Abstract
The identification of causal variants and mechanisms underlying complex disease traits in humans is important for the progress of human disease genetics; this requires finding strategies to detect functional regulatory variants in disease-relevant cell types. To achieve this, we collected genetic and transcriptomic data from the aortic endothelial cells of up to 157 donors and four epigenomic phenotypes in up to 44 human donors representing individuals of both sexes and three major ancestries. We found thousands of expression quantitative trait loci (eQTLs) at all ranges of effect sizes not detected by the Gene-Tissue Expression Project (GTEx) in human tissues, showing that novel biological relationships unique to endothelial cells (ECs) are enriched in this dataset. Epigenetic profiling enabled discovery of over 3,000 regulatory elements whose activity is modulated by genetic variants that most frequently mutated ETS, AP-1, and NF-kB binding motifs, implicating these motifs as governors of EC regulation. Using CRISPR interference (CRISPRi), allele-specific reporter assays, and chromatin conformation capture, we validated candidate enhancer variants located up to 750 kb from their target genes, VEGFC, FGD6, and KIF26B. Regulatory SNPs identified were enriched in coronary artery disease (CAD) loci, and this result has specific implications for PECAM-1, FES, and AXL. We also found significant roles for EC regulatory variants in modifying the traits pulse pressure, blood protein levels, and monocyte count. Lastly, we present two unlinked SNPs in the promoter of MFAP2 that exhibit pleiotropic effects on human disease traits. Together, this supports the possibility that genetic predisposition for complex disease is manifested through the endothelium.
Keywords: genomics, epigenetics, association mapping, endothelial cells, complex disease
Introduction
Over the past decade, genome-wide association studies (GWASs) for complex disease traits have established that roughly 90% of the detectable signals reside in the non-protein-coding genome. This suggests that a considerable proportion of genetic risk is conferred through perturbations of gene regulation.1 Compared to protein-coding variants, identification of the underlying mechanisms affecting complex disease through regulation is challenging because it often requires contextual information about regulatory elements, target genes, operational cell types, tissues, and organ systems. It is therefore valuable that large consortia such as the Encyclopedia of DNA Elements (ENCODE),2 the Roadmap Epigenomics Project,3 the Gene-Tissue Expression Project (GTEx),4 and the 1000 Genomes Project5 are providing the scientific community with atlases for genetic, epigenetic, and gene expression profiles of numerous human tissues and cell lines.
A major challenge remains, however: non-coding gene regulatory elements, particularly enhancer elements, are frequently cell-type specific. Given that tissues are composed of multiple cell types, their regulatory profiles reflect the weighted sum of all composite cell types. Numerous single-cell sequencing studies underscore this fact by demonstrating that tissues are more heterogeneous than has previously been appreciated.6 The implication for the identification of functional non-coding variants is that cell-restricted enhancer profiles may be diluted and appear as noise if the cell type is rare, or may not be appreciated as truly cell-type specific. For these reasons, it is important to empirically measure gene expression and epigenetic profiles in pure cell populations or in single cells. In addition, it is particularly useful to know the identities of lineage-determining transcription factors (LDTFs) that define different cell types. LDTFs, also called “master regulators” or “pioneering factors,” establish and maintain cell-type identity7 and prime cell-type-specific responsiveness to new signals.8,9 We and others have shown that the identities of LDTFs can be ascertained from enriched DNA motifs in epigenomic profiles of pure cell types.8,10 Importantly, knowing these factors and cognate binding motifs improves predictions for fine-mapping functional non-coding genetic variants that effect enhancer function, target-gene regulation, and signal-dependent changes to enhancer activation when specific cell types encounter varied exposures.9,10 In this study, we measure binding of the ETS-related gene ERG, as a LDTF in endothelial cells (ECs).
ECs are a nearly ubiquitous yet dynamic cell type in the human body. They regulate vascular tone and mediate an anti-thrombotic surface that lines arteries, veins, and capillaries. ECs also participate in the onsets and progressions of most complex diseases. Their activation and dysfunction are associated with common disease pathologies including atherosclerosis, hypertension, and respiratory diseases.11,12
In the current study, we performed epigenetic and gene expression mapping in ECs originating from aortic explants in up to 157 different human donors from three ancestral populations including both sexes. We considered the epigenetic profiles as locus-specific quantitative traits, and tested genotypes that are associated with differences in epigenetics and gene expression in cis, under both normal and pro-inflammatory conditions. This design enabled identification of thousands of expression quantitative trait loci (eQTLs) and hundreds to thousands of epigenetic molecular quantitative trait loci (molQTLs). Many variants associated with both epigenetic and expression traits, providing direct evidence for thousands of functional regulatory SNPs in ECs. To prioritize variants in this set to those that predispose individuals to complex diseases, we intersected mol/eQTL co-mapped SNPs with disease loci from GWASs and identified several common variants linked to altered regulatory function, gene expression, and disease. While this study serves as a valuable resource for research in vascular biology and is a proof-of-principle study for the utility of epigenetic quantitative trait locus (QTL) analysis in a pure cell type for functional human genetics, it represents only a foundational step on the path toward comprehensively understanding the complement of diverse regulatory programs that vary in the human genome.
Material and Methods
Cell Culture and Collection
Human aortic endothelial cells (HAECs) were isolated from de-identified deceased heart donor aortic trimmings at the University of California Los Angeles Hospital as described previously.13 Donor cells from up to 53 individuals were expanded and used for chromatin immunoprecipitation with sequencing (ChIP-seq), RNA sequencing (RNA-seq), and assay for transposase-accessible chromatin with sequencing (ATAC-seq) assays. Cells were treated prior to harvest for 4 h with 10 ng/mL human recombinant IL-1B protein or no additional protein.
High-throughput Sequencing
For RNA-seq, total RNA was extracted and polyA was selected ahead of library construction using previously described methods.14 Cells used for ChIP-seq were fixed with 1% formaldehyde or 1% formaldehyde and 2 nM disuccinimidyl glutarate according to previously described methods.14 ATAC-seq was performed according to the originally published protocol15 with an added size selection of 175–225 bp on Tris-borate-EDTA gels. Sequence libraries were prepared as previously described14 and sequenced on an Illumina HiSeq 4000. For quality control of all sequencing assays, samples were removed from further analysis if: (1) the sample contained less than three million unique mapped reads, (2) the sample had an average of six or more duplicate tags per site, (3) the sample was an extreme outlier in principal-component analysis (PCA), or (4) if the RNA-based genotyping at heterozygous loci could not replicate the genotyped-based identity on file for that individual through the use of hierarchical clustering of variant call format (VCF) files.
Microarray data of was provided from previous publications16,13 on 157 HAEC donors. Subsets of the same donors’ cells were grown again and used for RNA-seq, ATAC-seq, and ChIP-seq in this study. Microarrays were used to examine gene expression in 157 donors in untreated conditions and 156 donors in Oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (oxPAPC) treated conditions (see Supplemental Methods). These are publicly available at NCBI GEO database: GSE30169, GSE139377.
Mapping and Processing
The sequencing data were mapped utilizing Bowtie217 with default parameters. Mapping bias correction and duplicate read removal were implemented using the software package WASP.18 The correction resulted in a mapped binary alignment map (BAM) file for use in allele-specific analysis.
Genotyping
Genomic DNA was isolated from HAECs and genotyped according to the Affymetrix Genome-wide Human SNP Array 6.0 assay protocol and is available in the database of Genotypes and Phenotypes (dbGaP; see Web Resources). The image data were processed as described previously13 for determining the specific hybridizing signal for each SNP call and copy-number detection. IMPUTE219,20 was used to impute genotypes utilizing all populations from the 1000 Genomes Project reference panel. Genotypes were called for imputed SNPs with allelic R2 values greater than 0.9.
VCF File Preparation
To reduce multiple testing and to avoid false positives, VCF files were restricted using vcftools to only include genetic variants a minor allele frequencies of at least 0.05.21 This was performed for each assay treatment set individually to account for the differences in donor numbers passing the quality control measures from above.
Covariate Discovery
Because the HAECs were de-identified, biological sex and ethnicity were determined from genotyping data. Ancestry was determined through the use of PCA clusters using genotypes of HAEC donors and 1000 Genomes Project individuals of known ancestry. Biological sex was determined in PLINK22 based on heterozygosity on X chromosomes. To avoid spurious associations based on population stratification, and to power discovery of cis-acting genetic variants, we applied PEER to remove known and hidden systemic signals from normalized gene expression measured by RNA-seq and microarray (see Supplemental Methods).
eQTL Analysis
eQTL analysis was done via linear regression in MatrixeQTL. MatrixeQTL was run using a reads per kilobase million (RPKM)-normalized expression matrix generated in HOMER (see Supplemental Methods), a VCF file, and a covariate file containing biological sex, unique total tag counts, the top four principal components (PCs) from a PCA performed on the genotypes to account for ancestry, and either fifteen factors (RNA-seq) or thirty factors (microarray) discovered by PEER. SNPs were tested against a gene’s expression if they were in cis (i.e., within 1Mb of the gene). Results were restricted to a gene-level Benjamini-Hochberg false discovery rate (FDR) of less than 5% (see Supplemental Methods for more).
