Significance
The KRAS oncogene is frequently mutated in human cancers, and cancer-associated mutations are constitutively activated, which stimulates RAF kinase activity at the plasma membrane. Membrane-anchored KRAS engages the RAS-binding domain (RBD) of RAF, and the adjacent cysteine-rich domain (CRD) interacts with the plasma membrane via a poorly defined mechanism. Studying this multidomain, membrane-dependent interaction has been a major challenge; hence, assembly of the full KRAS–RAF complex on the membrane is not well-understood. We determined NMR-based structures of a KRAS:RBD–CRD complex on a lipid bilayer, revealing how multivalent, dynamic protein–protein and protein–lipid interactions stabilize the complex in two states and how perturbation of this equilibrium by a cancer-associated CRAF mutation promotes CRAF activation, potentially via KRAS dimerization.
Keywords: KRAS, protein membrane interactions, RAF kinase, cysteine-rich domain (CRD), RAS-binding domain (RBD)
Abstract
Membrane anchoring of farnesylated KRAS is critical for activation of RAF kinases, yet our understanding of how these proteins interact on the membrane is limited to isolated domains. The RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF engage KRAS and the plasma membrane, unleashing the kinase domain from autoinhibition. Due to experimental challenges, structural insight into this tripartite KRAS:RBD–CRD:membrane complex has relied on molecular dynamics simulations. Here, we report NMR studies of the KRAS:CRAF RBD–CRD complex. We found that the nucleotide-dependent KRAS–RBD interaction results in transient electrostatic interactions between KRAS and CRD, and we mapped the membrane interfaces of the CRD, RBD–CRD, and the KRAS:RBD–CRD complex. RBD–CRD exhibits dynamic interactions with the membrane through the canonical CRD lipid-binding site (CRD β7–8), as well as an alternative interface comprising β6 and the C terminus of CRD and β2 of RBD. Upon complex formation with KRAS, two distinct states were observed by NMR: State A was stabilized by membrane association of CRD β7–8 and KRAS α4–α5 while state B involved the C terminus of CRD, β3–5 of RBD, and part of KRAS α5. Notably, α4–α5, which has been proposed to mediate KRAS dimerization, is accessible only in state B. A cancer-associated mutation on the state B membrane interface of CRAF RBD (E125K) stabilized state B and enhanced kinase activity and cellular MAPK signaling. These studies revealed a dynamic picture of the assembly of the KRAS–CRAF complex via multivalent and dynamic interactions between KRAS, CRAF RBD–CRD, and the membrane.
The prevalence of activating KRAS and BRAF mutations in human cancers has spurred intense interest in studying the structure, function, and biology of both proteins (1, 2), as well as the mechanisms by which KRAS activates RAF kinases. There are three RAF kinase paralogs (A-, B-, and CRAF), which are each comprised of several conserved domains, including the RAS-binding domain (RBD) and cysteine-rich domain (CRD), together known as conserved region 1 (CR1), as well as the kinase domain (CR3), and a Ser/Thr-rich region (CR2) involved in regulation (3, 4). Inactive monomeric RAF in the cytoplasm is autoinhibited by interactions between the N-terminal (N-term) RBD–CRD region and the kinase domain (Fig. 1A). RAF can be activated by membrane-anchored RAS-guanosine triphosphate (GTP), which binds the RBD, disrupting autoinhibition and recruiting RAF to the membrane, where the CRD forms additional interactions with phospholipids (2). Structural studies of full-length RAF are challenging due to difficulties with producing and purifying the protein, as well as sample heterogeneity resulting from partial phosphorylation and long intrinsically disordered regions. Structures of the CRAF CRD (5), RBD, and RBD in complex with HRAS (6, 7), as well as active homo- and heterodimeric RAF kinase domains, have been solved (3); however, despite the wealth of structures, key details about the assembly of RAS-GTP and the intact CR1 region on the membrane, and how this promotes dimerization, remain unknown. It has been reported that an interaction between CRD and RAS is critical for the activation of RAF (8, 9) although it is not clear how these domains interact, particularly when both are associated with the membrane. Various RAS:CRD interfaces have been proposed (10), involving the farnesyl moiety (11, 12), Switch II (13), N26/V45 (9), and/or residues 23 to 30 (12) of RAS, and β7–8 (secondary structures have been numbered continuously in RBD–CRD) (SI Appendix, Fig. S1D) (11) or the C-terminal (C-term) region (14) of CRD. The potential binding sites, as well as the nucleotide dependence of the interaction, remain controversial. The CRD is an atypical C1 domain that has been shown to interact with membranes enriched in phosphatidylserine (PS) (15, 16); however, its phospholipid-binding site has not been precisely determined, nor has the effect of lipid composition on membrane binding. These questions have been interrogated in a recent series of high-quality molecular dynamics (MD) simulations (17–19); however, these investigations have not converged on a consensus, and experimental observations are required to support a data-driven model.
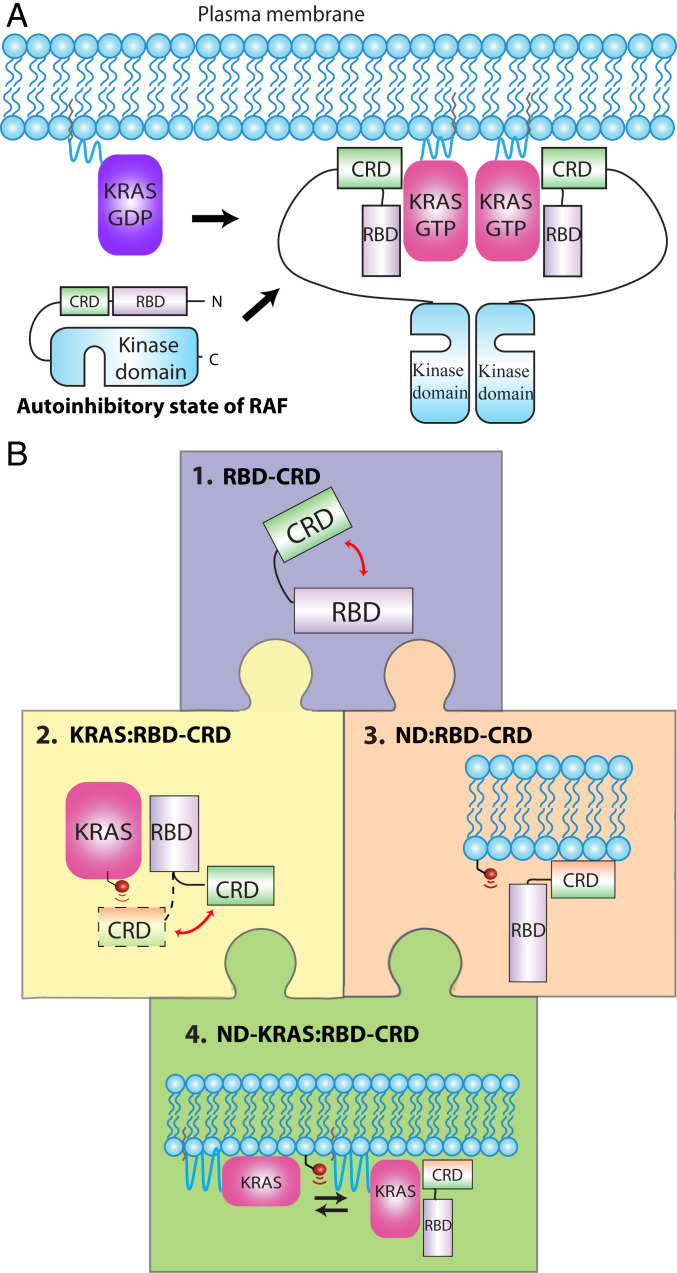
Fig. 1.
