Abstract
In this report, a series of novel piperidine-substituted thiophene[3,2-d]pyrimidine derivatives were designed to explore the hydrophobic channel of the non-nucleoside reverse transcriptase inhibitors binding pocket (NNIBP) by incorporating an aromatic moiety to the left wing of the lead K-5a2. The newly synthesized compounds were evaluated for anti-HIV potency in MT-4 cells and inhibitory activity to HIV-1 reverse transcriptase (RT). Most of the synthesized compounds exhibited broad-spectrum activity toward wild-type and a wide range of HIV-1 strains carrying single non-nucleoside reverse transcriptase inhibitors (NNRTI)-resistant mutations. Especially, compound 26 exhibited the most potent activity against wild-type and a panel of single mutations (L100I, K103N, Y181C, Y188L and E138K) with an EC50 ranging from 6.02 to 23.9 nmol/L, which were comparable to those of etravirine (ETR). Moreover, the RT inhibition activity, preliminary structure–activity relationship and molecular docking were also investigated. Furthermore, 26 exhibited favorable pharmacokinetics (PK) profiles and with a bioavailability of 33.8%. Taken together, the results could provide valuable insights for further optimization and compound 26 holds great promise as a potential drug candidate for the treatment of HIV-1 infection.
Keywords: HIV-1; NNRTIs; NNIBP; Thiophene[3,2-d]pyrimidine; Hydrophobic channel
Graphical abstract
Compound 26 exhibited potent activity against wild-type and a panel of single mutations with an EC50 ranging from 6.02 to 23.9 nmol/L. Furthermore, 26 exhibited favorable PK profiles and with a bioavailability of 33.8%. These results demonstrated that 26 holds great promise as a potential HIV-1 drug candidate.
1. Introduction
Acquired immune deficiency syndrome (AIDS) is caused by human immunodeficiency virus (HIV), which could enter the host cell and eventually leads to significant weakening of the host immune system. In the life cycle of HIV-1, reverse transcriptase (RT) is responsible for transcribing the single-stranded RNA genome to a double-stranded DNA, and it is a crucial target for development of novel anti-AIDS drugs1. Among the HIV-1 RT inhibitors, non-nucleoside reverse transcriptase inhibitors (NNRTIs), due to their unique antiviral potency, high specificity and low toxicity, gained a definitive position in highly active antiretroviral therapy (HAART) regimens used to treat HIV-12, 3, 4.
Nevirapine (NVP), delavirdine (DLV), and efavirenz (EFV) were approved as the first-generation NNRTIs for AIDS therapy5. However, their low genetic barrier results in the emergence of drug-resistant mutants rapidly. K103N and Y181C are the two most prevalent mutations in vivo, which elicited frequently resistance to NVP and EFV6. Etravirine (1, ETR) and rilpivirine (2, RPV), both of which belong to the diarylpyrimidine (DAPY) family, were approved as the second-generation NNRTIs in 2008 and 2011 by U. S. Food and Drug Administration (FDA), respectively5. Although they could effectively inhibit most of the RT-resistant mutations caused by the first-generation NNRTIs, they generally failed to suppress the most refractory mutations E138K and RES056 (K103N + Y181C)7, 8. In addition, the patients treated with second-generation NNRTIs were frequently reported with hypersensitivity reactions and other adverse effects9, 10. Therefore, new NNRTIs that offer a combination of improved potency against these mutants, a favorable profile of safety and tolerability is urgently needed11, 12, 13.
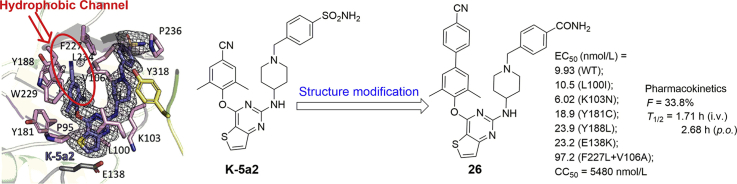
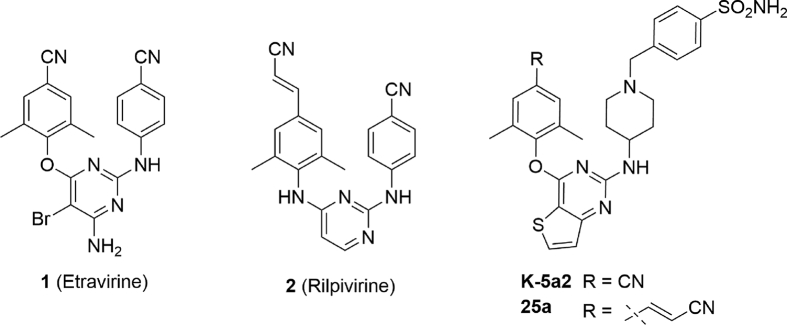
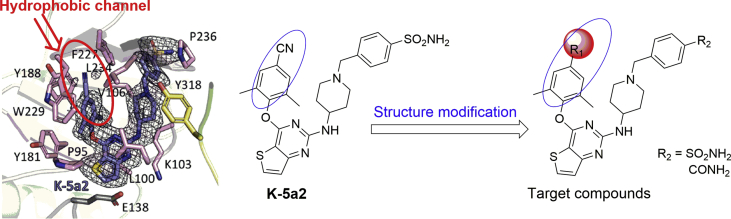
Previous research efforts in our lab led to the discovery of two potent HIV-1 NNRTIs, named as K-5a2 and 25a (Fig. 1), exhibiting excellent activities against drug-resistant mutations compared to that of ETR14, 15. Both compounds share a thiophene[3,2-d]pyrimidine central scaffold and a flexible extended right wing with piperidine-linked benzenesulfonamide structure, with the difference that K-5a2 retains the 4-cyano-2,6-dimethylphenyl structure of ETR in the left wing while 25a features the 4-cyanovinyl-2,6-dimethylphenyl structure of RPV in its left wing. The co-crystal structures of wild-type (WT) and mutant HIV-1 RT in complex with K-5a2 and 25a indicated that they displayed potent activity against WT and a panel of NNRTI-resistant mutant HIV-1 strains due to conformational flexibility and positional adaptability of the inhibitors, and the network of main chain hydrogen bonds and hydrophobic interactions developed between the inhibitors and NNIBP. Especially, the hydrophobic interactions between the left wing of K-5a2 and the hydrophobic channel surrounded by Tyr181, Tyr188, Phe227, Trp229, and Leu234 were critical for the enzyme–inhibitor complex formation. Although the 4-cyano-2,6-dimethylphenyl structure of K-5a2 projected into the hydrophobic channel, it contacted with Tyr188 and Trp229 only and did not develop an effective π–π interaction with the highly conserved residue Phe227. As depicted in Fig. 2, we can learn that the hydrophobic channel still has enough space to accommodate structurally diverse groups16, 17, 18.
Figure 1.
Chemical structures of second-generation NNRTI drugs and thiophene[3,2-d]pyrimidine lead K-5a2 and 25a.
Figure 2.
Design of the novel thiophene[3,2-d]pyrimidine derivatives.
In the present work, we paid our attention to explore the structure–activity relationship (SARs) of this underexplored region in the NNIBP by introducing different aromatic structures, including thiophene, furfuran and substituted benzene ring, to the left wing of K-5a2, with the aim to develop novel potent NNRTIs with improved drug resistance profiles. The synthetic routes, anti-HIV activity, preliminary SARs and modeling analysis of these novel thiophene[3,2-d]pyrimidine derivatives will be discussed.
2. Results and discussion
2.1. Chemistry
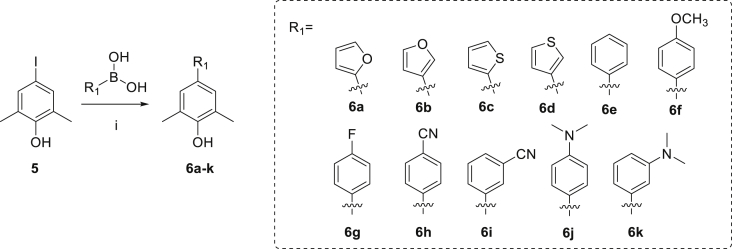
Scheme 1, Scheme 2 illustrate the general synthetic routes of the intermediates 6a–k and target compounds 11–32. Firstly, Suzuki coupling of various boronic acid derivatives with commercially available 4-iodo-2,6-dimethylphenol (5), in DMF/H2O solution using Pd(PPh3)4 as catalyst, provided the intermediates 6a–k (Scheme 1). Compounds 6a–k were followed by a nucleophilic substitution with 2,4-dichlorothieno[3,2-d]pyrimidine (7) in the presence of potassium carbonate to afford the intermediates 8a–k, respectively. Then, the intermediates 9a–k were obtained by Buchwald-Hartwig cross coupling reactions of 8a–k with 4-(tert-butoxycarbonyl)-aminopiperidine in the presence of BINAP and PdCl2(PPh3)2. Removing the Boc protecting group of 9a–k by trifluoroacetic acid (TFA) furnished the key intermediates 10a–k, which were reacted with 4-(bromomethyl)benzenesulfonamide or 4-(chloromethyl)benzamide to yield the target compounds 11–32 (Scheme 2).
Scheme 1.
Synthesis of 6a–k. Reagents and conditions: (i) Pd(PPh3)4, K2CO3, DMF, H2O, 100 °C.
Scheme 2.
Synthesis of the target compounds 11–32. Reagents and conditions: (i) 6a–k, DMF, K2CO3, r.t.; (ii) BINAP, PdCl2(PPh3)2, Cs2CO3, N-(tert-butoxycarbonyl)-4-aminopiperidine, 1,4-dioxane, 120 °C; (iii) TFA, DCM, r.t.; (iv) 4-(bromomethyl) benzenesulfonamide or 4-(chloromethyl) benzamide, DMF, K2CO3, r.t.
2.2. In vitro anti-HIV activities
The newly synthesized derivatives 11–32 were firstly evaluated for their antiviral potency against WT HIV-1 (IIIB) and a HIV-2 strain (ROD) in MT-4 cell cultures. NVP, EFV, ETR and the lead compound K-5a2 were selected as control. The values of EC50 (50% effective concentration), CC50 (50% cytotoxic concentration), and SI (selectivity index, CC50/EC50 ratio) of the target compounds are summarized in Table 1.
Table 1.
Anti-HIV-1 (IIIB) and HIV-2 (ROD) activity, cytotoxicity and SI of target compounds 11–32.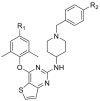
| Compd. | R1 | R2 | EC50 (nmol/L)a |
CC50 (μmol/L)b | SIc |
||
|---|---|---|---|---|---|---|---|
| IIIB | ROD | IIIB | ROD | ||||
| 11 |  |
SO2NH2 | 17.7 ± 4.83 | 4148 ± 1785 | 13.5 ± 3.94 | 762 | 3 |
| 12 |  |
CONH2 | 93.3 ± 82.8 | 25,289 ± 18,400 | 28.9 ± 15.6 | 310 | 1 |
| 13 |  |
SO2NH2 | 63.6 ± 11.4 | 6805 ± 1225 | 15.6 ± 4.97 | 246 | 2 |
| 14 |  |
CONH2 | 57.5 ± 12.7 | 3181 ± 832 | 15.8 ± 7.04 | 275 | 5 |
| 15 | 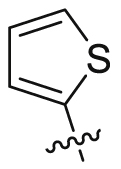 |
SO2NH2 | 49.5 ± 14.8 | 5111 ± 1707 | 21.5 ± 0.89 | 435 | 4 |
| 16 |  |
CONH2 | 203 ± 51.5 | >20,535 | ≥20.5 | ≥101 | NAd |
| 17 | 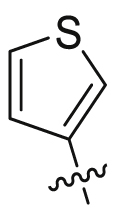 |
SO2NH2 | 31.6 ± 17.7 | 4802 ± 916 | 15.4 ± 5.86 | 489 | 3 |
| 18 |  |
CONH2 | 240 ± 94.3 | >17,044 | 17.0 ± 12.9 | 71 | <1 |
| 19 | 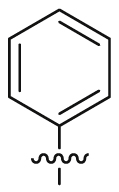 |
SO2NH2 | 43.8 ± 15.6 | 6135 ± 272 | 21.3 ± 1.26 | 487 | 3 |
| 20 |  |
CONH2 | 47.7 ± 11.8 | 3842 ± 1765 | 17.5 ± 6.18 | 367 | 5 |
| 21 | 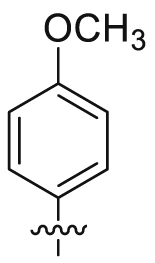 |
SO2NH2 | 61.1 ± 10.3 | >16,635 | 16.6 ± 4.65 | 272 | <1 |
| 22 |  |
CONH2 | 52.6 ± 11.4 | 5035 ± 685 | 13.4 ± 5.94 | 256 | 3 |
| 23 | 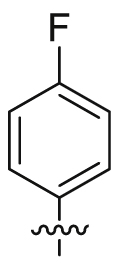 |
SO2NH2 | 11.3 ± 2.07 | 3283 ± 1255 | 16.9 ± 4.77 | 1492 | 5 |
| 24 |  |
CONH2 | 12.8 ± 2.73 | ≥5655 | 6.80 ± 3.13 | 532 | <1 |
| 25 | 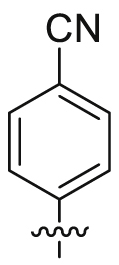 |
SO2NH2 | 17.0 ± 5.78 | 5560 ± 42.3 | 5.31 ± 2.56 | 312 | 1 |
| 26 |  |
CONH2 | 9.93 ± 1.27 | >5488 | 5.48 ± 2.12 | 552 | <1 |
| 27 | 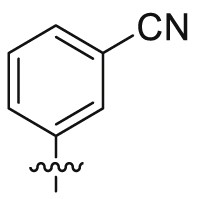 |
SO2NH2 | 13.3 ± 2.60 | 4916 ± 2247 | 14.0 ± 8.28 | 1054 | 3 |
| 28 |  |
CONH2 | 23.4 ± 16.9 | >4169 | 4.16 ± 1.75 | 178 | <1 |
| 29 | 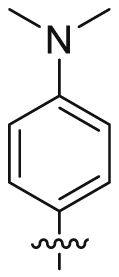 |
SO2NH2 | 79.2 ± 48.7 | >18,381 | 18.3 ± 4.50 | 232 | <1 |
| 30 |  |
CONH2 | 176 ± 26.4 | >23,535 | 23.5 ± 0.38 | 134 | <1 |
| 31 | 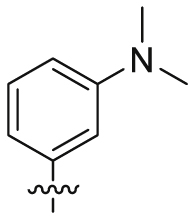 |
SO2NH2 | 882 ± 323 | >110,175 | 110 ± 1.46 | 125 | <1 |
| 32 |  |
CONH2 | 163 ± 57.8 | 4659 ± 824 | 19.8 ± 4.47 | 121 | 4 |
| NVP | – | – | 186 ± 59.0 | >9510 | >9.51 | >51 | NAd |
| EFV | – | – | 5.25 ± 1.98 | >5330 | >6.33 | >1205 | NAd |
| ETR | – | – | 5.03 ± 1.68 | >4594 | >4.59 | >913 | NAd |
| K-5a2e | – | – | 1.4 ± 0.4 | ≥227,890 | ≥227 | >159,101 | NAd |
EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytotoxicity, as determined by the MTT method. A smaller EC50 values means that the compound has greater activity.
