Abstract
Z-DNA and Z-RNA are left-handed double helix nucleic acid structures with poorly understood biological function1–3. Z-DNA binding protein 1 (ZBP1, also known as DAI or DLM-1) is a nucleic acid sensor containing two Zα domains that bind Z-DNA4,5 and Z-RNA6–8. ZBP1 mediates host-defence against certain viruses6,7,9–14 by sensing viral nucleic acids6,7,10. RIPK1 deficiency or mutation of its RIP homotypic interaction motif (RHIM) triggers ZBP1-dependent necroptosis and inflammation in mice15,16, however, the mechanisms inducing ZBP1 activation in the absence of viral infection remain elusive. Here we show that Zα-dependent sensing of endogenous ligands induces ZBP1-mediated perinatal lethality in mice expressing RIPK1 with mutated RHIM (Ripk1mR/mR) and skin inflammation in mice with epidermis-specific RIPK1 deficiency (RIPK1E-KO), as well as colitis in mice with intestinal epithelial-specific FADD deficiency (FADDIEC-KO). Consistently, functional Zα domains were required for ZBP1-induced necroptosis in fibroblasts that express RIPK1 with mutated RHIM or were treated with caspase inhibitors. Moreover, inhibition of nuclear export triggered Zα-dependent activation of RIPK3 in the nucleus resulting in cell death, suggesting that ZBP1 may recognise Z-form nucleic acids (Z-NA) in the nucleus. We found that ZBP1 constitutively bound cellular double stranded RNA (dsRNA) in a Zα-dependent manner. Furthermore, endogenous retroelement (ERE)-derived complementary reads were detected in epidermal RNA, suggesting that ERE-derived dsRNA may act as Zα domain ligand triggering ZBP1 activation. Collectively, our results provide evidence that sensing of endogenous Z-NA by ZBP1 triggers RIPK3-dependent necroptosis and inflammation, which could underlie the development of chronic inflammatory conditions particularly in patients with mutations in the RIPK1 and CASPASE-8 genes17–20.
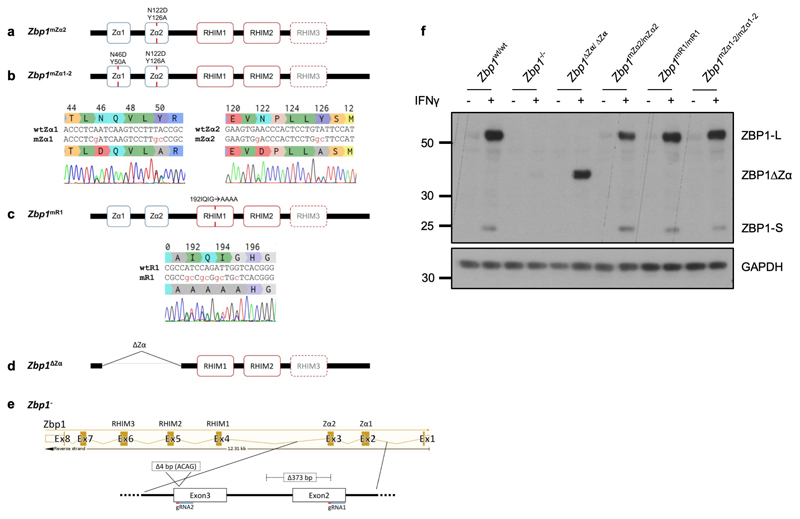
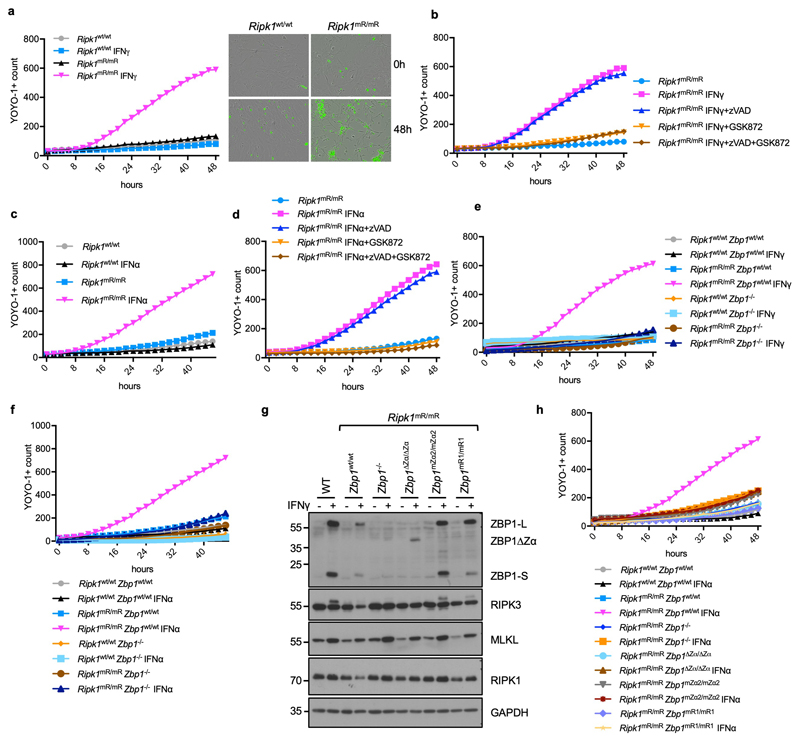
To study the physiological role of Z-NA sensing by ZBP1, we generated knock-in mice expressing ZBP1 with deletion (Zbp1∆Zα/∆Zα) or mutation of both Zα domains (Zbp1mZα1-2/mZα1-2) or Zα2 alone (Zbp1mZα2/mZα2), substituting key amino acids (Zα1: N46D, Y50A; Zα2: N122D, Y126A) that are essential for Z-NA binding4 (Extended Data Fig. 1). Moreover, to assess the role of RHIM1, which was shown to mediate ZBP1 signalling9,10,21,22, we generated Zbp1mR1/mR1 mice expressing ZBP1 with four amino acid substitutions disrupting its core RHIM sequence (192IQIG to 192AAAA) (Extended Data Fig. 1). These mutant ZBP1 mouse strains were viable and did not show any apparent abnormalities. Immunoblot analysis of IFNγ-stimulated lung fibroblasts (LFs) confirmed the expression of both full length (ZBP1-L) and the short form of ZBP1 (ZBP1-S), which is produced by alternative splicing and contains the Zα domains but lacks the C-terminal part containing the RHIMs, in Zbp1mZα2/mZα2, Zbp1mZα1-2/mZα1-2 and Zbp1mR1/mR1 mice, as well as the truncated ZBP1∆Zα protein in Zbp1∆Zα/∆Zα mice (Extended Data Fig. 1).
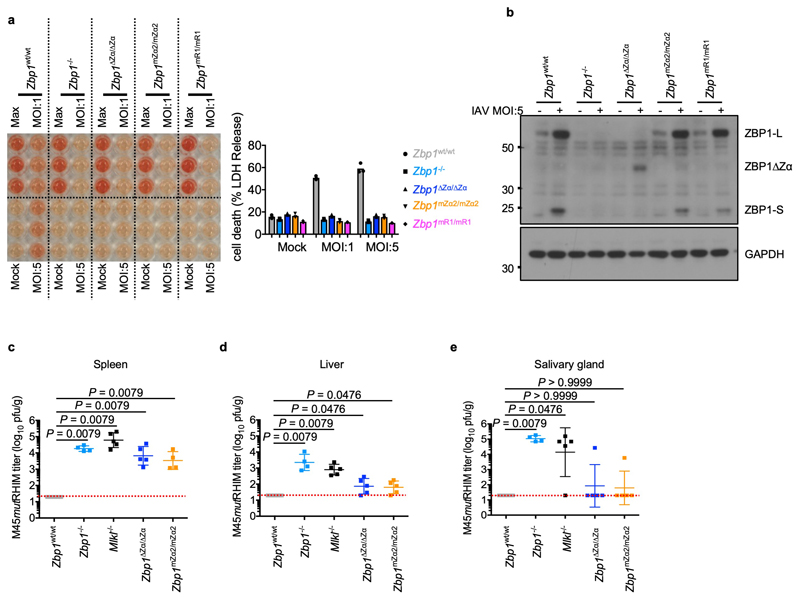
Previous studies showed that Zα-dependent ZBP1 signalling triggers cell death in response to infection with influenza A virus (IAV) or murine cytomegalovirus expressing M45 protein with mutation of the RHIM domain (MCMV-M45mutRHIM)6,7,10. Consistently, IAV-induced cell death was abrogated in LFs from Zbp1-/-, Zbp1∆Zα/∆Zα, Zbp1mZα2/mZα2 and Zbp1mR1/mR1 mice (Extended Data Fig. 2a, b). Furthermore, in agreement with earlier studies6,13, MCMV-M45mutRHIM could establish productive infection in Zbp1∆Zα/∆Zα, Zbp1mZα2/mZα2, Zbp1-/- and Mlkl-/- but not in wild type (WT) mice (Extended Data Fig. 2c, d). However, most Zbp1∆Zα/∆Zα and Zbp1mZα2/mZα2 mice displayed lower viral titres in spleen and liver five days after infection and did not contain detectable live virus in salivary gland after 14 days, suggesting that they could control the virus better compared to Zbp1-/- and Mlkl-/- mice (Extended Data Fig. 2c-e). Therefore, Zα-dependent ZBP1 activation is important for restricting MCMV-M45mutRHIM early during infection, however, Zα-independent functions contribute to the full clearance of the virus during later stages.