Assembly of the KRAS:CRAF complex on the membrane surface and experimental approach for structural studies of bilayer-anchored KRAS in complex with RBD–CRD. (A) assembly of a KRAS:CRAF signaling complex on the membrane upon activation of KRAS. KRAS-GTP binding to the RBD recruits CRAF to the plasma membrane, and the CRD interacts with the surface. This relieves autoinhibition and promotes dimerization of the kinase domain. (B) “Jigsaw puzzle” approach to determining structures of KRAS in complex with RBD–CRD on lipid bilayer. 1, Potential interdomain interactions within the 15-kDa dual RBD and CRD construct were assessed through CSPs (SI Appendix, Figs. S1 and S2). 2, Engagement of RBD–CRD by KRAS in solution was characterized using multiple techniques, including CSP, PRE, and NOEs (Fig. 2 and SI Appendix, Figs. S3–S12). 3, RBD–CRD interactions with the surface of membranes of different lipid composition were characterized using nanodiscs with a PRE-tagged lipid (SI Appendix, Figs. S13–S15). KRAS:membrane interactions were reported previously (21, 22). 4, Finally, the KRAS:RBD–CRD complex was oriented on the membrane surface using lipid-PRE restraints (Figs. 3 and 4 and SI Appendix, Figs. S16–S24).
In this study, we produced a RAF construct comprising the tandem RBD and CRD and characterized how this dual domain interacts with KRAS on the surface of a membrane (Fig. 1B). To build a structural model, we examined how the pieces of this puzzle fit together, starting by combining pairs of pieces, which could be observed by NMR with high sensitivity. We then examined the entire tripartite complex comprised of RBD–CRD, KRAS, and the membrane. Detection of this large complex by NMR was challenging; however, several restraints were obtained using a variety of isotopic labeling schemes. We then fit all these pieces together to assemble the puzzle (Fig. 1) and build a structural model of KRAS in complex with RBD–CRD on the membrane.
Results
Purification and Characterization of the CRAF RBD–CRD Dual Domain.
The RBD and CRD are both folded domains connected by only four residues (DHVP), but it is not known whether they are closely associated or independent. While structures of RBD and CRD have been determined in isolation, no structure of the tandem RBD–CRD domain was available because it was a challenge to produce. After several rounds of optimization, we were able to purify an RBD–CRD construct comprised of CRAF residues 55 to 187 (SI Appendix, SI Materials and Methods). We assigned 90% of the backbone resonances of isolated RBD and CRD domains (SI Appendix, Fig. S1 A and B) and compared these chemical shifts to those of the RBD–CRD dual domain (SI Appendix, Fig. S2D). The comparison of the 1H-15N heteronuclear single quantum coherence (HSQC) spectra revealed that separating the RBD and CRD induced only minimal chemical shift perturbations, suggesting that the two domains are relatively independent even though they are connected via a short sequence (SI Appendix, Fig. S2 C and D). Not surprisingly, the largest chemical shift differences were observed in sequences flanking the linkage between RBD and CRD, as well as the C-term of CRD (SI Appendix, Fig. S2D), which is spatially proximal to the N-term linkage (SI Appendix, Fig. S1E).
Interaction of the RBD–CRD Domain with KRAS in Solution.
With this RBD–CRD dual domain construct in hand, we proceeded to investigate how it interacts with KRAS, first in solution (i.e., in the absence of a membrane). To build structures of the KRAS:RBD–CRD complex, we first characterized the interactions formed between each pair of components within the complex and then compared these with data obtained from the whole complex. Addition of KRAS-GMPPNP to 15N-labeled tandem RBD–CRD induced the expected chemical shift perturbations (CSPs) in RBD peaks, particularly in β2 and α1, and also broadened a number of CRD peaks, especially in the C-term region (SI Appendix, Fig. S3). KRAS-GMPPNP did not induce appreciable CSP in isolated 15N CRD, indicative of an RBD-dependent interaction between KRAS and CRD (SI Appendix, Fig. S3). As expected, the RBD–CRD interaction with KRAS remained nucleotide-dependent as KRAS-GDP did not appreciably perturb the spectrum of RBD–CRD (SI Appendix, Fig. S4A). Likewise, farnesylated KRAS-GDP did not perturb RBD–CRD (SI Appendix, Fig. S4B) whereas farnesylated HRAS-GDP was reported to perturb CRAF CRD resonances (11, 12), potentially due to different titration ratios, buffer conditions, or RAS/RAF isoforms. These spectra were collected with 1:1 stoichiometry in 450 mM NaCl at pH 5.5, which was necessary to stabilize RBD–CRD, although this buffer condition may reduce the strength of electrostatic interactions in the complex. The interaction between RAS and CRAF RBD was previously shown to be enhanced by electrostatic complementarity, which is reduced with increasing ionic strength (20). Similarly, the high salt may weaken potential interactions between KRAS and CRD (21) and is likely to reduce the affinity of the polybasic C-term region of KRAS and the CRD for the membrane, which contains acidic phospholipids (22–24). Thus, we compared the interaction between KRAS and RBD–CRD in a physiological buffer condition (SI Appendix, Fig. S5) to that in the optimized NMR buffer (SI Appendix, Fig. S3C). In physiological conditions, the solubility of RBD–CRD was poor, and addition of the active farnesylated KRAS-GTPγS induced extensive broadening of RBD–CRD amides and methyls (SI Appendix, Fig. S5). Nevertheless, the major sites exhibiting broadening are β2 and α1 of RBD and the C terminus of CRD, which is partially consistent with SI Appendix, Fig. S3E, and suggests that the major sites of interaction (β2 and α1) appear to be retained.
