Abstract
The expression of desaturases is higher in many types of cancer, and despite their recognized role in oncogenesis, there has been no research on the expression of desaturases in glioblastoma multiforme (GBM). Tumor tissue samples were collected during surgery from 28 patients (16 men and 12 women) diagnosed with GBM. The effect of necrotic conditions and nutritional deficiency (mimicking conditions in the studied tumor zones) was studied in an in vitro culture of human brain (glioblastoma astrocytoma) U-87 MG cells. Analysis of desaturase expression was made by qRT-PCR and the immunohistochemistry method. In the tumor, the expression of stearoyl–coenzyme A desaturase (SCD) and fatty acid desaturases 2 (FADS2) was lower than in the peritumoral area. The expression of other desaturases did not differ in between the distinguished zones. We found no differences in the expression of SCD, fatty acid desaturases 1 (FADS1), or FADS2 between the sexes. Necrotic conditions and nutritional deficiency increased the expression of the studied desaturase in human brain (glioblastoma astrocytoma) U-87 MG cells. The obtained results suggest that (i) biosynthesis of monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA) in a GBM tumor is less intense than in the peritumoral area; (ii) expressions of SCD, SCD5, FADS1, and FADS2 correlate with each other in the necrotic core, growing tumor area, and peritumoral area; (iii) expressions of desaturases in a GBM tumor do not differ between the sexes; and (iv) nutritional deficiency increases the biosynthesis of MUFA and PUFA in GBM cells.
Keywords: glioblastoma multiforme, brain tumor, stearoyl–CoA desaturase, fatty acid desaturase, fatty acid
1. Introduction
Glioblastoma multiforme (GBM) is one of the most common primary brain tumors. It is also the most invasive and undifferentiated glioma and the most malignant tumor based on the histopathological criteria defined by the World Health Organization (WHO). It is characterized by high mortality despite surgical intervention, chemotherapy, and radiotherapy. The overall survival rate and 5-year postoperative survival are very low, at 8.1 months and 9.8%, respectively [1,2].
A promising direction of research into possible therapies for GBM is the metabolism of fatty acids in cancer cells [3,4]. Particularly interesting is the action of desaturases, enzymes forming double bonds in fatty acids, which can be divided into those with the highest catalytic activity for saturated fatty acids (stearoyl–CoA desaturase (SCD) and stearoyl-CoA desaturase 5 (SCD5)) or for unsaturated fatty acids (fatty acid desaturase 1 (FADS1), fatty acid desaturase 2 (FADS2), and fatty acid desaturase 3 (FADS3)).
SCD converts saturated fatty acids (SFA) into monounsaturated fatty acids (MUFA) [5], performing Δ9 desaturation in fatty acids with chain lengths of 12 to 19 carbons [6], with the highest specificity for stearic acid. The expression of this enzyme is highest in the adipose tissue, liver, and brain [7,8]. This enzyme plays an important role in lipid metabolism [9], and its expression is strictly regulated by hormones that are essential for the regulation of metabolism and by nutrients themselves [10]. Its expression and activity is elevated in cases of obesity [11,12,13] and insulin resistance [14]. SCD also participates in the synthesis of fatty acids, the structural elements of cell membranes, and therefore, its expression increases in rapidly dividing cells [15]. SCD expression is elevated in the tumors of many cancers, including esophageal cancer [16,17], hepatocellular adenoma and carcinoma [16,18], colorectal cancer [16,17], breast cancer [17,19,20], gastric cancer [17], thyroid cancer [17,21], lung adenocarcinoma [19,22], prostate cancer [19,20], and clear-cell renal cell carcinoma [23]. Importantly, the increased tumor expression of SCD is associated with worse prognosis.
SCD5 is the second Δ9-desaturase isoform found in humans; it also occurs in most fish, amphibians, reptiles, birds, and mammals except Muridae [24]. Expression of this enzyme is highest in the brain, particularly in the fetal stage, and in the pancreas, adrenal gland, and ovaries [7,8]. The properties of SCD5 are similar to those of SCD. However, an experiment on murine neuroblastoma Neuro2a cells showed that SCD5 exhibited substrate specificity only for palmitic acid but not for stearic acid [25], in contrast to observations in A375 melanoma and 4T1 breast cancer cells, where SCD5 did desaturate stearic acid [26]. Unlike SCD, SCD5 may not be responsible for increasing fatty acid synthesis during cell proliferation [26]. A change in the expression of SCD5 alters the composition of phospholipids in biological membranes, increasing the synthesis of phosphatidylcholine and cholesterolesters [25]. SCD5 is also known to play an important role in oncogenesis in breast cancer [26,27], and the expression of this enzyme increases in anaplastic thyroid carcinoma [21]. On the other hand, SCD5 acts as a tumor suppressor in melanoma, with the cancer reducing its expression [28].
FADS1 and FADS2 are responsible for the biosynthetic pathways that produce long-chain polyunsaturated fatty acids (PUFA). The expression of these desaturases is highest in the liver, brain, and adrenal glands [8,29,30]. They participate in the synthesis of arachidonic acid from γ-linolenic acid and linoleic acid [31,32,33]. By producing arachidonic acid, FADS1 and FADS2 participate in the production of prostaglandins and thus play an important role in inflammation and associated neoplastic processes [34,35]. For this reason, the expression of FADS1 and FADS2 is increased in some cancers, e.g., Lewis lung carcinoma [35], melanoma [35], colon cancer [36], hepatocellular carcinoma [37], and breast cancer [34]. FADS1 and FADS2 also participate in the biosynthetic pathway for n-3 family long-chain PUFA, resulting in the formation of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) from α-linolenic acid. These fatty acids exhibit anti-inflammatory [38] and anti-cancer properties [39,40,41].
The function of FADS3 is unclear; the gene encodes a protein with a structure similar to that of other desaturases. Its expression is highest in fatty tissue and placenta, and it is moderate in the brain [8]. FADS3 does not directly catalyze any reactions in the synthetic pathways for DHA or arachidonic acid or in the formation of MUFA from SFA [42]. Although it has displayed Δ13-desaturase activity for trans-vaccenic acid in transfected COS-7 cells and rat hepatocytes [42], those results have been contested [43]. The function of FADS3 remains unclear, although it appears that the expression of this gene supports the synthesis of arachidonic acid and DHA in the liver and brain by regulating the expression of FADS1 and FADS2 [43].
Although the expression of desaturases is increased in many types of cancer and despite their recognized role in oncogenesis, there has been no research on the role of desaturases in the formation of GBM. Therefore, the aim of this study was to analyze the mRNA expression of desaturase genes—SCD, SCD5, FADS1, FADS2, and FADS3—in GBM in three tumor zones: necrotic core, growing tumor area, and peritumoral area. The intention was to analyze differences in the expression of individual desaturases between the three tumor zones.
2. Materials and Methods
2.1. Patient Samples
The material used in the present study was obtained from patients with brain tumors diagnosed by neuroimaging (magnetic resonance imaging (MRI) or computed tomography (CT)), who were hospitalized in the Department of Neurosurgery and Pediatric Neurosurgery of Pomeranian Medical University in Szczecin, Poland. The present project and archiving of material was initiated in 2014 by the Department of the Biochemistry and the Department of Neurosurgery and Pediatric Neurosurgery of Pomeranian Medical University in Szczecin and concerned the engagement of purinergic receptors in GBM progression. The project was accepted by the local bioethical commission (KB-0012/96/14), and the study was conducted in accordance with the Declaration of Helsinki.
Tumor tissue samples were collected during surgery from 28 patients (16 males and 12 females) diagnosed with central nervous system (CNS) tumor and GBM (Table 1). Patients presented with symptoms resulting from increased intracranial pressure such as dizziness and nausea, and those resulting from local tumor growth, which included sensory and motor disorders as well as disorders of higher nervous functions (Table 2).
Table 1.
The statistical characteristics of the study group.
| N | Mean | Standard Deviation | Median | Minimum | Maximum | First Quartile | Third Quartile | Interquartile Range | |
|---|---|---|---|---|---|---|---|---|---|
| Age at surgery | 24 | 60.7 | 12.5 | 64 | 36 | 81 | 54 | 68.5 | 14.5 |
| Weight | 24 | 84 | 19 | 89 | 55 | 130 | 67.5 | 95 | 27.5 |
| Height | 23 | 172 | 12 | 172 | 147 | 196 | 163 | 182 | 19 |
| Body mass index (BMI) | 23 | 28.7 | 4.8 | 27.9 | 21.5 | 38.9 | 24.7 | 31.9 | 7.2 |
| Physical activity | 21 | 3.05 | 1.16 | 3 | 1 | 4 | 3 | 4 | 1 |
| Limitation of physical activity caused by disease | 21 | 2.10 | 0.83 | 2 | 1 | 3 | 1 | 3 | 2 |
| Limitation of cognitive abilities | 21 | 2.19 | 0.87 | 2 | 1 | 3 | 1 | 3 | 2 |
N: number of patients included in the analysis. Physical activity, limitation of physical activity, and limitation of cognitive abilities were calculated on the basis of the levels indicated in the questionnaires. Physical activity: everyday—4/a few times a week—3/rarely—2/almost never—1. Limitation of physical activity: none—1/partial—2/considerable—3. Limitation of cognitive abilities: none—1/partial—2/considerable—3.
Table 2.
Inclusion and exclusion criteria. GBM: glioblastoma multiforme.
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Age 18+ | Age below 18 |
| Central nervous system tumor confirmed by radiological examination | Histopathological diagnosis other than GBM |
| Surgical and anesthetic qualification for surgery | |
| Histopathological diagnosis of biopsied tumor mass: GBM |
Each patient was recommended for neurosurgery following the radiological diagnosis of CNS tumors. After qualifying for surgery, patients underwent a standard anesthetic procedure (general anesthesia with endotracheal intubation). During the neuronavigation procedure, craniotomy and tumor resection were performed according to the classical method (bone removal and dura incision, tumor visualization, resection, biopsy for histopathological and molecular examination, closure of the dura, bone restoration in some patients, subcutaneous tissue and skin closure in some patients). The range of resection was determined by the extent of the tumor and its topography.
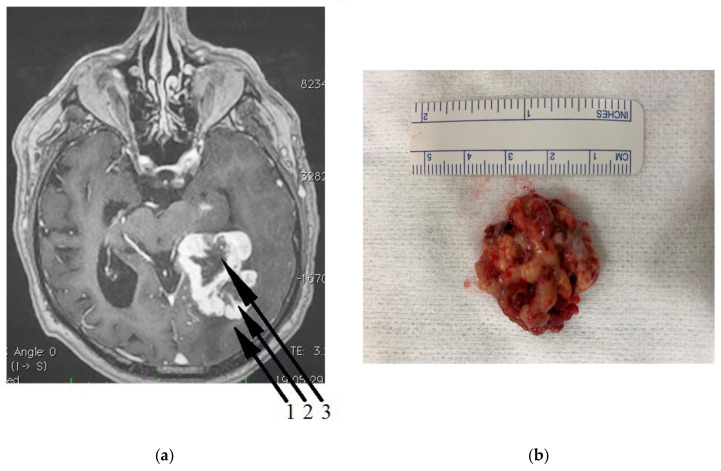
Clinical–radiological morphology made it possible to distinguish three tumor zones commonly used in the literature and clinical practice: the necrotic core (NC) is usually located in the central part of the tumor, the growing tumor area (GTA) (surrounding the necrotic core), and the peritumoral area (PA): a buffer zone between the tumor and healthy tissue, with individual foci of infiltration (Figure 1) [44].
Figure 1.
(a) Magnetic resonance imaging (MRI) scan of a patient with diagnosed glioblastoma multiforme (GBM). The particular zones 1,2,3 are visible on the tumor cross-section. Zone 1—the peritumoral area, Zone 2—the growing tumor area, Zone 3—the necrotic core. (b) GBM tissue obtained from temporal lobe. The tumor was removed by neuronavigation-guided craniotomy. II gyrus cortex was incised over a 5 cm section from the polar end and the central part of the tumor excised, followed by careful and fine debridement with an ultrasonic surgical aspirator.
