Abstract
Chromatin-associated architectural proteins are part of a fundamental support system for cellular DNA-dependent processes and can maintain/modulate the efficiency of DNA replication, transcription, and DNA repair. Interestingly, prognostic outcomes of many cancer types have been linked with the expression levels of several of these architectural proteins. The High Mobility Group Box (HMGB) architectural protein family has been well studied in this regard. The differential expression levels of HMGB proteins and/or mRNAs and their implications in cancer etiology and prognosis present the potential of novel targets that can be explored to increase the efficacy of existing cancer therapies. HMGB1, the most studied member of the HMGB protein family, has pleiotropic roles in cells including an association with nucleotide excision repair, base excision repair, mismatch repair, and DNA double-strand break repair. Moreover, the HMGB proteins have been identified in regulating DNA damage responses and cell survival following treatment with DNA-damaging agents, and as such, may play roles in modulating the efficacy of chemotherapeutic drugs by modulating DNA repair pathways. Here, we discuss the functions of HMGB proteins in DNA damage processing, and their potential roles in cancer etiology, prognosis and therapeutics.
Keywords: High mobility group box (HMGB) proteins, drug resistance, cisplatin, DNA damage repair, cancer prognosis
Introduction
The HMGB proteins are chromatin-associated DNA binding proteins that can bend DNA (1,2). Such a function is fundamental for efficient genome organization and execution of DNA replication, DNA repair, and transcription (3). Cancer cells often have higher levels of DNA replication and DNA repair (compared to normal cells) to support their growth and survival; thus, these ubiquitously expressed proteins may play important roles in cancer etiology at a very basic level. Because of their ubiquitous presence and lack of enzymatic function, they have not gained much attention as therapeutic targets in cancer therapy. However, more recent studies have suggested an association between expression levels of the HMGB proteins in various cancers. In this review, we discuss potential roles of the HMGB proteins in cancer etiology, and in the response to and repair of chemotherapeutic DNA-damaging agents.
The chromatin associated HMGB proteins, HMGB1, HMGB2, HMGB3, and HMGB4, were identified more than 40 years ago (4) [reviewed in (5)]. The HMGB proteins contain two conserved box domains; Box A, which facilitates DNA binding, and Box B, which assists in binding and bending DNA. In addition, HMGB1, HMGB2, and HMGB3 contain a C-terminal acidic tail that is lacking in HMGB4 (6). The C-terminal acidic tail has been shown to fold in between the two basic box domains and can regulate DNA binding and bending (7–9). Because HMGB4 does not contain a C-terminal acidic tail, its mode of interaction with DNA may differ from the other members of the family, and thus here we will focus on HMGB1–3.
HMGB proteins are largely localized in the nucleus with some cytoplasmic and extracellular distribution (10), which is in part regulated by posttranslational modifications (11). HMGB1 and HMGB2 have been shown to bind structurally distorted DNA in a sequence-independent manner and can topologically modify the DNA by introducing supercoiling and stabilizing DNA loops (12,13). Through DNA bending, HMGB1 has been shown to facilitate recruitment of other proteins to their DNA targets (14–16). Interestingly, some chemotherapeutic drug-induced DNA distortions/lesions are also high-affinity targets of HMGB1 (17,18), and perhaps of HMGB2 and HMGB3. Of relevance to cancer therapy, HMGB binding to structurally distorted DNA can modulate the processing of chemotherapeutic drug-induced DNA damage in cancer cells (19–25). Of note, the functions of HMGB1 have been the most studied among the HMGB family members and it is evident that HMGB1 has pleiotropic roles in cells, which will be discussed in a later section. In addition, several of the HMGB proteins have been found to play roles in various aspects of inflammation and immunity (26), though here we will focus on DNA damage responses and DNA repair in cancer etiology, prognosis, and therapeutic outcome.
Cancer etiology and prognosis correlate with HMGB mRNA and protein levels
With the accessibility of cancer genome databases, many gene products are emerging as potential cancer-associated genes/proteins. For example, an inverse relationship has been found between HMGB expression levels and overall survival of cancer patients. Correlations of the HMGB proteins with cancer etiology and prognosis have been supported by studies that measured the HMGB expression levels via immunohistochemistry and RT-qPCR from patient samples (27–29). In a meta-analysis of 18 studies with 11 different tumor types, including gastric cancer, colorectal cancer, hepatocellular carcinoma, pancreatic cancer, nasopharyngeal carcinoma, head and neck squamous-cell carcinoma (HNSCC), esophageal cancer, malignant pleural mesothelioma, bladder cancer, prostate cancer, and cervical carcinoma, higher HMGB1 protein levels indicated poor overall survival and poor progression-free survival (27). A similar correlation was demonstrated in ovarian cancer (28) and in these cancers, HMGB1 was found to upregulate the ERK1/2, MAPK, NF-kβ and Akt signaling pathways via its binding to the receptor for advanced glycation end products (RAGE), contributing to the reprogramming of cancer cells. HMGB1 upregulation in squamous cell carcinoma has been shown to promote chromatin modification and increase the phosphorylation of CHK1 to activate DNA damage responses (DDR), contributing to radio-resistance (29). Although targeting of HMGB1 for cancer therapy is a matter of debate due to its pleiotropic roles in cells (30), it has shown promising results in prostate cancer cells (31) and in HNSCC (32). Similar to HMGB1, HMGB2 expression levels appear to be positively correlated with HNSCC (33), breast cancer (34,35), colorectal cancer (36), gastric cancer (37), and ovarian cancer (38), largely by modulating the glycolytic pathway. In colorectal cancer cells, HMGB2 was found to modulate DNA damage repair in a TP53 dependent manner (36). HMGB3 follows a similar trend of poor prognosis with high expression levels in lung cancer (39), NSCLC (39,40), colorectal cancer (41), urinary bladder cancer (42), gastric adenocarcinoma (43), breast cancer (44), and in esophageal squamous cell carcinoma (ESCC) (45). We have shown that siRNA targeting of HMGB3 sensitizes chemoresistant human ovarian cancer cells to low doses of cisplatin by attenuating the ATR/CHK1 DNA damage checkpoint signaling pathway (46). Although it is not fully understood why the overexpression of HMGB proteins correlates with poor prognosis, based on their known roles in facilitating DNA repair, one could speculate that their overexpression may contribute to more efficient removal of chemotherapeutic DNA adducts. These observations are summarized in Table 1.
