Abstract
Hepatocellular carcinoma (HCC) is the most common subtype of liver cancer, and assessing its histopathological grade requires visual inspection by an experienced pathologist. In this study, the histopathological H&E images from the Genomic Data Commons Databases were used to train a neural network (inception V3) for automatic classification. According to the evaluation of our model by the Matthews correlation coefficient, the performance level was close to the ability of a 5-year experience pathologist, with 96.0% accuracy for benign and malignant classification, and 89.6% accuracy for well, moderate, and poor tumor differentiation. Furthermore, the model was trained to predict the ten most common and prognostic mutated genes in HCC. We found that four of them, including CTNNB1, FMN2, TP53, and ZFX4, could be predicted from histopathology images, with external AUCs from 0.71 to 0.89. The findings demonstrated that convolutional neural networks could be used to assist pathologists in the classification and detection of gene mutation in liver cancer.
Subject terms: Cancer imaging, Hepatocellular carcinoma, Cancer models
Introduction
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer-related mortality and is currently the main cause of liver-related death, leading to more than one million deaths annually worldwide1–3. Over several decades, substantial progress had been made in the understanding of HCC risk factors, epidemiology, and molecular pathogenesis. The early detection of HCC increases the chance of curative therapies in high overall survival. Unfortunately, most HCC patients are diagnosed at the intermediate to late-stage, which significantly decreases the overall survival4. Various predominant clinical risk factors for the development of HCC have been defined, including alcohol abuse, cirrhosis, metabolic syndrome, and hepatitis B and/or C virus infection5–8. However, multiple genetic alternation and signaling cascades also have a great influence on tumor progression and overall survival9.
The understanding of HCC molecular pathogenesis has been significantly improved over the past decade10. The development of genomic analysis has identified the major drivers that are responsible for cancer development and progression. HCC has been reported to have around 40 genomic aberrations, some of which are deemed as drivers. Several frequent HCC genomic alternations have been identified, including mutations in the CTNNB1 (β-catenin WNT pathway activation), TP53, telomere reverse transcriptase (telomere maintenance), AT-rich interaction domain 1A (ARID1A; chromatin remodeling), mammalian target of rapamycin signaling, RAS signaling, oxidative stress pathway activation, and aberrations in DNA methylation11. Previous studies have reported that the heterogeneity of HCC at both molecular and histological levels are correlated with gene mutations and oncogenic pathways12. The mutually exclusive CTNNB1 (40%) and TP53 (21%) mutations have been identified as two major groups of HCC according to its distinct phenotype. CTNNB1 mutated HCC is generally well-differentiated and large, with pseudoglandular and microtrabecular patterns, and lacks inflammatory infiltrates; whereas TP53 mutated HCC is generally poor-differentiated, with compact patterns, frequent vascular invasion, and pleomorphic, multinucleated cells13. The deeper understandings of the HCC phenotypes are essential for improving targeted therapies and clinical translation.
Pathologists could provide limited information regarding cancer reorganization from normal liver tissue and assess its histopathological grade via visual inspection, but it still lacks the underlying biological differences in HCC gene mutations associated with overall survival. The recent advances in artificial intelligence (AI) provided a novel way to assist clinicians to classify medical information and images14–17. Recently, Lin et al.18 used multiphoton microscopy with deep learning in the automated classification of HCC differentiation. Furthermore, Li et al.19 combined extreme learning machine with multiple convolutional neural network methods for nuclei grading in HCC. The development of graphics processing units allows the possibility to train a more complex neural network to satisfy the requirement of accomplishing complex visual recognition tasks, such as distinguishing tumors from normal tissue slides and classifying subtypes of tumors20,21. To the best of our knowledge, a previous study by Coudray et al.20 utilized the deep convolutional neural network on histopathological images to automatically classify the type and subtype of lung tumors. In addition, a promising result for the classification of colorectal22,23 and breast tumors24 using deep learning was also reported. Therefore, deep-learning models could be used to assist pathologists to effectively detect gene mutations and cancer subtypes. However, it remains unclear whether deep learning can be applied to solid tumors, especially for HCC. In addition, advances in AI tools in digital pathology have resulted in an increased demand for predictive assays in frozen slides that enable the selection and stratification of patients for additional treatment during surgery25.
Herein, based on the inception V3 network developed by Google26 and some packaging code from Coudray et al.20 via EASY DL platform and whole-slide images (WSIs) of H&E stained liver tissue, we have established a model to classify liver tissue and predict certain gene mutations. The model was externally validated by an independent cohort.