The use of neuronavigation helped map the tumor and determine the topography of the zones. The results of 1.5 and 3 Tesla MRI were entered into the computer station of a neuronavigation device and used during the operation to determine the position of surgical instruments in relation to cancer tissue with the help of a television camera. The camera monitored the surgical movement in relation to the radiological image to a precision of 2–3 mm. This allowed safe and reliable resection in places where the image of the operating microscope was uncertain and the macroscopic tumor boundaries were difficult to distinguish. Neuronavigation during biopsy and craniotomy allowed the material to be extracted from the three separate zones. Each sample was subjected to histopathological examination to confirm the criteria of grade IV brain tumor defined by the WHO: IDH mutation, 1p19q codeletion, and MGMT gene promoter methylation.
2.2. Immunohistochemistry
Samples of the brain tumor (glioblastoma) were fixed in 4% buffered formalin then washed with absolute ethanol (3 times over 3 h), absolute ethanol with xylene (1:1) (twice over 1 h), and xylene (3 times over 20 min). Then, the tissues were saturated with liquid paraffin for 3 h and embedded in paraffin blocks. Using a microtome (Microm HM340E), 3–5 µm serial sections were taken, placed on polysine histological slides (Thermo Scientific, UK; cat. no. J2800AMNZ), deparaffinized in xylene, and then rehydrated in decreasing concentrations of ethanol to be used for immunohistochemical staining. In order to expose the epitopes, the sections were boiled twice in a microwave oven (700 W for 4 min and 3 min) in 10 mM citrate buffer (pH 6.0). Once cooled and washed with phosphate buffer saline (PBS), the endogenous peroxidase was blocked by a 3% solution of perhydrol in methanol, and then the slides were incubated for 30 min. at room temperature (RT) with primary antibodies (anti-SCD, monoclonal mouse antibody from Invitrogen, cat. no.: MA5-27542, final dilution 1:100; and anti-FADS2 polyclonal rabbit antibody from Invitrogen, cat. no.: PA5-48353, final dilution 1:50 according to the manufacturer’s datasheet). To visualize the antigen–antibody complex, a Dako LSAB+System-HRP was used (DakoCytomation, Code K0679) based on the reaction of avidin–biotin–horseradish peroxidase with diaminobenzidine (DAB) as a chromogen, according to the included staining procedure instructions. Sections were washed in distilled water, counterstained with hematoxylin, and prepped for mounting. For a negative control, specimens were processed in the absence of primary antibodies. Positive staining was defined microscopically (Leica DM5000B, Wetzlar, Germany) by the visual identification of brown pigmentation.
2.3. Cell Culture and Treatment
Human brain cells (glioblastoma astrocytoma, U-87 MG cell line) from the European Collection of Authenticated Cell Cultures (ECACC) were cultured in eagle’s minimum essential medium (EMEM), (Sigma-Aldrich, Poznań, Poland) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Gibco Limited, Poznań Poland), 2 mM l-glutamine, 1 mM sodium pyruvate (Sigma-Aldrich, Poznań, Poland), 1% non-essential amino acids (Sigma-Aldrich, Poznań, Poland), 100 U/mL penicillin (Gibco Limited, Poznań, Poland), and 100 µg/mL streptomycin (Gibco Limited, Poznań, Poland), at 37 °C in a humidified atmosphere of 95% air and 5% CO2. The U-87 MG cells were seeded in 6-well plates at a density of 40,000 cells/cm2 in full medium. After 72 h incubation (70%–80% confluence), cells were washed three times with pre-warmed phosphate buffer saline (PBS) solution (37 °C). Next, the cells were cultured for 24 h under three different conditions (control, nutrient deficiency, and necrotic). The control cells were suspended in full medium, the starved cells were grown in medium with a low concentration of l-glutamine (0.2 mM) and without sodium pyruvate (volume supplemented with PBS). For the induction of necrotic conditions, cells were incubated in medium supplemented with 200 µM of H2O2. After 24 h of incubation, U-87 MG cells were trypsinized (0.25% trypsin–ethylenediaminetetraacetic acid (EDTA) solution, Sigma-Aldrich, Poznań, Poland) from the plate. After centrifuging (25 °C, 300 G, 5 min), the supernatant was discarded and the obtained cell pellet was used for RNA analysis.
In vitro studies were performed to analyze the influence of necrotic factors on the expression of desaturases. The growing GBM tumor area contained numerous and very small necrotic niches [45,46,47,48]. Around these areas, the intensive proliferation of neoplastic cells occurs. The surroundings of these areas were also poorly vascularized, resulting in a local nutritional deficiency in the GBM cells. In order to investigate the influence of necrotic factors on the expression of the desaturases in the GBM cells in this study, the U-87 MG cells were treated with 200 µM of H2O2, which is a necrotic factor commonly used in in vitro tests [49,50]. In addition, to better reflect the conditions in which GBM cells live, we studied the effect of nutritional deficiency on the expression of desaturases in U-87 MG cells. These cells grew in medium with a low concentration of l-glutamine (0.2 mM) and without sodium pyruvate. However, there was still 1.0 g/L (5.5 mM) of glucose in the medium. Under these conditions, the concentration of substances such as mineral salts, vitamins, other amino acids or growth factors were not changed. The reduction in the concentration of the selected substances significantly reduced the concentration of substrates for fatty acid production, because in the GBM cells, glucose is converted into pyruvate, then into acetyl-CoA, and then further into fatty acids. Glutamine is converted into acetyl-CoA and then into fatty acids [51,52]. In other words, we reduced the amount of substrate for the production of fatty acids and tested the expression of the tested desaturases. This model better represents the necrotic zones in the GBM tumor than the use of toxic substances that cause necrosis.
2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Quantitative analysis of mRNA expression of FADS1, FADS2, FADS3, SCD, and SCD5 genes was performed by two-step reverse transcription PCR (RT-PCR). Total RNA was extracted from 50–100 mg tissue samples using an RNeasy Lipid Tissue Mini Kit (Qiagen) and, for the in vitro study, from 300,000 cells using an RNeasy Mini Kit. cDNA was prepared from 1 μg of total cellular RNA in 20 μL of reaction volume using a FirstStrand cDNA synthesis kit and oligo-dT primers (Fermentas). Quantitative assessment of mRNA levels was performed by real-time RT-PCR using an ABI 7500Fast instrument with Power SYBR Green PCR Master Mix reagent (Applied Biosystems). Real-time conditions were as follows: 95 °C (15 s), 40 cycles at 95 °C (15 s), and 60 °C (1 min). According to melting point analysis, only one PCR product was amplified under these conditions. Each sample was analyzed in two technical replicates, and mean Ct values were used for further analysis. The relative quantity of a target, normalized to the levels of endogenous controls glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-2 macroglobulin (B2M) genes, was calculated as the fold difference (2^dCt) and further processed using statistical analysis. Data were presented as tumor tissue absolute expression.
The GAPDH reference gene was selected because it is considered a suitable control in research on the expression of various genes in GBM [53,54,55]. Additionally, we used also a second reference gene: B2M [54,55].
The following primer pairs were used: (5′-TCA TGG GTG TGA ACC ATG AGA A-3′ and 5′-GGC ATG GAC TGT GGT CAT GAG-3′) for GAPDH, 5′-AAT GCG GCA TCT TCA AAC CT-3′ and 5′-TGA CTT TGT CAC AGC CCA AGA TA-3′) for B2M, (5′-CCA ACT GCT TCC GCA AAG AC 3′ and 5′-GCT GGT GGT TGT ACG GCA TA-3′) for FADS1, (5′-TGA CCG CAA GGT TTA CAA CAT-3′ and 5′-AGG CAT CCG TTG CAT CTT CTC-3′) for FADS2, (5′-GGA GTC ATC CGT CGA GTA TGG-3′ and 5′-GGG CCG ATC AGG AAG AAG T-3′) for FADS3, (5′-TTC CTA CCT GCA AGT TCT ACA CC-3′ and 5′-CCG AGC TTT GTA AGA GCG GT-3′) for SCD, and (5′-TGC GAC GCC AAG GAA GAA AT-3′ and 5′-CCT CCA GAC GAT GTT CTG CC-3′) for SCD5.
2.5. Statistical Methods
The expression values of the desaturase genes were calculated in relation to the expression of two reference genes: B2M and GAPDH. The relative expression values for the three zones (necrotic core, growing tumor area, and peritumoral area) in each patient and their ratios were calculated, e.g., growing tumor area/necrotic core as a ratio of relative expression in growing tumor area to relative expression in necrotic core. The distribution of expression values significantly differed from the normal distribution (Shapiro–Wilk test), and therefore statistical analysis was based on non-parametric tests: the Mann–Whitney U-test for comparisons between groups of patients, Wilcoxon signed-rank test for comparisons between zones of tumor, and the Spearman rank correlation coefficient for the analysis of correlations between the expression of the tested genes in the three zones. The median and quartile values were given as descriptive statistics in tables and graphs. The statistical significance threshold was p < 0.05. Calculations were performed using Statistica 13 software.
3. Results
3.1. The Expression of SCD and FADS2 Was Lower in GBM Growing Tumor Area than in Peritumoral Area
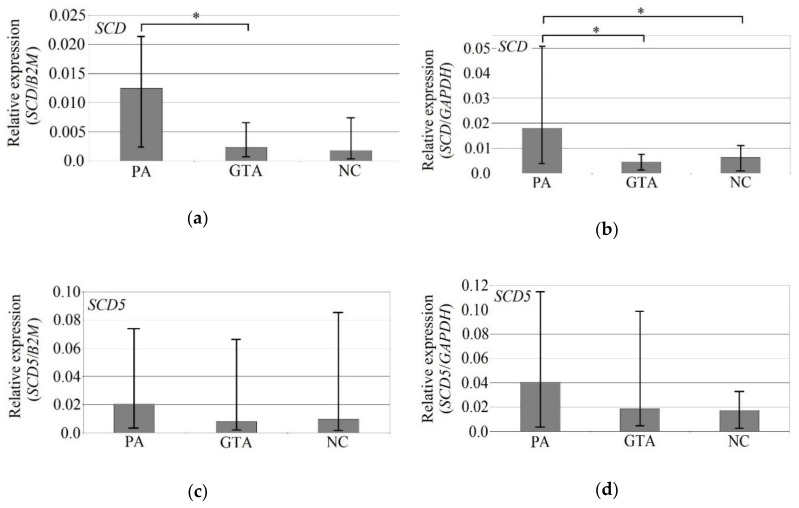
The expression of SCD in GBM was significantly lower in the growing tumor area compared to the peritumoral area and in the necrotic core compared to the peritumoral area (Figure 2). Using the GAPDH gene as reference, in the peritumoral area, we observed a significantly higher (almost 4 times) expression of SCD than in the growing tumor area (p = 0.015) and almost 3 times higher expression than in necrotic core (p = 0.019). Using the B2M gene as reference, the expression of SCD in the peritumoral area was more than 5 times higher (p = 0.005) than in the growing tumor area. However, the expression of B2M is significantly altered in the GBM tumor, and it is significantly different from that in the brain [54,55]. For this reason, all data using this reference gene should be analyzed with caution.
Figure 2.
mRNA expression of stearoyl–CoA desaturase (SCD) (a,b) and SCD5 (c,d) depending on the location in GBM, using B2M and GAPDH genes as reference. The graphs show the median mRNA expression of a given desaturase. The error bars are the first and third quartiles. * - statistically significant difference in the expression of a given desaturase between the tumor area according to the Wilcoxon signed-rank test (p < 0.05). PA—peritumoral area (n = 22); GTA—growing tumor area (n = 19); NC—necrotic core (n = 21).
The expression of brain-specific SCD5 was also lower in the growing tumor area versus the peritumoral area and in the necrotic core versus the peritumoral area, but they were not statistically significantly for either of the reference genes.
