Abstract
The type VI secretion system (T6SS) is a toxic effector delivery apparatus widely distributed in Gram-negative bacteria. The opportunistic pathogen Pseudomonas aeruginosa encodes three T6SSs, namely H1-, H2-, and H3-T6SS. Each T6SS possesses its own effectors and their roles are not yet fully understood. Here, we report that an H3-T6SS deletion mutant PAO1(ΔclpV3) significantly affected the virulence-related phenotypes including pyocyanin production, biofilm formation, proteolytic activity, and motilities. Most interestingly, the expression of T3SS genes was markedly affected, indicating a link between H3-T6SS and T3SS. RNA-Sequencing was performed to globally identify the genes differentially expressed when H3-T6SS was inactivated and the results obtained correlated well with the observed phenotypes. Interestingly, the expressions of T2SS, T3SS, H2-T6SS, and H3-T6SS were all significantly decreased, while H1-T6SS was increased in the PAO1(ΔclpV3) strain. We also observed that the intracellular concentration of secondary messenger cAMP was reduced in PAO1(ΔclpV3), and the c-di-GMP level was also decreased as indicated by the decreased cdrA reporter activity. Finally, by using a Galleria mellonella infection model, we show that H3-T6SS plays a key role in the pathogenicity of P. aeruginosa in vivo. Overall, our study highlights the unique connection of H3-T6SS in P. aeruginosa with T3SS, pyocyanin production, biofilm formation and in vivo pathogenicity.
Keywords: Pseudomonas aeruginosa, secretion system, ClpV3, virulence factors, signal molecules
Introduction
Pseudomonas aeruginosa is an important human pathogen capable of growing in a wide range of environmental conditions (Arai, 2011). On the list of antibiotic-resistant “priority pathogens” published by World Health Organization, P. aeruginosa is classified as one of the critical pathogens that pose the greatest threat to human health (Willyard, 2017). P. aeruginosa especially affects patients who are immunocompromised or suffer with burn wounds, urinary tract infections, and cystic fibrosis (Mittal et al., 2009; Migiyama et al., 2016; Stefani et al., 2017). P. aeruginosa has an arsenal of virulence factors and pathogenic mechanisms, such as toxic protein secretion systems, quorum sensing systems (QS), biofilm formation, and antibiotic resistance (Lau et al., 2005; Bleves et al., 2010; Singh et al., 2017). Among these, the contact-dependent type VI secretion system (T6SS) directly translocates toxic effectors into prokaryotic or eukaryotic target cells (Ho et al., 2014; Jiang et al., 2014; Cianfanelli et al., 2016).
The structure of T6SS consists of a phage like tail as well as several sub-complexes that secrets toxins into target cells in a one-step manner (Bleves et al., 2010). Thirteen essential genes are conserved in all T6SSs (Filloux et al., 2008). These proteins named from TssA to TssM. TssJ, TssL, and TssM make up the membrane core complex that serves as a platform for baseplate assembly and a T6SS docking station (Durand et al., 2012; Logger et al., 2016). The T6SS baseplate complex is composed of TssE, TssF, TssG, and TssK, which facilitates the correct assembly of inner tubes and contractile sheath (Zoued et al., 2013; Brunet et al., 2015). Type VI secretion system sheath consists of two contractile proteins, TssB and TssC, forming tubular structures (Zoued et al., 2016; Gallique et al., 2017) while TssA forms a dodecamer complex, connecting the sheath structure to the membrane complex (Planamente et al., 2016). ClpV is a cytoplasmic AAA+ ATPase protein and is an essential component of T6SS (Schlieker et al., 2005). In P. aeruginosa, T6SS effector delivery is driven by ATP hydrolysis which generates the force for toxin secretion (Corbitt et al., 2018). Once the sheath is contracted, ClpV recognizes and interacts with TssC to disassemble the sheath, therefore, TssB and TssC can be reused for a new round of translocation. Planamente et al. (2016) demonstrated that ClpV interacts with TssA, suggesting ClpV is not only responsible for TssBC sheath disassembly, but is also involved in recycling other T6SS components.
There are three T6SSs in P. aeruginosa: H1-T6SS, H2-T6SS, and H3-T6SS. The expression of T6SSs in P. aeruginosa is regulated by the QS system (Lesic et al., 2009). There are several QS systems in P. aeruginosa, two N-acyl-homoserine lactone based QS systems (las and rhl systems) and one quinolone PQS system (pqs). The expression of H1-T6SS is negatively regulated by both las and pqs QS systems, while the expression of H2- and H3-T6SS is positively regulated by las, rhl, and pqs (Lesic et al., 2009; Sana et al., 2013). The RNA-binding post-transcriptional regulator RsmA represses all the T6SS clusters in P. aeruginosa (Allsopp et al., 2017). Extrinsic environmental factors play an important role in shaping pathogenesis and iron availability regulates a wealth of genes via ferric uptake regulator Fur (Pasqua et al., 2017). Iron has been shown to reduce the expression of H2- and H3-T6SSs (Sana et al., 2012, 2013; Lin et al., 2017).
The T6SSs in P. aeruginosa is not only involved in competition against other bacteria, but also survival, colonization and full virulence against the host (Sana et al., 2016). H1-T6SS is the most studied and only associated with killing prokaryotic cells, while H2- and H3-T6SS have been shown to target both bacterial and host cells but are poorly understood. H1-T6SS translocates at least seven effectors, Tse1-7, to kill prokaryotic preys (Pissaridou et al., 2018), whereas H2- and H3-T6SS target both bacterial and host cells, but only a few effectors have been identified. Recently, the role of the T6SSs has been shown to extend beyond the delivery of toxic effectors (Lin et al., 2017; Han et al., 2019). However, in depth studies on their roles are lacking. PldA, PldB, Tle3, Azu, and TplE are injected through H2-T6SS (Moore et al., 2013; Berni et al., 2019; Han et al., 2019; Wettstadt et al., 2019), while TseF is secreted by H3-T6SS (Lin et al., 2017). Azu, a Cu2+ binding protein, is secreted by H2-T6SS in P. aeruginosa for Cu2+ acquisition (Han et al., 2019). TseF dependent on H3-T6SS directly interacts with PQS and is incorporated with outer membrane vesicles (OMVs) for iron acquisition (Lin et al., 2017). Multiple T6SSs empower bacteria to better adapt and survive in the complicated polymicrobial communities not just limited to translocating toxic effectors into prey cells. Studies on T6SSs in other bacteria suggest a far more complex role. The T6SS-4 from Yersinia pseudotuberculosis is involved in the transportation of Zinc to permit the bacteria to survive oxidative stress as well as host immunity (Wang et al., 2015). Weber et al. (2009) have shown that the T6SS proteins from Vibrio anguilarum regulate expression of the stress response regulator RpoS and the quorum sensing regulator VanT, and suggest that T6SS could also function as a signal sensing system as well (Weber et al., 2009). A recent study demonstrated that the T6SS-4 in Burkholderia thailandensis plays a role in accumulating intracellular concentration of manganese through the Mn2+ binding effector (TseM) under oxidative stress (Si et al., 2017). Matthey et al. (2019) have shown that V. cholerae are able to acquire free DNA from their surroundings in a T6SS-dependent manner suggesting a role in anti-microbial resistance.
In this study, we constructed a clpV3 deletion mutant in P. aeruginosa PAO1 and found that the virulence factors (pyocyanin production, biofilm formation, proteolytic activity, motility, and T3SS) were significantly affected by the impairment of H3-T6SS. Furthermore, we performed RNA-Sequencing to compare the globally transcriptional profiles between PAO1 and PAO1(ΔclpV3) and identified 311 differentially expressed genes (DEGs). The effect of H3-T6SS on in vivo pathogenicity was also examined by using a Galleria mellonella infection model.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa and Escherichia coli strains were grown in LB broth and agar plates at 37°C. The concentrations of antibiotic used were as follows: for E. coli, tetracycline (12.5 μg/ml), ampicillin (100 μg/ml), and kanamycin (50 μg/ml), and for P. aeruginosa, tetracycline (70 μg/ml), carbenicillin (250 μg/ml), and trimethoprim (300 μg/ml). For construction of P. aeruginosa mutant PAO1(ΔclpV3), tetracycline at 300 μg/ml in Pseudomonas isolation agar (PIA) was used for specific selection.
TABLE 1.