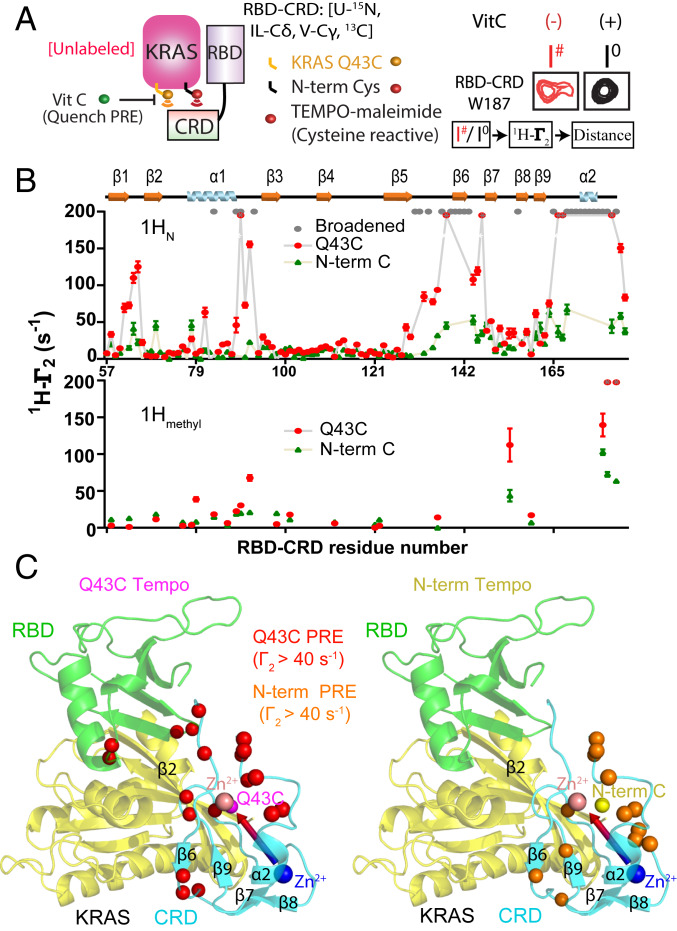
To map the binding site of RBD–CRD on KRAS, we compared the CSP induced by CRAF RBD versus RBD–CRD on uniformly 15N- and 13C-methyl Ile, Leu, and Val (ILV) labeled KRAS-GMPPNP and investigated whether the presence of CRD caused any additional perturbations. The RBD induced CSP on the KRAS β1–3 and α1–2 regions (SI Appendix, Fig. S6) whereas RBD–CRD caused larger chemical shift changes and more broadening of peaks from a KRAS region comprising α1, β2, and α5 (SI Appendix, Fig. S7), suggesting this surface interacts with the CRD when KRAS binds the tandem domain. To investigate whether these CSP result from a direct interaction versus structural/dynamical perturbations, we conjugated maleimide-2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO) spin labels at each of two cysteine residues introduced in the beta-sheet region of KRAS (an N-term Cys preceding Met1 near β1, and a Q43C substitution in β2) and analyzed the paramagnetic relaxation enhancement (PRE) effect of these spin labels on 15N amides and 13C ILV methyls of RBD–CRD (Fig. 2). PRE tags in either position on KRAS caused strong peak broadening and high 1H transverse relaxation rates (1H-Γ2) in the region near β6 and the C terminus of CRD (Fig. 2B), but Q43C induced higher overall PRE, indicating it is more proximal to CRD. The Cys43 spin label generated strong PRE on RBD β1-β2 and α1-β3 loops, consistent with the structure of HRAS in complex with RBD (Fig. 2 B and C), and also affected a loop between β9 and α2, suggesting that this surface of CRD is near KRAS β2 (Fig. 2C). The peak intensity ratios obtained from these PRE experiments were converted to 1H-Γ2 (SI Appendix, Fig. S8A) and distance restraints (SI Appendix, Fig. S8B). To obtain additional restraints, we performed nuclear Overhauser effect (NOE) analyses of KRAS:RBD, as well as KRAS:RBD–CRD complexes (SI Appendix, Fig. S9), and identified three intermolecular NOEs between RBD and KRAS in both complexes (SI Appendix, Fig.S9 C and D). These NOE, PRE, and CSP data (SI Appendix, Tables S1 and S2) were converted to 67 distance restraints (details described in SI Appendix, SI Materials and Methods), and high ambiguity driven protein–protein DOCKing (HADDOCK) software was used to generate NMR-driven structural models of KRAS:RBD and KRAS:RBD–CRD complexes in solution (SI Appendix, Fig. S10) (25). The RBD:KRAS contacts are conserved in both models and are consistent with the available crystal structure of the RBD:HRAS complex (7). Analysis of the KRAS:RBD models identified a large cluster comprising 98% of solutions while the KRAS:RBD–CRD models formed two clusters in which the RBD was fixed relative to KRAS, but the CRD adopted two different orientations. This is consistent with weak and transient interactions between KRAS and CRD (SI Appendix, Figs. S10 and S11), which is supported by conformity analysis of the PRE restraints (SI Appendix, Fig. S12), the lack of interaction between KRAS and CRD in the absence of RBD, and the lack of observable NOE between KRAS and CRD in the KRAS:RBD–CRD complex, as well as the observation that KRAS broadens specific CRD peaks in this complex, suggesting fast/intermediate chemical exchange.
Fig. 2.
Site-directed spin-labeling and PRE reveal an RBD-dependent interaction between KRAS and the CRD domain of CRAF. (A) Schematic diagram of spin-labeled KRAS and the resulting PRE on RBD–CRD. TEMPO-maleimide was covalently linked to one of two cysteine residues introduced to KRAS near switch I (Q43C) or at the N terminus (Cys0). Interaction with RBD–CRD caused PRE-induced broadening of peaks from proximal residues (e.g., W187, Right). I#/Io is the ratio of peak heights before and after quenching the TEMPO spin label with Vitamin C (Vit C). (B) Estimated 1HN-Γ2 rates of KRAS Q43C-TEMPO (red) or N-term-TEMPO (green) on 15N, 13C Ile-Cδ, Val-Cγ, Leu-Cδ, Met-Cε-RBD–CRD amide, and methyl resonances. Residues broadened beyond detection after adding KRAS (even in the absence of TEMPO) are colored gray. Residues broadened beyond detection in the presence of TEMPO are colored in red with a gray asterisk, and their 1HN-Γ2 rates are assigned to a fixed value of 200. (C) Structural model of KRAS in complex with RBD–CRD. The positions of Q43C and the N-term Cys (Cys0) in KRAS are shown in pink and yellow, respectively. Residues exhibiting strong PRE from TEMPO tags attached to Cys43 or N-term are colored red or orange, respectively.
Interaction between RBD–CRD and the Membrane.