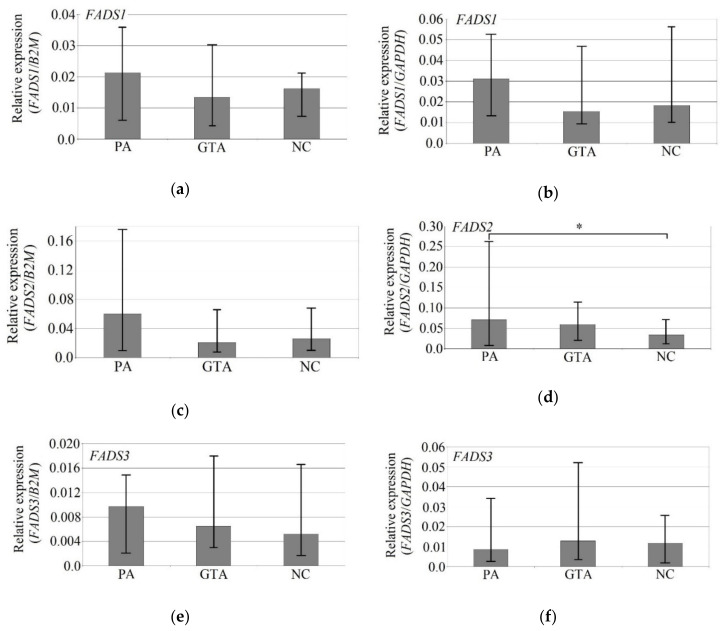
Similar to SCD, FADS1 expression was lower in the growing tumor area compared to the peritumoral area and in the necrotic core compared to the peritumoral area (Figure 3). However, the observed differences were not statistically significant in relation to either of the reference genes. The expression of FADS2 was also lower in the growing tumor area versus the peritumoral area and in the necrotic core versus the peritumoral area, but the difference was statistically significant only when using GAPDH as reference: FADS2 expression in the peritumoral area was 2 times higher than in the necrotic core (p = 0.027).
Figure 3.
mRNA expression of FADS1 (a,b), FADS2 (c,d), and FADS3 (e,f) depending on the location in GBM, using B2M and GAPDH genes as reference. The graphs show the median mRNA expression of a given desaturase. The error bars are the first and third quartiles. * - statistically significant difference in the expression of a given desaturase between the tumor area according to the Wilcoxon signed-rank test (p < 0.05). PA—peritumoral area (n = 22); GTA—growing tumor area (n = 19); NC—necrotic core (n = 21).
FADS3 expression measured against B2M reference gene was lower in the growing tumor area versus the peritumoral area and in the necrotic core versus the peritumoral area, but—similarly to previous genes—the observed differences were not statistically significant. The expression of FADS3 measured against the GAPDH reference gene was higher in the growing tumor area versus the peritumoral area and in the necrotic core versus the peritumoral area, but again the differences were statistically insignificant.
3.2. Desaturase Expression Positively Correlated within and between Individual GBM Zones Glucose
Some positive correlations were found between the expression of desaturases in each of the studied tumor zones.
In the growing tumor area, FADS1, FADS2, SCD, and SCD5 expression positively correlated when using the B2M gene as reference (Table 3). However, the expression of B2M is significantly altered in the GBM tumor, and it is significantly different from that in the brain [54,55]. For this reason, all data using this reference gene should be analyzed with caution.
Table 3.
Correlations between the mRNA expression of all studied desaturase genes in all three tumor zones, using B2M gene as reference.
| Studied Gene/ Location |
FADS1 | FADS2 | FADS3 | SCD | SCD5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTA | NC | PA | GTA | NC | PA | GTA | NC | PA | GTA | NC | PA | GTA | NC | PA | ||
| FADS1 | GTA | 1.00 | ||||||||||||||
| NC | 0.26 | 1.00 | ||||||||||||||
| PA | 0.48 * | 0.15 | 1.00 | |||||||||||||
| FADS2 | GTA | 0.77 * | 0.09 | 0.51 | 1.00 | |||||||||||
| NC | 0.15 | 0.58 * | 0.33 | 0.48 | 1.00 | |||||||||||
| PA | 0.22 | 0.10 | 0.48 * | 0.50 * | 0.53 * | 1.00 | ||||||||||
| FADS3 | GTA | 0.10 | −0.11 | 0.21 | 0.18 | 0.08 | 0.40 | 1.00 | ||||||||
| NC | 0.00 | 0.52 * | −0.01 | 0.14 | 0.21 | 0.37 | 0.40 | 1.00 | ||||||||
| PA | −0.10 | −0.03 | 0.43 * | 0.12 | 0.23 | 0.37 | 0.71 * | 0.29 | 1.00 | |||||||
| SCD | GTA | 0.58 * | 0.16 | 0.13 | 0.68* | 0.50 | 0.40 | 0.22 | 0.17 | 0.25 | 1.00 | |||||
| NC | 0.35 | 0.63 * | 0.04 | 0.38 | 0.45 * | 0.07 | 0.12 | 0.59 * | 0.09 | 0.30 | 1.00 | |||||
| PA | 0.20 | 0.35 | 0.46 * | 0.46 | 0.68 * | 0.74 * | 0.35 | 0.49 * | 0.39 | 0.64 * | 0.40 | 1.00 | ||||
| SCD5 | GTA | 0.65 * | −0.07 | 0.48 * | 0.82 * | 0.35 | 0.52 * | 0.03 | −0.05 | −0.05 | 0.56 * | 0.20 | 0.57 * | 1.00 | ||
| NC | 0.30 | 0.66 * | 0.37 | 0.51 * | 0.80 * | 0.65 * | 0.05 | 0.42 | 0.25 | 0.52 * | 0.63 * | 0.76 * | 0.48 | 1.00 | ||
| PA | 0.47 * | 0.21 | 0.55* | 0.68 * | 0.62 * | 0.70 * | 0.16 | 0.08 | −0.02 | 0.24 | 0.22 | 0.59 * | 0.65 * | 0.71 * | 1.00 | |
The values of Spearman’s rank correlation coefficients are given. * statistically significant correlation of the expression of two genes/locations (p < 0.05). GTA—growing tumor area; NC—necrotic core; PA—peritumoral area.
When using GAPDH as a reference gene, the results were similar, although no correlation was found between the expression of SCD and SCD5 (Table 4). FADS3 expression positively correlated only with SCD expression, albeit only when the GAPDH gene was used as reference.
Table 4.
Correlations between the mRNA expression of all studied desaturase genes in all three tumor zones, using GAPDH gene as reference.
| Studied Gene/ Location |
FADS1 | FADS2 | FADS3 | SCD | SCD5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTA | NC | PA | GTA | NC | PA | GTA | NC | PA | GTA | NC | PA | GTA | NC | PA | ||
| FADS1 | GTA | 1.00 | ||||||||||||||
| NC | 0.44 | 1.00 | ||||||||||||||
| PA | 0.22 | 0.31 | 1.00 | |||||||||||||
| FADS2 | GTA | 0.59 * | −0.31 | −0.20 | 1.00 | |||||||||||
| NC | 0.01 | 0.13 | 0.11 | −0.07 | 1.00 | |||||||||||
| PA | 0.16 | −0.26 | 0.52 * | 0.13 | 0.45 | 1.00 | ||||||||||
| FADS3 | GTA | 0.47 * | 0.14 | −0.01 | 0.29 | −0.16 | −0.01 | 1.00 | ||||||||
| NC | 0.13 | 0.57 * | −0.13 | −0.20 | −0.18 | −0.09 | 0.36 | 1.00 | ||||||||
| PA | −0.16 | 0.19 | 0.53 * | −0.53 * | 0.02 | 0.31 | 0.26 | 0.32 | 1.00 | |||||||
| SCD | GTA | 0.78 * | 0.30 | 0.10 | 0.67 * | 0.18 | 0.14 | 0.55 * | 0.12 | −0.13 | 1.00 | |||||
| NC | 0.14 | 0.44 | −0.06 | 0.10 | 0.24 | −0.14 | 0.02 | 0.26 | 0.11 | 0.13 | 1.00 | |||||
| PA | 0.35 | −0.09 | 0.44 | 0.37 | 0.50 * | 0.66 * | 0.22 | −0.17 | 0.32 | 0.51 * | 0.00 | 1.00 | ||||
| SCD5 | GTA | 0.65 * | −0.21 | 0.09 | 0.78 * | −0.05 | 0.20 | 0.11 | −0.44 | −0.55 * | 0.46 | 0.02 | 0.38 | 1.00 | ||
| NC | 0.19 | 0.03 | 0.03 | 0.13 | 0.69 * | 0.48 * | −0.09 | −0.19 | 0.00 | 0.15 | 0.40 | 0.45 | 0.14 | 1.00 | ||
| PA | 0.34 | −0.19 | 0.56 * | 0.30 | 0.33 | 0.72* | −0.01 | −0.35 | 0.03 | 0.04 | 0.02 | 0.42 | 0.62 * | 0.40 | 1.00 | |
The values of Spearman’s rank correlation coefficients are given. * statistically significant correlation of the expression of two genes/locations (p < 0.05). GTA—growing tumor area; NC—necrotic core; PA—peritumoral area.
The expression of desaturases also correlated in the necrotic core. Using the B2M gene as reference, SCD, SCD5, FADS1, and FADS2 expression were positively correlated in this zone. However, FADS3 expression positively correlated only with SCD and FADS1. Using GAPDH as a reference gene, positive correlations were found only for FADS2 versus SCD5, and FADS1 versus FADS3.
Similar correlations were found in the peritumoral area. Using the B2M gene as reference, positive correlations were found between SCD, SCD5, FADS1, and FADS2. FADS3 expression positively correlated only with FADS1 expression. Similar results were obtained using the second reference gene, GAPDH. The expression of FADS1, FADS2, FADS3, and SCD5 were positively correlated, while FADS1 versus SCD and SCD versus SCD5 were not. Again, FADS3 expression positively correlated with FADS1 expression only.
Some positive correlations were also found in the expression of individual desaturases between the studied structures. Taking B2M as the reference gene, correlations were observed between the growing tumor area and peritumoral area for all studied desaturase genes. There was also a positive correlation for FADS2 and SCD5 expression between the necrotic core and peritumoral area. However, when using the GAPDH gene as reference, correlations between the growing tumor area and peritumoral area were found only for SCD and SCD5.
When using the reference gene GAPDH, there were very few correlations between the GBM zones. A negative correlation was found between the expression of FADS2 in the growing tumor area and the expression of FADS3 in the peritumoral area; a positive correlation between FADS2 expression in the necrotic core and SCD in the peritumoral area; and a positive correlation between FADS2 expression in the peritumoral area and SCD5 expression in the necrotic core. At the same time, FADS3 expression in the peritumoral area negatively correlated with SCD5 expression in the growing tumor area.
There were many more correlations of different desaturase expressions between the studied tumor zones with respect to the B2M reference gene, as shown in detail in the table below. Most were between SCD5 and FADS2 expression. Correlations in the expression of these two genes were found between all structures except FADS2 expression in the necrotic core versus SCD5 expression in the growing tumor area.
3.3. No Significant Differences between the Sexes in the Expression of Desaturases
The expression of SCD and SCD5, the enzymes responsible for SFA desaturation, did not differ significantly between the sexes in the growing tumor area or peritumoral area (Table 5). Although the expression of SCD in the necrotic core was 6 times higher in men, the relationship was statistically insignificant (p = 0.09 for B2M, p = 0.105 for GAPDH).
Table 5.
mRNA expression of SCD and SCD5 by sex.
| Sex | Female | Male | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Studied Gene/Reference Gene/Location | Median | 1st Quartile | 3rd Quartile | N | Median | 1st Quartile | 3rd Quartile | N | p |
| SCD/ B2M | |||||||||
| GTA | 0.0023 | 0.0013 | 0.0064 | 9 | 0.0009 | 0.0004 | 0.0033 | 9 | 0.233 |
| NC | 0.0004 | 0.0002 | 0.0017 | 8 | 0.0026 | 0.0004 | 0.0193 | 12 | 0.090 |
| PA | 0.0063 | 0.0018 | 0.0207 | 8 | 0.0104 | 0.0019 | 0.0202 | 12 | 1.000 |
| GTA/NC | 3.5421 | 1.0997 | 9.4235 | 8 | 0.2712 | 0.0410 | 1.3336 | 7 | 0.083 |
| SCD/GAPDH | |||||||||
| GTA | 0.0044 | 0.0008 | 0.0107 | 9 | 0.0043 | 0.0017 | 0.0060 | 9 | 0.965 |
| NC | 0.0012 | 0.0005 | 0.0036 | 8 | 0.0078 | 0.0010 | 0.0125 | 12 | 0.105 |
| PA | 0.0092 | 0.0021 | 0.0470 | 8 | 0.0186 | 0.0046 | 0.0468 | 12 | 0.758 |
| SCD5/ B2M | |||||||||
| GTA | 0.0079 | 0.0017 | 0.0432 | 10 | 0.0078 | 0.0016 | 0.0088 | 9 | 0.568 |
| NC | 0.0016 | 0.0006 | 0.0102 | 9 | 0.0121 | 0.0036 | 0.0854 | 12 | 0.201 |
| PA | 0.0054 | 0.0021 | 0.0423 | 9 | 0.0235 | 0.0038 | 0.0654 | 13 | 0.664 |
| GTA/NC | 3.5979 | 1.3722 | 5.6384 | 9 | 0.3880 | 0.1523 | 0.9700 | 7 | 0.063 |
| SCD5/GAPDH | |||||||||
| GTA | 0.0090 | 0.0026 | 0.0199 | 10 | 0.0299 | 0.0046 | 0.0967 | 9 | 0.327 |
| NC | 0.0065 | 0.0012 | 0.0177 | 9 | 0.0235 | 0.0081 | 0.0328 | 12 | 0.320 |
| PA | 0.0085 | 0.0024 | 0.0502 | 9 | 0.0629 | 0.0090 | 0.1460 | 13 | 0.117 |
GTA—growing tumor area; NC—necrotic core; PA—peritumoral area. GTA/NC—the ratio of expression in growing tumor area to expression in necrotic core.