CC50: concentration required to reduce the viability of mock-infected cell cultures by 50%, as determined by the MTT method. A larger CC50 values means that the compound has lower toxicity.
SI: selectivity index, the ratio of CC50/EC50.
NA: stands for≥1 or<1.
Data were reported in ref. 14. –Not applicable.
As depicted in Table 1, all the target compounds exhibited effective potency against WT HIV-1 with EC50 values ranging from 9.93 to 882 nmol/L. Among them, most compounds demonstrated EC50 values of 9.93–93.3 nmol/L, which were superior to that of the control drug NVP (EC50 = 186 nmol/L). Especially, compound 26 proved to be the most potent inhibitor against HIV-1 (IIIB) with an EC50 value of 9.93 nmol/L, which was comparable to that of EFV (EC50 = 5.25 nmol/L) and ETR (EC50 = 5.03 nmol/L), but slightly weak to that of K-5a2 (EC50 = 1.4 nmol/L). Besides, compounds 11, 23, 24, 25 and 27 were also endowed with promising anti-HIV-1 potencies (EC50 = 17.7, 11.3, 12.8, 17.0 and 13.3 nmol/L, respectively) and moderate cytotoxicity (CC50 = 13.5, 16.9, 6.80, 5.31 and 14.0 μmol/L, respectively). In the case of HIV-2 (ROD), all the compounds showed weaker activity or lost activity.
Preliminary investigation of the SARs revealed that the nature of the substituent group of the left wing (R1) and right wing (R2) have a significant effect on their antiviral potency. Detailed comparison of the activities of 11–18 suggested that the substitution position of thiophene and furfuran ring have little effect on their activity toward WT HIV-1 strain. However, a phenyl ring seems more favorable to the activity than a thiophene or furfuran ring at the R1 position when comparing the activity of 11–18 (EC50 = 17.7–240 nmol/L) and 19, 20 (EC50 = 43.8–47.7 nmol/L), which could be explained by the stronger stacking interaction between the phenyl moiety and resides in the hydrophobic channel of NNIBP. Elaboration with a F atom or CN substituents at the para-position of the phenyl ring yielded 23–26 with a highly effective potency of 9.93–17.0 nmol/L, while OCH3 or N(CH3)2 substituents at this position (21, 22, 29, 30, EC50 = 52.6–176 nmol/L) resulted in a decreased activity, suggesting that the antiviral activity is very sensitive to the kind of the substituents. Furthermore, by comparing the activity of 25–28 with 29–32, we concluded that the position of CN and N(CH3)2 substituents has little effect on the activity. In addition, the effect of the type of substituent (SO2NH2 and CONH2) in the right wing on the antiviral activity was not obvious.
All the compounds were further evaluated against a panel of clinically relevant NNRTIs-resistant single-mutant strains (L100I, K103N, Y181C, Y188L, E138K) and double-mutant strains F227L + V106A, RES056. The results were depicted in Table 2. Moreover, the SI and RF (fold-resistance factor, EC50 mutant strain/EC50 WT strain) were summarized in Table 3. In general, the compounds activity to mutant strains exhibits a similar trend to their potency to WT HIV-1 strain. Compound 26 demonstrated the most active potency toward all the single-mutant HIV-1 strain, including L100I, K103N, Y181C, Y188L and E138K (EC50 = 10.5, 6.02, 18.9, 23.9 and 23.2 nmol/L, respectively), its activity was superior to that of NVP and EFV, and comparable to that of ETR. Interestingly, all the target compounds displayed lower fold resistance (FR, ratio of EC50 against mutant strain/EC50 against WT strain) values toward K103N than other mutant strains. Most compounds were demonstrated with improved activity against K103N mutation compared to WT strain. For example, 26 displayed the most effective activity against K103N (EC50 = 6.02 nmol/L, FR = 0.6), being more potent than its activity to WT HIV-1 (EC50 = 9.93 nmol/L). For double-mutant HIV-1 strain F227L + V106A, compounds 23 and 27 were proved to be the most potent inhibitors and exhibited EC50 values of 46.6 and 49.2 nmol/L, respectively, being far more potent than NVP (EC50 > 9510 nmol/L) and EFV (EC50 = 268 nmol/L), and comparable to ETR (EC50 = 14.2 nmol/L). In the case of RES056, which is the most challenging double mutation emerging in HIV patients, compounds 25 and 26 provided the greatest potency with EC50 values of 103 and 125 nmol/L respectively, being superior to EFV (EC50 = 265 nmol/L), but slightly inferior to ETR (EC50 42.5 nmol/L) and K-5a2 (EC50 = 30.6 nmol/L).
Table 2.
Activity against mutant HIV-1 strains of target compounds 11–32.
| Compd. | EC50 (nmol/L)a |
||||||
|---|---|---|---|---|---|---|---|
| L100I | K103N | Y181C | Y188L | E138K | F227L + V106A | RES056 | |
| 11 | 89.3 ± 23.0 | 20.7 ± 2.75 | 63.5 ± 8.75 | 53.4 ± 2.15 | 49.9 ± 0.35 | 85.7 ± 9.71 | 565 ± 22.5 |
| 12 | 414 ± 37.9 | 129 ± 5.87 | 511 ± 77.2 | 307 ± 58.2 | 287 ± 4.98 | 1108 ± 269 | 2903 ± 119 |
| 13 | 186 ± 29.4 | 62.9 ± 10.4 | 197 ± 16.1 | 133 ± 3.35 | 193 ± 14.8 | 358 ± 18.4 | 1194 ± 184 |
| 14 | 262 ± 75.0 | 74.8 ± 2.93 | 180 ± 33.4 | 222 ± 30.5 | 177 ± 2.93 | 525 ± 21.1 | 813 ± 177 |
| 15 | 97.9 ± 62.4 | 23.1 ± 2.56 | 91.7 ± 18.2 | 106 ± 43.6 | 124 ± 88.7 | 56.7 ± 16.2 | 999 ± 151 |
| 16 | 731 ± 274 | 184 ± 90.2 | 731 ± 198 | 619 ± 3.59 | 662 ± 58.4 | 1092 ± 226 | 10,011 ± 1066 |
| 17 | 76.0 ± 17.2 | 17.9 ± 1.51 | 63.2 ± 16.1 | 62.0 ± 5.83 | 52.5 ± 11.3 | 56.8 ± 3.85 | 932 ± 88.4 |
| 18 | 925 ± 286 | 171 ± 56.9 | 741 ± 154 | 628 ± 49.6 | 739 ± 318 | 854 ± 186 | 4515 ± 1447 |
| 19 | 344 ± 117 | 59.9 ± 6.01 | 167 ± 24.5 | 121 ± 9.19 | 107 ± 18.8 | 200 ± 10.4 | 1713 ± 473 |
| 20 | 416 ± 87.9 | 102 ± 2.50 | 196 ± 54.6 | 205 ± 13.6 | 146 ± 12.5 | 395 ± 5.14 | 1733 ± 5.51 |
| 21 | 169 ± 2.91 | 57.6 ± 0.67 | 157 ± 5.61 | 146 ± 44.7 | 114 ± 9.20 | 122 ± 59.9 | 1563 ± 1280 |
| 22 | 268 ± 103 | 58.4 ± 7.14 | 158 ± 27.3 | 110 ± 3.57 | 149 ± 42.0 | 297 ± 39.8 | 1123 ± 353 |
| 23 | 45.4 ± 5.26 | 11.7 ± 1.03 | 44.8 ± 5.26 | 51.8 ± 8.47 | 30.9 ± 5.26 | 46.6 ± 15.5 | 388 ± 76.2 |
| 24 | 61.1 ± 9.35 | 11.7 ± 1.09 | 36.1 ± 1.21 | 51.4 ± 9.60 | 34.8 ± 10.6 | 56.6 ± 13.7 | 575 ± 120 |
| 25 | 27.4 ± 4.41 | 13.2 ± 0.33 | 30.8 ± 0.11 | 53.6 ± 3.50 | 44.4 ± 3.62 | 59.9 ± 10.2 | 103 ± 3.39 |
| 26 | 10.5 ± 4.56 | 6.02 ± 0.84 | 18.9 ± 2.28 | 23.9 ± 2.40 | 23.2 ± 8.64 | 97.2 ± 25.3 | 125 ± 39.3 |
| 27 | 24.6 ± 4.97 | 8.56 ± 1.69 | 32.4 ± 4.07 | 54.7 ± 2.26 | 34.8 ± 9.28 | 49.2 ± 8.26 | 347 ± 75.9 |
| 28 | 50.2 ± 10.3 | 20.0 ± 1.20 | 51.6 ± 4.32 | 73.4 ± 5.16 | 41.9 ± 4.56 | 157 ± 6.60 | 402 ± 41.9 |
| 29 | 247 ± 4.50 | 97.2 ± 3.07 | 260 ± 30.2 | 269 ± 30.0 | 183 ± 19.4 | 140 ± 1.31 | 1682 ± 388 |
| 30 | 1115 ± 543 | 173 ± 46.3 | 585 ± 33.3 | 556 ± 57.4 | 245 ± 173 | 1253 ± 804 | >23,535 |
| 31 | 2401 ± 256 | 685 ± 0.21 | 2300 ± 413 | 1992 ± 46.5 | 979 ± 185 | 2157 ± 298 | 29,610 ± 4817 |
| 32 | 1135 ± 99.2 | 212 ± 7.34 | 626 ± 39.2 | 552 ± 45.6 | 198 ± 34.2 | 783 ± 196 | 4298 ± 1542 |
| NVP | 1403 ± 828 | 7818 ± 556 | >9510 | >9510 | 115 ± 24.5 | >9510 | >9510 |
| EFV | 46.5 ± 9.11 | 111 ± 35.4 | 6.33 ± 1.06 | 306 ± 53.9 | 5.12 ± 1.28 | 268 ± 48.7 | 265 ± 47.9 |
| ETR | 6.35 ± 1.77 | 3.13 ± 0.42 | 12.2 ± 2.63 | 17.3 ± 5.53 | 8.07 ± 2.52 | 14.2 ± 5.14 | 42.5 ± 15.1 |
| K-5a2b | 3.4 ± 0.6 | 2.9 | 3.2 ± 0.4 | 3.0 ± 0.1 | 2.9 | 4.2 ± 1.2 | 30.6 ± 12 |
EC50: concentration of compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytotoxicity, as determined by the MTT method.
Data were reported in Ref. 14.
Table 3.
SI and RF values of target compounds 11–32.
| Compd. | SI (RF)a |
||||||
|---|---|---|---|---|---|---|---|
| L100I | K103N | Y181C | Y188L | E138K | F227L + V106A | RES056 | |
| 11 | 151 (5.0) | 651 (1.2) | 213 (3.6) | 253 (3.0) | 271 (2.8) | 158 (4.8) | 24 (31.9) |
| 12 | 70 (4.4) | 224 (1.4) | 57 (5.5) | 94 (3.3) | 101 (3.1) | 26 (11.9) | 10 (31.1) |
| 13 | 84 (2.9) | 249 (1.0) | 79 (3.1) | 117 (2.1) | 81 (3.0) | 44 (5.6) | 13 (18.8) |
| 14 | 60 (4.6) | 212 (1.3) | 88 (3.1) | 71 (3.9) | 89 (3.1) | 30 (9.1) | 19 (14.1) |
| 15 | 220 (2.0) | 932 (0.5) | 235 (1.9) | 203 (2.1) | 173 (2.5) | 380 (1.1) | 22 (20.2) |
| 16 | ≥28 (3.6) | ≥111 (0.9) | ≥28 (3.6) | ≥33 (3.0) | ≥31 (3.3) | ≥19 (5.4) | ≥2 (49.2) |
| 17 | 203 (2.4) | 865 (0.6) | 245 (2.0) | 249 (2.0) | 295 (1.7) | 272 (1.8) | 17 (29.5) |
| 18 | 18 (3.8) | 99 (0.7) | 23 (3.1) | 27 (2.6) | 23 (3.1) | 20 (3.6) | 4 (18.8) |
| 19 | 62 (7.9) | 356 (1.4) | 127 (3.8) | 176 (2.8) | 200 (2.4) | 106 (4.6) | 12 (39.1) |
| 20 | 42 (8.7) | 171 (2.1) | 89 (4.1) | 85 (4.3) | 120 (3.1) | 44 (8.3) | 10 (36.3) |
| 21 | 98 (2.8) | 289 (0.9) | 105 (2.6) | 114 (2.4) | 145 (1.9) | 136 (2.0) | 11 (25.6) |
| 22 | 50 (5.1) | 230 (1.1) | 85 (3.0) | 122 (2.1) | 90 (2.8) | 45 (5.7) | 12 (21.4) |
| 23 | 372 (4.0) | 1441 (1.0) | 377 (4.0) | 326 (4.6) | 547 (2.7) | 363 (4.1) | 43 (34.3) |
| 24 | 111 (4.8) | 578 (0.9) | 189 (2.8) | 132 (4.0) | 195 (2.7) | 120 (4.4) | 12 (44.9) |
| 25 | 194 (1.6) | 403 (0.8) | 173 (1.8) | 99 (3.2) | 120 (2.6) | 89 (3.5) | 51 (6.1) |
| 26 | 521 (1.1) | 910 (0.6) | 290 (1.9) | 229 (2.4) | 236 (2.3) | 56 (9.8) | 44 (12.6) |
| 27 | 569 (1.9) | 1639 (0.6) | 432 (2.4) | 256 (4.1) | 402 (2.6) | 285 (3.7) | 40 (26.1) |
| 28 | 83 (2.1) | 208 (0.9) | 81 (2.2) | 57 (3.1) | 99 (1.8) | 26 (6.7) | 10 (17.2) |
| 29 | 74 (3.1) | 189 (1.2) | 71 (3.3) | 68 (3.4) | 100 (2.3) | 131 (1.8) | 11 (21.2) |
| 30 | 21 (6.3) | 135 (1.0) | 40 (3.3) | 42 (3.2) | 96 (1.4) | 19 (7.1) | <1 (>133) |
| 31 | 46 (2.7) | 161 (0.8) | 48 (2.6) | 55 (2.3) | 112 (1.1) | 51 (2.4) | 4 (33.5) |
| 32 | 17 (6.9) | 93 (1.3) | 32 (3.8) | 36 (3.4) | 100 (1.2) | 25 (4.8) | 5 (26.2) |
| NVP | >7 (7.5) | >1 (42) | NA (>51.1) | NA (>51) | >82 (0.6) | NA (>51.1) | NA (>51.1) |
| EFV | >136 (8.8) | >57 (21) | >1000 (1.2) | >21 (58) | >1237 (1.0) | >24 (51.1) | >24 (51.0) |
| ETR | >723 (1.3) | >1463 (0.6) | >376 (2.4) | >264 (3.5) | >569 (1.6) | >322 (2.8) | >108 (8.4) |
RF is the ratio of EC50 (resistant viral strain)/EC50 (wild-type viral strain).
