Abstract
Microtubule affinity-regulating kinase (MARK4) plays a key role in Alzheimer’s disease (AD) development as its overexpression is directly linked to increased tau phosphorylation. MARK4 is a potential drug target of AD and is thus its structural features are employed in the development of new therapeutic molecules. Donepezil (DP) and rivastigmine tartrate (RT) are acetylcholinesterase (AChE) inhibitors and are used to treat symptomatic patients of mild to moderate AD. In keeping with the therapeutic implications of DP and RT in AD, we performed binding studies of these drugs with the MARK4. Both DP and RT bound to MARK4 with a binding constant (K) of 107 M−1. The temperature dependency of binding parameters revealed MARK−DP complex to be guided by static mode while MARK−RT complex to be guided by both static and dynamic quenching. Both drugs inhibited MARK4 with IC50 values of 5.3 μM (DP) and 6.74 μM (RT). The evaluation of associated enthalpy change (ΔH) and entropy change (ΔS) implied the complex formation to be driven by hydrogen bonding making it seemingly strong and specific. Isothermal titration calorimetry further advocated a spontaneous binding. In vitro observations were further complemented by the calculation of binding free energy by molecular docking and interactions with the functionally-important residues of the active site pocket of MARK4. This study signifies the implications of AChE inhibitors, RT, and DP in Alzheimer’s therapy targeting MARK4.
Keywords: acetylcholinesterase inhibitors, drug design and discovery, kinase inhibitors, molecular modeling and docking, isothermal titration calorimetry, Alzheimer’s disease, MARK4
1. Introduction
Cellular functions of the human body are governed by protein phosphorylation and dephosphorylation by protein kinases and phosphatases [1,2]. The human genome encodes many protein kinases, among them many are considered as potential targets for drug design and development. Mutations and/or altered expression of these kinases are often associated with the cause and progression of diseases [3]. More than 25 kinase-targeting drugs have been approved by the US Food and Drug Administration (FDA) and many hundreds are still in the phase of clinical trials [4].
Microtubule affinity-regulating kinase (MARK4) is a member of Ser/Thr kinase involved in various biological functions including microtubule-dependent transport, microtubule dynamics, and cell polarity [5,6,7]. MARK4 is exploited as a potential drug target for cancer, neurodegenerative disorders, and metabolic disorders highlighting its clinical significance [8,9,10]. Many studies described the importance of MARK4 in obesity, diabetes, Alzheimer’s diseases (AD), and metastatic breast carcinomas [11,12,13]. The highest expression of MARK4 is found in the brain, kidney, and testes [14,15] and variation in its level leads to the altered Akt, mTOR [16], Wnt, and NF-κB [12] signaling and thereby causes associated diseases. The design and development of potential MARK4 inhibitors are of great significance to address cancer, diabetes, and AD therapy [17,18].
The progression of AD is linked with the hyperphosphorylated tau in the brain where the role of MARK4 has been well-established [19]. Phosphorylation of tau by MARK at the Ser262 site leads to detachment of tau from microtubules and consequently allows it to be phosphorylated by other kinases [20,21]. A study reported that methylene blue decreased MARK4-mediated tau phosphorylation in a dose-dependent manner in the 293T culture of Drosophila [22]. The overexpressed MARK4 is directly linked to defected synapses and dendritic spines [23]. Numerous studies suggested the potential of MARK4 as one of the best targets in Alzheimer’s therapy [22] as well as other neurodegenerative diseases [14,24]. Owing to its remarkable role in neurodegenerative disorders, MARK4 presents a novel therapeutic target, and hence, recently many studies have reported MARK4 inhibitors that can be used to treat MARK4-directed diseases. [19,25]. In light of all these important roles of MARK4, it is currently considered an attractive drug target especially for AD and some of the associated cancers.
Structure-based drug design is the best approach to identify bioactive leads with high specificity and affinity [26]. Exploring the interaction mechanisms of therapeutics and potential drugs with the proteins or target tissues is essential for pharmaceutical industries [27,28,29,30,31]. Studying protein−drug interaction is an essential and major step in pharmacological profiling. Drug−protein interactions are important to study as the binding of a ligand/inhibitor to protein affects its pharmacokinetics [32]. At present, acetylcholinesterase (AChE) inhibitors, rivastigmine tartrate (RT), and donepezil (DP) are in use to treat symptomatic patients of mild to moderate AD. RT is a carbamate inhibitor of AChE approved by the FDA for the treatment of mild to moderate AD in adults [33]. It improves the patient’s condition in all three major domains: cognitive function, global function, and behavior [34]. RT may prevent AD progression by preferential processing of amyloid precursor protein (APP) by α-secretase, preventing it from BACE1 [35]. DP is another AChE inhibitor, a piperidine-based reversible inhibitor, that is approved for first-line treatment of AD [36].
Post ligand binding to a protein, the structure and functionality are affected thus making it important to study drug−protein interactions. The role of MARK4 is well established in the case of AD and both RT and DP are used in AD treatment thereby providing a rationale to study the binding of these drugs with the MARK4. A detailed investigation of the binding of RT and DP with the MARK4 will be useful to understand molecular insights into the therapeutic mechanism. Such analysis could further strengthen our understanding to discover hidden targeting to improve effective therapeutic strategy.
In the present study, the binding mechanism and efficacy of DP and RT with MARK4 were investigated by spectroscopic, calorimetric, and cell-free enzyme assay complemented by molecular docking.
2. Material and Methods
2.1. Materials
Both drugs RT and DP were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Unless stated, all the chemicals were procured from Sigma-Aldrich Co. (St. Louis, MO, USA). Other reagents were analytical grade, procured from local suppliers.
2.2. Expression and Purification of MARK4
Human MARK4 was cloned, expressed, and purified as per our published protocol [37,38]. The quality of purified protein was assessed by kinase assay and purity was checked by SDS-PAGE. MARK4 protein was confirmed with the help of Western blot using specific primary antibodies [39].
2.3. Kinase Assay for Enzyme Activity
The activity of MARK4 was measured using standard malachite green (BIOMOL® reagent, Enzo Life Sciences) microtitre-plate assay using previously-published protocols [17,40]. MARK4 (4 μM) with increasing concentrations of ATP and assay buffer (20 mM Tris-HCl, pH 8.0, and 100 mM NaCl) were incubated for 15–20 min at 25 °C. Then, 100 μL of Biomol Green reagent was added to terminate the reaction followed by incubation for 20 min for color development. A multiplate ELISA reader was used to measure the absorbance of each well at 620 nm.
ATPase inhibition assay of MARK4 was performed in the presence of increasing concentrations (0–20 μM) of DP and RT. Initially, MARK4 (4 μM) was pre-incubated with increasing concentrations of ligands at room temperature for 60 min in a 96-well plate. Subsequently, 200 μM of freshly-prepared ATP was mixed to the reaction mixture and incubated for 15–20 min at 25 °C. At the end of this time, BIOMOL® reagent was added and kept for 15–20 min. The intensity of color was spectrophotometrically measured at 620 nm. The kinase activity of MARK4 was quantified and plotted as percent inhibition of DP and RT compared to the activity of native MARK4 considered as a reference of 100%.
