Significance
Enteric pathogens successfully colonize the host intestinal environment by regulating the expression of their metabolic and virulence genes. A high-throughput screen identified the arginine sensor ArgR as a regulator of virulence expression in enterohemorrhagic Escherichia coli. Here we show that ArgR senses arginine fluctuations and regulates the virulence of enterohemorrhagic E. coli and Citrobacter rodentium, both in vitro and during murine infections. Our work highlights the importance of gut metabolite sensing in affecting the disease outcome and vulnerability to pathogens.
Keywords: EHEC, arginine, ArgR
Abstract
Microbiota, host and dietary metabolites/signals compose the rich gut chemical environment, which profoundly impacts virulence of enteric pathogens. Enterohemorrhagic Escherichia coli (EHEC) engages a syringe-like machinery named type-III secretion system (T3SS) to inject effectors within host cells that lead to intestinal colonization and disease. We previously conducted a high-throughput screen to identify metabolic pathways that affect T3SS expression. Here we show that in the presence of arginine, the arginine sensor ArgR, identified through this screen, directly activates expression of the genes encoding the T3SS. Exogenously added arginine induces EHEC virulence gene expression in vitro. Congruently, a mutant deficient in arginine transport (ΔartP) had decreased virulence gene expression. ArgR also augments murine disease caused by Citrobacter rodentium, which is a murine pathogen extensively employed as a surrogate animal model for EHEC. The source of arginine sensed by C. rodentium is not dietary. At the peak of C. rodentium infection, increased arginine concentration in the colon correlated with down-regulation of the host SLC7A2 transporter. This increase in the concentration of colonic arginine promotes virulence gene expression in C. rodentium. Arginine is an important modulator of the host immune response to pathogens. Here we add that arginine also directly impacts bacterial virulence. These findings suggest that a delicate balance between host and pathogen responses to arginine occur during disease progression.
Enteric pathogens integrate sensing and responding to multiple metabolites/signals within the gastrointestinal tract to regulate expression of key virulence genes. This surveillance allows for the spatial–temporal control of virulence gene expression, preventing unnecessary energy expenditure. Enterohemorrhagic Escherichia coli (EHEC) colonizes the human colon causing bloody diarrhea that may lead to hemolytic-uremic syndrome (HUS) (1, 2). EHEC virulence determinants include the locus of enterocyte effacement (LEE) pathogenicity island that encodes a type III secretion system (T3SS), and the potent Shiga toxin (Stx2a) that is responsible for HUS. EHEC utilizes the T3SS to translocate multiple effectors into colonocytes promoting cytoskeletal rearrangement and attaching and effacing (AE) lesion formation (1). The LEE is organized in five major operons LEE1 to LEE5. Transcription of all LEE genes is under the control of a master regulator, Ler, encoded within the LEE1 operon (1) (Fig. 1A). Transcription of ler is controlled by several host and microbiota-derived signals such as epinephrine, norepinephrine, succinate, fucose, serine, cysteine, and ethanolamine among others (3).
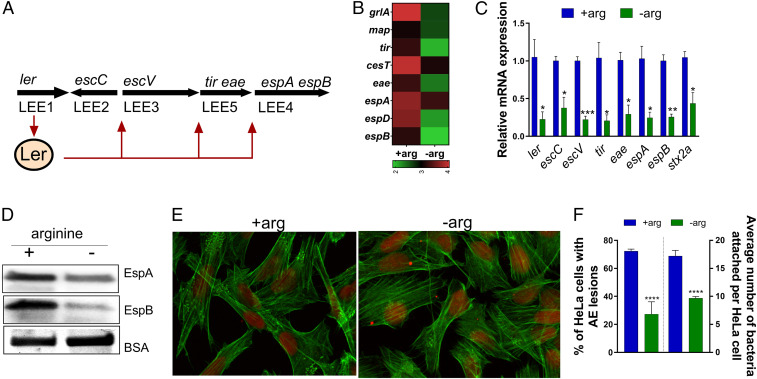
Fig. 1.
Arginine induces EHEC virulence in vitro. (A) Schematic of the LEE and the representative genes used in this work. The LEE1 operon encodes ler that activates expression of all five LEE operons. (B) Heat map showing the effect of arginine on LEE-encoded genes from WT EHEC grown in the presence (+arg) or absence (−arg) of 482 µM arginine. (C) qRT-PCR of LEE-encoded genes and Stx2a. (D) Western blot analysis of secreted proteins EspB and EspA from WT EHEC grown in the absence or presence of arginine. Experiments in B–D were performed in microaerophilic conditions using low-glucose DMEM, and samples were harvested in late log phase. (E) Fluorescein actin staining analysis. HeLa cells were infected with WT EHEC and incubated in the presence or absence of arginine for 4 h postinfection. HeLa cells were stained with FITC-phalloidin to visualize actin (green) and propidium iodide to stain for bacteria and nuclei (red). Pedestals were visualized as green dots. Pedestals were enumerated per cell (60×). (F) The percentage of infected cells and the number of pedestals per infected cell were quantified (n = 3). Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To dissect the metabolic pathways that influence virulence of EHEC, we previously performed a high-throughput screen (HTS) that identified the arginine sensor ArgR, and the transmembrane arginine importer ArtP as possible regulators of EHEC virulence gene expression (4). ArgR is a transcription factor encoded by the argR gene that inhibits the transcription of multiple genes including the arginine biosynthesis regulon, the arginine transport genes, as well as its own transcription (5, 6). ArgR also functions as a coactivator of the astCADBE operon encoding the arginine succinyltransferase pathway for arginine catabolism (7). Therefore, the biosynthesis, catabolism, and uptake of arginine are tightly regulated by ArgR. Biochemical and structural analyses have shown that the ArgR monomer consists of two domains separated by a protease-accessible linker: An N-terminal DNA-binding domain and a C-terminal arginine-binding domain (8). l-Arginine binding is required for ArgR oligomerization (9, 10). Mechanistically, in the presence of arginine, two trimers of ArgR dimerize forming a hexamer, which can bind to whole ARG boxes present in promoter regions, while trimeric ArgR still binds to half of the ARG box in the absence of l-arginine (11).