Bacterial strains and plasmids used in this study.
| Bacterial strain or plasmid | Relevant characteristics/sequence | Source |
| E. coli strains | ||
| DH5α | F– φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk–, mk+) phoA supE44 λ– thi–1 gyrA96 relA1 | Invitrogen |
| SM10-λ pir | Mobilizing strain, RP4 integrated in the chromosome; Knr | Simon et al., 1983 |
| P. aeruginosa strains | ||
| PAO1 | Wild type, lab strain | Holloway et al., 1994 |
| PAO1(ΔclpV3) | ClpV3 knockout mutant of PAO1 | This study |
| Plasmids | ||
| pMS402 | Expression reporter plasmid carrying the promoterless luxCDABE; Kmr Tmpr | Duan et al., 2003 |
| CTX-6.1 | Integration plasmid origins of plasmid mini-CTX-lux; Tcr | Kong et al., 2013 |
| pRK2013 | Broad-host-range helper vector; Tra+, Kmr | Ditta et al., 1980a |
| pEX18Tc | oriT+ sacB+ gene replacement vector with multiple-cloning site from pUC18; Tcr | Hoang et al., 1998 |
| pAK1900 | E. coli-P. aeruginosa shuttle cloning vector, Ampr | Sharp et al., 1996 |
| pEX18Tc-clpV3up | pEX18Tc carrying the upstream fragment of clpV3 | This study |
| pEX18Tc-clpV3up+dw | pEX18Tc carrying the upstream and downstream fragment of clpV3 | This study |
| pAK-clpV3 | pAK1900 with a 2774 bp fragment of clpV3 between KpnI and HindIII; Ampr, Cbr | This study |
| CTX-phzA1 | Integration plasmid, CTX6.1 with a fragment of pKD-phzA1 containing phzA1 promoter region and luxCDABE gene; Kmr, Tmpr, Tcr | Guo et al., 2016 |
| CTX-phzA2 | Integration plasmid, CTX6.1 with a fragment of pKD-phzA2 containing phzA2 promoter region and luxCDABE gene; Kmr, Tmpr, Tcr | Guo et al., 2016 |
| pKD -exoY | pMS402 containing exoY promoter region, Kmr, Tmpr | Guo et al., 2016 |
| pKD -exsD | pMS402 containing exsD promoter region, Kmr, Tmpr | Kong et al., 2013 |
| pKD-cdrA | pMS402 containing cdrA promoter region, Kmr, Tmpr | Bhagirath et al., 2018 |
Construction of P. aeruginosa PAO1(ΔclpV3) Mutant
To generate the clpV3 deletion mutant PAO1(ΔclpV3), the pEX18Tc sucrose counter selection system was used for unmarked deletion of clpV3 gene as described previously (Hoang et al., 1998; Hmelo et al., 2015). Briefly, the upstream and downstream fragment of clpV3 were amplified by PCR with the primers clpV3-up-S/clpV3-up-AS and clpV3-dw-S/clpV3-dw-AS, respectively (Table 2). The upstream fragment firstly was cloned into the vector pEX18Tc treated with the same restriction enzyme yielding pEX18Tc-clpV3up. Following double restriction enzyme digestion (BamHI and HindIII) as well as downstream fragment, these two products were ligated to generate pEX18Tc-clpV3up+dw. The PAO1(ΔclpV3) was obtained by means of tri-parental mating as described previously (Ditta et al., 1980b). In brief, overnight cultures of the donor strain E. coli containing the plasmid pEX18Tc-clpV3up+dw, the helper E. coli strain containing pRK2013 and the recipient PAO1 were collected and re-suspended in PBS. The bacteria were mixed in a ratio of 2:2:1 and then spotted onto LB agar plates. After culturing at 37°C overnight, the bacteria were scraped off and re-suspended in 500 μl of LB. The diluted suspensions were spread on PIA plates containing tetracycline at 300 μg/ml to select for merodiploids. After first crossover, the grown colony was streaked on no salt LB plate containing 10% sucrose to select for double crossover. The resultant clpV3 knockout mutant was verified by PCR with the primers C-clpV3-S/C-clpV3-AS and designated as PAO1(ΔclpV3).
TABLE 2.
Primers used in this study.
| Primer | Sequence (5′→3′) | Restriction site |
| clpV3-up-S | TAGGAATTCGCCCTATGCCTACCAGGAA | EcoRI |
| clpV3-up-AS | TAAGGATCCGCAGGTGCTCGATCTCTACG | BamHI |
| clpV3-dw-S | TACGGATCCTGGTGGTGGACTTCAGGAAC | BamHI |
| clpV3-dw-AS | CCCAAGCTTTTGCTTTCTTCGCTTGTGAA | HindIII |
| C-clpV3-S | GGTGGAAAGCCTGCTCGACGAC | |
| C-clpV3-AS | GCGAGGATCCTTTGCCACTTGG | |
| pAK-clpV3-S | TATGGTACCGACCTGGATTGTCGCCTGA | KpnI |
| pAK-clpV3-AS | CAGAAGCTTTCTTCGCTTGTGAATGGCAC | HindIII |
| rpsL-F | TCTGACCAACGGTTTCGAGG | |
| rpsL-R | GCCCGGAAGGTCCTTTACAC | |
| rsmA-F | GACGGTACTGGGTGTCAAAGGGAAC | |
| rsmA-R | CTCTTGATCTTTCTCTTTCTGGATGCG | |
| exoS-F | GCATCAGGTAATGAGCGAGGTCG | |
| exoS-R | GGCTGTCTGCCCAGGTACTTTTCC | |
| phzA1-F | CGGTCAGCGGTACAGGGAAACA | |
| phzA1-R | CGAACAGGCTGTGCCGCTGTA | |
| phzA2-F | GCGAGAGTACCAACGGTTGAAAGG | |
| phzA2-R | GAACAGGCTGTGCCGCTGTAAC | |
| lasA-F | CGCCATCCAACCTGATGCAAT | |
| lasA-R | CGTAGGACGCATCGAAGGACGA | |
| lasR-F | CTGTACCCAGAGCGTACTGCCGA | |
| lasR-R | CGGCATGGTCAGCCCATACAC |
Underlined are restriction site sequences.
Measurement of Pyocyanin Production
Supernatants from 18 h incubation bacterial culture were collected to extract and quantify the pyocyanin production according to the previously described methods (Essar et al., 1990). Briefly, 5 ml of the culture supernatant was fully mixed with 3 ml of chloroform, then the chloroform layer was transferred to a new tube containing 1 ml of 0.2 N HCl. After centrifugation at 4500 g for 10 min, the top layer was transferred to cuvette to measure its absorbance at 520 nm. The concentrations obtained, expressed as micrograms of pyocyanin produced per milliliter of culture supernatant, were calculated by multiplying the extinction coefficient of 17.072 at 520 nm.
Measurement of Promoter Activities
The procedures of the lux-based reporter assays were described previously (Duan et al., 2003; Bhagirath et al., 2017). In brief, bacteria were incubated overnight in LB broth followed by sub-inoculating into fresh medium to OD600 of 0.2 and cultivated for an additional 3 h before use as inoculants. The cultures were inoculated into 96-well plates with transparent bottom in triplicates in a ratio of 5 μl of inoculum to 95 μl of fresh medium. 50 μl of filter-sterilized mineral oil (Sigma Aldrich) was added on top to prevent evaporation during the assay. Luminescence (counts per second, cps) was measured every 30 min for 24 h in a Synergy H4 Multimode Microplate Reader (BioTek). Bacterial growth was monitored at the same time by measuring OD600. The level of gene expression was normalized to bacterial growth and is presented as cps/OD600.
Assays for Biofilm Formation
Biofilm production was quantified as previously described (O’Toole, 2011). Cells from overnight cultures were diluted at 1:100 into M63 minimal medium supplemented with magnesium sulfate, glucose and casamino acids, then inoculated in 96-well polystyrene microtiter plates (Costar) and grown at 37°C for 24 h. After incubation, the cells were discarded, and the plate was gently submerged in a small tub of water to remove unattached cells and media components. A 125 μl volume of 0.1% crystal violet was added to each well and staining was allowed for 20 min at room temperature. Wells were rinsed three times with distilled water, and 125 μl of 30% acetic acid in water was added to dissolve the remaining crystal violet. A 100 μl portion of this solution was transferred to a new plate, and the absorbance was measured at 550 nm (OD550).
Measurement of Proteolytic Activity
Skim milk proteolysis was determined through the use of agar plate assays as described in the previous study with minor modification (Gupta et al., 2009). One microliter of cells from overnight culture were inoculated on LB plate containing 2% skim milk and grown at 37°C for overnight. Zones of clearance surrounding the bacterial colonies indicate proteolytic activity and the sizes of the zones were measured.
Swarming and Swimming Motility Examination
Bacterial motility activities were assessed as described previously (Rashid and Kornberg, 2000). Medium used for swarming assay consisted of 8 g/l nutrient broth, 5 g/l glucose and 0.5% (wt/vol) agar. For swimming assay, the medium contained 10 g/l tryptone, 5 g/l NaCl and 0.3% agar. For swarming and swimming motilities, bacteria were spotted onto plates as a 1 μl of aliquot taken directly from overnight LB cultures. After inoculation, photographs were acquired with the Fusion FX7 Vilber Lourmat Imaging machine.
Quantification of Intracellular cAMP Levels
Intracellular concentration of cAMP was quantified by using Cyclic AMP Select ELISA kit (Cayman Chemical, United States). Overnight culture of 8 OD600 units were pelleted by centrifugation and mixed with 500 μl of 0.1 N HCl. Following 30 s of sonication, the supernatants were collected by centrifugation at 1000 g for 10 min and transferred to a fresh tube. According to the manufacturer’s protocol, the standards and samples were prepared and loaded into the 96-well supplied plate. After incubation and development of the plate, wavelengths between 405 and 420 nm were read. The concentration of each sample was determined by using the equation obtained from the standard curve plot.
Complementation of the clpV3 Knockout Mutant
For the complementation experiments, The E. coli-P. aeruginosa shuttle vector pAK1900 was used (Sharp et al., 1996). clpV3 gene was generated by PCR with the primers pAK-clpV3-S/pAK-clpV3-AS listed in Table 2. This PCR amplified fragment was cloned into PAK1900 and the resultant plasmid pAK-clpV3 was transformed into PAO1(ΔclpV3) by electroporation.
