Abstract
Enhanced brown adipose tissue (BAT) mass and activity have been demonstrated to promote the expenditure of excess stored energy and reduce prevalence of obesity. Cold is known as a potent stimulator of BAT and activates BAT primarily through the β3-adrenergic-cAMP signaling. Here, we performed RNA-sequencing to identify differential miRNAs in mouse BAT upon cold exposure and a total of 20 miRNAs were validated. With the treatment of CL-316,243 (CL) and forskolin (Fsk) in mouse and human differentiated brown adipocyte cells in vitro, miR-23b-5p, miR-133a-3p, miR-135-5p, miR-491-5p, and miR-150-3p expression decreased and miR-455-5p expression increased. Among these deferentially expressed miRNAs, miR-23b-5p expression was differentially regulated in activated and aging mouse BAT and negatively correlated with Ucp1 expression. Overexpression of miR-23b-5p in the precursor cells from BAT revealed no significant effects on lipid accumulation, but diminished mitochondrial function and decreased expression of BAT specific markers. Though luciferase reporter assays did not confirm the positive association of miR-23b-5p with the 3′UTRs of the predicted target Ern1, miR-23b-5p overexpression may affect brown adipocyte thermogenic capacity mainly through regulating genes expression involving in lipolysis and fatty acid β-oxidation pathways. Our results suggest that miRNAs are involved in cold-mediated BAT thermogenic activation and further acknowledged miR-23b-5p as a negative regulator in controlling thermogenic programs, further providing potential molecular therapeutic targets to increase surplus energy and treat obesity.
Keywords: brown adipose, cold stimulation, miR-23b-5p, lipolysis, β-oxidation
Introduction
Obesity and the associated disorders such as type 2 diabetes, hypertension, and cardiovascular disease are steadily increasing worldwide. It is now recognized that at least two distinct types of adipose tissue exist in mammals. White adipose tissue (WAT) is specialized for the energy storage in form of triglyceride. In contrast, brown adipose tissue (BAT) possesses a large amount of uncoupling protein 1 (Ucp1) in mitochondria (1). Experiments in mouse models have demonstrated that the activation of thermogenesis in fat could produce a healthier metabolic phenotype with traits such as whole-body energy metabolism, improved glucose homeostasis, and weight loss (2, 3). Moreover, functional BAT was discovered in adult humans (4, 5) and its activation was shown to have an inverse correlation with BMI and central obesity (6). In this light, the control of energy expenditure through the activation and/or formation of BAT represents as a promising strategy for combating obesity and associated metabolic abnormalities.
A critical physiological stimulus for augmenting the thermogenesis of BAT in mice and humans is cold stress. After exposure to a cold environment, activated BAT acquired a more significant thermogenic identity, as shown by increased glucose metabolism (7, 8) and accelerated lipid metabolism (9, 10) in both mice and humans. It has been demonstrated that mouse brown fat is activated by β3-adrenergic stimulation downstream of cold exposure (11), illustrating functional regulation by cAMP signaling in adipose thermogenesis. A similar response to cAMP signaling was also identified in human BAT in recent studies (12). Extensive studies revealed that the critical transcriptional regulators that respond to cold such as Atf4 (13), Zfp516 (14), and Irf4 (15) and upstream signaling molecules such as PKA-ASK1-p38 (16) and JAK–STAT (17) that precisely control the transcriptional system have been well-identified in brown adipocyte differentiation and cold-mediated thermogenic activation. Other types of cold-regulated adipokines such as FGF21 (18) and apelin (19) have been also demonstrated to regulate Ucp1 expression and adaptive thermogenesis. Identification of regulators and elucidation of underlying molecular mechanisms might aid in full comprehension of the metabolic potential of brown adipocytes.
MicroRNAs (miRNAs) are an abundant class of 21–24 nucleotide noncoding RNAs that control diverse biological processes. Only in recent years, several miRNAs were found to associate with transcription factors that switch brown adipogenesis and thermogenesis on or off. A BAT-enriched miRNAs cluster, miR-193b-365 cluster, was first demonstrated as a key regulator of BAT development (20). Subsequently, miR-196a (21) and miR-203 (22) were identified to have the ability to induce brown abiogenesis from white fat progenitor cells and miR-378 was found to function as a key regulatory component underlying the classical BAT-specific expansion and obesity resistance (23). Conversely, the miRNAs miR-133 (24), miR-155, (25) and miR-27a (26) have been classified as negative regulators for brown fat differentiation and thermogenesis. Although rodent studies strongly corroborated the importance of miRNAs involved in BAT function, the significance of miRNAs in humans brown adipocytes is just beginning to be revealed. miR-26b (27) and let-7i (28) were confirmed to play a role in the conversion of white adipocytes to brown adipocytes. Our previous studies also found that a BAT-enriched miRNA, miR-199a-3p, exerted an inhibitory role in brown adipocyte function (29). Thus, the elucidation of miRNAs responsiveness to cold and its downstream β3-adrenergic-cAMP signaling in classical brown adipocytes requires further research.
Herein, we performed global miRNA-sequencing to identify miRNAs that are important for cold-induced BAT activation. Temperature-regulated miRNAs in mouse brown adipocytes of mice showed comparable expression patterns to human brown adipocytes exposed to β3-adrenergic-cAMP signaling stimulation, indicating conserved regulation in human brown adipocytes. We further identified miR-23b-5p as a negative regulator that controls a brown adipocyte thermogenic gene program through reformed lipolysis and fatty acid β-oxidation pathways. Together, our observations show cold-responsive miRNAs in mouse BAT and close regulation in human brown adipocytes, providing further potential molecular therapeutic targets to combat obesity.
Materials and methods
Animal maintenance, cold exposure experiments, and CL-316, 243 administration in vivo
Male C57BL/6J mice at the age of 6 weeks, purchased from the Model Animal Research Center of Nanjing University, were randomly divided into two groups: one group was kept at room temperature (RT, 26 ± 2°C) and the other group was kept at much lower temperature for the cold exposure treatment (4°C) (n = 12 per group). The duration of the cold adaption was maintained for 7 days and interscapular BAT (iBAT) samples were removed for miRNA sequencing as well as qPCR detection. Another group of mice (also 6 weeks of age) were injected intraperitoneally (i.p) daily with saline or CL-316,243 (1 mg/kg, Sigma-Aldrich) (n = 12 per group). After 1 week of treatment, iBAT were harvested for qPCR analysis. Mice at the specific ages (1, 2, 5, 8, and 10 months) were humanely destroyed and their iBAT were harvested for qPCR detection (n = 6 per age). Our experiments involving mice were performed in accordance with the Use of Experimental Animals protocols published by the Ethics Committee at Nanjing Medical University.
Ethics statement
This work was carried out with approval from the Human Research Ethics Committee of Nanjing Maternity and Child Health Care Institute (permit number [2015]110). Human fetal adipose tissues were acquired from the interscapular areas of aborted fetuses after parental donors signed a written informed consent form.
Small RNA library construction, sequencing, and differential miRNAs analysis
Total RNAs of mouse BAT (n = 3 per group) were extracted using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol. A small RNA library was constructed and sequencing was carried out by RIBOBIO Co. Ltd (Guangzhou, China) in accordance with the manufacturer’s instructions. Briefly, small RNAs ranging from 18 nt to 30 nt in size were purified with gel and ligated to the 3′ adaptor and 5′adaptor. Subsequently, ligation products were purified, reverse transcribed, and amplified. The purified cDNA library was used for cluster generation on Illumina’s Cluster Station (Illumina, San Diego, CA, USA) and sequencing was run on Illumina HiSeq 2500 (Illumina). Raw sequencing reads were obtained using related Illumina’s analysis software. Up- or downregulated miRNAs were screened up by |log2 (Fold Change)| ≥1 as well as P value test <0.05.
Cell isolation, culture, and induction of adipogenesis
Mouse adipose tissue SVF preparation and culture
The mouse fat progenitor cells derived from stromal vascular fraction (SVF) were isolated from the iBAT of 4-week-old C57Bl/6J mice using a standard procedure according to our prior study (30). Mouse brown pre-adipocytes were induced via differentiation medium containing 10% FBS/DMEM (Gibco), 1% penicillin/streptomycin solution (Gibco), 0.5 mM IBMX, 0.5 mM dexamethasone, 0.125 mM indomethacin, 860 nM insulin, 1 nM T3, and 1 μM rosiglitazone (these regents were obtained from Sigma-Aldrich). IBMX, dexamethasone, indomethacin, and rosiglitazone were wiped out 2 days later and cells were incubated with a maintenance medium containing 10% FBS DMEM (Gibco), 1% antibiotics (Gibco), 850 nM insulin (Sigma-Aldrich), and 1 nM T3 (Sigma-Aldrich) for 6–8 days until mature lipids appeared.