l-Arginine is an important amino acid essential for protein synthesis and plays an equally important role in stress conditions. l-Arginine acts as a precursor molecule for the biosynthesis of others amino acids, polyamines, and nitric oxide, and it also serves as a source of nitrogen upon its degradation via distinct catabolic routes (12). Arginine uptake in E. coli is mediated by three transport systems of the ATP-binding cassette transporter family (6, 13). These import systems are encoded by two gene clusters, artPIQM-artJ and argT-hisJQMP, which are repressed by ArgR in E. coli. These import systems diverge in substrate specificity and affinity for arginine, and display differences in the regulation of their own synthesis and activity (5). The importer encoded by artPIQM-artJ has the highest affinity for arginine (Kd 0.4 μM), where ArtP is an ATP-binding protein present in the membrane (5).
Here, we show that exogenous l-arginine up-regulates the expression of LEE-encoded genes and Shiga toxin (Stx) via ArgR in the enteric pathogens EHEC and Citrobacter rodentium (surrogate murine infection model). Disruption of l-arginine uptake systems in EHEC also decreases the expression of their virulence determinants. Moreover, we also show that C. rodentium infection increases the l-arginine levels in the colon that correlates with increased pathogenesis. In summary, our findings add another node to the complex metabolic interactions that intersect with virulence gene regulation to promote enteric disease.
Results
EHEC Virulence Is Increased by Exogenous l-Arginine.
Our previous HTS screen identified the ArgR transcription factor that is an arginine sensor, and ArtP that is a transmembrane arginine importer as involved in LEE gene regulation (4). To confirm whether arginine affects the expression of virulence genes, we performed RNA-sequencing (RNA-seq; data deposited at the European Nucleotide Archive, accession no. PRJEB35294) in EHEC grown in low-glucose DMEM under microaerophilic conditions [the optimal in vitro condition for LEE gene expression in EHEC (14)] with or without l-arginine. We observed that the presence of l-arginine increased the expression of LEE virulence genes in EHEC (Fig. 1 B–D), as well as non-LEE–encoded genes (SI Appendix, Figs. S1A and S3B). l-Arginine augments the expression of LEE-encoded (ler, escC, escV, tir, eae, espA, and espB) and non-LEE–encoded (stx2a, nleA, espN, espW, espX, espY, espR, espJ, espK, espM) genes (Fig. 1 B and C and SI Appendix, Figs. S1A and S3B). Congruent with the transcription effect of l-arginine on LEE genes, l-arginine also increased the secretion of both EspA and EspB proteins (Fig. 1D). Importantly, EHEC growth remains unaffected by l-arginine under these conditions (SI Appendix, Fig. S1B). This suggests that l-arginine–dependent LEE regulation is not due to a metabolic defect caused by the absence of l-arginine. Because LEE gene expression is necessary for AE lesion formation, we next assessed the role of l-arginine on AE lesion formation. AE lesions are characterized by remodeling of the epithelial cell cytoskeleton and formation of a pedestal-like structure beneath the bacteria (15). In accordance with the LEE-expression phenotype, l-arginine enhanced AE lesion formation and increased attachment of bacteria to HeLa cells (Fig. 1 E and F). l-Arginine also increased LEE gene expression in less permissive in vitro conditions (DMEM high glucose under microaerophilic conditions) (SI Appendix, Fig. S1C). Overall, these data indicate that arginine increases the virulence gene expression and AE lesion formation in EHEC.