Results
The distribution of WSIs and tiles
There were 491 WSIs of H&E stained liver tissue from the Genomic Data Commons portal (GDC-portal, https://portal.gdc.cancer.gov/), including 402 WSIs of HCC and 89 WSIs of normal liver tissue. The information on histopathological grade was not available in 19 of 402 WSIs of HCC. According to the histopathological grade, they were then sorted into well (G1, n = 55), moderate (G2, n = 187), and poor group (G3/G4, n = 141) in the remaining 383 WSIs of HCC. A total of 387 WSIs of HCC with corresponding gene mutation information were available. Besides, 67 WSIs of HCC with histopathological grade and related gene mutation information and 34 WSIs of normal liver tissue were selected from Sir Run-Run Shaw Hospital (SRRSH). After each WSI was cropped into small “Tiles”, there are 119,596 “Tiles” (HCC vs. normal liver tissue, 87,422 vs. 32,174), 84,149 “Tiles” with histopathological grade (well vs. moderate vs. poor, 14,713 vs. 41,370 vs. 28,066) and 86,323 “Tiles” with corresponding gene mutation information. The distribution of WSIs and tiles was summarized in Table 1.
Table 1.
The distribution of patients, histopathological images/WSIs, and tiles in each subset.
| Patients | Histopathological images | Tiles | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tr | Te | IV | EV | Tr | Te | IV | EV | Tr | Te | IV | EV | |
| Normal and HCC | ||||||||||||
| HCC | 208 | 41 | 128 | 67 | 225 | 47 | 130 | 67 | 41,578 | 8157 | 24,294 | 13,393 |
| Normal | 53 | 9 | 27 | 34 | 53 | 9 | 27 | 34 | 12,614 | 2204 | 9493 | 7863 |
| Histopathological grade | ||||||||||||
| Well (G1) | 31 | 7 | 14 | 17 | 33 | 8 | 14 | 17 | 6967 | 1893 | 2654 | 3199 |
| Moderate (G2) | 98 | 17 | 60 | 38 | 106 | 20 | 61 | 38 | 18,754 | 3862 | 10,953 | 7801 |
| Poor (G3/G4) | 69 | 14 | 48 | 12 | 76 | 16 | 49 | 12 | 13,701 | 3189 | 8783 | 2393 |
| CTNNB1 mutation | ||||||||||||
| Yes | 60 | 13 | 26 | 21 | 63 | 15 | 26 | 21 | 11,283 | 3218 | 5329 | 4120 |
| No | 142 | 29 | 96 | 46 | 153 | 32 | 98 | 46 | 28,437 | 6321 | 18,342 | 9273 |
| FMN2 mutation | ||||||||||||
| Yes | 31 | 7 | 9 | 10 | 32 | 7 | 9 | 10 | 6335 | 1632 | 2143 | 2736 |
| No | 171 | 35 | 113 | 57 | 184 | 40 | 115 | 57 | 34,103 | 7963 | 20,754 | 10,657 |
| TP53 mutation | ||||||||||||
| Yes | 64 | 14 | 42 | 20 | 68 | 14 | 43 | 20 | 12,537 | 2873 | 7794 | 4341 |
| No | 138 | 28 | 80 | 47 | 148 | 33 | 81 | 47 | 26,521 | 6359 | 16,646 | 9052 |
| ZFX4 mutation | ||||||||||||
| Yes | 35 | 5 | 20 | 15 | 36 | 6 | 20 | 15 | 7273 | 1468 | 3892 | 3224 |
| No | 167 | 37 | 102 | 52 | 180 | 41 | 104 | 52 | 33,219 | 7845 | 19,233 | 10,169 |
Tr training subset, Te test subset, IV internal validation subset, EV external validation subset.
Deep learning framework
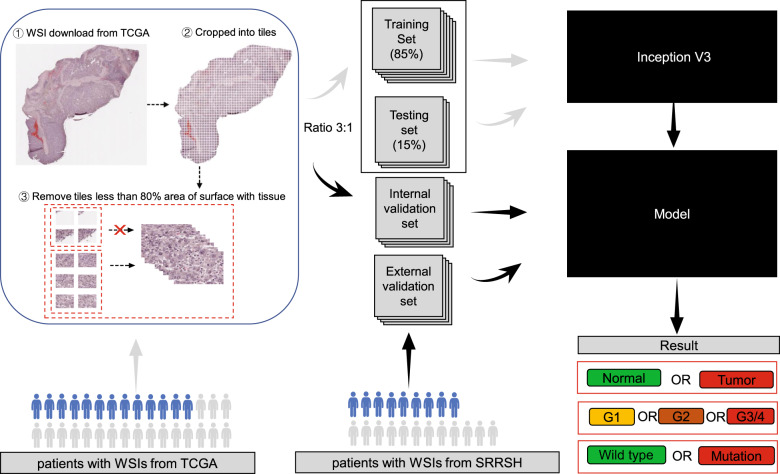
Patients from GDC-portal were selected and identified as the primary cohort. Based on a random split-sample approach, a total of 377 patients were then randomly divided into a training cohort (consisting of testing cohort) and an internal validation cohort with a ratio of 3:1. In addition, 67 patients from our medical center were identified as an external validation cohort. All WSIs were cropped into multiple small “tiles” at a magnification of 20×. Finally, the training and testing set consisting of a large collection of tiles were used to train a neural network (inception V3) for the classification of liver tissue via EASY DL. The internal and external validation was performed by the remaining tiles from internal and external validation sets, respectively (Fig. 1).