Human adipose tissue SVF preparation and culture
Primary pre-adipocytes were isolated from human fetal interscapular BAT as described by Lei Sun et al. (31). These cells were sustained in a pre-adipocyte medium containing 5% fetal bovine serum, 1% pre-adipocyte growth supplement, and 1% penicillin/streptomycin solution (PAM; Sciencell Research Laboratories, Carlsbad, CA, USA) supplied with 20 ng/mL hbFGF (R&D) and 16 nM ascorbic acid (Sigma-Aldrich). The differentiation program was performed according to our previous study (32).
Lentivirus production and infection
Lentiviral constructs for overexpression and negative control vectors were acquired from GenePharma Inc. (Shanghai, China). A detailed transfected procedure was performed according to manufacturer’s instructions. The pre-adipocytes at the 30–40% confluence were infected with the negative control lentivirus (NC, viral titre, 1 × 108 TU/mL) or miR-23b-5p lentivirus (miR-23b-5p, viral titre, 1 × 108 TU/mL) in 1 mL DMEM culture medium mixed with 5 µg/mL Polybrene (Sigma-Aldrich). After a 24-h infection, the pre-adipocytes were placed in a fresh medium. Adipogenic induction was performed until the cells reached confluency.
CL 316, 243 (CL), and forskolin (Fsk) treatment
To induce thermogenesis, fully differentiated mouse or human brown adipocytes were incubated in serum-free medium for 6 h followed by treatment with Fsk or CL in a final concentration of 10 µM in DMEM complete medium for 4 h according to our previous report (29).
Oil Red O staining and triacylglycerol (TG) content measurement
The fully differentiated cells were fixed in 4% formaldehyde for 15 min and then stained with 0.2% Oil Red O (Sigma) working solution for 30 min at 37°C. The stained cells were rinsed three times with distilled water and observed under the Observer D1 microscope (Carl Zeiss). Intracellular triglycerides content was measured by adopting a triglyceride assay kit (Applygen, Beijing, China) and normalizing the total protein content of the samples, obtained by BCA Protein Assay kit (Pierce).
Mitochondrial DNA abundance and Mitotracker Red staining
Mature adipocytes were scraped and total DNA was extracted using a QIAamp DNA minikit (QIAGEN). To quantify mitochondrial DNA content, quantitative real-time PCR was performed and the primers for mouse mtCox1 and mtNd1 as the mitochondrial genome and 18s as the nuclear genome were described in Supplementary Table 1 (see section on supplementary materials given at the end of this article). For MitoTracker Red staining, alive adipocytes were incubated with MitoTraker Red probe (Invitrogen) at a final concentration of 50 nM for 30 min in darkness at 37°C and imaged with the Observer D1 microscope (Carl Zeiss).
RNA extraction, reverse transcription, and quantitative real-time PCR
Adipose tissues from mice or cell cultures were collected using Trizol reagent (Invitrogen) at the indicated time. RNeasy Mini Kits (Qiagen) were utilized to extract and purify the total RNA in accordance with the manufacturer’s protocol. One microgram of the total RNA from every sample was reverse transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Applied Biosystems). miRNA levels were measured with the Bulge-LoopTM qPCR Primer Set (RIBOBIO, Guangzhou, China). Quantitative real-time PCRs were carried out using SYBR green technique (Applied Biosystems) on an Applied Biosystems ViiA 7 system (Applied Biosystems) according to the manufacturer’s instructions. The expression levels of miRNAs or mRNAs, which were normalized to U6 or Ppia expression, were analyzed by 2−ΔΔCt method. The sequences of primers refer to in the Supplementary Table 1.
Protein extraction and Western blot
Cell cultures were washed with ice cold PBS and extracted with RIPA lysis buffer (Beyotime Biotechnology, NanTong, China) which was supplemented with protease inhibitor cocktail (Roche) and phenylmethylsulfonyl fluoride (Beyotime) at 4°C with gentle shaking. Proteins were loaded onto an 8% to 12% SDS-PAG for electrophoresis, transferred to PVDF membranes, and immunoblotted with specific primary antibodies as following: rabbit polyclonal UCP1 (Abcam; Cat. No. ab10983), rabbit polyclonal AMPKa (Abcam; Cat. No. 3759), monoclonal rabbit HSL (Cell Signaling Technology; Cat. No. 18381), monoclonal rabbit ATGL (Cell Signaling Technology; Cat. No. 2439), polyclonal rabbit GLUT4 (Abcam; Cat. No. 18381), monoclonal rabbit ACCSL1 (Cell Signaling Technology; Cat. No. 3658), monoclonal rabbit ACSL1 (Cell Signaling Technology; Cat. No. 9189), monoclonal mouse ACADM (Santa Cruz; Cat. No. 65030), and monoclonal mouse ACADS (Santa Cruz; Cat. No. 65953) and the secondary antibodies were horse radish peroxidase-conjugated goat anti-rabbit IgG and goat anti-mouse IgG from Beijing Zhong Shan Biotechnology CO (Beijing, China).
Measurement of oxygen consumption and lipolysis capacity
Mouse brown pre-adipocytes were seeded in XF24 cell culture plates (Seahorse Biosciences, North Billerica, MA), transfected, and differentiated as previously described (33). Oxygen consumption rate (OCR) of differentiated cells was assessed using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience) as previously described (33). To detect lipolysis efficacy, differentiated adipocytes from NC and miR-23b-5p overexpressing groups were starved in DMEM medium (Gibco) without serum for 4 h. Adipocytes were then incubated with or without Fsk (10 µM) in DMEM medium without phenol red (Gibco) and serum containing 2% free fatty acid BSA (Equitech-Bio, Kerrville, TX, USA). After a 1-h incubation period, the culture medium was collected for glycerol measurements (Jiangcheng Bioengineering Insititute, Nanjing, China). The glycerol release was normalized to the total protein content of cell cultures, attained using the BCA Protein Assay kit (Pierce).
Cell immunofluorescent staining
At the designed time, adipocytes from the NC and miR-23b-5p overexpression groups were washed by PBS twice, fixed in 4% paraformaldehyde for 15 min, and permeabilized with 0.25% Triton X-100 in PBS (PBST) for another 20 min. The fixed cells were kept in blocking solution, 2.5% BSA in PBST for 30 min at 37°C, followed by UCP1 antibody (Abcam) incubation overnight at 4°C. Then the cells were incubated with secondary antibodies conjugated with Alexa Fluor 546 (Invitrogen) for 1 h at 37°C. After the cell’s nuclei were stained with DAPI (Invitrogen) for 5 min, cells were mounted with microscope slides and visualized under the fluorescence microscope Imager A2 (Carl Zeiss).
Bioinformatics predictions and assay of luciferase activity
TargetScan (http://www.targetscan.org) was used to predict the target sequences for miR-23b-5p. The 3′UTR region of Ern1 containing two predicted miR-23b-5p binding sites named WT1 and WT2 or corresponding mutant seed sequences named MUT1 and MUT2 were amplified via PCR and inserted into XhoI and NotI sites of psiCHECK-2 plasmid (Promega). The target sequence of miR-23b-5p within the endoplasmic reticulum to nucleus signaling 1 (Ern1) 3′UTR-1 was mutated from GGAACCA to CCTTGGA and 3′UTR-2 AGGAACC to ACCTTGG. 3T3-L1 cells (104 per well) were transfected with 100 ng of plasmid per well (96-well plate) for 16 h followed by a miR-23b-5p lentivirus infection (1 µL per well, viral titre, 1 × 108 TU/mL). After a 48-h infection, firefly and renilla luciferase activities were analyzed using the dual-luciferase reporter assay system (Promega). The renila luciferase activity was normalized to firefly luciferase activity and the results were plotted as a proportion of the control.
Statistical analysis
Data were analyzed with Graphpad Prism 5 and expressed as mean ± s.d. Unpaired two-tailed t-test and one-way ANOVA were used for most comparison. Correlations and differences between miR-23b-5p and Ucp1 expression level were analyzed using the Pearson’s correlation coefficient. P < 0.05 was considered significant and was indicated in the figures.