To determine the similarity between the eQTL results from the RNA-seq datasets and the microarray datasets, pairwise comparisons were made using the effect sizes of significant SNPs in one set, which were graphically compared to the effect sizes of the same SNPs in the second dataset regardless of significance in the second set.
ChIP-seq and ATAC-seq QTL Analysis
molQTL mapping analysis was performed using the software package RASQUAL,23 which incorporates allele-specific mapping information at heterozygous SNPs. The molQTL analysis using RASQUAL23 was run with VCF files containing allele-specific counts, a RPKM-normalized tag matrix, and the covariates: sex, unique total tag counts, and the first four genotype-generated PCs to adjust for ancestry. SNPs were tested for association against normalized tag counts at epigenetic peaks if they were within the boundaries of the peak. The results were filtered using a per-site FDR of 5%.
GWAS Comparison
The enrichment of eQTLs in CAD GWAS data was done by comparison of observed, experimental overlap between HAEC eQTL SNPs with the CAD GWAS SNPs and results from 1,000 random permutations’ significant CAD SNPs with HAEC eQTLs. Significance was assessed using Fisher’s Exact Test. For more details, see Supplemental Methods. The R package “coloc” was also used to verify the colocalization of the CAD GWAS SNPs and the eQTLs found in this study. This was run using crude p values, minor allele frequencies, and sample numbers using the command coloc.abf().
Motif Enrichment and Motif Mutation Analysis
Motif enrichment analysis in sequences underlying regulatory elements was performed in HOMER using findMotifsGenome.pl. Motif mutations were detected when the local sequence was altered by alleles of a SNP such that one allele dropped the match to the motif’s position weight matrix (PWM) below the motif detection threshold that is defined in the HOMER motif database.8 Significance testing for effects of motif mutations on epigenetic trait pi values (from RASQUAL) was performed using unpaired two-tailed t tests assuming unequal variance (e.g., Figures 3C and 4C–D. Pi values that deviate from 0.5 indicate allele-specific effects.
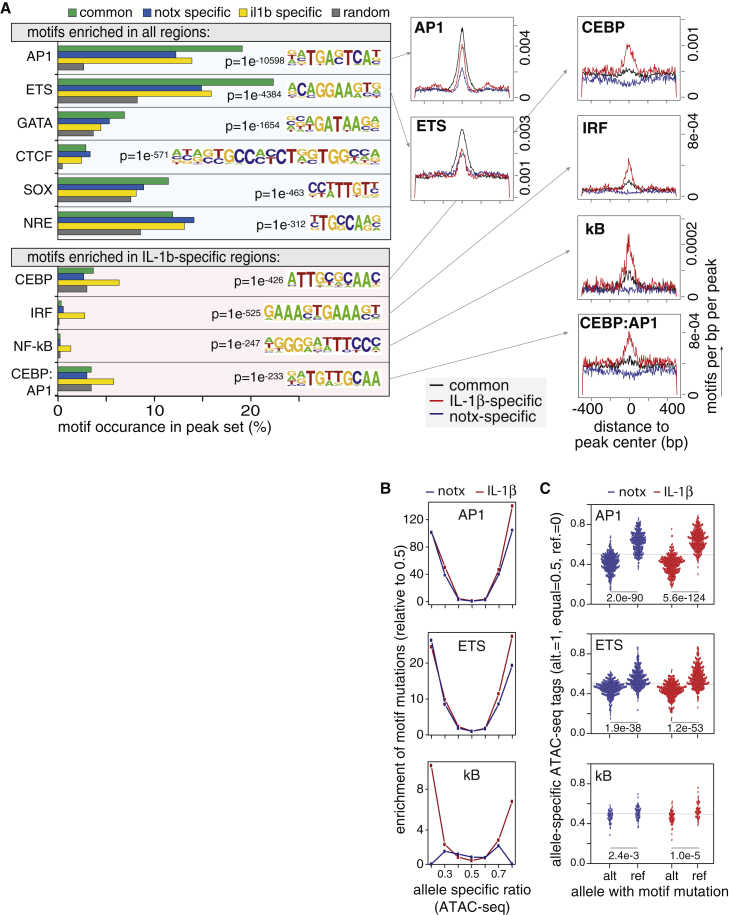
Figure 3.
Motif Enrichment Analysis of EC Regulatory Elements and molQTLs
(A) Discovery of TF binding motifs enriched in all regulatory elements (top), or those specific to IL-1b treatment (bottom). p values for enrichment are compared to random background (top), or untreated (bottom)., Respective densities of these motifs are shown to the right relative to the summit of the ATAC-seq-defined peak.
(B) Y-axes are enrichment of motif mutations in bins of allele-specific ATAC-seq tags on the x-axes for each motif indicated in notx (blue) and IL-1b (red) datasets as normalized to 0.5 (equal tags on ref versus alt). x values of 0 = exclusive openness on ref.; values of 1 = exclusive openness on the alt.
(C) Instances of motif mutations created by alt. or ref. alleles (x axis) show expected effects on ratios of allele-specific accessibility (y axis) for AP-1, ETS, and kB motif mutations in notx (blue) and IL-1b-treated (red) ATAC-seq datasets. p values are from an unpaired two-tailed t test assuming unequal variance.
Figure 4.
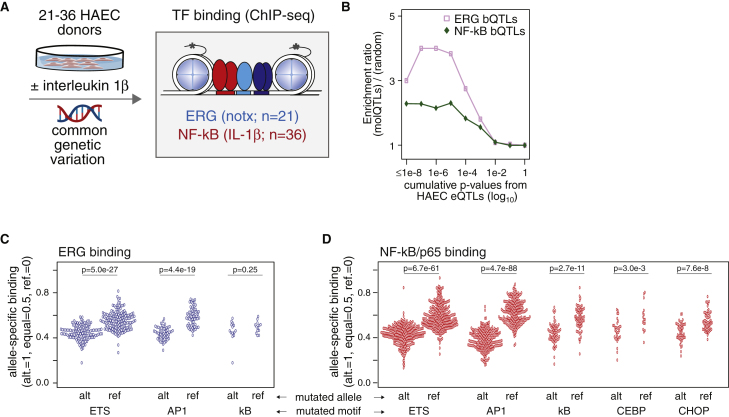
Transcription Factor Binding QTL Analysis
(A) Schematic of ChIP-seq collection and sample size for binding QTL analysis.
(B) Enrichment of bQTLs in cumulative p value bins for RNA-seq eQTLs of corresponding treatment.
(C) Allele-specific ratios (y axis) for ERG binding by motif mutations in ETS, AP-1, and NF-kB motifs (x axis).
(D) Allele-specific ratios (y axis) for NF-kB binding by motif mutations in ETS, AP-1, NF-kB, CEBP, and CHOP motifs (x axis). p values are from an unpaired two-tailed t test assuming unequal variance.
Biological Validation
For the dual luciferase reporter assay, 198 bp fragments of the enhancer regions were cloned into Addgene plasmid #99297 (see Web Resources),24 which was co-transfected into telomerase-immortalized human aortic endothelial cells (teloHAECs) with the control vector pGL4.75 (Promega), which encodes the luciferase gene hRluc (Renilla reniformis). Luciferase activity was measured 48 h post-transfection. The data are presented proportional to the control vector. Three independent experiments with four technical replicates were performed. Intra-haplotype or haplotype-control statistical analyses were performed with two-tailed t test.
For the CRISPR interference (CRISPRi) assay, a fusion protein of catalytically dead Cas9 (dCas9) fused to KRAB repressor protein (addGene cat#46911) was produced in HAECs via transfection of in vitro transcripts. 8 h post-transfection, cells were lysed for RNA collection, cDNA synthesis, and analysis via quantitative polymerase chain reaction (qPCR). A guide RNA sequence targeted to a previously identified endothelial enhancer in PLPP3 rs17114036 locus was used as a positive control.
Results
eQTL Mapping in HAECs
We measured gene expression through the use of RNA-seq in Human Aortic Endothelial Cell (HAEC; EC for short) primary cultures from 53 individuals’ ECs in two different environments: control (untreated) and pro-inflammatory cytokine interleukin 1 beta (IL-1b) treated (Table S1; with an average per-individual unique mapped tag read count of 12,120,267). We also utilized microarray-based expression data from our previous study in which 157 EC donors were cultured at low passage with and without treatment with pro-inflammatory oxidized phospholipid (oxPL)13 To identify genetic variants that are associated with gene expression, we performed eQTL mapping using Matrix eQTL on a total of four EC datasets: microarray-determined expression in untreated (notx) and oxPL-treated ECs, and RNA-seq-determined expression in notx and IL-1b-treated ECs (overview in Figure 1A and Table 1). RNA-seq and microarray eQTL results were each utilized throughout this study as deemed appropriate for the questions asked. Notably, 5,784 more transcripts were tested for RNA-seq-based eQTL analysis than for microarray due to the unbiased nature of RNA-seq-based eQTL analysis in sampling mRNAs independent of genomic build.