To study interactions between proteins and the membrane surface by NMR, we previously developed a nanodisc PRE assay, whereby the paramagnetic ion Gd3+ can be incorporated in the nanodiscs to identify residues proximal to the bilayer surface through PRE-induced broadening of their resonances (26, 27). We used this method to map the sites of membrane interaction in RBD, CRD (SI Appendix, Fig. S13), and the dual RBD–CRD domain (SI Appendix, Fig. S14). As expected, in isolation, CRD exhibited stronger overall PRE from the membrane than RBD (SI Appendix, Fig. S13A). The PRE effect was severe on strand β6 and also prominent on β7–9 and the C terminus of CRD. In the RBD, strand β2 experienced modest PRE. The strong PRE probes on the CRD were used as restraints in HADDOCK to model its interactions with nanodiscs. Among 200 solutions, CRD was found in two orientations: a major “Perpendicular 1” cluster (84%) and a minor “Parallel 1” cluster (15%), designated by the orientation of the vector connecting the two zinc ions relative to the bilayer surface (SI Appendix, Fig. S13 B and C). In the Perpendicular 1 cluster, CRD interacts with the lipids through β6, β7, and β8, consistent with the “canonical” membrane interface previously identified by mutagenesis (15, 16) and MD simulations (17–19), whereas Parallel 1 involves an adjacent surface.
In comparison to either domain in isolation, the RBD–CRD dual domain exhibited higher overall PRE from the membrane (SI Appendix, Fig. S14A), consistent with a recent report demonstrating that linking the domains enhanced membrane affinity (28). When the RBD and CRD were linked, the whole RBD and a C-term region of the CRD experienced stronger PRE whereas the PRE on the CRD β-strands, particularly β8, was reduced. These data suggest that CRD recruits RBD to the membrane where RBD-membrane contacts pull the CRD into a new orientation. These PRE data were used as distance restraints in HADDOCK simulations, which identified three orientations of the CRD, whereas the RBD orientation was highly variable (SI Appendix, Fig. S14B), which promotes productive encounters with RAS (29). Among 200 solutions, the Perpendicular 1 orientation of CRD involving β7–8 was still observed, but the size of the cluster was reduced (14.5%). The major cluster was a distinct “Perpendicular 2” orientation (56%) that interacts with the bilayer through the opposite surface of CRD comprising β9–10 and the N terminus. A “Parallel 2” orientation, mediated by α2-β6, formed another minor cluster (21%) (SI Appendix, Fig. S14C). The drastic reduction of PRE in CRD β8 in the dual domain implies that the Perpendicular 1 and Parallel 1 orientations, which share the β7–8 interface, become less favored as RBD-membrane interactions stabilize the Perpendicular 2 and Parallel 2 orientations.
PS has been proposed to be a critical lipid component for membrane association of CRD (30) and KRAS (31–33), as well as activation of RAF kinase activity (34). To study the PS dependence of RBD–CRD membrane binding, we examined how the concentration of 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS, 0 to 20%) affects the RBD–CRD PRE profile (SI Appendix, Fig. S15A). Increasing concentrations of DOPS enhanced the overall PRE on RBD–CRD. Specifically, PRE was enhanced on β2 in RBD and the “canonical” Perpendicular1 interface in CRD (β6–9); however, PRE on the C terminus of CRD (Parallel interface) was reduced. These effects are likely mediated by the distribution of hydrophobic and charged residues throughout the construct (SI Appendix, Fig. S15A). In addition to PS, the plasma membrane also contains other types of anionic lipids, and their interactions with KRAS have been characterized in “lipid strip” assays (35, 36); thus, we further investigated how addition of phosphatidic acid (PA) and phosphatidylinositol 4,5-bisphosphate (PIP2) affect interactions with RBD–CRD (SI Appendix, Fig. S15B). The three lipids (PA, PS, and PIP2) were associated with similar overall RBD–CRD PRE profiles; however, PRE was weaker from nanodiscs containing PA, which has a shorter head group, whereas a concentration of only 4% of the more anionic PIP2 led to PRE comparable to that of 20% PA. To our surprise, the properties of the lipid acyl chains also affected RBD–CRD interactions with membrane. A comparison of the PRE from nanodiscs containing symmetric DOPS to that from the asymmetric, more saturated and shorter POPS revealed that RBD–CRD membrane binding was promoted by the longer, less saturated acyl chain of DOPS.
NMR-Driven Structure and Dynamics of the KRAS:RBD–CRD Complex on the Membrane.
Our data show that the RAS-binding site (β2) of CRAF RBD was involved in interactions between RBD–CRD and the membrane, and we previously showed that maleimide-conjugated KRAS (MC-KRAS) can adopt an “occluded” orientation in which its RBD-binding site (Switch I) is blocked by the membrane (26, 27); thus, we sought to investigate the structure of a KRAS:RBD–CRD complex on the membrane surface. This model was built using our solution structure of KRAS:RBD–CRD combined with PRE-derived distance restraints between the proteins and the membrane. To examine KRAS interactions with a membrane when bound to RBD–CRD, we used our previously described MC-KRAS nanodisc system (26, 27); however, addition of RBD–CRD broadened most of the 13C-methyl resonances of ILV-labeled KRAS beyond detection, even in the absence of PRE (SI Appendix, Fig. S16A). This indicates that the relatively dynamic motion of MC-KRAS on the membrane was restrained by complexation with RBD–CRD, and its tumbling rate was significantly reduced in this large (∼150 kDa) complex. To overcome this issue, we expressed uniformly deuterated 13C-methyl ILV KRAS using media comprising D2O and deuterated glucose (37, 38), which enabled detection of most of the 13C-methyl resonances (SI Appendix, Fig. S16B). The similarity between this spectrum and that of the complex in solution (SI Appendix, Fig. S7B) suggested that the overall conformation of the MC-KRAS:RBD–CRD complex was preserved upon binding to the membrane.