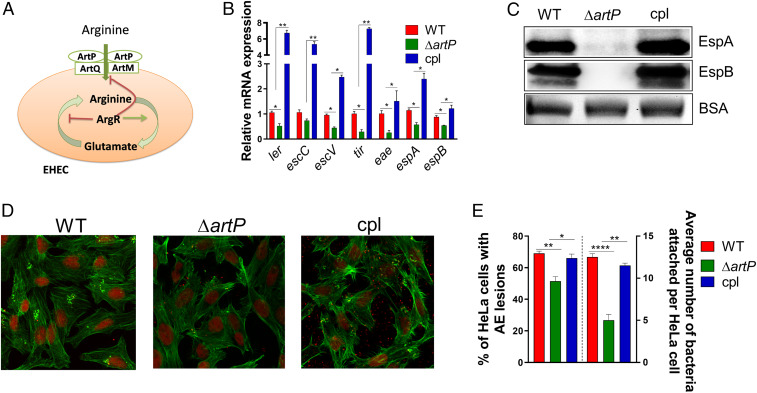
In E. coli, l-arginine is imported by binding to three periplasmic proteins, and is subsequently transported into the cell by two different transmembrane complexes (Fig. 2A). ArtP is a transmembrane protein necessary for the function of both arginine import systems (5). To investigate whether exogenous arginine regulates virulence gene expression in EHEC, we generated an artP mutant. Deletion of artP decreases LEE gene expression (ler, escC, escV, tir, eae, espA, and espB), and these phenotypes are rescued upon complementation (Fig. 2B). The overall decrease in transcription of LEE-encoded T3SS genes is corroborated with the decrease in the levels of the secreted proteins EspA and EspB (Fig. 2C), and decreased AE lesion formation in ΔartP EHEC (Fig. 2 D and E). Of note, EHEC growth was unaffected by deletion of artP (SI Appendix, Fig. S2A). These data further indicate that ArtP-dependent LEE regulation is not due to a metabolic defect, but to the lack of import of exogenous arginine (SI Appendix, Fig. S2A).
Fig. 2.
Arginine transporter ATP-binding protein artP regulates EHEC virulence in vitro. (A) Schematic of the arginine transmembrane transporter systems in E. coli. (B) qRT-PCR analysis to compare the expression of select virulence genes and (C) Western blot analysis of secreted proteins EspB and EspA from WT, ΔartP, and cpl EHEC (complemented with artP gene in pACYC184 vector). Experiments in B and C were performed in microaerophilic conditions using low-glucose DMEM with arginine, and samples were harvested in late log phase. (D) For fluorescein actin staining analysis, HeLa cells were infected with WT, ΔartP and complemented artP EHEC (clp) and incubated in low-glucose DMEM for 4 h postinfection in the presence of arginine. Then, the cells were stained with FITC-phalloidin to visualize actin (green) and propidium iodide to stain for bacteria and nuclei (red). Pedestals were visualized as green puncta. Pedestals were enumerated for each field (60×). (E) The percentage of infected cells and the number of pedestals per infected cell were quantified (n = 3). Error bars represent SEM. *P < 0.05, **P < 0.01, ****P < 0.0001.
In the absence of exogenous l-arginine, the arginine intracellular demand is likely met by the endogenous biosynthesis of arginine. The arginine biosynthetic pathway in E. coli requires eight enzymatic steps (12). To understand the relevance of endogenous arginine biosynthesis for EHEC, we generated a ΔargH mutant. The argH gene encodes the last enzyme in the arginine biosynthetic pathway. As predicted, the EHEC ΔargH can only grow in the presence of exogenous arginine (SI Appendix, Fig. S2 B and C), suggesting that ΔargH is auxotrophic for arginine. Taken together, these data suggest that exogenous arginine is not necessary for growth in WT EHEC, but up-regulates virulence.
ArgR Is a Direct Activator of LEE-Encoded Gene Expression.
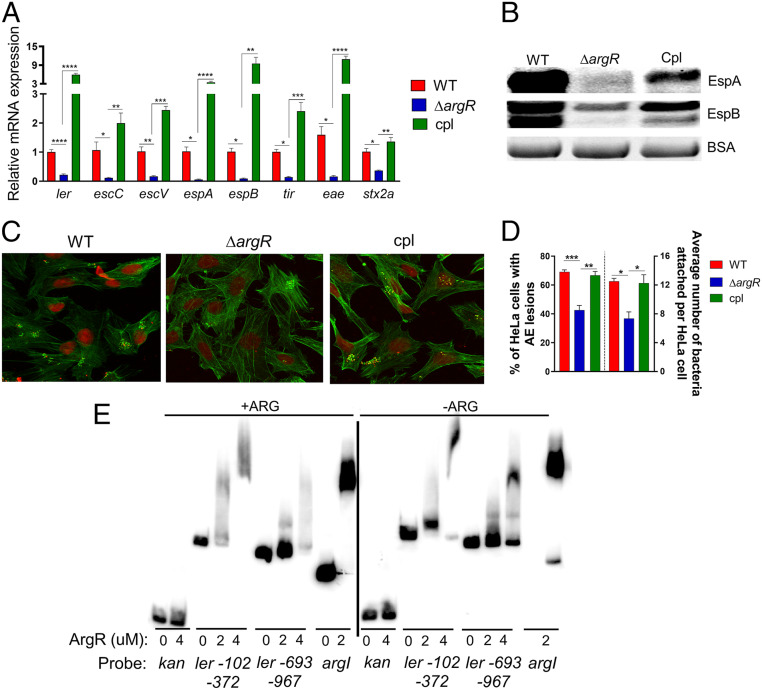
Next, we sought to investigate the role of the ArgR transcription factor in regulating EHEC virulence gene expression in response to arginine. Although the effects of ArgR on arginine metabolism are well described (6), its effect on modulating the virulence of pathogenic E. coli has not yet been explored. In the presence of arginine, ΔargR EHEC has no growth defect (SI Appendix, Fig. S3A). However, under these conditions, ΔargR had decreased LEE and nleA (non-LEE–encoded effector) gene expression (Fig. 3A and SI Appendix, Fig. S3B), reduced secretion of EspA and EspB (Fig. 3B), and AE lesion formation (Fig. 3 C and D) compared to WT and complemented strains. Of note, WT, ΔargR and complemented strains of EHEC grown in the absence of arginine showed no difference in LEE gene expression (SI Appendix, Fig. S3 C and D). Furthermore, WT grown in the absence of arginine expresses similar levels of EspB when compared to ΔargR grown with arginine (SI Appendix, Fig. S3E). These data suggest that ArgR induces LEE expression in vitro in an arginine-dependent manner.
