Anal. Chem, 2009, 81, 9507–9511. [DOI] [PubMed] [Google Scholar]
- 5.Schoch RB, Ronaghi M and Santiago JG, Lab Chip, 2009, 9, 2145–2152. [DOI] [PubMed] [Google Scholar]
- 6.Persat A and Santiago JG, Anal. Chem, 2011, 83, 2310–2316. [DOI] [PubMed] [Google Scholar]
- 7.Persat A, Chivukula RR, Mendell JT and Santiago JG, Anal. Chem, 2010, 82, 9631–9635. [DOI] [PubMed] [Google Scholar]
- 8.Eid C, Garcia-Schwarz G and Santiago JG, Analyst, 2013, 138, 3117–3120. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Schwarz G and Santiago JG, Anal. Chem, 2012, 84, 6366–6369. [DOI] [PubMed] [Google Scholar]
- 10.Stellwagen NC and Stellwagen E, J. Chromatogr. A, 2009, 1216, 1917–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everaerts FM, Beckers JL and E TP, Isotachophoresis: Theory, Instrumentation and Applications, Elsevier, 2011. [Google Scholar]
- 12.Inano K, Tezuka S, Miida T and Okada M, Ann. Clin. Biochem, 2000, 37 (Pt 5), 708–716. [DOI] [PubMed] [Google Scholar]
- 13.Kuriyama K, Shintaku H and Santiago JG, Electrophoresis, 2015, 36, 1658–1662. [DOI] [PubMed] [Google Scholar]
- 14.Marshall LA, Han CM and Santiago JG, Anal. Chem, 2011, 83, 9715–9718. [DOI] [PubMed] [Google Scholar]
- 15.Han CM, Katilius E and Santiago JG, Lab Chip, 2014, 14, 2958–2967. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian Parimalam S, Oguchi Y, Abdelmoez MN, Tsuchida A, Ozaki Y, Yokokawa R, Kotera H and Shintaku H, Anal. Chem, 2018, 90, 12512–12518. [DOI] [PubMed] [Google Scholar]
- 17.Khnouf R, Shore S, Han CM, Henderson JM, Munro SA, McCaffrey AP, Shintaku H and Santiago JG, Anal. Chem, 2018, 90, 12609–12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X and Mortazavi A, Genome Biol, 2016, 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter JA, Spacek DV and Snyder MP, Mol. Cell, 2015, 58, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, Stanton R, Rigo F, Guttman M and Yeo GW, Nat. Methods, , DOI: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ and Yu G, J. Biol. Chem, 2011, 286, 1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY and Segal E, Nature, 2010, 467, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingolia NT, Ghaemmaghami S, Newman JRS and Weissman JS, Science, 2009, 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingolia NT, Brar GA, Rouskin S, McGeachy AM and Weissman JS, Nat. Protoc, 2012, 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cenik C, Cenik ES, Byeon GW, Grubert F, Candille SI, Spacek D, Alsallakh B, Tilgner H, Araya CL, Tang H, Ricci E and Snyder MP, Genome Res, 2015, 25, 1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazzini AA, Lee MT and Giraldez AJ, Science, 2012, 336, 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGlincy NJ and Ingolia NT, Methods, , DOI: 10.1016/j.ymeth.2017.05.028. [DOI] [Google Scholar]
- 28.Chou H-J, Donnard E, Gustafsson HT, Garber M and Rando OJ, Mol. Cell, , DOI: 10.1016/j.molcel.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cameron V and Uhlenbeck OC, Biochemistry, 1977, 16, 5120–5126. [DOI] [PubMed] [Google Scholar]
- 30.Beis I and Newsholme EA, Biochem. J, 1975, 152, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dard-Dascot C, Naquin D, d’Aubenton-Carafa Y, Alix K, Thermes C and van Dijk E, BMC Genomics, 2018, 19, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fishman A, Light D and Lamm AT, Genome Biol, 2018, 19, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vadnere M, Amidon G, Lindenbaum S and Haslam JL, Int. J. Pharm, 1984, 22, 207–218. [Google Scholar]
- 34.Markham R and Smith JD, Biochem. J, 1952, 52, 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin M, EMBnet.journal, 2011, 17, 10–12. [Google Scholar]
- 36.Rodriguez JM, Maietta P, Ezkurdia I, Pietrelli A, Wesselink J-J, Lopez G, Valencia A and Tress ML, Nucleic Acids Res, 2013, 41, D110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R and Hubbard TJ, Genome Res, 2012, 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall LA, Rogacs A, Meinhart CD and Santiago JG, J. Chromatogr. A, 2014, 1331, 139–142. [DOI] [PubMed] [Google Scholar]
- 39.Lilley DM, Bhattacharyya A and McAteer S, Biotechnol. Genet. Eng. Rev, 1992, 10, 379–401. [DOI] [PubMed] [Google Scholar]
- 40.Gerashchenko MV and Gladyshev VN, Nucleic Acids Res, DOI: 10.1093/nar/gkw822. [DOI] [Google Scholar]
- 41.Lahens NF, Kavakli IH, Zhang R, Hayer K, Black MB, Dueck H, Pizarro A, Kim J, Irizarry R, Thomas RS, Grant GR and Hogenesch JB, Genome Biol, 2014, 15, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.