Figure 1.
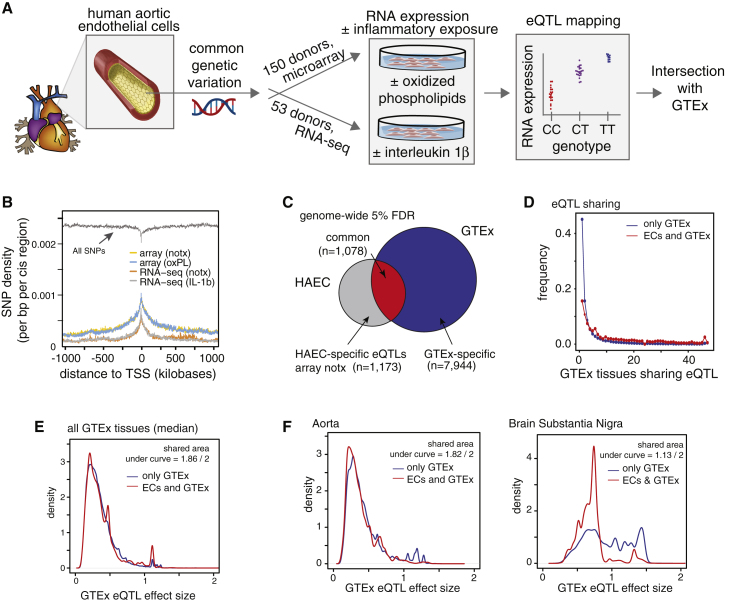
eQTL Analysis in ECs and Comparison to GTEx eQTLs
(A) Pipeline of eQTL analysis.
(B) Density plot of SNP localization relative to transcriptional start sites (TSSs).
(C) Venn diagram of unique eQTL transcripts in HAECs (5% FDR), shared between HAECs and any GTEx tissue, or unique to GTEx. All eQTLs were tested in both datasets.
(D) Histogram of eQTL sharing across GTEx tissues (number of tissues with eQTL on x axis) for two sets: eQTLs only in GTEx (blue) or in GTEx and HAECs (red).
(E) Histogram of the median GTEx eQTL effect sizes across all tissues for the array notx eQTL dataset.
(F) Histogram of median GTEx eQTL effect sizes in two exemplar comparisons: aorta and substantia nigra.
Table 1.
Summary of eQTL Analysis in Endothelial Cells
| IL-1B | oxPL | Number of Transcripts with cis-Variation at MAF > 5% Tested for Association | Number of Significant SNP-Transcript Pairs | Number of Lead Significant SNP-Transcript Pairs (R2 < 0.6) | Unique Transcripts with ≥ 1 eQTL | % of Transcripts Tested with ≥ 1 eQTL | % of Variants Tested with Significant eQTL |
|---|---|---|---|---|---|---|---|
| RNA-seq | |||||||
| - | - | 15,045 | 180,956 | 9,446 | 1,666 | 11.07% | 2.01% |
| + | - | 14,790 | 207,954 | 10,767 | 1,804 | 12.20% | 2.35% |
| Array | |||||||
| - | - | 11,878 | 571,161 | 43,619 | 3,887 | 20.90% | 4.19% |
| - | + | 11,878 | 568,262 | 43,330 | 3,795 | 20.41% | 4.12% |
| Total | |||||||
| N/A | N/A | 20,187 | 725,845 | NA | 4,911 | 24.33% | 7.45% |
Transcripts were associated with SNPs in cis (1 Mb from gene) for each expression quantitative trait locus (eQTL) dataset (row). These datasets were used to discover functional regulatory variants in this study. Significance is defined by locus-wide Benjamini-Hochberg false discovery rate correction. IL-1b—interleukin 1 beta. N/A—not applicable. oxPL—oxidized phospholipid. MAF—minor allele frequency. RNA-seq—RNA sequencing.
Focusing on cis-eQTLs, called eQTLs hereafter, discovered at 5% locus-wide false discovery (within 1 Mb of promoters), we observed depletion of SNPs tested at transcription start sites (TSS) and enrichment of significant eQTLs near TSSs and in gene bodies (Figure 1B, Figure S1B); this result demonstrates that gene bodies are enriched for functional SNPs in this EC population. Using eQTL effect sizes, which reflect the direction and magnitude of allelic effects on associated genes, we found high concordance in eQTLs called across our datasets (correlation p < 2 × 10-16, Figure S1C and S1D); this concordance demonstrated that EC eQTL results were robust to technical differences in platform and culture batch.
Half of Endothelial eQTLs are Not in GTEx
With data from 46 human tissues and two cell lines, GTEx is currently the largest compendium of eQTLs.4 GTEx does not include pure EC samples, so we sought to compare our most statically powered EC eQTLs (the locus- and genome-wide corrected, array-based notx set from 157 people) with GTEx tissue eQTLs. We found that about one-half of EC eQTLs were present in at least one GTEx tissue, and most EC eQTLs were evident in less than 10 GTEx tissues (Figure 1C and 1D). Because sample number and genotypic effect size in part determine statistical power to detect eQTLs, we compared whether the eQTL effect size showed any difference in magnitude between eQTLs common to GTEx and ECs versus GTEx-only eQTLs. We found some variation in shared effect sizes between eQTLs unique to GTEx versus shared with ECs (Figure 1E, values in Table S2), but found no trend suggesting that our EC eQTLs were selectively enriched in large GTEx effect sizes. Instead, we found similar distributions of effect sizes overall with two examples of the extremes in Figure 1F. These data show that shared GTEx/EC eQTLs had similar effect sizes in the aortic artery to those unique to GTEx, whereas shared eQTL effects in the substantia nigra brain region were smaller relative to eQTLs specific to this brain region. Comparison of effect sizes measured in ECs revealed no global differences in magnitude between eQTLs shared with GTEx and EC-only sets (Figure S1E). Together, these data show that our dataset uncovered about 1,000 EC eQTLs that were not detected in tissue samples, and the data also demonstrate that genotypic effects in ECs are not merely a subset of large or small effects in tissues.
Epigenetic Profiles Are Genetically Regulated by cis-Variants and Enriched for eQTLs
Given the prevalence of non-coding eQTL SNPs (Figure S1B), which is suggestive of perturbations to gene regulatory function, we mapped variants that perturb regulatory function in human ECs (see Figure 2A for an overview of our strategy). To locate regulatory elements and quantify their activity, we performed two epigenomic assays: (1) ChIP-seq for acetylation of lysine 27 on histone H3 (H3K27ac), which is a robust and quantitative marker of active regulatory elements at both promoters and enhancers,25 and (2) the assay for ATAC-seq15 that provides highly resolved positions of open, transcription factor (TF)-bound chromatin. These sequencing assays had average per-individual unique mapped reads of 13,956,326 and 11,547,593, respectively (Table S1). Regulatory elements were defined as loci with an ATAC-seq peak and adjacent H3K27ac signal in at least one individual across EC donors in untreated notx (n = 44 donors) or IL-1b-treated (n = 43 donors) conditions. This resulted in 109,817 regulatory elements common to both treatments, 46,263 specific to untreated, and 49,874 specific to IL-1b-treated ECs (Figure 2B 5% FDR).
Figure 2.
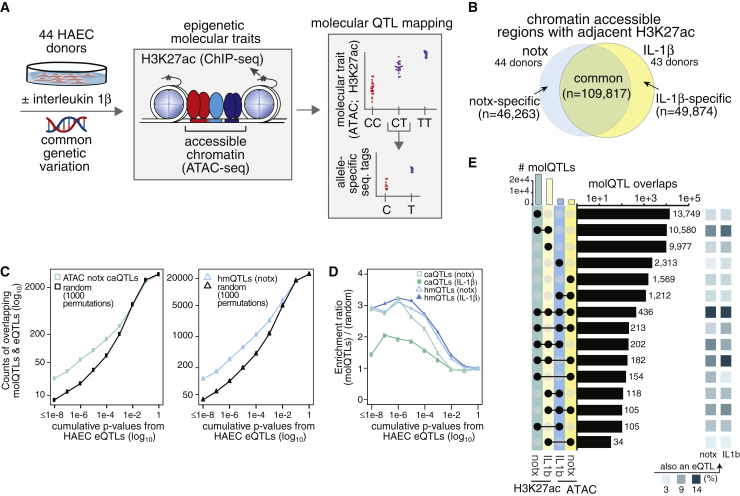
Regulatory Element Identification and molQTL Analysis
(A) Pipeline of chromatin accessibility and histone modification QTL analysis.