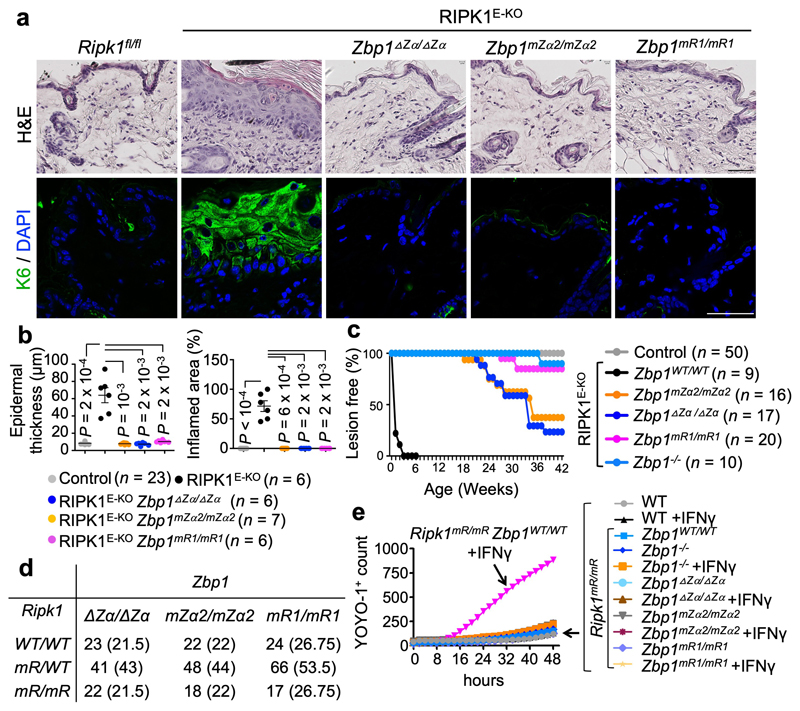
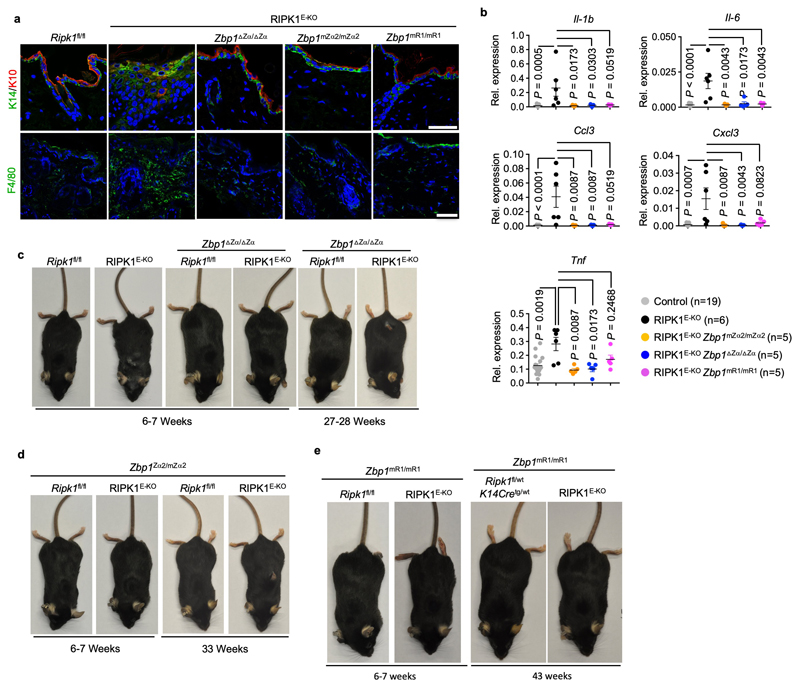
To address whether Zα-dependent sensing of endogenous ligands is required for ZBP1-mediated responses in the absence of virus infection, we crossed Zbp1∆Zα/∆Zα and Zbp1mZα2/mZα2 with RIPK1E-KO mice, which develop skin inflammation due to ZBP1-RIPK3-MLKL-dependent keratinocyte necroptosis16,23. In contrast to RIPK1E-KO mice, which developed inflammatory skin lesions with marked epidermal hyperplasia, upregulation of keratin 6 (K6) and K10, myeloid cell infiltration and upregulation of inflammatory cytokines and chemokines, and needed to be culled by 4-7 weeks of age, RIPK1E-KO Zbp1∆Zα/∆Zα and RIPK1E-KO Zbp1mZα2/mZα2 mice remained healthy at this age (Fig. 1a-c, and Extended Data Fig. 3a-d). RIPK1E-KO Zbp1mR1/mR1 mice also did not develop skin inflammation (Fig. 1a-c and Extended Data Fig. 3a, b, e) revealing an essential role for RHIM1. However, most RIPK1E-KO Zbp1∆Zα/∆Zα and RIPK1E-KO Zbp1mZα2/mZα2 mice developed mild skin lesions between 18 - 42 weeks (Fig. 1c and Extended Data Fig. 3c, d), in contrast to RIPK1E-KO Zbp1mR1/mR1 but also RIPK1E-KO Zbp1-/- mice, which were produced using a newly generated C57Bl/6N Zbp1-/- strain to fully match the genetic background of the ZBP1 mutants (Extended Data Fig. 1), which remained healthy with only 15% of these animals showing very mild skin lesions at the age of 40 weeks (Fig. 1c and Extended Data Fig. 3e). Therefore, whereas Zα domain-dependent sensing of endogenous ligands triggers ZBP1-mediated keratinocyte necroptosis and skin inflammation in young RIPK1E-KO mice, ZBP1 also exhibits Zα-independent functions that contribute to skin lesion development in older animals, consistent with our findings in MCMV-M45mutRHIM infection.
Figure 1. ZBP1 causes skin inflammation in RIPK1E-KO mice and perinatal lethality in Ripk1mR/mR mice by Zα-dependent and -independent mechanisms.
a, Representative images of skin sections from 6 - 7 week-old mice with the indicated genotypes stained with haematoxylin & eosin (H&E) or anti-keratin 6 (K6) antibodies and DAPI (DNA stain). RIPK1E-KO, n=6 for H&E and n≥5 for K6; RIPK1E-KO Zbp1mZα2/mZα2, n=9 for H&E and n≥3 for K6; RIPK1E-KO Zbp1∆Zα/∆Zα, n=15 for H&E and n≥3 for K6; RIPK1E-KO Zbp1mR1/mR1, n=10 for H&E and n≥3 for K6. Scale bars, 50 μm. b, Graphs depicting microscopic quantification of mean epidermal thickness (Epi. th.) and inflamed skin area in 6 - 7 week-old mice. Dots represent individual mice. Mean ± s.e.m are shown. P value by two-sided nonparametric Mann–Whitney test. c, Kaplan-Meier plot depicting lesion-free survival of mice with the indicated genotypes. Control mice include Ripk1FL/FL mice that do not express K14-Cre, or Ripk1FL/WT K14-Cre mice, with WT or mutated Zbp1 alleles. d, Table showing the numbers of weaned offspring from Ripk1mR/wt parents that were homozygous for the respective Zbp1 alleles. Expected numbers for mendelian inheritance are shown in brackets. e, Cell death measured by YOYO-1 uptake in MEFs with the indicated genotypes treated with IFNγ (1,000 u ml−1). Graph shows mean values from technical triplicates (n = 3) from one representative out of three independent experiments.
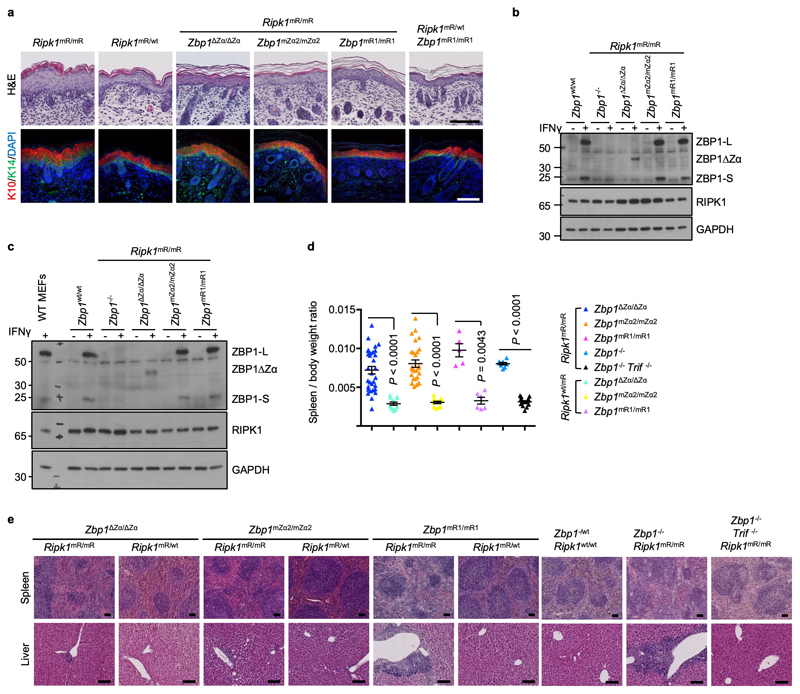
To address whether perinatal lethality of Ripk1mR/mR mice15,16 requires Zα-dependent sensing of endogenous ligands and RHIM1-dependent signalling, we crossed them with Zbp1∆Zα/∆Zα, Zbp1mZα2/mZα2 and Zbp1mR1/mR1 mice. Ripk1mR/mR Zbp1∆Zα/∆Zα, Ripk1mR/mR Zbp1mZα2/mZα2 and Ripk1mR/mR Zbp1mR1/mR1 mice were viable (Fig. 1d), did not develop epidermal hyperplasia (Extended Data Fig. 4a), and reached adulthood without showing macroscopic signs of pathology at least up to the age of one year. Immunoblot analysis of IFNγ-stimulated LFs and keratinocytes confirmed that the mutant ZBP1 proteins were expressed similarly to WT ZBP1, with the exception of the truncated ZBP1∆Zα that was detected at lower levels indicating that it may exhibit reduced stability (Extended Data Fig. 4b,c). Ripk1mR/mR Zbp1∆Zα/∆Zα, Ripk1mR/mR Zbp1mZα2/mZα2, Ripk1mR/mR Zbp1mR1/mR1 as well as Ripk1mR/mR Zbp1-/- mice examined at the age of 6 months or older showed splenomegaly and mild inflammatory infiltrates in the liver, which depended on TRIF as they were not observed in triple mutant Ripk1mR/mR Zbp1-/- Trif-/- mice (Extended Data Fig. 4d,e). Consistent with our in vivo findings, IFNγ or IFNα stimulation induced ZBP1-mediated necroptosis in mouse embryonic fibroblasts (MEFs) from Ripk1mR/mR but not WT mice (Fig. 1e, Extended Data Fig. 5a-f), which depended on intact Zα domains and RHIM1 (Fig. 1e and Extended Data Fig. 5g,h). Collectively, Zα-dependent sensing of endogenous ligands and RHIM1-mediated signalling by ZBP1 causes perinatal lethality in Ripk1mR/mR mice and IFN-induced necroptosis in Ripk1mR/mR cells.