Results
Global profiling identified differentially expressed miRNAs during iBAT activation by cold stimulation
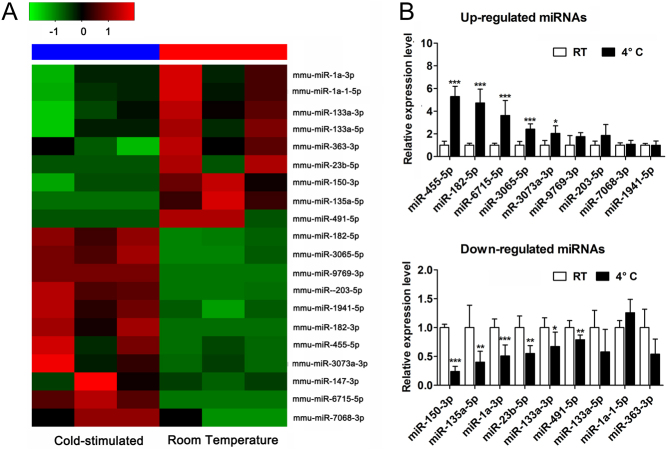
To identify the miRNAs differentially expressed in activated BAT via cold stimulation, we exposed 6-week-old male mice to a cold (4°C) or room temperature (26 ± 2°C, termed RT) environment for 7 days. To ascertain BAT activation, H&E staining was used to evaluate morphological change between RT and 4°C groups (Supplementary Fig. 1A). UCP1 in BAT was further confirmed at the protein level by immunohistochemical staining (Supplementary Fig. 1B). We visualized the deeper color of BAT in 4°C groups compared to RT groups. Additionally, smaller droplet and increased UCP1 content were observed in 4°C groups compared to RT groups. With the usage of Illumina HiSeqTM 2500 sequencing technology, the differentially expressed miRNAs were identified in the iBAT between cold and room temperature exposure. Based on the selection criteria (|fold change| ≥1, P < 0.05), a total of 82 miRNAs were detected that significantly changed in the interscapular BAT after cold exposure: 38 miRNAs were identified to be upregulated after cold exposure, while 44 miRNAs were downregulated (summarized reads information was shown in Supplementary Table 2). This defined a total of 20 miRNAs for further validation (summarized reads >0.8), 11 upregulated miRNAs and 9 downregulated miRNAs. Hierarchical clustering of 20 miRNAs was depicted in Fig. 1A. However, most cold-dependent differentially expressed miRNAs we identified have never been described in BAT. The results showed that miR-455-5p, miR-182-5p, miR-6715-5p, miR-3065-5p, and miR-3073a-3p were upregulated in mouse BAT upon cold stimulation, whereas miR-150-3p, miR-135a-5p, miR-1a-3p, miR-23b-5p, miR-133a-3p, and miR-491-5p expression decreased (Fig. 1B). However, miR-9769-3p, miR-203-5p, miR-7068-3p, miR-1a-1-5p, miR-363-3p, miR-1941-5p, and miR-133a-5p expression showed no significant difference after cold exposure. Another two miRNAs miR-147-3p and miR-182-3p did not detect CT value. We also detected miRNAs expression in ingWAT from mice exposed to a cold (4°C) or RT (26 ± 2°C). Among these cold responsive miRNAs in BAT, most miRNAs presented consistent expression patterns in cold-stimulated ingWAT (Supplementary Fig. 2A, B, C, D, E, F, G, H, I, J, K, L, M, N, and O).
Figure 1.
Global analysis of miRNAs regulated during mouse BAT activation by cold expoure. (A) Heatmap of miRNA sequencing results from the samples treated with cold exposure or room temperature for 7 days (n = 3 respectively for RT or 4°C groups). Red bars indicate higher expression (compared with value 1), and green bars indicate lower expression (compared with value −1). Black bars indicate the median expression between red and green bars. (B) Relative expression level of the selected miRNAs upregulated or downregulated in mouse BAT after cold exposure were validated by real-time PCR (n = 12 for each group). U6 expression was used as internal control for miRNA expression analysis. Values are the means ± s.d. *P < 0.05; **P < 0.01; ***P < 0.001.
The effect of β3-adrenergic-cAMP signaling stimulation on cold-responsive miRNAs expression in the differentiated mouse and human brown adipocytes
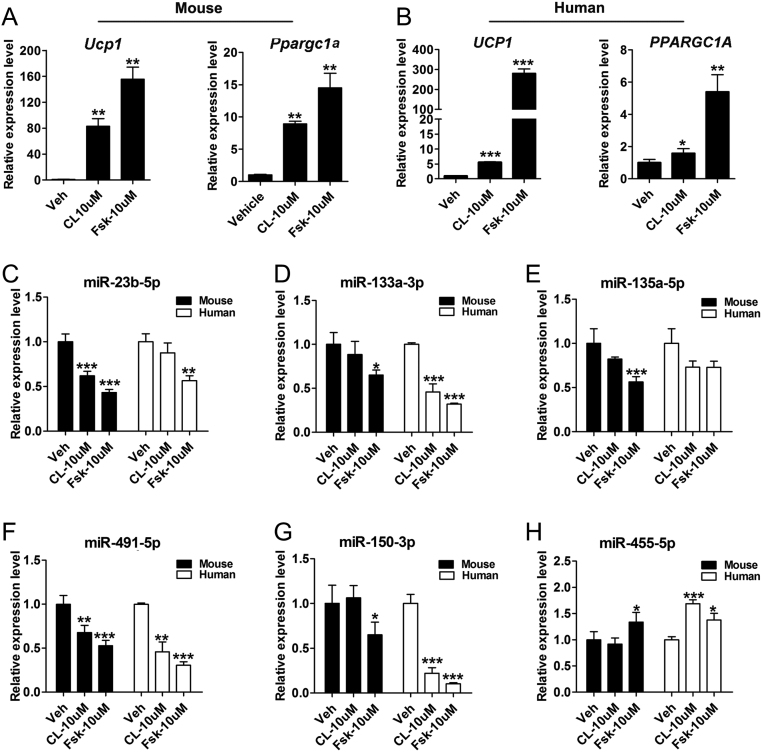
To mimic the regulatory role of β3-adrenergic-cAMP signaling in miRNAs expression of BAT upon prolonged cold exposure, we employed β3-ARs agonist CL 316, 243 (CL), and cAMP-inducing agent Fsk to analyze the expression of cold-responsive miRNAs. Treatment of primary brown pre-adipocytes after full differentiation (day 8) with CL (10 μM) and Fsk (10 μM) for 4 h showed the enhancement of thermogenic program via significant upregulation of Ucp1/UCP1 and peroxisome proliferative activated receptor gamma coactivator 1 alpha Ppargc1a/PPARGC1A) mRNA expression (Fig. 2A and B). Of note, Fsk administration has a much stronger effect on the transcription of thermogenic genes. After confirming successful stimulation, we selected 11 cold-responsive miRNAs for further investigation: the upregulated miRNAs of miR-455-5p, miR-182-5p, miR-6715-5p, miR-3065-5p, and miR-3073a-3p and the downregulated miRNAs of miR-150-3p, miR-135a-5p, miR-1a-3p, miR-23b-5p, miR-133a-3p, and miR-491-5p. We first confirmed these 11 miRNAs expression in mouse differentiated brown adipocytes and identified miR-23b-5p and miR-491-3p were down-regulated by both β3-adrenergic stimuli. The expression levels of miR-133a-3p, miR-135a-5p, and miR-150-3p decreased and miR-455-5p expression was upregulated by Fsk stimulation (Fig. 2C, D, E, F, G and H). These results highlight that β3-adrenergic signaling is essential for the regulation of cold sensitive-miRNAs (miR-23b-5p and miR-491-3p), whereas others (miR-455-5p, miR-133a-3p, miR-150-3p, and miR-135a-5p) are only sensitive to intracellular cAMP signaling. Except for miR-135-5p, these selected miRNAs showed constant expression pattern in human brown adipocytes (Fig. 2C, D, E, F, G and H). As some selected miRNAs (miR-455-5p, miR-133a-3p, and miR-150-3p) have been reported to be induced by cold exposure and play significant roles in BAT function (34, 35, 36), we focused our study on miR-23b-5p and miR-491-3p for further characterization.
Figure 2.