(B) Venn diagram of the number of regulatory elements identified in notx ECs and IL-1B treated ECs using ATAC-seq-defined open chromatin with adjacent H3K27ac marks.
(C) Numbers of untreated caQTLs (left) and hmQTLs (right) in RNA-seq EC eQTLs, for datasets of corresponding treatment, are plotted in blue for increasing cumulative p value thresholds (x axis) with empirical estimates of random samplings by 1,000 permutations (black).
(D) Enrichment scores for caQTLs and hmQTLs as a function of cumulative significance thresholds (x), calculated by dividing expected by random values in c. molQTL enrichment is calculated in the RNA-seq eQTL dataset of corresponding treatment.
(E) Upset plot showing co-mapping of molQTL SNPs between caQTL and hmQTL datasets with proportions of eQTLs for RNA-seq datasets are shown by colored boxes to right.
Next, we performed QTL analysis to identify molQTLs across our HAEC population. These molQTLs reflect non-random associations between genotype and the quantitative abundance of ATAC or H3K27ac ChIP sequence tags in the immediate cis region (within the peaks). Using the program RASQUAL, which combines terms for both allele-specific reads in heterozygotes and diploid genotypic effects across individuals,23 we identified thousands of molQTLs (Table 2). Variants associated with differential abundance in ATAC-seq data are termed chromatin accessibility QTLs (caQTLs), and those in H3K27ac ChIP-seq are termed histone modification QTLs (hmQTLs). Between 2,130 and 3,415 regulatory elements were significantly associated (FDR < 5%) with an underlying cis variant, reflecting precise instances whereby the presumed activities of regulatory elements are modulated by common genetic variation in human ECs (Table 2).
Table 2.
Summary of molQTL Analysis in Endothelial Cells
| IL-1B | # Peaks with cis-Variation MAF > 5% Tested for Association | Significant molQTLs (SNP-Peak Pairs) | Unique Peaks with ≥ 1 Significant molQTL | % Peaks Tested with ≥ 1 Significant molQTL | % Variants Tested Underlying Significant molQTLs |
|---|---|---|---|---|---|
| H3K27ac ChIP-seq | |||||
| - | 84,820 | 25,621 | 2,620 | 3.81% | 6.52% |
| + | 91,950 | 21,634 | 2,130 | 2.96% | 5.61% |
| ATAC-seq | |||||
| - | 435,081 | 3,905 | 2,815 | 3.46% | 3.62% |
| + | 390,257 | 4,704 | 3,415 | 4.88% | 4.97% |
| ERG ChIP-seq | |||||
| - | 69,342 | 557 | 354 | 1.40% | 1.52% |
| p65 ChIP-seq | |||||
| + | 154,714 | 5,791 | 3,742 | 6.19% | 6.41% |
Pileups of unique mapped tags at regulatory elements (peaks) per donor were used as quantitative traits in quantitative trait locus (QTL) mapping, and associations were termed molecular QTLs (molQTLs). molQTL results are shown for each epigenetic assay (rows), with significance defined by 5% false discovery at the locus level using RASQUAL software. IL-1b—interleukin 1 beta. MAF—minor allele frequency. H3K27ac— acetylation of lysine 27 on histone H3. ChIP-seq— chromatin immunoprecipitation with sequencing. ATAC-seq—assay for transposase-accessible chromatin with sequencing.
Co-mapping analysis revealed a significant enrichment of molQTL variants that were also eQTLs (Figure 2C), with H3K27ac hmQTLs being ∼3 times and caQTLs ∼2 times more likely to also have expression associations (p < 1 × 10-8) than by random expectation (Figure 2D). We observed many different combinations of overlap among molQTLs, with variants underlying hmQTLs in both datasets (n = 10,580) and variants at hmQTL+caQTLs in all datasets (n = 436) as most likely to also be eQTLs (Figure 2E). We interpret these data to mean that allelic perturbations affecting H3K27ac signals are most predictive of eQTLs, and this interpretation is consistent with a model whereby this post-translational modification closely reflects productive transcription.
Motif Mutation Analysis Identifies Genetic Variants Whose Alleles Confer cis-Regulatory Function
With the eventual goal of identifying putative causal non-coding variants, and because TF motif mutations are a powerful means to identify causal variants, we performed de novo motif enrichment analysis8 to investigate which TF motifs are enriched in EC regulatory elements. Consistent with our previous report in a single EC donor,14 we found the AP-1, ETS, and GATA DNA binding motifs were significantly enriched across the HAEC epigenetic landscape in all subsets of regulatory elements (subsets: notx-specific, IL-1b-specific, and common; Figure 3A, top). Additionally, we found that CEBP, IRF, NF-kB, and CEBP:AP-1 cognate motifs were enriched in regulatory elements gained upon IL-1b treatment (Figure 3A, bottom). Consistent with previous work,14 this suggests that TFs from these families regulate dynamic expression changes downstream of IL-1b signaling.
For these TF motifs, we tested the hypothesis that SNPs whose alleles differentiate between a “match” or a “mutation” of the motif should be enriched in elements that exhibit allele-specific molecular traits. Indeed, we found that SNPs whose alleles mutate AP-1, ETS, and NF-kB motifs were significantly enriched in the extremes of ATAC-seq allele-specific ratios, where 0.5 is an equal number of reads from both alleles in heterozygotes, and 0.0 and 1.0 are ATAC-seq reads that map exclusively to reference or alternative alleles, respectively (Figure 3B and 3C). Motif mutations to the kB motif were only enriched in allele-specific ATAC-seq regions after IL-1b treatment, and not in untreated cells; because NF-kB is only present in the nucleus after IL-1b treatment, this result provides confirmation that the motif mutation approach is able to capture functional “mutations.”
Binding QTL Analysis Reveals SNPs and Motifs that Affect DNA Binding by the EC Lineage-Determining Factor ERG and Signal-Dependent Factor NF-kB
With insights afforded by molQTL mapping, we collected ChIP-seq data for two TFs of particular importance to ECs: (1) the ETS-related gene, ERG, and (2) NF-kB using ChIP for the RelA/p65 subunit (Figure 4A). These sequencing assays had per-individual average unique mapped read counts of 9,220,510 and 10,109,014 respectively (Table S1). ERG was selected for ChIP-seq because it is one of the predominant binders to the ETS motif that is prevalent in endothelial enhancers14 (Figure 2A). Further, ERG is expressed predominantly in ECs, and is an essential gene for vascular development.14,26, 27, 28 We and others have shown that ERG upregulates quintessential EC-specific genes (e.g., NOS3, PECAM1, and VWF), and it directly or indirectly represses pro-inflammatory genes (e.g., IL-8, CCL2, and IL1B).14,29,30 The rationale for measuring NF-kb’s binding profile across donors included its role as a master pro-inflammatory TF downstream of many signals, including IL-1 signaling; we did this to inform interpretation of many diseases with a vascular inflammatory component.14
Using RASQUAL, we performed TF-binding QTL (bQTL) analysis for ERG in untreated ECs from 21 EC donors and NF-kB in IL-1b treated ECs from 36 donors. We discovered 354 and 3,742 binding regions that were significantly associated with at least one cis-variant (Table 2; 5% FDR). We reason that fewer ERG bQTLs were discovered relative to NF-kB based on lesser sample size, and therefore statistical power. Still, we observed enrichment of ERG and NF-kB bQTLs at eQTL loci (Figure 4B); this suggests that functional non-coding variants are enriched in our bQTL sets.
To fine-map functional regulatory variants and identify particular TF motifs whose mutation corresponds with diminished binding, we analyzed the relationship between ERG’s allelic binding ratios and the presence of motif mutations. ERG binding in untreated conditions, as expected, was influenced most by mutations to its respective ETS binding motif (Figure 4C, p = 5 × 10-27). More interestingly, ERG binding was also affected by mutations to the AP-1 motif (p = 4.4 × 10-19), whereas ERG binding was not affected by mutations in the kB motif (p = 0.25, Figure 4C). We interpret this to mean that ERG’s binding is influenced not only by the sequence it binds, but also by the coordinated binding of other TFs in the local chromatin landscape. Results from all tested motifs are in Table S3.
For NF-kB binding after IL-1b treatment, we found many TF motifs (e.g., ETS, AP-1, kB, CEBP, and CHOP), in which mutated alleles corresponded to less NF-kB binding relative to the non-mutated allele (all p < 3 × 10-3, Figure 4D). This demonstrates that, while differences in NF-kB binding are significantly affected by mutations in the kB motif, these instances are ∼4 times less frequent in the genome relative to differential NF-kB binding corresponding to ETS motif mutations, and ∼3 times less frequent than AP-1 motif mutations. This observation builds upon our previous report, which used a similar approach in macrophages taken from inbred strains of mice;9 however, the current study is a qualitative advancement in that we utilized allele-specific binding in heterozygous human cells in an outbred population. Together, these findings strongly support a model whereby signal-induced TFs like NF-kB bind chromatin with patterns that depend on previously established patterns via cell lineage-determining TFs (reviewed by Romanoski et al.31).