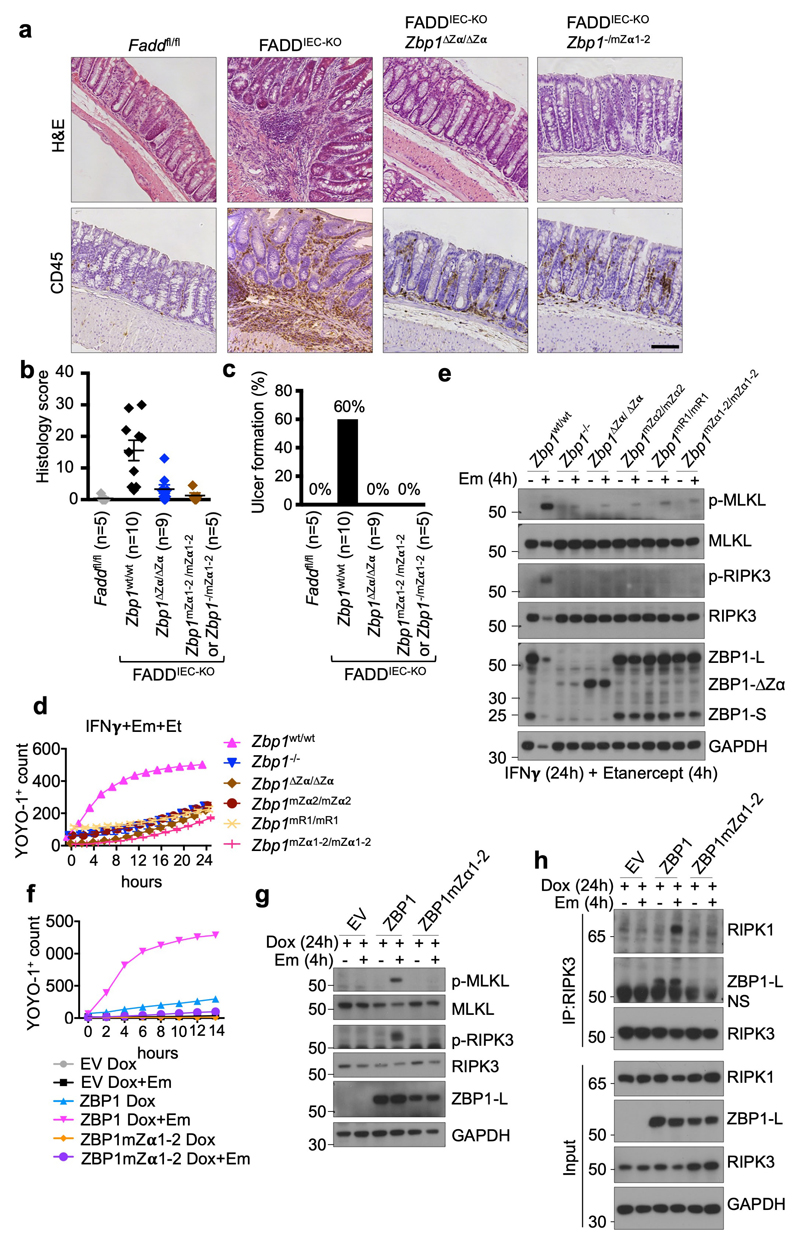
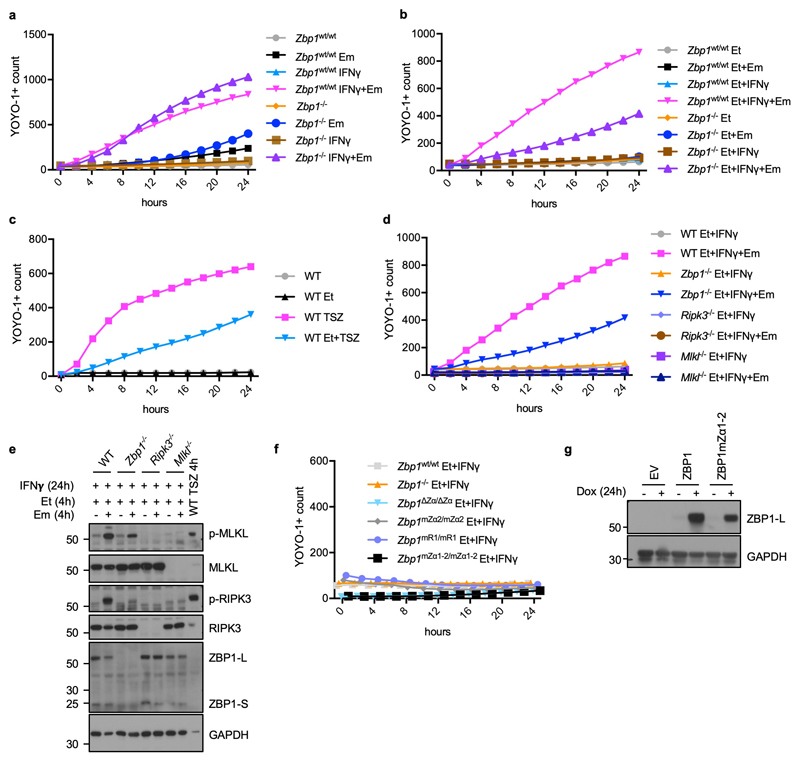
To address whether sensing of endogenous Zα domain ligands by ZBP1 also triggers necroptosis and inflammation also in the presence of intact RIPK1, we assessed the potential role of Zα-dependent ZBP1 activation in mice with intestinal epithelial cell (IEC)-specific knockout of FADD (FADDIEC-KO mice), which develop colitis due to RIPK3-dependent IEC necroptosis24. Indeed, deletion or mutation of the two Zα domains inhibited colitis development in FADDIEC-KO mice, as shown by strongly reduced immune cell infiltration, absence of ulcer formation and an overall normal colon tissue architecture in FADDIEC-KO Zbp1∆Zα/∆Zα and FADDIEC-KO Zbp1mZα1-2/mZα1-2 mice (Fig. 2a-c). These results suggested that inhibition of FADD-caspase-8 signalling sensitizes cells to Zα-dependent ZBP1-mediated necroptosis. To assess this in a cellular system, we stimulated WT or ZBP1-deficient MEFs with IFNγ in the presence of the caspase inhibitor emricasan (IDN-6556) and found that caspase inhibition sensitized cells to IFNγ-induced cell death, which, however, was not affected by ZBP1-deficiency (Extended Data Fig. 6a). We reasoned that autocrine TNF signalling could contribute to IFNγ-induced cell death and repeated this experiment in the presence of the TNF inhibitor etanercept. Indeed, ZBP1-deficient cells were strongly protected from IFNγ+emricasan-induced death when autocrine TNF signalling was inhibited (Fig 2d and Extended Data Fig. 6b). The residual cell death observed in ZBP1-deficient MEFs was likely due to incomplete TNF neutralisation, as similar etanercept treatment did not fully prevent TNF-induced necroptosis (Extended Data Fig. 6c). IFNγ+emricasan+etanercept (IFNγ+Em+Et)-induced cell death correlated with RIPK3 and MLKL phosphorylation and was prevented by RIPK3 or MLKL deficiency, showing that this treatment induced necroptosis (Fig. 2e and Extended Data Fig. 6d,e). LFs from Zbp1∆Zα/∆Zα, Zbp1mZα1-2/mZα1-2, Zbp1mZα2/mZα2 and Zbp1mR1/mR1 mice were protected from IFNγ+Em+Et-induced activation of RIPK3 and MLKL and cell death (Fig. 2d, e and Extended Data Fig. 6f), showing that Zα-dependent sensing of endogenous ligands triggers ZBP1-RIPK3-MLKL-mediated necroptosis when caspases are inhibited. Furthermore, doxycycline-induced expression of WT ZBP1 but not ZBP1mZα1-2 in immortalised MEFs (iMEFs) induced strong phosphorylation of RIPK3 and MLKL and cell death in the presence of emricasan (Fig. 2f, g and Extended Data Fig. 6g), demonstrating that ZBP1 expression in the absence of IFN stimulation was sufficient to trigger necroptosis in a Zα-dependent manner when caspase-8 was inhibited. These findings implied that caspase-8 might negatively regulate ZBP1-mediated RIPK3 activation by cleaving components of the ZBP1-RIPK3 complex, as suggested by studies in L929 cells25, similarly to its function in inhibiting TNF-induced necroptosis. We reasoned that RHIM-dependent recruitment of RIPK1 could inhibit activation of the ZBP1-RIPK3 complex by recruiting caspase-8 via FADD and examined whether RIPK1 is recruited to the ZBP1-RIPK3 complex in the presence of emricasan. Indeed, we found that RIPK1 co-immunoprecipitated with RIPK3 in cells overexpressing ZBP1 only in the presence of emricasan (Fig. 2h). Moreover, ZBP1mZα1-2 did not interact with RIPK3 (Fig. 2h), suggesting that Z-NA sensing promotes RHIM-dependent binding of ZBP1 to RIPK3. Collectively, our studies showed that Zα-dependent sensing of endogenous ligands triggers ZBP1-dependent activation of RIPK3-MLKL-mediated necroptosis when caspase-8 is inhibited, a mechanism that critically contributes to colitis development in FADDIEC-KO mice.
Figure 2. Zα-dependent Z-NA sensing induces ZBP1-mediated necroptosis and colitis when FADD-caspase-8 function is inhibited.
a-c, Representative images of colon sections stained with H&E or immunostained for CD45 (a), and graphs depicting histological colitis score (b) and incidence of ulcer formation (c) from 10 week-old mice with the indicated genotypes. Scale bar = 100μm. Dots represent individual mice. Mean ± s.e.m are shown. P value by two-sided nonparametric Mann–Whitney test. d, e, Cell death measured by YOYO-1 uptake (d) and immunoblot analysis of total lysates (e) in LFs with the indicated genotypes treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), etanercept (50 μg ml−1) and emricasan (5 μM). f-h, Cell death measured by YOYO-1 uptake (f), and immunoblot analysis of total lysates (g) and anti-RIPK3 immunoprecipitates (f) in iMEFs expressing doxycycline (Dox)-induced Flag (EV), Flag-tagged ZBP1 or Flag-tagged ZBP1mZα1-2 stimulated with combinations of Dox (1 μg ml−1) (24 h pre-treatment) and emricasan (5 μM). Data are representative of two (e, g, h), four (d) and three (f) independent experiments. Graphs in d and f show mean from technical triplicates (n = 3). GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.
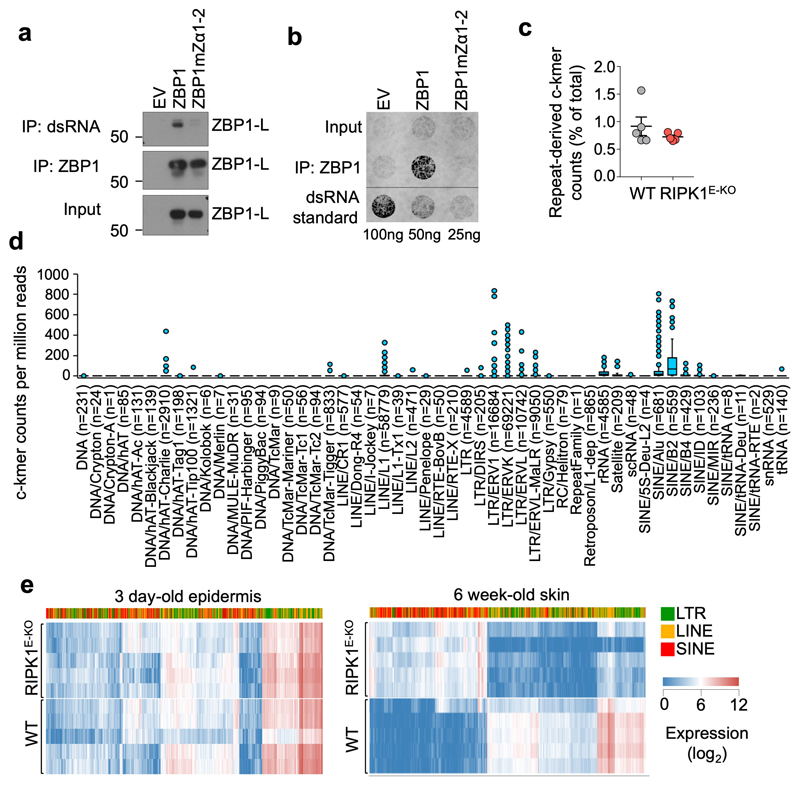
Previous studies suggested that ZBP1 senses viral but also endogenous RNA through its Zα domains6,7. To assess whether ZBP1 binds cellular dsRNA via its Zα domains we immunoprecipitated dsRNA from iMEFs expressing WT ZBP1 or ZBP1mZa1-2 using the dsRNA-specific J2 antibody, followed by immunoblotting with anti-ZBP1 antibodies. We found that WT ZBP1, but not ZBP1mZa1-2, co-immunoprecipitated with dsRNA (Fig. 3a), suggesting that ZBP1 binds to dsRNA in a Zα-dependent manner. To confirm this interaction, we immunoprecipitated ZBP1 from the same cells and assessed the presence of dsRNA in the immunoprecipitates using the J2 antibody on a dot-blot assay. As shown in Fig. 3b, a strong signal for dsRNA was detected in immunoprecipitates from cells expressing WT ZBP1 but not ZBP1mZa1-2. Together, these experiments showed that ZBP1 binds to endogenous dsRNA via its Zα domains, implicating dsRNA as a putative ligand triggering ZBP1-mediated necroptosis and inflammation in our mouse models.