2.4. Fluorescence Measurements
To study the binding affinity of DP and RT with MARK4, the fluorescence emission spectrum was recorded using the Jasco spectrofluorometer (FP-6200) and analyzed as per previous studies [41,42,43]. The observed fluorescence intensities were corrected for the inner filter effect [44]. Obtained data were fitted into the Stern−Volmer and modified Stern−Volmer equations to estimate Stern-Volmer constant (Ksv), binding constant (Ka), and the number of binding sites (n).
2.5. Thermodynamics of the Complex
Equation (1) (van’t Hoff equation) [45] was used to find the thermodynamic parameters such as changes in enthalpy, entropy, and Gibbs free energy associated with the reaction.
| (1) |
K is the obtained binding constant, ΔH is the associated enthalpy change and ΔG is the associated free Gibbs energy change. ΔS denotes the associated entropy change with reaction, and R is the universal gas constant.
The slope of the van’t Hoff plot (LnK on Y-axis against 1/T on the X-axis) gives the value of −ΔH/R, and the intercept giving the value of ΔS/R.
2.6. Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) measurements were carried out on a VP-ITC microcalorimeter (MicroCal, Inc, GE, MicroCal, Northampton, MA, USA) at 25 °C. All the solutions were degassed in thermovac for 20 min to remove air bubbles. For sample preparation, recombinant MARK4 was extensively dialyzed in 20 mM Tris buffer, pH 8.5. The sample cell was filled with 20 µM MARK4 and the syringe was filled with 100 µM DP and 200 µM RT. We used standard protocol [46,47] to run the ITC in which the first injection was a false injection of 2 μL followed by injections of 10 μL of the ligands at a span of 260 s intervals into the sample cell. The stirring rate was maintained at 320 rpm. The obtained titration data were analyzed by Origin 8.0 software to estimate thermodynamic parameters such as stoichiometry of binding (n), enthalpy change (ΔH), an association constant (Ka).
2.7. Molecular Docking
Atomic coordinates of human MARK4 were taken from the Protein Data Bank (PDB ID: 5ES1, [48]) and refined through the Swiss PDB Viewer and the MGL tools [49]. Hydrogen atoms were added to the polar groups in the protein along with the Kollman charges using the Molecular Graphics Laboratory Tools. The docking was performed using AutoDock Vina with a blind search space for both DP and RT with the exhaustiveness of 8. The binding affinity and interaction of DP and RT towards MARK4 were studied while using PyMOL and Discovery Studio.
2.8. Statistical Analysis
All the experiments were performed in triplicate and the data obtained have been expressed in mean ± standard error of the mean (SEM).
3. Results
3.1. Fluorescence-Based Binding Studies
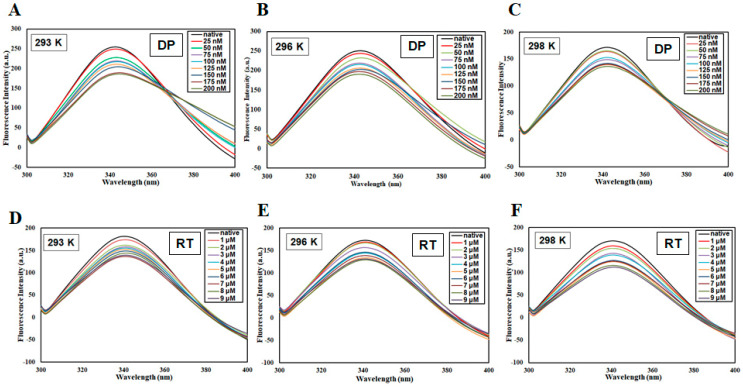
The intrinsic fluorescence of protein due to tryptophan is often used to get an insight into the ligand binding to the protein because binding often leads to changes in the surrounding environment around tryptophan residues which corresponds to observed changes in fluorescence. The recombinant MARK4 was expressed and purified to carry out all binding studies [50]. Figure 1 shows the fluorescence emission spectra of MARK4 in the presence of DP (0–200 nM) and RT (0–9 µM) at three different temperatures. For both the compounds, the fluorescence intensity decreased with a subsequent increase in ligand concentrations. This decrease in fluorescence (fluorescence quenching) is indicative of a stable complex formation between protein and ligand [51]. This data was mathematically expressed to estimate associated binding parameters, Stern−Volmer constant (Ksv), binding constant (K), and the number of the binding site (n).
Figure 1.
Steady-state fluorescence of MARK4 in the presence of DP (0−200 nM) at (A) 293 K, (B) 296 K, and (C) 298 K, and RT (0−9 µM) at (D) 293 K, (E) 296 K, and (F) 298 K. The concentration of MARK4 was 4 μM. The protein was excited at 280 nm and emission was recorded in the range of 300−400 nm.
Fluorescence quenching can be static or dynamic or a combination of both [52,53,54]. The mode of quenching operative for a particular protein−ligand interaction can be obtained by analyzing the temperature dependency of the binding parameters since it can differentiate between static and dynamic modes of quenching [52]. Thus, to have an insight into the operative quenching mechanism, fluorescence measurements were carried out at three different temperatures.
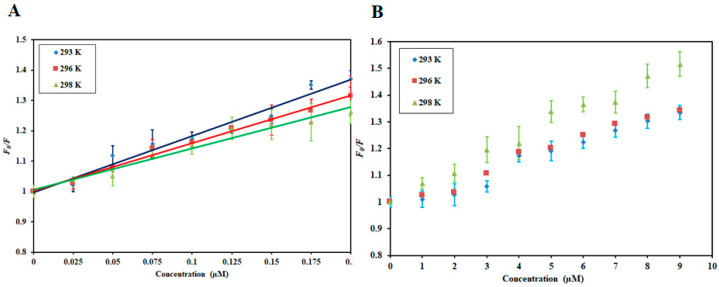
Stern−Volmer (SV) plots of F0/F on the Y-axis and [DP] and [RT] on the X-axis is shown in Figure 2. Ksv was obtained from the slope of this plot at a fixed intercept after linear regression using the Stern−Volmer equation (Equation (2)).
| (2) |
Figure 2.
Stern−Volmer plots of (A) MARK4−DP and (B) MARK4−RT at three different temperatures. The temperatures used in the study were 293 K, 296 K, and 298 K.
The SV plot of combined quenching (both static and dynamic) is characteristically an upward curvature while the linear Stern−Volmer plot depicts that only one mode of quenching was operative. The SV plot for MARK4−DP was linear while for MARK4−RT interaction there was a slight positive deviation implying that this interaction is guided by combined quenching. The linear plot and upward curvature are not enough to define the quenching type. Hence, the variation in Ksv values as a function of temperature was further analyzed. In the static quenching, there was a decrease in Ksv values while in the dynamic quenching Ksv increased with a corresponding increase in temperature [55]. Table 1 depicts the Ksv values obtained for MARK4−DP and MARK4−RT interactions. For MARK4−DP interaction, Ksv values decrease with increasing temperature suggesting that MARK4−DP complex formation is guided by static mode of quenching. However, an opposite effect was observed for MARK4−RT interaction where an increase in Ksv values was observed with increasing temperature implying MARK4−RT interaction to be guided by a dynamic mode of quenching. Additionally, the value of the bimolecular quenching rate constant, Kq, affirmed it. Equation (3) uses Ksv and τo (average integral fluorescence lifetime of tryptophan) (2.7 × 10−9 s) [56] to find biomolecular quenching rate constant (Kq)
| (3) |
Table 1.