Table 1.
Association of HMGB proteins with cancer outcome.
| Protein | Cancer outcome | DNA damage processing modulation | Ref. |
|---|---|---|---|
| HMGB1 | • Gastric cancer • Hepatocellular cancer • HNSCC • Nasopharyngeal cancer • Colorectal cancer • ESCC • Mesothelioma • Bladder cancer • Prostate cancer • Cervical cancer • Ovarian cancer • NSCLC |
• Promotes error-free repair via NER • Binds with high affinity to chemotherapeutic DNA lesions • Positive cooperative binding with NER damage recognition proteins on DNA lesions • Assists in recruitment of XPA to damaged DNA • Introduces negative supercoiling preferentially to damaged DNA • HMGB1 depletion increases sensitivity to DNA crosslinking agents • HMGB1 depletion results in increased DSBs |
27–31 54–64 84–105 |
| HMGB2 | • HNSCC • Breast cancer • Colorectal cancer • Ovarian cancer • Gastric cancer |
• Increases γ-H2AX foci formation after damage, and delays repair | 33–38 |
| HMGB3 | • Ovarian cancer • NSCLC • Colorectal cancer • Gastric cancer • ESCC • Urinary bladder cancer |
• HMGB3 depletion downregulates the ATR/CHK1 signaling pathway | 39 – 46 |
HNSCC = Head and neck squamous cell carcinoma, NSCLC = Non-small cell lung cancer, ESCC = Esophageal squamous cell carcinoma
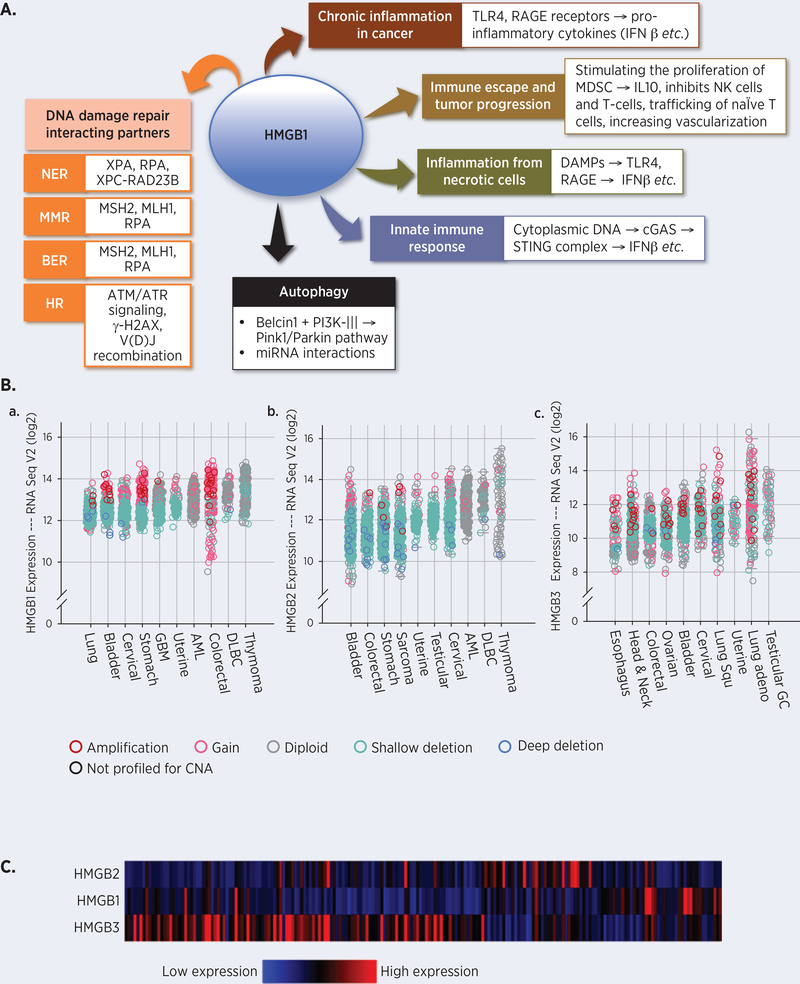
An analysis of HMGB mRNA expression levels and copy number alterations using the cBioPortal (47,48) in patient cancer samples from 32 TCGA PanCancer Atlas studies indicated a 4- to 16-fold increase in their mRNA levels when compared to the surrounding non-cancerous tissues (49). We plotted the top 10 HMGB1–3 expressing cancers based on their median mRNA expression levels (Figure 1A) (50–52), which revealed that not all HMGB proteins are equally overexpressed in each cancer type. In addition, the expression of different HMGB mRNAs varied among the individual patient samples within the same cancer type. For example, mRNA expression analysis from 398 ovarian serous cystadenocarcinoma patient samples indicated a selective, non-overlapping expression pattern of HMGB1, HMGB2, and HMGB3 (Figure 1B) (52).