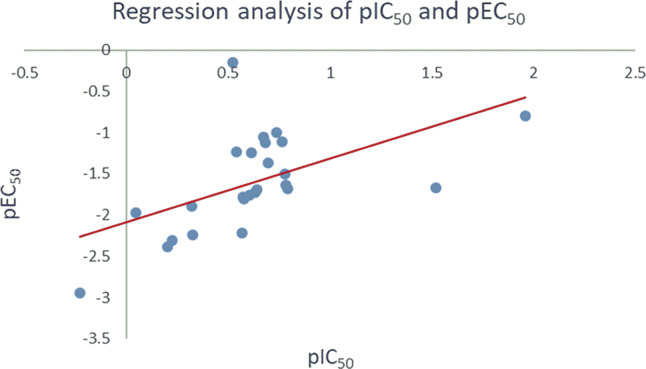
With the aim to validate the binding target of these novel piperidine-substituted thiophene[3,2-d]pyrimidine derivatives, their inhibitory effects on WT HIV-1 RT were investigated. As shown in Table 4, most compounds exhibited effective inhibitory activity (IC50 = 0.161–0.624 μmol/L) to HIV-1 RT, being comparable to the lead K-5a2 (IC50 = 0.300 μmol/L), superior to the reference drug NVP (IC50 = 1.220 μmol/L) and inferior to ETR (IC50 = 0.011 μmol/L). Compound 31, the weakest inhibitor of WT HIV-1 strain in cell culture (EC50 = 882 nmol/L), exhibited the lowest binding-affinity for WT HIV-1 RT (IC50 = 1.681 μmol/L). In general, the enzyme inhibitory activity (IC50) of these novel synthesized compounds was consistent with their anti-HIV-1 (IIIB) activity (EC50), and there was a good linear relationship between them (Fig. 3). All the results indicated that these novel synthesized piperidine-substituted thiophene[3,2-d]pyrimidine derivatives are classical NNRTIs.
Table 4.
Inhibitory activity against WT HIV-1 RT of target compounds 11–32.
| Compd. | IC50 (μmol/L)a | Compd. | IC50 (μmol/L)a |
|---|---|---|---|
| 11 | 0.243 ± 0.037 | 22 | 0.233 ± 0.028 |
| 12 | 0.901 ± 0.045 | 23 | 0.211 ± 0.015 |
| 13 | 0.265 ± 0.059 | 24 | 0.171 ± 0.010 |
| 14 | 0.249 ± 0.001 | 25 | 0.288 ± 0.026 |
| 15 | 0.227 ± 0.046 | 26 | 0.182 ± 0.016 |
| 16 | 0.595 ± 0.044 | 27 | 0.207 ± 0.006 |
| 17 | 0.167 ± 0.014 | 28 | 0.202 ± 0.020 |
| 18 | 0.624 ± 0.055 | 29 | 0.477 ± 0.009 |
| 19 | 0.164 ± 0.005 | 30 | 0.473 ± 0.059 |
| 20 | 0.161 ± 0.014 | 31 | 1.681 ± 0.056 |
| 21 | 0.268 ± 0.052 | 32 | 0.271 ± 0.016 |
| NVP | 1.220 ± 0.337 | ETRb | 0.011 ± 0.000 |
| EFVb | 0.030 ± 0.000 | K-5a2b | 0.300 ± 0.060 |
IC50: inhibitory concentration of test compound required to inhibit biotin deoxyuridine triphosphate (biotindUTP) incorporation into WT HIV-1 RT by 50%.
The data were obtained from the same laboratory with the same method (Prof. Erik De Clercq, Rega Institute for Medical Research, KU Leuven, Belgium).
Figure 3.
Regression analysis of pIC50 and pEC50 values for the newly synthesized derivatives.
2.3. Molecular modelling studies
Classical molecular dynamics (MD) simulations and MM/GBSA free energy calculations were performed to identify the molecular determinants responsible of the activity profile of the most effective inhibitor 26 bound to the WT and the mutated forms of HIV-1 RT. To this end, the X-ray crystal structure of the WT HIV-1 RT (PDB code: 6c0n), and its K103N (PDB code: 6c0o) and RES056 (K103N/Y181C) (PDB code: 6c0r) variants were used16. The co-crystallized NNRTIs K-5a2 was used as template to place 26 into the inhibitor's binding site, according to the close similarity between the crystallographic and simulated ligands.
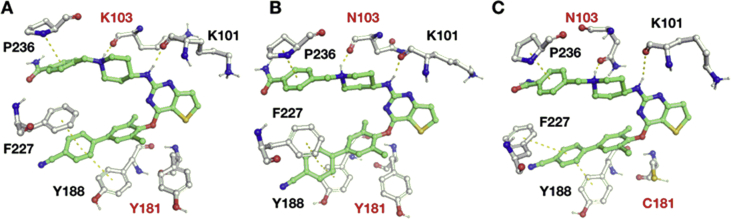
As depicted in Fig. 4, compound 26 adopts a horseshoe-like conformation in the NNIBP, which was commonly observed in most DAPY NNRTIs. The complexes of 26 with WT, K103N or RES056 RT display the following common interactions: (i) the newly introduced biphenyl structure is stably accommodated into the hydrophobic channel and stabilized by π-stacking interactions with Y/C181, Y188, F227, and W229; (ii) the thiophene ring of 26 is projected towards the NNIBP entrance channel surrounded by V179 and E138; and (iii) the piperidine-linked benzamide structure of the right wing occupies the tolerant region I surrounded by K/N103, K104, V106, and F227.
Figure 4.
Final geometries for 26 in complexs with WT (A), K103N (B) and RES056 (C) RT. Hydrogen-bonding interactions for the –NH guanidine group and the protonated nitrogen atom with K101 and K103 of the protein as well as the π-stacking with F227 and Y188 are also highlighted.
The stability of the three protein–ligand complexes formed by 26 with the WT HIV-1 RT, mutant HIV-1 K103N and RES056 RT was analysed by means of 100 ns MD simulations. Final geometries for the three simulated systems are shown in Fig. 4. All the simulated systems were stable during MD simulation, as demonstrated by the root-mean square deviation (RMSD) values (Supporting Information Fig. S1A, B and C) for both the ligand and the residues that shape the NN binding site.
Accordingly to the binding mode, the –NH guanidine group of the ligand is hydrogen-bonded to the backbone oxygen of K101 with average (K101)C O┄HN (26) distances of 2.4 ± 0.5, 2.3 ± 0.5 and 3.2 ± 0.5 Å respectively for the WT RT, and its single and double mutated variants (Fig. S1D, E and F) calculated along the last 30 ns of the MD trajectory. The protonated pyrimidine nitrogen of 26 is also involved in hydrogen-bonding with the backbone oxygen of K/N103 with average (K/N103)C O┄HN (26) distances of 2.5 ± 0.5, 2.3 ± 0.5 and 4.1 ± 0.6 Å respectively for the WT, singly and doubly mutated species (Fig. S1D, E and F).
In the hydrophobic channel of NNIBP, the benzonitryl moiety of 26 stacks against Y188 and F227 (Fig. 4) with average distances of (Y188/P227) 4.4 ± 0.6/3.3 ± 0.6 Å for WT RT, 3.9 ± 0.4/3.4 ± 0.6 Å for the K103N variant, and 4.5 ± 0.7/4.7 ± 0.7 Å for the K103N/Y181C mutated enzyme (Fig. S1G, H and I). The involvement of F227 is allowed by the rearrangement of its side chain during the second part of the MD simulations.
As noted in the Experimental Section of this work, a similar (EC50 = 9.93 and 6.02 nmol/L) inhibitory activity was found by 26 in the WT and the single mutant K013N, whereas it was 20-fold less potent (EC50 = 125 nmol/L) against the double mutant RES056. This last data could be explained by the gradual displacement of the ligand within the NNIBP observed during the MD simulation. This gradual displacement allowed the insertion of a water molecule with a consequent weakening of the (K101)C O┄HN (26) hydrogen–bonding interaction (Fig. S1F). Furthermore, the benzonitrile moiety slightly deviates from the optimal orientation required for the π-stacking interaction with Y188 and F227 in the hydrophobic channel. All these factors would contribute to the reduced activity of 26 against RES056. Indeed, the structural destabilization observed for RES056 in complex with 26 was confirmed by MM/GBSA free energy calculations performed on the last 30 ns for the previously generated MD trajectories. In this regard, binding affinities (ΔGbind) of –73.4 and –74.1 kcal/mol were obtained respectively for the WT HIV-1 RT and the K103N mutant (Table 5), whereas a binding free energy of –65.9 kcal/mol was observed for 26 in the NNIBP of the double mutant RES056.
Table 5.
MM/GBSAa calculations for the three MD simulated systems. Values are expressed in kcal/mol.
| System | EvdW | Eelect | ΔGgas | EGB | ESURF | ΔGsolv | ΔGbind |
|---|---|---|---|---|---|---|---|
| WT+26 | −87.2 ± 4.3 | −2.9 ± 8.8 | −90.1 ± 10.8 | 26.2 ± 8.8 | −9.6 ± 0.2 | 16.6 ± 8.7 | −73.4 ± 4.7 |
| K103N + 26 | −88.2 ± 3.3 | −39.3 ± 9.2 | −127.4 ± 9.8 | 62.0 ± 9.3 | −9.5 ± 0.2 | 52.5 ± 9.3 | −74.1 ± 3.9 |
| RES056 + 26 | −3.1 ± 3.4 | −35.2 ± 13.3 | −118.2 ± 14.4 | 62.1 ± 13.1 | −9.6 ± 0.2 | 52.4 ± 13.0 | −65.9 ± 3.8 |
EvdW: ligand–protein van der Waals interaction energy; Eelect: ligand–protein electrostatic interaction energy; ΔGgas = EvdW + Eelect; EGB: Generalized-Born electrostatic component of the solvation free energy; ESURF: Solvent-accessible surface component of the solvation free energy; ΔGsolv = EGB + ESURF; ΔGbind = ΔGgas+ΔGsolv.
A total of 100 snapshots taken from the last 30 ns of the MD trajectories for the three simulated complexes were considered.
2.4. In vivo pharmacokinetics (PK) study and acute toxicity assessment
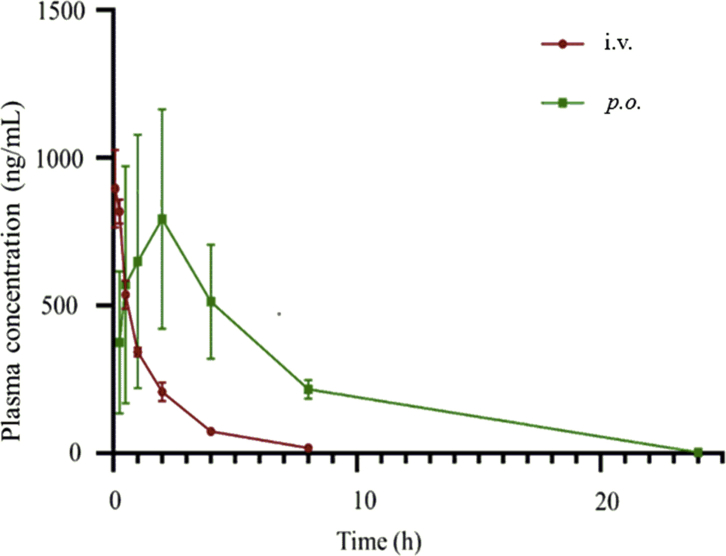
Our study was approved by the ethics committee of Cheeloo College of Medicine, Shandong University (Jinan, China). All procedures performed in studies were in accordance with the ethical standards of the institution or practice at which the studies were conducted. The in vivo pharmacokinetic profile of compound 26 was examined in Wistar rat PK model (Fig. 5 and Table 6). After a single 2 mg/kg i.v. dose, 26 was characterized by a modest clearance (CL = 25.0 mL/min/kg), a short terminal half-life (t1/2 = 1.71 h) and a maximum concentration (Cmax) of 941 ng/mL. When administered at 20 mg/kg orally, compound 26 was rapidly absorbed with a Tmax of 2.00 h, a favorable half-life of 2.68 h, and a Cmax of 762 ng/mL. The oral bioavailability (F) was 33.8%, which was sufficiently enough for a drug candidate.
Figure 5.
The plasma concentration–time profiles of 26 in Wistar rat following p.o. administration (20 mg/kg) and i.v. administration (2 mg/kg).
Table 6.