Fig. 1. Deep-learning framework for training and evaluating the model to classify and predict mutation.
Patients from TCGA were randomly divided into training cohorts (training and test) and internal validation cohort. Some patients had multiple virtual slides, and each slide was sliced into smaller “tiles”. The training, test, and internal and external validation sets were made up of multiple tiles from related cohorts. Model selection was done based on the performance in the test set. After learning and selection, the model was applied to tiles in the internal and validation sets to assess their performances.
Performance of classification
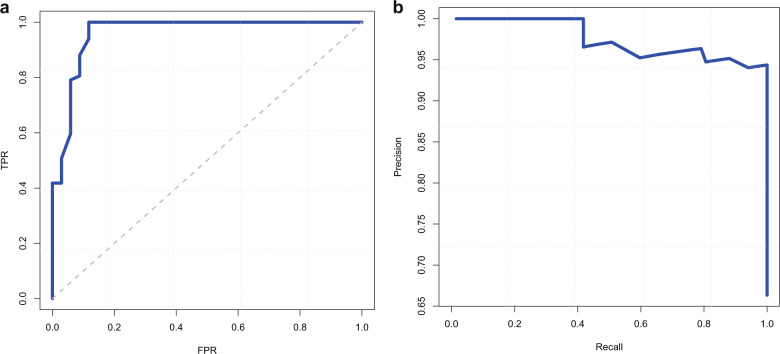
The high-performance level of our models at recognizing tumors from normal liver tissue (AUC = 0.961; 95% CI 0.939–0.981) was observed in the validation set (Fig. 2a). Based on the class-imbalanced problem, the precision-recall curves (PR-curves) and Matthews correlation coefficient (MCC) were also used to evaluate its performance (Fig. 2b). The MCC was up to 0.82 for benign or malignant classification, and 0.738 for assessing histopathological grade (well, moderate, or poor). Compared to three pathologists with 2-year, 5-year, and 10-year experience in respective, the performances of our classifiers nearly reached the ability of pathologists with 5-year experience (Table 2).
Fig. 2. The performance of the model at automated recognizes tumors from normal liver tissue.
a The receiver operating characteristic curve. TPR represents true positive rate, and FPR represents false positive rate. b Precision-recall curve.
Table 2.
The performance of our models and pathologists’ ability for classification.
| Classifiers | Performance | Our models | Pathologists with different years’ experience | ||
|---|---|---|---|---|---|
| 2-year | 5-year | 10-year | |||
| Normal vs. tumor | Accuracy | 0.960 | 0.911 | 0.970 | 0.990 |
| Precision | 0.945 | 0.926 | 0.957 | 0.985 | |
| Recall | 1.000 | 0.940 | 1.000 | 1.000 | |
| F1-score | 0.971 | 0.933 | 0.978 | 0.993 | |
| MCC | 0.912 | 0.799 | 0.934 | 0.977 | |
| Well (G1) vs. moderate(G2) vs. poor (G3/G4) | Accuracya | 0.896 | 0.851 | 0.910 | 0.955 |
| Precisiona | 0.879 | 0.831 | 0.869 | 0.944 | |
| Recalla | 0.771 | 0.758 | 0.807 | 0.895 | |
| Micro F1-score | 0.820 | 0.754 | 0.836 | 0.914 | |
| MCCa | 0.738 | 0.637 | 0.764 | 0.882 | |
MCC Matthews correlation coefficient.
aAverage value.