Figure 3. The ZBP1 Zα domains bind cellular dsRNA likely derived from endogenous retroelements.
a, b, Immunoblot analysis of J2 anti-dsRNA mAb immunoprecipitates, anti-ZBP1 immunoprecipitates and total lysates (input) (a) and dot blot analysis performed with J2 anti-dsRNA mAb on RNA isolated from anti-ZBP1 immunoprecipitates or total cell lysates (input) (b) from iMEFs expressing doxycycline-inducible Flag (EV), Flag-tagged ZBP1 or Flag-tagged ZBP1mutZα1-2 stimulated with Dox (1 μg ml−1) for 24 h. c, Proportion of repeat-derived complementary kmer (c-kmer) counts in total kmer counts in stranded RNA-seq data from the epidermis of 3 day-old WT (n=5) and RIPK1E-KO mice (n=5). Dots represent individual mice. Mean ± s.e.m are shown. d, Box plots of repeat-derived c-kmer counts, normalised by sequencing depth, according to the repeat class they associate with. Counts represent the mean value of all WT (n=5) and RIPK1E-KO mice (n=5) from c. Counts of the least abundant c-kmer of each complementary pair are shown. Numbers in brackets denote the total number of unique c-kmers identified for each repeat class. Box plots show the upper and lower quartiles, centre lines show the median, whiskers represent the 1.5x interquartile range and individual points represent outliers. e, Heatmap showing expression of EREs in the epidermis of 3 day-old or skin of 6 week-old WT and RIPK1E-KO mice. Only EREs that were differentially expressed by ≥10-fold (q≤0.01) between 6 week-old WT and RIPK1E-KO mice are shown. Each row represents an individual mouse. Data are representative of four (a) and two (b) independent experiments. For gel source data, see Supplementary Fig. 1.
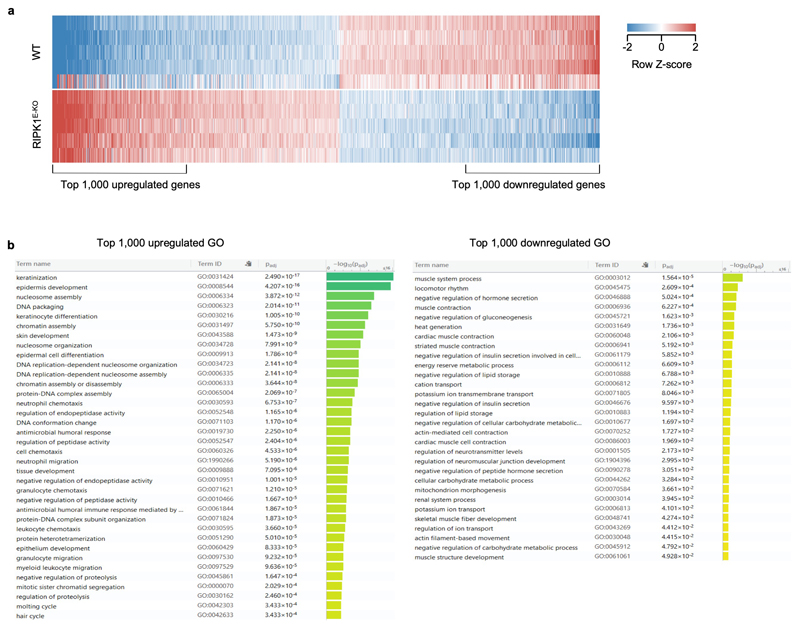
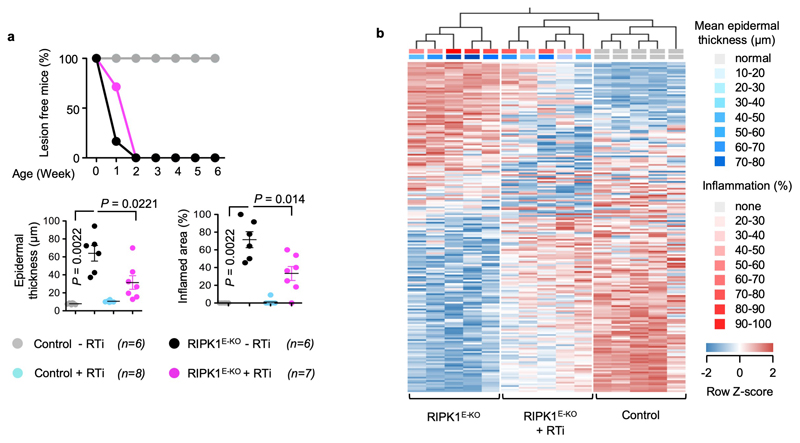
The ZBP1 Zα domains are structurally similar to the Zα domain of ADAR15, which was shown to bind Z-RNA8. ADAR1 edits dsRNA from endogenous retroelements (EREs), particularly from inverted Alu repeats of the short interspersed nuclear elements (SINE) class, thus preventing their recognition by MDA5 and activation of interferon signalling26. An ADAR1 mutation predicted to disrupt Zα domain binding to Z-NA (P193A) found in patients with Aicardi-Goutières Syndrome was shown to decrease ADAR1 editing efficiency27, suggesting that Zα-dependent binding of ADAR1 to ERE-derived Z-RNAs facilitates their editing. ERE-derived Z-RNA could therefore provide a ligand for the ZBP1 Zα domains. To test whether ERE-derived dsRNA could be present in epidermal keratinocytes in vivo, we examined the complementarity of reads in stranded RNA-seq data from the epidermis of 3 day-old WT or RIPK1E-KO mice, before the onset of the inflammatory skin lesions. Whilst not all complementary RNA reads in the RNA-seq libraries would necessarily have been double-stranded, any dsRNA present in the samples at the time of isolation should have both strands represented in the stranded libraries. Complementarity was calculated using a kmer-based method comparing counts of repeat-derived kmers and their reverse complements. Repeat-derived complementary kmer (c-kmer) counts amassed to ~0.8% of all kmer counts in both WT and RIPK1E-KO keratinocytes (Fig. 3c). Indicative of overall expression, the highest counts of uniquely mapped c-kmers were observed for B2 and Alu SINEs, followed by long interspersed nuclear elements (LINE)/L1 and long terminal repeat (LTR) elements, with these ERE groups accounting for ~87% of all repeat-mapped c-kmer counts (Fig. 3d). Although ERE expression was comparable between WT and RIPK1E-KO mice before the onset of inflammation, skin biopsies from RIPK1E-KO mice at 6 weeks of age exhibited marked deregulation of ERE expression compared to WT littermates, with strong upregulation of SINEs and downregulation of LTR elements (Fig. 3e). This was accompanied by extensive changes in the expression of genes involved in inflammation, myeloid cell migration and epidermal development (Extended Data Fig. 7a, b), consistent with the inflammatory hyperplastic skin lesions observed at this age. Therefore, EREs, and particularly B2 and Alu SINEs, are expressed in healthy epidermis and show the highest potential to form dsRNA, which could act as Zα domain ligands inducing ZBP1-mediated necroptosis and skin inflammation in RIPK1E-KO mice. ERE deregulation could also contribute to the upregulation of ZBP1 expression observed in the epidermis of RIPK1E-KO mice16 by activating MDA5-MAVS and/or cGAS-STING signalling. Moreover, the Zα domain also binds DNA-RNA hybrids28, which could conceivably be generated during reverse transcription of EREs providing additional ligands for ZBP1. Consistent with this notion, treatment of RIPK1E-KO mice with a combination of reverse transcriptase inhibitors (RTi) partially ameliorated the skin lesions (Extended Data Fig. 8a,b), supporting that EREs likely contribute to skin inflammation in RIPK1E-KO mice.
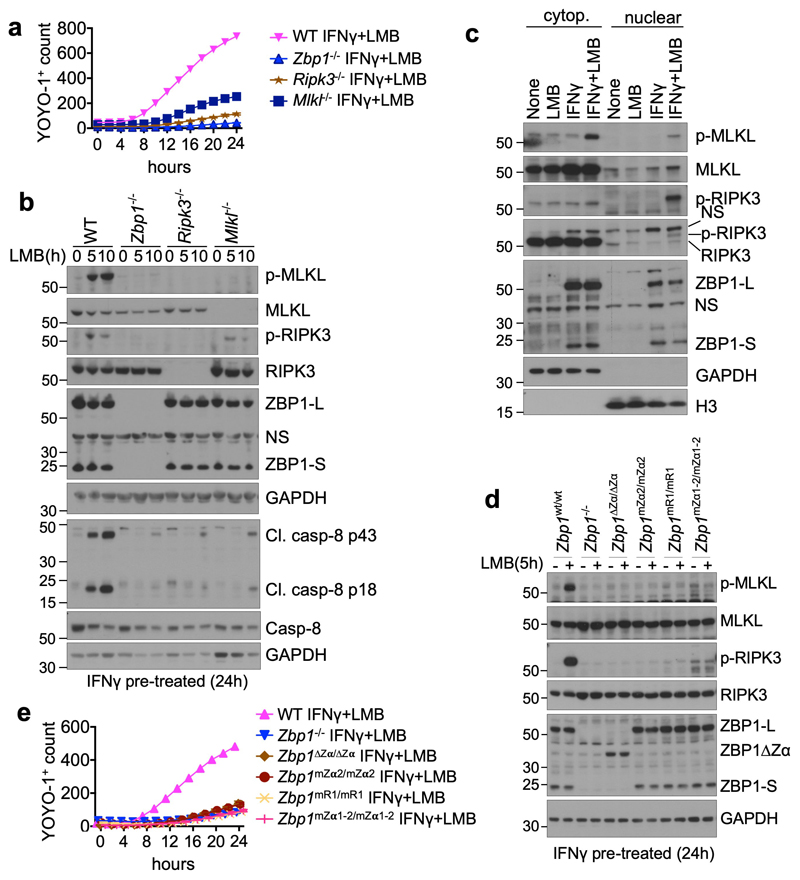
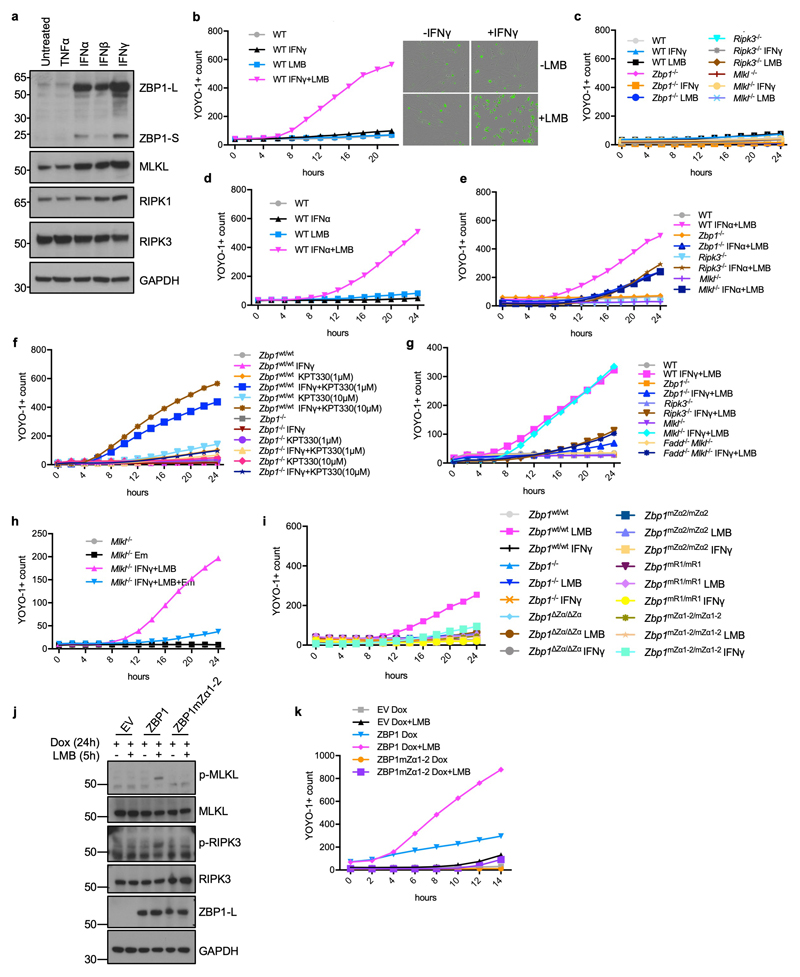
Transcription of viral genomes in the nucleus was suggested to trigger ZBP1-mediated cell death6,9. To assess whether sensing of endogenous Z-NA may trigger ZBP1 activation in the nucleus we pre-stimulated WT MEFs with interferons for 24 hours to induce the expression of ZBP1 (Extended Data Fig. 9a), followed by treatment with leptomycin B (LMB) to inhibit nuclear export. LMB treatment of IFNγ- or IFNα-pre-stimulated MEFs caused ZBP1- and RIPK3-dependent cell death, which was partially inhibited by MLKL deficiency (Fig. 4a and Extended Data Fig. 9b-e). Treatment of IFNγ pre-stimulated cells with KPT330, another nuclear export inhibitor29, also induced cell death in WT but not Zbp1-/- LFs (Extended Data Fig. 9f). Immunoblot analysis of protein extracts from MEFs pre-stimulated with IFNγ and treated with LMB for 5 and 10 hours revealed phosphorylation of RIPK3 and MLKL but also cleavage of caspase-8 in WT but not in Zbp1-/- MEFs (Fig. 4b). Emricasan treatment or FADD deficiency prevented IFN+LMB-induced death of MLKL-deficient cells, showing that this treatment induces both apoptosis and necroptosis (Extended Data Fig. 9g, h). Immunoblot analysis of cytoplasmic and nuclear extracts from IFNγ pre-stimulated cells that were treated for 5 hours with LMB revealed that most of ZBP1, RIPK3 and MLKL were found in the cytosolic fraction (Fig. 4c). Only small amounts of these proteins were detected in the nucleus, which did not increase further by LMB treatment suggesting that blocking nuclear export did not induce their nuclear accumulation (Fig. 4c). However, we detected strong phosphorylation of RIPK3 and more weakly of MLKL in the nucleus of IFNγ+LMB-treated cells, suggesting that ZBP1 activates RIPK3 in the nucleus (Fig. 4c). Zα-dependent sensing of endogenous ligands was required for ZBP1 activation, as IFNγ+LMB-induced phosphorylation of RIPK3 and MLKL and cell death was prevented in Zbp1∆Zα/∆Zα, Zbp1mZα2/mZα2 and Zbp1mZα1-2/mZα1-2, but also in Zbp1mR1/mR1 cells (Fig. 4d, e and Extended Data Fig. 9i). Furthermore, LMB treatment induced RIPK3 and MLKL phosphorylation and cell death in cells expressing doxycycline-inducible WT ZBP1 but not ZBP1mZα1-2 (Extended Data Fig. 9j,k). Taken together, these results showed that ZBP1 senses endogenous Z-NA and activates RIPK3 in the nucleus inducing cell death. While the precise mechanism by which LMB treatment triggers ZBP1 activation remains unclear, it is possible that nuclear export blockade causes accumulation of Zα ligands, such as ERE-derived dsRNA, in the nucleus. Alternatively, nuclear export inhibition may protect the ZBP1-RIPK3 complex from the negative regulatory function of the cytosolic RIPK1-FADD-Caspase-8 machinery.