Pharmacokinetic profile of 26a.
| Subject | t1/2 | Tmax | Cmax | AUC0–last | AUC0–inf | CL | F |
|---|---|---|---|---|---|---|---|
| (h) | (h) | (ng/mL) | (h⋅ng/mL) | (h⋅ng/mL) | (mL/min/kg) | (%) | |
| 26 (i.v.)b | 1.71 ± 0.24 | – | 941 ± 181 | 1290 ± 62.7 | 1336 ± 53.9 | 25.0 ± 1.04 | – |
| 26 (p.o.)c | 2.68 ± 0.20 | 2.00 ± 0.32 | 762 ± 296 | 4359 ± 1570 | 4761 ± 1354 | – | 33.8 |
PK parameters (mean ± SD, n = 3).
Dosed i.v. at 2 mg/kg.
Dosed p.o. at 20 mg/kg –Not applicable.
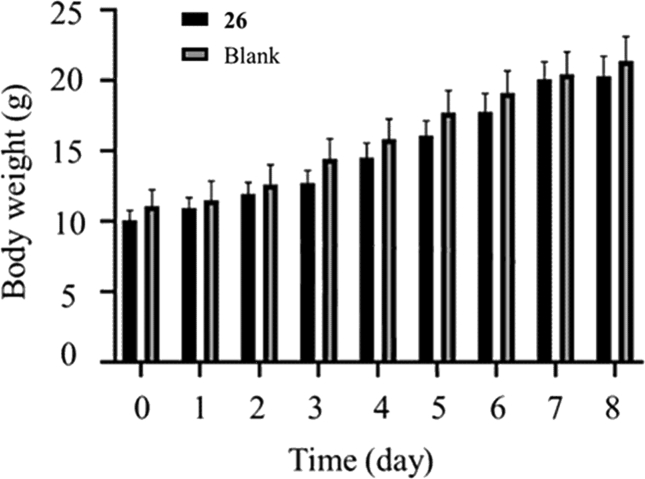
Furthermore, 26 was performed acute toxicity experiment in Kunming mice. As depicted in Fig. 6, no mortality or poisoning symptom was found after given the mice with single oral doses of 2000 mg/kg. Besides, there have no significant weight and behaviors changes compared to the control group during the post-treatment period of 8 days.
Figure 6.
The relative body weight changes of Kunming mice in different groups (26 and blank).
3. Conclusions
In conclusion, we designed and synthesized a novel series of piperidine-substituted thiophene[3,2-d]pyrimidine derivatives by incorporating an aromatic moiety to interact with the hydrophobic channel of NNIBP. These compounds showed effective potency against WT and most of NNRTIs-resistant HIV-1 strain, especially to the challenging K103N mutation. Compound 26 turns out as the most potent inhibitor with EC50 values of 9.93, 10.5, 6.02, 18.9, 23.9 and 23.2 nmol/L against WT, L100I, K103N, Y181C, Y188L and E138K strains, respectively, being comparable to those of ETR. Furthermore, 26 also exerted stronger binding affinity for WT HIV-1 RT with an IC50 value of 0.182 μmol/L. Molecular simulation provides a reasonable explanation why 26 exhibited different activity against a panel of HIV-1 mutant strains. Notably, 26 exhibited favorable PK profiles, with a moderate clearance in rats and its bioavailability is up to 33.8%. Taken together, the results will contribute to explore the hydrophobic channel of NNIBP for developing novel potent NNRTIs and compound 26 holds great promise as a potential drug candidate for the treatment of HIV-1 infection.
4. Experimental
4.1. Chemistry
All melting points were determined on a micro melting point apparatus (YRT-3, Tianjin, China) and are uncorrected. 1H NMR and 13C NMR spectra were recorded in CDCl3 or DMSO-d6 on a Bruker AV-400 spectrometer (Zurich, Switzerland) with tetramethylsilane (TMS) as the internal standard. A G1313A Standard LC Autosampler (Agilent, Palo Alto, CA, USA) was used to collect samples for measurement of mass spectra. All reactions were monitored by thin layer chromatography (TLC), and spots were visualized with iodine vapor or by irradiation with UV light. Flash column chromatography was performed on columns packed with silica gel (200–300 mesh, Qingdao, China). Solvents were of reagent grade and were purified by standard methods when necessary.
4.1.1. General procedure for the preparation of intermediates 6a–k
4-Iodo-2,6-dimethylphenol (5, 1.00 mmol) and boric acid substituents (1.20 mmol) were dissolved in DMF (10 mL), then Pd(PPh3)4 (0.05 mmol) and 2 mol/L Na2CO3 aqueous solution (2.00 mmol) were added. The reaction mixture was stirred under N2 at 100 °C for 6–10 h (monitored by TLC). The solution was cooled to room temperature and 10 mL of water was added. The aqueous layer was extracted with EtOAc, then the organic phase was dried over Na2SO4, filtered and evaporated under reduced pressure gave the intermediates 6a–k, which were conducted next step without further purification.
4.1.2. General procedure for the preparation of intermediates 8a–k
A reaction mixture of 6a–k (10 mmol), 2,4-dichlorothiopheno[3,2-d]pyrimidine (2.1 g, 10 mmol) and potassium carbonate (1.7 g, 12 mmol) in DMF (30 mL) was stirred at room temperature for 2–4 h (monitoring with TLC), then the mixture was poured into water (100 mL). The precipitated white solid was collected by filtration, washed with water, and recrystallized in DMF-H2O to provide the desired compounds 8a–k.
4.1.2.1. 2-Chloro-4-(4-(furan-2-yl)-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidine (8a)
White solid, Yield 82%, m.p.: 175–177 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.57 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.78 (d, J = 1.8 Hz, 1H, C3-furan-H), 7.67 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.57 (s, 2H, C3,C5-Phʹ-H), 6.97 (d, J = 3.3 Hz, 1H, C5-furan-H), 6.63 (dd, J = 3.4, 1.8 Hz, 1H, C4-furan-H), 2.13 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 165.0, 163.7, 152.8, 148.3, 143.5, 140.3, 131.5, 129.1, 124.3, 124.1, 115.7, 112.6, 106.6, 16.4. ESI-MS: m/z 357.2 [M+H]+, 379.4 [M+Na]+. C18H13ClN2O2S (356.04).
4.1.2.2. 2-Chloro-4-(4-(furan-3-yl)-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidine (8b)
White solid, Yield 84%, m.p.: 163–165 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.00 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.57–7.56 (m, 2H), 7.52 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.28 (s, 2H, C3,C5-Phʹ-H), 7.13–7.12 (m, 1H), 2.20 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.5, 163.8, 163.7, 161.2, 156.8, 148.5, 138.5, 136.9, 136.6, 131.0, 128.7, 128.6, 127.5, 124.1, 115.7, 115.5, 16.6. ESI-MS: m/z 357.2 [M+H]+. C18H13ClN2O2S (356.04).
4.1.2.3. 2-Chloro-4-(2,6-dimethyl-4-(thiophen-2-yl)phenoxy)thieno[3,2-d]pyrimidine (8c)
White solid, Yield 92%, m.p.: 143–145 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.98 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.50 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.38 (s, 2H, C3,C5-Phʹ-H), 7.31 (dd, J = 3.6, 1.2 Hz, 1H, C3-thiophen-H), 7.28 (dd, J = 5.1, 1.1 Hz, 1H, C5-thiophen-H), 7.08 (dd, J = 5.1, 3.6 Hz, 1H, C4-thiophen-H), 2.18 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.6, 163.8, 148.5, 143.5, 136.9, 132.7, 131.3, 128.0, 126.4, 124.9, 124.0, 123.3, 16.5. ESI-MS: m/z 373.2 [M+H]+. C18H13ClN2OS2 (372.02).
4.1.2.4. 2-Chloro-4-(2,6-dimethyl-4-(thiophen-3-yl)phenoxy)thieno[3,2-d]pyrimidine (8d)
White solid, Yield 90%, m.p.: 140–142 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.58 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.91 (dd, J = 3.0, 1.4 Hz, 1H, C4-thiophen-H), 7.69–7.66 (m, 2H), 7.61 (d, J = 1.4 Hz, 1H, C5-thiophen-H), 7.59 (s, 2H, C3,C5-Ph′-H), 2.13 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 165.0, 163.7, 155.7, 148.1, 141.1, 140.2, 133.9, 131.2, 127.5, 127.0, 126.7, 124.1, 121.6, 115.8, 39.7, 16.5. ESI-MS: m/z 373.1 [M+H]+, 395.2 [M+Na]+. C18H13ClN2OS2 (372.02).
4.1.2.5. 2-Chloro-4-((3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidine (8e)
White solid, Yield 93%, m.p.: 150–151 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.59 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.72 (d, J = 7.3 Hz, 2H, C2,C6-Phʹʹ-H), 7.68 (d, J = 5.4, 1H, C7-thienopyrimidine-H), 7.53 (s, 2H, C3,C5-Phʹ-H), 7.49 (t, J = 7.5 Hz, 2H, C3,C5-Phʹʹ-H), 7.40 (d, J = 7.5 Hz, 1H, C4-Phʹʹ-H), 2.16 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 165.0, 164.9, 163.7, 163.3, 155.7, 148.6, 140.3, 139.8, 138.8, 137.7, 133.6, 131.2, 129.3, 127.9, 127.2, 124.1, 115.9, 92.2, 16.5. ESI-MS: m/z 367.2 [M+H]+, 389.2 [M+Na]+. C20H15ClN2OS (366.06).
4.1.2.6. 2-Chloro-4-((4′-methoxy-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidine (8f)
White solid, Yield 94%, m.p.: 150–151 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.97 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.52 (s, 2H, C2,C6-Phʹʹ-H), 7.50 (d, J = 5.6 Hz, 1H, C7-thienopyrimidine-H), 7.30 (s, 2H, C3,C5-Phʹ-H), 6.99–6.91 (m, 2H, C3,C5-Phʹʹ-H), 3.85 (s, 3H, CH3), 2.19 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.5, 163.9, 159.2, 148.1, 139.1, 136.9, 132.9, 130.9, 128.1, 127.2, 124.0, 114.1, 55.3, 16.6. ESI-MS: m/z 397.2 [M+H]+, 419.2 [M+Na]+. C21H17ClN2O2S (396.07).
4.1.2.7. 2-Chloro-4-((4′-fluoro-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidine (8g)
White solid, Yield 89%, m.p.: 126–128 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.98 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.74–7.68 (m, 2H, C2,C6-Phʹʹ-H), 7.51 (d, J = 5.6 Hz, 1H, C7-thienopyrimidine-H), 7.31 (s, 2H, C3,C5-Phʹ-H), 6.72–6.70 (m, 2H, C3,C5-Phʹʹ-H), 2.17 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.5, 163.8, 156.8, 148.0, 143.7, 138.5, 136.9, 131.1, 130.7, 127.6, 126.3, 125.7, 124.0, 115.4, 108.9, 16.5. ESI-MS: m/z 385.2 [M+H]+, 407.3 [M+Na]+. C20H14ClFN2OS (384.05).
4.1.2.8. 4'-((2-Chlorothieno[3,2-d]pyrimidin-4-yl)oxy)-3′,5′-dimethyl-[1,1′-biphenyl]-4-carbonitrile (8h)
White solid, Yield 85%, m.p.: 131–133 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.59 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.95 (s, 4H, Phʹʹ-H), 7.68 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.64 (s, 2H, C3, C5-Phʹ-H), 2.16 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 165.0, 163.6, 149.6, 144.2, 140.3, 136.8, 133.2, 131.6, 128.1, 128.0, 124.2, 119.3, 110.5, 16.5. ESI-MS: m/z 392.2 [M+H]+. C21H14ClN3OS (391.05).
4.1.2.9. 4'-((2-Chlorothieno[3,2-d]pyrimidin-4-yl)oxy)-3′,5′-dimethyl-[1,1′-biphenyl]-3-carbonitrile (8i)
White solid, Yield 82%, m.p.: 142–144 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.03 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.86 (t, J = 1.7 Hz, 1H, C4-Phʹʹ-H), 7.82 (dt, J = 7.9, 1.6 Hz, 1H, C6-Phʹʹ-H), 7.66–7.62 (m, 1H, C2-Phʹʹ-H), 7.56 (d, J = 7.8 Hz, 1H, C5-Phʹʹ-H), 7.53 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.31 (s, 2H, C3,C5-Phʹ-H), 2.22 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.6, 163.6, 149.3, 141.7, 137.0, 136.9, 131.5, 131.4, 130.7, 129.5, 127.6, 124.1, 118.8, 112.9, 16.6. ESI-MS: m/z 392.2 [M+H]+, 414.2 [M+Na]+. C21H14ClN3OS (391.05).
4.1.2.10. 4'-((2-Chlorothieno[3,2-d]pyrimidin-4-yl)oxy)-N,N,3′,5′-tetramethyl-[1,1′-biphenyl]-4-amine (8j)
White solid, Yield 88%, m.p.: 137–138 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.95 (d, J = 5.5 Hz, 1H, C6-thienopyrimidine-H), 7.51 (d, J = 8.9 Hz, 2H, C2,C6-Phʹʹ-H), 7.49 (d, J = 5.6 Hz, 1H, C7-thienopyrimidine-H), 7.30 (s, 2H, C3,C5-Phʹ-H), 6.81 (d, J = 8.8 Hz, 2H, C3,C5-Phʹʹ-H), 3.00 (s, 6H, NCH3), 2.18 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.6, 164.0, 156.8, 150.0, 147.6, 139.5, 136.9, 130.8, 128.3, 127.6, 126.7, 124.0, 115.4, 112.7, 76.8, 40.6, 16.7. ESI-MS: m/z 410.2 [M+H]+. C22H20ClN3OS (409.10).
4.1.2.11. 4'-((2-Chlorothieno[3,2-d]pyrimidin-4-yl)oxy)-N,N,3′,5′-tetramethyl-[1,1′-biphenyl]-3-amine (8k)
White solid, Yield 84%, m.p.: 130–132 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.98 (d, J = 5.5 Hz, 1H, C6-thienopyrimidine-H), 7.51 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.34 (s, 2H, C3,C5-Phʹ-H), 7.32–7.24 (m, 1H), 6.99–6.91 (m, 2H), 6.75 (dd, J = 8.3, 2.5 Hz, 1H, C5-Phʹʹ-H), 3.03 (s, 6H, NCH3), 2.20 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.5, 163.9, 156.8, 150.9, 148.4, 141.4, 140.6, 136.9, 130.8, 129.4, 127.8, 124.0, 115.8, 115.5, 111.7, 111.5, 40.7, 16.6. ESI-MS: m/z 410.2 [M+H]+, 422.2 [M+Na]+. C22H20ClN3OS (409.10).