The effect of β3-adrenergic-cAMP signaling stimulation on cold-responsive miRNAs expression in brown adipocytes after differentiation. Primary mouse and human brown pre-adipocytes after full differentiation (day 10) were treated with CL (10 μM) and Fsk (10 μM) for 4 h. (A and B) qPCR analysis of the thermogenic genes in mouse and human brown adipocytes. (C, D, E, F, G, and H) qPCR analysis of the selected miRNAs in mouse and human brown adipocytes. Peptidylprolyl isomerase A (Ppia) or U6 expression was used as internal control for mRNA or miRNA expression analysis, respectively. Values are the means ± s.d. of three separate experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
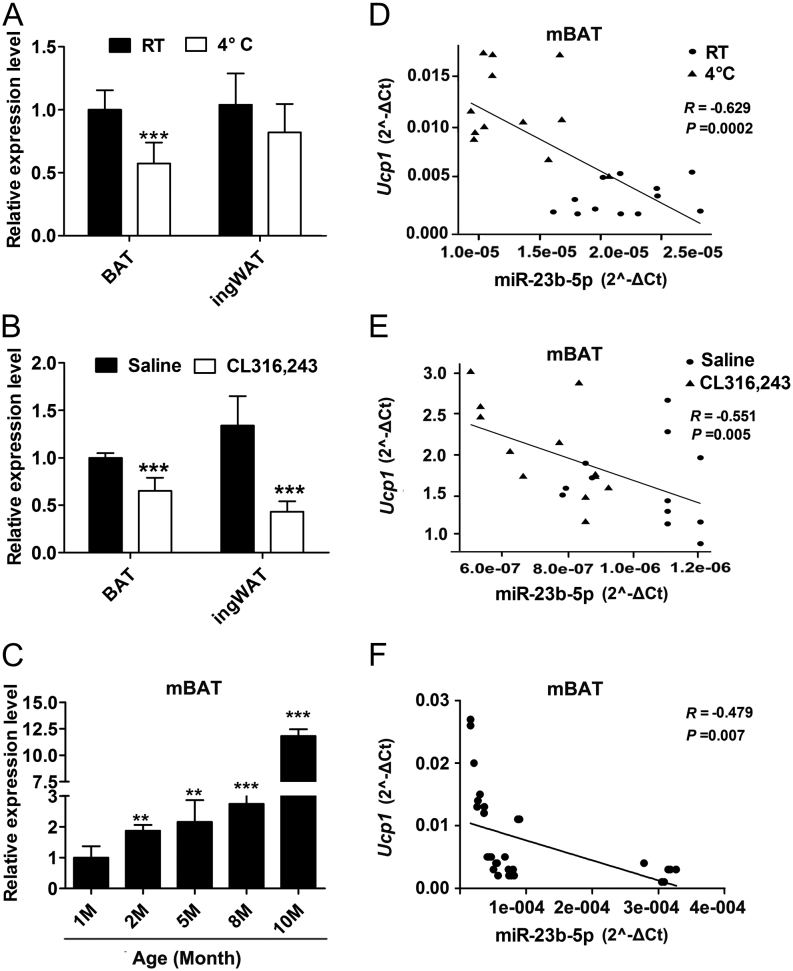
miR-23b-5p expression is differentially regulated in activated and aging mouse BAT and negatively correlated with Ucp1 expression
To elicit the role of selected miR-23b-5p and miR-491-3p in BAT function, we detected their expression in different adipose depot including BAT, ingWAT (inguinal depot), and epiWAT (epididymal depot), and in the activated or aging mouse BAT and further analyzed their expression levels with Ucp1 in BAT. The results showed miR-23b-5p was relatively abundant in skeletal muscle than BAT, ingWAT, and epiWAT (as shown in Supplementary Fig. 3). miR-23b-5p expression in BAT was significantly downregulated to 50% after the cold exposure in vivo (Figs 1B and 3A). Specifically, the responsiveness of miR-23b-5p upon cold stimulation showed no significance in ingWAT (Fig. 3A). After the β3-adrenergic agonist (CL-316,243) stimulation in vivo, miR-23b-5p’s expression level reduced remarkably both in BAT and ingWAT (Fig. 3B). As aging has been identified as a strong negative factor for BAT mass and activity in rodents and humans (37, 38), we detected miR-23b-5p expression in BAT obtained from mice of different ages and observed a progressively increased expression of miR-23b-5p (Fig. 3C). Additionally, miR-23b-5p expression was very negatively correlated with BAT activity, as measured by Ucp1 mRNA expression (Fig. 3D, E and F). However, we did not find the specific expression pattern of miR-491-3p in these models (as shown in Supplementary Fig. 4). In summary, these observations imply the potential role of miR-23b-3p in regulating BAT function and became the candidate miRNA for functional evaluation.
Figure 3.
miR-23b-5p expression was differentially regulated in the activated and the aging BAT. (A) miR-23b-5p expression in BAT and WAT between room temperature (RT) and cold stimulation (4°C) groups were evaluated by real-time PCR (n = 12 per group). (B) miR-23b-5p expression in BAT and WAT of mice intraperitoneally administered with β3-adrenergic agonist CL-316,243 (1 mg/kg body weight) were detected by real-time PCR (n = 12 per group). (C) Relative abundance of miR-23b-5p in BAT of age group 1-month- (1M), 2-month- (2M), 5-month- (5M), 8-month- (8M), and 10-month-old (10M) male C57BL6 mice was determined by real-time PCR (n = 6 per time point). (D, E, and F) Correlation between the mRNA expression of miR-23b-5p and Ucp1 in the activated (upper and middle) and aging (lower) BAT. Ppia or U6 expression was used as internal control for mRNA or miRNA expression analysis respectively. Values are the means ± s.d. *P < 0.05; **P < 0.01; ***P < 0.001.
The role of miR-23b-5p in brown adipocytes differentiation and thermogenic programs
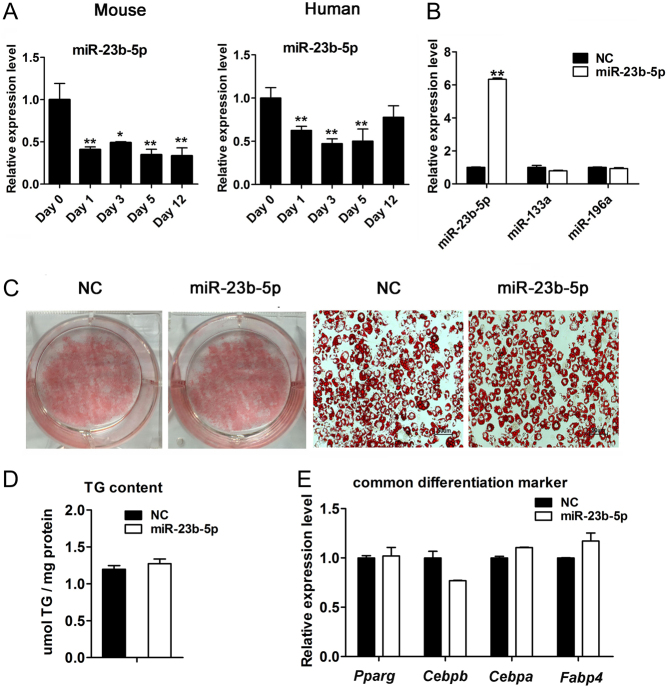
As the differentiation time increases, miR-23b-5p expression significantly deceased at various time points (day 0, day 1, day 3, day 5, and day 12) following induction of differentiation in mouse and human brown adipocytes (Fig. 4A), indicating a potential role in brown adipocyte adipogenesis. We also detected the expression levels of miR-150, miR-491-3p, miR-135a-5p, and miR-133a-3p during the differentiation periods as shown in Supplementary Fig. 5. To characterize the effect of miR-23b-5p on brown adipocytes differentiation, we forced miR-23b-5p expression via lentivirus vectors in SVF and induced these cell cultures to mature brown adipocytes (differentiation day 10). The abundance of miR-23b-5p was elevated in miR-23b-5p overexpressing cells almost 6-fold compared to NC-infected cells by real-time PCR (Fig. 4B). Furthermore, the expression levels of miR-133a-3p and miR-196-3p, which play well-known roles in modulating brown adipocyte function, were not influenced by miR-23b-5p overexpression. Oil Red O staining and triglyceride (TG) content revealed that miR-23b-5p overexpression has a weak effect on lipid accumulation as shown in Fig. 4C and D. Consistent with these results, forced expression of miR-23b-5p did not alter the mRNA expression levels of common differentiation markers including peroxisome proliferator activated receptor gamma (Pparg), CCAAT/enhancer binding protein beta (Cebpb), CCAAT/enhancer binding protein alpha (Cebpa), and fatty acid binding protein 4 (Fabp4) (Fig. 4E).
Figure 4.