Figure 4. Inhibition of nuclear export triggers Zα-dependent ZBP1-mediated cell death.
a, e, Cell death analysed by YOYO-1 uptake in MEFs (a) or LFs (e) from mice with the indicated genotypes treated with IFNγ (1,000 u ml−1) (24 h pre-treatment) and LMB (1 ng ml−1 for MEFs and 5 ng ml−1 for LFs). b, c, d, Immunoblot analysis of total lysates from MEFs (b) or LFs (d) with the indicated genotypes and of nuclear and cytosolic extracts from WT MEFs (c) treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment) and LMB (1 ng ml−1 for MEFs and 5 ng ml−1 for LFs). Data information: data are representative of four (a), two (b, d) or five (c, e) independent experiments. Graphs in a and e show mean of technical triplicates (n = 3). GAPDH and histone 3 (H3) were used as loading controls and cytosolic and nuclear fraction markers respectively for immunoblot analysis. For gel source data, see Supplementary Fig. 1.
Although the existence of Z-DNA and Z-RNA has been known for many years, their physiological role has remained poorly understood1–3. This is largely because of the lack of reliable tools allowing the detection and functional analysis of Z-NA in living cells and organisms. Studying the function of the Zα protein domains that sense them is currently the only means to interrogate the functional roles of Z-NA. Here we provide experimental evidence supporting that sensing of endogenous Z-NA via its Zα domains activates ZBP1-mediated cell death and inflammation. Whilst the exact nature of the endogenous Z-NA that are sensed by ZBP1 remains elusive at present, our results suggest that ERE-derived dsRNA is the most likely ligand triggering ZBP1 activation. Together, our findings support a model whereby ZBP1 constitutively binds cellular z-RNA via its Zα domains, but RIPK3 activation is normally inhibited by the RIPK1-FADD-dependent recruitment of caspase-8 that cleaves components of the complex such as RIPK1 and RIPK3 (Extended Data Fig. 10). Zα-mediated activation of ZBP1 signalling may therefore contribute to the pathogenesis of autoinflammatory conditions in patients with mutations in the RIPK1 and CASPASE-8 genes, who show no or partial response to anti-TNF treatment17–20. Collectively, our studies suggest that Zα-dependent sensing of endogenous Z-NA triggers ZBP1-mediated cell death and inflammation, which could be relevant for the pathogenesis of inflammatory diseases.
Methods
Mice
Ripk1fl/fl (23) and Faddfl/fl (24), K14-cre 30, Triffl/fl (23). Zbp1 mutant mice were generated in C57BL6/N genetic background as previously described16,31. For Zbp1ΔZα mice a short-guide RNA (sgRNA) (5’GAACGACGACACCACCCAG3’) upstream of Zα1-encoding exon2 and a sgRNA (5’ATCAAGATTAGGTCACCTA3’) downstream of Zα2-encoding exon3 were in vitro transcribed and co-injected into fertilized wt oocytes together with Cas9 protein and mRNA. For Zbp1mZα2 mice, a sgRNA (5’ TTCTCATGGAATACAGGAGT 3’) targeting the Zα domain-encoding exon 3 was co-injected into fertilized wt oocytes with Cas9 protein and mRNA as well as a long ssODN (5’GACGTGAGTGGTAGATCTTCCACGTCTGTCCGTCATAGCTCAGAAGGTGCTTATTTCTCATGGAAgcCAGGAGTGGGTcCACTTCTTTGGCTGTCGTCATTCCCAGAGCCTTGGCGATGTGCAGGGCCCTGTGAGGCCCATTGG3’) harbouring the desired mutations (indicated in lowercase letters). To mutate RHIM1 of ZBP1 a sgRNA (5’ ATTCCCGTGACCAATCTGGA 3’) targeting the RHIM-encoding exon 4 was co-injected into fertilized wt oocytes together with Cas9 protein and mRNA, as well as a long ssODN (5’CAGCCAGGACCCTCCTCTTACCTGGCTCA CCACAGGCTTTCTCTCTTACTATGACATTCCCGTGAgCAgcCgcGgcGGCGTTTGAATTGGCAATGGAGATGTGGCTGTTGGCTCCTTGTTGGCAGATCATGTTGACCGGAT3’) with the desired mutations (indicated in lowercase letters). To generate the combined Zbp1mZα1-2 mutant mice, a sgRNA (5‘GGCGGTAAAGGACTTGATTG3‘) targeting the Zα1 domain was co-injected together with Cas9 protein and mRNA, as well as a ssODN harbouring the desired mutations (indicated in lowercase letters) (5’CAGCCCCGCCTATGCTCCATGTTGCAGGCTCTGGGGAGGACACTCTGTCCTCCTTCTTCAGGCGGgcAAGGACTTGATcGAGGGTTTTCTTGGGCACTTGGCATTTCTTCACCAGCTGGCCAATCTTCACAGGGCCGCCGTCAT3’) in zygotes produced by in vitro fertilization of wt oocytes with sperm from Zbp1mZα2/mZα2 mice. After confirmation of the correct mutations by genomic DNA sequencing analysis, founder mice carrying the targeted mutations were backcrossed to C57BL/6N mice to establish independent mouse-lines. We further generated a new Zbp1 knockout mouse line in a pure C57BL/6N background to avoid potential problems stemming from a mixed background. Since all exons of murine Zbp1 are in the same frame we could not cause a frameshift that generates a stop codon by deleting an exon. We therefore targeted both exons 2 (sgRNA1: 5’CAGGTGTTGAGCGATGACGG3’) and 3 (sgRNA2: 5’TGAGCTATGACGGACAGACG3’) in order to generate a fusion exon that would cause a frameshift. Mice with a fusion exon however showed a truncated protein, resembling the size of the ZBP1ΔZα protein indicating exon skipping (data not shown). The functional Zbp1 knockout allele chosen for further experiments was found to have lost 373bp starting in exon2 and spanning into the 3’ intron as well as a 4bp deletion at the gRNA target site in exon3. These mice were backcrossed to eventually generate a Zbp1-/- colony. Lack of ZBP1 expression in cells from homozygous mice was confirmed by immunoblotting. Mice were maintained at the SPF animal facilities of the Institute for Genetics and the CECAD Research Center of the University of Cologne, or of the University of Texas Health Science Center at San Antonio (UTHSCSA). All animal procedures were conducted in accordance with national and institutional guidelines and protocols were approved by the responsible local authorities in Germany (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, Germany) and in Texas (Institutional Animal Care and Use Committee at UTHSCSA). Animals requiring medical attention were provided with appropriate care and were sacrificed when they developed macroscopically visible skin lesions to minimize suffering. No other exclusion criteria existed. Calculations to determine group sizes were not performed. Mice of the indicated genotype were assigned at random to groups and experiments were unblinded. Both male and female mice are included in all groups as the phenotypes studied in this manuscript were not affected by the mouse gender.
Antiretroviral drug treatment
WT (n=8) and RIPK1E-KO (n=7) littermate mice were given reverse transcriptase inhibitors (RTi) every day in drinking water starting from birth for a period of 6 weeks. A combination of Emtricitabine/Tenofovir (Aliud Pharma GmbH) and Nevirapine (Ratiopharm GmbH) were dissolved in drinking water at the following concentration: Emtricitabin, 660 μM; Tenofovir, 314 μM; Nevirapine at 375 μM. The RTi-containing water was changed once every week.