4.1.3. General procedure for the preparation of intermediates 10a–k
PdCl2(PPh3)2 (0.07 g, 0.1 mmol) and BINAP (0.06 g, 0.1 mmol) were dissolved in dry dioxane (20 mL), and then tert-butyl(4-aminophenyl)carbamate (0.50 g, 2.4 mmol), intermediates 8a–k (0.01 mmol) and Cs2CO3 (0.97 g, 3.0 mmol) were added to the solution. The reaction mixture was stirred under N2 at 120 °C for 10–12 h (monitoring with TLC). After cooling to room temperature, the solvent was evaporated under reduce pressure, and EtOAc was added to the residue. The organic layer was extracted three times with water, then the organic phase was dried over Na2SO4, filtered and evaporated under reduced pressure gave the intermediates 9a–k, which was used directly in the next step without further purification. Then the intermediates 9a–k (2.0 mmol) and TFA (1.50 mL, 20 mmol) were dissolved in DCM (4 mL), the mixed solution was stirred for 3–6 h (monitored by TLC) at room temperature. Then the solution was alkalized to pH 9 with saturated NaHCO3 and extracted with DCM (3 × 5 mL). The organic phase was dried over anhydrous Na2SO4, filtered, concentrated and purified by flash column chromatography to give the intermediates 10a–k.
4.1.3.1. 4-(4-(Furan-2-yl)-2,6-dimethylphenoxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10a)
White solid, Yield 56%, m.p.: 150–152 °C. 1H NMR (400 MHz, chloroform-d) δ 9.20 (s, 1H, NH), 7.78 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.45 (d, J = 1.7 Hz, 1H, C3-furan-H), 7.42 (s, 2H, C3,C5-Ph′-H), 7.21 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.62 (d, J = 3.3 Hz, 1H, C5-furan-H), 6.46 (dd, J = 3.4, 1.8 Hz, 1H, C4-furan-H), 5.25 (s, 1H, NH), 3.87 (s, 1H), 3.33–3.30 (m, 2H), 2.87 (s, 2H), 2.16 (s, 6H), 2.11–2.02 (m, 2H), 1.75–167 (s, 2H). 13C NMR (100 MHz, chloroform-d) δ 163.7, 159.6, 153.3, 148.7, 142.1, 135.2, 131.4, 128.7, 123.9, 122.9, 111.7, 107.4, 105.0, 46.3, 42.7, 28.5, 16.5. ESI-MS: m/z 421.2 [M+H]+. C23H24N4O2S (420.16).
4.1.3.2. 4-(4-(Furan-3-yl)-2,6-dimethylphenoxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10b)
White solid, Yield 50%, m.p.: 148–150 °C. 1H NMR (400 MHz, chloroform-d) δ 8.23 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.92–7.91 (m, 2H), 7.83 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.46 (s, 2H, C3,C5-Phʹ-H), 7.42–7.41 (m, 1H), 5.32 (s, 1H, NH), 3.88 (s, 1H), 3.30–3.27 (m, 2H), 2.87–2.85 (m, 2H), 2.21 (s, 6H), 2.03–1.71 (m, 4H). 13C NMR (100 MHz, chloroform-d) δ 165.2, 164.7, 163.9, 161.5, 155.9, 147.6, 138.7, 136.1, 135.7, 131.6, 128.7, 126.8, 124.9, 115.7, 115.2, 46.4, 42.2, 28.7, 16.6. ESI-MS: m/z 421.2 [M+H]+, 438.3 [M+NH4]+. C23H24N4O2S (420.16).
4.1.3.3. 4-(2,6-Dimethyl-4-(thiophen-2-yl)phenoxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10c)
White solid, Yield 47%, m.p.: 161–162 °C. 1H NMR (400 MHz, chloroform-d) δ 7.92 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.48 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.32 (s, 2H, C3,C5-Phʹ-H), 7.31 (dd, J = 3.6, 1.1 Hz, 1H, C3-thiophen-H), 7.27 (dd, J = 5.2, 1.1 Hz, 1H, C5-thiophen-H), 7.10 (dd, J = 5.2, 3.6 Hz, 1H, C4-thiophen-H), 5.32 (s, 1H, NH), 3.88 (s, 1H), 3.30–3.27 (m, 2H), 2.86–2.85 (m, 2H), 2.18 (s, 6H), 2.03–1.71 (m, 4H). 13C NMR (100 MHz, chloroform-d) δ 164.6, 163.8, 148.5, 143.5, 136.9, 132.7, 131.3, 128.0, 126.4, 124.9, 124.0, 123.3, 46.3, 42.5, 28.6, 16.5. ESI-MS: m/z 437.2 [M+H]+. C23H24N4OS2 (436.14).
4.1.3.4. 4-(2,6-Dimethyl-4-(thiophen-3-yl)phenoxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10d)
White solid, Yield 57%, m.p.: 157–159 °C. 1H NMR (400 MHz, chloroform-d) δ 8.52 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.87 (dd, J = 3.1, 1.5 Hz, 1H, C4-thiophen-H), 7.68–7.62 (m, 2H), 7.61 (d, J = 1.5 Hz, 1H, C5-thiophen-H), 7.58 (s, 2H, C3,C5-Phʹ-H), 5.31 (s, 1H, NH), 3.88 (s, 1H), 3.29–3.28 (m, 2H), 2.86–2.83 (m, 2H), 2.13 (s, 6H), 2.01–1.98 (m, 2H), 1.74–1.52 (m, 2H). 13C NMR (100 MHz, chloroform-d) δ 165.2, 163.5, 155.9, 148.6, 143.6, 141.3, 140.0, 133.4, 131.2, 127.8, 127.0, 126.4, 123.9, 121.6, 116.0, 46.3, 39.7, 28.6, 16.5. ESI-MS: m/z 437.2 [M+H]+, 454.6 [M+NH4]+. C23H24N4OS2 (436.14).
4.1.3.5. 4-((3,5-Dimethyl-[1,1′-biphenyl]-4-yl)oxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10e)
White solid, Yield 46%, m.p.: 153–155 °C. 1H NMR (400 MHz, chloroform-d) δ 7.79 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.59 (d, J = 7.2 Hz, 2H, C2,C6-Phʹʹ-H), 7.43 (t, J = 7.6 Hz, 2H, C3,C5-Phʹ-H), 7.34–7.32 (m, 3H), 7.21 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 5.04 (s, 1H, NH), 3.89 (s, 1H), 3.32–3.30 (m, 2H), 2.92–2.87 (m, 2H), 2.20 (s, 6H), 2.16–2.05 (m, 2H), 1.77–1.69 (m, 2H). 13C NMR (100 MHz, chloroform-d) δ 177.8, 167.9, 163.6, 159.8, 155.2, 149.0, 144.4, 140.4, 139.0, 135.0, 132.1, 131.3, 128.7, 127.0, 123.1, 115.0, 107.5, 46.3, 42.7, 28.6, 16.6. ESI-MS: m/z 431.3 [M+H]+, 453.4 [M+Na]+. C25H26N4OS (430.18).
4.1.3.6. 4-((4′-Methoxy-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10f)
White solid, Yield 67%, m.p.: 158–160 °C. 1H NMR (400 MHz, chloroform-d) δ 7.77 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.54–7.47 (m, 2H, C2,C6-Phʹʹ-H), 7.27 (s, 2H, C3,C5-Phʹ-H), 7.21 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.96 (d, J = 2.7 Hz, 2H), 5.01 (s, 1H, NH), 3.84 (s, 1H), 3.83 (s, 3H), 3.32–3.31 (m, 2H), 2.95–2.87 (m, 2H), 2.18 (s, 6H), 2.12–2.04 (m, 4H). 13C NMR (100 MHz, chloroform-d) δ 164.7, 163.6, 159.8, 159.1, 148.5, 138.6, 135.0, 133.0, 131.2, 128.0, 127.3, 126.8, 123.1, 114.2, 55.3, 42.8, 28.5, 16.6. ESI-MS: m/z 461.3 [M+H]+, 478.2 [M+NH4]+. C26H28N4O2S (460.19).
4.1.3.7. 4-((4′-Fluoro-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10g)
White solid, Yield 48%, m.p.: 145–147 °C. 1H NMR (400 MHz, chloroform-d) δ 7.84 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.72–7.66 (m, 2H, C2,C6-Phʹʹ-H), 7.43 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.32 (s, 2H, C3,C5-Phʹ-H), 6.71–6.68 (m, 2H, C3,C5-Phʹʹ-H), 5.03 (s, 1H, NH), 3.86 (s, 1H), 3.30 (s, 2H), 2.90–2.86 (m, 2H), 2.17 (s, 6H), 2.12–1.45 (m, 4H). 13C NMR (100 MHz, chloroform-d) δ 167.8, 164.3, 163.4, 156.8, 148.2, 143.7, 138.7, 137.0, 130.7, 130.1, 127.6, 126.0, 125.7, 124.0, 115.1, 109.0, 46.5, 42.3, 28.2, 16.4. ESI-MS: m/z 449.2 [M+H]+, 471.3 [M+Na]+. C25H25FN4OS (448.17).
4.1.3.8. 3′,5′-Dimethyl-4'-((2-(piperidin-4-ylamino)thieno[3,2-d]pyrimidin-4-yl)oxy)-[1,1′-biphenyl]-4-carbonitrile (10h)
White solid, Yield 60%, m.p.: 157–159 °C. 1H NMR (400 MHz, chloroform-d) δ 8.52 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.95–7.92 (m, 4H), 7.65 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 7.62 (s, 2H, C3,C5-Ph’-H), 5.02 (s, 1H, NH), 3.85 (s, 1H), 3.30–3.19 (m, 2H), 2.89–2.85 (m, 2H), 2.16 (s, 6H), 2.09–1.32 (m, 4H). 13C NMR (100 MHz, chloroform-d) δ 172.3, 167.3, 165.1, 163.2, 150.7, 144.1, 139.2, 137.0, 133.0, 131.2, 128.4, 128.0, 124.5, 120.5, 110.7, 46.6, 42.4, 28.1, 16.5. ESI-MS: m/z 456.2 [M+H]+, 473.4 [M+NH4]+. C26H25N5OS (455.18).
4.1.3.9. 3′,5′-Dimethyl-4'-((2-(piperidin-4-ylamino)thieno[3,2-d]pyrimidin-4-yl)oxy)-[1,1′-biphenyl]-3-carbonitrile (10i)
White solid, Yield 55%, m.p.: 171–173 °C. 1H NMR (400 MHz, chloroform-d) δ 8.06 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.89–7.88 (m, 1H, C4-Phʹʹ-H), 7.84 (dt, J = 7.8, 1.6 Hz, 1H, C6-Phʹʹ-H), 7.69–7.64 (m, 1H, C2-Phʹʹ-H), 7.57 (d, J = 7.8 Hz, 1H, C5-Phʹʹ-H), 7.52 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.30 (s, 2H, C3,C5-Phʹ-H), 5.01 (s, 1H, NH), 3.86 (s, 1H), 3.30–3.19 (m, 2H), 2.88–2.85 (m, 2H), 2.18 (s, 6H), 2.03–1.88 (m, 2H), 1,67–1.51 (m, 2H). 13C NMR (100 MHz, chloroform-d) δ 167.3, 164.2, 162.7, 149.03, 141.4, 137.4, 137.0, 131.9, 131.4, 130.8, 129.9, 126.8, 124.4, 119.3, 113.2, 46.4, 42.1, 28.5, 16.6. ESI-MS: m/z 456.2 [M+H]+, 473.6 [M+NH4]+. C26H25N5OS (455.18).
4.1.3.10. 4-((4'-(Dimethylamino)-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10j)
White solid, Yield 63%, m.p.: 170–171 °C. 1H NMR (400 MHz, chloroform-d) δ 7.98 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.52 (d, J = 8.6 Hz, 2H, C2,C6-Phʹʹ-H), 7.51 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.32 (s, 2H, C3,C5-Phʹ-H), 6.82 (d, J = 8.8 Hz, 2H, C3,C5-Phʹʹ-H), 5.01 (s, 1H, NH), 3.84 (s, 1H), 3.33–3.31 (m, 2H), 3.02 (s, 6H, NCH3), 2.18 (s, 6H), 2.09–1.61 (m, 6H). 13C NMR (100 MHz, chloroform-d) δ 167.2, 164.3, 163.8, 156.0, 150.3, 147.1, 140.8, 136.2, 130.6, 128.3, 127.2, 126.7, 124.7, 115.4, 112.8, 46.3, 42.7, 40.6, 28.4, 16.7. ESI-MS: m/z 474.2 [M+H]+, 491.2 [M+NH4]+. C27H31N5OS (473.22).
4.1.3.11. 4-((3'-(Dimethylamino)-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)-N-(piperidin-4-yl)thieno[3,2-d]pyrimidin-2-amine (10k)
White solid, Yield 50%, m.p.: 166–167 °C. 1H NMR (400 MHz, chloroform-d) δ 7.83 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.50 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 7.32 (s, 2H, C3,C5-Ph′-H), 7.31–7.25 (m, 1H), 6.98–6.92 (m, 2H), 6.72 (dd, J = 8.8, 2.4 Hz, 1H, C5-Phʹʹ-H), 5.00 (s, 1H, NH), 3.83 (s, 1H), 3.32–3.30 (m, 2H), 3.03 (s, 6H, NCH3), 2.86–1.83 (m, 2H), 2.20 (s, 6H), 2.12–1.54 (m, 4H). 13C NMR (100 MHz, chloroform-d) δ 167.6, 164.2, 163.5, 156.4, 150.5, 148.6, 141.3, 139.6, 136.2, 130.8, 129.8, 127.1, 124.0, 115.7, 115.5, 112.0, 111.2, 46.5, 42.4, 40.6, 28.3, 16.6. ESI-MS: m/z 474.3 [M+H]+, 491.2 [M+NH4]+. C27H31N5OS (473.22).