The expression of PUFA-processing desaturases did not differ between the sexes (Table 6). In men, FADS1 expression was elevated in the necrotic core (p = 0.155 for B2M) and reduced in the peritumoral area (p = 0.133), but not statistically significantly. In men, the ratio of FADS1 expression in the necrotic core to its expression in the peritumoral area (NC/PZ) was significantly 3 times higher than in women (p = 0.021, for B2M). There were no statistically significant sex-related differences in the expression of FADS2, in contrast to FADS3; in women in the peritumoral area, FADS3 expression was 4 times greater than in men (p = 0.021, for B2M). However, taking into account the second reference gene, GAPDH, these differences were statistically insignificant (50% higher expression, p = 0.526).
Table 6.
mRNA expression of FADS1, FADS2, and FADS3, by sex.
| Sex | Female | Male | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Studied Gene/Reference Gene/Location | Median | 1st Quartile | 3rd Quartile | N | Median | 1st Quartile | 3rd Quartile | N | p |
| FADS1/ B2M | |||||||||
| GTA | 0.0175 | 0.0051 | 0.0271 | 10 | 0.0065 | 0.0039 | 0.0143 | 9 | 0.369 |
| NC | 0.0128 | 0.0069 | 0.0175 | 9 | 0.0208 | 0.0054 | 0.0402 | 12 | 0.155 |
| PA | 0.0277 | 0.0155 | 0.0357 | 9 | 0.0088 | 0.0021 | 0.0306 | 13 | 0.133 |
| NC/PA | 0.4949 | 0.2562 | 0.6068 | 8 | 1.7582 | 0.4924 | 4.4001 | 11 | 0.021 * |
| FADS1/GAPDH | |||||||||
| GTA | 0.0121 | 0.0050 | 0.0204 | 10 | 0.0394 | 0.0112 | 0.0591 | 9 | 0.142 |
| NC | 0.0115 | 0.0035 | 0.0485 | 9 | 0.0183 | 0.0116 | 0.0548 | 12 | 0.477 |
| PA | 0.0340 | 0.0084 | 0.0497 | 9 | 0.0286 | 0.0050 | 0.0513 | 13 | 0.867 |
| FADS2/ B2M | |||||||||
| GTA | 0.0205 | 0.0086 | 0.1085 | 10 | 0.0153 | 0.0044 | 0.0392 | 9 | 0.414 |
| NC | 0.0179 | 0.0075 | 0.0468 | 9 | 0.0260 | 0.0024 | 0.1177 | 12 | 0.670 |
| PA | 0.0836 | 0.0130 | 0.2199 | 9 | 0.0448 | 0.0079 | 0.1201 | 13 | 0.193 |
| FADS2/GAPDH | |||||||||
| GTA | 0.0482 | 0.0116 | 0.0644 | 10 | 0.0485 | 0.0184 | 0.1061 | 9 | 0.744 |
| NC | 0.0303 | 0.0066 | 0.0493 | 9 | 0.0301 | 0.0112 | 0.1144 | 12 | 0.887 |
| PA | 0.0163 | 0.0096 | 0.2084 | 9 | 0.0867 | 0.0057 | 0.2571 | 13 | 0.973 |
| FADS3/ B2M | |||||||||
| GTA | 0.0086 | 0.0053 | 0.0178 | 10 | 0.0029 | 0.0019 | 0.0057 | 9 | 0.060 |
| NC | 0.0020 | 0.0013 | 0.0048 | 9 | 0.0073 | 0.0017 | 0.0242 | 12 | 0.201 |
| PA | 0.0112 | 0.0096 | 0.0255 | 9 | 0.0030 | 0.0010 | 0.0094 | 13 | 0.021 * |
| NC/PA | 0.2007 | 0.1072 | 0.2872 | 8 | 3.1901 | 0.5512 | 9.2635 | 11 | 0.013 * |
| FADS3/GAPDH | |||||||||
| GTA | 0.0100 | 0.0026 | 0.0280 | 10 | 0.0205 | 0.0032 | 0.0474 | 9 | 0.568 |
| NC | 0.0068 | 0.0008 | 0.0226 | 9 | 0.0111 | 0.0029 | 0.0257 | 12 | 0.356 |
| PA | 0.0103 | 0.0022 | 0.0358 | 9 | 0.0067 | 0.0032 | 0.0255 | 13 | 0.526 |
*—statistically significant difference between two groups according to the Mann–Whitney U test (p < 0.05). GTA—growing tumor area; NC—necrotic core; PA—peritumoral area; NC/PA—the ratio of expression in necrotic core to expression in peritumoral area.
3.4. The Immunolocalization of SCD and FADS2 Desaturases in Glioblastoma Multiforme Tumor
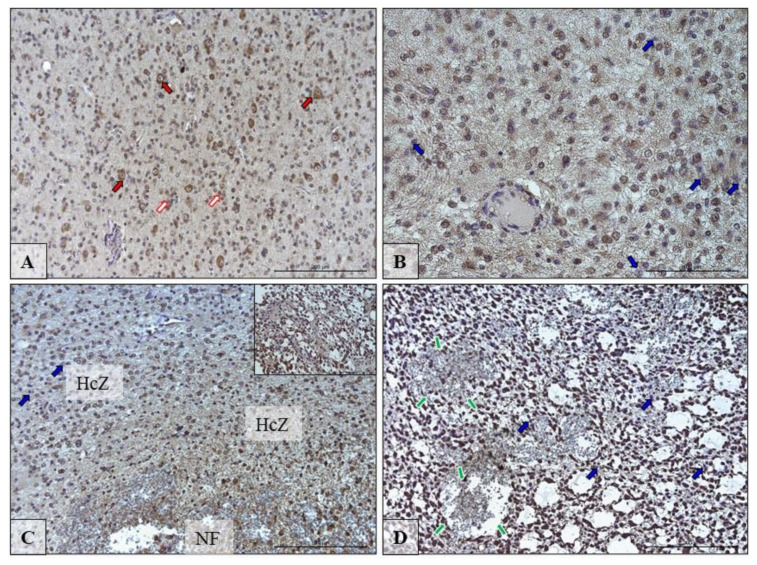
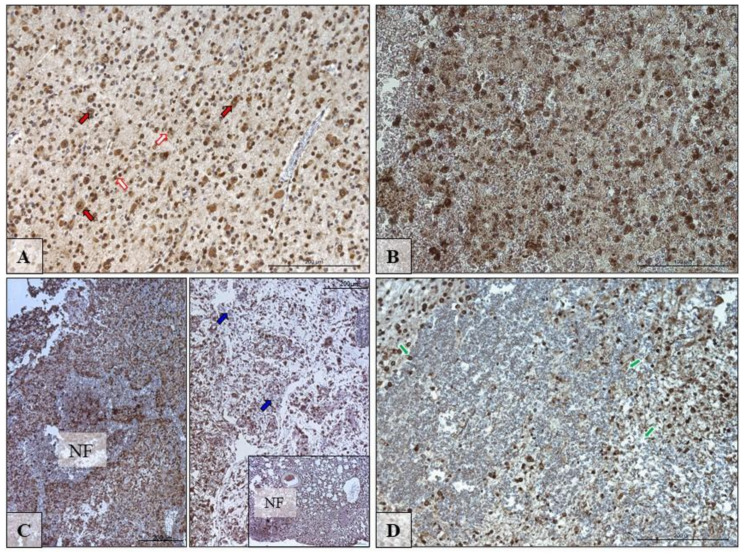
Figure 4 and Figure 5 show the immunolocalization of SCD and FADS2. The nerve cells and glia cells show the high expression of detected desaturases: SCD and FADS2 (Figure 4A and Figure 5A; red and white arrows).
Figure 4.
Representative microphotographs showing the protein expression of desaturase SCD in neurons and glia cells in the peritumoral area (A; red and white arrows) and in cells in the growing tumor area (B; blue arrows) necrotic core (C,D; blue arrows) of brain tumors from patients diagnosed with glioblastoma. Please note that on microphotography of GTA/NC, there are many pathologically changed regions of the brain tissue that indicate brain cell necrosis envelope by hypercellular zones (HcZ), with additional microvascular hyperplasia (green arrows). There are microphotographs from different objective (obj. magnification: A,C,D ×20, B ×40); scale bar are 200 µm or 100 µm.
Figure 5.
Representative microphotographs showing the protein expression of desaturase FADS2 in neurons and glia cells in the peritumoral area (A; red and white arrows) and in cells in the growing tumor area (B) necrotic foci (NF) (C,D; blue arrows) of brain tumors from patients diagnosed with glioblastoma. Please note that on microphotography of GTA/NC, there are many pathologically changed regions of the brain tissue that indicate brain cell necrosis envelope by hypercellular zones (HcZ), with additional microvascular hyperplasia (green arrows). There are microphotographs from different objective (obj. magnification: A, ×20, C ×20, B,D ×40); scale bars are 200 µm or 100 µm.
The pathological features observed in the GBM (glioblastoma) are necrotic foci (NF) surrounded by hypercellular zone (HcZ; pseudopalisades, Figure 4C and Figure 5C) and also was present specialized form of angiogenesis—microvascular hyperplasia (Figure 4D and Figure 5D; green arrows). In these pathological regions, not all glia cells expressed highly SDC and FADS2 (Figure 4B–D and Figure 5C; blue arrows).
3.5. The Effect on Necrotic Factors on the Expression of Desaturases in U-87 MG Cells
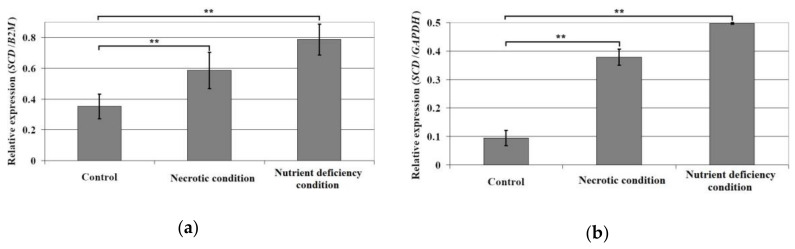
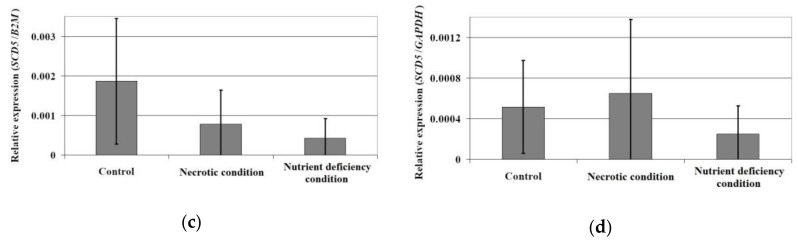
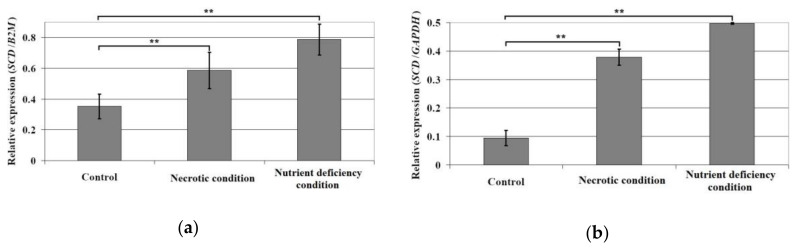
Nutritional deficiency increased the expression of SCD (Figure 6). The expression of this gene in U-87 MG cells was statistically significantly 5 times (p = 0.004) and 2 times (p = 0.004) higher than for GAPDH and B2M, respectively. Oxidative stress increased the expression of SCD by 50% (compared to B2M, p = 0.004) and 3 times (compared to GAPDH, p = 0.004). Neither of these factors resulted in a statistically significant change in the expression of SCD5 (p > 0.05).