Histological analysis, Immunostaining and quantification of skin sections
Skin tissues were embedded in paraffin or snap frozen in OCT compound. Antigen retrieval for paraffin sections was performed in citrate buffer, pH6 for the skin sections. Anti-F4/80 (clone A3-1, MCA497G, BIO-RAD), anti-Keratin 14 (MA5-11599, Invitrogen), anti-Keratin 6 (905701, Biolegend), anti-Keratin 10 (905401, Biolegend) were used for the staining. Alexa-488, Alexa-594 and Alexa-633 fluorescence conjugated secondary antibodies were used for detection. F4/80 staining was performed on cryo sections. All sections were counterstained with DAPI. Quantification of epidermal thickness was performed by measurement of epidermal thickness in five optical fields per section. In each field, four measurements were performed. Percentage of inflamed area was determined as the percentage of inflamed versus total number of optical fields at 20x on individual skin sections. Assessment of tissue pathology was performed in a blinded fashion. All images were acquired using either a Zeiss Meta 710 confocal or PerkinElmer Spinning Disc confocal microscope for fluorescent images and brightfield images were acquired using a Leica SCN400 slide scanner or a Leica DM5500 B microscope.
Quantitative RT-PCR
Total RNA from skin tissue was extracted with Trizol Reagent (Life Technologies) and RNeasy Columns (Qiagen) and cDNA was prepared with Superscript III cDNA-synthesis Kit (Life Technologies). qRT–PCR of Il1-b, Il-6, Tnf, Cxcl3 and Ccl3 genes was performed with TaqMan probes (Life Technologies). HPRT was used as a reference gene. Data were analysed according to the ∆CT method.
Histopathological analysis of colon sections
Mouse colon sections were prepared and stained as previously described 32. Primary antibody for IHC was anti-CD45 (14-0451-85, eBioscience). Histopathological evaluation was performed on H&E-stained swiss roll colon sections in a blinded fashion as described previously33. Histopathology scores are composed of four parameters: epithelial hyperplasia, epithelial cell death, epithelial injury and quantity and localization of tissue inflammation (parameter scores; severity 0-3). Additionally, the fraction of affected tissue (area factor) was scored and multiplied with the respective parameter score (1=0-25% area affected; 2=25-50%; 3=50-75%, 4=75-100%). One section per mouse was scored. Ulcer formation was defined as loss of epithelial barrier integrity associated with infiltrating immune cells within the mucosa and/or submucosa.
Subcellular fractionation
Primary MEFs were directly lysed in hypotonic buffer (50 mM Tris-HCl, pH 8.0, 2 mM EDTA, 10% glycerol, and 0.1% Nonidet P-40) for 10 minutes on ice, followed by at 2,300 g centrifugation at 4°C for 5 minutes. The supernatants were collected as cytoplasmic extracts. The pellets were washed three times with hypotonic buffer and lysed in 20 mM HEPES-KOH, pH 7.6, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol, PhosSTOP phosphatase inhibitors (Roche), and complete protease inhibitors (Roche) for 30 minutes on ice. Cell lysates were centrifuged for 10 minutes at 13,200 rpm at 4°C, and the supernatants were collected as nuclear extracts.
Immunoblotting, immunoprecipitation and RNA dot blot
Antibodies against the following proteins were used for western blot analysis: RIPK3 (ADI-905-242-100, Enzo), RIPK1 (610459, BD), ZBP1 (AG-20B-0010, Adipogen or custom-made, Eurogentec), MLKL (MABC604, Millipore), GAPDH (NB300-221, Novus), p-RIPK3 (57220, Cell Signaling Technology), p-MLKL (37333, Cell Signaling Technology), Caspase-8 (4790, Cell Signaling Technology), cleaved Caspase-8 (8592, Cell Signaling Technology), H3 (ab1791, Abcam), donkey anti-rabbit IgG-HRP (NA934, GE Healthcare), sheep anti-mouse IgG-HRP (NA931, GE Healthcare) and goat anti-rat lgG-HRP (112-035-003, Jackson Immuno Research). The signals were detected by Amersham ECL Western Blotting Detection Reagent (RPN2106, GE Healthcare), SuperSignal West Pico PLUS Chemiluminescent substrate (34580, Thermo) or SuperSignal West Femto Maximum Sensitivity substrate (34095, Thermo). The membranes were re-probed after incubation in Restore Western Blot stripping buffer (21063, Thermo). To check MLKL and RIPK3 phosphorylation, total lysates of MEFs treated with TNF (VIB Protein Service Facility, Ghent), birinapant (2597, BioCat) and Z-VAD-FMK (N-1560, Bachem) were used as positive control. GAPDH was used as a loading control and cytosolic fraction marker and histone 3 (H3) was used as a loading control and nuclear fraction marker in immunoblots.
In the immunoprecipitation and dot blot experiments, Flag tag, Flag-tagged WT ZBP1 and Flag-tagged ZBP1mZa1-2 (N46D, Y50A, N122D, Y126A) were subcloned into lentiviral pCW-Cas9 (a gift from Eric Lander & David Sabatini (Addgene plasmid # 50661)) to replace Cas9 coding sequence. Immortalized wild-type MEFs (via serial passaging) were transduced with recombinant lentiviruses and were selected by 4 μg mL-1 puromycin (P8833, Sigma). Cell lysates were prepared in immunoprecipitation buffer (20 mM Tris pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, PhosSTOP phosphatase inhibitors (Roche), complete protease inhibitors (Roche), 100 U ml-1 RNAse inhibitor (M0314S, NEB)). In order to co-immunoprecipitate dsRNA-interacting proteins or RIPK3-interacting proteins, dsRNA or RIPK3 immunoprecipitation was performed by incubating the cell lysates with J2 anti-dsRNA mAb (10010200, Scicons) or anti-RIPK3 mAb and protein G Dynabeads (10004D, Life Technologies). For the dot blot experiment, cell lysates were incubated with homemade rabbit anti-ZBP1 antibody and protein G Dynabeads were subsequently washed 6 times in IP buffer and eluted in elution buffer (100 mM Tris.HCl pH8.0; 10 mM EDTA; 1% SDS; 100 mM DTT; 0.5 M NaCl) supplemented with 50 μg proteinase K at 56°C for 1 h. Supernatant was collected from the beads and RNA was subsequently extracted using the standard Trizol (15596018, Thermo Fisher) protocol using Glycoblue (AM9516, Thermo Fisher) as co-precipitant. The isolated RNA was dissolved in RNAse free H2O and measured. For the Dot Blot, RNA samples were mixed 1:1 with 20x SSC (V4261, Promega) and spotted onto a positively charged Nylon membrane (RPN82B, GE Healthcare) that was soaked in H2O and briefly pre-incubated in 10x SSC. Subsequently RNA was crosslinked to the membrane using UVC light followed by complete drying of the membrane. For detection of dsRNA the membrane was shortly soaked in H2O and blocked for 30 min in blocking buffer (927-40000, Licor) followed by incubation with the dsRNA specific J2 antibody. Blots were developed using fluorescently labelled secondary antibodies and the Odyssey imaging system (Licor).
Cell death assays
Primary or immortalised MEFs or LFs were seeded in 96 well plates (1 × 104 cells per well) one day before treatment. On the day of the experiment, indicated amounts of recombinant murine TNF (VIB Protein Service Facility, Ghent), IFNα (752804, Biolegend), IFNβ (97265.25, Biomol), IFNγ (12343537, ImmunoTools), Z-VAD-FMK (N-1560, Bachem), GSK’872 (5303890001, Sigma), emricasan (S7775, Selleckchem) or etanercept (Enbrel®, Amgen) were added to cells. Cell death assays were performed using the IncuCyte bioimaging platform (Essen); two to four images per well were captured, analysed and averaged. Cell death was measured by the incorporation of YOYO™-1 Iodide (491/509) (Y3601, Thermo). In the IFNγ+emricasan+etanercept or Dox+emricasan experiments, the cells were pre-treated with IFNγ or Dox for 24 h, then the medium was replaced with medium containing emricasan with or without etanercept and cell death was assessed using Incucyte. In the LMB (L2913, Sigma) or KPT-330 (S7252, Selleckchem) experiments, the cells were pretreated with IFNα, IFNγ or Dox for 24 h, then the medium was replaced with medium containing leptomycin B or KPT-330 and cell death was assessed in IncuCyte. LDH activity in the supernatant was used to evaluate IAV-induced cell death. LDH measurement was performed using a CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (Promega) according to the manufacturer’s protocol. All values represent the percentage of LDH release compared to a maximum lysis control (1% Triton X-100-lysed cells).
Virus infections
Influenza A virus strain A/PR/8/34 (H1N1, PR8) was obtained from ATCC. The virus was propagated in allantoic cavity of 9- to 11-day old embryonated SPF chicken eggs and viral titres were enumerated by standard plaque assays. Lung fibroblasts were infected with PR8 virus (MOI: 1 or MOI: 5) in serum free medium for 2 h. Complete DMEM medium was replaced after 2 h. Total cell lysates were harvested at 8 h after infection for immunoblotting analysis and cell supernatants were harvested at 24 h after infection for LDH release analysis. For MCMV infection, infections and organ titres as well as construction and propagation of the BAC-derived parental WT K181 and M45mutRHIM viruses were performed as previously described 34.
Repeat region annotation, RNA-seq read mapping and counting
Repetitive regions were annotated as previously described 35. Briefly, the mouse genome (GRCm38.78) was masked using RepeatMasker (RepeatMasker v.4.09, repeatmasker.org) configured with nhmmer 36 in sensitive mode using the Dfam 2.0 library (v150923). RepeatMasker annotates LTR and internal regions separately, complicating the summation of reads spanning these divides. Tabular outputs were, therefore, parsed to merge adjacent annotations for the same element and to produce a gene transfer format (GTF) file compatible with popular read-counting programs (Supplementary file 1). Read pairs were aligned with HISAT2 37 and primary, stranded, mappings counted with featureCounts (Subread 38) using standard GTFs for annotated genes and the curated RepeatMasker GTFs for repeat regions. For accuracy and to prevent ambiguity, only reads that could be uniquely assigned to a single feature were counted. Those remaining were normalized to account for variable sequencing depth between samples using DESeq2 39. All downstream differential expression analyses and visualization were carried out using Qlucore Omics Explorer 3.3 (Qlucore, Lund, Sweden).
Quantitation of complementary RNA-seq reads
Direct, non-canonical, kmers (k = ⌈log42.7e9⌉ = 16, the smallest k such that the majority of kmers are found uniquely) were counted for adapter-trimmed, quality-filtered stranded reads with the exact kmer counting tool KMC v3.1.1 40. First-in-pair reads were counted directly and merged with counts from reverse-complemented second-in-pair reads using KMC to form a total direct count. By matching using their canonical (lexicographically lowest) representation, kmers where both the forward and reverse-complement had been observed within the dataset were selectively retained. The queryRepeats script (RepeatMasker v.4.09, repeatmasker.org) was used to extract Mus musculus repetitive elements from the Dfam3.1 database 41 which were broken into their constitutive canonical kmers and filtered such that only kmers uniquely identifying a particular repeat family were retained. 1,073,365 kmers annotated with their corresponding repeat families were retained in the subsequent database, which was used to screen the previously counted kmers. kmers were converted into complementary kmers using shell commands and Python code (Supplementary file 2). Counts of repeat-associated c-kmers were normalised by sequencing depth (kmers per million reads) and the counts of the least abundant c-kmer of each pair was used for the calculations.