Overexpression of miR-23b-5p did not affect the lipid accumulation during brown pre-adipocytes differentiation. The brown pre-adipocytes were transfected with the NC and miR-23b-5p overexpressing lentivirus and exposed to the induction medium and formed mature lipid on days 8–10. (A) Expression level of miR-23b-5p during mouse and human brown adipocytes differentiation program. (B) qPCR analyses of miR-23b-5p, miR-133a-3p, and miR-196a-3p expression level after miR-23b-5p overexpression in mouse brown adipocytes. (C) Lipid accumulation was detected by Oil Red O staining and representative image of staining was shown. Scale bar, 50 μm. (D) The triacylglycerol content was quantified to detect lipid accumulation between NC and miR-23b-5p overexpression groups and relative to total protein concentration. (E) Common differentiation-related genes expression were detected by qPCR and normalized to Ppia expression. Values are the means ± s.d. of three separated experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
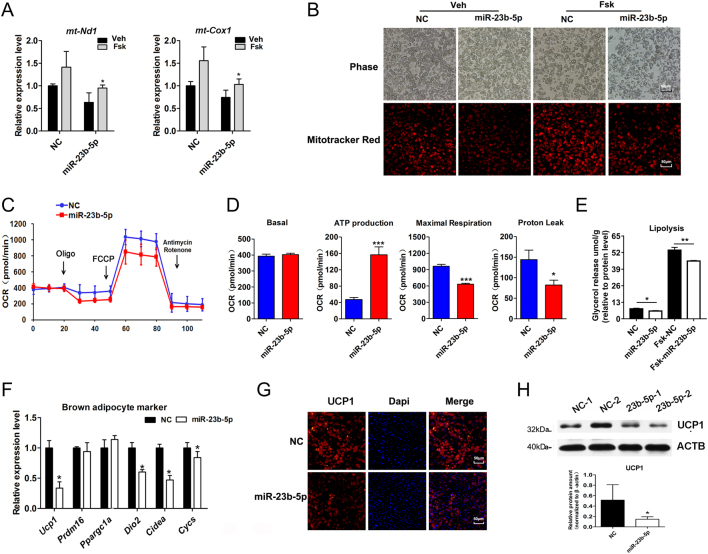
We further verified whether miR-23b-5p affects mitochondria mass by assessing mitochondrial DNA copy number (mNd1 and mCox1) after miR-23b-3p overexpression. It was notable that mNd1 and mCox1 expression was suppressed in miR-23b-5p-expressing adipocytes compared with NC groups under thermogenic status induced by Fsk stimulation (Fig. 5A), suggesting lower levels of mitochondrial amounts in thermogenic status. Indeed, MitoTraker Red staining also confirmed the attenuated abundance of mitochondria after miR-23b-5p overexpression at the basal status as well as thermogenic propensity via Fsk stimulation (Fig. 5B). To address the functional changes in cellular metabolism of miR-23b-5p-overexpressing brown adipocytes, we studied real-time bioenergetic kinetics on a Seahorse extracellular analyzer (Fig. 5C). Importantly, measurements of OCR indicated that miR-23b-5p-overexpressing cells had a lower rate of maximal respiratory capacity as well as oxygen consumption depended on proton leaks (Fig. 5D). However, basal OCR was not affected (Fig. 5D). Lipolysis levels were decreased in miR-23b-5p overexpression groups under the basal and Fsk stimulation (Fig. 5E). Further, mRNA expression of brown adipocyte marker genes including Ucp1, Dio2, Cidea, and Cycs were lower in miR-23b-5p-expressing adipocytes (Fig. 5F). UCP1 protein levels evaluated via immunofluorescent staining and Western blot were significantly declined after miR-23b-5p overexpression (Fig. 5G and H). As such, lentivirus-mediated miR-23b-5p expression could decrease mitochondrial content and function and lead to the decline of brown adipocyte specific gene expression.
Figure 5.
Upregulation of miR-23b-5p expression inhibited brown adipocytes thermogenic program. (A) Mitochondrial DNA content following with vehicle (Veh) or Fsk (10 μM) for 1 h, calculated by comparison of Nd1 and Cox1 expression determined by qPCR. (B) Phase-contrast and MitoTracker (Red) fluorescence images of live cultured brown adipocytes (red) following with vehicle (veh) or Fsk (10 μM) for 4 h. Scale bar, 50 μm. (C) Representative metabolic flux curves of the oxygen consumption rate (OCR). Brown adipocytes from NC and miR-23b-5p groups were treated with respiratory inhibitors including oligomycin (Oligo), FCCP, and antimycin A plus rotenone (A&R) to dissect the various parts of the respiration program. (D) Quantitation of the OCRs including basal respiration, ATP production, maximal respiration, and Proton leak. Data were analyzed between NC and miR-23b-5p overexpression groups using Student’s t-test. n = 5 wells per group. (E) The release of glycerol content was normalized to the total amount of protein for each sample in NC and miR-23b-5p overexpression groups under the basal and Fsk stimulation. Data show the mean of three replicates for each experiment. (F) Brown adipocytes enriched genes including Ucp1, Ppargc1a, Dio2, Cidea, and Cycs were detected by qPCR and normalized to Ppia expression. (G) Representative images of UCP1 (red) and Dapi (blue) in NC and miR-23b-5p overexpression brown adipocytes. Scale bar: 50 μm. (H) Immunoblot of UCP1 (predicted in 32 kDa) expression in the NC and miR-23b-5p overexpression brown adipocytes. ACTB was used as the internal control. Values are the means ± s.d. of three separate experiments. *P < 0.05; **P < 0.01; ***P < 0.001 vs the NC groups.
Overexpression of miR-23b-5p inhibited the thermogenic program through lipolysis and fatty acid β-oxidation pathways
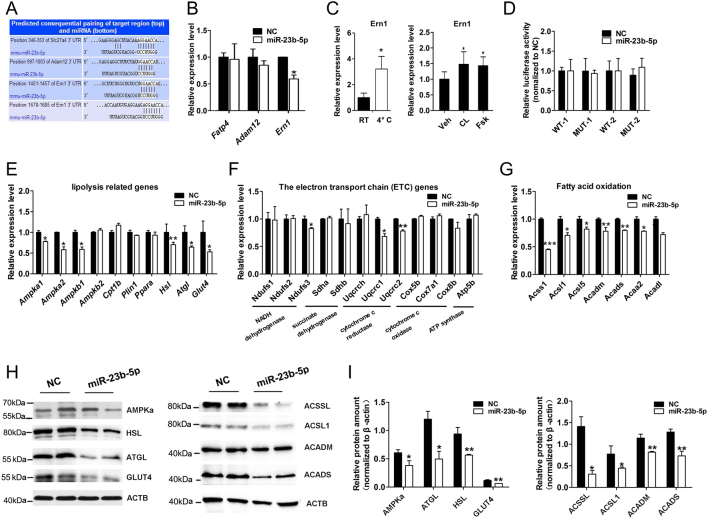
Based on the miRNA target prediction analysis (www.targetscan.org), target genes of miR-23b-5p with the matched sites in the 3′UTR region were selected (Fig. 6A). Fatty acid transporter 4 (Fatp4), a disintegrin, metallopeptidase domain 12 (Adam12), and endoplasmic reticulum to nucleus signaling 1 (Ern1) were selected for further validation among all the potential targets and these genes were identified to be involved in fat metabolism (39, 40, 41). We further detected the expression of these potential targets by qPCR in miR-23b-5p over-expressing adipocytes and results showed a significant decrease of Ern1 (Fig. 6B). Ern1 expression was increased by CL and Fsk stimulation (Fig. 6C). However, luciferase reporter assays showed that miR-23b-5p overexpression did not alter the activity of the reporter construct harboring WT (WT1 and WT2) or mutant (MUT1 and MUT2) 3′UTRs (Fig. 6D).
Figure 6.
Overexpression of miR-23b-5p impaired brown adipocytes thermogenesis mainly through decreasing lipolysis-and β-oxidation-related genes expression. (A) TargetScan predicted miR-23b-5p binds the 3′ UTR of Slc27a4 (Fatp4), Adam12, and Ern1. (B) The relative expression of targets genes after miR-23b-5p overexpression in mouse brown adipocytes. (C) Ern1 expression level upon cold exposure in BAT and its expression level in the brown adipocytes treated with CL (10 µM) and Fsk (10 μM) for 4 h. (D) The psi-CHECK2 luciferase reporter plasmids containing Ern1 WT (WT1 and WT2) 3′UTR or mutant 3′UTR (MUT1 and MUT2) were transfected into mouse 3T3-L1 cells (six replicates per group) for 16 h followed by miR-23b-5p or NC lentivirus infection. After 48 h infection, cells were harvested and renilla luciferase activity was analyzed using the dual-luciferase reporter Assay System and normalized to firefly luciferase activity. (E, F, and G) The relative expression of lipolysis, the electron transport chain (ETC)-, fatty acid oxidation-related genes involved in brown adipocytes thermogenesis were detected in NC and miR-23b-5p overexpression groups by qPCR and normalized to Ppia expression. (H and I) The relative protein amount of AMPKa (predicted in 63 kDa), HSL (predicted in 81–83 kDa), ATGL (predicted in 54 kDa), GLUT4 (predicted in 55 kDa), ACCSL1 (predicted in 78 kDa), ACSL1 (predicted in 78 kDa), ACADM (predicted in 45 kDa), and ACADS (predicted in 43 kDa) among NC and miR-23b-5p overexpression groups. Values are the means ± s.d. of three separated experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
The thermogenic capacity of brown adipocytes relies on proper mitochondrial biogenesis and lipid metabolism. Recent studies identified several BAT pathways required for brown adipocyte thermogenesis and such genes involving in the respiratory chain/oxidative phosphorylation system (OxPhos) (42), lipolysis (43), and fatty acid β-oxidation (FAO) (44, 45). Next, we analyzed whether any of these genes were affected by miR-23b-5p overexpression, thus participating in sustaining thermogenesis. qPCR results showed expression levels of these genes decreased after miR-23b-5p overexpression including AMP-activated protein kinase alpha 1 catalytic subunit (Ampka1), AMP-activated protein kinase alpha 2 catalytic subunit (Ampka2), AMP-activated non-catalytic subunit beta 1 (Ampkb2), hormone sensitive lipase (Hsl), patatin-like phospholipase domain containing 2 (Atgl), and facilitated glucose transporter 4 (Glut4), all of which were involved in lipolysis (Fig. 6E). Similarly, characteristic mitochondrial FAO enzymes related genes in BAT compared to WAT including acyl-coenzyme A (acyl-CoA) synthetase short-chain family member 1 (Acss1), acyl-CoA synthetase-1 (Acsl1), acyl-CoA synthetase-5 (Acsl5), medium chain acyl-CoA dehydrogenase (Acadm), short chain acyl-CoA dehydrogenase (Acads), long-chain acyl-CoA dehydrogenase (I), and acetyl-CoA acyltransferase 2 (Acaa2) revealed a notable decrease after miR-23b-5p overexpression (Fig. 6G). However, these genes associated with the OxPhos system were similar between NC and miR-23b-5p overexpression groups. Moreover, only a few OxPhos genes’ expression including NADH dehydrogenase (ubiquinone) Fe-S protein 3 (Ndufs3), ubiquinol cytochrome c reductase core protein 1 (Uqcrc1), and ubiquinol cytochrome c reductase core protein 2 (Uqcrc2) were downregulated in miR-23b-5p overexpressing cells (Fig. 6F). Western blot further revealed that miR-23b-5p-transduced cells showed reduced protein expression when involved in lipolysis and fatty acid β-oxidation pathways (Fig. 6H and I).