4.1.4. General procedure for the preparation of the target compounds 11–32
Compounds 10a–k (1.0 mmol), substituted benzyl chloride (bromine) (1.1 mmol) and anhydrous K2CO3 (1.2 mmol) were dissolved in anhydrous DMF (10 mL). The reaction mixture was stirred at room temperature for 4–8 h (monitored by TLC). The solvent was removed under reduced pressure, and then water was added. Extracted with ethyl acetate (3 × 10 mL), and the organic phase was dried over anhydrous Na2SO4, then purified by flash column chromatography and recrystallized from EA/PE to give the target compounds 11–32.
4.1.4.1. 4-((4-((4-(4-(Furan-2-yl)-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (11)
White solid, Yield 77%, m.p.: 158–160 °C. 1H NMR (400 MHz, chloroform-d) δ 7.83 (d, J = 8.0 Hz, 2H, C2,C6-Phʹʹ-H), 7.72 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.48 (d, J = 1.7 Hz, 1H, C3-furan-H), 7.40 (s, 4H), 7.15 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.62 (d, J = 3.3 Hz, 1H, C5-furan-H), 6.49 (dd, J = 3.4, 1.8 Hz, C4-furan-H), 5.44–5.24 (m, 2H, SO2NH2), 5.16 (s, 1H, NH), 3.47 (s, 2H, N–CH2), 2.69–2.65 (m, 2H), 2.15 (s, 6H), 2.07–1.21 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.0, 163.5, 160.3, 153.6, 149.0, 143.9, 142.0, 140.7, 134.8, 131.6, 129.5, 128.5, 126.3, 123.9, 123.0, 111.7, 104.8, 62.3, 52.1, 48.3, 31.8, 16.6. HRMS m/z C30H31N5O4S2: Calcd. 589.1817, Found 590.1895 [M+H]+. HPLC purity: 96.83%.
4.1.4.2. 4-((4-((4-(4-(Furan-2-yl)-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (12)
White solid, Yield 79%, m.p.: 167–168 °C. 1H NMR (400 MHz, chloroform-d) δ 7.84 (d, J = 8.1 Hz, 2H, C2,C6-Phʹʹ-H), 7.72 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.46 (d, J = 1.7 Hz, 1H, C3-furan-H), 7.42–7.40 (m, 4H), 7.15 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 6.62 (d, J = 3.2 Hz, 1H, C5-furan-H), 6.48 (dd, J = 3.2, 1.8 Hz, C4-furan-H), 6.07–5.88 (m, 2H, CONH2), 5.16 (s, 1H, NH), 3.47 (s, 2H, N–CH2), 2.69–2.65 (m, 2H), 2.15 (s, 6H), 2.07–1.21 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.1, 163.5, 160.2, 153.6, 149.1, 143.7, 142.2, 140.9, 134.4, 131.6, 129.7, 128.5, 126.3, 123.0, 114.3, 111.7, 104.7, 62.4, 52.1, 48.3, 31.8, 16.6. HRMS m/z C31H31N5O3S: Calcd. 553.2148, Found 554.2225 [M+H]+. HPLC purity: 97.63%.
4.1.4.3. 4-((4-((4-(4-(Furan-3-yl)-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (13)
White solid, Yield 81%, m.p.: 154–156 °C. 1H NMR (400 MHz, chloroform-d) δ 7.83 (d, J = 8.1 Hz, 2H), 7.74 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.56 (d, J = 5.4 Hz, 1H), 7.53 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 7.40 (d, J = 8.0 Hz, 2H), 7.16 (d, J = 7.1 Hz, 1H), 7.12 (d, J = 8.6 Hz, 1H), 5.23 (s, 2H, SO2NH2), 5.00 (s, 1H, NH), 3.47 (s, 2H, N–CH2), 2.70 (d, J = 10.9 Hz, 2H), 2.18 (s, 6H), 2.11–1.33 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.0, 160.4, 149.1, 144.1, 140.6, 137.7, 136.8, 134.7, 131.5, 129.4, 128.6, 127.0, 126.3, 123.0, 115.7, 62.3, 52.3, 31.9, 16.6. HRMS m/z C30H31N5O4S2: Calcd. 589.1817, Found 590.1886 [M+H]+. HPLC purity: 97.30%.
4.1.4.4. 4-((4-((4-(4-(Furan-3-yl)-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (14)
White solid, Yield 83%, m.p.: 182–185 °C. 1H NMR (400 MHz, chloroform-d) δ 7.74 (t, J = 2.7 Hz, 2H), 7.72 (s, 1H), 7.56–7.54 (m, 2H), 7.34 (d, J = 7.8 Hz, 2H), 7.27 (d, J = 2.1 Hz, 2H), 7.19 (d, J = 5.4 Hz, 1H), 7.13 (t, J = 8.6 Hz, 1H), 6.08–5.82 (m, 2H, CONH2), 4.93 (d, J = 8.0 Hz, 1H, NH), 3.47 (s, 2H, N–CH2), 2.72 (d, J = 11.3 Hz, 2H), 2.19 (s, 6H), 2.07–1.84 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.2, 165.1, 161.2, 160.4, 149.2, 143.0, 137.7, 136.8, 134.6, 132.0, 131.5, 129.1, 128.6, 128.5, 127.3, 127.0, 123.2, 115.7, 115.4, 62.6, 52.3, 32.0, 16.6. HRMS m/z C31H31N5O3S: Calcd. 553.2148, Found 554.22195 [M+H]+. HPLC purity: 98.82%.
4.1.4.5. 4-((4-((4-(2,6-Dimethyl-4-(thiophen-2-yl)phenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (15)
White solid, Yield 69%, m.p.: 138–140 °C. 1H NMR (400 MHz, chloroform-d) δ 7.95 (d, J = 8.2 Hz, 2H, C2,C6-Phʹʹ-H), 7.76 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.42–7.40 (m, 4H), 7.32 (dd, J = 3.5, 1.2 Hz, 1H, C3-thiophen-H), 7.29 (dd, J = 5.2, 1.2 Hz, 1H, C5-thiophen-H), 7.19 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 7.03 (dd, J = 5.2, 3.6 Hz, 1H, C4-thiophen-H), 5.41–5.30 (m, 2H, SO2NH2), 5.11 (s, 1H, NH), 3.47 (s, 2H, N–CH2), 2.69–2.65 (m, 2H), 2.12 (s, 6H), 2.01–1.43 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.2, 163.8, 160.4, 153.6, 149.2, 143.5, 142.4, 140.7, 134.2, 131.6, 130.2, 128.7, 126.3, 123.9, 123.1, 111.7, 104.2, 52.1, 48.3, 31.8, 28.6, 16.6. HRMS m/z C30H31N5O3S3: Calcd. 605.1589, Found 606.1656 [M+H]+. HPLC purity: 92.90%.
4.1.4.6. 4-((4-((4-(2,6-Dimethyl-4-(thiophen-2-yl)phenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (16)
White solid, Yield 78%, m.p.: 148–150 °C. 1H NMR (400 MHz, chloroform-d) δ 7.92 (d, J = 8.2 Hz, 2H, C2,C6-Phʹʹ-H), 7.79 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.40–7.38 (m, 4H), 7.31 (dd, J = 3.5, 1.2 Hz, 1H, C3-thiophen-H), 7.29–7.28 (m, 1H), 7.19 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.07 (dd, J = 5.1, 3.4 Hz, 1H, C4-thiophen-H), 6.03 (s, 2H, CONH2), 5.10 (s, 1H, NH), 3.48 (s, 2H, N–CH2), 2.70–2.68 (m, 2H), 2.11 (s, 6H), 1.98–1.40 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.3, 163.2, 160.8, 152.9, 148.7, 143.1, 142.7, 140.7, 134.6, 131.2, 130.2, 128.4, 126.3, 124.0, 123.1, 111.2, 104.2, 52.1, 48.5, 31.8, 28.2, 16.6. HRMS m/z C31H31N5O2S2: Calcd. 569.1919, Found 570.1987 [M+H]+. HPLC purity: 99.90%.
4.1.4.7. 4-((4-((4-(2,6-Dimethyl-4-(thiophen-3-yl)phenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (17)
White solid, Yield 86%, m.p.: 168–170 °C. 1H NMR (400 MHz, chloroform-d) δ 7.83 (d, J = 8.0 Hz, 2H, C2,C6-Phʹʹ-H), 7.73 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.43 (d, J = 1.6 Hz, 1H), 7.41–7.36 (m, 4H), 7.31 (s, 2H, C3,C5-Phʹ-H), 7.16 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 5.16 (s, 2H, SO2NH2), 5.00 (s, 1H, NH), 3.46 (s, 2H, N–CH2), 2.69 (d, J = 11.2 Hz, 2H), 2.17 (s, 6H), 2.07–1.59 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.0, 163.5, 160.4, 148.9, 144.1, 141.8, 140.6, 134.7, 133.4, 131.5, 129.5, 126.4, 126.2, 123.0, 120.0, 62.3, 52.2, 48.4, 31.9, 16.6. HRMS m/z C30H31N5O3S3: Calcd. 605.1589, Found 606.1659 [M+H]+. HPLC purity: 96.62%.
4.1.4.8. 4-((4-((4-(2,6-Dimethyl-4-(thiophen-3-yl)phenoxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (18)
White solid, Yield 82%, m.p.: 152–154 °C. 1H NMR (400 MHz, chloroform-d) δ 7.73–7.71 (m, 3H), 7.47–7.42 (m, 1H), 7.42–7.36 (m, 2H), 7.33–7.31 (m, 4H), 7.18 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.06–6.04 (m, 2H, CONH2), 5.00 (d, J = 7.4 Hz, 1H, NH), 3.46 (s, 2H, N–CH2), 2.71 (d, J = 11.2 Hz, 2H), 2.18 (s, 6H) C, 2.11–1.31 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.3, 165.1, 163.5, 160.4, 143.0, 141.8, 134.6, 133.4, 132.0, 131.5, 129.1, 127.3, 126.4, 126.2, 123.1, 120.0, 62.6, 52.2, 48.5, 32.0, 16.6. HRMS m/z C31H31N5O2S2: Calcd. 569.1919, Found 570.1995 [M+H]+. HPLC purity: 96.84%.
4.1.4.9. 4-((4-((4-((3,5-Dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (19)
White solid, Yield 75%, m.p.: 130–132 °C. 1H NMR (400 MHz, chloroform-d) δ 7.78–7.70 (m, 3H), 7.63–7.60 (m, 2H), 7.44 (t, J = 7.5 Hz, 2H), 7.38–7.32 (m, 5H), 7.18 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 5.08 (s, 2H, SO2NH2), 4.97 (d, J = 7.4 Hz, 1H, NH), 3.46 (s, 2H, N–CH2), 2.71 (d, J = 11.2 Hz, 2H), 2.20 (s, 6H), 2.02–1.36 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.2, 165.1, 161.3, 149.2, 143.5, 140.7, 138.9, 134.6, 132.2, 131.4, 129.7, 128.4, 127.8, 127.2, 123.1, 110.5, 62.9, 52.3, 48.5, 32.2, 16.6. HRMS m/z C32H33N5O3S2: Calcd. 599.2025, Found 600.2100 [M+H]+. HPLC purity: 100.00%.
4.1.4.10. 4-((4-((4-((3,5-Dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (20)
White solid, Yield 88%, m.p.: 143–145 °C. 1H NMR (400 MHz, chloroform-d) δ 7.76–7.70 (m, 3H), 7.64–7.59 (m, 2H), 7.45 (t, J = 7.5 Hz, 2H), 7.39–7.30 (m, 5H), 7.18 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.01–6.00 (m, 2H, CONH2), 4.96 (d, J = 7.4 Hz, 1H, NH), 3.46 (s, 2H, N–CH2), 2.71 (d, J = 11.2 Hz, 2H), 2.20 (s, 6H), 2.07–1.02 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.2, 165.1, 160.5, 149.2, 143.0, 140.7, 138.7, 134.6, 132.0, 131.4, 129.1, 128.7, 127.3, 127.1, 123.1, 62.6, 52.3, 32.0, 16.6. HRMS m/z C33H33N5O2S: Calcd. 563.2355, Found 564.2426 [M+H]+. HPLC purity: 95.41%.
4.1.4.11. 4-((4-((4-((4′-Methoxy-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (21)
White solid, Yield 70%, m.p.: 138–140 °C. 1H NMR (400 MHz, chloroform-d) δ 7.82 (d, J = 8.3 Hz, 2H), 7.73 (d, J = 5.4 Hz, 1H, C6-thienopyrimidine-H), 7.53 (d, J = 8.6 Hz, 2H), 7.47–7.45 (m, 2H), 7.40 (d, J = 7.9 Hz, 2H), 7.16 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 7.02–6.94 (m, 2H), 5.01 (d, J = 7.2 Hz, 1H, NH), 3.48 (s, 2H, N–CH2), 2.70 (d, J = 12.3 Hz, 2H), 2.18 (s, 6H), 2.11–1.36 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.0, 163.5, 160.4, 159.1, 148.7, 144.1, 140.5, 138.3, 134.7, 133.2, 131.3, 129.5, 128.0, 127.3, 126.7, 126.4, 123.0, 114.1, 114.0, 62.3, 55.4, 52.3, 31.9, 16.6. HRMS m/z C33H35N5O4S2: Calcd. 629.2130, Found 630.2207 [M+H]+. HPLC purity: 92.74%.
4.1.4.12. 4-((4-((4-((4′-Methoxy-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (22)
White solid, Yield 74%, m.p.: 140–142 °C. 1H NMR (400 MHz, chloroform-d) δ 7.81 (d, J = 8.2 Hz, 2H), 7.72 (d, J = 5.3 Hz, 1H, C6-thienopyrimidine-H), 7.54 (d, J = 8.6 Hz, 2H), 7.47 (s, 2H), 7.42 (d, J = 7.9 Hz, 2H), 7.14 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 7.03–6.98 (m, 2H), 6.02–5.98 (m, 2H, CONH2), 5.01 (d, J = 7.2 Hz, 1H, NH), 3.48 (s, 2H, N–CH2), 2.70 (d, J = 12.1 Hz, 2H), 2.18 (s, 6H), 2.11–1.36 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.6, 163.2, 160.4, 159.6, 148.1, 143.6, 140.2, 138.7, 134.7, 133.5, 131.3, 129.5, 128.2, 127.3, 127.1, 126.4, 125.8, 123.0, 114.6, 62.2, 55.4, 52.8, 31.9, 16.6. HRMS m/z C34H35N5O3S: Calcd. 593.2461, Found 594.2532 [M+H]+. HPLC purity: 99.92%.