Stern−Volmer constants calculated for interactions of MARK4 with DP and RT at different temperatures.
| Temperature, K | Ksv (105 M−1) | Kq (1014 M−1 s−1) | R2 |
|---|---|---|---|
| MARK4−DP | |||
| 293 | 18.5 | 6.85 | 0.99 |
| 296 | 15.7 | 5.81 | 0.98 |
| 298 | 13.6 | 5.03 | 0.96 |
| MARK4−RT | |||
| Temperature, K | Ksv (104 M−1) | K q (1013 M−1 s−1) | R2 |
| 293 | 3.4 | 1.25 | 0.97 |
| 296 | 3.9 | 1.44 | 0.97 |
| 298 | 5.8 | 2.14 | 0.98 |
Ksv, Stern−Volmer constant; Kq, quenching rate constant.
The values of Kq for MARK4−DP interaction and MARK4−RT interaction (Table 1) were substantially higher than the maximum dynamic quenching constant (∼1010 M−1 s−1) [57], suggesting that both MARK4−DP and MARK4−RT interactions are guided by static mode of quenching. For MARK4−RT interaction, earlier-obtained Ksv values as a function of temperature suggested that dynamic mode guides MARK4−RT interaction. Hence, we concluded that MARK4−RT quenching is guided by a mixture of the static and dynamic modes of quenching. Figure S1 gives a clear idea of the type of quenching operative for specific protein−ligand interactions.
K, binding constant; n, number of the binding site; ΔG, change in Gibbs free energy; ΔS, change in entropy; ΔH, change in enthalpy.
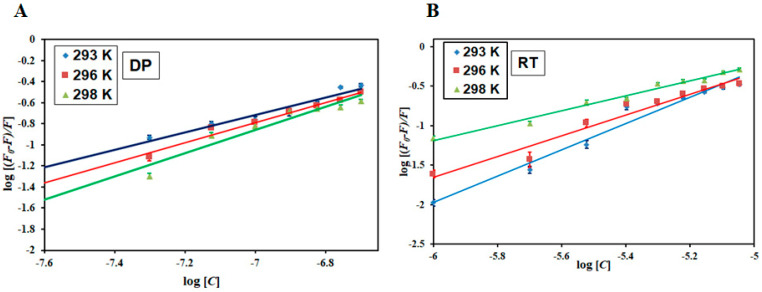
To get insights into other binding parameters, Equation (4) was used [58]. This equation gives binding constant (K) and the number of binding sites (n).
| (4) |
Figure 3 illustrates the experimental fitting obtained as per Equation (4) for MARK4−DP and MARK4−RT. The intercept of the plots gives the binding constant (K) and slope provides the number of binding sites (n). Table 2 lists the values of binding constants of MARK4−DP and MARK4−RT at three different temperatures. The obtained binding constants of DP and RT with the MARK4 indicate a strong affinity. The values of K decreased at high temperatures indicating the formation of a stable complex at a lower temperature. All these observations suggest a strong binding affinity of RT and DP to MARK4. Moreover, the MARK4−DP complex is governed by static mode and MARK4−RT complex by a combination of static and dynamic modes of quenching.
Figure 3.
Modified Stern−Volmer plots of (A) MARK4−DP and (B) MARK4−RT at three different temperatures.
Table 2.
Thermodynamic parameters obtained from fluorescence quenching studies carried out at different temperatures.
| Temperature (K) |
K 107 M−1 |
n | ΔG kcal mol−1 |
ΔS cal mol−1K−1 |
ΔH kcal mol−1 |
kcal mol−1 |
|---|---|---|---|---|---|---|
| MARK4 − DP | ||||||
| 293 | 9.4 | 1.66 | −10.70 | −432.749 | −137.5 | −126.79 |
| 296 | 0.9 | 1.311 | −9.40 | −128.09 | ||
| 298 | 0.002 | 0.1 | −8.54 | −128.95 | ||
| MARK4−R Τ | ||||||
| 293 | 10.5 | 1.66 | −10.74 | −455.78 | −144.23 | −133.48 |
| 296 | 0.16 | 1.311 | −8.46 | −135.76 | ||
| 298 | 0.002 | 0.94 | −6.18 | −138.04 | ||
3.2. Thermodynamic Analysis
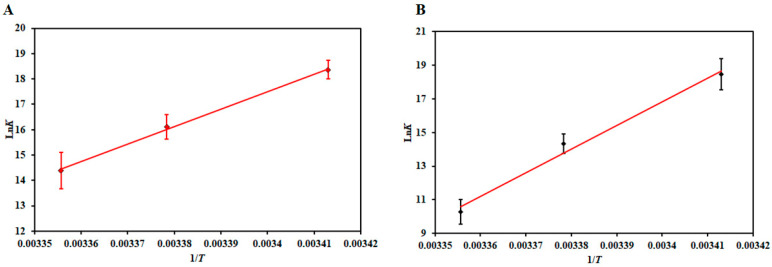
After estimating the binding parameters, the van’t Hoff equation (Equation (1)) gives the idea of different thermodynamic parameters of the complexes [52]. These values are an indicator of the prevalent driving forces associated with the binding. According to Ross and Subramaniam [59], the negative values of ΔH and ΔS suggest that this reaction is driven by van der Waals force and/or hydrogen bonding while the positive values of ΔH and ΔS suggest the reaction to be dominated by hydrophobic interactions. The negative ΔH and positive ΔS values indicate that electrostatic interactions drive the reaction [56]. Figure 4 shows the van’t Hoff plot of MARK4−DP and MARK4−RT with a linear fit of the obtained data points using Equation (1). The slope of this plot gives the value of −ΔH/R, and the intercept gives the value of ΔS/R. Table 2 lists the thermodynamic parameters estimated for MARK4−DP and MARK4−RT, respectively. The binding was spontaneous as evident from ΔG < 0 [60]. Moreover, the negative values of ΔH and ΔS (Table 2) obtained for both MARK4−DP and MARK4−RT suggest that the reaction is exothermic and driven by enthalpy, not entropy. The negative values of ΔH and ΔS further suggest prevalent forces driving the reactions are van der Waals and hydrogen bonding [59], thus making this process seemingly specific.
Figure 4.
Van’t Hoff plot for (A) MARK4−DP and (B) MARK4−RT interaction at three different temperatures.