Performance of mutation prediction
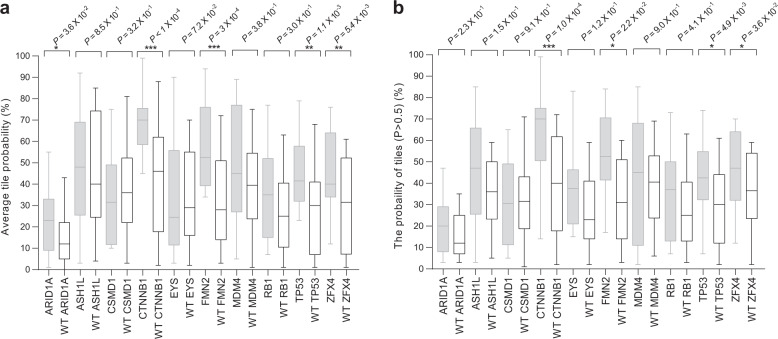
Our models were trained and validated based on the ten most significantly mutated genes to estimate the possibility of mutation. The performances, including accuracy, precision, and recall rate, F1-score, and MCC, were summarized in Table 3. In order to reduce heterogeneity, the performance was assessed both the average predicted probability on region (tiles)-level and the probability of predicted tile (P > 0.5) on slide-level in the external validation set. On the region(tiles)-level, we found that five of which, including ARID1A (P = 0.036), CTNNB1 (P < 0.0001), FMN2 (P = 0.0003), TP53 (P = 0.0011) and ZFX4 (P = 0.0054), showed significant differences between mutation and wild type group (Fig. 3a), with the area under the receiver operating characteristic curves (AUCs) from 0.71 to 0.89 in the external validation set. In addition, similar differences were observed on the slide-level, except for ARID1A (Fig. 3b). The per-slide AUCs after aggregation by average predicted probability and percentage of tiles with positive classification were listed in Table 4.
Table 3.
The performances of our models for gene mutation prediction.
| GENE | Accuracy | Precision | Recall | F1-score | MCC |
|---|---|---|---|---|---|
| ARID1A | 0.925 | 0.833 | 0.769 | 0.800 | 0.755 |
| ASH1L | 0.896 | 0.778 | 0.583 | 0.667 | 0.615 |
| CSMD1 | 0.910 | 0.714 | 0.556 | 0.625 | 0.581 |
| CTNNB1 | 0.910 | 0.895 | 0.810 | 0.850 | 0.788 |
| EYS | 0.925 | 0.800 | 0.500 | 0.615 | 0.596 |
| FMN2 | 0.925 | 0.727 | 0.800 | 0.762 | 0.719 |
| MDM4 | 0.925 | 0.750 | 0.429 | 0.545 | 0.532 |
| RB1 | 0.940 | 0.800 | 0.571 | 0.667 | 0.646 |
| TP53 | 0.925 | 0.895 | 0.850 | 0.872 | 0.820 |
| ZFX4 | 0.910 | 0.846 | 0.733 | 0.786 | 0.732 |
MCC Matthews correlation coefficient.
Fig. 3. Prediction of the ten most common mutated genes in liver cancer using our deep-learning model and histopathology images.
a comparison of the mutation and wild type in the distribution of the mutation probability in genes from tiles. b comparison of the mutation and wild type in the distribution of the mutation probability (Predicted P > 0.5) in each slide. P values were estimated with the two-tailed Mann–Whitney U-test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). For the two box plots, the middle line within the box represents the median; box limits represent 95% upper and lower quartiles; and whiskers represent the minima and maxima.
Table 4.
The performance of our models at mutation prediction in the external validation set.
| Mutations | Per-tile AUC | Per-slide AUC after aggregation by | |
|---|---|---|---|
| Average predicted probability | Percentage of positive tiles | ||
| CTNNB1 | 0.805 (0.759–0.851) | 0.898 (0.810–0.986) | 0.817 (0.713–0.922) |
| FMN2 | 0.727 (0.666–0.789) | 0.737 (0.613–0.861) | 0.838 (0.742–0.935) |
| TP53 | 0.736 (0.696–0.777) | 0.770 (0.650–0.890) | 0.715 (0.591–0.840) |
| ZFX4 | 0.720 (0.675–0.765) | 0.724 (0.591–0.858) | 0.751 (0.614–0.888) |
Discussion
In this study, the deep-learning classifiers displayed a high-level performance at recognizing cancer apart from normal liver tissue and assessing histopathological grade (well, moderate, or poor). The performances nearly reached the ability of pathologists with 5-year experience. Interestingly, the model found 9 out of 13 WSIs from our center with grading misclassified by at least a pathologist. Although the sensitivity and accuracy still need to be improved to be on par with a 10-year experience pathologist, it could be used to assist young pathologists at diagnosing with shorter learning curve period, faster speed, and higher accuracy. Moreover, the prediction of the four genes mutation (CTNNB1, FMN2, TP53, and ZFX4) is beyond the ability of pathologists.