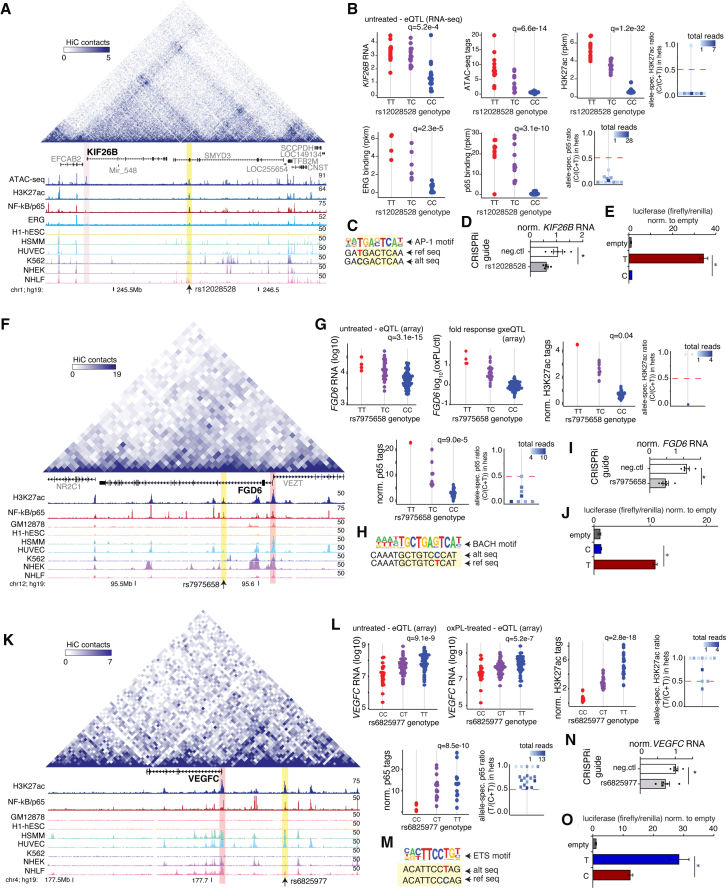
Identification of Enhancers and Functional Regulatory Variants for KIF26B, FGD6, and VEGFC
To examine the utility of our system’s genetic dataset, we cross-referenced EC eQTLs, molQTLs, and motif mutations to fine-map functional regulatory variants. In total, there were 2,818 instances of SNPs underlying all three data types. To exemplify these data, we focus on three examples: KIF26B, FGD6, and VEGFC (Figure 5).
Figure 5.
KIF26B, FGD6, and VEGFC Loci
(A) Browser-style track of KIF26B locus with 3D Hi-C data from HUVECs shown by heatmap above with epigenetic tracks shown below. Vertical yellow bar highlights enhancer-like region containing SNP of interest, with KIF26B promoter highlighted in pink. Cell types listed for H3K27ac are from E—NCODE. GM12878human B-lymphocyte-lymphoblastoid cell line; H1-hESC—human embryonic stem cells; HSMM—human skeletal muscle myoblasts; A549—human epithelial lung carcinoma-derived cell line; HUVEC—human umbilical vein endothelial cells; K562—human chronic myelogenous leukemia-derived cell line; NHEK—human epidermal keratinocytes; NHLF—human lung fibroblasts.
(B) The SNP shown in a is an eQTL for KIF26B, a molQTL for chromatin accessibility, H3K27ac (including allele-specific plot), ERG binding, and p65 binding (including allele-specific plot).
(C) rs12028528 mutates an AP-1 motif.
(D) Reduced KIF26B expression in HAECs treated with pooled gRNAs with CRISPR interference (CRISPRi) within 200 bp of rs12028528. n = 5. Expression measured by qPCR and normalized to GAPDH. Data show mean ± standard error of the mean. ∗p < 0.05, ∗∗∗p < 0.001 as determined from unpaired t test.
(E) The functional effect rs12028528 was replicated via luciferase assay in teloHAECs (n = 3 independent experiments; p < 0.05 by unpaired 2-tailed t test).
(F) FGD6 locus with SNP rs7975658 in an enhancer-like region (yellow) within FGD6 intron.
(G) Plots for eQTL for FGD6 in untreated and gene-by-environment eQTL (y axis = oxPL RNA − notx RNA for FGD6). This SNP is also an hmQTL with allele-specificity (within heterozygotes) and an NF-kB bQTL with allele-specificity.
(H) rs7975658 mutates a BACH motif.
(I) CRISPRi and guides at rs7975658 reduced FGD6 compared to control as in (D).
(J) Luciferase reporter assay in teloHAECs as in (E).
(K) VEGFC locus with SNP rs6825977 in an enhancer-like region (yellow), which loops with the VEGFC promotor (pink) based on high-throughput chromatin conformation capture (HiC) data.
(L) The SNP is an eQTL for VEGFC, an hmQTL with allele-specificity, and an NF-kB bQTL with allele-specificity.
(M) rs6825977mutates an ETS motif.
(N) CRISPRi with gRNA targeting rs6825977 reduced VEGFC RNA versus control as in (D).
(O) Luciferase reporter assay in teloHAECs as in (E).
As a first example, for KIF26B, RNA levels associated with genotypes at the SNP rs12028528, located ∼725kb downstream, in an intron of the neighboring gene SMYD3. Greater KIF26B expression associated with the T allele and less expression with C (Figure 5B). This SNP is within a chromatin-accessible, H3K27ac-positive regulatory element that is bound by ERG and NF-kB, and these quantitative measures each associate with genotypes at rs12028528 in the same direction as does KIF26B RNA (Figure 5B). Further analysis of 3D chromosome conformation capture Hi-C data from human umbilical vein ECs (HUVECs),32 shown in the triangular heatmap above the genomic tracks, revealed that this enhancer loops to physically interact with the genomic locus that contains the KIF26B promoter (Figure 5A, top). Motif mutation analysis revealed that the C allele of rs12028528 mutates an AP-1 motif, whereas the T allele maintains an intact motif (Figure 5C). Analysis of ENCODE H3K27ac ChIP-seq data in multiple cell types revealed that this enhancer is specific to endothelial cells (HUVECs) (Figure 5A). Using the CRISPRi approach (CRISPR/Cas9 + KRAB domain) to target the repressive machinery by guide RNA within 200 bp of rs12028528 resulted in ∼2 fold less KIF26B expression than did the non-targeted control (Figure 5D). Further, cloning of 198 bp of genomic sequence, centered on rs12028528, into a luciferase reporter and transfecting into ECs confirmed that the T allele-containing sequence had over 30 times greater enhancer activity than did the C-containing sequence (Figure 5E). KIF26B has been identified as a microtubule-associated kinesin protein that polarizes endothelial cells in response to sheer stress,33 and these data demonstrate that rs12028528 in a distal enhancer regulates its expression.
As a second example, at the FGD6 locus, shown in Figure 5F, the SNP rs7975658, located in the second intron of FGD6, is a significant eQTL for FGD6 and is also a molQTL for H3K27ac and NF-kB/p65 (Figure 5G). For each ‘omic assay, the T allele associates with greater expression, acetylation, and/or binding than does the C allele. Analysis of ENCODE data shows this enhancer-like element to be acetylated on H3K27 specifically in endothelial cells, suggesting cell-type specificity. Motif mutation analysis identified a BACH motif that is mutated by the C allele, consistent with the T allele having regulatory activity (Figure 5H). We validated enhancer activity from this locus on FGD6 through the use of CRSIPRi in ECs (Figure 5I). Lastly, we tested the allele-specific enhancer activity by using transient transfection of luciferase constructs into teloHAECs, and we found that only the T allele corresponded to enhancer function compared to both the empty vector (no enhancer) and the enhancer sequence with the C allele (Figure 5J).
As a third example, at the VEGFC locus, shown in Figure 5K, we found that SNP rs6825977 (∼100kb downstream of VEGFC) was both an eQTL for VEGFC and a molQTL for H3K27ac and NF-kB. The T allele corresponded to greater expression, acetylation, and binding (Figure 5L). Here, the T allele preserves an ETS motif, whereas the C allele mutates a key residue in the sequence (Figure 5M). Enhancer activity was confirmed using CRISPRi at this enhancer (Figure 5N), and was confirmed specifically for the sequence with the T allele, whereas the C allele did not enhance luciferase production above background (Figure 5O). Taken together, these data demonstrate how our unbiased genetics approach was able to identify functional regulatory variants and enhancers that direct target-gene expression in human ECs.