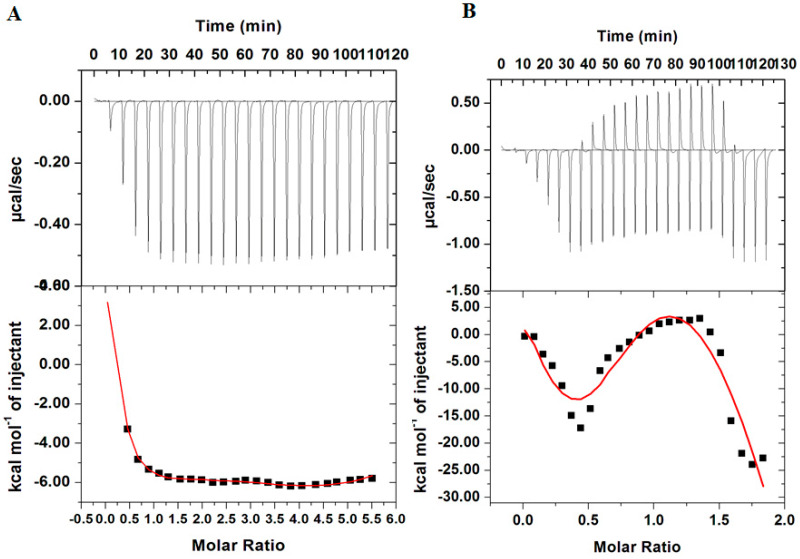
3.3. Isothermal Titration Calorimetry
To further validate the binding results obtained from fluorescence studies, ITC measurements were performed. Figure 5A shows an isotherm of MARK4 titrated with 100 µM DP while Figure 5B depicts a calorimetric profile obtained for MARK4 upon titrating with 200 µM RT. The upper panel corresponds to raw data obtained due to consecutive injection of the ligand into MARK4 with the bottom panel showing binding curves obtained after subtracting the dilution heat of both ligands and protein. The final figure was obtained using Micro Cal VP-ITC Origin 8.0., the obtained ITC isotherm advocates spontaneous binding of DP and RT with MARK4 with a significantly high binding affinity validating our fluorescence results. The thermodynamic parameters obtained for MARK4−DP interaction and MARK4−RT interaction are depicted in Table S1. Our ITC results are in agreement with fluorescence results as both methods suggested that both DP and RT have a strong binding affinity to the MARK4, despite a notable variation in the values of binding parameters being observed. These variations are due to the assumptions made in noncalorimetric approaches that ΔH does not depend on temperature [61,62]. Additionally, ITC measures a global change in property with fluorescence spectroscopy measuring only local changes around Tryptophan. These variations in thermodynamic parameters obtained from fluorescence spectroscopy and ITC are a common observation and has been reported in many protein−ligand interaction studies [52,56,63].
Figure 5.
(A) Isothermal titration calorimetric profile obtained upon titration of 100 µM DP into 20 µM MARK4. (B) Isothermal titration calorimetric profile obtained upon titration of 200 µM RT into 20 µM MARK4. The bottom panel shows resultant binding isotherm before the integration of peak area and normalization to yield a plot of molar enthalpy change against the ligand/protein ratio.
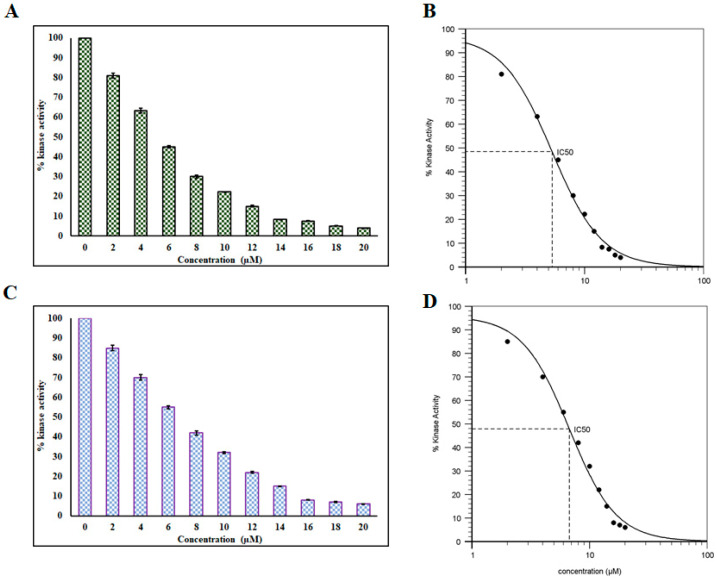
3.4. Enzyme Inhibition Assay
Enzyme inhibition assays were performed to see the effect of RT and DP on the kinase activity of MARK4 as described [64,65]. Interestingly, we observed a significant reduction in the kinase activity of MARK4 with increasing concentrations of DP (Figure 6A) and RT (Figure 6C), in a dose-dependent manner. IC50 was calculated as 5.3 µM (Figure 6B) and 6.74 µM (Figure 6D), for DP and RT, respectively. The obtained IC50 values were in the order of reported inhibitors of MARK4 [6,17,66], thus implying the significance of DP and RT as potential MARK4 inhibitors. These observations suggest DP and RT have the potential to inhibit MARK4 and may be implicated in AD therapy. Finally, DP and RT bind to the MARK4 and subsequently inhibit the kinase activity of MARK4, and thus offer a newer approach of targeting AD.
Figure 6.
(A) ATPase assay showing the effect of DP on the kinase activity of MARK4. (B) IC50 plot of DP obtained through the AAT Bioquest calculator [67]. (C) ATPase assay depicting the effect of RT on the kinase activity of MARK4. (D) IC50 plot of RT obtained through the AAT Bioquest calculator [67].
3.5. Molecular Docking
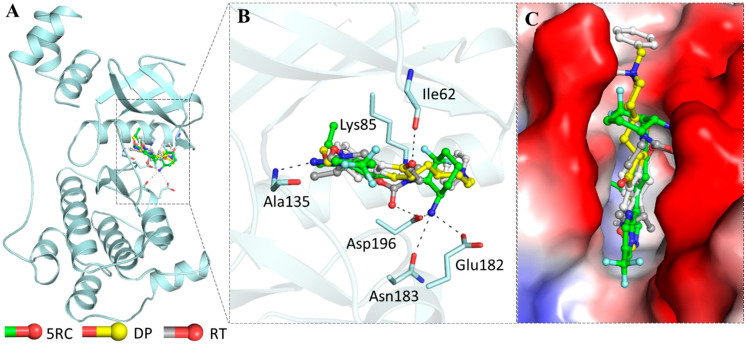
Both DP and RT were subjected to docking analyses which showed a considerable binding affinity towards MARK4 with the calculated binding free energies as −8.1 kcal/mol and −6.3 kcal/mol, respectively. The standard compound, the co-crystalized MARK4 pyrazolopyrimidine inhibitor, 5RC (~-[(1~,6~)-6-azanyl-2,2-bis(fluoranyl)cyclohexyl]-5-ethyl-4-[6-(trifluoromethyl)pyrazolo[1 ,5-a]pyrimidin-3-yl]thiophene-2-carboxamide) was also docked to estimate the binding affinity. The binding affinity of this ligand (5RC) was calculated as −9.8 kcal/mol indicating a higher strength of binding affinity than DP and RT. The docking analysis is in agreement with our fluorescence binding studies and ITC which revealed both compounds bind to MARK4 and subsequently inhibit its kinase activity. All possible docked conformers of DP and RT were analyzed to investigate their mode of interaction with the functionally-important residues of the binding pocket of MARK4. Here, we found that DP and RT are preferentially binding with the active residues of MARK4.