Fig. 3.
Arginine repressor ArgR is a direct activator of LEE gene expression in vitro. (A) qRT-PCR analysis to compare the expression of select virulence genes and (B) Western blot analysis of secreted proteins EspB and EspA from WT, ΔargR and cpl EHEC (complemented with argR gene on the pACYC184). Experiments in A and B were performed in microaerophilic conditions using low-glucose DMEM with arginine, and samples were harvested in late log phase. (C) For fluorescein actin staining analysis, HeLa cells were infected with WT, ΔargR and complemented argR EHEC (clp) and incubated in low-glucose DMEM with arginine for 4 h postinfection. Then, the cells were stained with FITC-phalloidin to visualize actin (green) and propidium iodide to stain for bacteria and nuclei (red). Pedestals were visualized as a green spot. Pedestals were enumerated for each field (60×). (D) The percentage of infected cells and the number of pedestals per infected cell were quantified (n = 3). (E) EMSA of His-tagged ArgR with argI promoter probe (positive control), kanamycin promoter (negative control), and segments of the EHEC LEE1 regulatory region numbered from the proximal transcriptional start site. Error bars represent SEs of means (SEM). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Stx is associated with exacerbated disease. The stx genes are encoded within the λ-phage late genes, and are expressed upon induction of the phage lytic cycle. In induction the SOS pathway promotes the initiation of the lytic cycle via overproduction of RecA (16, 17). We observed decreased expression of stx2A in ΔargR (Fig. 3A) compared to WT and complemented strains. This effect was associated with reduced recA gene expression in an arginine-dependent manner (SI Appendix, Figs. S1A and S3B).
The role of ArgR in the arginine biosynthetic pathway is well characterized. A single hexameric ArgR binds to a tandem pair of ARG box sequences in an arginine-dependent fashion (9). In EHEC, the majority of the LEE genes are organized in five major operons (LEE1–5) (Fig. 1A), and the first gene within LEE1 encodes the master regulator Ler of the entire LEE (Fig. 3E) (15). Electrophoretic mobility shift assays (EMSAs) with ArgR (in the presence or absence of arginine) and overlapping fragments of the LEE1 promoter region, showed that it interacts with the LEE1 fragments ranging from −102 to −372 bp and −693 to −967 bp upstream of the proximal transcription start site (Fig. 3E and SI Appendix, Fig. S4). An in silico search for the ARG box consensus sequence found potential binding sites within both regions of the shifted probe fragments of the ler regulatory region (10). As expected, ArgR binds to the argI promoter region (positive control), but not to the promoter region of the kanamycin gene (negative control) (Fig. 3E). Notably, the absence of arginine led to an incomplete shift with the argI and LEE1 promoter regions (Fig. 3E), suggesting that arginine may enhance the interactions between ArgR and these promoter regions. Taken together, our data suggest that arginine increases the expression of LEE encoded genes via ArgR.
ArgR Increases C. rodentium Virulence during Murine Infection.
To investigate the role of ArgR and the arginine transporter ArtP in the progression of mammalian disease, we employed the C. rodentium murine infection model. C. rodentium is extensively used as a surrogate for EHEC infections in mice. C. rodentium contains the LEE and forms AE lesions on murine colonic cells (18). We generated ΔartP and ΔargR strains of DBS770 C. rodentium (19). The DBS770 C. rodentium strain was engineered to produce Stx, and when inoculated into mice displays key aspects of EHEC disease in humans: Formation of AE lesions, Stx-dependent intestinal inflammation, kidney damage, and lethality (19). The disease caused by the DBS770 strain has been extensively documented (19). Furthermore, this strain is the most suitable for these studies in light of the fact that exogenous arginine induced Stx expression in vitro (Fig. 1C) via ArgR (Fig. 3A).