Figure 6.
The effect of necrotic condition or nutritional deficiency on the mRNA expression of SCD (a,b) and SCD5 (c,d). Human brain (glioblastoma astrocytoma) U-87 MG cell line cultured in eagle’s minimum essential medium (EMEM) with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, 1% non essential amino acids, 100 U/mL penicillin, and 100 µg/mL streptomycin. U-87 MG cells were seeded in 6-well plates. After 72 h incubation, cells were washed three times with phosphate-buffered saline (PBS) solution and cultured for 24 h under three different conditions (control, nutrient deficiency, and necrotic). Control cells were suspended in full medium, starved cells were grown in medium with a low concentration of l-glutamine (0.2 mM) and without sodium pyruvate. For the induction of necrotic conditions, cells were incubated in medium supplemented with 200 µM H2O2. After 24 h of incubation, the cells were trypsinized and analyzed using qRT-PCR. **—statistically significant difference in the expression of a given desaturase between the tumor area according to the Wilcoxon signed-rank test (p < 0.01).
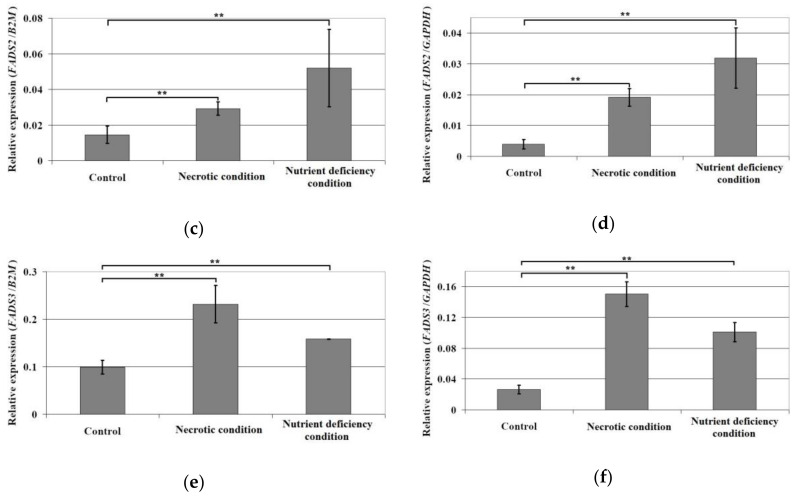
Both the nutritional deficiency and necrotic conditions increased the expression of PUFA desaturases in U-87 MG cells (Figure 7). Compared to GAPDH, nutritional deficiency increased the expression of FADS2 8 times (p = 0.004) and necrotic conditions 5 times (p = 0.004). Similar results were obtained in comparison to B2M (p = 0.004), albeit the increase in expression was two times smaller. Nutritional deficiency and necrotic conditions also increased the expression of FADS1 2 times compared to B2M (p = 0.004) and 5 times compared to GAPDH (p = 0.004). The results show that the studied conditions caused an increase in long-chain PUFA biosynthesis.
Figure 7.
Effect of the necrotic condition or nutritional deficiency on the mRNA expression of FADS1 (a,b), FADS2 (c,d), and FADS3 (e,f). Human brain (glioblastoma astrocytoma) U-87 MG cell line cultured in EMEM medium with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 1% non-essential amino acids, 100 U/mL penicillin, and 100 µg/mL streptomycin. U-87 MG cells were seeded in 6-well plates. After 72 h incubation, cells were washed three times with PBS solution. Next, cells were cultured for 24 h under three different conditions (control, nutrient deficiency, and necrotic). Control cells were suspended in full medium, while starved cells were grown in medium with low concentration of l-glutamine (0.2 mM) and without sodium pyruvate. For the induction of necrotic conditions, cells were incubated in medium supplemented with 200 µM H2O2. After 24 h of incubation, the cells were trypsinized and analyzed using qRT-PCR. **—statistically significant difference in the expression of a given desaturase between the tumor area according to the Wilcoxon signed-rank test (p < 0.01).
The studied conditions also increased the expression of FADS3. Compared to B2M, it increased twice (p = 0.004), while compared to GAPDH, it increased 5 times (necrotic conditions, p = 0.004) and 4 times (nutritional deficiency, p = 0.004).
4. Discussion
4.1. Analysis of SCD Expression in GBM Tumors
In our study of GBM tumors, the expression of the SCD gene—one that may play a significant role in the proliferation of rapidly dividing cells—was lower in the growing tumor area and necrotic core than in the peritumoral area. This stands in opposition to other reports. For example, the induction of proliferation through transformation using simian virus 40 caused the increased expression of this enzyme [15]. SCD expression is also increased in neoplasms and is associated with worse prognosis for patients [16,17,18,19,20,21,22,23,56].
Although our results regarding decreased SCD expression in GBM are not consistent with the results of most studies on other types of neoplasms, a reduction in SCD expression has also been observed in prostate cancer [57]. However, most importantly, our study was the first to examine SCD expression in GBM, and the discrepancies with the results of other studies may result from our comparing GBM tumors with brain tissue, where SCD expression is higher than in other organs [8]. It is noteworthy that lesions in the brain area with the subsequent loss of eloquent and silent neurological deficit is promoting neuroneogenesis based on stem cells as well as angio and glial cells’ growth hyperactivity [58]. The latter is to be taken under consideration analyzing the SCD unexpected level changes in peritumoral and tumor itself areas.
However, in GBM, the expression of SCD has a very significant clinical impact. Research on U87 MG cells showed that SCD expression was increased by the activation of epidermal growth factor receptor (EGFR) [59] and platelet-derived growth factor receptor (PDGFR) [60], which are receptors that are crucial for GBM development and are often activated in GBM tumors [22,61,62]. In another study, the increased expression of SCD in T98G and U87 MG glioma cells correlated with higher temozolomide (TMZ)-resistance, which contributed to the reduced survival of patients after TMZ-based chemotherapy [63]. Finally, in other cancers, SCD expression also positively correlates with the proliferation index, which is a factor associated with the lower survival of GBM patients [64]. There are no data that would link higher SCD expression to negative prognoses for glioma patients. Data published in the Human Protein Atlas (available from http://www.proteinatlas.org) show 3-year survival in 26% of patients with increased SCD expression in a tumor and only in 5% patients with lower SCD expression [65,66]; however, these data are statistically insignificant (p = 0.11).
In our study, the expression of SCD in the peritumoral area and in the growing tumor area did not differ between the sexes, although it was distinctly but statistically insignificantly higher in the necrotic core in men. This lack of statistically significant differences confirms the observations made by other authors researching the expression of SCD in the brain [67,68]. However, comparisons of SCD expression in the necrotic core between sexes should be performed on larger groups of patients than in our study. Further research is also required to investigate the potential influence of sex hormones on SCD expression and GBM cell proliferation. As androgen receptor activation enhances GBM cell proliferation [69,70]—and cell proliferation is closely related to SCD expression [15]—the activation of androgen receptors in men could increase SCD expression in GBM. In women, the activation of estrogen receptor β significantly improves patient prognosis [71,72,73]. The protein expression of SCD in neurons and glia cells of tumors showed pathological changes in the brain tissue, which indicated brain tissue necrosis.
4.2. Analysis of SCD5 Expression in GBM Tumors
In our study, the expression of SCD5 did not differ between the examined areas. However, the expression of the SCD5 gene showed a statistically insignificant decrease in the necrotic core in relation to the peritumoral area. However, due to the limited number of patients, the observed dependence requires confirmation with larger groups of patients.
SCD5 is an enzyme that is specific to the brain [8], with a well-known anti-cancer effect [26,28]. Although it does not influence proliferation intensity in melanoma, it does reduce melanoma dissemination and lung metastases by reducing intracellular acidification [26]. Experiments on Neuro2a cells showed that the increased expression of SCD5 may disturb signaling pathways relevant to GBM development, in particular by disrupting signal transmission from EGFR [25]. However, this effect of SCD5 has not been studied on any GBM model. If it turns out that SCD5 also disrupts signal transmission from EGFR in GBM, this would indicate its anti-cancer properties, since this receptor is essential for the induction of GBM cell proliferation [61,74].
In the aforementioned experiment on Neuro2a cells, SCD5 also disturbed the activation of the canonical Wnt pathway [25] and the increased expression of SCD5 caused activation of the non-canonical Wnt pathway, in which GBM may intensify the dissemination of cancer cells [25]. Activation of the canonical and non-canonical Wnt pathways is important for the functioning of cancer stem cells in GBM [75]. SCD5 may also have an oncogenic effect in other cancers, as shown on a breast cancer model, where it increased the viability of cancer cells [26,27]. SCD5 expression is also increased in anaplastic thyroid carcinoma, which may indicate that SCD5 has important functions in that cancer [21]. SCD5 is also likely to play an important role in neoplastic processes in GBM. Nevertheless, in glioma, SCD5 tumors may not be significant in tumor development. By The Human Protein Atlas (available from http://www.proteinatlas.org), 3-year survival of patients with high and low SCD5 gene expression is 11% and 4%, respectively, but the differences in these results are statistically insignificant (p = 0.18) [65,76].
In our study, the expression of SCD5 did not differ between male and female patients; however, the ratio of expression in the growing tumor area versus necrotic core was statistically insignificantly higher (p = 0.063) in women. As no studies have been conducted on sex-related differences in the expression of this enzyme in the brain or in cancers, it is very difficult to discuss these results. Due to the high expression of SCD5 in the brain, further studies are required to examine the regulation of its expression in healthy tissues and its significance for neoplastic cells, especially in GBM.
4.3. Expression of FADS1 and FADS2 in GBM Tumors
In our study, the expression of FADS2 in the necrotic core was significantly lower than in the peritumoral area. In addition, the expression of FADS1 in the growing tumor area was distinctly, although not statistically significantly, lower than in the peritumoral area. This stands in opposition to other reports. In other types of tumors, FADS1 and FADS2 exhibit oncogenic properties, for example in non-small-cell lung cancer, where the lower expression of FADS1 is associated with worse prognosis [77]. The oncogenic properties of FADS1 have also been shown in HCA-7 (colon cancer) [78] and BxPC-3 (pancreatic cancer cells) [78]. In turn, FADS2 displayed oncogenic properties in B16 melanoma [35], LLC Lewis lung carcinoma [35], MCF7 cell line (breast cancer) [34], and HT-29 human colon cancer cells [36]. In patients with esophageal squamous cell carcinoma, a lower expression of FADS1 was associated with improved prognosis [79]. Nevertheless, it seems that FADS1 and FADS2 in glioma tumors have anti-neoplastic properties. Based on The Human Protein Atlas (available from http://www.proteinatlas.org), the greater expression of FADS1 and FADS2 genes in a glioma increases the 3-year survival of patients (p = 0.0045 for FADS1, p = 0.030 for FADS2) [65,80,81].
In our study, the expression of FADS1 and FADS2 did not differ between sexes, which is consistent with literature on their expression in the brain [67,68,82]. This stands in opposition to other organs such as the liver [82,83], where differences were related to the effects of progesterone [84]. In our study, with B2M as a reference, we observed only a negligible elevation in FADS1 expression in the necrotic core in men and a statistically insignificantly elevation of expression of FADS1 in the peritumoral area in women. This resulted in a significantly higher ratio of FADS1 expression in the necrotic core to peritumoral area in men. Nevertheless, there are no detailed literature data concerning the influence of sex hormones on the aforementioned sex-related differences. The increased expression of FADS1 in the peritumoral area in women could be attributed to the influence of estradiol, which increased the expression of FADS1 in human neuroblastoma SH-SY5Y cells [85]. In the same study, the expression of FADS1 was reduced by testosterone. However, there is no data in the literature that could help explain the mechanisms underlying the influence of sex hormones on FADS1 expression in the necrotic core.