It is well known that Lys85 is a catalytically critical residue for MARK4 kinase function and plays an important role in ATP binding which forms close interactions with DP and RT. The binding pattern of DP and RT along with the standard MARK4 inhibitor with MARK4 is illustrated in Figure 7. We observed that both DP and RT interacted with the ATP-binding site of MARK4 and mimicked the pose of a co-crystallized MARK4 inhibitor (Figure 8B). Both compounds, along with standard inhibitor [48], bind into the deep cavity of MARK4 binding pocket (Figure 8C). A set of MARK4 ATP-binding site residues is interacting with DP and RT suggesting that these compounds can block the accessibility of ATP in the binding pocket of MARK4.
Figure 7.
Interactions of DP and RT with MARK4. (A) Cartoon representation showing the docked DP, RT, and standard inhibitor 5RC, interacting with the binding site of MARK4. (B) Zoomed view of the binding pocket and its interaction with DP, RT, and standard MARK4 inhibitor. (C) Potential surface view of MARK4 binding pocket occupied by DP, RT, and standard MARK4 inhibitor.
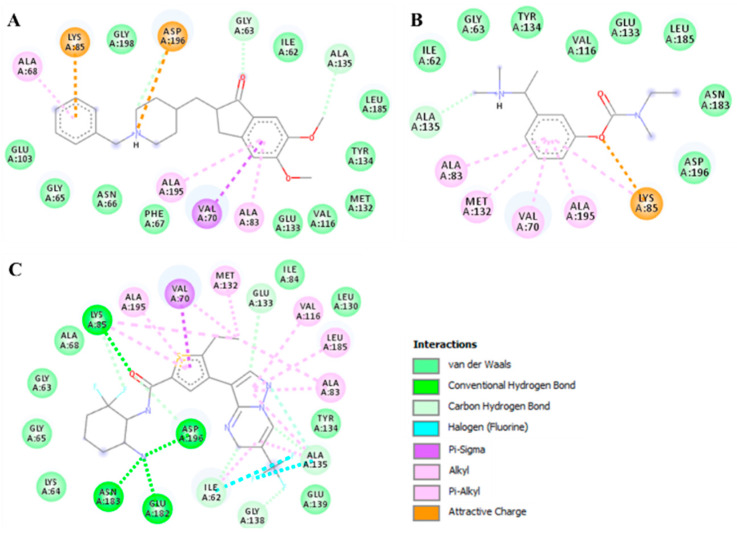
Figure 8.
2D structural representation of MARK4 residues interacting with (A) DP (B) RT, and (C) standard MARK4 inhibitor (5RC, [48]).
Both compounds, DP and RT were further explored to see their possible interactions with the active site residues of MARK4 and to infer possible mechanisms of inhibition. It was observed that the docked complexes of MARK4 with DP and RT were stabilized by several non-covalent interactions shared by the ATP-binding site (Lys85) and other functionally-active residues of MARK4 binding pocket. The interaction pattern for DP and RT was found to be similar to the standard MARK4 inhibitor (Figure 8). It is evident from Figure 8 that DP, RT, and the standard MARK4 inhibitor are interacting with the MARK4 ATP-binding site, Lys85. Both compounds are showing common interactions and interacting with the attractive charge towards Lys85. Specific interactions between the functionally-vital residues of MARK4 binding pocket and DP and RT indicate strong binding and their further implications as ATP-competitive inhibitors of MARK4.
4. Conclusions
MARK4 is being used as a drug target for the development of therapeutic molecules in Alzheimer’s therapy. Overexpression of MARK4 is directly linked to the tau hyperphosphorylation which is a key event in AD pathology thus highlighting the clinical significance of developing molecules with inhibitory potential against MARK4. This study reports the inhibitory potential of AChE inhibitors, DP, and RT, against MARK4. As we know that both DP and RT are AChE inhibitors that have been recommended for the symptomatic treatment of mild to moderate AD, inhibition of MARK4 by the same drugs indicating a common targeting during AD therapy. In addition, new molecules with a high binding affinity and selectivity to MARK4 may be designed by considering the chemical scaffold of DP and RT.
Acknowledgments
A.S. is thankful to DST-SERB for the award of the National Postdoctoral Fellowship (Grant (NPDF/2017/000745). M.F.A., A.H., and M.T.R. acknowledge the generous support from the Deanship of Scientific Research at King Saud University, Riyadh, Kingdom of Saudi Arabia, for funding his research group (Grant No. RGP-1441-150). S.A. thanks to the Indian Council of Medical Research for the Senior Research Fellowship (45/37/2019-BIO/BMS). The authors sincerely thank the DST for the FIST support (FIST program No. SR/FST/LSI-541/2012).
Supplementary Materials
The following can be found at https://www.mdpi.com/2218-273X/10/5/789/s1; Figure S1: Graphical representation of the operative mode of quenching for specific protein-ligand interaction; Table S1: Thermodynamic parameters obtained from ITC measurements.
Author Contributions
All the authors contributed to this manuscript. Conceptualization, A.S., S.A., and M.I.H.; methodology, S.A., A.S., and T.M.; software, M.F.A., G.M.H., T.M., and A.H. validation, M.T.R. and M.I.H.; formal analysis, S.A. and A.S.; investigation, S.A., A.I., and G.M.H.; resources, A.I. and M.I.H.; data curation, A.S., M.T.R., and A.H.; writing—original draft preparation, S.A., A.S., and M.I.H.; writing—review and editing, A.I. and M.I.H.; visualization, A.S., S.A., and T.M.; supervision, M.F.A. and M.I.H.; project administration, A.I. and M.I.H.; funding acquisition, M.I.H. and M.F.A. All authors have read and agreed to publish the current version of the manuscript.
Funding
This work is supported by the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India (Grant No. EMR/2015/002372). APC is paid by King Saud University, Riyadh, Kingdom of Saudi Arabia.
Conflicts of Interest
There are no conflicts to declare.