Figure 1.
A. Pleiotropic roles of HMGB1 in cells. HMGB1 has been found to be involved in DNA repair, chronic inflammation, immune responses, autophagy, DNA damage responses etc. Please see text for detailed description. B. Expression levels of HMGB mRNAs are higher in cancerous tissues when compared to the normal surrounding tissues as measured by Illumina RNA Seq V2 of cancer patient samples from 158 studies. The fold increase in expression levels as a function of mRNA in cancer samples are plotted in an increasing order of median values of the expression level. Samples with mutations are also identified in the same plot. The copy number and expression levels are not always linearly correlated. A gain in copy number can also be accompanied by missense mutations or small truncating mutations, resulting in lower expression levels in those particular samples. a, HMGB1 expression levels and copy number alterations data from 42,049 samples. The HMGB1 gene was mutated in 263 patients. b, HMGB2 expression levels and copy number alterations from 42,049 samples. The HMGB2 gene was mutated in 285 of the queried patients. c, HMGB3 expression levels and copy number alterations from the 42,049 samples. The HMGB3 gene is altered in 351 of the queried patients. The data was procured, analyzed, and the graphs were prepared using cBioPortal. The copy number level per gene was derived from copy number analysis algorithms. Amplification = many additional gene copies, local; Gain = a few additional copies, broad; Diploid = two complete sets of chromosomes; Shallow deletion = possible heterozygous deletion; Deep deletion = possible homozygous deletion. CNA = Copy Number Alteration; GBM = Glioblastoma; AML = Acute Myeloid Leukemia; DLBC = Diffuse large B cell lymphoma; Lung Squ = Lung squamous cell carcinoma; Testicular GC = Testicular Germ Cell. C. HMGB mRNAs are expressed differentially in different ovarian serous cystadenocarcinoma patients. Heat maps representing the mRNA levels from 398 patient samples were generated from the TCGA Next-Generation Clustered Heat Map (NG-CHM) Compendium (https://bioinformatics.mdanderson.org/TCGA/NGCHMPortal/).
HMGB proteins are involved in cellular responses to chemotherapeutic DNA-damaging agents
Cytotoxic therapy has been a mainstay of cancer treatment and these regimens often include DNA interstrand crosslink (ICL)-inducing platinum drugs (e.g. carboplatin, oxaliplatin, and cisplatin), DNA double-strand break (DSB)-inducing drugs (e.g. doxorubicin, mitoxantrone, etoposide, and PARP inhibitors), radiation therapy, alkylating agents (e.g. cyclophosphamide), and antimetabolites (e.g. gemcitabine, clofarabine, cytarabine) that induce DNA damage and repair synthesis and/or inhibit DNA synthesis. Such damage is processed by DNA repair proteins from different repair pathways (53). It has been found that the HMGB proteins play roles in several DNA repair mechanisms. Below we summarize the roles of HMGB1 in processing ICLs by different DNA repair pathways.
HMGB1 as a modulator of ICL processing by nucleotide excision repair (NER) proteins
The NER mechanism processes bulky DNA adducts, including those induced by many chemotherapeutic drugs, such as the ICL-inducing agents mentioned above. In NER, the damage-induced local distortion of the DNA is identified by the XPC-RAD23B protein complex in global-genome NER, and by stalled RNA polymerase II in transcription-coupled NER (54–58). In subsequent steps, a pre-initiation complex is formed by further recruitment of TFIIH, XPA, and RPA (59,60), and the DNA is unwound by the helicase activities of the XPB and XPD subunits of TFIIH. The open complex is stabilized by XPA binding on the single-stranded and double-stranded region of the DNA (61). In the stabilized open-complex, the lesion is removed by dual incision on one strand of the DNA surrounding the lesion. The ERCC1-XPF structure-specific nuclease complex cleaves the DNA on the 5ʹ-side of the lesion, and XPG cleaves the DNA on the 3ʹ-side of the lesion (62), releasing the nucleotides surrounding the lesion.
We found that HMGB1 functions as an NER co-factor, as it facilitated the error-free processing of lesions by the NER mechanism (63). It has been found that HMGB1 binds to 1,2 GG intrastrand cisplatin crosslinks (64) and site-specific psoralen ICLs with higher affinity compared to non-damaged DNA (18). Further, HMGB1 binds to ICLs in a positive cooperative fashion with the NER damage recognition complexes, XPC-RAD23B and XPA-RPA, to form higher order DNA-protein complexes (20), indicating its association with and facilitation of NER protein binding to DNA lesions. The physiological relevance of the binding interactions of HMGB1 with NER proteins on ICLs was studied by using cell viability and mutagenesis assays. In HMGB1 knockout mouse embryonic fibroblasts, induction of psoralen ICLs or UVC-mediated DNA adducts (canonical NER substrates) were more cytotoxic and mutagenic than in wild-type cells (63), in part, due to less efficient NER-mediated lesion processing (63). Similarly, in human osteosarcoma cells, HMGB1 promoted the error-free processing of psoralen ICLs by associating with the lesions and by assisting in the recruitment of XPA to the damaged sites (19). HMGB1 also induced more negative supercoiling preferentially in a damaged DNA substrate compared to a lesion-free DNA substrate (19), which may facilitate NER-mediated lesion removal.