The prediction of mutation based on histopathological H&E images using deep learning may have a positive influence on the diagnosis and treatment of patients with cancer given the importance of gene mutation21,27. For example, the mutations in CTNNB1 occurred at a relatively high frequency in HCC, with a high expression of the protein kinase human monopolar spindle 1 (hMps1/TTK), and TTK inhibitors regarded as one of the potential targeted drugs for CTNNB1 mutant HCC28–30. Interestingly, our models showed a high-performance level of predicting CTNNB1 mutation. The prediction of CTNNB1 mutation using deep learning may make a great contribution to select patients who are most likely to respond to TTK inhibitor targeted therapy.
Due to the unclear AI algorithmic data processing in a “black box”, developers and users do not know how computers arrive at conclusion, thereby making it difficult to find out the detail of evidence resulting in a conclusion31,32. Therefore, as a novel tool for diagnosis and treatment, AI should be validated against current quality standards to ensure clinical effectiveness and safety in clinical practice33,34. In this study, an independent database from our center was used to validate the performance of our models. It was demonstrated that convolutional neural networks could be used to assist in the classification and mutation prediction, based on histopathological H&E slides in liver cancer. However, the model still needs to be improved and validated by larger studies in the future. Even though it is impossible for AI to completely replace humans in practice nowadays, it is still a useful and effective tool to assist clinicians in dealing with repetitive work to provide important prognostic and therapeutic information. For example, mutation prediction could serve as pre-screening to improve cost-efficiency before immunohistochemistry or next-generation sequencing.
Overall, the study demonstrates that convolutional neural networks can predict histopathological grade and mutation in liver cancer. Although AI is likely to be a useful tool to assist surgeons and pathologists in classification of WSIs of HCC, the black box that how to get the conclusion is unclear and should be further studied. Besides, it is the first study to predict the gene mutation in HCC, meanwhile, internal and external validation cohorts were utilized to improve the accuracy of the model. In addition, the information on pathology and gene mutations may potentially be significant in applying the appropriate targeted therapy to HCC patients, thereby improving the performance of precision medicine.
The present study has several limitations to discuss. On the one hand, the size of the validation cohort is small. On the other hand, the model is not a complete replacement for pathologists’ examination, which included the diversity and heterogeneity of tissues that pathologists typically inspect (e.g., inflammation, necrosis, and blood vessels) and some clinical factors. Therefore, further validation of our model is necessary in a larger dataset with multiple centers and clinical factors or characteristics should be considered in further study. Moreover, EASY DL platform is exclusively available in Chinese which considerably limits the scope and audience targeted. To address the limitation, we provided the step-by-step instruction (figures and detailed English descriptions) for training deep-learning models via EASY DL, which was available at GitHub (https://github.com/drmaxchen-gbc/HCC-deep-learning/) named “How_to_use_EASY DL”.
In conclusion, our study demonstrated that the convolutional neural networks could assist pathologists in the classification of liver cancer and the detection of gene mutation. It also revealed that this method might be successfully adopted for other types of solid tumors.
Methods
Prepare histopathological tiles dataset of liver cancer
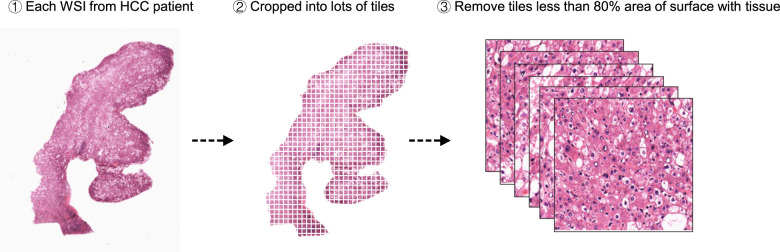
The frozen slide images and the corresponding cancer information were obtained from the GDC-portal (https://portal.gdc.cancer.gov/). On slide-level, there were 491 WSIs (HCC vs. normal liver tissue, 402 vs. 89), 383 WSIs of HCC with available histopathological grade (well vs. moderate vs. poor, 55 vs. 187 vs. 141) and 387 WSIs of HCC with corresponded gene mutation information. Besides, 67 WSIs of HCC with completed information and 34 WSIs of normal liver tissue were selected from Sir Run-Run Shaw Hospital. All WSIs should be cropped into multiple small “tiles” at a magnification of 20×. The majority of slides could be cropped into more than 200 “tiles” on region (tiles)-level (Supplementary Fig. 1). Each tile was saved as a JPG format by nonoverlapping 256 × 256-pixel windows. In order to avoid heterogeneity, each tile, where less than 80% of the surface was covered by tissue, should be removed (Fig. 4). Finally, the liver cancer tiles dataset consisted of four subsets, including the training, testing, internal validation, and external validation sets. The data in the training and internal validation cohorts from the Genomic Data Commons portal (https://portal.gdc.cancer.gov/) were publicly available without restriction, authentication or authorization. The independent external validation cohort we used consisted of slide images without identifiable information and all participants had provided written informed consent. Our study was approved by the SRRSH of Medicine Institutional Review Board (KY20181209-5).