References
- 1.Bononi A., Agnoletto C., De Marchi E., Marchi S., Patergnani S., Bonora M., Giorgi C., Missiroli S., Poletti F., Rimessi A. Protein kinases and phosphatases in the control of cell fate. Enzym. Res. 2011;2011:329098. doi: 10.4061/2011/329098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter T. Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 3.Hoda N., Naz H., Jameel E., Shandilya A., Dey S., Hassan M.I., Ahmad F., Jayaram B. Curcumin specifically binds to the human calcium–calmodulin-dependent protein kinase IV: Fluorescence and molecular dynamics simulation studies. J. Biomol. Struct. Dyn. 2016;34:572–584. doi: 10.1080/07391102.2015.1046934. [DOI] [PubMed] [Google Scholar]
- 4.Katopodis P., Chudasama D., Wander G., Sales L., Kumar J., Pandhal M., Anikin V., Chatterjee J., Hall M., Karteris E. Kinase Inhibitors and Ovarian Cancer. Cancers. 2019;11:1357. doi: 10.3390/cancers11091357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naz F., Anjum F., Islam A., Ahmad F., Hassan M.I. Microtubule affinity-regulating kinase 4: Structure, function, and regulation. Cell Biochem. Biophys. 2013;67:485–499. doi: 10.1007/s12013-013-9550-7. [DOI] [PubMed] [Google Scholar]
- 6.Naz F., Khan F.I., Mohammad T., Khan P., Manzoor S., Hasan G.M., Lobb K.A., Luqman S., Islam A., Ahmad F., et al. Investigation of molecular mechanism of recognition between citral and MARK4: A newer therapeutic approach to attenuate cancer cell progression. Int. J. Biol. Macromol. 2018;107:2580–2589. doi: 10.1016/j.ijbiomac.2017.10.143. [DOI] [PubMed] [Google Scholar]
- 7.Naz F., Shahbaaz M., Bisetty K., Islam A., Ahmad F., Hassan M.I. Designing New Kinase Inhibitor Derivatives as Therapeutics Against Common Complex Diseases: Structural Basis of Microtubule Affinity-Regulating Kinase 4 (MARK4) Inhibition. OMICS. 2015;19:700–711. doi: 10.1089/omi.2015.0111. [DOI] [PubMed] [Google Scholar]
- 8.Arash E.H., Shiban A., Song S., Attisano L. MARK4 inhibits Hippo signaling to promote proliferation and migration of breast cancer cells. EMBO Rep. 2017;18:420–436. doi: 10.15252/embr.201642455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naz F., Sami N., Naqvi A.T., Islam A., Ahmad F., Imtaiyaz Hassan M. Evaluation of human microtubule affinity-regulating kinase 4 inhibitors: Fluorescence binding studies, enzyme, and cell assays. J. Biomol. Struct. Dyn. 2017;35:3194–3203. doi: 10.1080/07391102.2016.1249958. [DOI] [PubMed] [Google Scholar]
- 10.Pardo O.E., Castellano L., Munro C.E., Hu Y., Mauri F., Krell J., Lara R., Pinho F.G., Choudhury T., Frampton A.E. miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep. 2016;17:570–584. doi: 10.15252/embr.201540970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian L., Wen A., Dong S., Yan P. Molecular Characterization of Microtubule Affinity-Regulating Kinase4 from Sus scrofa and Promotion of Lipogenesis in Primary Porcine Placental Trophoblasts. Int. J. Mol. Sci. 2019;20:1206. doi: 10.3390/ijms20051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z., Gan L., Chen Y., Luo D., Zhang Z., Cao W., Zhou Z., Lin X., Sun C. Mark4 promotes oxidative stress and inflammation via binding to PPARγ and activating NF-κB pathway in mice adipocytes. Sci. Rep. 2016;6:21382. doi: 10.1038/srep21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak G.A., Zhou Z., Silzer T.K., Barber R.C., Phillips N.R. Two-stage Bayesian GWAS of 9576 individuals identifies SNP regions that are targeted by miRNAs inversely expressed in Alzheimer’s and cancer. Alzheimers Dement. 2020;16:162–177. doi: 10.1002/alz.12003. [DOI] [PubMed] [Google Scholar]
- 14.Trinczek B., Brajenovic M., Ebneth A., Drewes G. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J. Biol. Chem. 2004;279:5915–5923. doi: 10.1074/jbc.M304528200. [DOI] [PubMed] [Google Scholar]
- 15.Tang E.I., Xiao X., Mruk D.D., Qian X.-J., Mok K.-W., Jenardhanan P., Lee W.M., Mathur P.P., Cheng C.Y. Microtubule affinity-regulating kinase 4 (MARK4) is a component of the ectoplasmic specialization in the rat testis. Spermatogenesis. 2012;2:117–126. doi: 10.4161/spmg.20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Guan K.-L. Microtubule-associated protein/microtubule affinity-regulating kinase 4 (MARK4) is a negative regulator of the mammalian target of rapamycin complex 1 (mTORC1) J. Biol. Chem. 2013;288:703–708. doi: 10.1074/jbc.C112.396903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan P., Queen A., Mohammad T., Smita, Khan N.S., Hafeez Z.B., Hassan M.I., Ali S. Identification of alpha-Mangostin as a Potential Inhibitor of Microtubule Affinity Regulating Kinase 4. J. Nat. Prod. 2019;82:2252–2261. doi: 10.1021/acs.jnatprod.9b00372. [DOI] [PubMed] [Google Scholar]
- 18.Voura M., Khan P., Thysiadis S., Katsamakas S., Queen A., Hasan G.M., Ali S., Sarli V., Hassan M.I. Probing the Inhibition of Microtubule Affinity Regulating Kinase 4 by N-Substituted Acridones. Sci. Rep. 2019;9:1676. doi: 10.1038/s41598-018-38217-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Annadurai N., Agrawal K., Dzubak P., Hajduch M., Das V. Microtubule affinity-regulating kinases are potential druggable targets for Alzheimer’s disease. Cell. Mol. Life Sci. 2017;74:4159–4169. doi: 10.1007/s00018-017-2574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drewes G., Ebneth A., Preuss U., Mandelkow E.M., Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger tnicrotubule disruption. Cell. 1997;89:297–308. doi: 10.1016/S0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 21.Mandelkow E.-M., Thies E., Trinczek B., Biernat J., Mandelkow E. MARK/PAR1 kinase is a regulator of microtubule-dependent transport in axons. J. Cell Biol. 2004;167:99–110. doi: 10.1083/jcb.200401085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun W., Lee S., Huang X., Liu S., Inayathullah M., Kim K.-M., Tang H., Ashford J.W., Rajadas J. Attenuation of synaptic toxicity and MARK4/PAR1-mediated Tau phosphorylation by methylene blue for Alzheimer’s disease treatment. Sci. Rep. 2016;6:34784. doi: 10.1038/srep34784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu W., Polepalli J., Wagh D., Rajadas J., Malenka R., Lu B. A critical role for the PAR-1/MARK-tau axis in mediating the toxic effects of Aβ on synapses and dendritic spines. Hum. Mol. Genet. 2012;21:1384–1390. doi: 10.1093/hmg/ddr576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenardhanan P., Mannu J., Mathur P.P. The structural analysis of MARK4 and the exploration of specific inhibitors for the MARK family: A computational approach to obstruct the role of MARK4 in prostate cancer progression. Mol. Biosyst. 2014;10:1845–1868. doi: 10.1039/C3MB70591A. [DOI] [PubMed] [Google Scholar]