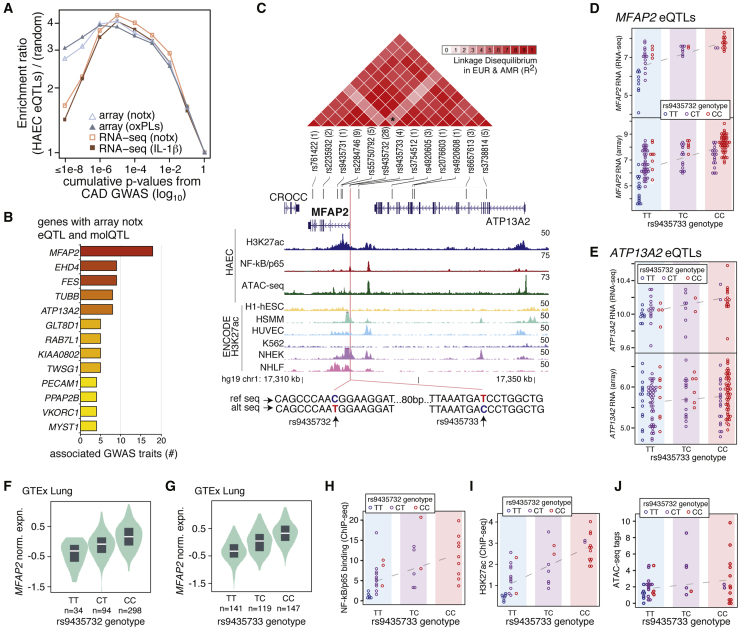
eQTLs are Enriched in CAD-Relevant Loci and molQTLs Refine Candidate Causal Variants
To investigate the utility of our datasets for fine-mapping functional variants underlying coronary artery disease (CAD), we cross-referenced EC eQTLs with summary statistics from the most recent meta-GWAS for CAD.34 We found that EC eQTLs are significantly enriched at CAD loci above randomly permuted rates of expected overlap (Figure 6A). EC eQTLs were enriched for both genome-wide significant CAD SNPs (p < 1 × 10-8), as well in CAD associations in the sub-genome-wide significant range (1 × 10-7 < p < 1 × 10-3). This finding strongly supports a model whereby the “mid-hanging fruit” in GWAS studies are functionally affecting biological pathways, and suggests that these effects for CAD are operating in part through ECs. Next, we cross-referenced significant molQTLs with the eQTL/CAD loci, resulting in a list of 18 variants associated with nine genes (Table S4).
Figure 6.
Enrichment of EC QTLs in CAD and Complex Disease GWAS
(A) Enrichment (y axis) of HAEC eQTLs at CAD loci (x axis) over permuted rates of random overlap for all HAEC eQTL datasets.
(B) HAEC eQTL genes co-mapping with a molQTL that are associated with multiple GWAS traits (x axis).
(C) Genomic region of MFAP2 locus. Linkage disequilibrium between GWAS-associated loci (top) epigenetic traits (bottom), and local sequence surrounding SNPs of interest, rs9435732 and rs9435733.
(D) eQTL plots for MFAP2 where each dot is an HAEC donor, colored bars along the x axis represent genotypes at rs9435733, and the colors of dots represent genotypes at rs9435732.
(E) Graph as in (D) for expression of ATP13A2.
(F and G) GTEx eQTLs in lung for MFAP2 replicates the direction of rs9435732 (F) and rs9435733 (G).
(H–J) molQTLs for rs9435733 for NF-kB binding (H), H3K27ac (I), and ATAC-seq (J), with dot colors representing genotypes at rs9435732.
Next, we compared the precision of this molQTL fine-mapping method to another recent method, coloc.35 Coloc uses summary statistics and allele frequencies to test whether a putative “causal” SNP signal underlying associations along one trait’s locus (e.g., eQTLs) are likely to also drive associations at that locus for another trait (e.g., GWAS). We applied coloc to the eight of the nine loci above which contained putative causal relationships (one transcript was not on the microarray). This resulted in confirmatory posterior probabilities (>0.821), indicating that our molQTL fine-mapping strategy was well adept at discovering functional regulatory variants.
The list of 18 variants includes the CAD-associated SNP rs17114036, with eQTL for PPAP2B/PLPP3 (Figure S2A). We have previously reported rs17114036 to be a functional intronic enhancer variant that modulates PPAP2B expression in a sheer stress-sensitive manner.36 Here, consistent with previous reports, we find that rs17114036 is also a molQTL for NF-kB/p65 binding with the G allele associated with greater binding and PPAP2B expression and CRISPRi confirming enhancer activity at this SNP (Figure S2B and Table S4).
Another notable gene among those with CAD, eQTL, and/or molQTL hits is PECAM1 (CD31), which is frequently used as a cell-surface marker to identify endothelial cells in tissues and is part of the mechano-sensitive complex that senses hemodynamic forces.37 The associated region on chromosome 17 is a replicated CAD locus,34,38 and loss-of-function experiments in mice result in decreased or increased atherosclerosis in relation to the branching arteries and innominate artery or aortic arch inner curve, respectively.39,40 These characteristics make PECAM1 an attractive positional candidate at this GWAS locus, one that would only be detectable in EC-enriched datasets. Our analysis identified six SNPs along this locus that are eQTLs and hmQTLs in both IL-1b and untreated H3K27ac datasets (Figure S2C and S2D and Table S4). All six SNPs are near the 3′ end of PECAM1 (three intronic, one in 3′ UTR, and two intergenic) with moderate linkage disequilibrium (LD) (R2 = 0.6–0.8). Further experimentation will be required to test for causality among this set.
Another interesting gene implicated by our CAD, eQTL, and molQTL analysis was FES. We detected three SNPs (rs1894400, rs35346340, and rs7497304) within FES that were eQTLs for FES as well as hmQTLs for H3K27ac after IL-1B treatment (Table S4). The nearby SNP rs12906125 was also a robust FES eQTL, a significant hmQTL, and a suggestive caQTL (Figure S2E). rs12906125 is in near-perfect LD with all three CAD SNPs but was not tested or reported in the CAD meta-GWAS (R2 > 0.99, using European and American combined LD structure, Figure S2F). rs12906125 is a promising positional candidate because, aside from its strong associations, it is located in the center of the nucleosome-free region at the center of the FES promoter, near the summit of chromatin accessibly and binding peak for ERG and NF-kB/p65. We previously demonstrated that these characteristics increase the probability that SNPs affect the activity of regulatory elements.9 In addition, this promoter is selectively marked by H3K27ac with nucleosome depletion selectively in HUVECs in ENCODE and in our HAEC data, but not other cell types; this selective marking suggests that this gene and its allelic regulation is likely restricted in its cell-type expression profile (Figure S2F). The potential functional effect of rs12906125 on FES expression is corroborated by significant eQTLs in GTEx in numerous tissues including artery aorta tissue and artery coronary tissue (Figure S2G). Lastly, because the FES promoter is bound by ERG, we hypothesized that it would be downregulated upon ERG knockdown, and this was confirmed by analysis of our previously published data (Figure S2H). Together, these data provide functional evidence that FES is a likely mediator of vascular health and that its function is likely modulated by the haplotype including rs12906125.
EC QTLs Underlie Multi-Trait Associations
Finally, given the ubiquity of ECs throughout tissues of the human body, we reasoned that our dataset would be valuable for fine-mapping regulatory variants that affect many different complex traits in addition to CAD. We therefore cross-referenced our set of overlapping eQTLs and molQTLs with summary data from the GWAS Catalog41 containing lead variants from 3,616 complex traits and diseases that span 3,551 studies.41 To test for enrichment, we generated empirical null distributions of expected overlap by 1,000 permutations of eQTL and molQTL genomic locations, and we counted the random occurrences of overlap with GWAS traits. The traits with significant eQTL and/or molQTL enrichment are shown in Table S6, and top traits include pulse pressure (p = 1.03 × 10-11), blood protein levels (p = 2.48 × 10-10), monocyte count (p = 1.77 × 10-6), and lung function by FEV1/FVC (p = 1.2 × 10-5). These data support the hypothesis that common genetic variation in the EC regulome modifies EC biology to affect these complex traits with implications in disease.
Next, to identify loci with pleiotropic effects, we tabulated loci harboring an EC eQTL, a molQTL, and multiple GWAS trait associations using all EC datasets combined. The top 13 genes with multiple GWAS associations in this analysis are shown in Figure 6B (a detailed list of overlaps is in Table S7). Based on these criteria, the MFAP2 gene locus demonstrated the most trait associations, including peak expiratory flow, lung function, and chronic obstructive pulmonary disease, as well as several anthropometric measures such as waist-to-hip ratio (Table S7).