RNA Isolation
Strains were grown overnight at 37°C followed by sub-culturing into fresh medium and grown to mid-exponential phase. Total RNA was extracted by TRIzol-based method (Life Technologies, CA, United States). In brief, the cultures were centrifuged at 12,000 g for 5 min. The supernatant was discarded, cell pellets were resuspended in 1 ml of TRIzol and then incubated at room temperature for 5 min to permit complete dissociation of nucleoproteins complex. Followed by adding 0.2 ml of chloroform into the tube and mix well, the samples were centrifuged (12,000 g at 4°C for 15 min) to form three layers. The upper aqueous phase was transferred to a fresh tube and added by 0.5 ml of cold isopropanol to precipitate RNA. After centrifugation, the supernatant was removed, and the pellet was suspended in 1 ml of 75% ethanol. Samples were centrifuged and the supernatant was discarded, air-dried RNA pellet was resolved in 50 μl of RNase-free Water.
RNA-Seq Library Construction and Sequencing
RNA integrity was measured using Bioanalyzer 2100 (Agilent, Santa Clara, CA, United States) and rRNA was removed from 1 mg of total RNA with Ribo-Zero Magnetic Gold Kit (Epicentre Biotechnologies, Madison, WI, United States). To construct the RNA-Seq library, TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA, United States) was used. rRNA-depleted RNA was fragmented into small pieces using Elute Prime Fragment Mix. First-strand cDNA was synthesized with First Strand Master Mix and Super Script II reverse transcriptase (Invitrogen, Carlsbad, CA, United States). Following purification by Agencourt RNAClean XP beads (Beckman Coulter, CA, United States), the second-strand cDNA library was synthesized using Second Strand Master Mix and dATP, dGTP, dCTP, dUTP mix. Purified fragmented cDNA was end repaired (30 min at 37°C) prior to ligating sequencing adapters. Amplified RNA-Seq libraries were purified by using AMPureXP Beads. The clustering of the index-coded samples was performed on a cBot Cluster Generation System following to the manufacturer’s instructions, and the sequencing was performed using the Illumina Hiseq TM 2500 platform with pair-end 150 base reads.
Bioinformatics Analysis
After RNA-Sequencing, the obtained raw data were filtered according to the following standards: (1) removing reads with ≥10% unidentified nucleotides (N); (2) removing reads with >50% bases having Phred quality scores of ≤20; (3) removing reads aligned to the barcode adapter using FASTP1. Quality trimmed reads were aligned using Bowtie2 (Langmead and Salzberg, 2012) (version 2.2.8) to the P. aeruginosa PAO1 reference genome to identify known genes and calculated gene expression by RSEM (Li and Dewey, 2011). The gene expression level was calculated and further normalized by using the fragments per kb of transcript per million (FPKM) mapped reads method to eliminate the influence of different gene lengths and amount of sequencing data on the calculation of gene expression. The edgeR package2 was used to identify DEGs across samples with fold changes ≥2 and a false discovery rate-adjusted P (q value) <0.05. Go terms and KEGG pathway were defined as being significantly enriched when the q value ≤0.05.
Synthesis of cDNA and Quantitative Real-Time PCR
cDNA was generated from 1 μg total RNA using a Quanta qScript cDNA Synthesis kit (Quanta BioSciences, MD, United States). rpsL was used as housekeeping gene control and the primers were listed in Table 2. The qPCR was performed using PowerUpTM SYBRTM Green Master Mix (Thermo Fisher Scientific) on an Eco Illumina real-time detection system (Montreal Biotech) under the following conditions: UDG activation at 50°C for 2 min, Dual-LockTM DNA polymerase initiation at 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Fold changes of gene expression levels were calculated according to the 2–ΔΔCq method. Each sample was measured in triplicate and repeated at least three times.
Galleria mellonella Killing Assays
The G. mellonella infection model is a widely-accepted animal model and the experiments were performed as previously described (Desbois and Coote, 2011). The larvae were stored in wood chips at 10°C and used within two weeks from shipment. Prior to inoculation into G. mellonella caterpillars, P. aeruginosa cells were washed twice with PBS and then diluted in PBS to a final concentration of 1000 CFU/ml. A 10 μl Hamilton syringe was used to inject 10 μl of bacterial suspension into G. mellonella via the last left proleg. The infected larvae were incubated in a static incubator in the dark at 30°C, the optimum temperature for insect growth and development (Brennan et al., 2002). The number of dead caterpillars was scored at each time point. Caterpillars were considered dead when they displayed no movement in response to touch.
Results
Inactivation of H3-T6SS Altered Multiple P. aeruginosa PAO1 Phenotypes
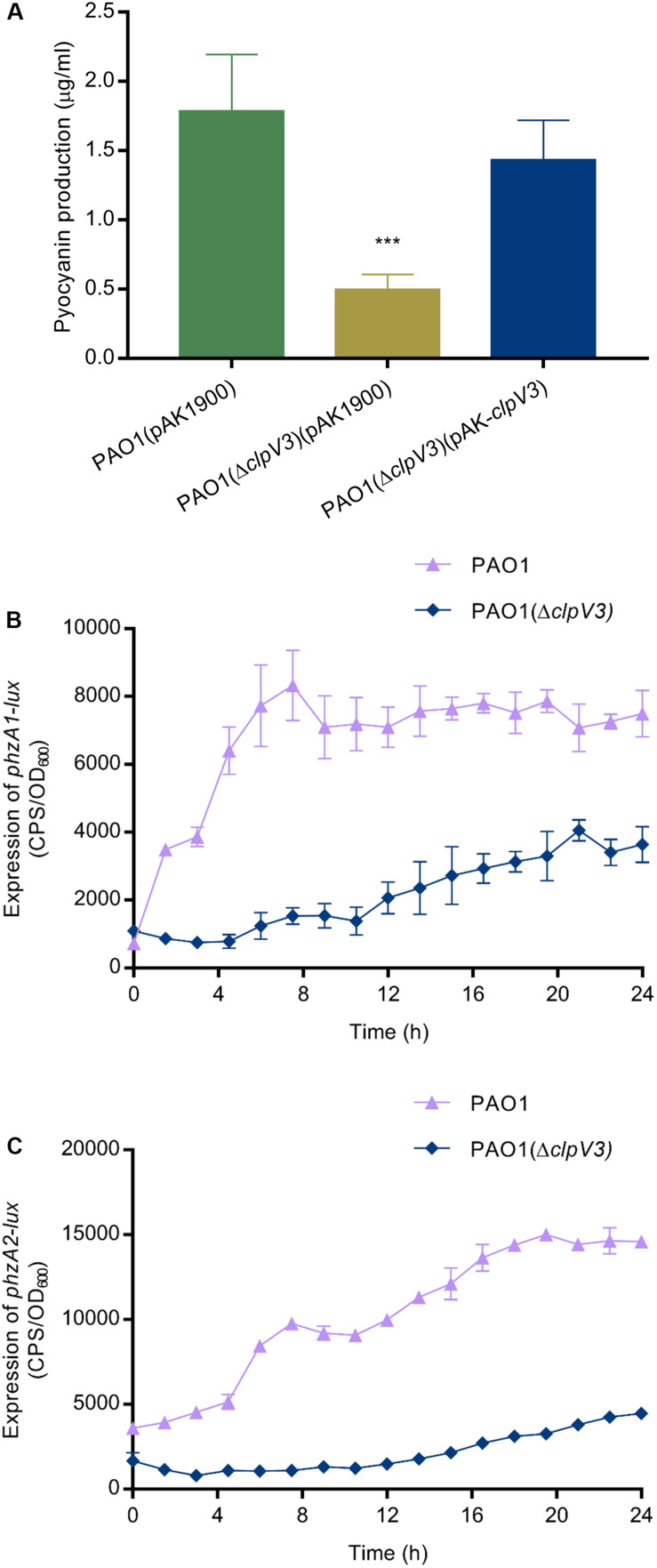
In light of the non-competition functions of T6SSs and the fact that H3-T6SS is poorly understood, we constructed an H3-T6SS mutant PAO1(ΔclpV3) and examined virulence-related phenotypical changes in the mutant. Deletion of clpV3 completely inactivates the function of H3-T6SS (Sana et al., 2013). We compared the virulence-related phenotypes between the wild type and the clpV3 knockout mutant. A striking difference that was easily observed was the color change of the bacterial cultures. Measurement of pyocyanin production indicated that when compared with the wild-type PAO1 pyocyanin was significantly reduced in PAO1(ΔclpV3). Overexpression of clpV3 gene in the PAO1(ΔclpV3) strain restored the pyocyanin production to the wild-type level (Figure 1A). Since two homologous operons are associated with synthesizing phenazine compounds in P. aeruginosa, phzA1B1C1D1G1 (phzA1) and phzA2B2C2D2G2 (phzA2) (Mavrodi et al., 2001), the reduced pyocyanin production in PAO1(ΔclpV3) strain led us to analyze the promoter activities of these two operons. As shown in Figures 1B,C, the phzA1 and phzA2 promoter activities in the PAO1(ΔclpV3) were 3-fold lower than in the wild-type PAO1, indicating the reduced pyocyanin production in the mutant was probably a result of the reduced transcription of these operons.
FIGURE 1.
Reduced pyocyanin production and decreased expression of phzA1 and phzA2 in PAO1(ΔclpV3). (A) Comparison of pyocyanin production in PAO1(ΔclpV3),the wild-type PAO1 and the complementation strain. The experiment was independently performed three times. Unpaired Student’s t-test was used to analyze the data. ***p < 0.001. (B,C) Reduced promoter activities of two pyocyanin synthesis gene clusters phzA1 and phzA2 in PAO1(ΔclpV3). The promoter activity is presented as light production (counts per second, cps) normalized to OD600. The experiment was independently performed three times. Error bars indicate standard deviations.