Discussion
Brown and beige adipocytes are specialized for burning chemical energy to produce heat. Considering its potential effects as a target of anti-obesity therapeutics, studies in the development and activity of BAT have recently received a considerable amount of attention. Most of reported miRNAs, which play significant roles in adipogenesis and metabolic diseases, were responsive to cold exposure or downstream hormones. Therefore, identification of cold-responsive miRNAs in BAT is more beneficial in order to describe the molecular regulatory mechanism underlying thermogenic function.
miRNA microarray profiling of BAT from cold-stimulated mice may offer a look at BAT activity-related miRNAs and present a snapshot of how BAT responds to thermogenic activation. For example, Trajkovski et.al identified miRNAs during a short cold stimulation (one day, 8°C) (46) and another report by Cong Tao et al. also investigated differentially expressed miRNAs in murine BAT and WAT in response to prolonged cold stimulation (10 days, 6°C) (47). Similar BAT phenotype by cold exposure was observed no matter in our study (7 days, 4°C) as shown in Supplementary Fig. 1 or Tao’s study (10 days, 6°C). Actually, we compared our small RNA-seq data as shown in Fig. 1 A and B with Tao’s and found that six miRNAs (6/27) overlapped in their sequencing, because we sequenced more miRNAs (6/82) and thus the overlapping rate (6/82) is relatively low. These overlapping miRNAs include miR-3074-5p, miR-196b-5p, miR-203-5p, miR-455-5p, miR-3073a-3p, and miR-150-3p. This relatively low overlapping rate may result from experimental error and length of exposure (7 days vs 10 days). The previous microarray results observed that several miRNAs including miR-133a-3p (24), miR-182-5p (48), miR-1a-3p (49), miR-135a-5p (50) and miR-150-3p (47) were also detected in our RNA-seq. RNA-sequencing has the advantage of quantifying low-abundance miRNAs without prior sequence knowledge (51), facilitating a more accurate determination and quantification of ‘unknown’ miRNAs expressed in BAT or miRNAs responsive to cold exposure. miR-23b-5p and miR-150-3p could be detected with the relatively low abundance in adipose tissues and mature adipocytes induced from primary cultures of SVF, while their expression patterns exert a significantly downregulated trend after cold exposure in Fig. 1B or pharmaceutical stimulation (CL and Fsk) in Fig. 2C and G. Additionally, we also found a higher abundance of miR-23b-5p in skeletal muscle compared to other adipose depot as shown in Supplementary Fig. 3. As described by Fei Yu et al., miR-23b-5p was upregulated in myocardial tissues from chronic heart failure and its overexpression in vivo showed a aggravated cardiac hypertrophy and dysfunction (52). This observation may indicate a potential effect of miR-23b-5p on differentiation or function of skeletal muscle, which possess a similar structure with cardiac muscle in some extent. This deserves our further exploration of miR-23b-5p in myotubes differentiation or metabolism.
While BAT from rodents and humans presents the differences in anatomic location, cell composition, and molecular signatures (53), the function of mitochondrial UCP1 was comparable in the two species from a physiological perspective (54). Importantly, the classical BAT in humans is mostly from fetuses or newborns and represents a rapid adrenergic stimulation resembling that in rodents (55). Revealed by Güller et al., a comparison of miRNA profiles between mouse and human BAT further validated an expression pattern of 145 miRNAs in both species (56). Our results suggested that miRNAs involved in cold-mediated mouse BAT thermogenic activation also could be detected in the human brown adipocytes and showed a similar responsiveness to β3-adrenergic-cAMP signaling as shown in Fig. 2C, D, E, F, G, and H. Characterization of miR-23b-5p expression during the preadipocytes differentiation course of mouse and human BAT was shown in Fig. 4A.
Activation of brown adipocytes is characterized by enhanced mitochondria numbers (57), oxidative respiration (58), and lipolysis capacities (59). We detected the previously mentioned indicators and the results revealed that miR-23b-5p overexpression led to decreased mitochondria content and impaired oxidative respiration and lipolysis, especially after Fsk stimulation as shown in Fig. 5. To attain the molecular regulatory mechanism underlying the effect of miR-23b-5p on BAT thermogenic programs, we screened several genes with matched sites in 3′UTR region and expression validations revealed that Ern1 could be the predicted target of miR-23b-5p as defined in Fig. 6B and C. However, the luciferase reporter experiments demonstrated that miR-23b-5p did not interact with the predicted target sites of the Ern1 transcripts as shown in Fig. 6D. Based on the previous study, the verified target of miR-23b-5p is high-mobility group box 2 (Hmgb2) in myocardial tissues (52). However, Hmgb2 was observed with pro-adipogenic effect in mesenchymal stem cells (60), which is not associated with the phenotype in mouse brown adipocytes as shown in Fig. 4C. We did not find targeting genes of miR-23b-5p through bioinformatics prediction and reported studies, but scanning these genes involved in several BAT pathways indirectly indicated that miR-23b-5p might regulate brown adipocyte thermogenic programs through lipolysis and fatty acid β-oxidation pathways as described in Fig. 6E, G, H and I. Previous research concerning knockout mice models revealed that lipolysis-related metabolism in adipose tissue was largely dependent on ATGL and HSL expression (61, 62). Specifically, ATGL had a profound effect on BAT phenotype and affected the expression levels of other lipolytic enzymes mainly in BAT rather than WAT. AMPK is a cellular sensor of energy homeostasis and has also been confirmed to be critical in the maintenance of mitochondrial function (63) as well as lipolytic capacity in BAT (61). In comparison with WAT, BAT is characterized by abundant FAO enzymes. First, fatty acids delivered to the mitochondria are activated to fatty acyl-CoAs by acyl-CoA synthetases. ACSL1 contributes 80% of total ACSL activity in adipose tissue, and knockout of ACSL1 expression in vivo showed a deficiency in FAO and remarkable cold intolerance (64). Among the acyl-CoA dehydrogenases (ACADL, ACADM, and ACADS), higher ACADM expression was associated with the increased UCP1 expression in brown-like adipocytes and promoted FAO capacity in brown adipocytes (65). Though we found evidence for miR-23b-5p in the modulation of brown adipocyte function, further research is needed to elucidate its direct association with key molecules such as Ampk, Atgl, and Acadm.
In summary, we performed RNA-sequencing to identify miRNAs in mouse iBAT under cold conditions and validated that cold-regulated miRNAs in mouse brown adipocytes showed a similar expression pattern to human brown adipocytes exposed to β3-adrenergic or cAMP signaling stimulation. Moreover, miR-23b-5p was demonstrated to be a negative regulator in controlling brown adipocyte thermogenic gene program. There are also existing some limitations in our study, such as functional evaluation after miR-23b-5p inhibition in brown adipocytes and elucidation of direct targets of miR-23b-5p. Further experiments are needed in the future. Together, our observations show cold-responsive miRNAs in mouse BAT and close regulation in human brown adipocytes, further providing potential molecular therapeutic targets to combatobesity.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81600687, 81671359, 81800772, and 81670773), Jiangsu Province Natural Science Foundation (BK20191126, BK20180146), The Subproject of Key Research and Development Program of China (Grant No. 2016YFC1000204-6), Jiangsu Provincial Medical Innovation Team (Grant No. CXTDA2017001), The ‘six talent peak’ High-level Talents Training Project of Jiangsu Province (Grant No. WSN-165), Nanjing Medical Science and Technique Development Foundation (Grant No. QRX17160, YKK17177), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (Grant No. JX22013535).