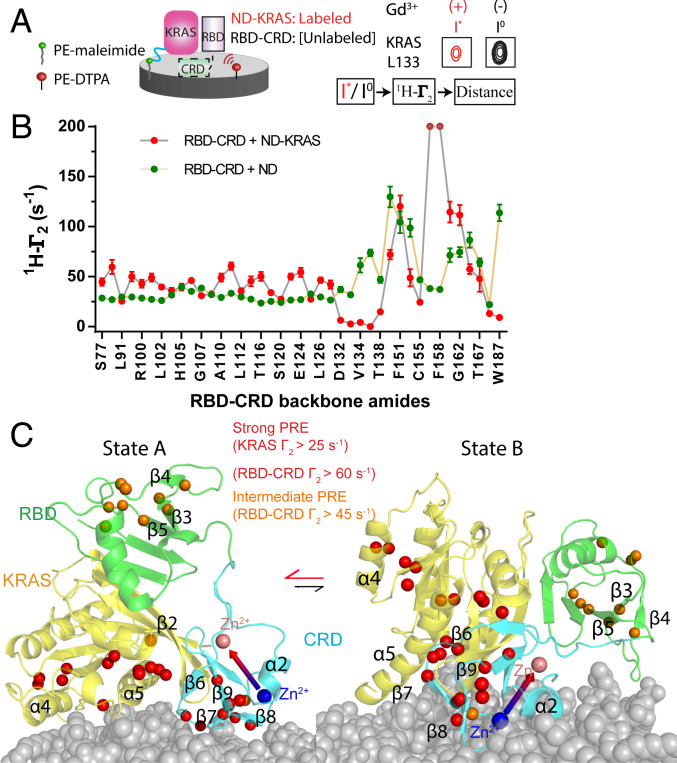
To map the protein surfaces of MC-KRAS:RBD–CRD that interact with the membrane for an NMR data-driven model, Gd3+ was incorporated into the nanodiscs (Fig. 3A). Addition of RBD–CRD to nanodisc-tethered MC-KRAS enhanced PRE on a region near α4-α5 (SI Appendix, Figs. S17C and S3C), consistent with stabilization of an orientation in which the RBD-binding site is exposed. A comparison of the PRE on 15N-labeled RBD–CRD induced by nanodiscs alone versus nanodiscs containing MC-KRAS indicated that the presence of KRAS enhanced membrane interactions with RBD β3–5 (opposite the Ras-binding site) and CRD β7–8 (Perpendicular1 interface), while reducing that of the region that links the RBD to the CRD as well as the C terminus of CRD (Fig. 3B and SI Appendix, Fig. S17B). To orient the complex on the membrane, two separate HADDOCK simulations were performed using high (1-I*/Io > 0.7) or intermediate (1-I*/Io > 0.6) PRE thresholds (SI Appendix, Fig. S18 and Table S3). The distribution of these residues is shown in Fig. 3C. In both cases, the resulting HADDOCK models exhibit a major cluster in which CRD adopts the “canonical” Perpendicular1 orientation that interacts with the membrane through β6–8. In this cluster, which we denote state “A,” MC-KRAS α4-α5 associates with the membrane surface while the loop region between β2–3 interacts with CRD and RBD is distal to the membrane (Fig. 3C). Interaction with MC-KRAS significantly broadened the line width of peaks from RBD–CRD in the presence of nanodiscs (SI Appendix, Fig. S14B and Fig. 3 B and C). The inclusion of intermediate-strength PRE probes in the HADDOCK restraints resulted in the appearance of an additional smaller cluster in which CRD adopted a parallel orientation, and a polybasic stretch in the RBD β3–4 loop (KGKKAR) contacted the membrane, leading to exposure of MC-KRAS α4-α5. This orientation was denoted state “B.” Conformity and violation analysis of strong and intermediate PRE restraints indicated that a two-conformer model (state A&B) was in good agreement with observed restraints (SI Appendix, Fig. S19). The KRAS in state A adopted an orientation very similar to our previously reported exposed orientation, except its α5 was slightly lifted away from membrane because of its interaction with CRD. State “B” resembles the semiexposed orientation, except the angle between α5 and the membrane is greater (more exposed) due to the existence of CRD.
Fig. 3.
Mapping the membrane association interface of the MC-KRAS:RBD–CRD complex. (A) Schematic diagram of the experimental system to determine membrane interfaces of MC-KRAS:RBD–CRD. Gd3+ was chelated on a lipid head group and incorporated into a nanodisc-bound lipid bilayer. The resulting PRE on KRAS and RBD–CRD (alone and in complex) identified residues proximal to the membrane surface (e.g., KRAS L133, Right). Functionalized lipids 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (PE)-N-diethylenetriaminepentaacetic acid (DTPA, red) and PE-maleimide (green) were used to chelate Gd3+ or crosslink KRAS, respectively. (B) Estimated 1HN-Γ2 rates induced by Gd3+ on amide resonances of RBD–CRD in the presence (red) or absence (green) of KRAS tethered to the nanodisc (ND). (C) Structural models of the KRAS:RBD–CRD complex on the membrane. Isotopic probes are shown in orange and red according to severity of broadening. State A represents a major cluster based on strong PRE restraints while state B represents an alternate conformation that satisfies restraints from intermediate PRE effects. Two CRD bound zinc ions are shown as large spheres colored pink or blue and connected by an arrow to indicate the orientation of CRD.
Cancer-Associated Mutation in CRAF RBD Stabilizes State B and Enhances Kinase Activity.
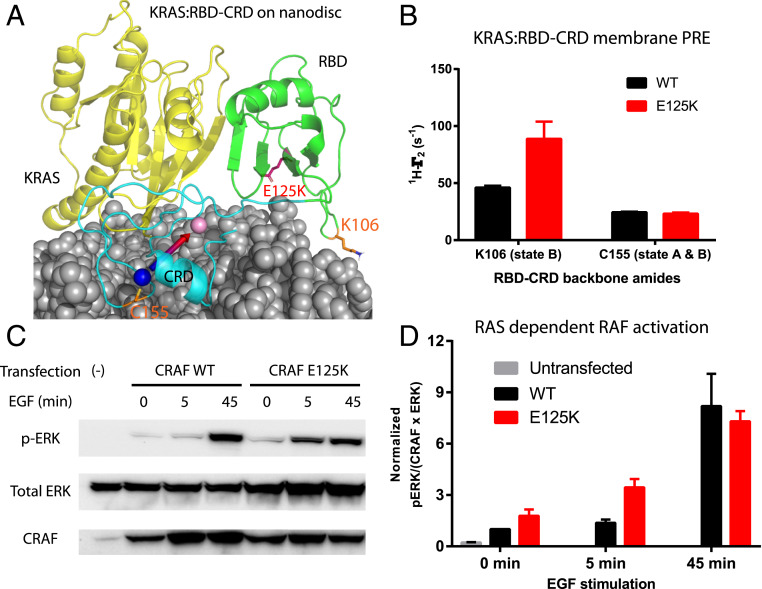
RAF kinases are frequently mutated in a number of cancers (39), and, while the kinase domain is affected most frequently, mutations have also been observed in RBD and CRD. A rare CRAF mutation detected in esophageal squamous cell carcinoma (E125K) (40, 41) is located in the RBD, and the resulting charge reversal would be predicted to stabilize state B of the complex through favorable electrostatic interactions with the anionic membrane surface (SI Appendix, Table S4 and Fig. 4A). We examined the PRE profile of the RBD–CRD E125K mutant in complex with MC-KRAS on nanodiscs and found that it indeed enhanced the PRE (1H-Γ2) of K106, consistent with a shift toward the state B orientation (Fig. 4B). This rare mutation has not been characterized in cells; thus, we assayed its kinase activity in HEK293T cells in the presence and absence of epidermal growth factor (EGF) stimulation to promote RAS activation (Fig. 4 C and D). Transient transfection of CRAF E125K caused more ERK phosphorylation in response to EGF stimulation than wild type (WT) (Fig. 4 C and D), even though both expressed at similar levels (SI Appendix, Fig. S20). Before it was identified in cancer, Kiel et al. engineered the E125K mutant in CRAF RBD to alter the electrostatic surface, but this mutation did not alter affinity for RAS in solution, as determined by isothermal titration calorimetry (42). This suggests the E125K-enhanced RAF kinase activation was not due to increased affinity for RAS; however, the correlation between elevated kinase activation and the population of the state B orientation of KRAS:CRAF is consistent with our model that stabilization of state B, which is compatible with KRAS dimerization, promoted more efficient RAF activation and enhanced signal transduction.