4.4. FADS3 Expression in GBM Tumors
In our study, the expression of FADS3 did not differ between the tumor and peritumoral area. FADS3 shows Δ13-desaturase activity for trans-vaccenic acid [42], meaning it forms conjugated linoleic acids, which act as anticancer agents by activating proliferator-activated receptor γ (PPARγ) [86,87]. Another function of FADS3 is to regulate the synthesis of arachidonic acid and DHA through changes in the expression of FADS1 and FADS2 [43]; however, a study on FADS3 knockout mice showed a significant age-dependent variability of n-6 and n-3 PUFA levels in the liver and brain. Nevertheless, the function of FADS3 in GBM is not clear and requires further research. However, data from The Human Protein Atlas (available from http://www.proteinatlas.org) show that FADS3 can enhance neoplastic processes in glioma. Patients who had a higher expression of the FADS3 gene in the tumor showed a lower 3-year survival (6%) than patients with a low expression of this gene (13%, p = 0.039) [65,88].
In our study, the expression of FADS3 in the peritumoral area was significantly higher in women. However, we cannot relate this observation to any literature data, as this paper is the first to study the difference in the expression of FADS3 between sexes. Although FADS3 is a gene located close to the relatively well-researched FADS1 and FADS2 [89], and at the transcription level it can possibly can be subject to regulation together with these genes, little is known about the regulation of FADS3 expression. In the cerebral cortex, the expression of FADS1 and FADS2 does not differ between sexes [67,68,82,83], while in the liver, the expression of FADS1 and FADS2 was higher in female rats [82,83] and human women [84], which is attributed to the action of progesterone. Nevertheless, it is not possible to determine whether the sex-related difference in FADS3 in our study was due to a difference existing in healthy brains or the simultaneous effect of sex hormones and signal molecules secreted by the GBM tumor.
4.5. The effect of Necrotic Factors on the Expression of Desaturases in U-87 MG Cells
The growth of GBM tissue is not accompanied by the development of blood vessels that supply the cancer cells with nutrients. This results in the formation of nutrient-deficient areas, which causes numerous small necrotic niches [45,46,47,48]. Around these areas, the intensive proliferation of neoplastic cells occurs.
According to our analysis, necrotic factors such as nutritional deficiency increased SCD expression but not SCD5. These results may explain the occurrence of intense GBM cell proliferation around the necrotic niche [45,46,47,48]. SCD expression is associated with the proliferation index [64]. Thus, necrotic factors increase GBM cell proliferation and thus SCD expression. In turn, SCD5 expression is not associated with cell proliferation [26]. Therefore, the expression of this gene should not be higher under nutritional deficiency, as confirmed by our results.
The examined factors (nutrient deficiency) also increased the expression of PUFA desaturation: FADS1, FADS2, and FADS3. Currently, there is no literature data showing the influence of oxidative stress, starvation, or nutritional deficiency on the expression of the discussed desaturases. It is only known that high-carbohydrate diets increase the protein expression of FADS2 (Δ6 desaturase) in the rat liver but do not affect the protein expression of FADS1 (Δ5 desaturase) [90]. Due to the lack of literature data, it is difficult to refer to the results of other studies.
4.6. Effect of Mutation in IDH1/2 Genes on the Expression of Desaturases
In glioma tumor cells, point mutations often occur in one of the alleles of the isocitrate dehydrogenase (IDH)1 gene in codon 132 or IDH2 gene in codon 172 [91]. The mutated IDH catalyzes the production of an oncometabolite, 2-hydroxyglutarate, from α-ketoglutarate. An inhibitor of α-ketoglutarate-dependent dioxygenases [92,93], this compound increases the methylation of histones and DNA. In addition, 2-hydroxyglutarate inhibits the activity of prolyl hydroxylases, which are enzymes that are involved in hypoxia inducible factor α subunits (HIF-α) degradation [94]. The frequency of IDH mutations depends on the type of GBM. Mutation in IDH2 occurs in only 4% of GBM cases and in II–III grade gliomas [95,96]. IDH1 mutation occurs in primary GBM in only 2% to 16% of cases, depending on the study cited [95,96,97]. In secondary GBM, the frequency of IDH1 mutation ranges from 50% to 85% depending on the study [96,97].
Mutation in IDH genes causes changes in the synthesis of fatty acids. Experiments on acute myeloid leukemia HL60 line have demonstrated that IDH1 mutation causes an increase in lipid anabolic fluxes [98] and significantly more MUFA and more SFA, albeit not statistically significantly (p = 0.07) [98]. In the same experiment, the level of PUFA in the studied cells did not differ between those with or without the IDH mutation (p = 0.6) [98]. The observed changes were confirmed on the glioma tumor with and without the IDH1 mutation. The IDH1 mutation causes an increase in SCD activity in the tumor [99]. The mutation increases EPA and docosapentaenoic acid (DPA), but it does not affect DHA nor linoleic acid, linolenic acid, and arachidonic acid [99]. This indicates an increase in FADS1 activity, although with no effect on arachidonic acid.
Biosynthesis 2-hydroxyglutarate may cause endoplasmic reticulum autophagy, which results in the degradation of enzymes involved in lipid synthesis [100]. Thus, it may reduce the protein expression of genes tested in this study.
The study cited above shows that the mutation of IDH1 in GBM results in an increase in SCD expression (increase in the amount of MUFA) and probably an increase in FADS1 expression, but it does not affect the expression of FADS2 (no effect on the overall PUFA level).
4.7. Conclusions
The obtained results suggest that (i) biosynthesis of MUFA and PUFA in a GBM tumor is less intense than in the peritumoral area; (ii) expressions of SCD, SCD5, FADS1, and FADS2 correlate with each other in the necrotic core, growing tumor area, and peritumoral area; (iii) expressions of desaturases in a GBM tumor do not differ between the sexes; and (iv) nutritional deficiency increases the biosynthesis of MUFA and PUFA in GBM cells.
Author Contributions
J.K.: literature search and review, manuscript writing; K.K.: manuscript writing; D.J.: data collection; D.S.: literature search and review; M.T.: data collection; P.K.: data collection; K.S.: data collection; I.G.: literature search and review; M.G.: literature search and review; A.K.-W.: data collection; B.W.: data collection; D.C.: founds collection; I.B.-B.: literature search and review, writing the manuscript, final acceptance of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the statutory budget of the Department of Biochemistry and Medical Chemistry Pomeranian Medical University in Szczecin, Poland.
Conflicts of Interest
The authors declare no conflict of interest
References
- 1.Stupp R., Hegi M.E., Mason W.P., Bent M.J.B.V.D., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Bélanger K., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Aulakh S., DeDeo M.R., Free J., Rosenfeld S.S., Quinones-Hinojosa A., Paulus A., Manna A., Manochakian R., Chanan-Khan A.A., Ailawadhi S. Survival trends in glioblastoma and association with treating facility volume. J. Clin. Neurosci. 2019;68:271–274. doi: 10.1016/j.jocn.2019.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uto Y. Recent progress in the discovery and development of stearoyl CoA desaturase inhibitors. Chem. Phys. Lipids. 2016;197:3–12. doi: 10.1016/j.chemphyslip.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Imamura K., Tomita N., Kawakita Y., Ito Y., Ono K., Nii N., Miyazaki T., Yonemori K., Tawada M., Sumi H., et al. Discovery of Novel and Potent Stearoyl Coenzyme A Desaturase 1 (SCD1) Inhibitors as Anticancer Agents. Bioorganic Med. Chem. 2017;25:3768–3779. doi: 10.1016/j.bmc.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Hodson L., Fielding B.A. Stearoyl-CoA desaturase: Rogue or innocent bystander? Prog. Lipid Res. 2013;52:15–42. doi: 10.1016/j.plipres.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Enoch H.G., Catalá A., Strittmatter P. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J. Biol. Chem. 1976;251:5095–5103. [PubMed] [Google Scholar]
- 7.Wang J., Yu L., Schmidt R.E., Su C., Huang X., Gould K., Cao G. Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem. Biophys. Res. Commun. 2005;332:735–742. doi: 10.1016/j.bbrc.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2013;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paton C.M., Ntambi J.M. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Metab. 2008;297:E28–E37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauvoisin D., Mounier C. Hormonal and nutritional regulation of SCD1 gene expression. Biochimie. 2011;93:78–86. doi: 10.1016/j.biochi.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Jiang G., Li Z., Liu F., Ellsworth K., Dallas-Yang Q., Wu M., Ronan J., Esau C., Murphy C., Szalkowski D., et al. Prevention of obesity in mice by antisense oligonucleotide inhibitors of stearoyl-CoA desaturase-1. J. Clin. Investig. 2005;115:1030–1038. doi: 10.1172/JCI200523962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyazaki M., Flowers M.T., Sampath H., Chu K., Otzelberger C., Liu X., Ntambi J.M. Hepatic Stearoyl-CoA Desaturase-1 Deficiency Protects Mice from Carbohydrate-Induced Adiposity and Hepatic Steatosis. Cell Metab. 2007;6:484–496. doi: 10.1016/j.cmet.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Yee J.K., Wahjudi P.N., Vega J., Lim S., Martin A., Patterson M.E., Cohen J.N., Mao C.S., Lee W.-N.P. Stearoyl-CoA desaturase enzyme 1 inhibition reduces glucose utilization for de novo fatty acid synthesis and cell proliferation in 3T3-L1 adipocytes. Metabolomics. 2013;9:809–816. doi: 10.1007/s11306-013-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobrzyn P., Jazurek M., Dobrzyn A. Stearoyl-CoA desaturase and insulin signaling—What is the molecular switch? Biochim. Biophys. Acta. 2010;1797:1189–1194. doi: 10.1016/j.bbabio.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Scaglia N., Igal R.A. Stearoyl-CoA Desaturase Is Involved in the Control of Proliferation, Anchorage-independent Growth, and Survival in Human Transformed Cells. J. Biol. Chem. 2005;280:25339–25349. doi: 10.1074/jbc.M501159200. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Ding S.-F., Habib N.A., Fermor B.F., Wood C.B., Gilmour R.S. Partial characterization of a cDNA for human stearoyl-CoA desaturase and changes in its mRNA expression in some normal and malignant tissues. Int. J. Cancer. 1994;57:348–352. doi: 10.1002/ijc.2910570310. [DOI] [PubMed] [Google Scholar]
- 17.Guo S., Wang Y., Zhou D., Li Z. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci. Rep. 2014;4:5959. doi: 10.1038/srep05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal S., Berk M., Alkhouri N., Partrick D.A., Fung J.J., Feldstein A. Stearoyl-CoA desaturase plays an important role in proliferation and chemoresistance in human hepatocellular carcinoma. J. Surg. Res. 2013;186:29–38. doi: 10.1016/j.jss.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roongta U.V., Pabalan J.G., Wang X., Fargnoli J., Henley B.J., Zhu J., Madireddi M.T., Lawrence R.M., Wong T.W., Rupnow B.A., et al. Cancer Cell Dependence on Unsaturated Fatty Acids Implicates Stearoyl-CoA Desaturase as a Target for Cancer Therapy. Mol. Cancer Res. 2011;9:1551–1561. doi: 10.1158/1541-7786.MCR-11-0126. [DOI] [PubMed] [Google Scholar]