Fig. 4. Strategy of preparing tiles dataset.
First, each WSI of liver tissue was selected from GDC-portal or SRRSH. Then, they were cropped into lots of tiles. Finally, the tiles less than 80% area of surface with tissue were removed, and the remaining tiles were used for further analysis.
Technical detail on frozen slides in the external validation cohort
The obtained specimens (e.g., liver tissues) were macroscopically examined, measured, sectioned through their longest axis, and then midsections were examined. The material was frozen at −28 °C, cut into 5–10 µm thick sections, Hematoxylin-Eosin (H&E) stained, and then analysed by pathologists with the light microscope. There were 67 out of 70 patients diagnosed as HCC and the related frozen slide were collected. Notably, normal liver tissues cannot be available in half of the obtained specimens, because normal liver tissues should be at least 2 cm away from tumors. Therefore, there were only 34 WSIs of normal liver tissues. In order to obtain digital pathology images, each slide was scanned at a magnification of 20× by using digital pathology scanner VS120 (Olympus).
Deep-learning with convolution neural networks
Typical convolutional neural networks contain several levels of convolution filters, pooling layers, and fully connected layers. In our study, we primarily used inception V3 architecture, which makes use of inception modules which are made from a spread of convolutions having different kernel sizes and a max-pooling layer. The initial five convolution nodes are combined with two max-pooling operations and followed by 11 stacks of inception modules. A fully connected layer to the end of the inception modules was then added to permit us to utilize the pre-trained model and finetune the parameters for our own task. Finally, a softmax layer was added as a classifier outputting a probability for every class, and the one with the highest probability was chosen as the predicted class.
We used the pre-trained model offered by TensorFlow and finetuned it using histopathological images. It was pre-trained on the ImageNet dataset and available at the TensorFlow-Slim image classification library (http://tensorflow.org). We initialized the parameters from the pre-trained model because pre-training can speed up the convergence of the network. Most importantly, it was difficult to train a deep network with a small number of images due to the massive number of network parameters.
Comparison with pathologists
One hundred and one WSIs of liver tissues without a label from the external validation cohort were used to test pathologist’s performance and compared with our model performance. All pathologists should report whether there is HCC, and if there is HCC, they should report histopathological grade via digital pathology images. The outcomes reported by six pathologists with 2-years, 5-years, and 10-years experience (two pathologists in each category) and our model were collected and analyzed by the R 3.6.0 (https://www.r-project.org). Cohen’s Kappa analysis was performed to assess inter-observer agreement. Good inter-operator agreements were observed in pathologists with 2-year experience (Kappa = 0.894; 95% CI, 0.837–0.944), pathologists with 5-year experience (Kappa = 0.933; 95% CI, 0.888–0.975), and pathologists with 5-year experience (Kappa = 0.967; 95% CI, 0.930–0.992).
Identification of significantly mutated genes
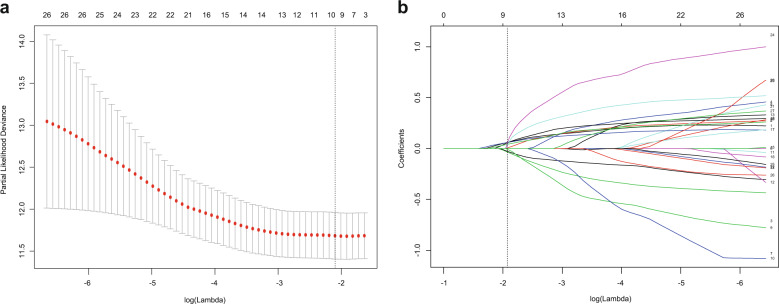
The gene mutation data for the matched patient sample were downloaded from the cancer genome atlas (TCGA). The gene mutated at least 10% of the available liver cancer samples were selected from the 283 cancer-related genes (Supplementary Fig. 2). The least absolute shrinkage and selection operator (LASSO) regression with a 10-fold cross-validation method was then performed to identify significant prognosis-related gene mutations by using R software packages (http://www.r-project.org). Finally, the ten most significant prognosis-related gene mutations, including ARID1A, ASH1L, CSMD1, CTNNB1, EYS, FMN2, MDM4, RB1, TP53, and ZFX4 were identified (Fig. 5).