Fig. 4.
CRAF kinase activity is enhanced by a cancer-associated mutation that stabilizes state B orientation of KRAS:RBD–CRD. (A) The cancer-associated mutation E125K (red), which is located in the state B membrane interface of RBD, is shown in the KRAS:RBD–CRD complex. Probes that report on state A and/or state B (K106 and C155) are colored orange. Nanodisc, KRAS, RBD, and CRD are colored in grey, yellow, green, and cyan, respectively. The two zinc ions in CRD are colored in blue or pink and connected by an arrow to illustrate the orientation. (B) Effect of the E125K substitution on the PRE profile of the KRAS:RBD–CRD complex on Gd3+-tagged nanodiscs. Enhanced Γ2 of K106 indicates stabilization of state B. Error bars are estimated from spectral noise in diamagnetic and paramagnetic samples. (C) Serum-starved human embryonic kidney 293T (HEK293T) cells without transfection or transfected with full-length CRAF WT or E125K were treated with 2 ng/mL EGF for 0, 5, and 45 min, and Western blotted for phosphorylated ERK (pERK), total ERK, and CRAF. (D) Quantification of ERK phosphorylation. The ratio of pERK/(ERK x CRAF) detected in each sample in B was normalized to the WT-CRAF sample at time 0 min. Error bars represent the SD from three independent experiments.
Discussion
The activation of the MAPK pathway is initiated by the recruitment of RAF kinases to the plasma membrane by RAS, releasing RAF autoinhibition and promoting dimerization of the kinase domain. However, it has been challenging to integrate the available structures of individual domains to elucidate the molecular and structural basis of activation; thus, more structural studies are needed to understand this complex process. Due to the challenges associated with experimental approaches to this system, extensive efforts have been invested in performing MD simulations, and, recently, three independent in silico studies were published on KRAS and RBD–CRD interactions (17–19). In a nutshell, our NMR-driven models agree with and share some similarities with certain subsets of these published MD models, but there are a number of key differences. All three groups acknowledged that the dominant membrane interface is β7–8 of CRD and the hypervariable region of KRAS, which were previously characterized biochemically (43, 16). Our NMR data-driven structures support these biochemical observations; however, details of the orientation and interaction of KRAS and RBD–CRD with the membrane surface differ from any of the reported MD models. The models from Buck’s group (18) predict that CRD β7–8 and KRAS α3-α4 form the major membrane interaction sites and that they would compete for membrane interaction and lead to a “tug of war” because they cannot engage the membrane simultaneously. In our state A model, MC-KRAS interactions with the membrane that occur through the α4-α5 surface are compatible with simultaneous engagement of CRD. However, an alternate competition was observed between these interfaces and the RBD β3–4 loop, which establishes the equilibrium between state A and B (Fig. 3C). Nussinov’s group (17) proposed highly dynamic models of KRAS:RBD–CRD on the membrane, all of which have KRAS α4-α5 exposed, similar to our state B. But these models contrast with our state B model, in that their CRD adopts multiple positions relative to KRAS, and RBD β3–4 does not contact the membrane in most solutions. Gnanakaran (19) built models that either included or excluded restraints between KRAS and BRAF CRD based on previous KRAS mutagenesis data (9). Interestingly, incorporation of this restraint produced models that are somewhat similar to our state A model, with simultaneous engagement of the membrane by MC-KRAS α4-α5 and the canonical CRD loops; however, the CRD is rotated by 70 ° relative to our model A and thus interacts with MC-KRAS through a different surface. Exclusion of this restraint produced models that resemble state B, except the CRD was further rotated by 180 ° (Fig. 3C).
The present NMR-driven structure of the MC-KRAS:RBD–CRD complex revealed an interface between MC-KRAS β2–3 and CRD β6–7, C-term, and the β9-α2 loop, which is dependent on KRAS–RBD interaction. This interface is consistent with a report that a V45E mutation on RAS β2 disrupted the binding of CRD and impaired RAS-induced RAF activation (9). Furthermore, sequence alignments of 150 homologous RAF kinase sequences show that CRD β6–7 and β9-α2 loop (residues 162 to 176), along with the RAS-binding site in RBD (residues 66 to 70), are the most highly conserved regions in RBD–CRD whereas the CRD C-term is more variable (SI Appendix, Fig. S21). The high conservation of the CRD β9-α2 loop underscores a functional significance of this structural element. The divergence of the CRD C terminus among RAFs may have evolved for isoform-specific functions. Interestingly, the autoinhibited state of RAF was recently solved by cryogenic electron microscopy, revealing that CRD adopts a core position where it interacts with both 14-3-3 and RAF kinase domain through its β6–7 and β9-α2 loop, respectively (SI Appendix, Fig. S21B) (44). This suggests that the RAS-interacting region of CRD is also involved in the proposed autoinhibitory mechanism, in which the engagement of KRAS:RBD–CRD on the membrane is coupled with release of the kinase domain for activation.
To characterize how multivalent interactions with the membrane may potentially enhance association of the complex with the bilayer surface, we measured the affinities of farnesylated KRAS, RBD–CRD, and farnesylated KRAS:RBD–CRD complex for immobilized nanodiscs using biolayer interferometry (BLI). The BLI data showed that the affinity of farnesylated KRAS and RBD–CRD for the membrane (dissociation constant [Kd] = 20 µM and 30 μM, respectively) was enhanced when they formed a complex (Kd = 10 µM) (SI Appendix, Fig. S22), suggesting cooperative assembly of the complex on the membrane. It has been shown that KRAS nanoclusters colocalize with regions of enriched PS on the membrane (45), which would favor CRD docking proximal to KRAS. Enhanced membrane affinity and local enrichment of these proteins may facilitate the formation of dimers and nanoclusters of RAS:RAF complexes.