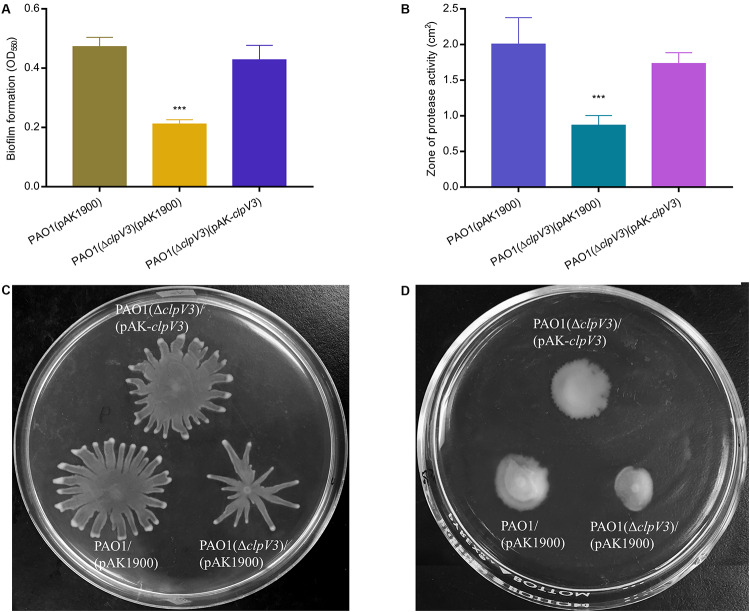
We further examined the effect of H3-T6SS on other virulence factors including proteolytic activity, biofilm formation and bacterial motility. Proteases in P. aeruginosa play two important roles: subverting host immune responses and mediating host-directed damage (Laarman et al., 2012). Biofilms function as a protective barrier and allow bacteria to survive from the antimicrobial agents and the host immune systems (Drenkard, 2003; Haussler and Parsek, 2010). Motility is a key feature of acute phase of infection and is regulated by RsmA among other regulators (Heurlier et al., 2004). As shown in Figure 2, the results obtained demonstrated that biofilm formation was significantly decreased in PAO1(ΔclpV3) as compared to the wild-type PAO1 or the complemented strain (Figure 2A) and so was protease production (Figure 2B). In addition, PAO1(ΔclpV3) exhibited altered swarming patterns characterized by less and short tendrils compared with the pattern of wild type or the complemented strain (Figure 2C and Supplementary Figure S1A). Inactivation of H3-T6SS resulted in reduced swimming motility as well (Figure 2D and Supplementary Figure S1B). These results clearly indicate that the functionality of H3-T6SS is intricately connected with key virulence factors in P. aeruginosa.
FIGURE 2.
Inactivation of H3-T6SS in P. aeruginosa alters numerous phenotypes. (A) Biofilm formation was evaluated in 96-well microtiter plates and our results showed that biofilm formation was significantly decreased in PAO1(ΔclpV3) compared with in PAO1 or the complementation strain. (B) PAO1(ΔclpV3) showed smaller proteolytic zone than PAO1, which indicated that deletion of clpV3 leads to producing less protease production. (C,D) Inactivation of H3-T6SS resulted in altered swarming motility and reduced swimming motility. The experiment was independently performed three times. Unpaired Student’s t-test was used to analyze the data. Error bars indicate standard deviations. ***p < 0.001.
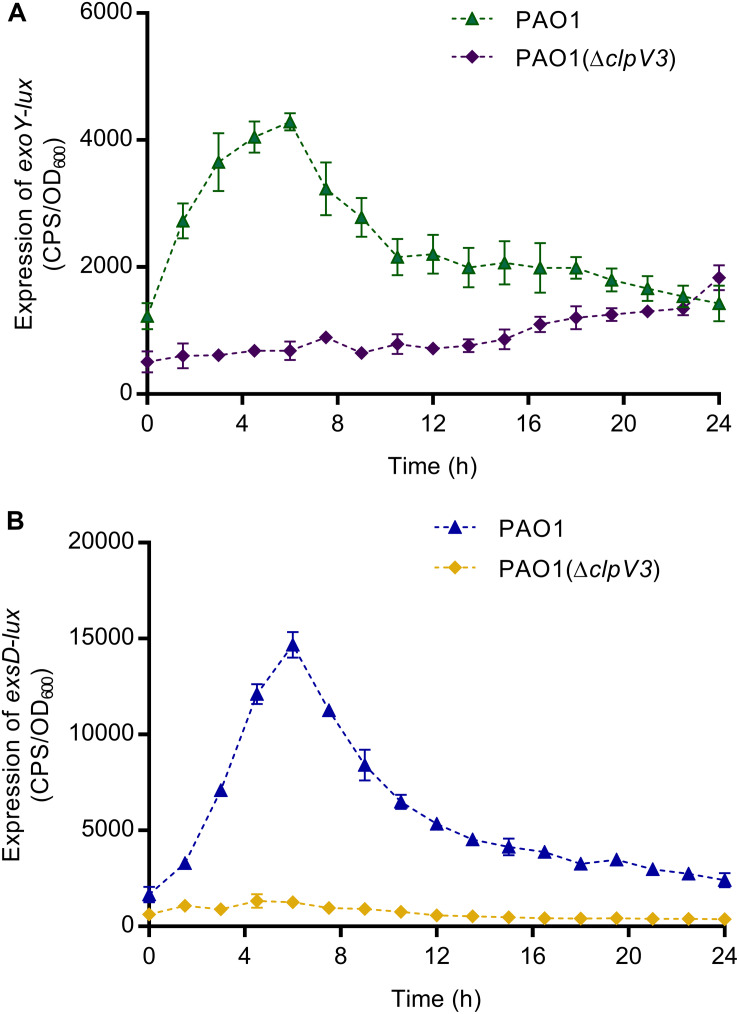
Deletion of clpV3 Resulted in Decreased Expression of exoY and exsD
Pseudomonas aeruginosa utilizes a complex type III secretion apparatus to inject effector proteins (ExoS, ExoY, ExoT, and ExoU) into the host cells (Yan et al., 2019). T3SS and T6SS have been shown to be oppositely regulated (Moscoso et al., 2011) and both can target host cells. To find out whether the expression of T3SS was also affected in PAO1(ΔclpV3), the promoter activities of T3SS genes, including effector (exoY) and the exsD-pscBCDEFGHIJKL operon were analyzed in PAO1 and PAO1(ΔclpV3) using the lux-based pKD-exoY and pKD-exsD reporters. As shown in Figures 3A,B, the promoter activities of exoY and exsD were significantly decreased in PAO1(ΔclpV3) when compared with those in the wild-type PAO1, indicating that inactivation of H3-T6SS caused a significant reduction of the expression of T3SS genes.
FIGURE 3.
The promoter activities of exoY (A) and exsD (B) were significantly decreased in PAO1(ΔclpV3) when compared with in wild-type PAO1. The results clearly indicate that the expression of T3SS was significantly reduced when H3-T6SS is inactivated.
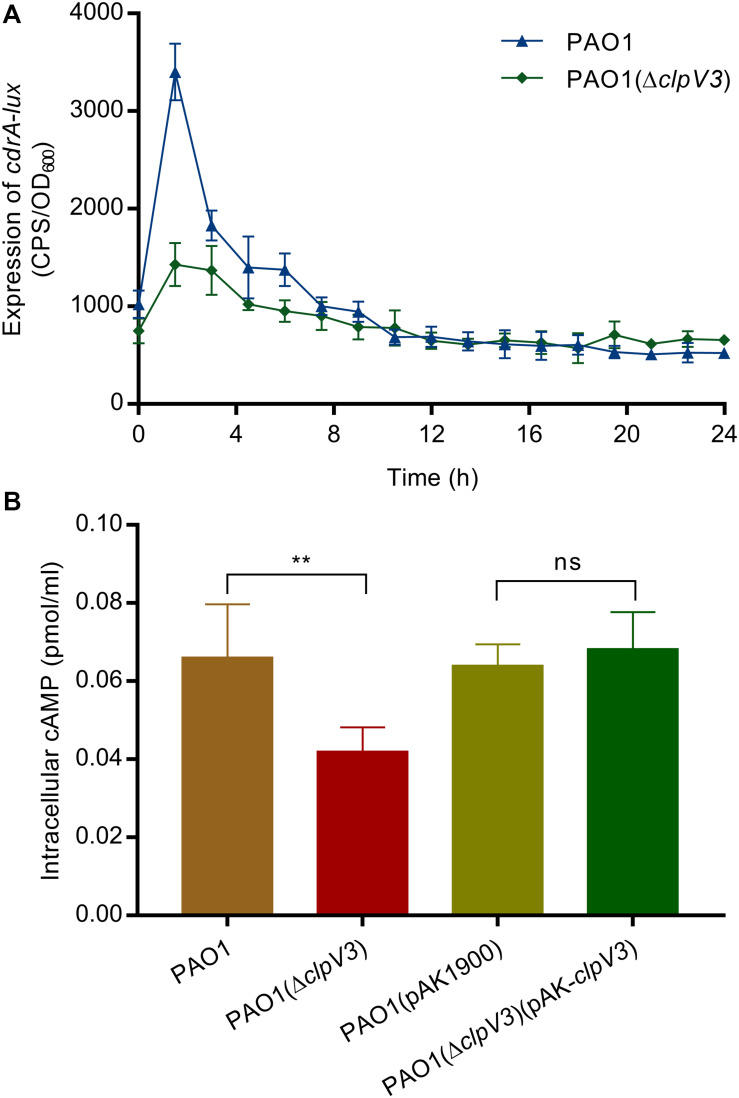
PAO1(ΔclpV3) Had Reduced Levels of Second Messengers c-di-GMP and cAMP
Signaling molecules cyclic-di-GMP (c-di-GMP) and cyclic AMP (cAMP) play an important role in virulence factor regulation in P. aeruginosa (Almblad et al., 2015). c-di-GMP is a second messenger used by P. aeruginosa and other bacteria to regulate the expression of genes associated with flagella motility, Type IV pili and biofilm initiation (Valentini and Filloux, 2019). To investigate if the observed phenotypical changes could be a result of altered c-di-GMP levels, we tested cdrA promoter activity in the clpV3 knockout mutant. The cdrA promoter activity levels have been shown to faithfully reflect the fluctuations in intracellular c-di-GMP levels (Rybtke et al., 2012). Quantifications of cdrA promotor activity in PAO1(ΔclpV3) and wild-type PAO1 were performed in triplicates and the data obtained are presented in Figure 4A. Compared with the wild-type PAO1, a decreased cdrA promoter activity was observed in PAO1(ΔclpV3), indicating a potential negative effect of H3-T6SS impairment on intracellular c-di-GMP levels. To determine if clpV3 deletion affected cAMP levels in P. aeruginosa, the concentration of intracellular cAMP was measured for PAO1 and PAO1(ΔclpV3) using the Cyclic AMP Select ELISA Kit (Cayman Chemical Company, USA). As shown in Figure 4B, the clpV3 mutant had significantly lower cellular cAMP than the wild-type PAO1, suggesting that the clpV3 or the functionality of H3-T6SS contributes to the balance of in vivo cAMP concentrations. The level of cAMP in PAO1(ΔclpV3) was restored to the wild-type level upon complementation of clpV3 on a plasmid. The decreased intracellular concentrations of the second messengers c-di-GMP and cAMP in the clpV3 deletion mutant may explain, at least partially, the altered virulence factor production in this H3-T6SS mutant observed.