HMGB1 as a modulator of ICL processing by mismatch repair (MMR) proteins
MMR repairs mismatches and small loops in DNA caused primarily by replication errors. In human cells the mismatch is recognized by the MSH2-MSH6 heterodimer and subsequently processed by the MLH1-PMS2 heterodimer, PCNA, 5ʹ→3ʹ EXO1 exonuclease, RPA, DNA polymerase δ, and DNA ligase I [reviewed in (65)]. Helix-distorting ICLs can also be recognized and processed, in part, by MMR proteins in human cells (66), as well as in cell-free extracts from Xenopus egg extracts (67). In Xenopus egg extracts that do not support replication and transcription, ICLs were identified by the MSH2-MSH6 heterodimer and subsequently excised by MLH1-PMS2 and EXO1 (67). In human cells, MSH2 plays a role in processing of psoralen ICLs (66). In addition, we have shown that MSH2-MSH3 can bind to site-directed psoralen ICLs with the NER proteins, XPC-RAD23B and XPA-RPA (68).
The capacity of HMGB1 to modulate the MMR process has been reported. HMGB1 was shown to interact with MSH2 and MLH1 to identify the mismatched bases (69). Additionally, HMGB1 can substitute for RPA in the EXOI-mediated excision step in MMR (70). However, results from studies using HMGB1 knockout mouse embryonic fibroblasts suggested that HMGB1 was dispensable for MMR (71). These studies indicated that HMGB1 may be involved in the MMR-associated removal of ICLs, although further studies are warranted for a conclusive understanding of the role(s) of HMGB1 in MMR-mediated ICL processing.
HMGB1 as a modulator of ICL processing by base excision repair (BER) proteins
BER removes endogenous DNA lesions by initiation of a glycosylase followed by 3ʹ→5ʹ apurinic/apyrimidinic endonuclease 1 (APE1)-mediated cleavage of the DNA backbone, and then subsequent strand displacement and synthesis of DNA by Polβ, and Ligase III, or in long-patch BER by Polβ, Fen1, Pol λ, PCNA, and Ligase I [reviewed in (72)]. In vitro studies have demonstrated that the BER glycosylases, NEIL1 and NEIL3, can excise a psoralen ICL from a three or four-stranded DNA structure (73), and APE1 can incise cisplatin ICL-containing DNA (74). There are also indications of epistatic roles of BER and MMR in the processing of cisplatin-induced DNA lesions (75). However, psoralen ICLs and cisplatin ICLs may be processed differently by BER proteins (74,76).
Interestingly, HMGB1 has been found to be associated with BER via mass spectrometry analyses, which identified associations of HMGB1 with BER intermediates in mouse embryonic fibroblast extracts. In addition, HMGB1 was found to stimulate the activities of APE1 and FEN1 (77,78).
HMGB1 as a modulator of ICL processing by DNA double-strand break (DSB) repair proteins
The repair of ICLs is complex and is thought to involve proteins from several DNA repair pathways, including NER, MMR, BER, translesion synthesis (TLS), homologous recombination (HR), and Fanconi Anemia (FA) proteins [reviewed in (53,79)]. The structure-specific endonucleases, ERCC1-XPF and FAN1, can assist in the resolution of ICLs (80–84). Subsequently, RAD51, RAD52, RAD54, RPA, BRCA1 and FA proteins can generate intermediate structures that can be resolved by the MUS81-EME1 complex (85–88). In addition to these proteins, multiple TLS polymerases can bypass “unhooked” ICLs, often in a mutagenic fashion (89–91). In S-phase, unrepaired DNA damage induced by many chemotherapeutic drugs, including ICLs, can stall replication forks (91–93). Such stalled replication forks can lead to fork collapse generating DSBs resulting in repair processing by homology-directed repair (HDR) (94,95).
How do the HMGB proteins modulate this complex process? Although detailed interactions are not clear, there are several studies that have associated HMGB1 and HMGB2 with DSB processing. In mouse brain tissues, accumulation of DSBs were inversely correlated with the expression of HMGB1, as assessed by immunostaining and immunoblotting (96). Similarly, HMGB1 depletion in MCF7 breast cancer cells resulted in inefficient repair of strand breaks caused by ionizing radiation due to downregulation of the ATM and ATR signaling pathways, and alterations in telomere-binding protein levels (97). HMGB1 depletion in ESCC cells impaired radiation-induced DNA damage repair as indicated by increased γ-H2AX foci and reduced PARP1 expression (98). Colorectal cancer cells were sensitized to radiation when HMGB2 was downregulated, likely due to unresolved DSBs as evidenced by prolonged γ-H2AX foci (36).
The role of HMGB1 and HMGB2 in facilitating V(D)J recombination is well documented. During V(D)J recombination, the RAG1 and RAG2 proteins bind to the conserved recombination signal sequences adjacent to the V, D, and J sequences and bring them together into a synaptic complex, which is then cleaved by the RAG1 protein (99–101). Although HMGB proteins are not essential for V(D)J recombination, they have been shown to enhance the affinity of the RAG complexes for DNA and increased the cleavage up to 100-fold by stabilizing the required DNA bending (26,102–106).
These studies clearly indicated an association of HMGB1 with various DNA damage processing pathways. However, the roles of HMGB2 and HMGB3 in processing DNA ICLs are not yet clearly defined. Because of the structural similarities between the HMGB1, HMGB2, and HMGB3 proteins and their abundance in cancer cells (Figure 1), further studies to determine the extent to which these proteins have functional redundancy or whether they have distinct and unique roles in the response to and the repair of genome-destabilizing chemotherapeutic and carcinogenic lesions may provide a better understanding of DNA lesion processing in cells.