Gene functional annotation
Pathway analyses were performed using g:Profiler (https://biit.cs.ut.ee/gprofiler) with genes ordered by the degree of differential expression. P values were estimated by hypergeometric distribution tests and adjusted by multiple testing correction using the g:SCS (set counts and sizes) algorithm, integral to the g:Profiler server42.
Statistical analysis
Data shown in graphs represent mean or mean ± s.e.m. If the data fulfilled the criteria for Gaussian distribution tested by column statistics, unpaired parametric t test with Welch’s correction was performed for statistical analysis. If not, nonparametric Mann–Whitney test was performed. All statistical tests listed in the figure legends were two-sided and performed using Graphpad Prism or Qlucore Omics Explorer 3.3. Exact P values are presented in the figures.
Extended Data
Extended Data Figure 1. Generation of mutant Zbp1 knock-in mice.
a-e, Schematic depicting the different Zbp1 mutant mice generated using CRISPR/Cas9-mediated gene targeting in C57BL/6N zygotes as indicated. To generate Zbp1mZα2 mice amino acids N122 and Y126 in Zα2 domain were substituted with D and A respectively (a). For Zbp1mZα1-2 mice both Zα1 (N46D and Y50A) and Zα2 (N122D, Y126A) were mutated (b). For Zbp1mR1 mice the residues of the core RHIM motif starting at amino acid position 192 (IQIG) were replaced by alanines (c). Sequencing traces of the desired mutations are shown in heterozygous mice. d, Zbp1ΔZα mice were generated by deleting exons two and three which contain both Zα domains (d). A new Zbp1- knockout mouse was generated by targeting both exons 2 and 3. An allele harboring a 373 bp deletion in exon 2 and the following intron as well as an additional 4 bp deletion in exon 3 was chosen as Zbp1 knockout allele (e). f, Immunoblot analysis of total lysates from LFs with the indicated genotypes stimulated with IFNγ (1,000 u ml−1) for 24 h. Data information: one representative out of two independent experiments shown (f). GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2. ZBP1 exhibits Zα-dependent and -independent functions in anti-viral defence.
a, Cell death analysed by measuring LDH release in supernatants of LFs with the indicated genotypes infected with IAV (MOI:1 or 5) for 24 h. Data show mean from technical triplicates (n = 3). b, Immunoblot analysis of total lysates from LFs with the indicated genotypes infected with IAV (MOI:5) for 8 h with the indicated antibodies. GAPDH was used as a loading control. c-e, Viral titres in spleen (c), liver (d) or salivary gland (e) from mice with the indicated genotypes 5 days (c, d) or 14 days (e) after intraperitoneal inoculation with 1 × 106 MCMV-M45mutRHIM. Graphs show values from individual mice as well as mean ± s.e.m. P values by two-sided nonparametric Mann–Whitney test. The dotted horizontal red line depicts the detection limit of the assay. Data information: data shown are representative of two independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3. ZBP1 causes keratinocyte necroptosis and skin inflammation in RIPK1E-KO mice by Zα-dependent and -independent mechanisms.
a, Representative images of skin sections from 6 - 7 week-old mice with the indicated genotypes immunostained with the indicated antibodies and DAPI (DNA stain). RIPK1E-KO (n ≥ 5); RIPK1E-KO Zbp1mZα2/mZα2 (n ≥ 3); RIPK1E-KO Zbp1∆Zα/∆Zα (n ≥ 3); RIPK1E-KO Zbp1mR1/mR1 (n ≥ 3). Scale bars, 50 μm. b, qRT-PCR analysis of the mRNA expression of the indicated cytokines and chemokines in RNA isolated from total skin from 6 - 7 week-old mice with the indicated genotypes. Each dot represents an individual mouse. Data show mean ± s.e.m. P value by two-sided nonparametric Mann–Whitney test. Control mice include Ripk1FL/FL mice that do not express K14-Cre, or Ripk1FL/WT K14-Cre mice, with WT or mutated Zbp1 alleles. c-e, Representative images of mice with the indicated genotypes at the indicated age. RIPK1E-KO (n = 6) and RIPK1E-KO Zbp1∆Zα/∆Zα (n = 17) (c); RIPK1E-KO Zbp1mZα2/mZα2 (n = 16) (d); RIPK1E-KO Zbp1mR1/mR1(n = 20) (e).
Extended Data Figure 4. ZBP1-mediated perinatal lethality in Ripk1mR/mR mice depends on Zα-dependent sensing of Z-NA and RHIM1-mediated signalling.
a, Representative pictures of skin sections from mice with the indicated genotypes stained with H&E or immunostained with antibodies against K10 and K14 and counterstained with DAPI visualising nuclear DNA. Skin sections from Ripk1mR/mR or Ripk1mR/wt E18.5 embryos are compared with skin from newborn (P1 to P3) double mutant mice. Scale bars = 100 μm. Ripk1mR/mR (n = 3), Ripk1mR/wt (n = 3), Ripk1mR/mR Zbp1∆Zα/∆Zα (n = 3), Ripk1mR/mR Zbp1mZα2/mZα2 (n = 3), Ripk1mR/mR Zbp1mR1/mR1 (n = 6), Ripk1mR/wt Zbp1mR1/mR1 (n = 3). b, c Immunoblot analysis of total lysates from primary LFs (b) and keratinocytes (c) with the indicated genotypes stimulated with IFNγ (1,000 u ml−1) for 24 h. d, Spleen to bodyweight ratio of mice with the indicated genotypes. Each dot represents an individual mouse. Data show mean ± s.e.m. P value by two-sided unpaired t-test or nonparametric Mann–Whitney test. e, Representative H&E stained sections from spleen and liver of mice with the indicated genotypes. Ripk1wt/wt Zbp1-/wt (n = 5), Ripk1mR/mR Zbp1-/- (n = 6), Ripk1mR/mR Zbp1-/- Trif-/- (n = 6), Ripk1mR/mR Zbp1∆Zα/∆Zα (n = 13), Ripk1mR/wt Zbp1∆Zα/∆Zα (n = 10), Ripk1mR/mR Zbp1mZα2/mZα2 (n = 21), Ripk1mR/wt Zbp1mZα2/mZα2 (n = 3), Ripk1mR/mR Zbp1mR1/mR1 (n = 4), Ripk1mR/wt Zbp1mR1/mR1 (n = 6). Representative data shown in panels b (n = 2) and c (n = 2). GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 5. IFNα and IFNγ induce death of Ripk1mR/mR cells by inducing Zα-dependent ZBP1 activation and RHIM1-mediated downstream signalling.
a, Cell death measured by YOYO-1 uptake in Ripk1wt/wt or Ripk1mR/mR MEFs treated with IFNγ (1,000 u ml−1) for 48 h and IncuCyte images of Ripk1wt/wt or Ripk1mR/mR MEFs before and 48 h after IFNγ (1,000 u ml−1) treatment. YOYO-1 staining is shown in green. b-f, h, Graphs depicting cell death assessment by YOYO-1 uptake in MEFs with the indicated genotypes treated with combinations of IFNγ (1,000 u ml−1), IFNα (50 ng ml−1), Z-VAD-FMK (20 μM) and GSK’872 (3 μM) for 48 h. g Immunoblot analysis of total lysates from MEFs with the indicated genotypes stimulated with IFNγ (1,000 u ml−1) for 24 h. Representative data in panel a (n = 7), b (n = 5), c (n = 3), d (n = 3), e (n = 7), f (n = 3), g (n = 2) and h (n = 3). Panels (a-f, h) show mean values from technical triplicates (n = 3). GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 6. IFNγ+emricasan in the presence of etanercept induces ZBP1 dependent cell death in MEFs.
a, b, Graphs depicting cell death assessment by YOYO-1 uptake in WT and ZBP1 KO MEFs treated with combinations of IFNγ (1,000 u ml−1, 24h pre-treatment), emricasan (em) (5 μM) and etanercept (et) (50 μg ml-1). c, Graph depicting cell death assessment by YOYO-1 uptake in WT MEFs treated with combinations of TNF (20 ng ml−1), birinapant (1 μM), Z-VAD-FMK (20 μM) and etanercept (50 μg ml-1). d, f, Graphs depicting cell death assessment by YOYO-1 uptake in MEFs (d) and LFs (f) with the indicated genotypes treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), etanercept (50 μg ml−1) and emricasan (5 μM). e, Immunoblot analysis of total lysates from MEFs with the indicated genotypes stimulated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), etanercept (50 μg ml−1) and emricasan (5 μM). Total lysate from WT MEFs treated with TNF (20 ng ml−1)+birinapant (1 μM)+Z-VAD-FMK (20 μM) (TSZ) for 4h was used as positive control for p-MLKL and p-RIPK3 detection. g, Immunoblot analysis of total lysates from iMEFs transduced with lentiviruses expressing Flag (EV), Flag-tagged ZBP1 or Flag-tagged ZBP1mZα1-2 stimulated with doxycycline (1 μg ml−1) for 24 h. Representative data in panel a (n = 6), b (n = 9), c (n = 3), d (n = 6), e (n = 3), f (n = 4) and g (n = 2). Panels (a, b, c, d, f) show mean values from technical triplicates (n = 3). Data shown in panel f serve as control for the data shown in Fig. 2d and come from the same experiment. GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 7. RIPK1 deficiency in keratinocytes causes extensive transcriptional changes in the skin.
a, Differential expression (≥ 2-fold, p ≤0.05) of 4,204 genes in skin biopsies from 6 week-old WT (n = 5) and RIPK1E-KO mice (n = 5). b, Functional annotation by gene ontology (GO) of the top n = 1,000 upregulated genes and the top n = 1,000 down regulated genes between the samples in a, performed with g:Profiler (https://biit.cs.ut.ee/gprofiler). P values were estimated by hypergeometric distribution tests and adjusted by multiple testing correction using the g:SCS (set counts and sizes) algorithm, integral to the g:Profiler server.
Extended Data Figure 8. Treatment with reverse transcriptase inhibitors ameliorates skin inflammation in RIPK1E-KO mice.
a, Kaplan-Meier plot depicting lesion-free survival (top panel) and microscopic quantification of mean epidermal thickness and inflamed skin area (bottom panels) of mice with the indicated genotypes that were treated or not with reverse transcriptase inhibitors (RTi) from birth till the age of 6 weeks. Each dot represents an individual mouse. Data show mean ± s.e.m. P value by two-sided nonparametric Mann–Whitney test. The data from untreated control and RIPK1E-KO mice are included also in Figure 1b, c. b, Hierarchical clustering of samples from control (n = 5) and RIPK1E-KO mice (n = 10) according to the expression of genes that are differentially expressed (≥ 1.75-fold, p ≤0.05, two-sided t-test, performed in Qlucore Omics Explorer 3.3) between untreated (n = 5) and RTi-treated RIPK1E-KO mice (n = 5). Each column corresponds to one mouse. The mean epidermal thickness and inflammation score for each mouse are indicated. Control mice include Ripk1FL/FL mice that do not express K14-Cre, or Ripk1FL/WT K14-Cre mice.