References
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews 2004. 84 277–359. ( 10.1152/physrev.00015.2003) [DOI] [PubMed] [Google Scholar]
- 2.Liu X, Zheng Z, Zhu X, Meng M, Li L, Shen Y, Chi Q, Wang D, Zhang Z, Li C, et al. Brown adipose tissue transplantation improves whole-body energy metabolism. Cell Research 2013. 23 851–854. ( 10.1038/cr.2013.64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanford KI, Middelbeek RJ, Townsend KL, An D, Nygaard EB, Hitchcox KM, Markan KR, Nakano K, Hirshman MF, Tseng YH, et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. Journal of Clinical Investigation 2013. 123 215–223. ( 10.1172/JCI62308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. New England Journal of Medicine 2009. 360 1509–1517. ( 10.1056/NEJMoa0810780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. New England Journal of Medicine 2009. 360 1518–1525. ( 10.1056/NEJMoa0808949) [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Zhang M, Xu M, Gu W, Xi Y, Qi L, Li B, Wang W. Brown adipose tissue activation is inversely related to central obesity and metabolic parameters in adult human. PLoS ONE 2015. 10 e0123795 ( 10.1371/journal.pone.0123795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao Q, Yadav R, Basse AL, Petersen S, Sonne SB, Rasmussen S, Zhu Q, Lu Z, Wang J, Audouze K, et al. Transcriptome profiling of brown adipose tissue during cold exposure reveals extensive regulation of glucose metabolism. American Journal of Physiology: Endocrinology and Metabolism 2015. 308 E380–E392. ( 10.1152/ajpendo.00277.2014) [DOI] [PubMed] [Google Scholar]
- 8.Orava J, Nuutila P, Lidell ME, Oikonen V, Noponen T, Viljanen T, Scheinin M, Taittonen M, Niemi T, Enerback S, et al. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metabolism 2011. 14 272–279. ( 10.1016/j.cmet.2011.06.012) [DOI] [PubMed] [Google Scholar]
- 9.Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. Brown adipose tissue activity controls triglyceride clearance. Nature Medicine 2011. 17 200–205. ( 10.1038/nm.2297) [DOI] [PubMed] [Google Scholar]
- 10.Chondronikola M, Volpi E, Borsheim E, Porter C, Saraf MK, Annamalai P, Yfanti C, Chao T, Wong D, Shinoda K, et al. Brown adipose tissue activation is linked to distinct systemic effects on lipid metabolism in humans. Cell Metabolism 2016. 23 1200–1206. ( 10.1016/j.cmet.2016.04.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner A, Kjellstedt A, Carreras A, Bottcher G, Peng XR, Seale P, Oakes N, Linden D. Activation of beta3-adrenoceptors increases in vivo free fatty acid uptake and utilization in brown but not white fat depots in high-fat-fed rats. American Journal of Physiology: Endocrinology and Metabolism 2016. 311 E901–E910. ( 10.1152/ajpendo.00204.2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cypess AM, Weiner LS, Roberts-Toler C, Franquet EE, Kessler SH, Kahn PA, English J, Chatman K, Trauger SA, Doria A, et al. Activation of human brown adipose tissue by a beta3-adrenergic receptor agonist. Cell Metabolism 2015. 21 33–38. ( 10.1016/j.cmet.2014.12.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Xia T, Du Y, Meng Q, Li H, Liu B, Chen S, Guo F. Effects of ATF4 on PGC1alpha expression in brown adipose tissue and metabolic responses to cold stress. Metabolism: Clinical and Experimental 2013. 62 282–289. ( 10.1016/j.metabol.2012.07.017) [DOI] [PubMed] [Google Scholar]
- 14.Dempersmier J, Sambeat A, Gulyaeva O, Paul SM, Hudak CS, Raposo HF, Kwan HY, Kang C, Wong RH, Sul HS. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Molecular Cell 2015. 57 235–246. ( 10.1016/j.molcel.2014.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell 2014. 158 69–83. ( 10.1016/j.cell.2014.04.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hattori K, Naguro I, Okabe K, Funatsu T, Furutani S, Takeda K, Ichijo H. ASK1 signalling regulates brown and beige adipocyte function. Nature Communications 2016. 7 11158 ( 10.1038/ncomms11158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi SY, Zhang W, Luk CT, Sivasubramaniyam T, Brunt JJ, Schroer SA, Desai HR, Majerski A, Woo MJAK. JAK2 promotes brown adipose tissue function and is required for diet- and cold-induced thermogenesis in mice. Diabetologia 2016. 59 187–196. ( 10.1007/s00125-015-3786-2) [DOI] [PubMed] [Google Scholar]
- 18.Lee P, Linderman JD, Smith S, Brychta RJ, Wang J, Idelson C, Perron RM, Werner CD, Phan GQ, Kammula US, et al. Irisin and FGF21 are cold-induced endocrine activators of brown fat function in humans. Cell Metabolism 2014. 19 302–309. ( 10.1016/j.cmet.2013.12.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Than A, He HL, Chua SH, Xu D, Sun L, Leow MK, Chen P. Apelin enhances brown adipogenesis and browning of white adipocytes. Journal of Biological Chemistry 2015. 290 14679–14691. ( 10.1074/jbc.M115.643817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nature Cell Biology 2011. 13 958–965. ( 10.1038/ncb2286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori M, Nakagami H, Rodriguez-Araujo G, Nimura K, Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biology 2012. 10 e1001314 ( 10.1371/journal.pbio.1001314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X, Zhang Z, Zeng T, Lim YC, Wang Y, Xie X, Yang S, Huang C, Xu M, Tao L, et al. cAMP-MicroRNA-203-IFNgamma network regulates subcutaneous white fat browning and glucose tolerance. Molecular Metabolism 2019. 28 36–47. ( 10.1016/j.molmet.2019.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan D, Mao C, Quattrochi B, Friedline RH, Zhu LJ, Jung DY, Kim JK, Lewis B, Wang YX. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nature Communications 2014. 5 4725 ( 10.1038/ncomms5725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nature Cell Biology 2012. 14 1330–1335. ( 10.1038/ncb2612) [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Siegel F, Kipschull S, Haas B, Frohlich H, Meister G, Pfeifer A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nature Communications 2013. 4 1769 ( 10.1038/ncomms2742) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Trajkovski M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism: Clinical and Experimental 2014. 63 272–282. ( 10.1016/j.metabol.2013.10.004) [DOI] [PubMed] [Google Scholar]
- 27.Karbiener M, Pisani DF, Frontini A, Oberreiter LM, Lang E, Vegiopoulos A, Mossenbock K, Bernhardt GA, Mayr T, Hildner F, et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 2014. 32 1578–1590. ( 10.1002/stem.1603) [DOI] [PubMed] [Google Scholar]
- 28.Giroud M, Karbiener M, Pisani DF, Ghandour RA, Beranger GE, Niemi T, Taittonen M, Nuutila P, Virtanen KA, Langin D, et al. Let-7i-5p represses Brite adipocyte function in mice and humans. Scientific Reports 2016. 6 28613 ( 10.1038/srep28613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Cao Y, Cui X, Wang X, Zhou Y, Huang F, Wang X, Wen J, Xie K, Xu P, et al. miR-199a-3p regulates brown adipocyte differentiation through mTOR signaling pathway. Molecular and Cellular Endocrinology 2018. 476 155–164. ( 10.1016/j.mce.2018.05.005) [DOI] [PubMed] [Google Scholar]
- 30.You LH, Zhu LJ, Yang L, Shi CM, Pang LX, Zhang J, Cui XW, Ji CB, Guo XR. Transcriptome analysis reveals the potential contribution of long noncoding RNAs to brown adipocyte differentiation. Molecular Genetics and Genomics 2015. 290 1659–1671. ( 10.1007/s00438-015-1026-6) [DOI] [PubMed] [Google Scholar]
- 31.Seiler SE, Xu D, Ho JP, Lo KA, Buehrer BM, Ludlow YJ, Kovalik JP, Sun L. Characterization of a primary brown adipocyte culture system derived from human fetal interscapular fat. Adipocyte 2015. 4 303–310. ( 10.1080/21623945.2015.1042192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, You L, Cui X, Li Y, Wang X, Xu P, Zhu L, Wen J, Pang L, Guo X, et al. Evaluation and optimization of differentiation conditions for human primary brown adipocytes. Scientific Reports 2018. 8 5304 ( 10.1038/s41598-018-23700-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You L, Zhou Y, Cui X, Wang X, Sun Y, Gao Y, Wang X, Wen J, Xie K, Tang R, et al. GM13133 is a negative regulator in mouse white adipocytes differentiation and drives the characteristics of brown adipocytes. Journal of Cellular Physiology 2018. 233 313–324. ( 10.1002/jcp.25878) [DOI] [PubMed] [Google Scholar]