- 25.Hayden E.Y., Putman J., Nunez S., Shin W.S., Oberoi M., Charreton M., Dutta S., Li Z., Komuro Y., Joy M.T., et al. Ischemic axonal injury up-regulates MARK4 in cortical neurons and primes tau phosphorylation and aggregation. Acta Neuropathol. Commun. 2019;7:135. doi: 10.1186/s40478-019-0783-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammad T., Khan F.I., Lobb K.A., Islam A., Ahmad F., Hassan M.I. Identification and evaluation of bioactive natural products as potential inhibitors of human microtubule affinity-regulating kinase 4 (MARK4) J. Biomol. Struct. Dyn. 2019;37:1813–1829. doi: 10.1080/07391102.2018.1468282. [DOI] [PubMed] [Google Scholar]
- 27.Beg A., Khan F.I., Lobb K.A., Islam A., Ahmad F., Hassan M.I. High throughput screening, docking, and molecular dynamics studies to identify potential inhibitors of human calcium/calmodulin-dependent protein kinase IV. J. Biomol. Struct. Dyn. 2019;37:2179–2192. doi: 10.1080/07391102.2018.1479310. [DOI] [PubMed] [Google Scholar]
- 28.Hassan M.I. Recent Advances in the Structure-Based Drug Design and Discovery. Curr. Top. Med. Chem. 2016;16:899–900. doi: 10.2174/1568026616999150918145640. [DOI] [PubMed] [Google Scholar]
- 29.Mohammad T., Arif K., Alajmi M.F., Hussain A., Islam A., Rehman M.T., Hassan I. Identification of high-affinity inhibitors of pyruvate dehydrogenase kinase-3: Towards therapeutic management of cancer. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1711810. [DOI] [PubMed] [Google Scholar]
- 30.Mohammad T., Batra S., Dahiya R., Baig M.H., Rather I.A., Dong J.J., Hassan I. Identification of High-Affinity Inhibitors of Cyclin-Dependent Kinase 2 Towards Anticancer Therapy. Molecules. 2019;24:4589. doi: 10.3390/molecules24244589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammad T., Siddiqui S., Shamsi A., Alajmi M.F., Hussain A., Islam A., Ahmad F., Hassan M.I. Virtual Screening Approach to Identify High-Affinity Inhibitors of Serum and Glucocorticoid-Regulated Kinase 1 among Bioactive Natural Products: Combined Molecular Docking and Simulation Studies. Molecules. 2020;25:823. doi: 10.3390/molecules25040823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ajmal M.R., Nusrat S., Alam P., Zaidi N., Khan M.V., Zaman M., Shahein Y.E., Mahmoud M.H., Badr G., Khan R.H. Interaction of anticancer drug clofarabine with human serum albumin and human α-1 acid glycoprotein. Spectroscopic and molecular docking approach. J. Pharm. Biomed. Anal. 2017;135:106–115. doi: 10.1016/j.jpba.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Pan Y., Xu X., Wang X. Rivastigmine blocks voltage-activated K+ currents in dissociated rat hippocampal neurons. Br. J. Pharmacol. 2003;140:907–912. doi: 10.1038/sj.bjp.0705503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson B., Samanta M.K., Santhi K., Kumar K.P.S., Paramakrishnan N., Suresh B. Poly (n-butylcyanoacrylate) nanoparticles coated with polysorbate 80 for the targeted delivery of rivastigmine into the brain to treat Alzheimer’s disease. Brain Res. 2008;1200:159–168. doi: 10.1016/j.brainres.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 35.Ray B., Maloney B., Sambamurti K., Kumar Karnati H., Nelson P.T., Greig N.H., Lahiri D.K. Rivastigmine modifies the α-secretase pathway and potentially early Alzheimer’s disease. Transl. Psychiatry. 2020;10:1–17. doi: 10.1038/s41398-020-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cecilia Rodrigues Simoes M., Pereira Dias Viegas F., Soares Moreira M., de Freitas Silva M., Maximo Riquiel M., Mattos da Rosa P., Rosa Castelli M., Henrique dos Santos M., Gomes Soares M., Viegas C. Donepezil: An important prototype to the design of new drug candidates for Alzheimer’s disease. Mini Rev. Med. Chem. 2014;14:2–19. doi: 10.2174/1389557513666131119201353. [DOI] [PubMed] [Google Scholar]
- 37.Naz F., Sami N., Islam A., Ahmad F., Hassan M.I. Ubiquitin-associated domain of MARK4 provides stability at physiological pH. Int. J. Biol. Macromol. 2016;93:1147–1154. doi: 10.1016/j.ijbiomac.2016.09.087. [DOI] [PubMed] [Google Scholar]
- 38.Naz F., Asad M., Malhotra P., Islam A., Ahmad F., Hassan M.I. Cloning, expression, purification and refolding of microtubule affinity-regulating kinase 4 expressed in Escherichia coli. Appl. Biochem. Biotechnol. 2014;172:2838–2848. doi: 10.1007/s12010-014-0733-5. [DOI] [PubMed] [Google Scholar]
- 39.Khan P., Idrees D., Moxley M.A., Corbett J.A., Ahmad F., von Figura G., Sly W.S., Waheed A., Hassan M.I. Luminol-based chemiluminescent signals: Clinical and non-clinical application and future uses. Appl. Biochem. Biotechnol. 2014;173:333–355. doi: 10.1007/s12010-014-0850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anwar S., Kar R.K., Haque M.A., Dahiya R., Gupta P., Islam A., Ahmad F., Hassan M.I. Effect of pH on the structure and function of pyruvate dehydrogenase kinase 3: Combined spectroscopic and MD simulation studies. Int. J. Biol. Macromol. 2020;147:768–777. doi: 10.1016/j.ijbiomac.2020.01.218. [DOI] [PubMed] [Google Scholar]
- 41.Gupta P., Mohammad T., Khan P., Alajmi M.F., Hussain A., Rehman M.T., Hassan M.I. Evaluation of ellagic acid as an inhibitor of sphingosine kinase 1: A targeted approach towards anticancer therapy. Biomed. Pharmacother. 2019;118:109245. doi: 10.1016/j.biopha.2019.109245. [DOI] [PubMed] [Google Scholar]
- 42.Khan N.S., Khan P., Ansari M.F., Srivastava S., Hasan G.M., Husain M., Hassan M.I. Thienopyrimidine-Chalcone Hybrid Molecules Inhibit Fas-Activated Serine/Threonine Kinase: An Approach To Ameliorate Antiproliferation in Human Breast Cancer Cells. Mol. Pharm. 2018;15:4173–4189. doi: 10.1021/acs.molpharmaceut.8b00566. [DOI] [PubMed] [Google Scholar]
- 43.Shamsi A., Al Shahwan M., Ahamad S., Hassan M.I., Ahmad F., Islam A. Spectroscopic, calorimetric and molecular docking insight into the interaction of Alzheimer’s drug donepezil with human transferrin: Implications of Alzheimer’s drug. J. Biomol. Struct. Dyn. 2020;38:1094–1102. doi: 10.1080/07391102.2019.1595728. [DOI] [PubMed] [Google Scholar]
- 44.Chi Z., Liu R. Phenotypic characterization of the binding of tetracycline to human serum albumin. Biomacromolecules. 2010;12:203–209. doi: 10.1021/bm1011568. [DOI] [PubMed] [Google Scholar]