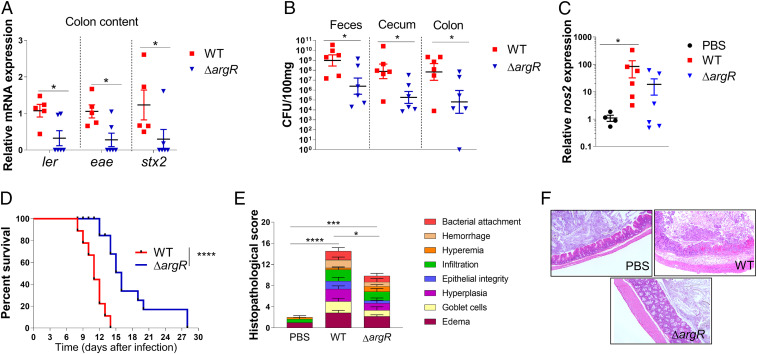
The ΔartP strain of DBS770 does not colonize the murine intestine, and consequently, mice infected with this mutant survive infection (SI Appendix, Fig. S5). These data suggest that ArtP is essential for C. rodentium gut colonization. Although ΔargR colonized the murine intestine, this strain presented decreased bacterial loads in stools, cecum, and colon tissues compared to WT (Fig. 4B). The attenuation of ΔargR can be attributed to decreased virulence gene expression in vivo (the LEE genes: ler and eae, and stx2a) (Fig. 4A). Furthermore, ΔargR presented decreased inflammation in the colon (Fig. 4 C, E, and F), and delayed mortality (Fig. 4D) compared to WT. The attenuated phenotype observed for ΔargR is not due to a growth defect, given that there is no difference in growth between ΔargR and WT DBS770 (SI Appendix, Fig. S6D). Furthermore, in vitro studies utilizing the DBS770 strain with exogenous arginine and ΔargR corroborated data obtained with EHEC (SI Appendix, Fig. S6). Exogenous arginine induced LEE expression (SI Appendix, Fig. S6 B and D) in C. rodentium. The ΔargR DBS770 strain also had reduced LEE expression (SI Appendix, Fig. S6 E and F) when compared to WT. Together, these results suggest that the arginine-dependent induction of virulence gene expression is conserved between EHEC and C. rodentium, and is important during murine infection.
Fig. 4.
ArgR increases C. rodentium virulence in vivo. (A) Expression of LEE gene and Stx2 from bacteria recovered from stools of C3H/HeJ mice infected with either WT or ΔargR C. rodentium 8 d postinfection. (B) CFUs recovered from cecum, colon, and stool samples of mice with infected either WT or ΔargR C. rodentium. (C) Expression of nos2 in the colon tissue obtained from C. rodentium or PBS-treated mice, 8 d postinfection. (D) Survival curve of mice infected with either WT or ΔargR C. rodentium. A total of n = 10 mice per group were used in this study. (E) Histological score and (F) representation (10×) of cecum 8 d after C. rodentium infection (n = 3). Each symbol represents an individual mouse. Error bars represent SEM. *P < 0.05, ***P < 0.001, ****P < 0.0001.
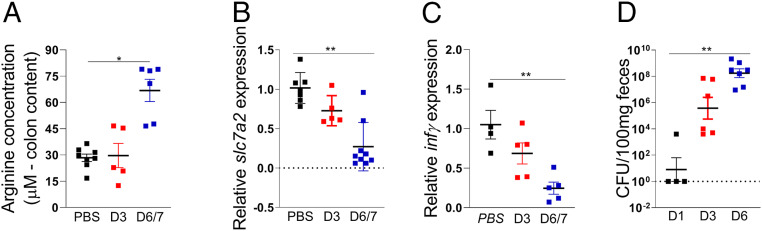
Inasmuch as ArgR-dependent LEE regulation requires arginine, we measured the intestinal levels of this amino acid in the colon of mice either infected or uninfected with C. rodentium at different time points. Arginine increased in the colon of infected mice 6/7 d postinfection compared to uninfected (PBS) animals (Fig. 5A). This increase correlated with the peak of infection, and enhanced C. rodentium colonization at 6 d postinfection (Fig. 5D), suggesting that arginine sensing by C. rodentium might dictate its expansion in the gut. This increase in arginine levels in the colon correlates with the decreased expression of the solute carrier family 7 member 2 (SLC7A2) that transports l-arginine into mammalian cells, and IFN γ, which activates expression of SLC7A2 (Fig. 5 B and C) (20). These data are opposite from a previous report (20) where expression of SLC7A2 was increased during C. rodentium infection. We note that in this previous study these measurements were taken at day 14 postinfection, when infection is at the resolution process in the less susceptible murine strain C57BL/6 (18, 20), while we investigated these parameters at the peak of infection in susceptible C3H/HeJ that succumb to death (18). It is worth noting that differences in susceptibility to C. rodentium infection between these murine strains are not associated with differences in Toll-like receptor 4 signaling, and are due to the more severe diarrhea caused by C. rodentium in C3H/HeJ mice (21, 22). Additionally, we also used different C. rodentium strains. Singh et al. (20) used C. rodentium strain DBS100, while we used DBS770. Both strains harbor the LEE, as EHEC, which is necessary to form AE lesions on enterocytes of mice leading to colonic hyperplasia and colitis (23, 24). We used the C. rodentium DBS770 strain engineered to produce Stx, which when inoculated into mice display key aspects of EHEC disease in humans: Formation of AE lesions, Stx-dependent intestinal inflammation, kidney damage, and lethality. The disease caused by the DBS770 strain has been extensively documented, and this model is a strong in vivo model for studying EHEC pathogenesis (4, 19, 25, 26). The source of arginine modulating C. rodentium virulence in the colon is not dictated by diet, given that mice fed chow with or without arginine had similar pathogen burdens (SI Appendix, Fig. S7A). Importantly, there was no difference in arginine levels obtained from the colons of mice fed chow with or without arginine (SI Appendix, Fig. S7B).
Fig. 5.
Arginine levels is incresead in colon content after C. rodentium infection. (A) Arginine measurements in colon contents, (B) slc7a2, and (C) infγ mRNA expression in colon tissue of mice in the third and sixth days after WT C. rodentium infection or not (PBS). (D) CFU in stools of mice in the first, third, and sixth days after WT C. rodentium infection. Each symbol represents an individual mouse. Error bars represent SEM. *P < 0.05, **P < 0.01.