- 34.Chou CF, Lin YY, Wang HK, Zhu X, Giovarelli M, Briata P, Gherzi R, Garvey WT, Chen CY. KSRP ablation enhances brown fat gene program in white adipose tissue through reduced miR-150 expression. Diabetes 2014. 63 2949–2961. ( 10.2337/db13-1901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nature Cell Biology 2012. 14 1330–1335. ( 10.1038/ncb2612) [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Guan M, Townsend KL, Huang TL, An D, Yan X, Xue R, Schulz TJ, Winnay J, Mori M, et al. MicroRNA-455 regulates brown adipogenesis via a novel HIF1an-AMPK-PGC1alpha signaling network. EMBO Reports 2015. 16 1378–1393. ( 10.15252/embr.201540837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald RB, Horwitz BA. Brown adipose tissue thermogenesis during aging and senescence. Journal of Bioenergetics and Biomembranes 1999. 31 507–516. ( 10.1023/a:1005404708710) [DOI] [PubMed] [Google Scholar]
- 38.Pfannenberg C, Werner MK, Ripkens S, Stef I, Deckert A, Schmadl M, Reimold M, Haring HU, Claussen CD, Stefan N. Impact of age on the relationships of brown adipose tissue with sex and adiposity in humans. Diabetes 2010. 59 1789–1793. ( 10.2337/db10-0004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenz LS, Marx J, Chamulitrat W, Kaiser I, Grone HJ, Liebisch G, Schmitz G, Elsing C, Straub BK, Fullekrug J, et al. Adipocyte-specific inactivation of acyl-CoA synthetase fatty acid transport protein 4 (Fatp4) in mice causes adipose hypertrophy and alterations in metabolism of complex lipids under high fat diet. Journal of Biological Chemistry 2011. 286 35578–35587. ( 10.1074/jbc.M111.226530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masaki M, Kurisaki T, Shirakawa K, Sehara-Fujisawa A. Role of meltrin {alpha} (ADAM12) in obesity induced by high-fat diet. Endocrinology 2005. 146 1752–1763. ( 10.1210/en.2004-1082) [DOI] [PubMed] [Google Scholar]
- 41.Asada R, Kanemoto S, Matsuhisa K, Hino K, Cui M, Cui X, Kaneko M, Imaizumi K. IRE1alpha-XBP1 is a novel branch in the transcriptional regulation of Ucp1 in brown adipocytes. Scientific Reports 2015. 5 16580 ( 10.1038/srep16580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pardo R, Blasco N, Vila M, Beiroa D, Nogueiras R, Canas X, Simo R, Sanchis D, Villena JA. EndoG Knockout mice show increased brown adipocyte recruitment in white adipose tissue and improved glucose homeostasis. Endocrinology 2016. 157 3873–3887. ( 10.1210/en.2015-1334) [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Fromme T, Schweizer S, Schottl T, Klingenspor M. Taking control over intracellular fatty acid levels is essential for the analysis of thermogenic function in cultured primary brown and brite/beige adipocytes. EMBO Reports 2014. 15 1069–1076. ( 10.15252/embr.201438775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metabolism 2009. 10 324–335. ( 10.1016/j.cmet.2009.08.014) [DOI] [PubMed] [Google Scholar]
- 45.Nakamura Y, Sato T, Shiimura Y, Miura Y, Kojima M. FABP3 and brown adipocyte-characteristic mitochondrial fatty acid oxidation enzymes are induced in beige cells in a different pathway from UCP1. Biochemical and Biophysical Research Communications 2013. 441 42–46. ( 10.1016/j.bbrc.2013.10.014) [DOI] [PubMed] [Google Scholar]
- 46.Liu W, Bi P, Shan T, Yang X, Yin H, Wang YX, Liu N, Rudnicki MA, Kuang S. miR-133a regulates adipocyte browning in vivo. PLoS Genetics 2013. 9 e1003626 ( 10.1371/journal.pgen.1003626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao C, Huang S, Wang Y, Wei G, Zhang Y, Qi D, Wang Y, Li K. Changes in white and brown adipose tissue microRNA expression in cold-induced mice. Biochemical and Biophysical Research Communications 2015. 463 193–199. ( 10.1016/j.bbrc.2015.05.014) [DOI] [PubMed] [Google Scholar]
- 48.Kim HJ, Cho H, Alexander R, Patterson HC, Gu M, Lo KA, Xu D, Goh VJ, Nguyen LN, Chai X, et al. MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes 2014. 63 4045–4056. ( 10.2337/db14-0466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. Journal of Cellular Physiology 2009. 218 444–449. ( 10.1002/jcp.21621) [DOI] [PubMed] [Google Scholar]
- 50.Chen C, Peng Y, Peng Y, Peng J, Jiang S. miR-135a-5p inhibits 3T3-L1 adipogenesis through activation of canonical Wnt/beta-catenin signaling. Journal of Molecular Endocrinology 2014. 52 311–320. ( 10.1530/JME-14-0013) [DOI] [PubMed] [Google Scholar]
- 51.Eipper-Mains JE, Eipper BA, Mains RE. Global approaches to the role of miRNAs in drug-induced changes in gene expression. Frontiers in Genetics 2012. 3 109 ( 10.3389/fgene.2012.00109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boureima OD, Ji H, Xu J, Li S, Ruan W, Xiao F, Yu F. Involvement of microRNA-23b-5p in the promotion of cardiac hypertrophy and dysfunction via the HMGB2 signaling pathway. Biomedicine and Pharmacotherapy 2019. 116 108977 ( 10.1016/j.biopha.2019.108977) [DOI] [PubMed] [Google Scholar]
- 53.Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homoe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J, et al. A classical brown adipose tissue mRNA signature partly overlaps with Brite in the supraclavicular region of adult humans. Cell Metabolism 2013. 17 798–805. ( 10.1016/j.cmet.2013.04.011) [DOI] [PubMed] [Google Scholar]
- 54.Porter C, Herndon DN, Chondronikola M, Chao T, Annamalai P, Bhattarai N, Saraf MK, Capek KD, Reidy PT, Daquinag AC, et al. Human and mouse brown adipose tissue mitochondria have comparable UCP1 function. Cell Metabolism 2016. 24 246–255. ( 10.1016/j.cmet.2016.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Franco A, Guasti D, Squecco R, Mazzanti B, Rossi F, Idrizaj E, Gallego-Escuredo JM, Villarroya F, Bani D, Forti G, et al. Searching for classical brown fat in humans: development of a novel human fetal brown stem cell model. Stem Cells 2016. 34 1679–1691. ( 10.1002/stem.2336) [DOI] [PubMed] [Google Scholar]
- 56.Guller I, McNaughton S, Crowley T, Gilsanz V, Kajimura S, Watt M, Russell AP. Comparative analysis of microRNA expression in mouse and human brown adipose tissue. BMC Genomics 2015. 16 820 ( 10.1186/s12864-015-2045-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan X, Wei G, You Y, Huang Y, Lee HJ, Dong M, Lin J, Hu T, Zhang H, Zhang C, et al. Rutin ameliorates obesity through brown fat activation. FASEB Journal 2017. 31 333–345. ( 10.1096/fj.201600459RR) [DOI] [PubMed] [Google Scholar]
- 58.Schilperoort M, van Dam AD, Hoeke G, Shabalina IG, Okolo A, Hanyaloglu AC, Dib LH, Mol IM, Caengprasath N, Chan YW, et al. The GPR120 agonist TUG-891 promotes metabolic health by stimulating mitochondrial respiration in brown fat. EMBO Molecular Medicine 2018. 10 e8047 ( 10.15252/emmm.201708047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mottillo EP, Bloch AE, Leff T, Granneman JG. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) alpha and delta in brown adipocytes to match fatty acid oxidation with supply. Journal of Biological Chemistry 2012. 287 25038–25048. ( 10.1074/jbc.M112.374041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee D, Taniguchi N, Sato K, Choijookhuu N, Hishikawa Y, Kataoka H, Morinaga H, Lotz M, Chosa E. HMGB2 is a novel adipogenic factor that regulates ectopic fat infiltration in skeletal muscles. Scientific Reports 2018. 8 9601 ( 10.1038/s41598-018-28023-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morak M, Schmidinger H, Riesenhuber G, Rechberger GN, Kollroser M, Haemmerle G, Zechner R, Kronenberg F, Hermetter A. Adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) deficiencies affect expression of lipolytic activities in mouse adipose tissues. Molecular and Cellular Proteomics 2012. 11 1777–1789. ( 10.1074/mcp.M111.015743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metabolism 2011. 13 739–748. ( 10.1016/j.cmet.2011.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mottillo EP, Desjardins EM, Crane JD, Smith BK, Green AE, Ducommun S, Henriksen TI, Rebalka IA, Razi A, Sakamoto K, et al. Lack of adipocyte AMPK exacerbates insulin resistance and hepatic steatosis through brown and beige adipose tissue function. Cell Metabolism 2016. 24 118–129. ( 10.1016/j.cmet.2016.06.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellis JM, Li LO, Wu PC, Koves TR, Ilkayeva O, Stevens RD, Watkins SM, Muoio DM, Coleman RA. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metabolism 2010. 12 53–64. ( 10.1016/j.cmet.2010.05.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gan L, Liu Z, Chen Y, Dan L, Feng F, Liu G, Sun C. alpha-MSH and Foxc2 promote fatty acid oxidation through C/EBPbeta negative transcription in mice adipose tissue. Scientific Reports 2016. 6 36661 ( 10.1038/srep36661) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a