HMGB3 inhibition as a potential neoadjuvant therapy
Recently, we have discovered that siRNA-mediated depletion of HMGB3 can sensitize chemoresistant human ovarian cancer cells to cisplatin treatment (46). This sensitization, among other possibilities, could be due to less efficient cisplatin-DNA adduct removal (Mukherjee & Vasquez, unpublished observation) in the absence of HMGB3. Therefore, the idea of a small molecule that can selectively inhibit the DNA binding properties of HMGB3 resulting in less efficient chemotherapeutic lesion removal has potential for increasing chemotherapeutic efficacy. However, HMGB1, HMGB2 and HMGB3 have similar sequences and domain structures, which may present a challenge in developing HMGB3-specific small molecules that inhibit only the DNA binding function of HMGB3. Although HMGB3 has not yet been implicated in modulating pro-inflammatory cytokine functions, HMGB1 has been shown to act as a pro-inflammatory cytokine/mediator by interacting with TLR (Toll-like receptors) (107), RAGE receptors (108), and by stimulating the synthesis of Myeloid Derived Suppressor Cells (MDSCs) (109), all of which contribute to chronic inflammation in a tumor microenvironment [reviewed in (110)]. These pro-inflammatory properties of HMGB1 are mediated through the B-Box (111), which also possess DNA binding properties. Therefore, at least for HMGB1, a small molecule that interacts with the B-Box to inhibit its DNA binding properties may also inhibit its pro-inflammatory functions, which might increase the efficacy of chemotherapy by reducing the cancer-associated chronic inflammation burden.
For HMGB1, this approach has a significant paradox. In a non-tumor microenvironment, HMGB1 is released from necrotic cells and acts as a proinflammatory cytokine and induces the immune system to protect against infections (112). Free cytosolic DNA resulting from intracellular pathogens can bind HMGB1 and stimulates the activity of cyclic GMP-AMP (cGAMP) synthase (cGAS), and subsequently the stimulator of interferon genes (STING) complex to induce synthesis of proinflammatory cytokines (113). Loss of the pro-inflammatory cytokine functions of HMGB1 in a non-tumor microenvironment may increase the risk of infection and lead to autophagy deficiency (114), contributing to inflammation. In addition to its involvement in inflammation, HMGB1 has antitumor roles mediated by its interactions with tumor suppressors, such as RB (115) and TP53 (116). Further, HMGB1 mediates genomic stability by facilitating error-free DNA repair (19,63), and maintaining chromosomal telomere length via interactions with Topo IIα (117). Interestingly, unlike HMGB1, HMGB3 is not ubiquitously expressed and has not yet been shown to have pro-inflammatory cytokine properties. Although careful evaluation is warranted, HMGB3 may provide a more favorable target for neoadjuvant therapy than HMGB1. There are many described strategies to inhibit HMGB1, which include recombinant A-Box-mediated inhibition of RAGE receptor-mediated signaling, small molecule inhibitors derived from naturally-occurring molecules and steroids, anti-HMGB1 antibodies, peptide and protein inhibitors, and oligonucleotide-based inhibitors [reviewed in (118)]. Although these approaches show promise, they are not yet under clinical evaluation. Reports of the development of inhibitors or strategies to inhibit HMGB3 binding to DNA are lacking. Although there are structural similarities between the different HMGB proteins, there are also differences between them that may allow for specific targeting. For example, in cells HMGB1, HMGB2, and HMGB3 can be targeted distinctly with siRNAs, and there are antibodies that interact with separate and distinct epitopes of HMGB1, HMGB2, and HMGB3 (46). In addition, depending on the cancer type, not all HMGB proteins may be expressed equally (Figure 1C). Therefore, targeting the DNA binding function of HMGB3 may be possible without significantly disrupting the functions of other HMGB proteins, though further studies are warranted.
Conclusions
The involvement of the HMGB proteins in various DNA damage repair processing pathways, such as NER, BER, MMR and HR are evident. We have recently shown that siRNA-mediated depletion of HMGB1, HMGB2, or HMGB3 in human osteosarcoma cells increases their sensitivity to chemotherapeutic ICL-inducing agents. In addition, we have shown that targeting HMGB3 can sensitize chemoresistant ovarian cancer cells to lower doses of cisplatin by modulating the response to and repair of cisplatin-DNA adducts. Whether these effects of HMGB3 depletion are specific for ovarian cancer cells or if they can be extended to other cell types needs to be experimentally determined. Moreover, the extent to which HMGB1 and/or HMGB2 play similar roles in cisplatin sensitivity is not known; thus, we are currently investigating this possibility. Nonetheless, the HMGB proteins, when targeted individually or in combination, may open new possibilities for therapeutic intervention to treat recurrent and/or chemoresistant cancers and may increase survival and improve the quality of life for cancer patients.