Four variants were associated with expression of MFAP2. Three of these variants (rs9435731, rs2284746, and rs9435733) are in high LD with each other (R2 = 15,42) and the alternate alleles are associated with higher expression of MFAP2 (Figure 6C, LD shown by heatmap in red triangles). These three variants are associated with the lung function traits listed above. The fourth variant, rs9435732, is not in high LD with the rest (max R2 = 0.245) and is associated with waist-to-hip ratio. The reference allele (C) for this variant is associated with greater MFAP2 expression. Figure 6C shows the epigenetic landscape in ECs and ENCODE cell types across the MFAP2 and ATP13A2 locus that includes many regulatory elements with variable activity profiles across cell types. Whereas MFAP2 and ATP13A2 both have eQTLs at rs9435731, rs2284746, and rs9435733, the ATP13A2 eQTLs are much less significant in the EC dataset than is MFAP2 (Figure 6D and 6E). Further, rs9435732 is not an eQTL for ATP13A2 in ECs, supporting the likelihood that MFAP2 is the functional gene that mediates trait differences through ECs. Lastly, MFAP2 eQTLs at rs9435732/rs9435733 are replicated in GTEx lung tissue in the same direction we observe in ECs (Figure 6F and 6G).
Given our observation that two sets of alleles (the three SNPs and rs9435732) are independently associated with MFAP2 expression as well as distinct sets of traits by GWAS, we tested whether combinations of alleles at this locus further discriminated MFAP2 expression relative to either set alone. Indeed, we found that within genotypes of rs9435733, genotypes at rs9435732 further discriminated MFAP2 levels (Figure 6D) but not ATP13A2 levels (Figure 6E). Interestingly, both of these SNPs are located in the promoter of MFAP2 (Figure 6C, pink vertical line), 96 bp apart and with significant relationships to NF-kb/p65 binding and H3K27ac levels (Figure 6H and 6I; all with FDR < 5%) and ATAC-seq (Figure 6J; all with FDR < 8%) in the direction consistent with gene expression. Taken together, these data provide robust functional evidence that SNPs along the haplotypes created by these MFAP2 promoter SNPs modify risk for pulmonary and anthropometric traits by modulating MFAP2 expression in ECs. Further experimentation will be required to understand which of these variants are necessary and sufficient for the epigenetic and transcriptomic differences observed.
Discussion
In this study, we identified non-coding, common genetic variants that modify levels of target-gene expression in a single cell type that is critical for health. The design was to propagate genetically diverse human ECs in low-passage, model disease microenvironments through the use of pro-inflammatory stimuli, and to quantitatively measure genome-wide features of gene expression and epigenetic state. By analysis of enriched motifs at EC regulatory elements, we provide genome-wide evidence that mutations in ETS, AP-1, and NF-kB motifs by common alleles perturb activities of regulatory function. We demonstrated the utility of our integrated analysis and the accuracy of our approach through experimental validation of a handful of candidate regulatory SNPs, modification of enhancer activity, and target-gene expression (KIF26B, VEGFC, FGD6) for regulatory elements up to 750kb away from their targets. Intersection of these data with public catalogs, including GTEx, ENCODE, and the GWAS Catalog led to additional discoveries, including that roughly half of EC eQTLs were not evident in GTEx tissues, and that EC eQTLs and molQTLs are enriched at disease loci for several disease traits (CAD, lung function, pulse pressure, and blood protein levels) with some loci (MFAP2) harboring pleiotropic effects. The thousands of eQTLs and molQTLs detected here serve as a resource such that investigators may query significant regulatory relationships for specific genes or loci of interest. Lastly, while this work is a qualitative advancement toward functional annotation of the non-coding genome, much work remains to be done toward this goal. These topics are discussed below.
Delineation of the cell and tissue-type patterns of operational regulatory elements is a challenging requirement toward annotating the functional non-coding genome. It will require a combination of experimental and computational approaches to discern epigenetic and expression states of single cell types that comprise human tissues during health and disease. Here, the comparison of eQTLs between ECs and GTEx showed that eQTLs collected from a single cell type enabled discovery of about 1,000 transcripts with novel eQTLs not evident in GTEx at all ranges of effect sizes (Figure 1). This suggests that eQTLs present in a constituent cell type of tissue is often below the limit of detection. We recognize that some of the eQTLs in this study could result from genes whose expression is exaggerated in culture, for example by proliferation or exposure to serum, but this cannot fully explain the differences, because two cell lines in GTEx (Epstein-Barr virus (EBV)-transformed lymphocytes and cultured fibroblasts) were propagated in similar conditions, and their profiles are distinct from ECs. Together, this finding indicates that a large proportion of functional, common variants regulating cell-specific target genes remain to be identified, and that this can be achieved through eQTL analysis of single cell types.
Perhaps one of the most exciting promises of molQTL mapping is that, in conjunction with eQTLs, it enables fine mapping of causal regulatory variants. Our study is among the first to demonstrate this in human cells, and to our knowledge, the first to do this in ECs. We provided three examples in Figure 5: KIF26B, FGD6, and VEGFC, where experimentation confirmed the predicted function of non-coding SNPs in gene regulation through enhancers up to 750kb distant from the genes. While effective, validation using traditional experimental approaches (like ours) is rate limiting. Application of high-throughput assays, such as massively parallel reporter assays (MPRAs) and multiplexed CRISPR applications, will accelerate the rate of validation for predicted non-coding variants. It will be important, however, that these techniques be applied in the primary cell types from which QTLs were generated, because only these cell types will contain the relevant complement of TFs needed to achieve cell-appropriate regulation.
To quantify how many eQTLs we were able to “explain” with molQTLs in this study, we found that about 10% of EC eQTLs are in linkage with molQTLs (R2 > 0.8). As similar studies are published, it will be important for the community to estimate this value, because it leads to estimates of the proportion of expression differences between people that stem from common regulatory variants in the proximal genomic landscape. Analogous to the “missing heritability” paradigm in GWAS, this value reflects the proportion of expression differences we can explain using single genetic loci. It is difficult to know from our data how well this estimate reflects the effects of true causal regulatory variants, but we provide here non-exclusive scenarios that explain possibilities. It is possible this value is an under-estimate of the true causal SNP set and that nearly every expression trait has an underlying causal variant. Explanations for why we cannot observe them relate to statistical power due to sample size and technical sensitivity in quantifying epigenetic traits. Quantitation of RNA transcripts through the use of array or RNA-seq spans a large dynamic range, whereas epigenetic assays like ATAC-seq are semiquantitative because they average binary on-off states of proteins accessing DNA on two chromosomes per cell. Single-cell approaches, or assay developments that improve sensitivity, performed with larger sample numbers, should improve molQTL detection. In addition, our study might have missed informative regulatory marks simply because these marks were not included in the design. Repressive marks, for example, could help identify modulating regulatory SNPs. It is also possible, and worth considering, that the “proportion explained” value is a somewhat accurate reflection of loci harboring true causal regulatory variants, and that gene expression is genetically regulated by yet undiscovered mechanisms that are not reflected in the understood regulome of the cell. Such higher-dimensional possibilities may include roles for combinatorial and/or hierarchical relationships among regulatory elements, 3D chromatin structure, histone positioning, or biophysical constraints, among others. Nonetheless, our study confirms the utility of molQTL mapping and provides a foundation for future studies.
One of the most exciting findings in our study was the potentially causal role for unlinked SNPs in the promoter of MFAP2 to modify human disease. This locus associates with multiple GWAS traits including lung function and waist-to-hip ratio (Figure 6). While not fully understood, MFAP2 encodes a secreted protein that has been shown to bind microfibrils in the extracellular matrix.43, 44, 45, 46 MFAP2 has been shown to bind active transforming growth factor beta, and bone morphogenic proteins to modulate downstream signaling in paracrine cells of elastic tissues. Mfap2−/− knockout mice exhibit increased body fat, weight, size, altered wound healing in bone and skin, osteopenia, bone fractures, bleeding diathesis, and increased metabolic dysfunction. Recently, MFAP2 was implicated in regulating EC functions through sequestration of the endothelial-specific protein EGFL7, which controls various endothelial functions including repression of endothelial-derived lysyl oxidase (LOX), adhesion molecule expression, and Notch signaling.47 Exactly how MFAP2 modifies traits in humans has yet to be elucidated. Nonetheless, its eQTLs at rs9435733 and rs9435732 are reproducible, as they are evident in multiple GTEx tissues. Our data support the possibility that these variants primarily regulate MFAP2 in ECs, rather than the neighboring gene ATP13A2, which could be perturbed in other cell types. Further, our molecular data demonstrate that haplotypes across rs9435733 and rs9435732 better discriminate MFAP2 expression in ECs than either variant does alone (Figure 6D). While future work is required to establish the molecular explanations for how these variants perturb MFAP2 function and modify disease risk, these data support a model whereby this gene is controlled by at least two regulatory variants and the likelihood that this locus has pleiotropic effects on human complex disease.