Our structural studies, which were performed using an average of one KRAS per nanodisc, revealed that the MC-KRAS:RBD–CRD complex adopted two orientations on the membrane mediated by either KRAS α4–5 and CRD β7–8 (state A), or KRAS α5 and CRD α2/RBD β3–5 (state B). Recent studies have proposed that KRAS signaling is promoted by the formation of KRAS dimers on the membrane surface, and α4-α5 has been proposed to mediate dimerization, based on superresolution fluorescence microscopy (46), molecular simulations (47, 48), crystal packing (49), NMR PRE assay (50), and mutagenesis (51). In our state A MC-KRAS:RBD–CRD model, α4-α5 interacts with the membrane surface, which is incompatible with α4-α5–mediated KRAS dimerization. By contrast, in state B, the proposed α4-α5 dimerization interface is fully solvent-exposed and accessible for interaction with a second molecule of KRAS. To investigate the effect of increasing density of the KRAS:RBD–CRD complex on its orientation on nanodiscs, we performed experiments in which we added unlabeled farnesylated KRAS:RBD–CRD complex to labeled MC-KRAS tethered to a Gd3+-tagged nanodisc and bound to RBD–CRD. Although NMR detection was challenging, we observed a notable reduction of PRE on KRAS α4-α5 (reduction of state A) and significant enhancement of PRE on KRAS β1–3 (enrichment of state B) (SI Appendix, Fig. S23), indicating that increased density of the KRAS:RBD–CRD complex led to a state B-like orientation. The β4 region of RBD (106KGKKAR111 in CRAF) is the major interface that stabilizes state B, and, interestingly, this sequence varies among RAF paralogs (66KGRKTV71 in ARAF; 202DGEKKP207 in BRAF) (SI Appendix, Fig. S1D). CRAF has the most basic β4 region (net charge +4), and it is preceded by a three-residue insertion (HEH) relative to ARAF/BRAF whereas BRAF β4 is the most neutral (net charge 0) in nature (SI Appendix, Fig. S1D). The sequence divergence in this region may dictate how each RAF paralog interacts with the membrane, as well as the population and “residence time” of state B. Interestingly, a number of rare mutations in this β4-α2 region have been reported in the Catalogue of Somatic Mutations (COSMIC) and The Cancer Genome Atlas (TCGA) database (SI Appendix, Table S4) in both BRAF and CRAF, which increase the net positive charge in the β4-α2 region (SI Appendix, Fig. S24). We speculate that these alterations would augment the interactions between β4-α2 and the membrane, thereby enriching state B and promoting KRAS homodimerization.
In summary, our studies have characterized the interaction between KRAS and RBD–CRD in solution and revealed two NMR-visible conformational states of KRAS in complex with RBD–CRD on the membrane. The two states of KRAS:RBD–CRD on the membrane share similar protein:protein interfaces but differ in their membrane interactions, which dictate the orientation of both KRAS and RBD–CRD relative to the membrane surface. State B is fully compatible for KRAS homodimerization through the α4-α5 interface whereas state A is not. Increasing the number of KRAS:RBD–CRD complexes on the nanodisc promoted a shift toward state B, suggesting KRAS clustering on lipid bilayers stabilizes state B. Further, a cancer-associated mutation in RBD that increases the positive charge on the membrane interface of state B both stabilized that state and promoted kinase activity. Dimerization of the RAF kinase domain is likely to play additional roles in the assembly of the activated signaling complex. If future studies can overcome the technical challenges associated with preparing sufficient quantities of full-length RAF and detection of NMR signals from a much larger complex, further structural mechanisms underlying the KRAS-dependent activation of RAF may be revealed, along with new therapeutic strategies.
Materials and Methods
Preparation of proteins and nanodiscs, NMR data collection and analysis, cell transfection, Western blot, BLI assays, and RAF sequence analysis are fully described in SI Appendix, SI Materials and Methods.
All docking simulations were performed using high ambiguity driven biomolecular DOCKing (HADDOCK, version 2.2) software (25). The starting structure of full-length KRAS4B GMPPNP was derived from PDB ID 4DST. The complex formed by KRAS4B and RBD–CRD in solution was modeled in HADDOCK using restraints derived from intermolecular NOEs, CSPs, and PREs (SI Appendix, Table S2). A distance range of 2 to 6 Å and 2 to 5 Å was applied to CSP and membrane-PRE–based ambiguous interaction restraints (AIRs), respectively, and a distance range of 2 to 5.5 Å was applied to all NOE-based unambiguous restraints in HADDOCK. For TEMPO PRE experiments, two classes of distance restraints were used for structure calculation. Peaks with an intensity ratio <0.5, but still detectable in the paramagnetic spectra, were converted to restraints of the calculated distances ±4 Å. Severely broadened peaks not detectable in the paramagnetic spectra were converted to restraints with no lower bound and a target distance estimated from the noise in the spectrum plus an upper bound of 4 Å. The conversion from intensity ratio to calculated distance is described in SI Appendix, SI Materials and Methods (29, 52–54). Due to the lack of protons in the HADDOCK-derived structural model, the proton–proton NOEs were converted to distance restraints between the adjacent carbon atoms of KRAS4B and RBD–CRD. The TEMPO PRE data were converted to distance restraints between KRAS4B Cys43 or the N-term Cys0 γ-position atoms and the carbon/nitrogen atoms of RBD–CRD methyls/amides. In HADDOCK simulations, the flexible dual domain RBD–CRD was treated as two independent molecules with connectivity restraints between them, which consist of two files and was first proposed by Bonvin’s group (55). The first file was used during rigid body docking (it0) and semiflexible refinement (it1) to keep RBD C termini (H133) and CRD N termini (V134) within 10 Å. A real peptide distance of 1.3 Å was imposed to RBD C termini and CRD N termini in the second file (H-bond restraint file) to restore connectivity. To keep Zn2+ coordination in CRD, 0 to 2 Å unambiguous distance restraints were defined between zinc atoms and its coordinating atoms (H139, C165, C168, and C184 for one zinc, and C152, C155, C176, and H173 for the other zinc in CRD). The docking protocol comprised a 3,000 rigid-body docking stage, and the top 200 HADDOCK scored structures were further refined using semiflexible simulated annealing, followed by water refinement.
Data Availability.
The atomic coordinates and structures of the MC-KRAS:RBD–CRD complex on nanodisc have been deposited in the Protein Data Bank, https://www.rcsb.org/ under the accession code 6PTS (state A) and 6PTW (state B). Materials and reagents are available upon request.
Supplementary Material
Acknowledgments
This work was supported by Cancer Research Society (Canada) Grant 14014, Canadian Cancer Society Research Institute Grant 703209, the Princess Margaret Cancer Foundation, and Canadian Institutes of Health Research Foundation Grant 410008598. Z.F. acknowledges the Connaught Fund for a Connaught International Scholarship. We thank Dominic Esposito (National Cancer Institute RAS Initiative) for kindly providing the baculovirus expression system for production of fully processed KRAS; Carl Virtanen and Zhibin Lu for providing access to the Princess Margaret Computational Biology Resource Center cluster and technical assistance; and Alexandre Bonvin (Utrecht University) for HADDOCK support. NMR spectrometers were funded by the Canada Foundation for Innovation and supported by the Princess Margaret Cancer Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.L.C. is a guest editor invited by the Editorial Board.
Data deposition: Two structures have been deposited in the Protein Data Bank (PDB ID codes 6PTS [state A] and 6PTW [state B]). State A and state B are two conformations of KRAS:RBD–CRD on nanodiscs.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914076117/-/DCSupplemental.