4.1.4.13. 4-((4-((4-((4′-Fluoro-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (23)
White solid, Yield 83%, m.p.: 138–140 °C. 1H NMR (400 MHz, chloroform-d) δ 7.84 (d, J = 8.1 Hz, 2H), 7.74–7.71 (m, 2H), 7.50–7.47 (m, 2H), 7.41 (d, J = 8.0 Hz, 2H), 7.21 (s, 2H), 7.16 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.70 (d, J = 1.8 Hz, 1H), 5.08 (s, 2H, SO2NH2), 4.95 (d, J = 7.5 Hz, 1H, NH), 3.48 (s, 2H, N–CH2), 2.70 (d, J = 11.3 Hz, 2H), 2.15 (s, 6H), 1.90–1.40 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.0, 160.4, 148.7, 144.1, 143.7, 140.5, 138.3, 134.7, 131.6, 129.9, 129.5, 127.1, 126.6, 126.4, 125.9, 125.8, 123.0, 108.9, 62.3, 52.2, 31.9, 16.5. HRMS m/z C32H32FN5O3S2: Calcd. 617.1931, Found 618.2008 [M+H]+. HPLC purity: 97.39%.
4.1.4.14. 4-((4-((4-((4′-Fluoro-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (24)
White solid, Yield 88%, m.p.: 132–135 °C. 1H NMR (400 MHz, chloroform-d) δ 7.75–7.73 (m, 4H), 7.50 (d, J = 1.7 Hz, 2H), 7.35 (d, J = 7.9 Hz, 2H), 7.22 (s, 2H), 7.18 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.70 (s, 1H), 5.98–5.96 (m, 2H, CONH2), 4.92 (d, J = 7.5 Hz, 1H, NH), 3.48 (s, 2H, N–CH2), 2.72 (d, J = 11.3 Hz, 2H), 2.16 (s, 6H), 1.93–1.43 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.2, 165.1, 163.4, 160.4, 148.7, 143.6, 138.3, 134.6, 132.0, 131.6, 129.9, 129.2, 127.3, 126.0, 125.8, 123.2, 108.9, 62.6, 52.2, 32.0, 16.5. HRMS m/z C33H32FN5O2S: Calcd. 581.2261, Found 582.2370 [M+H]+. HPLC purity: 91.70%.
4.1.4.15. 4-((4-((4-((4′-Cyano-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (25)
White solid, Yield 66%, m.p.: 148–150 °C. 1H NMR (400 MHz, chloroform-d) δ 7.79–7.62 (m, 7H), 7.35–7.32 (m, 4H), 7.19 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 5.07 (s, 2H, SO2NH2), 4.90 (d, J = 7.4 Hz, 1H, NH), 3.47 (s, 2H, N–CH2), 2.72 (d, J = 11.2 Hz, 2H), 2.23 (s, 6H), 2.10–1.40 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.4, 164.8, 163.7, 161.6, 150.2, 145.8, 136.3, 134.2, 132.5, 132.1, 129.0, 127.6, 127.3, 119.0, 112.4, 62.6, 52.4, 32.1, 16.7. HRMS m/z C33H32N6O3S2: Calcd. 624.1977, Found 625.2050 [M+H]+. HPLC purity: 96.65%.
4.1.4.16. 4-((4-((4-((4′-Cyano-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (26)
White solid, Yield 74%, m.p.: 156–158 °C. 1H NMR (400 MHz, chloroform-d) δ 7.77–7.71 (m, 7H), 7.36–7.33 (m, 4H), 7.20 (d, J = 5.4 Hz, 1H), 6.11–5.90 (m, 2H), 4.88 (d, J = 7.4 Hz, 1H, NH), 3.47 (s, 2H, N–CH2), 2.72 (d, J = 11.3 Hz, 2H), 2.21 (s, 6H), 1.90–1.42 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.1, 165.2, 163.2, 160.4, 150.3, 145.1, 136.5, 134.6, 132.5, 132.0, 129.0, 127.6, 127.3, 127.2, 119.0, 110.7, 62.6, 52.3, 32.0, 16.7. HRMS m/z C34H32N6O2S: Calcd. 588.2307, Found 589.2382 [M+H]+. HPLC purity: 97.28%.
4.1.4.17. 4-((4-((4-((3′-Cyano-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (27)
White solid, Yield 74%, m.p.: 140–143 °C. 1H NMR (400 MHz, chloroform-d) δ 7.86 (d, J = 1.5 Hz, 1H), 7.82 (dd, J = 7.6, 1.6 Hz, 1H), 7.76–7.74 (m, 3H), 7.62 (dd, J = 7.8, 1.5 Hz, 1H), 7.57 (d, J = 7.8 Hz, 1H), 7.37 (d, J = 7.6 Hz, 2H), 7.30 (s, 2H), 7.20 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 5.09 (s, 2H, SO2NH2), 4.90 (d, J = 7.2 Hz, 1H, NH), 3.47 (s, 2H, N–CH2), 2.73 (d, J = 11.2 Hz, 2H), 2.21 (s, 6H), 2.11–1.39 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.1, 165.7, 159.3, 150.7, 142.2, 141.6, 136.3, 134.7, 132.3, 131.4, 130.8, 129.6, 129.2, 127.4, 127.0, 123.2, 119.0, 112.9, 62.7, 52.3, 32.1, 16.7. HRMS m/z C33H32N6O3S2: Calcd. 624.1977, Found 625.2045 [M+H]+. HPLC purity: 98.93%.
4.1.4.18. 4-((4-((4-((3′-Cyano-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (28)
White solid, Yield 83%, m.p.: 140–143 °C. 1H NMR (400 MHz, chloroform-d) δ 7.88 (d, J = 1.6 Hz, 1H), 7.83 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.74 (m, 3H), 7.64 (dd, J = 7.6, 1.5 Hz, 1H), 7.57 (d, J = 7.8 Hz, 1H), 7.35 (d, J = 7.8 Hz, 2H), 7.30 (s, 2H), 7.20 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 5.98–5.96 (m, 2H, CONH2), 4.90 (d, J = 7.2 Hz, 1H, NH), 3.48 (s, 2H, N–CH2), 2.73 (d, J = 11.2 Hz, 2H), 2.21 (s, 6H), 2.11–1.39 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.2, 165.2, 160.4, 150.0, 142.9, 141.9, 136.3, 134.7, 132.1, 131.4, 130.6, 130.5, 129.6, 129.1, 127.4, 127.1, 123.2, 118.9, 112.9, 62.6, 52.3, 32.0, 26.9, 16.7. HRMS m/z C34H32N6O2S: Calcd. 588.2307, Found 589.2378 [M+H]+. HPLC purity: 100.00%.
4.1.4.19. 4-((4-((4-((4'-(Dimethylamino)-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (29)
White solid, Yield 66%, m.p.: 172–175 °C. 1H NMR (400 MHz, chloroform-d) δ 7.84–7.70 (m, 3H), 7.48 (d, J = 2.0 Hz, 2H), 7.43 (d, J = 8.0 Hz, 2H), 7.21 (s, 2H), 7.16 (d, J = 5.4 Hz, 1H, C7-thienopyrimidine-H), 6.72 (d, J = 1.8 Hz, 2H), 5.08 (s, 2H, SO2NH2), 5.01 (s, 1H, NH), 3.46 (s, 2H, N–CH2), 3.02 (s, 6H, NCH3), 2.71 (d, J = 11.2 Hz, 2H), 2.18 (s, 6H), 2.09–1.42 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.0, 160.4, 148.7, 144.1, 143.7, 140.5, 138.3, 134.7, 131.6, 129.9, 129.5, 127.1, 126.6, 125.9, 123.0, 108.9, 52.2, 46.9, 42.7, 40.3, 28.4, 16.5. HRMS m/z C34H38N6O3S2: Calcd. 642.2447, Found 643.2524 [M+H]+. HPLC purity: 92.28%.
4.1.4.20. 4-((4-((4-((4'-(Dimethylamino)-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (30)
White solid, Yield 69%, m.p.: 141–143 °C. 1H NMR (400 MHz, chloroform-d) δ 7.84 –7.73 (m, 3H), 7.46 (d, J = 2.0 Hz, 2H), 7.41 (d, J = 8.0 Hz, 2H), 7.20 (s, 2H), 7.16 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 6.74 (d, J = 2.0 Hz, 2H), 5.98 (s, 2H, CONH2), 5.00 (s, 1H, NH), 3.48 (s, 2H, N–CH2), 3.02 (s, 6H, NCH3), 2.72 (d, J = 11.2 Hz, 2H), 2.18 (s, 6H), 2.04–1.40 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 165.4, 160.9, 148.7, 144.5, 143.7, 140.2, 138.3, 134.4, 132.2, 130.0, 129.2, 127.5, 126.6, 126.0, 125.9, 123.5, 109.2, 52.0, 46.9, 42.9, 39.7, 28.1, 16.5. HRMS m/z C35H38N6O2S: Calcd. 606.2777, Found 607.2852 [M+H]+. HPLC purity: 96.74%.
4.1.4.21. 4-((4-((4-((3'-(Dimethylamino)-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzenesulfonamide (31)
White solid, Yield 76%, m.p.: 138–140 °C. 1H NMR (400 MHz, chloroform-d) δ 7.86–7.82 (m, 3H), 7.76–7.74 (m, 2H), 7.61 (dd, J = 7.6, 1.5 Hz, 1H), 7.54 (d, J = 7.6 Hz, 1H), 7.35–7.34 (m, 2H), 7.30 (s, 2H), 7.21 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 5.06 (s, 2H, SO2NH2), 5.03 (s, 1H, NH), 3.42 (s, 2H, N–CH2), 3.02 (s, 6H, NCH3), 2.70 (d, J = 11.2 Hz, 2H), 2.16 (s, 6H), 2.03–1.36 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.7, 165.0, 158.9, 150.7, 142.6, 141.6, 137.5, 134.6, 132.3, 131.1, 130.4, 129.2, 127.8, 127.2, 123.1, 119.3, 113.0, 111.8, 62.9, 52.5, 46.9, 42.9, 39.7, 28.1, 16.6. HRMS m/z C34H38N6O3S2: Calcd. 642.2447, Found 643.2524 [M+H]+. HPLC purity: 94.44%.
4.1.4.22. 4-((4-((4-((3'-(Dimethylamino)-3,5-dimethyl-[1,1′-biphenyl]-4-yl)oxy)thieno[3,2-d]pyrimidin-2-yl)amino)piperidin-1-yl)methyl)benzamide (32)
White solid, Yield 74%, m.p.: 133–135 °C. 1H NMR (400 MHz, chloroform-d) δ 7.86–7.81 (m, 3H), 7.76–7.73 (m, 2H), 7.61–7.59 (m, 1H), 7.53 (d, J = 7.6 Hz, 1H), 7.32–7.31 (m, 2H), 7.29 (s, 2H), 7.21 (d, J = 5.3 Hz, 1H, C7-thienopyrimidine-H), 5.97 (s, 2H, CONH2), 5.01 (s, 1H, NH), 3.42 (s, 2H, N–CH2), 3.02 (s, 6H, NCH3), 2.72 (d, J = 11.2 Hz, 2H), 2.16 (s, 6H), 2.00–1.41 (m, 7H). 13C NMR (100 MHz, chloroform-d) δ 169.7, 165.0, 159.2, 142.6, 141.7, 137.5, 134.2, 132.3, 131.6, 130.4, 129.2, 127.3, 123.1, 120.5, 113.0, 111.2, 62.9, 52.4, 46.9, 43.2, 39.3, 28.2, 16.6. HRMS m/z C35H38N6O2S: Calcd. 606.2777, Found 607.2852 [M+H]+. HPLC purity: 95.29%.
4.2. In vitro anti-HIV activities assays
The anti-HIV activity and cytotoxicity of the novel synthesized compounds were evaluated using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method in MT-4 cell cultures as described previously19, 20. Firstly, stock solutions (10 × final concentration) of the compounds were added in 25 μL volumes to two series of triplicate wells with the aim to allow simultaneous evaluation of their effects on mock- and HIV-infected cells. With a Biomek 3000 robot (Beckman Instruments, Fullerton, CA, USA), serial 5-fold dilutions of the test compounds were made directly in flat-bottomed 96-well microtiter trays, including untreated control HIV-1 and mock-infected cell samples for each sample. Wild-type and mutant HIV-1 strains stock (50 μL at 100–300 CCID50) or culture medium was added to either the infected or mock-infected wells of the microtiter tray. Mock-infected cells were used to evaluate the effect on uninfected cells to assess compounds cytotoxicity. Exponentially growing MT-4 cells were centrifuged for 5 minat 1000 rpm (Eppendorf 5424, Hamburg, Germany) and then supernatant was discarded. The MT-4 cells were resuspended at 6 × 105 cells/mL, and 50 μL aliquots were transferred to the microtiter tray wells. At 5 days after infection, the viability of mock- and HIV-infected cells was determined spectrophotometrically by means of MTT assay.
The MTT assay is based on the reduction of yellow-colored MTT (Acros Organics, Geel, Belgium) by mitochondrial dehydrogenase of metabolically active cells to form a blue-purple formazan. The absorbances were read in an eight-channel computer-controlled photometer at the wavelengths of 540 and 690 nm. All data were calculated using the median optical density (OD) value of three wells. The EC50 was defined as the concentration of the test compound affording 50% protection from viral cytopathogenicity. The CC50 was defined as the compound concentration that reduced the absorbance (OD540) of mock-infected cells by 50%.
4.3. Recombinant HIV-1 RT inhibitory assays
The HIV-1 RT inhibition assay was tested with an RT assay kit (Roche, Basel, Switzerland)21. Briefly, the reaction mixture containing HIV-1 RT enzyme, reconstituted template and viral nucleotides [digoxigenin (DIG)-dUTP, biotin-dUTP and dTTP] in the incubation buffer with or without inhibitors was incubated for 1 hat 37 °C. Then, the reaction mixture was transferred to a streptavidin-coated microtitre plate (MTP) and incubated for 2 hat 37 °C. The biotin-labeled dNTPs were incorporated into the cDNA chain in the presence of RT. The unbound dNTPs were washed with washing buffer, and anti-DIG-POD was added to the MTPs.