In conclusion, we present an integrative analysis of the effects of common genetic variation on human endothelial cell molecular traits. The findings presented here support the likelihood that numerous novel biological relationships are present in this dataset, and this will serve as a useful resource to accelerate discovery in the research community.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants to C.E.R. (R00HL123485 and R01HL147187), as well as by the following training fellowships: T32ES007091-36A1 (L.K.S and A.C.C.), American Heart Association 20PRE35200195 (L.K.S.), T32ES007091-36A1 (A.E.S.), and the Achievement Rewards for College Scientists (ARCS) Foundation in Phoenix (A.C.C.). Additional support was provided to Y.F. by R01HL138223 and R01HL136765. M.U.K. and group members AT, KÕ, TÖ, and M.L.R. were supported by grants from the Academy of Finland (287478, 319324, and 314554 to M.U.K.), the European Research Council (ERC) research and innovation program (802825), the Finnish Foundation for Cardiovascular Research, the Sigrid Jusélius Foundation, and the Jane and Aatos Erkko Foundation. M.L.R. was also supported by personal grants from the Finnish Foundation for Cardiovascular Research, the Finnish Cultural Foundation, and the Finnish Diabetes Research Foundation. I.S. and A.T. were supported by the University of Eastern Finland Doctoral Program in Molecular Medicine.
Published: May 21, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.04.008.
Accession Numbers
The accession numbers for the next-generation sequencing data reported in this paper are NCBI GEO database: GSE30169, GSE139377.
Web Resources
1000 Genomes Project, https://www.internationalgenome.org/
Addgene global plasmid repository, https://www.addgene.org/
Database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap/
Encyclopedia of DNA Elements (ENCODE), https://www.encodeproject.org/
Gene-Tissue Expression Project (GTEx), https://gtexportal.org/home/
Roadmap Epigenomics Project, http://www.roadmapepigenomics.org/
Supplemental Data
References
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium G.T., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A. A global reference for human genetic variation. Nature 526. 2015:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinz S., Romanoski C.E., Benner C., Glass C.K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinz S., Romanoski C.E., Benner C., Allison K.A., Kaikkonen M.U., Orozco L.D., Glass C.K. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F., Glass C.K. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajendran P., Rengarajan T., Thangavel J., Nishigaki Y., Sakthisekaran D., Sethi G., Nishigaki I. The vascular endothelium and human diseases. Int. J. Biol. Sci. 2013;9:1057–1069. doi: 10.7150/ijbs.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimbrone M.A., Jr., García-Cardeña G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romanoski C.E., Lee S., Kim M.J., Ingram-Drake L., Plaisier C.L., Yordanova R., Tilford C., Guan B., He A., Gargalovic P.S. Systems genetics analysis of gene-by-environment interactions in human cells. Am. J. Hum. Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan N.T., Whalen M.B., Stolze L.K., Hadeli N.K., Lam M.T., Springstead J.R., Glass C.K., Romanoski C.E. Transcriptional networks specifying homeostatic and inflammatory programs of gene expression in human aortic endothelial cells. eLife. 2017;6:e22536. doi: 10.7554/eLife.22536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romanoski C.E., Che N., Yin F., Mai N., Pouldar D., Civelek M., Pan C., Lee S., Vakili L., Yang W.P. Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ. Res. 2011;109:e27–e41. doi: 10.1161/CIRCRESAHA.111.241869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Geijn B., McVicker G., Gilad Y., Pritchard J.K. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis G., R Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumasaka N., Knights A.J., Gaffney D.J. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muerdter F., Boryń L.M., Woodfin A.R., Neumayr C., Rath M., Zabidi M.A., Pagani M., Haberle V., Kazmar T., Catarino R.R. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat. Methods. 2018;15:141–149. doi: 10.1038/nmeth.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayaraj P., Le Bras A., Mitchell N., Kondo M., Juliao S., Wasserman M., Beeler D., Spokes K., Aird W.C., Baldwin H.S., Oettgen P. Erg is a crucial regulator of endocardial-mesenchymal transformation during cardiac valve morphogenesis. Development. 2012;139:3973–3985. doi: 10.1242/dev.081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han R., Pacifici M., Iwamoto M., Trojanowska M. Endothelial Erg expression is required for embryogenesis and vascular integrity. Organogenesis. 2015;11:75–86. doi: 10.1080/15476278.2015.1031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lathen C., Zhang Y., Chow J., Singh M., Lin G., Nigam V., Ashraf Y.A., Yuan J.X., Robbins I.M., Thistlethwaite P.A. ERG-APLNR axis controls pulmonary venule endothelial proliferation in pulmonary veno-occlusive disease. Circulation. 2014;130:1179–1191. doi: 10.1161/CIRCULATIONAHA.113.007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah A.V., Birdsey G.M., Peghaire C., Pitulescu M.E., Dufton N.P., Yang Y., Weinberg I., Osuna Almagro L., Payne L., Mason J.C. The endothelial transcription factor ERG mediates Angiopoietin-1-dependent control of Notch signalling and vascular stability. Nat. Commun. 2017;8:16002. doi: 10.1038/ncomms16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah A.V., Birdsey G.M., Randi A.M. Regulation of endothelial homeostasis, vascular development and angiogenesis by the transcription factor ERG. Vascul. Pharmacol. 2016;86:3–13. doi: 10.1016/j.vph.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanoski C.E., Link V.M., Heinz S., Glass C.K. Exploiting genomics and natural genetic variation to decode macrophage enhancers. Trends Immunol. 2015;36:507–518. doi: 10.1016/j.it.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillabert-Gourgues A., Jaspard-Vinassa B., Bats M.L., Sewduth R.N., Franzl N., Peghaire C., Jeanningros S., Moreau C., Roux E., Larrieu-Lahargue F. Kif26b controls endothelial cell polarity through the Dishevelled/Daam1-dependent planar cell polarity-signaling pathway. Mol. Biol. Cell. 2016;27:941–953. doi: 10.1091/mbc.E14-08-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Harst P., Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krause M.D., Huang R.T., Wu D., Shentu T.P., Harrison D.L., Whalen M.B., Stolze L.K., Di Rienzo A., Moskowitz I.P., Civelek M. Genetic variant at coronary artery disease and ischemic stroke locus 1p32.2 regulates endothelial responses to hemodynamics. Proc. Natl. Acad. Sci. USA. 2018;115:E11349–E11358. doi: 10.1073/pnas.1810568115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzima E., Irani-Tehrani M., Kiosses W.B., Dejana E., Schultz D.A., Engelhardt B., Cao G., DeLisser H., Schwartz M.A. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 38.Coronary Artery Disease (C4D) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 39.Harry B.L., Sanders J.M., Feaver R.E., Lansey M., Deem T.L., Zarbock A., Bruce A.C., Pryor A.W., Gelfand B.D., Blackman B.R. Endothelial cell PECAM-1 promotes atherosclerotic lesions in areas of disturbed flow in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2008;28:2003–2008. doi: 10.1161/ATVBAHA.108.164707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goel R., Schrank B.R., Arora S., Boylan B., Fleming B., Miura H., Newman P.J., Molthen R.C., Newman D.K. Site-specific effects of PECAM-1 on atherosclerosis in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2008;28:1996–2002. doi: 10.1161/ATVBAHA.108.172270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinbaum J.S., Broekelmann T.J., Pierce R.A., Werneck C.C., Segade F., Craft C.S., Knutsen R.H., Mecham R.P. Deficiency in microfibril-associated glycoprotein-1 leads to complex phenotypes in multiple organ systems. J. Biol. Chem. 2008;283:25533–25543. doi: 10.1074/jbc.M709962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craft C.S., Pietka T.A., Schappe T., Coleman T., Combs M.D., Klein S., Abumrad N.A., Mecham R.P. The extracellular matrix protein MAGP1 supports thermogenesis and protects against obesity and diabetes through regulation of TGF-β. Diabetes. 2014;63:1920–1932. doi: 10.2337/db13-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craft C.S., Broekelmann T.J., Mecham R.P. Microfibril-associated glycoproteins MAGP-1 and MAGP-2 in disease. Matrix Biol. 2018;71-72:100–111. doi: 10.1016/j.matbio.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Craft C.S., Broekelmann T.J., Zou W., Chappel J.C., Teitelbaum S.L., Mecham R.P. Oophorectomy-induced bone loss is attenuated in MAGP1-deficient mice. J. Cell. Biochem. 2012;113:93–99. doi: 10.1002/jcb.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villain G., Lelievre E., Broekelmann T., Gayet O., Havet C., Werkmeister E., Mecham R., Dusetti N., Soncin F., Mattot V. MAGP-1 and fibronectin control EGFL7 functions by driving its deposition into distinct endothelial extracellular matrix locations. FEBS J. 2018;285:4394–4412. doi: 10.1111/febs.14680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.