Acknowledgements
This work was supported by NIH/NCI grants to K.M.V. (CA193124 and CA093729) to KMV. The results shown here are in whole or in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
References
- 1.Cubenas-Potts C, Corces VG. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett 2015;589:2923–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McBryant SJ, Adams VH, Hansen JC. Chromatin architectural proteins. Chromosome Res 2006;14:39–51 [DOI] [PubMed] [Google Scholar]
- 3.Almeida R, Fernandez-Justel JM, Santa-Maria C, Cadoret JC, Cano-Aroca L, Lombrana R, et al. Chromatin conformation regulates the coordination between DNA replication and transcription. Nat Commun 2018;9:1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem 1973;38:14–9 [DOI] [PubMed] [Google Scholar]
- 5.Stros M HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 2010;1799:101–13 [DOI] [PubMed] [Google Scholar]
- 6.Read CM, Cary PD, Crane-Robinson C, Driscoll PC, Norman DG. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res 1993;21:3427–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair RH, Horn AE, Pazhani Y, Grado L, Goodrich JA, Kugel JF. The HMGB1 C-Terminal Tail Regulates DNA Bending. J Mol Biol 2016;428:4060–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Zeng M, Wang W, Tang J. The HMGB1 acidic tail regulates HMGB1 DNA binding specificity by a unique mechanism. Biochem Biophys Res Commun 2007;360:14–9 [DOI] [PubMed] [Google Scholar]
- 9.Stott K, Watson M, Howe FS, Grossmann JG, Thomas JO. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J Mol Biol 2010;403:706–22 [DOI] [PubMed] [Google Scholar]
- 10.Muller S, Ronfani L, Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med 2004;255:332–43 [DOI] [PubMed] [Google Scholar]
- 11.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J 2003;22:5551–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 1994;10:94–100 [DOI] [PubMed] [Google Scholar]
- 13.Stros M, Reich J. Formation of large nucleoprotein complexes upon binding of the high-mobility-group (HMG) box B-domain of HMG1 protein to supercoiled DNA. Eur J Biochem 1998;251:427–34 [DOI] [PubMed] [Google Scholar]
- 14.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol 1998;18:4471–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell 1998;1:1011–9 [DOI] [PubMed] [Google Scholar]
- 16.Stros M, Kucirek M, Sani SA, Polanska E. HMGB1-mediated DNA bending: Distinct roles in increasing p53 binding to DNA and the transactivation of p53-responsive gene promoters. Biochim Biophys Acta Gene Regul Mech 2018;1861:200–10 [DOI] [PubMed] [Google Scholar]
- 17.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 1999;399:708–12 [DOI] [PubMed] [Google Scholar]
- 18.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry 2005;44:4188–95 [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee A, Vasquez KM. HMGB1 interacts with XPA to facilitate the processing of DNA interstrand crosslinks in human cells. Nucleic Acids Res 2016;44:1151–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange SS, Reddy MC, Vasquez KM. Human HMGB1 directly facilitates interactions between nucleotide excision repair proteins on triplex-directed psoralen interstrand crosslinks. DNA Repair (Amst) 2009;8:865–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange SS, Vasquez KM. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol Carcinog 2009;48:571–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park S, Lippard SJ. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry 2011;50:2567–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung Y, Lippard SJ. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry 2003;42:2664–71 [DOI] [PubMed] [Google Scholar]
- 24.McA’Nulty MM, Lippard SJ. The HMG-domain protein Ixr1 blocks excision repair of cisplatin-DNA adducts in yeast. Mutat Res 1996;362:75–86 [DOI] [PubMed] [Google Scholar]
- 25.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci U S A 1994;91:10394–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandke P, Vasquez KM. Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: Implications in DNA repair and immune responses. DNA Repair (Amst) 2019;83:102701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu T, Zhang W, Yang G, Li H, Chen Q, Song R, et al. HMGB1 overexpression as a prognostic factor for survival in cancer: a meta-analysis and systematic review. Oncotarget 2016;7:50417–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado LR, Moseley PM, Moss R, Deen S, Nolan C, Spendlove I, et al. High mobility group protein B1 is a predictor of poor survival in ovarian cancer. Oncotarget 2017;8:101215–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Yi J, Tao L, Huang G, Chu X, Song H, et al. Wnt signaling induces radioresistance through upregulating HMGB1 in esophageal squamous cell carcinoma. Cell Death Dis 2018;9:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang R, Zhang Q, Zeh HJ 3rd, Lotze MT, Tang D. HMGB1 in cancer: good, bad, or both? Clin Cancer Res 2013;19:4046–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gnanasekar M, Kalyanasundaram R, Zheng G, Chen A, Bosland MC, Kajdacsy-Balla A. HMGB1: A Promising Therapeutic Target for Prostate Cancer. Prostate Cancer 2013;2013:157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menger L, Vacchelli E, Adjemian S, Martins I, Ma Y, Shen S, et al. Cardiac glycosides exert anticancer effects by inducing immunogenic cell death. Sci Transl Med 2012;4:143ra99. [DOI] [PubMed] [Google Scholar]
- 33.Syed N, Chavan S, Sahasrabuddhe NA, Renuse S, Sathe G, Nanjappa V, et al. Silencing of high-mobility group box 2 (HMGB2) modulates cisplatin and 5-fluorouracil sensitivity in head and neck squamous cell carcinoma. Proteomics 2015;15:383–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu D, Li J, Wei J, Zhang Z, Luo Y, Tan H, et al. HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun Signal 2018;16:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redmond AM, Byrne C, Bane FT, Brown GD, Tibbitts P, O’Brien K, et al. Genomic interaction between ER and HMGB2 identifies DDX18 as a novel driver of endocrine resistance in breast cancer cells. Oncogene 2015;34:3871–80 [DOI] [PubMed] [Google Scholar]