After incubation for additional 1 h at 37 °C, the DIG-labeled dNTPs incorporated in cDNA were bound to the anti-DIG-POD antibody. The unbound anti-DIG-PODs were washed out and the peroxide substrate (ABST) solution was added to the MTPs. The absorbance of the sample was determined at OD405 using a microtiter plate ELISA reader (Multiskan Sky, Waltham, MA, USA). The percentage inhibitory activity of RT inhibitors was calculated using the following Eq. (1):
| Inhibition (%)=(OD value with RT but without inhibitors–OD value with RT and inhibitors)/(OD value with RT and inhibitors–OD value without RT and inhibitors) | (1) |
The IC50 values correspond to the concentrations of the inhibitors required to inhibit biotin-dUTP incorporation by 50%.
4.4. Molecular modelling studies
4.4.1. Models preparation
The X-ray crystal structure of HIV-1 WT RT (PDB code: 6c0n), HIV-1 K103N RT (PDB code: 6c0o) and HIV-1 RES056 RT (PDB code: 6c0r), respectively corresponding to the X-ray crystallographic structure of the WT, K103N and RES056 RT in complex with the NNRTIs K-5a2 were chosen to build up the initial models for compound 26. The atom–atom pair fitting procedure implemented in PyMOL V1.7 (https://pymol.org/) was applied to manually dock compound 26 into the NNRTIs binding site, according to the high molecular similarity of the crystallographic and simulated ligands.
4.4.2. Molecular dynamics simulations
Amber16 was used to explore the stability of compound 26 into the NNRTI binding site of the three simulated HIV-1 RT models by means of unbiased MD simulations. The Amber ff99SB-ILDN force field22 was used for the protein and protonation states at physiological pH were optimized in accordance to the pKa values estimated by Propka23, 24. The general Amber force field (GAFF)25. was used to parameterize the ligand and partial charges were derived at the B3LYP/6-31G(d) level of theory with Gaussian 0926, after preliminary optimization of the molecular structure, by using the restrained electrostatic potential (RESP) fitting method implemented in Antechamber.
The complex was then solvated with a truncated octahedral (TIP3P) water box with a layer of 20 Å and neutralized by adding Cl– counterions. The position of the conserved water molecule (HOH 817 in 6c0n, 824 in 6c0o and 736 in 6c0r), which mediates H-bonding interactions between the protonated pyrimidine nitrogen of the ligand and the protein backbone oxygen atoms of K/N103 and P236, was retained during generation of the stating complexes although it was revealed to be not stable along the simulation.
A stepwise minimization involving firstly all hydrogen atoms, then water molecules, and finally all the system with a maximum number of minimization cycles of 10,000 (the first 2000 by using the steepest descent method and the rest with conjugate gradient) for the latter stage was applied to the three pre-generated complexes. Prior to MD simulation, heating and equilibration of the system from 0 to 300 K was accomplished in six steps, the first being performed at constant volume and the rest at constant pressure. Harmonic restraints with a force constant of 10 kcal/mol/Å were applied during heating to some crucial interactions involving compound 26, the conserved water molecule and HIV-1 RT during equilibration in order to avoid artefactual structural changes on the pre-generated binding mode. These restraints were gradually reduced at 5 kcal/mol/Å during the 5 ns of equilibration and totally eliminated in the first 10 ns of MD production. The SHAKE algorithm was applied to constrain bonds involving hydrogen atoms. Periodic boundary conditions at constant volume were imposed on the systems during the MD simulations. Cut-off for the non-bonded interactions was set to 10 Å. The electrostatic interactions beyond the cut-off within the periodic box were computed by applying the Particle Mesh Ewald (PME) method. Langevin dynamics with a collision frequency of 1.0/ps was applied for temperature regulation during the heating. A total of 100 ns of unrestrained MD simulation at constant volume and temperature (300 K) using the weak-coupling algorithm with a time constant of 10.0 were run for the three pre-equilibrated systems. The time step for saving of trajectory was set to 2 ps.
Finally, trajectories for the unrestrained part of the three MD runs were generated and analysed by using the CPPTRAJ module of Amber. A total of 20,000 frames were collected for each simulated system. Complexes stability during MD simulation was evaluated by means of RMSD analysis for the NN binding site (residues 8 Å around the ligand), and for the ligand itself. The stability of some relevant ligand–protein interactions was also analysed.
4.4.3. Binding free energy calculations
The contributions to the binding free energy were calculated for the three complexes according to the GB method implemented in the MMGBSA.py module of Amber [S14]. The method allows calculating the free energy (G) of each species (ligand, protein and complex) as the sum of enthalpic (gas-phase) (Egas), solvation (Gsolv) and entropy (S) terms, according to Eq. (2):
| (2) |
where Eint, Eelect and EvdW are the internal, Coulomb, and van der Waals energy terms and account, respectively, for bonded, polar, and non-polar energy contributions in the gas-phase, Gsolv,pol is the polar contribution to solvation free energy evaluated by using the Generalized-Born solvation method, and Gsolv,n-pol accounts for non-polar contributions to solvation free energy and was computed linearly to the solvent-accessible surface area (SASA; Eq. (3)).
| (3) |
where γ is the surface tension (set to 0.0072 kcal/mol/Å) and b is a correction term which is assumed to be zero in the present calculation.
All these contributions were calculated for complex, receptor and ligand and the binding free energy (Gbind) was evaluated as the difference for the three species (Eq. (4))
| (4) |
where accounts for the average value for each species (x: complex, receptor, ligand) determined for an ensemble of 100 snapshots taken from the last 30 ns of MD trajectory of the complexes within the framework of the single-trajectory approach. The vibrational entropy term (S) was not determined.
Acknowledgments
We gratefully acknowledge financial support from the Key Project of NSFC for International Cooperation (No.81420108027, China), the National Natural Science Foundation of China (NSFC Nos. 81273354, 81573347, 81903453), Young Scholars Program of Shandong University (YSPSDU No. 2016WLJH32, China), Shandong Provincial Natural Science Foundation (ZR2019BH011, China), China Postdoctoral Science Foundation (2018M640641, 2019T120596), Key research and development project of Shandong Province (No. 2017CXGC1401, China) and KU Leuven (GOA 10/014, Belgium). The technical assistance of Mr. Kris Uyttersprot and Mrs. Kristien Erven, for the HIV experiments is gratefully acknowledged. T.G. thanks the Spanish Government (MINECO Project SAF2017-881074-R, AEI/FEDER, UE), and Generalitat de Catalunya (2017SGR1746, Spain) for the financial support. The Barcelona Supercomputer Center (BSC) is also acknowledged for providing access to supercomputation resources.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Author contributions
Dongwei Kang, Peng Zhan and Xinyong Liu conceived the project. Dongwei Kang, Da Feng, Fenju Wei and Yanying Sun finished the compounds synthesis and structure confirmation. Dongwei Kang, Jinmi Zou, Tong Zhao and Samuel Desta designed the pharmacokinetics and acute toxicity experiment. Dongwei Kang and Boshi Huang performed the data analysis. Tiziana Ginex performed the molecular modeling study. Eeik De Clercq and Christophe Pannecouque performed the activity test. Peng Zhan and Xinyong Liu provided the resources, supervision and funding assistance.
All authors critically evaluated the manuscript prior to submission.
Conflicts of interest
The authors have no conflicts of interest to declare.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.08.013.
Contributor Information
Peng Zhan, Email: zhanpeng1982@sdu.edu.cn.
Xinyong Liu, Email: xinyongl@sdu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shattock R.J., Warren M., McCormack S., Hankins C.A. AIDS. Turning the tide against HIV. Science. 2011;333:42–43. doi: 10.1126/science.1206399. [DOI] [PubMed] [Google Scholar]
- 2.Guo H., Zhuang X., Qian K., Sun L., Wang X., Li H., et al. Prodrug design, synthesis and pharmacokinetic evaluation of (3'R,4'R)-3-hydroxymethyl-4-methyl-3',4'-di-O-(S)-camphanoyl-(+)-cis-khellactone. Acta Pharm Sin B. 2012;2:213–219. doi: 10.1016/j.apsb.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhan P., Chen X., Li D., Fang Z., de Clercq E., Liu X. HIV-1 NNRTIs: structural diversity, pharmacophore similarity, and impliations for drug design. Med Res Rev. 2013;33(Suppl 1):E1–E72. doi: 10.1002/med.20241. [DOI] [PubMed] [Google Scholar]
- 4.Bec G., Meyer B., Gerard M.A., Steger J., Fauster K., Wolff P., et al. Thermodynamics of HIV-1 reverse transcriptase in action elucidates the mechanism of action of non-nucleoside inhibitors. J Am Chem Soc. 2013;135:9743–9752. doi: 10.1021/ja4018418. [DOI] [PubMed] [Google Scholar]
- 5.Namasivayam V., Vanangamudi M., Kramer V.G., Kurup S., Zhan P., Liu X., et al. The journey of HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) from lab to clinic. J Med Chem. 2019;62:4851–4883. doi: 10.1021/acs.jmedchem.8b00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyrer C., Pozniak A. HIV drug resistance—an emerging threat to epidemic control. N Engl J Med. 2017;377:1605–1607. doi: 10.1056/NEJMp1710608. [DOI] [PubMed] [Google Scholar]
- 7.Xu H.T., Asahchop E.L., Oliveira M., Quashie P.K., Quan Y., Brenner B.G., et al. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J Virol. 2011;85:11300–11308. doi: 10.1128/JVI.05584-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang B., Chen W., Zhao T., Li Z., Jiang X., Ginex T., et al. Exploiting the tolerant region I of the non-nucleoside reverse transcriptase inhibitor (NNRTI) binding pocket: discovery of potent diarylpyrimidine-typed HIV-1 NNRTIs against Wild-Type and E138K mutant virus with significantly improved water solubility and favorable safety profiles. J Med Chem. 2019;62:2083–2098. doi: 10.1021/acs.jmedchem.8b01729. [DOI] [PubMed] [Google Scholar]
- 9.Li D., Zhan P., de Clercq E., Liu X. Strategies for the design of HIV-1 non-nucleoside reverse transcriptase inhibitors: lessons from the development of seven representative paradigms. J Med Chem. 2012;55:3595–3613. doi: 10.1021/jm200990c. [DOI] [PubMed] [Google Scholar]
- 10.Kang D., Wang Z., Chen M., Feng D., Wu G., Zhou Z., et al. Discovery of potent HIV-1 non-nucleoside reverse transcriptase inhibitors by exploring the structure–activity relationship of solvent-exposed regions I. Chem Biol Drug Des. 2019;93:430–437. doi: 10.1111/cbdd.13429. [DOI] [PubMed] [Google Scholar]
- 11.Zhan P., Pannecouque C., de Clercq E., Liu X. Anti-HIV drug discovery and development: current innovations and future trends. J Med Chem. 2016;59:2849–2878. doi: 10.1021/acs.jmedchem.5b00497. [DOI] [PubMed] [Google Scholar]
- 12.Kang D., Zhang H., Wang Z., Zhao T., Ginex T., Luque F.J., et al. Identification of dihydrofuro[3,4-d]pyrimidine derivatives as novel HIV-1 non-nucleoside reverse transcriptase inhibitors with promising antiviral activities and desirable physicochemical properties. J Med Chem. 2019;62:1484–1501. doi: 10.1021/acs.jmedchem.8b01656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo X., Huo Z., Kang D., Wu G., Zhou Z., Liu X., et al. Current insights into anti-HIV drug discovery and development: a review of recent patent literature (2014–2017) Expert Opin Ther Pat. 2018;28:299–316. doi: 10.1080/13543776.2018.1438410. [DOI] [PubMed] [Google Scholar]
- 14.Kang D., Fang Z., Li Z., Huang B., Zhang H., Lu X., et al. Targeting the hydrophobic channel of NNIBP: discovery of nove. J Med Chem. 2016;59:7991–8007. doi: 10.1021/acs.jmedchem.6b00738. [DOI] [PubMed] [Google Scholar]
- 15.Kang D., Fang Z., Huang B., Lu X., Zhang H., Xu H., et al. Structure-based optimization of thiophene[3,2-d]pyrimidine derivatives as potent HIV-1 non-nucleoside reverse transcriptase inhibitors with improved potency against resistance-associated variants. J Med Chem. 2017;60:4424–4443. doi: 10.1021/acs.jmedchem.7b00332. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y., Kang D., Nguyen L.A., Smithline Z.B., Pannecouque C., Zhan P., et al. Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors. eLife. 2018;7 doi: 10.7554/eLife.36340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z., Liu T., Wu G., Kang D., Fu Z., Wang Z., et al. Targeting the hydrophobic channel of NNIBP: discovery of novel 1,2,3-triazole-derived diarylpyrimidines as novel HIV-1 NNRTIs with high potency against wild-type and K103N mutant virus. Org Biomol Chem. 2019;17:3202–3217. doi: 10.1039/c9ob00032a. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z., Liu T., Kang D., Huo Z., Wu G., Daelemans D., et al. Discovery of novel diarylpyrimidines as potent HIV-1 NNRTIs by investigating the chemical space of a less explored “hydrophobic channel”. Org Biomol Chem. 2018;16:1014–1028. doi: 10.1039/c7ob02828h. [DOI] [PubMed] [Google Scholar]
- 19.Pauwels R., Balzarini J., Baba M., Snoeck R., Schols D., Herdewijn P., et al. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 20.Pannecouque C., Daelemans D., de Clercq E. Tetrazolium-based colorimetric assay for the detection of HIV replication inhibitors: revisited 20 years later. Nat Protoc. 2008;3:427–434. doi: 10.1038/nprot.2007.517. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki K., Craddock B.P., Okamoto N., Kano T., Steigbigel R.T. Poly A-linked colorimetric microtiter plate assay for HIV reverse transcriptase. J Virol Methods. 1993;44:189–198. doi: 10.1016/0166-0934(93)90054-u. [DOI] [PubMed] [Google Scholar]
- 22.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Søndergaard C.R., Olsson M.H., Rostkowski M., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 25.Olsson M.H., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.