- 45.Shamsi A., Ahmed A., Khan M.S., Husain F.M., Amani S., Bano B. Investigating the interaction of anticancer drug temsirolimus with human transferrin: Molecular docking and spectroscopic approach. J. Mol. Recognit. 2018;31:e2728. doi: 10.1002/jmr.2728. [DOI] [PubMed] [Google Scholar]
- 46.Gupta P., Mohammad T., Dahiya R., Roy S., Noman O.M.A., Alajmi M.F., Hussain A., Hassan M.I. Evaluation of binding and inhibition mechanism of dietary phytochemicals with sphingosine kinase 1: Towards targeted anticancer therapy. Sci Rep. 2019;9:18727. doi: 10.1038/s41598-019-55199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan P., Rahman S., Queen A., Manzoor S., Naz F., Hasan G.M., Luqman S., Kim J., Islam A., Ahmad F., et al. Elucidation of Dietary Polyphenolics as Potential Inhibitor of Microtubule Affinity Regulating Kinase 4: In silico and In vitro Studies. Sci. Rep. 2017;7:9470. doi: 10.1038/s41598-017-09941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sack J.S., Gao M., Kiefer S.E., Myers J.E., Jr., Newitt J.A., Wu S., Yan C. Crystal structure of microtubule affinity-regulating kinase 4 catalytic domain in complex with a pyrazolopyrimidine inhibitor. Acta Cryst. F Struct. Biol. Commun. 2016;72:129–134. doi: 10.1107/S2053230X15024747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naqvi A.A.T., Jairajpuri D.S., Noman O.M.A., Hussain A., Islam A., Ahmad F., Alajmi M.F., Hassan M.I. Evaluation of pyrazolopyrimidine derivatives as microtubule affinity regulating kinase 4 inhibitors: Towards therapeutic management of Alzheimer’s disease. J. Biomol. Struct. Dyn. 2019:1–16. doi: 10.1080/07391102.2019.1666745. [DOI] [PubMed] [Google Scholar]
- 50.Parveen I., Khan P., Ali S., Hassan M.I., Ahmed N. Synthesis, molecular docking and inhibition studies of novel 3-N-aryl substituted-2-heteroarylchromones targeting microtubule affinity regulating kinase 4 inhibitors. Eur. J. Med. Chem. 2018;159:166–177. doi: 10.1016/j.ejmech.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 51.Albani J.R. Structure and Dynamics of Macromolecules: Absorption and Fluorescence Studies. Elsevier; Amsterdam, The Netherlands: 2011. [Google Scholar]
- 52.Khan S.N., Islam B., Yennamalli R., Sultan A., Subbarao N., Khan A.U. Interaction of mitoxantrone with human serum albumin: Spectroscopic and molecular modeling studies. Eur. J. Pharm. Sci. 2008;35:371–382. doi: 10.1016/j.ejps.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Mariam J., Dongre P., Kothari D. Study of interaction of silver nanoparticles with bovine serum albumin using fluorescence spectroscopy. J. Fluoresc. 2011;21:2193. doi: 10.1007/s10895-011-0922-3. [DOI] [PubMed] [Google Scholar]
- 54.Van de Weert M., Stella L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J. Mol. Struct. 2011;998:144–150. doi: 10.1016/j.molstruc.2011.05.023. [DOI] [Google Scholar]
- 55.Anand U., Jash C., Boddepalli R.K., Shrivastava A., Mukherjee S. Exploring the mechanism of fluorescence quenching in proteins induced by tetracycline. J. Phys. Chem. B. 2011;115:6312–6320. doi: 10.1021/jp2008978. [DOI] [PubMed] [Google Scholar]
- 56.Rehman M.T., Shamsi H., Khan A.U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharm. 2014;11:1785–1797. doi: 10.1021/mp500116c. [DOI] [PubMed] [Google Scholar]
- 57.Kang J., Liu Y., Xie M.-X., Li S., Jiang M., Wang Y.-D. Interactions of human serum albumin with chlorogenic acid and ferulic acid. Biochim. Biophys. Acta (BBA) Gen. Subj. 2004;1674:205–214. doi: 10.1016/j.bbagen.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 58.Shamsi A., Ahmed A., Bano B. Probing the interaction of anticancer drug temsirolimus with human serum albumin: Molecular docking and spectroscopic insight. J. Biomol. Struct. Dyn. 2018;36:1479–1489. doi: 10.1080/07391102.2017.1326320. [DOI] [PubMed] [Google Scholar]
- 59.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 60.Cahyana Y., Gordon M.H. Interaction of anthocyanins with human serum albumin: Influence of pH and chemical structure on binding. Food Chem. 2013;141:2278–2285. doi: 10.1016/j.foodchem.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe H., Tanase S., Nakajou K., Maruyama T., Kragh-Hansen U., Otagiri M. Role of Arg-410 and Tyr-411 in human serum albumin for ligand binding and esterase-like activity. Biochem. J. 2000;349:813–819. doi: 10.1042/bj3490813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aneja B., Queen A., Khan P., Shamsi F., Hussain A., Hasan P., Rizvi M.M.A., Daniliuc C.G., Alajmi M.F., Mohsin M., et al. Design, synthesis & biological evaluation of ferulic acid-based small molecule inhibitors against tumor-associated carbonic anhydrase IX. Bioorg. Med. Chem. 2020:115424. doi: 10.1016/j.bmc.2020.115424. [DOI] [PubMed] [Google Scholar]
- 63.Zaidi N., Ajmal M.R., Rabbani G., Ahmad E., Khan R.H. A comprehensive insight into binding of hippuric acid to human serum albumin: A study to uncover its impaired elimination through hemodialysis. PLoS ONE. 2013;8:e71422. doi: 10.1371/journal.pone.0071422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dahiya R., Mohammad T., Roy S., Anwar S., Gupta P., Haque A., Khan P., Kazim S.N., Islam A., Ahmad F., et al. Investigation of inhibitory potential of quercetin to the pyruvate dehydrogenase kinase 3: Towards implications in anticancer therapy. Int. J. Biol. Macromol. 2019;136:1076–1085. doi: 10.1016/j.ijbiomac.2019.06.158. [DOI] [PubMed] [Google Scholar]
- 65.Gupta P., Khan F.I., Roy S., Anwar S., Dahiya R., Alajmi M.F., Hussain A., Rehman M.T., Lai D., Hassan M.I. Functional implications of pH-induced conformational changes in the Sphingosine kinase 1. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020;225:117453. doi: 10.1016/j.saa.2019.117453. [DOI] [PubMed] [Google Scholar]
- 66.Aneja B., Khan N.S., Khan P., Queen A., Hussain A., Rehman M.T., Alajmi M.F., El-Seedi H.R., Ali S., Hassan M.I., et al. Design and development of Isatin-triazole hydrazones as potential inhibitors of microtubule affinity-regulating kinase 4 for the therapeutic management of cell proliferation and metastasis. Eur. J. Med. Chem. 2019;163:840–852. doi: 10.1016/j.ejmech.2018.12.026. [DOI] [PubMed] [Google Scholar]
- 67.Bioquest, A. Inc., Quest Graph™ IC50 Calculator. [(accessed on 23 April 2020)];2019 Available online: https://www.aatbio.com/tools/ic50-calculator20.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.