Discussion
The gastrointestinal environment is complex and profoundly impacted by the membership of the resident microbiota, host genetics, diet, and enteric diseases. Bacterial enteric pathogens evolved intricate regulatory systems to integrate multiple cues toward exquisite control of the expression of their virulence repertoire (3, 27). The significance of this molecular circuitry is conspicuous in EHEC, which colonizes the densely populated colon with a remarkable low infectious dose (50 CFUs) (1, 3).
EHEC integrates metabolism and virulence at multiple levels, with nutrients also serving as signals to modulate virulence gene expression (3, 4). We previously performed a comprehensive HTS screen to map metabolic pathways that impacted virulence expression (with emphasis on LEE gene expression) in EHEC (4). This screen pulled ArgR, the arginine sensor that controls arginine utilization and biosynthesis, and the ArtP arginine importer (5, 6), as potential regulators of LEE gene expression. Here we show that exogenous arginine enhances LEE and Stx expression in both EHEC and C. rodentium through the ArgR transcription factor (Figs. 1–3 and SI Appendix, Figs. S3–S6). The impact of arginine in EHEC/C. rodentium virulence is not due to its role in metabolism, given that no growth defects exist between ΔargR, ΔartP, and WT strains. C. rodentium infection increases the concentration of arginine in the colon, correlated with the down-regulation of the mammalian SLC7A2 importer, leading to increased pathogen loads and disease (Figs. 4 and 5). This is in contrast to a report that during infection with C. rodentium expression of SLC7A2 is up-regulated (20). A notable difference between these two studies is that ours investigates the role of arginine in C. rodentium pathogenesis at the peak of infection, in a very susceptible host, while Singh et al. (20) assess these parameters at the onset of disease resolution, in a less susceptible host. Arginine is known to be present in the colon content, and is absorbed by the intestinal epithelial cells where it can lead to the production of nitrogen oxide (NO) (25, 28). In fact, arginine modulates the host immune responses changing host susceptibility to infections (29). Taking these data together, we find that fluctuations of arginine concentrations in the intestine during infection impact the onset and resolution of disease.
The presence or absence of dietary arginine did not impact the colonic levels of this amino acid, nor pathogen burden (SI Appendix, Fig. S7). Our studies investigated the role of dietary arginine in the peak of C. rodentium infection in a susceptible murine strain. However, a previous report suggested that supplementation of arginine in the diet decreased pathogen burden, in a less susceptible murine strain at a later time point, close to the resolution of the disease (30). Interestingly, in both our and the previous studies the levels of arginine in the colon remained the same regardless of its presence or concentration in the diet (SI Appendix, Fig. S7) (30). However, arginine dietary differences impacted the levels of this amino acid in the serum under homeostasis, and its levels were enhanced in both the colon and serum upon the onset of inflammation due to dextran sulfate sodium treatment (30). The combination of these studies suggests that arginine orchestrate a complex interaction between pathogen and host responses, and arginine dietary supplementation to enhance gut health grants further research.
Materials and Methods
Bacterial Strains, Plasmids, and Growth and Culture Condition.
All strains and plasmids used in this study are listed in SI Appendix, Table S1. Bacterial cultures were grown in low glucose DMEM (Gibco) with or without 482 µM arginine in microaerophilic conditions. For microaerophilic conditions, bacterial cultures were grown standing in the 37 °C incubator. The cultures were harvested in late logarithmic growth phase unless stated otherwise. HeLa cells were routinely cultured in high-glucose DMEM (4.5 g/L glucose), 10% FBS, and penicillin plus streptomycin plus glutamine (PSG) mixture.
Recombinant DNA Techniques.
All primers used for qRT-PCR, mutant generation, and plasmid construction can be found in SI Appendix, Table S2. Knockout strains were constructed using the λ-red method (31). Plasmid pACYC184 (New England Biolabs) was used as complementation vector.
RNA-Seq Library Preparation and Analysis.
Briefly, RNAs extracted from three biological replicates were used to perform RNA-seq experiments. Sequencing was run at the University of Texas Medical Center Next Generation Sequencing core. RNA libraries were prepared using Epicentre Bacteria Kit. RNA libraries were run on an Illumina HiSEq. 2500 sequencer with SE-50. To analyze the data, Microbial Ecology software package (v1.91) (32, 33) was used. Reads were mapped to the E. coli O157:H7 strain EDL 933 genome (National Center for Biotechnology Information NZ_CP008957.1). The data analysis was conducted using the software DESeq2. Statistical significance was calculated using Student’s t test followed by false-discovery rate (Benjamini–Hochberg) correction. A P value of less than 0.05 was considered significant. (RNA-seq data have been deposited at the European Nucleotide Archive, accession no. PRJEB35294)
qRT-PCR.
RNA from three biological replicates was extracted using the RiboPure bacterial isolation kit according to the manufacturer’s protocols (Ambion). qRT-PCR was performed as follows. Briefly, 2 μg of diluted extracted RNA was converted to cDNA with addition of superscript, random primers, DTT, and dNTPs. Validated Primers (SI Appendix, Table S2) and SYBR Green were added to the cDNA and the mix run in Quantstudio 6 flex (Applied Biosystems). Data were collected using QuantStudio Real-Time PCR Software v1.3, normalized to endogenous rpoA levels, and analyzed using the comparative critical threshold (CT) method. For all of the in vitro experiments, error bars indicate SD. A P value of less than 0.05 was considered significant.