FIGURE 4.
Intracellular concentration of secondary messengers (c-di-GMP and cAMP) were significantly reduced when clpV3 gene was deleted. (A) PAO1(ΔclpV3) exhibited decreased cdrA promoter activity when compared with that of the wild-type PAO1, indicating a clear effect of H3-T6SS on intracellular c-di-GMP levels. (B) cAMP concentration was quantified in wild-type PAO1, PAO1(ΔclpV3) mutant and its complimented strain by using the Cyclic AMP Select ELISA Kit. Lower expressed cAMP was detected in clpV3 deletion mutant compared with either the wild type or its complemented strain. Unpaired Student’s t-test was used to analyze the data, and the means and standard deviations are shown. **p < 0.01, ns, not significant.
Transcriptional Profiling of the clpV3 Deletion Mutant
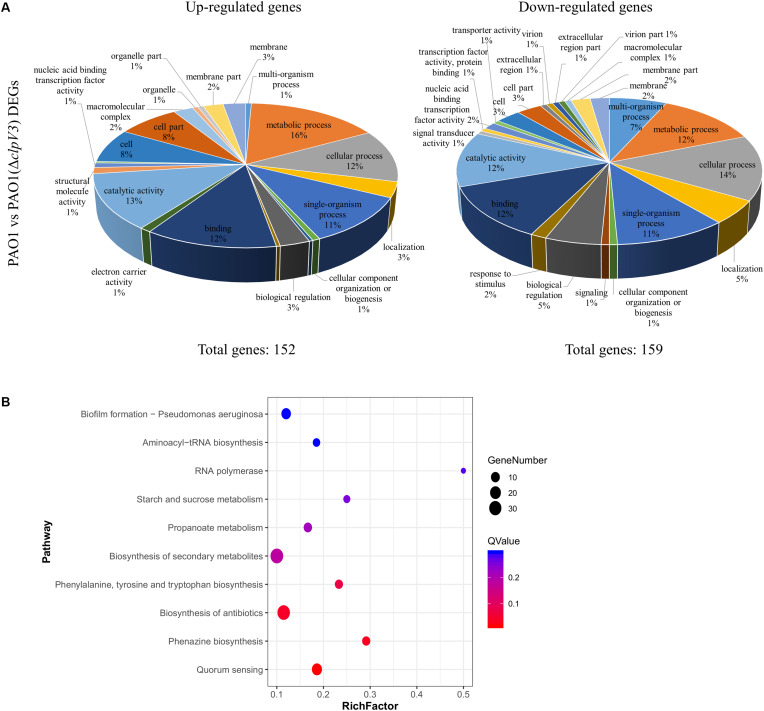
It was apparent that the H3-T6SS mutant produced a reduced amount of virulence factors, such as pyocyanin. In order to determine globally the effect of clpV3 inactivation, we performed RNA sequencing. The mRNAs were extracted from the culture grown in LB medium at the mid-exponential phase and the transcriptional profiles of PAO1(ΔclpV3) and wild-type PAO1 were compared. We found 311 significantly affected genes that were differentially expressed in these two strains. Hundred and fifty-two genes were up-regulated in clpV3 deletion mutant and 159 down-regulated. The DEGs were assigned to 29 functional groups based on sequence homology. The significantly enriched (q < 0.05) gene ontology (GO) categories among these DEGs are shown in Figure 5A and Supplementary Table S1. Comparing the expression patterns between PAO1 and PAO1(ΔclpV3), we were able to identify DEGs participating in quorum sensing, phenazine biosynthesis, biosynthesis of antibiotics, phenylalanine, tyrosine, and tryptophan biosynthesis, biosynthesis of secondary metabolism, and biofilm formation (Figure 5B).
FIGURE 5.
Comparison of the global transcriptomes of PAO1 and PAO1(Δclpv3) by RNA-Seq. (A) Totally 311 genes were significantly differentially expressed in PAO1(ΔclpV3) compared to wild-type PAO1 with absolute fold change >2 and FDR <0.05. Pie charts showing Gene Ontology analysis of the differentially expressed genes outlined in the PAO1 vs PAO1(ΔclpV3), subdivided into up-regulated (left) and down-regulated (right) groups. (B) The top 10 of significant KEGG enrichments between PAO1 and PAO1(ΔclpV3).
It is observable that in PAO1(ΔclpV3) the expression of genes related with T2SS, T3SS, and pyocyanin synthesis were significantly down regulated. The specific virulence factors and regulatory pathways are grouped and listed in Supplementary Table S2. Such results are in agreement with the observation of reduced pyocyanin production, proteolytic activity, and lower promoter activities of T3SS in the mutant. Interestingly, all three T6SSs in P. aeruginosa were differently expressed when clpV3 was deleted. The expression of both H2- and H3-T6SS was significantly decreased, compared with the wild-type PAO1, while the expression of H1-T6SS was significantly increased in PAO1(ΔclpV3). In addition, the RNA level of the post-transcriptional regulator RsmA which positively regulates T3SS and negatively regulates T6SSs was also lower in PAO1(ΔclpV3). Although RsmA activity is regulated mostly by small RNAs, such as RsmY and RsmZ, the altered transcription level of rsmA could also result in changed functionality of this regulator, The reduced expression of T3SS and increased H1-T6SS could, therefore, be due to the lower rsmA expression, at least partially. A previous study has shown that QS positively regulates H2- and H3-T6SS, but negatively regulates H1-T6SS (Lesic et al., 2009). We noted that the expression of several transcriptional regulators including LasR, VqsR, MexL, MvaT, and PauR were significantly decreased, while PmpR, CgrC, CgrB, PtxS, AntR, NarL, and Zur were significantly increased in the H3-T6SS mutant (Supplementary Table S2). The regulators that showed changed expression in the PAO1(ΔclpV3) potentially resulted in the lower expression of H2- and H3-T6SS, T2SS and T3SS and higher expression of H1-T6SS, which were also shown by the RNA-Sequencing analysis. Taken together, these results demonstrated that H3-T6SS impairment not only affected other T6SSs but also several genes involved in virulence and metabolic regulation in P. aeruginosa.