References
- 1.Prior I. A., Lewis P. D., Mattos C., A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavoie H., Therrien M., Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 16, 281–298 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Rajakulendran T., Sahmi M., Lefrançois M., Sicheri F., Therrien M., A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Hu J. et al., Allosteric activation of functionally asymmetric RAF kinase dimers. Cell 154, 1036–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mott H. R. et al., The solution structure of the Raf-1 cysteine-rich domain: A novel ras and phospholipid binding site. Proc. Natl. Acad. Sci. U.S.A. 93, 8312–8317 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L., Hofer F., Martin G. S., Kim S. H., Structural basis for the interaction of Ras with RalGDS. Nat. Struct. Biol. 5, 422–426 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Fetics S. K. et al., Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada T. et al., The strength of interaction at the Raf cysteine-rich domain is a critical determinant of response of Raf to Ras family small GTPases. Mol. Cell. Biol. 19, 6057–6064 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu C. D. et al., Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J. Biol. Chem. 270, 30274–30277 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Brtva T. R. et al., Two distinct Raf domains mediate interaction with Ras. J. Biol. Chem. 270, 9809–9812 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Williams J. G. et al., Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem. 275, 22172–22179 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Thapar R., Williams J. G., Campbell S. L., NMR characterization of full-length farnesylated and non-farnesylated H-Ras and its implications for Raf activation. J. Mol. Biol. 343, 1391–1408 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Drugan J. K. et al., Ras interaction with two distinct binding domains in Raf-1 may be required for Ras transformation. J. Biol. Chem. 271, 233–237 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Daub M. et al., The RafC1 cysteine-rich domain contains multiple distinct regulatory epitopes which control Ras-dependent Raf activation. Mol. Cell. Biol. 18, 6698–6710 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh S. et al., The cysteine-rich region of raf-1 kinase contains zinc, translocates to liposomes, and is adjacent to a segment that binds GTP-ras. J. Biol. Chem. 269, 10000–10007 (1994). [PubMed] [Google Scholar]
- 16.Improta-Brears T., Ghosh S., Bell R. M., Mutational analysis of Raf-1 cysteine rich domain: Requirement for a cluster of basic aminoacids for interaction with phosphatidylserine. Mol. Cell. Biochem. 198, 171–178 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Li S., Jang H., Zhang J., Nussinov R., Raf-1 cysteine-rich domain increases the affinity of K-Ras/Raf at the membrane, promoting MAPK signaling. Structure 26, 513–525.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z. L., Prakash P., Buck M., A “Tug of War” maintains a dynamic protein-membrane complex: Molecular dynamics simulations of C-Raf RBD-CRD bound to K-Ras4B at an anionic membrane. ACS Cent. Sci. 4, 298–305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travers T. et al., Molecular recognition of RAS/RAF complex at the membrane: Role of RAF cysteine-rich domain. Sci. Rep. 8, 8461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Block C., Janknecht R., Herrmann C., Nassar N., Wittinghofer A., Quantitative structure-activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat. Struct. Biol. 3, 244–251 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Schreiber G., Haran G., Zhou H. X., Fundamental aspects of protein-protein association kinetics. Chem. Rev. 109, 839–860 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez G. A., Daniotti J. L., Electrical properties of plasma membrane modulate subcellular distribution of K-Ras. FEBS J. 274, 2210–2228 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Ben-Tal N., Honig B., Miller C., McLaughlin S., Electrostatic binding of proteins to membranes. Theoretical predictions and experimental results with charybdotoxin and phospholipid vesicles. Biophys. J. 73, 1717–1727 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulgrew-Nesbitt A. et al., The role of electrostatics in protein-membrane interactions. Biochim. Biophys. Acta 1761, 812–826 (2006). [DOI] [PubMed] [Google Scholar]
- 25.van Zundert G. C. P. et al., The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Mazhab-Jafari M. T. et al., Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. U.S.A. 112, 6625–6630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Z. et al., Inhibition of K-RAS4B by a unique mechanism of action: Stabilizing membrane-dependent occlusion of the effector-binding site. Cell Chem. Biol. 25, 1327–1336.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Lakshman B. et al., Quantitative biophysical analysis defines key components modulating recruitment of the GTPase KRAS to the plasma membrane. J. Biol. Chem. 294, 2193–2207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang C., Iwahara J., Clore G. M., Visualization of transient encounter complexes in protein-protein association. Nature 444, 383–386 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Colón-González F., Kazanietz M. G., C1 domains exposed: From diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta 1761, 827–837 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y. et al., Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168, 239–251.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeung T. et al., Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y. et al., SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 349, 873–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansen J. M. et al., Inhibition of prenylated KRAS in a lipid environment. PLoS One 12, e0174706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao S. et al., K-Ras G-domain binding with signaling lipid phosphatidylinositol (4,5)-phosphate (PIP2): Membrane association, protein orientation, and function. J. Biol. Chem. 294, 7068–7084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjee A., Jang H., Nussinov R., Gaponenko V., The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr. Opin. Struct. Biol. 36, 10–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bax A., Ikura M., Kay L. E., Barbato G., Spera S., Multidimensional triple resonance NMR spectroscopy of isotopically uniformly enriched proteins: A powerful new strategy for structure determination. Ciba Found. Symp. 161, 108–119, discussion 119–135 (1991). [DOI] [PubMed] [Google Scholar]
- 38.Gardner K. H., Kay L. E., The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 27, 357–406 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Holderfield M., Deuker M. M., McCormick F., McMahon M., Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 14, 455–467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tate J. G. et al., COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawada G. et al., Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology 150, 1171–1182 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Kiel C. et al., Improved binding of raf to Ras.GDP is correlated with biological activity. J. Biol. Chem. 284, 31893–31902 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock J. F., Paterson H., Marshall C. J., A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63, 133–139 (1990). [DOI] [PubMed] [Google Scholar]
- 44.Park E. et al., Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 575, 545–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y. et al., Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 34, 862–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nan X. et al., Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prakash P. et al., Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers. Sci. Rep. 7, 40109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R., Membrane-associated Ras dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem. J. 473, 1719–1732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer-Smith R. et al., Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 13, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee K. Y. et al., Two distinct structures of membrane-associated homodimers of GTP- and GDP-bound KRAS4B revealed by paramagnetic relaxation enhancement. Angew. Chem. Int. Ed. Engl., 10.1002/anie.202001758 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ambrogio C. et al., KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Battiste J. L., Wagner G., Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39, 5355–5365 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Iwahara J., Clore G. M., Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440, 1227–1230 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Iwahara J., Tang C., Marius Clore G., Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 184, 185–195 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karaca E., Bonvin A. M., A multidomain flexible docking approach to deal with large conformational changes in the modeling of biomolecular complexes. Structure 19, 555–565 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structures of the MC-KRAS:RBD–CRD complex on nanodisc have been deposited in the Protein Data Bank, https://www.rcsb.org/ under the accession code 6PTS (state A) and 6PTW (state B). Materials and reagents are available upon request.