Fig. 5. Prognosis-related mutated genes selection using the least absolute shrinkage and selection operator (LASSO) Cox regression model.
a Selection of the super parameter λ in the LASSO model via 10-fold cross-validation based on the minimum standard. The optimal λ value of 0.122. b Shown here is a coefficient section view plotted against the log(λ) magnitude. The optimal λ corresponding to ten non-zero coefficients were obtained where the vertical line was drawn.
Training deep-learning network
Pathological diagnosis was the primary endpoint of interest for the classifier that recognizes tumors from normal liver tissue and the assessment of the histopathological grade. The status of gene mutation (mutation or wild type), based on the next-generation sequencing results, was the primary prerequisite in the classifier of mutation prediction. The model’s training strategy was based on an easy-to-use platform called EASY DL (https://ai.baidu.com/easydl/) that uses PaddlePaddle deep learning framework V3.0 created by Baidu Brain AI technology, inception V3 network developed by Google, and packaging code form Coudray20 and co-workers. The training set was used for training, and the testing set was used to evaluate the performances, finetune those parameters, and improve the models. A final model was selected according to the results of the testing set, where the F1-scores as a stopping rule. Notably, the subsets were grouped based on HCC patients rather than the WSIs. This method could maximize the size of the training set and avoid training and testing on tiles originating from the same human subjects. Thereby preventing the classifier from relying on intra-subject correlations between samples and resulting in inflated estimates of accuracy. In order to reduce selection bias, the performance of our model was then validated in the internal and external validation sets.
Statistical analysis
The ten most common and prognostic mutated genes were identified using the LASSO Cox regression model, and any differences of overall survival were evaluated by the Kaplan–Meier method with a log-rank test. The performance of those models was evaluated with F1-scores, MCC, and AUC. The F1-scores, ranging from 1 (perfect) to 0 (bad), is the harmonic average of the precision and recall21. MCC ranges from 1 (perfect) to −1 (bad). In addition, the probability of gene mutation was estimated and compared using the two-tailed Mann–Whitney U-tests. A P value of less than 0.05, was considered as statistical significance.
Supplementary information
Acknowledgements
We would like to thank the EASY DL team and Hangzhou Xixuan Health technology Co., Ltd. for their assistance in training our models. Thanks to Y.C., J.H.H., S.J.L., F.Y. and all our colleagues for their assistance in this study. This abstract of the study was presented at The International Liver Congress TM 2019 (EASL 2019) as Late-Breaker poster, in Vienna, Austria, on April 11–13, 2019. This work was supported by the Opening Fund of Engineering Research Center of Cognitive Healthcare of Zhejiang Province (No.2018KFJJ09), Zhejiang Medical Health Science and Technology Project (No.2016133597), and National Natural Science Foundation of China (No.81827804).
Author contributions
M.Y.C., J.S.C., W.T., H.Y., and B.Z. were involved in the study design, data collection and analysis, and drafted the paper; H.P.Z., S.J., and Q.J.M. collected and checked data; M.Y.C., J.S.C., X.J.C., and W.T. revised the paper; X.J.C. designed, supervised the study; and all authors wrote the paper.
Data availability
The slide images and the corresponding cancer information were uploaded from the Genomic Data Commons portal (https://portal.gdc.cancer.gov/) and were in whole or in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). These data were publicly available without restriction, authentication, or authorization. The datasets for the independent cohorts generated and/or analyzed during the current study are available from the corresponding author (X.J.C.) upon reasonable request and through collaborative investigations.
Code availability
The codes that were used to train and validate the deep-learning model in the manuscript are available at https://github.com/drmaxchen-gbc/HCC-deep-learning. It also used other open-source codes (inception V3), which were available at https://github.com/openslide/openslide-python.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hong Yu, Email: 3195016@zju.edu.cn.