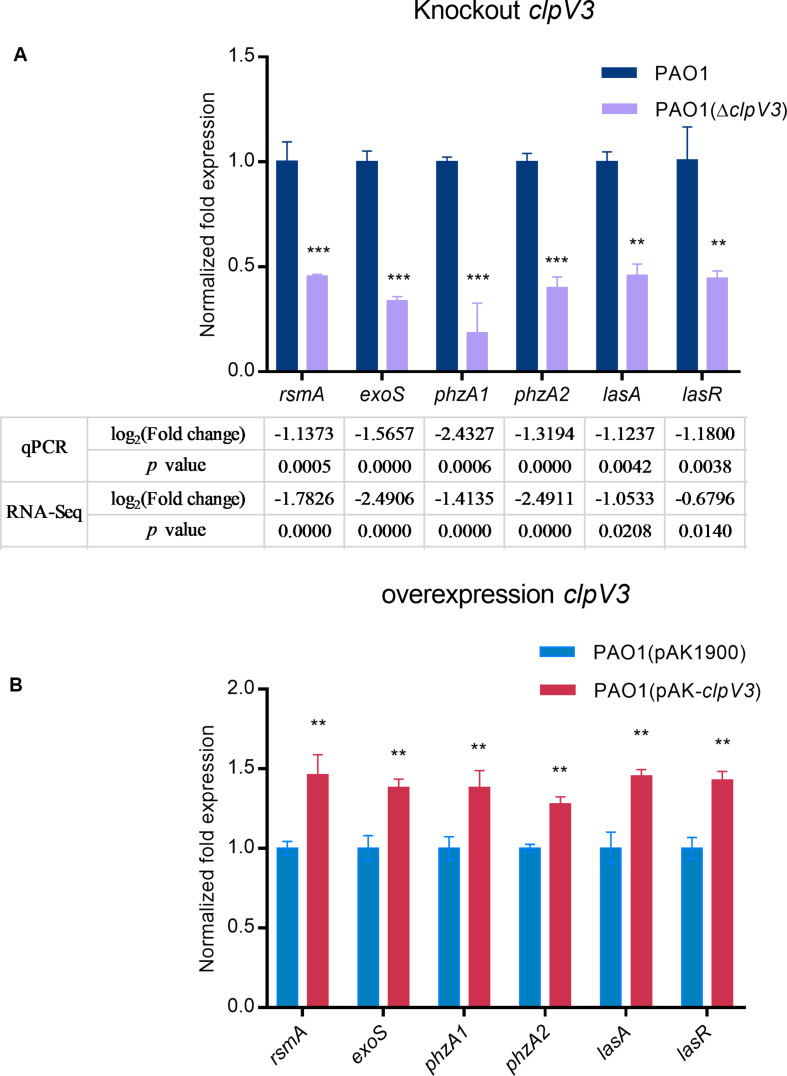
Confirmation of the RNA-Sequencing Results With Selected Genes
RT-qPCR was carried out with selected genes to verify the RNA-Sequencing data. cDNA was prepared using the same RNA that was used for RNA-Sequencing. Genes selected for validation included rsmA, exoS, phzA1, phzA2, lasA, and lasR. Gene expression relative to housekeeping gene (rpsL) levels was calculated and shown in Figure 6A. The results showed decreased expression of these genes, in agreement with the RNA-Seq results. Furthermore, we constructed a clpV3 overexpression strain and measured the expression of these previous tested genes. As shown in Figure 6B, the expression of these genes was significantly increased when clpV3 was overexpressed. P. aeruginosa PAO1 strain carrying an empty vector PAO1(pAK1900) was used as a control in the comparison.
FIGURE 6.
qPCR results after knocking down clpV3 (A) and after overexpression of clpV3 in PAO1 (B). Assays were performed in triplicate. Statistical analysis was performed by unpaired Student’s t-test. ***p < 0.001; **p < 0.01.
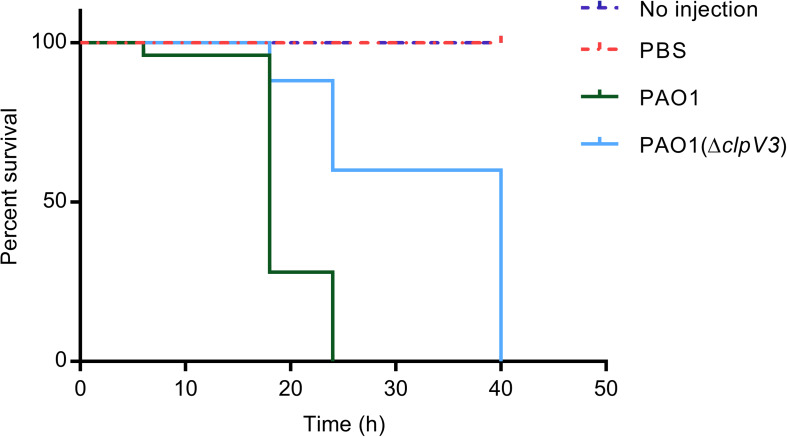
The Effect of clpV3 Deletion on P. aeruginosa PAO1 Virulence in vivo
To verify the effect of the H3-T6SS on P. aeruginosa PAO1 pathogenicity in vivo, we used a G. mellonella infection model to examine the pathogenicity of PAO1(ΔclpV3). The relative survival rates of the infected G. mellonella larvae (n = 25) were compared between the clpV3 mutant and the wild-type PAO1. As shown in Figure 7, the PAO1(ΔclpV3) infected larvae exhibited significantly increased survival rate compared with those infected with the wild-type PAO1. The decreased pathogenicity can be attributed to the decreased expression of virulence factors, such as pyocyanin production, T3SS, H2- and H3-T6SS. Our results, taken collectively, shown that H3-T6SS plays an important role in P. aeruginosa pathogenicity in vivo and may serve as a potential therapeutic target against its infection.
FIGURE 7.
The effect of H3-T6SS on the pathogenicity of P. aeruginosa in vivo. The relative survival rates of the infected G. mellonella larvae were compared. PAO1(ΔclpV3) exhibited significantly increased survival rate compared with the wild-type PAO1. The experiment was performed three times on different days.
Discussion
T6SS is a macromolecular weapon that is present in more than 25% of Gram-negative bacteria (Bingle et al., 2008). Multiple distinct T6SSs have been identified in a given bacterium including P. aeruginosa, suggesting T6SS’ versatile roles or specificities for particular niches. Distinct T6SSs might be activated in response to different environmental cues and hence have the extended functions beyond the delivery of toxic effectors (Bernard et al., 2010).
The results we obtained showed that deletion of clpV3 in P. aeruginosa, which abolishes the functionality of H3-T6SS (Sana et al., 2013), dramatically affected phenotypes associated with pathogenicity. This is the first report that H3-T6SS affects virulence factors and biofilm formation in P. aeruginosa, which supports a role of H3-T6SS beyond the function of a weapon in competition against other microorganisms and/or killing host cells. In the clpV3 deletion mutant, multiple virulence-related phenotypes were altered which included pyocyanin production, proteolytic activity, motilities, T3SS, and biofilm formation. Interestingly all of them were significantly reduced when H3-T6SS was inactivated by deletion of clpV3.
Among the virulence factors affected, pyocyanin is used by P. aeruginosa for interspecies competition and combating the host immune systems. It kills other competitors residing in the same niche and is able to elicit host responses by inactivating catalases known to protect against reactive oxygen species (ROS; Wilson et al., 1988; Ozyurek et al., 2011). Motility (swarming and swimming) is a key feature of P. aeruginosa during the acute infection phase and is also strongly associated with P. aeruginosa pathogenesis, as it enables colonization of different environments, attachment to surfaces, and formation of biofilm (Arora et al., 2005). Swarming is characterized as coordinated group activity and the motility pattern is dependent of the surfactant rhamnolipids (Caiazza et al., 2005; Tremblay et al., 2007). RhlAB are involved in the synthesis of rhamnolipids (Deziel et al., 2003), and the expression of rhlA and rhlB was reduced by 4.51- and 3.20-fold, respectively observed in PAO1(ΔclpV3) compared with that in wild-type PAO1 by RNA-Sequencing analysis (Supplementary Table S1). The changed motility in clpV3 mutant is in agreement with the recent reported study that deletion of clpV1 decreased swarming motility (Chen et al., 2020). Alkaline protease in P. aeruginosa is not only important for its virulence but is also important for immune evasion, though the exact mechanism is yet unknown (Laarman et al., 2012). We observed a decrease in proteolytic activity in clpV3 mutant compared to the wild-type PAO1 strain. Thus, the inactivation of H3-T6SS not only abolished the function of H3-T6SS as a weapon against competitors and/or the host but also impaired other traits that are involved in such actions against competitors or the hosts.
Another important consequence of inactivation of H3-T6SS is the decreased biofilm formation. Biofilms function as both a structural scaffold and protective barrier to prevent access of antimicrobial agents and host immune identification (Drenkard, 2003; Haussler and Parsek, 2010). A recent study done by Chen et al. (2020) found that the H1-T6SS, H3-T6SS and lasR were expressed at higher levels in strong biofilm forming clinical isolates than in non-biofilm forming groups. Our observation that the clpV3 mutant formed less biofilms than the wild type is in agreement with the reported findings suggesting a clinical relevance of the affected biofilm formation by H3-T6SS in P. aeruginosa.
T3SS is a system mainly used for interacting with eukaryotic host cells. T3SS and T6SS systems are reversibly regulated by the RsmA regulatory pathway (Brencic and Lory, 2009; Moscoso et al., 2011). The observation in our study that the expression of T3SS related genes (exoY and exsD-pscBCDEFGHIJKL) was significantly decreased in the clpV3 mutant is the first report of a direct effect of T6SS on T3SS expression. Although common regulators have been reported such as RsmA that regulate both T6SS and T3SS, to our knowledge, no effect of T6SS has been reported to cause a change in T3SS.
Secondary messengers (cAMP and c-di-GMP) play important roles in regulating multiple phenotypical characteristics, and c-di-GMP has been shown to regulate multiple phenotypes including motility and biofilm formation (Moscoso et al., 2011). We measured these two secondary messengers and found a decreased level of cAMP in the clpV3 mutant. This decreased cAMP level in clpV3 mutant could explain the lower expression of T3SS because the genes involved in this secretion system are regulated by Vfr coupled with cAMP (Marsden et al., 2016). The other secondary messenger molecule c-di-GMP is capable of regulating the life styles of bacteria and controlling many key virulence factors (Valentini and Filloux, 2019). As compared to PAO1, the clpV3 deletion mutant demonstrated a decrease in cdrA promoter activity, suggesting a reduced c-di-GMP level in the mutant. The decreased c-di-GMP signal molecules could hypothetically provide an explanation about the reduced virulence factors including pyocyanin production, proteolytic activity and biofilm formation (Hengge, 2009; Lo et al., 2016; Chang, 2017).