Western Blotting.
Secreted proteins were isolated as previously described (34) from cultures grown to the same OD600 in DMEM at 37 °C. A total of 10 μg BSA was added to secreted proteins as a loading control. Proteins were separated on a 5 to 20% SDS polyacrylamide gel, transferred to a polyvinylidene fluoride (PVDF) membrane, and blocked with 5% milk in PBS containing 0.05% Tween (PBST). Membranes were probed with either anti-EspB or anti-EspA primary antibody and then incubated with a secondary rabbit antibody conjugated to streptavidin-horseradish peroxidase. Enhanced chemiluminescence reagent was added, and the membranes were developed using the ChemiDoc touch imaging system (software 1.0.0.15) with Image Lab 5.2.1 software for image display.
Fluorescein Actin Staining Assays.
For the fluorescein actin staining assays, HeLa cells were grown overnight to about 70 to 80% confluence at 37 °C and 5% CO2 on coverslips in wells containing high-glucose (4.5 g/L) DMEM. Prior to infection, fresh low-glucose DMEM lacking antibiotics replaced overnight cell medium. Then, HeLa cells were infected with late log-phase bacterial cultures with equal CFU for 4 h. HeLa cells were infected with a multiplicity of infection of 100. After 4 h, the coverslips were washed, fixed, and permeabilized. The samples were treated with FITC-labeled phalloidin to visualize actin accumulation and propidium iodide to visualize bacterial DNA and HeLa nuclei. The coverslips were then mounted on slides and imaged with a confocal microscope (Zeiss LSM 880 confocal/multiphoton) at 60× resolution. The number of pedestals per HeLa cell was quantified.
Protein Purification and EMSA.
ArgR was cloned downstream to the arabinose operon promoter (pBAD) via Gibson cloning (SI Appendix, Table S2) to create an N-terminal His-tagged construct. This was transformed into Top10 cells and later into BL21 cells. His-tagged ArgR was purified through Ni-NTA column according to the manufacturer’s instructions. For EMSA, DNA probes were prepared by PCR (see SI Appendix, Table S2 for primers) from genomic templates. Probes were purified by gene electrophoresis and labeled with 32P γ-ATP by T4 PNK (New England Biolabs). Labeled probes were further purified by Qiagen PCR purification kit. EMSA reactions were prepared as protein diluted into a 3× EMSA buffer (10 mM Tris⋅HCl [pH 7.4], 5.0 mM MgCl2, 250 mM KCl, 2.5 mM CaCl2, 0.5 mM DTT, 2.5% [v/v] glycerol, 10 mM l-arginine) and incubated at 37 °C for 30 min. The EMSA reaction products were ran on 5% polyacrylamide gels in 1× TBE buffer. Gels were dried onto filter paper and exposed to phosphorimager screens and assessed on a GE Typhoon scanner.
Animal Experiments.
SPF female C3H/HeJ mice between 4 and 6 wk of age were used. Mice were infected by oral gavage of 108 CFU of DBS770, ΔargR or ΔartP mutants in PBS or PBS alone. Mice were checked daily for survival. Stools and tissues were collected for analysis of CFU, inflammation, and LEE gene expression after infection on different days. At the indicated time points, mice were killed, and the colon tissue and content were collected. The tissue was washed in PBS twice to remove any residual fecal content. The content and tissues were snap-frozen in liquid nitrogen. The feces from infected mice were collected in tubes. The tubes were snap-frozen in liquid nitrogen and stored in −80 °C until use. RNA was isolated from individual mice fecal pellets using RNeasy Power Microbiome kit (Qiagen) as per the manufacturer’s instructions. qRT-PCR was performed as described earlier using Quantstudio 6 flex (Applied Biosystems). rpoA was used as an internal control for C. rodentium. Significance was determined either by unpaired t test or Mann–Whitney U test, as stated in the figure legends. For diet experiment, mice were given an arginine-plus (12.1 g/kg) or an arginine-minus chow. Research involving animals has been approved by the University of Texas Southwestern Institutional Animal Care and Use Committee.
Arginine Measurements.
Colon content from a standard arginine-plus (12.1 g/kg), or an arginine-minus diet fed mice were collected on first, third, and sixth day after C. rodentium infection. Colon contents were vortexed for 2 min in a 10-fold volume of PBS:protease inhibitor (10 mL PBS+17 μL protease inhibitor − 10× weight of tissue in grams = volume of PBS:protease inhibitor added; total homogenate volume = 10× weight of tissue). The samples were spun twice, and supernatants were collected. An aliquot of each colon content supernatant was pooled and diluted 10× in methanol and centrifuged for protein and amino acid precipitation. After, the pellets were dried down in a vacuum centrifuge. The dried pellets were resuspended in water in the initial volume. The resuspended pellet was then transferred to an HPLC vial with insert and analyzed by LC-MS/MS. The Qtrap 6500 + analytical conditions used were: Buffer A: ddH20 + 0.1% formic acid; buffer B: ACN + 0.1% formic acid; column: Luna Phenomenex 5 µ C18 100A − 100 × 4.6 mm; gradient conditions: 0 to 1.9 min 3% B; 2 to 3.5 min 100% B; 3.6 to 4 min 3% B; flow rate: 1.5 L/min; ion source/gas parameters: CUR = 35, CAD = 9, IS = 4500, TEM = 700, GS1 = 60, GS2 = 60. 13C6 15N4 Butyl-Arginine; Butyl-Arginine.