- 20.Peck B., Schug Z.T., Zhang Q., Dankworth B., Jones D.T., Smethurst E., Patel R., Mason S., Jiang M., Saunders R., et al. Inhibition of fatty acid desaturation is detrimental to cancer cell survival in metabolically compromised environments. Cancer Metab. 2016;4:6. doi: 10.1186/s40170-016-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Von Roemeling C., Marlow L., Pinkerton A.B., Crist A., Miller J., Tun H.W., Smallridge R.C., Copland J.A. Aberrant lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl CoA desaturase 1 as a novel therapeutic target. J. Clin. Endocrinol. Metab. 2015;100:E697–E709. doi: 10.1210/jc.2014-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J., Fan X.-X., He J., Pan H., Li R.-Z., Huang L., Jiang Z., Yao X.-J., Liu L., Leung E.L.-H., et al. SCD1 is associated with tumor promotion, late stage and poor survival in lung adenocarcinoma. Oncotarget. 2016;7:39970–39979. doi: 10.18632/oncotarget.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Xu Y., Zhu L., Zou Y., Kong W., Dong B., Huang J., Chen Y., Xue W., Huang Y., et al. High Expression of Stearoyl-CoA Desaturase 1 Predicts Poor Prognosis in Patients with Clear-Cell Renal Cell Carcinoma. PLoS ONE. 2016;11:e0166231. doi: 10.1371/journal.pone.0166231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro L.F.C., Wilson J.M., Gonçalves O., Galante-Oliveira S., Rocha E., Cunha I. The evolutionary history of the stearoyl-CoA desaturase gene family in vertebrates. BMC Evol. Biol. 2011;11:132. doi: 10.1186/1471-2148-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinner D.I., Kim G.J., Henderson G.C., Igal R.A. StearoylCoA Desaturase-5: A Novel Regulator of Neuronal Cell Proliferation and Differentiation. PLoS ONE. 2012;7:e39787. doi: 10.1371/journal.pone.0039787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellenghi M., Puglisi R., Pedini F., De Feo A., Felicetti F., Bottero L., Sangaletti S., Errico M.C., Petrini M., Gesumundo C., et al. SCD5-induced oleic acid production reduces melanoma malignancy by intracellular retention of SPARC and cathepsin B. J. Pathol. 2015;236:315–325. doi: 10.1002/path.4535. [DOI] [PubMed] [Google Scholar]
- 27.Angelucci C., D’Alessio A., Iacopino F., Proietti G., Di Leone A., Masetti R., Sica G. Pivotal role of human stearoyl-CoA desaturases (SCD1 and 5) in breast cancer progression: Oleic acid-based effect of SCD1 on cell migration and a novel pro-cell survival role for SCD5. Oncotarget. 2018;9:24364–24380. doi: 10.18632/oncotarget.25273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puglisi R., Bellenghi M., Pontecorvi G., Gulino A., Petrini M., Felicetti F., Bottero L., Mattia G., Carè A. SCD5 restored expression favors differentiation and epithelial-mesenchymal reversion in advanced melanoma. Oncotarget. 2018;9:7567–7581. doi: 10.18632/oncotarget.24085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho H.P., Nakamura M., Clarke S.D. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 30.Cho H.P., Nakamura M.T., Clarke S.D. Cloning, expression, and nutritional regulation of the mammalian Delta-6 desaturase. J. Biol. Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 31.De Antueno R., Knickle L.C., Smith H., Elliot M.L., Allen S.J., Nwaka S., Winther M.D. Activity of human Delta5 and Delta6 desaturases on multiple n-3 and n-6 polyunsaturated fatty acids. FEBS Lett. 2001;509:77–80. doi: 10.1016/S0014-5793(01)03135-0. [DOI] [PubMed] [Google Scholar]
- 32.D’Andrea S., Guillou H., Jan S., Catheline D., Thibault J.-N., Bouriel M., Rioux V., Legrand P. The same rat Delta6-desaturase not only acts on 18- but also on 24-carbon fatty acids in very-long-chain polyunsaturated fatty acid biosynthesis. Biochem. J. 2002;364:49–55. doi: 10.1042/bj3640049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J.Y., Kothapalli K.S., Brenna J.T. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr. Opin. Clin. Nutr. Metab. Care. 2016;19:103–110. doi: 10.1097/MCO.0000000000000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park W.J., Kothapalli K.S.D., Lawrence P., Brenna J.T. FADS2 Function Loss at the Cancer Hotspot 11q13 Locus Diverts Lipid Signaling Precursor Synthesis to Unusual Eicosanoid Fatty Acids. PLoS ONE. 2011;6:e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He C., Qu X., Wan J.-B., Rong R., Huang L., Cai C., Zhou K., Gu Y., Qian S.Y., Kang J.X. Inhibiting Delta-6 Desaturase Activity Suppresses Tumor Growth in Mice. PLoS ONE. 2012;7:e47567. doi: 10.1371/journal.pone.0047567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen-Petrik M.B., McEntee M., Johnson B.T., Obukowicz M.G., Masferrer J., Zweifel B., Chiu C.H., Whelan J. Selective inhibition of Delta-6 desaturase impedes intestinal tumorigenesis. Cancer Lett. 2002;175:157–163. doi: 10.1016/S0304-3835(01)00715-7. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y., Zhu X., Zhu J., Liao S., Tang Q., Liu K., Guan X., Zhang J., Feng Z. Identification of differential expression of genes in hepatocellular carcinoma by suppression subtractive hybridization combined cDNA microarray. Oncol. Rep. 2007;18:943–951. doi: 10.3892/or.18.4.943. [DOI] [PubMed] [Google Scholar]
- 38.Allam-Ndoul B., Guénard F., Barbier O., Vohl M.-C. A Study of the Differential Effects of Eicosapentaenoic Acid (EPA) and Docosahexaenoic Acid (DHA) on Gene Expression Profiles of Stimulated Thp-1 Macrophages. Nutrients. 2017;9:424. doi: 10.3390/nu9050424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsson S.C., Kumlin M., Ingelman-Sundberg M., Wolk A. Dietary long-chain n−3 fatty acids for the prevention of cancer: A review of potential mechanisms. Am. J. Clin. Nutr. 2004;79:935–945. doi: 10.1093/ajcn/79.6.935. [DOI] [PubMed] [Google Scholar]
- 40.Das M., Das S. Identification of cytotoxic mediators and their putative role in the signaling pathways during docosahexaenoic acid (DHA)-induced apoptosis of cancer cells. Apoptosis. 2016;21:1408–1421. doi: 10.1007/s10495-016-1298-2. [DOI] [PubMed] [Google Scholar]
- 41.Kim S., Jing K., Shin S., Jeong S., Han S.H., Oh H., Yoo Y.S., Han J., Jeon Y.J., Heo J.Y., et al. ω3-polyunsaturated fatty acids induce cell death through apoptosis and autophagy in glioblastoma cells: In vitro and in vivo. Oncol. Rep. 2018;39:239–246. doi: 10.3892/or.2017.6101. [DOI] [PubMed] [Google Scholar]
- 42.Rioux V., Pédrono F., Blanchard H., Duby C., Boulier-Monthéan N., Bernard L., Beauchamp E., Catheline D., Legrand P. Trans-vaccenate is Δ13-desaturated by FADS3 in rodents. J. Lipid Res. 2013;54:3438–3452. doi: 10.1194/jlr.M042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J.Y., Qin X., Liang A., Kim E., Lawrence P., Park W.J., Kothapalli K.S.D., Brenna J.T. Fads3 modulates docosahexaenoic acid in liver and brain. Prostaglandins Leukot. Essent. Fat. Acids. 2017;123:25–32. doi: 10.1016/j.plefa.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemée J.-M., Com E., Clavreul A., Avril T., Quillien V., De Tayrac M., Pineau C., Menei P. Proteomic analysis of glioblastomas: What is the best brain control sample? J. Proteom. 2013;85:165–173. doi: 10.1016/j.jprot.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 45.Gorin F.A., Harley W., Schnier J., Lyeth B., Jue T. Perinecrotic glioma proliferation and metabolic profile within an intracerebral tumor xenograft. Acta Neuropathol. 2004;107:235–244. doi: 10.1007/s00401-003-0803-1. [DOI] [PubMed] [Google Scholar]
- 46.Schiffer D., Mellai M., Annovazzi L., Caldera V., Piazzi A., Denysenko T., Melcarne A. Stem Cell Niches in Glioblastoma: A Neuropathological View. BioMed Res. Int. 2014;2014:725921. doi: 10.1155/2014/725921. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Nigim F., Cavanaugh J., Patel A.P., Curry W.T., Esaki S.-I., Kasper E.M., Chi A.S., Louis D.N., Martuza R.L., Rabkin S.D., et al. Targeting Hypoxia-Inducible Factor 1α in a New Orthotopic Model of Glioblastoma Recapitulating the Hypoxic Tumor Microenvironment. J. Neuropathol. Exp. Neurol. 2015;74:710–722. doi: 10.1097/NEN.0000000000000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishii A., Kimura T., Sadahiro H., Kawano H., Takubo K., Suzuki M., Ikeda E. Histological Characterization of the Tumorigenic “Peri-Necrotic Niche” Harboring Quiescent Stem-Like Tumor Cells in Glioblastoma. PLoS ONE. 2016;11:e0147366. doi: 10.1371/journal.pone.0147366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ali M.A., Kandasamy A.D., Fan X., Schulz R. Hydrogen peroxide-induced necrotic cell death in cardiomyocytes is independent of matrix metalloproteinase-2. Toxicol. In Vitro. 2013;27:1686–1692. doi: 10.1016/j.tiv.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 50.Zhao L., Lin H., Chen S., Chen S., Cui M., Shi D., Wang B., Ma K., Shao Z. Hydrogen peroxide induces programmed necrosis in rat nucleus pulposus cells through the RIP1/RIP3-PARP-AIF pathway. J. Orthop. Res. 2018;36:1269–1282. doi: 10.1002/jor.23751. [DOI] [PubMed] [Google Scholar]
- 51.Maher E.A., Marin-Valencia I., Bachoo R.M., Mashimo T., Raisanen J., Hatanpaa K.J., Jindal A., Jeffrey F.M., Choi C., Madden C., et al. Metabolism of [U-13C]glucose in human brain tumorsin vivo. NMR Biomed. 2012;25:1234–1244. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ta N.L., Seyfried T.N. Influence of Serum and Hypoxia on Incorporation of [14C]-d-Glucose or [14C]-l-Glutamine into Lipids and Lactate in Murine Glioblastoma Cells. Lipids. 2015;50:1167–1184. doi: 10.1007/s11745-015-4075-z. [DOI] [PubMed] [Google Scholar]
- 53.Said H.M., Hagemann C., Anacker J., Schömig B., Vince G., Flentje M., Roosen K., Vordermark D. GAPDH is not regulated in human glioblastoma under hypoxic conditions. BMC Mol. Biol. 2007;8:55. doi: 10.1186/1471-2199-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kreth S., Heyn J., Grau S., Kretzschmar H.A., Egensperger R., Kreth F.W. Identification of valid endogenous control genes for determining gene expression in human glioma. Neuro-Oncology. 2010;12:570–579. doi: 10.1093/neuonc/nop072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grube S., Göttig T., Freitag D., Ewald C., Kalff R., Walter J. Selection of suitable reference genes for expression analysis in human glioma using RT-qPCR. J. Neuro-Oncology. 2015;123:35–42. doi: 10.1007/s11060-015-1772-7. [DOI] [PubMed] [Google Scholar]
- 56.Holder A., Gonzalez-Angulo A.M., Chen H., Akcakanat A., Anh K.-, Symmans W.F., Pusztai L., Hortobagyi G.N., Mills G.B., Meric-Bernstam F. High stearoyl-CoA desaturase 1 expression is associated with shorter survival in breast cancer patients. Breast Cancer Res. Treat. 2012;137:319–327. doi: 10.1007/s10549-012-2354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore S., Knudsen B., True L.D., Hawley S., Etzioni R., Wade C., Gifford D., Coleman I.M., Nelson P.S. Loss of stearoyl-CoA desaturase expression is a frequent event in prostate carcinoma. Int. J. Cancer. 2005;114:563–571. doi: 10.1002/ijc.20773. [DOI] [PubMed] [Google Scholar]
- 58.Zhang R.L., Chopp M., Roberts C., Liu X., Wei M., Nejad-Davarani S., Wang X., Zhang Z.G. Stroke Increases Neural Stem Cells and Angiogenesis in the Neurogenic Niche of the Adult Mouse. PLoS ONE. 2014;9:e113972. doi: 10.1371/journal.pone.0113972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo D., Prins R.M., Dang J., Kuga D., Iwanami A., Soto H., Lin K.Y., Huang T.T., Akhavan D., Hock M.B., et al. EGFR Signaling Through an Akt-SREBP-1-Dependent, Rapamycin-Resistant Pathway Sensitizes Glioblastomas to Antilipogenic Therapy. Sci. Signal. 2009;2:ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma D., Nutt C.L., Shanehsaz P., Peng X., Louis D.N., Kaetzel D.M. Autocrine Platelet-Derived Growth Factor–Dependent Gene Expression in Glioblastoma Cells Is Mediated Largely by Activation of the Transcription Factor Sterol Regulatory Element Binding Protein and Is Associated with Altered Genotype and Patient Survival in Human Brain Tumors. Cancer Res. 2005;65:5523–5534. doi: 10.1158/0008-5472.can-04-2582. [DOI] [PubMed] [Google Scholar]
- 61.Andersson U., Guo D., Malmer B., Bergenheim A., Henriksson R. Epidermal growth factor receptor family (EGFR, ErbB2?4) in gliomas and meningiomas. Acta Neuropathol. 2004;108:135–142. doi: 10.1007/s00401-004-0875-6. [DOI] [PubMed] [Google Scholar]
- 62.Phillips J.J., Aranda D., Ellison D.W., Judkins A.R., Croul S.E., Brat D.J., Ligon K.L., Horbinski C., Venneti S., Zadeh G., et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol. 2013;23:565–573. doi: 10.1111/bpa.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai S., Yan Y., Xu Z., Zeng S., Qian L., Huo L., Li X., Sun L., Gong Z. SCD1 Confers Temozolomide Resistance to Human Glioma Cells via the Akt/GSK3β/β-Catenin Signaling Axis. Front. Pharmacol. 2018;8:960. doi: 10.3389/fphar.2017.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gzell C., Wheeler H., Huang D., Gaur P., Chen J., Kastelan M., Back M., Information P.E.K.F.C. Proliferation Index Predicts Survival after Second Craniotomy within 6 Months of Adjuvant Radiotherapy for High-grade Glioma. Clin. Oncol. 2016;28:215–222. doi: 10.1016/j.clon.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 65.Uhlén M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 66.Expression of SCD in Glioma—The Human Protein Atlas. [(accessed on 14 January 2020)]; Available online: https://www.proteinatlas.org/ENSG00000099194-SCD/pathology/glioma.