Extended Data Figure 9. Inhibition of nuclear export triggers Zα-dependent ZBP1-mediated cell death.
a, Immunoblot analysis of WT MEFs treated with TNFα (20 ng ml−1), IFNα (50 ng ml−1), IFNβ (50 ng ml−1) or IFNγ (1,000 u ml−1) for 24 h. b, Cell death measured by YOYO-1 uptake in WT MEFs treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment) and LMB (1 ng ml−1). IncuCyte images of WT MEFs before and after treatment with LMB for 24 h. YOYO-1 staining is shown in green. c-e, Graphs depicting cell death assessment by YOYO-1 uptake in MEFs with the indicated genotypes treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), IFNα (50 ng ml−1) (24 h pre-treatment) and LMB (1 ng ml−1). f, g, i, Graphs depicting cell death assessment by YOYO-1 uptake in LFs with the indicated genotypes treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), KPT-330 (1 μM or 10 μM) (f) and LMB (5 ng ml−1) (g, i). h, Graphs depicting cell death assessment by YOYO-1 uptake in Mlkl-/- MEFs treated with combinations of IFNγ (1,000 u ml−1) (24 h pre-treatment), LMB (1 ng ml−1) and emricasan (5 μM). j, Immunoblot analysis of total lysates from iMEFs transduced with lentiviruses expressing Flag (EV), Flag-tagged ZBP1 or Flag-tagged ZBP1mZα1-2 stimulated with combinations of Dox (1 μg ml−1) (24 h pre-treatment) and LMB (5 ng ml−1). k, Graphs depicting cell death assessment by YOYO-1 uptake in iMEFs transduced with lentiviruses expressing Flag (EV), Flag-tagged ZBP1 or Flag-tagged ZBP1mZα1-2 stimulated with combinations of Dox (1 μg ml−1) (24 h pre-treatment) and LMB (5 ng ml−1). Dox, doxycycline. Representative data in panel a (n = 2), b (n = 6), c (n = 6), d (n = 6), e (n = 6), f (n = 4), g (n = 4), h (n = 4), i (n = 5), j (n = 2) and k (n = 3). Panels (b-i, k) show mean values from technical triplicates (n = 3). Data shown in panels c and i serve as controls for the data shown in Fig. 4a and 4e respectively, and come from the same experiments. GAPDH was used as a loading control for immunoblot analysis. For gel source data, see Supplementary Fig. 1.
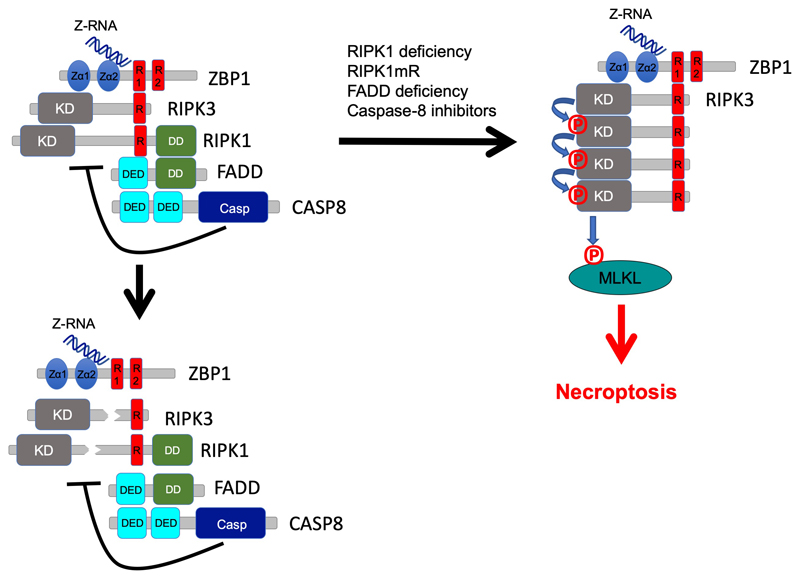
Extended Data Figure 10. Schematic model depicting the regulation of ZBP1-mediated activation of RIPK3-MLKL-dependent necroptosis by RIPK1 and caspase-8.
Sensing of endogenous cellular Z-RNA by its Zα domains activates ZBP1 inducing its interaction with RIPK3, but cell death is inhibited due to negative regulation by RIPK1 and caspase-8. RIPK1 inhibits ZBP1-induced activation of RIPK3 by FADD-mediated recruitment of caspase-8, which cleaves components of the complex such as RIPK1 and RIPK3. In cells lacking RIPK1 or expressing RIPK1 with mutated RHIM, in FADD-deficient cells as well as in cells treated with caspase inhibitors, Zα-dependent sensing of endogenous Z-RNA activates ZBP1 that strongly engages RIPK3 triggering MLKL-dependent necroptosis.
Supplementary Material
Acknowledgements
We thank E. Gareus, J. Kuth, B. Kühnel, E. Stade, C. Uthoff-Hachenberg and J. von Rhein for technical assistance, B. Zevnik and the CECAD Transgenic Core Unit for the generation of mutant ZBP1 knock-in mice and A. Schauss and the CECAD Imaging Facility for microscopy support. We also thank A. Athanasiadis for valuable discussions. Research reported in this publication was supported by funding from the European Research Council (Grant Agreement No. 787826), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; projects SFB829 (Project No. 73111208), SFB1218 (Project No. 269925409), SFB1399 (Project No. 413326622), SFB1403 (Project No. 414786233), PA 1476/8-1 (Project No. 411102043), and CECAD (project no. 390661388)), and the Federal Ministry of Education and Research (BMBF, e:med project InCa, Grant No. 01ZX1901A) to M.P., and by the Francis Crick Institute (FC001099) and the Wellcome Trust (102898/B/13/Z) to G.K.. J.L. and R.O.E. were supported by postdoctoral fellowships from the Alexander von Humboldt Foundation.
Footnotes
Data availability
The original RNA sequencing data are uploaded and available at the Gene Expression Omnibus (GEO) under accession (GSE143955). Source Data for Figs. 1–4 and Extended Data Figs. 2-6, 8 and 9 are provided with the paper.
Code availability
A gene transfer format (GTF) of repetitive region annotations for the mouse genome (GRCm38.78), with the adjacent annotations for the same element merged is included in Supplementary Data File 1. Specific shell commands executed and Python code to convert kmers into complementary kmers are included in Supplementary Data File 2.
Author contributions
H. J., L.W., S.K., R.S. and M.P. conceived the study and designed the experiments. H. J., L.W., S.K., R.S., J.L. R.O.E., A.F., R.L. and G.R.Y. performed and analysed experiments. W.J.K., G.K. and M.P. supervised the experiments. H. J., L.W., S.K., R.S., G.K., W.J.K. and M.P. interpreted data and wrote the paper.
Declaration of competing interests
The authors declare the following competing interests: M.P. received consulting and speaker fees from Genentech, GSK, Boehringer Ingelheim and Sanofi.
References
- 1.Wang AH, et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 2.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 3.Herbert A. Z-DNA and Z-RNA in human disease. Commun Biol. 2019;2:7. doi: 10.1038/s42003-018-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha SC, et al. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc Natl Acad Sci U S A. 2008;105:20671–20676. doi: 10.1073/pnas.0810463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat Struct Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 6.Maelfait J, et al. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. 2017;36:2529–2543. doi: 10.15252/embj.201796476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thapa RJ, et al. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe. 2016;20:674–681. doi: 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Placido D, Brown BA, 2nd, Lowenhaupt K, Rich A, Athanasiadis A. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H, et al. Species-independent contribution of ZBP1/DAI/DLM-1-triggered necroptosis in host defense against HSV1. Cell Death Dis. 2018;9:816. doi: 10.1038/s41419-018-0868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sridharan H, et al. Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis. EMBO Rep. 2017;18:1429–1441. doi: 10.15252/embr.201743947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koehler H, et al. Inhibition of DAI-dependent necroptosis by the Z-DNA binding domain of the vaccinia virus innate immune evasion protein, E3. Proc Natl Acad Sci U S A. 2017;114:11506–11511. doi: 10.1073/pnas.1700999114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuriakose T, et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. 2016;1 doi: 10.1126/sciimmunol.aag2045. aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Upton JW, Kaiser WJ, Mocarski ES. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe. 2012;11:290–297. doi: 10.1016/j.chom.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels BP, et al. The Nucleotide Sensor ZBP1 and Kinase RIPK3 Induce the Enzyme IRG1 to Promote an Antiviral Metabolic State in Neurons. Immunity. 2019;50:64–76 e64. doi: 10.1016/j.immuni.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton K, et al. RIPK1 inhibits ZBP1-driven necroptosis during development. Nature. 2016;540:129–133. doi: 10.1038/nature20559. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, et al. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 2016;540:124–128. doi: 10.1038/nature20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehle AS, et al. Intestinal Inflammation and Dysregulated Immunity in Patients With Inherited Caspase-8 Deficiency. Gastroenterology. 2019;156:275–278. doi: 10.1053/j.gastro.2018.09.041. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, et al. Human RIPK1 deficiency causes combined immunodeficiency and inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2019;116:970–975. doi: 10.1073/pnas.1813582116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuchet-Lourenco D, et al. Biallelic RIPK1 mutations in humans cause severe immunodeficiency, arthritis, and intestinal inflammation. Science. 2018;361:810–813. doi: 10.1126/science.aar2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchiyama Y, et al. Primary immunodeficiency with chronic enteropathy and developmental delay in a boy arising from a novel homozygous RIPK1 variant. J Hum Genet. 2019;64:955–960. doi: 10.1038/s10038-019-0631-3. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser WJ, Upton JW, Mocarski ES. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J Immunol. 2008;181:6427–6434. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebsamen M, et al. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dannappel M, et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014;513:90–94. doi: 10.1038/nature13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welz PS, et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 25.Yang D, et al. ZBP1 mediates interferon-induced necroptosis. Cell Mol Immunol. 2019 doi: 10.1038/s41423-019-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad S, et al. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell. 2018;172:797–810 e713. doi: 10.1016/j.cell.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannion NM, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae S, et al. Energetics of Z-DNA binding protein-mediated helicity reversals in DNA, RNA, and DNA-RNA duplexes. J Phys Chem B. 2013;117:13866–13871. doi: 10.1021/jp409862j. [DOI] [PubMed] [Google Scholar]
- 29.Etchin J, et al. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol. 2013;161:117–127. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hafner M, et al. Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis. 2004;38:176–181. doi: 10.1002/gene.20016. [DOI] [PubMed] [Google Scholar]
- 31.Troder SE, et al. An optimized electroporation approach for efficient CRISPR/Cas9 genome editing in murine zygotes. PLoS One. 2018;13:e0196891. doi: 10.1371/journal.pone.0196891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlantis K, et al. NEMO Prevents RIP Kinase 1-Mediated Epithelial Cell Death and Chronic Intestinal Inflammation by NF-kappaB-Dependent and -Independent Functions. Immunity. 2016;44:553–567. doi: 10.1016/j.immuni.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adolph TE, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upton JW, Kaiser WJ, Mocarski ES. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe. 2010;7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Attig J, Young GR, Stoye JP, Kassiotis G. Physiological and Pathological Transcriptional Activation of Endogenous Retroelements Assessed by RNA-Sequencing of B Lymphocytes. Frontiers in microbiology. 2017;8:2489. doi: 10.3389/fmicb.2017.02489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler TJ, Eddy SR. nhmmer: DNA homology search with profile HMMs. Bioinformatics (Oxford, England) 2013;29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kokot M, Dlugosz M, Deorowicz S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics (Oxford, England) 2017;33:2759–2761. doi: 10.1093/bioinformatics/btx304. [DOI] [PubMed] [Google Scholar]
- 41.Hubley R, et al. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016;44:D81–89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raudvere U, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.