- 36.Shin YJ, Kim MS, Kim MS, Lee J, Kang M, Jeong JH. High-mobility group box 2 (HMGB2) modulates radioresponse and is downregulated by p53 in colorectal cancer cell. Cancer Biol Ther 2013;14:213–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui G, Cai F, Ding Z, Gao L. HMGB2 promotes the malignancy of human gastric cancer and indicates poor survival outcome. Hum Pathol 2019;84:133–41 [DOI] [PubMed] [Google Scholar]
- 38.Li H, Zhang H, Wang Y. Centromere protein U facilitates metastasis of ovarian cancer cells by targeting high mobility group box 2 expression. Am J Cancer Res 2018;8:835–51 [PMC free article] [PubMed] [Google Scholar]
- 39.Hayes DC, Secrist H, Bangur CS, Wang T, Zhang X, Harlan D, et al. Multigene real-time PCR detection of circulating tumor cells in peripheral blood of lung cancer patients. Anticancer Res 2006;26:1567–75 [PubMed] [Google Scholar]
- 40.Song N, Wang B, Feng G, Duan L, Yuan S, Jia W, et al. Knockdown of high mobility group box 3 impairs cell viability and colony formation but increases apoptosis in A549 human non-small cell lung cancer cells. Oncol Lett 2019;17:2937–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Chang Y, Zhang J, Lu Y, Zheng L, Hu Y, et al. HMGB3 promotes growth and migration in colorectal cancer by regulating WNT/beta-catenin pathway. PLoS One 2017;12:e0179741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Cai Y, Zhao H, Xu Z, Sun Q, Luo M, et al. Overexpression of HMGB3 protein promotes cell proliferation, migration and is associated with poor prognosis in urinary bladder cancer patients. Tumour Biol 2015;36:4785–92 [DOI] [PubMed] [Google Scholar]
- 43.Guo S, Wang Y, Gao Y, Zhang Y, Chen M, Xu M, et al. Knockdown of High Mobility Group-Box 3 (HMGB3) Expression Inhibits Proliferation, Reduces Migration, and Affects Chemosensitivity in Gastric Cancer Cells. Med Sci Monit 2016;22:3951–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu J, Xu T, Huang QH, Zhang CM, Chen HY. HMGB3 silence inhibits breast cancer cell proliferation and tumor growth by interacting with hypoxia-inducible factor 1alpha. Cancer Manag Res 2019;11:5075–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J, Zou Z, Gao J, Zhang H, Lin Z, Zhang Y, et al. Increased expression of HMGB3: a novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol 2015;8:345–52 [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee A, Huynh V, Gaines K, Reh WA, Vasquez KM. Targeting the High-Mobility Group Box 3 Protein Sensitizes Chemoresistant Ovarian Cancer Cells to Cisplatin. Cancer Res 2019;79:3185–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400–16 e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540–56 e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018;173:291–304 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasquez KM. Targeting and processing of site-specific DNA interstrand crosslinks. Environ Mol Mutagen 2010;51:527–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clement FC, Camenisch U, Fei J, Kaczmarek N, Mathieu N, Naegeli H. Dynamic two-stage mechanism of versatile DNA damage recognition by xeroderma pigmentosum group C protein. Mutat Res 2010;685:21–8 [DOI] [PubMed] [Google Scholar]
- 55.Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, et al. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. EMBO J 2009;28:2387–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trego KS, Turchi JJ. Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry 2006;45:1961–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 1985;40:359–69 [DOI] [PubMed] [Google Scholar]
- 58.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 1987;51:241–9 [DOI] [PubMed] [Google Scholar]
- 59.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J 1997;16:6559–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J 1997;16:625–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol 2006;13:278–84 [DOI] [PubMed] [Google Scholar]
- 62.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J 2009;28:1111–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci U S A 2008;105:10320–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malina J, Kasparkova J, Natile G, Brabec V. Recognition of major DNA adducts of enantiomeric cisplatin analogs by HMG box proteins and nucleotide excision repair of these adducts. Chem Biol 2002;9:629–38 [DOI] [PubMed] [Google Scholar]
- 65.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res 2008;18:85–98 [DOI] [PubMed] [Google Scholar]
- 66.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep 2005;6:551–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kato N, Kawasoe Y, Williams H, Coates E, Roy U, Shi Y, et al. Sensing and Processing of DNA Interstrand Crosslinks by the Mismatch Repair Pathway. Cell Rep 2017;21:1375–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J, Jain A, Iyer RR, Modrich PL, Vasquez KM. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res 2009;37:4420–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J Biol Chem 2004;279:20935–40 [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, et al. Reconstitution of 5’-directed human mismatch repair in a purified system. Cell 2005;122:693–705 [DOI] [PubMed] [Google Scholar]
- 71.Genschel J, Modrich P. Functions of MutLalpha, replication protein A (RPA), and HMGB1 in 5’-directed mismatch repair. J Biol Chem 2009;284:21536–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin PR, Couve S, Zutterling C, Albelazi MS, Groisman R, Matkarimov BT, et al. The Human DNA glycosylases NEIL1 and NEIL3 Excise Psoralen-Induced DNA-DNA Cross-Links in a Four-Stranded DNA Structure. Sci Rep 2017;7:17438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kothandapani A, Dangeti VS, Brown AR, Banze LA, Wang XH, Sobol RW, et al. Novel role of base excision repair in mediating cisplatin cytotoxicity. J Biol Chem 2011;286:14564–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kothandapani A, Sawant A, Dangeti VS, Sobol RW, Patrick SM. Epistatic role of base excision repair and mismatch repair pathways in mediating cisplatin cytotoxicity. Nucleic Acids Res 2013;41:7332–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Couve S, Mace-Aime G, Rosselli F, Saparbaev MK. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J Biol Chem 2009;284:11963–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, et al. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell 2007;27:829–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y, Prasad R, Wilson SH. HMGB1: roles in base excision repair and related function. Biochim Biophys Acta 2010;1799:119–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol 2010;45:23–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol 2004;24:5776–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, et al. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol Cell Biol 2009;29:6427–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang R, Persky NS, Yoo B, Ouerfelli O, Smogorzewska A, Elledge SJ, et al. DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science 2014;346:1127–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hodskinson MR, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, et al. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell 2014;54:472–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene 2015;569:153–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, et al. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol 2000;20:2446–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J 2006;25:4921–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J 2003;22:6137–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, et al. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell 2001;8:1117–27 [DOI] [PubMed] [Google Scholar]
- 89.Roy U, Scharer OD. Involvement of translesion synthesis DNA polymerases in DNA interstrand crosslink repair. DNA Repair (Amst) 2016;44:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu X, Zhang N, Vasquez K, Barton M, Legerski R. Repair of psoralen interstrand cross-links in Xenopus laevis egg extracts is highly mutagenic. Biochem Biophys Res Commun 2005;336:69–75 [DOI] [PubMed] [Google Scholar]
- 91.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 2008;134:969–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cortez D Replication-Coupled DNA Repair. Mol Cell 2019;74:866–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol 2015;22:242–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6:1670–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol 2011;18:500–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Enokido Y, Yoshitake A, Ito H, Okazawa H. Age-dependent change of HMGB1 and DNA double-strand break accumulation in mouse brain. Biochem Biophys Res Commun 2008;376:128–33 [DOI] [PubMed] [Google Scholar]
- 97.Ke S, Zhou F, Yang H, Wei Y, Gong J, Mei Z, et al. Downregulation of high mobility group box 1 modulates telomere homeostasis and increases the radiosensitivity of human breast cancer cells. Int J Oncol 2015;46:1051–8 [DOI] [PubMed] [Google Scholar]
- 98.Di X, He G, Chen H, Zhu C, Qin Q, Yan J, et al. High-mobility group box 1 protein modulated proliferation and radioresistance in esophageal squamous cell carcinoma. J Gastroenterol Hepatol 2019;34:728–35 [DOI] [PubMed] [Google Scholar]
- 99.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet 2011;45:167–202 [DOI] [PubMed] [Google Scholar]
- 100.Tonegawa S Somatic generation of antibody diversity. Nature 1983;302:575–81 [DOI] [PubMed] [Google Scholar]
- 101.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol 2009;16:499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J 1997;16:2665–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lovely GA, Brewster RC, Schatz DG, Baltimore D, Phillips R. Single-molecule analysis of RAG-mediated V(D)J DNA cleavage. Proc Natl Acad Sci U S A 2015;112:E1715–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ciubotaru M, Trexler AJ, Spiridon LN, Surleac MD, Rhoades E, Petrescu AJ, et al. RAG and HMGB1 create a large bend in the 23RSS in the V(D)J recombination synaptic complexes. Nucleic Acids Res 2013;41:2437–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thwaites DT, Carter C, Lawless D, Savic S, Boyes JM. A novel RAG1 mutation reveals a critical in vivo role for HMGB1/2 during V(D)J recombination. Blood 2019;133:820–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim MS, Chuenchor W, Chen X, Cui Y, Zhang X, Zhou ZH, et al. Cracking the DNA Code for V(D)J Recombination. Mol Cell 2018;70:358–70 e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Das N, Dewan V, Grace PM, Gunn RJ, Tamura R, Tzarum N, et al. HMGB1 Activates Proinflammatory Signaling via TLR5 Leading to Allodynia. Cell Rep 2016;17:1128–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol 2010;28:367–88 [DOI] [PubMed] [Google Scholar]
- 109.Parker KH, Sinha P, Horn LA, Clements VK, Yang H, Li J, et al. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res 2014;74:5723–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gorgulho CM, Romagnoli GG, Bharthi R, Lotze MT. Johnny on the Spot-Chronic Inflammation Is Driven by HMGB1. Front Immunol 2019;10:1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li J, Kokkola R, Tabibzadeh S, Yang R, Ochani M, Qiang X, et al. Structural basis for the proinflammatory cytokine activity of high mobility group box 1. Mol Med 2003;9:37–45 [PMC free article] [PubMed] [Google Scholar]
- 112.Rovere-Querini P, Capobianco A, Scaffidi P, Valentinis B, Catalanotti F, Giazzon M, et al. HMGB1 is an endogenous immune adjuvant released by necrotic cells. EMBO Rep 2004;5:825–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andreeva L, Hiller B, Kostrewa D, Lassig C, de Oliveira Mann CC, Jan Drexler D, et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 2017;549:394–8 [DOI] [PubMed] [Google Scholar]
- 114.Zhang YG, Zhu X, Lu R, Messer JS, Xia Y, Chang EB, et al. Intestinal epithelial HMGB1 inhibits bacterial infection via STAT3 regulation of autophagy. Autophagy 2019;15:1935–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol Sin 2007;28:1957–67 [DOI] [PubMed] [Google Scholar]
- 116.Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res 2012;72:1996–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stros M, Bacikova A, Polanska E, Stokrova J, Strauss F. HMGB1 interacts with human topoisomerase IIalpha and stimulates its catalytic activity. Nucleic Acids Res 2007;35:5001–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Musumeci D, Roviello GN, Montesarchio D. An overview on HMGB1 inhibitors as potential therapeutic agents in HMGB1-related pathologies. Pharmacol Ther 2014;141:347–57 [DOI] [PubMed] [Google Scholar]