Data Availability Statement.
The RNA-seq data are deposited at the European Nucleotide Archive, accession no. PRJEB35294. All materials and detailed protocols are available upon request to the corresponding author (vanessa.sperandio@utsouthwestern.edu).
Supplementary Material
Acknowledgments
This study was supported by NIH Grants AI053067, AI05135, AI077613, and AI114511.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (ENA) database, https://www.ebi.ac.uk/ena (accession no. PRJEB35294).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919683117/-/DCSupplemental.
References
- 1.Kaper J. B., Nataro J. P., Mobley H. L., Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Bäumler A. J., Sperandio V., Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535, 85–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner N. C. A., Connolly J. P. R., Roe A. J., Control freaks-signals and cues governing the regulation of virulence in attaching and effacing pathogens. Biochem. Soc. Trans. 47, 229–238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pifer R., Russell R. M., Kumar A., Curtis M. M., Sperandio V., Redox, amino acid, and fatty acid metabolism intersect with bacterial virulence in the gut. Proc. Natl. Acad. Sci. U.S.A. 115, E10712–E10719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldara M., Minh P. N., Bostoen S., Massant J., Charlier D., ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J. Mol. Biol. 373, 251–267 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Charlier D., Bervoets I., Regulation of arginine biosynthesis, catabolism and transport in Escherichia coli. Amino Acids 51, 1103–1127 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Lu C. D., Houghton J. E., Abdelal A. T., Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. J. Mol. Biol. 225, 11–24 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Sunnerhagen M., Nilges M., Otting G., Carey J., Solution structure of the DNA-binding domain and model for the complex of multifunctional hexameric arginine repressor with DNA. Nat. Struct. Biol. 4, 819–826 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Torres Montaguth O. E., Bervoets I., Peeters E., Charlier D., Competitive repression of the artPIQM operon for arginine and ornithine transport by arginine repressor and leucine-responsive regulatory protein in Escherichia coli. Front. Microbiol. 10, 1563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho S., et al. , The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli. Nucleic Acids Res. 43, 3079–3088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherney L. T., Cherney M. M., Garen C. R., Lu G. J., James M. N., Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis. J. Mol. Biol. 384, 1330–1340 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Charlier D., Glansdorff N., Biosynthesis of arginine and polyamines. Ecosal Plus 1, 10.1128/ecosalplus.3.6.1.10 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Biemans-Oldehinkel E., Doeven M. K., Poolman B., ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580, 1023–1035 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Carlson-Banning K. M., Sperandio V., Catabolite and oxygen regulation of enterohemorrhagic Escherichia coli virulence. MBio 7, e01852-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens M. P., Frankel G. M., The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol. spectrum 2, EHEC-0007-2013 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Wagner P. L., et al. , Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183, 2081–2085 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neely M. N., Friedman D. I., Functional and genetic analysis of regulatory regions of coliphage H-19B: Location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28, 1255–1267 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Mullineaux-Sanders C., et al. , Citrobacter rodentium-host-microbiota interactions: Immunity, bioenergetics and metabolism. Nat. Rev. Microbiol. 17, 701–715 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Mallick E. M., et al. , A novel murine infection model for Shiga toxin-producing Escherichia coli. J. Clin. Invest. 122, 4012–4024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh K., et al. , The L-arginine transporter solute carrier family 7 member 2 mediates the immunopathogenesis of attaching and effacing bacteria. PLoS Pathog. 12, e1005984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borenshtein D., et al. , Diarrhea as a cause of mortality in a mouse model of infectious colitis. Genome Biol. 9, R122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borenshtein D., McBee M. E., Schauer D. B., Utility of the Citrobacter rodentium infection model in laboratory mice. Curr. Opin. Gastroenterol. 24, 32–37 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Mundy R., et al. , Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol. Microbiol. 48, 795–809 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Deng W., et al. , Dissecting virulence: Systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 101, 3597–3602 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis M. M., et al. , The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16, 759–769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A., Sperandio V., Indole signaling at the host-microbiota-pathogen interface. MBio 10, e01031-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A., Ellermann M., Sperandio V., Taming the beast: Interplay between gut small molecules and enteric pathogens. Infect. Immun. 87, e00131-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundberg J. O., Weitzberg E., Gladwin M. T., The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7, 156–167 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Fritz J. H., Arginine cools the inflamed gut. Infect. Immun. 81, 3500–3502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh K., et al. , Dietary arginine regulates severity of experimental colitis and affects the colonic Microbiome. Front. Cell. Infect. Microbiol. 9, 66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso J. G., et al. , QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vázquez-Baeza Y., Pirrung M., Gonzalez A., Knight R., EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2, 16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarvis K. G., et al. , Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U.S.A. 92 (17), 7996–8000, 10.1073/pnas.92.17.7996 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are deposited at the European Nucleotide Archive, accession no. PRJEB35294. All materials and detailed protocols are available upon request to the corresponding author (vanessa.sperandio@utsouthwestern.edu).