In an effort to globally investigate the effect of ClpV3, RNA-sequencing was performed to compare the DEGs between PAO1 and PAO1(ΔclpV3). The transcriptomic data revealed that a total of 311 genes were differentially expressed between the two strains. In a clpV3 mutant, lower expression of genes related to pyocyanin synthesis, T2SS, T3SS, H2-T6SS, H3-T6SS was observed while an increased expression of H1-T6SS was also noted. These results correlate well with the observed phenotypic changes in the mutant. In addition, a number of transcriptional or post-transcriptional regulators were differentially expression, including rsmA, vqsR, mexL, mvaT, pauR, pmpR, cgrC, cgrB, ptxS, antR, narL, and zur. The down-regulated lasR could explain the decreased expression of genes related to H2-T6SS, H3-T6SS, T2SS and pyocyanin, and an increase in the H1-T6SS (Venturi, 2006; Lesic et al., 2009). Other virulence factors affected by H3-T6SS can either directly or indirectly attributed to these regulators. ClpV is a cytoplasmic AAA+ ATPase protein and it is recently found to be localized at discrete and dynamic foci in the cell (Basler and Mekalanos, 2012; Lennings et al., 2019), suggesting additional roles beyond providing energy for T6SSs for this protein. The energy intensive function of T6SS depends on such an ATPase. It is tempting to speculate that, once clpV3 is deleted, the balance of ATP pool of the cell might be disrupted. ATP dependent synthesis of signaling molecules such as cAMP, as well as the quorum sensing signal molecules might have been affected as a result, which in turn would affect the downstream of the regulatory pathways, contributing to the altered pathogenicity associated phenotypes including biofilm formation, proteolytic activity, pyocyanin production, swarming and swimming motilities. T3SS is an ATP-dependent protein secretion system. T3SS activity is energy intensive and potentially influenced by cellular energy level; therefore, any changes in the cellular ATP pool could theoretically affect this system.
As multiple virulence factors changed significantly in the H3-T6SS mutant, we examined the relevance of such changes in vivo using a well-established G. mellonella infection model. G. mellonella larvae were infected by both the wild-type PAO1 and PAO1(ΔclpV3) and the survival rates were compared. The significantly decreased mortality rate in PAO1(ΔclpV3), when compared to the PAO1, confirmed that H3-T6SS plays an important role in the in vivo pathogenicity of P. aeruginosa. The lower expressed virulence including T3SS, pyocyanin production, and elastase may account for the decreased mortality. Because it is possible that H3-T6SS may be directly involved in killing the host, more studies are required to differentiate the direct contributions of the impairment of H3-T6SS in the decreased pathogenicity against G. mellonella.
Collectively, the results obtained in this study suggest that H3-T6SS in P. aeruginosa plays far more diverse roles and affects many more genes rather than just those related to T6SS, revealing a surprisingly complex connection of this system with other secretions systems and pathogenicity. These results add to the limited pool of knowledge in this field and suggest that T6SS might be a potential therapeutic target against P. aeruginosa infections.
Data Availability Statement
RNA-seq data can be found in the NCBI database, the data was assigned a accession number of GSE148116.
Author Contributions
KD directed and designed the research. YL, PZ, and LC conducted the experiments. YL and AB analyzed the data. YL and KD wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-05864-2019). The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01096/full#supplementary-material
References
- Allsopp L. P., Wood T. E., Howard S. A., Maggiorelli F., Nolan L. M., Wettstadt S., et al. (2017). RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114 7707–7712. 10.1073/pnas.1700286114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almblad H., Harrison J. J., Rybtke M., Groizeleau J., Givskov M., Parsek M. R., et al. (2015). The cyclic AMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by cyclic di-GMP. J. Bacteriol. 197 2731–2731. 10.1128/JB.00493-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H. (2011). Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2:103. 10.3389/fmicb.2011.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S. K., Neely A. N., Blair B., Lory S., Ramphal R. (2005). Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 73 4395–4398. 10.1128/IAI.73.7.4395-4398.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M., Mekalanos J. J. (2012). Type 6 secretion dynamics within and between bacterial cells. Science 337 815–815. 10.1126/science.1222901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C. S., Brunet Y. R., Gueguen E., Cascales E. (2010). Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192 3850–3860. 10.1128/JB.00370-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berni B., Soscia C., Djermoun S., Ize B., Bleves S. (2019). A type VI secretion system trans-kingdom effector is required for the delivery of a novel antibacterial toxin in Pseudomonas aeruginosa. Front. Microbiol. 10:1218. 10.3389/fmicb.2019.01218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagirath A. Y., Pydi S. P., Li Y., Lin C., Kong W., Chelikani P., et al. (2017). Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect. Dis. 3 162–175. 10.1021/acsinfecdis.6b00153 [DOI] [PubMed] [Google Scholar]
- Bhagirath A. Y., Somayajula D., Li Y., Duan K. (2018). CmpX affects virulence in Pseudomonas aeruginosa through the Gac/Rsm signaling pathway and by modulating c-di-GMP levels. J. Membr. Biol. 251 35–49. 10.1007/s00232-017-9994-6 [DOI] [PubMed] [Google Scholar]
- Bingle L. E., Bailey C. M., Pallen M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11 3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Bleves S., Viarre V., Salacha R., Michel G. P., Filloux A., Voulhoux R. (2010). Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300 534–543. 10.1016/j.ijmm.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Brencic A., Lory S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72 612–632. 10.1111/j.1365-2958.2009.06670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M., Thomas D. Y., Whiteway M., Kavanagh K. (2002). Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 34 153–157. 10.1111/j.1574-695X.2002.tb00617.x [DOI] [PubMed] [Google Scholar]
- Brunet Y. R., Zoued A., Boyer F., Douzi B., Cascales E. (2015). The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 11:e1005545. 10.1371/journal.pgen.1005545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza N. C., Shanks R. M., O’Toole G. A. (2005). Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187 7351–7361. 10.1128/JB.187.21.7351-7361.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. Y. (2017). Surface sensing for biofilm formation in Pseudomonas aeruginosa. Front. Microbiol. 8:2671. 10.3389/fmicb.2017.02671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zou Y., Kronfl A. A., Wu Y. (2020). Type VI secretion system of Pseudomonas aeruginosa is associated with biofilm formation but not environmental adaptation. Microbiologyopen 9:e991. 10.1002/mbo3.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli F. R., Monlezun L., Coulthurst S. J. (2016). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24 51–62. 10.1016/j.tim.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Corbitt J., Yeo J. S., Davis C. I., LeRoux M., Wiggins P. A. (2018). Type VI secretion system dynamics reveals a novel secretion mechanism in Pseudomonas aeruginosa. J. Bacteriol. 200 e744–e717. 10.1128/JB.00744-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois A. P., Coote P. J. (2011). Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 66 1785–1790. 10.1093/jac/dkr198 [DOI] [PubMed] [Google Scholar]
- Deziel E., Lepine F., Milot S., Villemur R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149(Pt 8), 2005–2013. 10.1099/mic.0.26154-0 [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. (1980a). Broad host range DNA cloning system for Gram-negative bacteria - construction of a gene bank of Rhizobium-Meliloti. Proc. Natl. Acad. Sci. U.S.A. 77 7347–7351. 10.1073/pnas.77.12.7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. (1980b). Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 77 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E. (2003). Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5 1213–1219. 10.1016/j.micinf.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Duan K., Dammel C., Stein J., Rabin H., Surette M. G. (2003). Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50 1477–1491. 10.1046/j.1365-2958.2003.03803.x [DOI] [PubMed] [Google Scholar]
- Durand E., Zoued A., Spinelli S., Watson P. J., Aschtgen M. S., Journet L., et al. (2012). Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J. Biol. Chem. 287 14157–14168. 10.1074/jbc.M111.338731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essar D. W., Eberly L., Hadero A., Crawford I. P. (1990). Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172 884–900. 10.1128/jb.172.2.884-900.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A., Hachani A., Bleves S. (2008). The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154(Pt 6), 1570–1583. 10.1099/mic.0.2008/016840-0 [DOI] [PubMed] [Google Scholar]
- Gallique M., Bouteiller M., Merieau A. (2017). The type VI secretion system: a dynamic system for bacterial communication? Front. Microbiol. 8:1454. 10.3389/fmicb.2017.01454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Wu Q., Bai D., Liu Y., Chen L., Jin S., et al. (2016). Potential use of dimethyl sulfoxide in treatment of infections caused by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60 7159–7169. 10.1128/AAC.01357-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Gobble T. R., Schuster M. (2009). GidA posttranscriptionally regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 191 5785–5792. 10.1128/JB.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Wang T., Chen G., Pu Q., Liu Q., Zhang Y., et al. (2019). A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 15:e1008198. 10.1371/journal.ppat.1008198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler S., Parsek M. R. (2010). Biofilms 2009: new perspectives at the heart of surface-associated microbial communities. J. Bacteriol. 192 2941–2949. 10.1128/JB.00332-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Heurlier K., Williams F., Heeb S., Dormond C., Pessi G., Singer D., et al. (2004). Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186 2936–2945. 10.1128/jb.186.10.2936-2945.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmelo L. R., Borlee B. R., Almblad H., Love M. E., Randall T. E., Tseng B. S., et al. (2015). Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10 1820–1841. 10.1038/nprot.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B. T., Dong T. G., Mekalanos J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15 9–21. 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 77–86. 10.1016/s0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Romling U., Tummler B. (1994). Genomic mapping of Pseudomonas aeruginosa PAO. Microbiology 140(Pt 11), 2907–2929. [DOI] [PubMed] [Google Scholar]