- 67.Liu Y., Jandacek R., Rider T., Tso P., McNamara R.K. Elevated delta-6 desaturase (FADS2) expression in the postmortem prefrontal cortex of schizophrenic patients: Relationship with fatty acid composition. Schizophr. Res. 2009;109:113–120. doi: 10.1016/j.schres.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McNamara R.K., Liu Y. Reduced expression of fatty acid biosynthesis genes in the prefrontal cortex of patients with major depressive disorder. J. Affect. Disord. 2010;129:359–363. doi: 10.1016/j.jad.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu X., Jiang Y., Wei W., Cong P., Ding Y., Xiang L., Wu K. Androgen receptor signaling regulates growth of glioblastoma multiforme in men. Tumor Biol. 2014;36:967–972. doi: 10.1007/s13277-014-2709-z. [DOI] [PubMed] [Google Scholar]
- 70.Bao D., Cheng C., Lan X., Xing R., Chen Z., Zhao H., Sun J., Wang Y., Niu C., Zhang B., et al. Regulation of p53wt glioma cell proliferation by androgen receptor-mediated inhibition of small VCP/p97-interacting protein expression. Oncotarget. 2017;8:23142–23154. doi: 10.18632/oncotarget.15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X., Wang L., Chen J., Ling Q., Wang H., Li S., Li L., Yang S., Xia M., Jing L. Estrogen receptor β agonist enhances temozolomide sensitivity of glioma cells by inhibiting PI3K/AKT/mTOR pathway. Mol. Med. Rep. 2014;11:1516–1522. doi: 10.3892/mmr.2014.2811. [DOI] [PubMed] [Google Scholar]
- 72.Cao L., Qu D., Wang H., Zhang S., Jia C., Shi Z., Wang Z., Zhang J., Ma J. Toosendanin Exerts an Anti-Cancer Effect in Glioblastoma by Inducing Estrogen Receptor β- and p53-Mediated Apoptosis. Int. J. Mol. Sci. 2016;17:1928. doi: 10.3390/ijms17111928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sareddy G.R., Li X., Liu J., Viswanadhapalli S., Garcia L., Gruslova A., Cavazos D., Garcia M., Strom A.M., Gustafsson J.-A., et al. Selective Estrogen Receptor β Agonist LY500307 as a Novel Therapeutic Agent for Glioblastoma. Sci. Rep. 2016;6:24185. doi: 10.1038/srep24185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang P.H., Xu A., White F.M. Oncogenic EGFR Signaling Networks in Glioma. Sci. Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 75.Zuccarini M., Giuliani P., Ziberi S., Carluccio M., Di Iorio P., Caciagli F., Ciccarelli R. The Role of Wnt Signal in Glioblastoma Development and Progression: A Possible New Pharmacological Target for the Therapy of This Tumor. Genes. 2018;9:105. doi: 10.3390/genes9020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Expression of SCD5 in Glioma—The Human Protein Atlas. [(accessed on 14 January 2020)]; Available online: https://www.proteinatlas.org/ENSG00000145284-SCD5/pathology/glioma.
- 77.Wang N., Lin Y., Gao B., Yan S., Wu H., Li Y., Wu Q., Wei Y.-C. Reduced Expression of FADS1 Predicts Worse Prognosis in Non-Small-Cell Lung Cancer. J. Cancer. 2016;7:1226–1232. doi: 10.7150/jca.15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X., Xu Y., Wang T., Shu D., Guo P., Miskimins K., Qian S.Y. Inhibition of cancer migration and invasion by knocking down delta-5-desaturase in COX-2 overexpressed cancer cells. Redox Biol. 2017;11:653–662. doi: 10.1016/j.redox.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du Y., Yan S.-M., Gu W.-Y., He F., Huang L.-Y., Li M., Yuan Y., Chen R.-H., Zhong Q., Li M.-Z., et al. Decreased Expression of FADS1 Predicts a Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Asian Pac. J. Cancer Prev. 2015;16:5089–5094. doi: 10.7314/APJCP.2015.16.12.5089. [DOI] [PubMed] [Google Scholar]
- 80.Expression of FADS1 in Glioma—The Human Protein Atlas. [(accessed on 14 January 2020)]; Available online: https://www.proteinatlas.org/ENSG00000149485-FADS1/pathology/glioma.
- 81.Expression of FADS2 in Glioma—The Human Protein Atlas. [(accessed on 14 January 2020)]; Available online: https://www.proteinatlas.org/ENSG00000134824-FADS2/pathology/glioma.
- 82.Kitson A.P., Smith T.L., Marks K.A., Stark K.D. Tissue-specific sex differences in docosahexaenoic acid and Δ6-desaturase in rats fed a standard chow diet. Appl. Physiol. Nutr. Metab. 2012;37:1200–1211. doi: 10.1139/h2012-103. [DOI] [PubMed] [Google Scholar]
- 83.Extier A., Langelier B., Perruchot M.-H., Guesnet P., Van Veldhoven P.P., Lavialle M., Alessandri J.-M. Gender affects liver desaturase expression in a rat model of n−3 fatty acid repletion. J. Nutr. Biochem. 2010;21:180–187. doi: 10.1016/j.jnutbio.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 84.Sibbons C.M., Brenna J.T., Lawrence P., Hoile S.P., Clarke-Harris R., Lillycrop K.A., Burdge G.C. Effect of sex hormones on n-3 polyunsaturated fatty acid biosynthesis in HepG2 cells and in human primary hepatocytes. Prostaglandins Leukot. Essent. Fat. Acids. 2013;90:47–54. doi: 10.1016/j.plefa.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Extier A., Perruchot M.-H., Baudry C., Guesnet P., Lavialle M., Alessandri J.-M. Differential effects of steroids on the synthesis of polyunsaturated fatty acids by human neuroblastoma cells. Neurochem. Int. 2009;55:295–301. doi: 10.1016/j.neuint.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 86.Maggiora M., Bologna M., Ceru M.P., Possati L., Angelucci A., Cimini A., Miglietta A., Bozzo F., Margiotta C., Muzio G., et al. An overview of the effect of linoleic and conjugated-linoleic acids on the growth of several human tumor cell lines. Int. J. Cancer. 2004;112:909–919. doi: 10.1002/ijc.20519. [DOI] [PubMed] [Google Scholar]
- 87.Cimini A., Cristiano L., Colafarina S., Benedetti E., Di Loreto S., Festuccia C., Amicarelli F., Canuto R.A., Cerù M.P. PPARgamma-dependent effects of conjugated linoleic acid on the human glioblastoma cell line (ADF) Int. J. Cancer. 2005;117:923–933. doi: 10.1002/ijc.21272. [DOI] [PubMed] [Google Scholar]
- 88.Expression of FADS3 in Glioma—The Human Protein Atlas. [(accessed on 14 January 2020)]; Available online: https://www.proteinatlas.org/ENSG00000221968-FADS3/pathology/glioma.
- 89.Marquardt A., Stöhr H., White K., Weber B.H. cDNA Cloning, Genomic Structure, and Chromosomal Localization of Three Members of the Human Fatty Acid Desaturase Family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 90.Drag J., Goździalska A., Knapik-Czajka M., Gawedzka A., Gawlik K., Jaśkiewicz J. Effect of high carbohydrate diet on elongase and desaturase activity and accompanying gene expression in rat’s liver. Genes Nutr. 2017;12:2. doi: 10.1186/s12263-017-0551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaminska B., Czapski B., Guzik R., Król S.K., Gielniewski B. Consequences of IDH1/2 Mutations in Gliomas and an Assessment of Inhibitors Targeting Mutated IDH Proteins. Molecules. 2019;24:968. doi: 10.3390/molecules24050968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chowdhury R., Yeoh K.K., Tian Y.-M., Hillringhaus L., Bagg E., Rose N., Leung I.K.H., Li X.S., Woon E.C.Y., Yang M., et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.-H., Ito S., Yang C., Wang P., Xiao M.-T., et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koivunen P., Lee S., Duncan C., Lopez G., Lu G., Ramkissoon S., Losman J.A., Joensuu P., Bergmann U., Gross S., et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cykowski M.D., Allen R.A., Fung K.-M., Harmon M.A., Dunn S.T. Pyrosequencing of IDH1 and IDH2 Mutations in Brain Tumors and Non-neoplastic Conditions. Diagn. Mol. Pathol. 2012;21:214–220. doi: 10.1097/PDM.0b013e31825d802b. [DOI] [PubMed] [Google Scholar]
- 96.Deng L., Xiong P., Luo Y., Bu X., Qian S., Zhong W., Lv S. Association between IDH1/2 mutations and brain glioma grade. Oncol. Lett. 2018;16:5405–5409. doi: 10.3892/ol.2018.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mellai M., Piazzi A., Caldera V., Monzeglio O., Cassoni P., Valente G., Schiffer D. IDH1 and IDH2 mutations, immunohistochemistry and associations in a series of brain tumors. J. Neuro-Oncology. 2011;105:345–357. doi: 10.1007/s11060-011-0596-3. [DOI] [PubMed] [Google Scholar]
- 98.Stuani L., Riols F., Millard P., Sabatier M., Batut A., Saland E., Viars F., Tonini L., Zaghdoudi S., Linares L.K., et al. Stable Isotope Labeling Highlights Enhanced Fatty Acid and Lipid Metabolism in Human Acute Myeloid Leukemia. Int. J. Mol. Sci. 2018;19:3325. doi: 10.3390/ijms19113325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou L., Wang Z., Hu C., Zhang C., Kovatcheva-Datchary P., Yu D., Liu S., Ren F., Wang X., Li Y., et al. Integrated Metabolomics and Lipidomics Analyses Reveal Metabolic Reprogramming in Human Glioma with IDH1 Mutation. J. Proteome Res. 2018;18:960–969. doi: 10.1021/acs.jproteome.8b00663. [DOI] [PubMed] [Google Scholar]
- 100.Viswanath P., Radoul M., Izquierdo-García J.L., Ong W.Q., Luchman H.A., Cairncross J.G., Huang B., Pieper R.O., Phillips J.J., Ronen S.M. 2-Hydroxyglutarate-Mediated Autophagy of the Endoplasmic Reticulum Leads to an Unusual Downregulation of Phospholipid Biosynthesis in Mutant IDH1 Gliomas. Cancer Res. 2018;78:2290–2304. doi: 10.1158/0008-5472.CAN-17-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]