- Jiang F., Waterfield N. R., Yang J., Yang G., Jin Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15 600–610. 10.1016/j.chom.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Kong W., Chen L., Zhao J., Shen T., Surette M. G., Shen L., et al. (2013). Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 88 784–797. 10.1111/mmi.12223 [DOI] [PubMed] [Google Scholar]
- Laarman A. J., Bardoel B. W., Ruyken M., Fernie J., Milder F. J., van Strijp J. A., et al. (2012). Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J. Immunol. 188 386–393. 10.4049/jimmunol.1102162 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau G. W., Hassett D. J., Britigan B. E. (2005). Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 13 389–397. 10.1016/j.tim.2005.05.011 [DOI] [PubMed] [Google Scholar]
- Lennings J., Mayer C., Makhlouf M., Brotz-Oesterhelt H., Schwarz S. (2019). Polar localization of the ATPase ClpV-5 occurs independent of type VI secretion system apparatus proteins in Burkholderia thailandensis. BMC Res. Notes 12:109. 10.1186/s13104-019-4141-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesic B., Starkey M., He J., Hazan R., Rahme L. G. (2009). Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155 2845–2855. 10.1099/mic.0.029082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. S., Zhang W. P., Cheng J. L., Yang X., Zhu K. X., Wang Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. 10.1038/ncomms14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y. L., Shen L., Chang C. H., Bhuwan M., Chiu C. H., Chang H. Y. (2016). Regulation of motility and phenazine pigment production by FliA is cyclic-di-GMP dependent in Pseudomonas aeruginosa PAO1. PLoS One 11:e0155397. 10.1371/journal.pone.0155397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logger L., Aschtgen M. S., Guerin M., Cascales E., Durand E. (2016). Molecular dissection of the interface between the type VI secretion tssm cytoplasmic domain and the TssG baseplate component. J. Mol. Biol. 428 4424–4437. 10.1016/j.jmb.2016.08.032 [DOI] [PubMed] [Google Scholar]
- Marsden A. E., Intile P. J., Schulmeyer K. H., Simmons-Patterson E. R., Urbanowski M. L., Wolfgang M. C., et al. (2016). Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 198 1442–1450. 10.1128/JB.00049-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthey N., Stutzmann S., Stoudmann C., Guex N., Iseli C., Blokesch M. (2019). Neighbor predation linked to natural competence fosters the transfer of large genomic regions in Vibrio cholerae. eLife 8:e48212. 10.7554/eLife.48212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi D. V., Bonsall R. F., Delaney S. M., Soule M. J., Phillips G., Thomashow L. S. (2001). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183 6454–6465. 10.1128/JB.183.21.6454-6465.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migiyama Y., Yanagihara K., Kaku N., Harada Y., Yamada K., Nagaoka K., et al. (2016). Pseudomonas aeruginosa bacteremia among immunocompetent and immunocompromised patients: relation to initial antibiotic therapy and survival. Jpn. J. Infect. Dis. 69 91–96. 10.7883/yoken.JJID.2014.573 [DOI] [PubMed] [Google Scholar]
- Mittal R., Aggarwal S., Sharma S., Chhibber S., Harjai K. (2009). Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J. Infect. Public Health 2 101–111. 10.1016/j.jiph.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Moore M. L., Stokes K. L., Hartert T. V. (2013). The impact of viral genotype on pathogenesis and disease severity: respiratory syncytial virus and human rhinoviruses. Curr. Opin. Immunol. 25 761–768. 10.1016/j.coi.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso J. A., Mikkelsen H., Heeb S., Williams P., Filloux A. (2011). The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13 3128–3138. 10.1111/j.1462-2920.2011.02595.x [DOI] [PubMed] [Google Scholar]
- O’Toole G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47:2437. 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozyurek S. B., Gur S. D., Bilkay I. S. (2011). Production of pyocyanin pigment from Pseudomonas aeruginosa strains and investigation of the antimicrobial effect of pyocyanin on other microorganisms. Curr. Opin. Biotechnol. 22 S113–S113. 10.1016/j.copbio.2011.05.363 [DOI] [Google Scholar]
- Pasqua M., Visaggio D., Lo Sciuto A., Genah S., Banin E., Visca P., et al. (2017). Ferric uptake regulator Fur is conditionally essential in Pseudomonas aeruginosa. J. Bacteriol. 199:e00472-17. 10.1128/JB.00472-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pissaridou P., Allsopp L. P., Wettstadt S., Howard S. A., Mavridou D. A. I., Filloux A. (2018). The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc. Natl. Acad. Sci. U.S.A. 115 12519–12524. 10.1073/pnas.1814181115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planamente S., Salih O., Manoli E., Albesa-Jove D., Freemont P. S., Filloux A. (2016). TssA forms a gp6-like ring attached to the type VI secretion sheath. EMBO J. 35 1613–1627. 10.15252/embj.201694024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M. H., Kornberg A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97 4885–4890. 10.1073/pnas.060030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtke M. T., Borlee B. R., Murakami K., Irie Y., Hentzer M., Nielsen T. E., et al. (2012). Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78 5060–5069. 10.1128/AEM.00414-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana T. G., Berni B., Bleves S. (2016). The T6SSs of Pseudomonas aeruginosa strain PA01 and their effectors: beyond bacterial-cell targeting. Front. Cell. Infect. Microbiol. 6:61. 10.3389/fcimb.2016.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana T. G., Hachani A., Bucior I., Soscia C., Garvis S., Termine E., et al. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J. Biol. Chem. 287 27095–27105. 10.1074/jbc.M112.376368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana T. G., Soscia C., Tonglet C. M., Garvis S., Bleves S. (2013). Divergent control of two type VI secretion systems by RpoN in Pseudomonas aeruginosa. PLoS One 8:e76030. 10.1371/journal.pone.0076030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C., Zentgraf H., Dersch P., Mogk A. (2005). ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol. Chem. 386 1115–1127. 10.1515/BC.2005.128 [DOI] [PubMed] [Google Scholar]
- Sharp R., Jansons I. S., Gertman E., Kropinski A. M. (1996). Genetic and sequence analysis of the cos region of the temperate Pseudomonas aeruginosa bacteriophage, D3. Gene 177 47–53. 10.1016/0378-1119(96)00268-5 [DOI] [PubMed] [Google Scholar]
- Si M., Zhao C., Burkinshaw B., Zhang B., Wei D., Wang Y., et al. (2017). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. U.S.A. 114 E2233–E2242. 10.1073/pnas.1614902114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1 784–791. [Google Scholar]
- Singh M., Yau Y. C. W., Wang S., Waters V., Kumar A. (2017). MexXY efflux pump overexpression and aminoglycoside resistance in cystic fibrosis isolates of Pseudomonas aeruginosa from chronic infections. Can. J. Microbiol. 63 929–938. 10.1139/cjm-2017-0380 [DOI] [PubMed] [Google Scholar]
- Stefani S., Campana S., Cariani L., Carnovale V., Colombo C., Lleo M. M., et al. (2017). Relevance of multidrug-resistant Pseudomonas aeruginosa infections in cystic fibrosis. Int. J. Med. Microbiol. 307 353–362. 10.1016/j.ijmm.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Tremblay J., Richardson A. P., Lepine F., Deziel E. (2007). Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9 2622–2630. 10.1111/j.1462-2920.2007.01396.x [DOI] [PubMed] [Google Scholar]
- Valentini M., Filloux A. (2019). Multiple roles of c-di-GMP signaling in bacterial pathogenesis. Annu. Rev. Microbiol. 73 387–406. [DOI] [PubMed] [Google Scholar]
- Venturi V. (2006). Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30 274–291. 10.1111/j.1574-6976.2005.00012.x [DOI] [PubMed] [Google Scholar]
- Wang T., Si M., Song Y., Zhu W., Gao F., Wang Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. 10.1371/journal.ppat.1005020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B., Hasic M., Chen C., Wai S. N., Milton D. L. (2009). Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 11 3018–3028. 10.1111/j.1462-2920.2009.02005.x [DOI] [PubMed] [Google Scholar]
- Wettstadt S., Wood T. E., Fecht S., Filloux A. (2019). Delivery of the Pseudomonas aeruginosa phospholipase effectors PldA and PldB in a VgrG- and H2-T6SS-dependent manner. Front. Microbiol. 10:1718. 10.3389/fmicb.2019.01718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willyard C. (2017). The drug-resistant bacteria that pose the greatest health threats. Nature 543:15. 10.1038/nature.2017.21550 [DOI] [PubMed] [Google Scholar]
- Wilson R., Sykes D. A., Watson D., Rutman A., Taylor G. W., Cole P. J. (1988). Measurement of Pseudomonas-aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56 2515–2517. 10.1128/Iai.56.9.2515-2517.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Hu S., Ma N., Song P., Liang Q., Zhang H., et al. (2019). Regulatory effect of DNA topoisomerase I on T3SS activity, antibiotic susceptibility and quorum-sensing-independent pyocyanin synthesis in Pseudomonas aeruginosa. Int. J. Mol. Sci. 20:1116. 10.3390/ijms20051116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoued A., Durand E., Bebeacua C., Brunet Y. R., Douzi B., Cambillau C., et al. (2013). TssK Is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J. Biol. Chem. 288 27031–27041. 10.1074/jbc.M113.499772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoued A., Durand E., Brunet Y. R., Spinelli S., Douzi B., Guzzo M., et al. (2016). Priming and polymerization of a bacterial contractile tail structure. Nature 531 59–63. 10.1038/nature17182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data can be found in the NCBI database, the data was assigned a accession number of GSE148116.