Xiujun Cai, Email: srrsh_cxj@zju.edu.cn.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41698-020-0120-3.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, et al. Cancer statistics for Hispanics/Latinos, 2018. Cancer J. Clin. 2018;68:425–445. doi: 10.3322/caac.21494. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Kudo M, et al. Brivanib as adjuvant therapy to transarterial chemoembolization in patients with hepatocellular carcinoma: a randomized phase III trial. Hepatology. 2014;60:1697–1707. doi: 10.1002/hep.27290. [DOI] [PubMed] [Google Scholar]
- 5.Sayiner Mehmet, Golabi Pegah, Younossi Zobair M. Disease Burden of Hepatocellular Carcinoma: A Global Perspective. Digestive Diseases and Sciences. 2019;64(4):910–917. doi: 10.1007/s10620-019-05537-2. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi VK, et al. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb. Pathog. 2019;128:184–194. doi: 10.1016/j.micpath.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Torres HA, et al. The oncologic burden of hepatitis C virus infection: a clinical perspective. Cancer J. Clin. 2017;67:411–431. doi: 10.3322/caac.21403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandenbulcke H, et al. Alcohol intake increases the risk of HCC in hepatitis C virus-related compensated cirrhosis: a prospective study. J. Hepatol. 2016;65:543–551. doi: 10.1016/j.jhep.2016.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Rao CV, Asch AS, Yamada HY. Frequently mutated genes/pathways and genomic instability as prevention targets in liver cancer. Carcinogenesis. 2017;38:2–11. doi: 10.1093/carcin/bgw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juengpanich, S. et al. Role of cellular, molecular, and tumor microenvironment in hepatocellular carcinoma: possible targets and future directions in the Regorafenib Era. Int. J. Cancer.10.1002/ijc.32970 (2020). [DOI] [PubMed]
- 11.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 12.Nault JC, Villanueva A. Intratumor molecular and phenotypic diversity in hepatocellular carcinoma. Clin. Cancer Res. 2015;21:1786–1788. doi: 10.1158/1078-0432.CCR-14-2602. [DOI] [PubMed] [Google Scholar]
- 13.Calderaro J, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 2017;67:727–738. doi: 10.1016/j.jhep.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Q, et al. Grading of hepatocellular carcinoma using 3D SE-DenseNet in dynamic enhanced MR images. Comput. Biol. Med. 2019;107:47–57. doi: 10.1016/j.compbiomed.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Weston AD, et al. Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology. 2019;290:669–679. doi: 10.1148/radiol.2018181432. [DOI] [PubMed] [Google Scholar]
- 16.Yi F, Huang J, Yang L, Xie Y, Xiao G. Automatic extraction of cell nuclei from H&E-stained histopathological images. J. Med. Imaging. 2017;4:027502. doi: 10.1117/1.JMI.4.2.027502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing F, Xie Y, Yang L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans. Med. Imaging. 2016;35:550–566. doi: 10.1109/TMI.2015.2481436. [DOI] [PubMed] [Google Scholar]
- 18.Lin, H. et al. Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J. Biophoton.10.1002/jbio.201800435 (2019). [DOI] [PubMed]
- 19.Li S, Jiang H, Pang W. Joint multiple fully connected convolutional neural network with extreme learning machine for hepatocellular carcinoma nuclei grading. Comput. Biol. Med. 2017;84:156–167. doi: 10.1016/j.compbiomed.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Coudray N, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darcy AM, Louie AK, Roberts LW. Machine learning and the profession of medicine. JAMA. 2016;315:551–552. doi: 10.1001/jama.2015.18421. [DOI] [PubMed] [Google Scholar]
- 22.Skrede OJ, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 23.Kather JN, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehteshami Bejnordi B, et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 2018;31:1502–1512. doi: 10.1038/s41379-018-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019;16:703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2818–2826 (2015).
- 27.Agarwal R, Narayan J, Bhattacharyya A, Saraswat M, Tomar AK. Gene expression profiling, pathway analysis and subtype classification reveal molecular heterogeneity in hepatocellular carcinoma and suggest subtype specific therapeutic targets. Cancer Genet. 2017;216–217:37–51. doi: 10.1016/j.cancergen.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Zaman GJR, et al. TTK inhibitors as a targeted therapy for CTNNB1 (beta-catenin) mutant cancers. Mol. Cancer Ther. 2017;16:2609–2617. doi: 10.1158/1535-7163.MCT-17-0342. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Liao W, Yuan Q, Ou Y, Huang J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget. 2015;6:34309–34320. doi: 10.18632/oncotarget.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang XD, et al. Expression and function analysis of mitotic checkpoint genes identifies TTK as a potential therapeutic target for human hepatocellular carcinoma. PLoS ONE. 2014;9:e97739. doi: 10.1371/journal.pone.0097739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietz, R. L. & Pantanowitz, L. The future of anatomic pathology: deus ex machina? J. Med. Artif. Intell.2, 4 (2019).
- 32.Tizhoosh HR, Pantanowitz L. Artificial intelligence and digital pathology: challenges and opportunities. J. Pathol. Inf. 2018;9:38. doi: 10.4103/jpi.jpi_53_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maddox TM, Rumsfeld JS, Payne PRO. Questions for artificial intelligence in health care. JAMA. 2019;321:31–32. doi: 10.1001/jama.2018.18932. [DOI] [PubMed] [Google Scholar]
- 34.Stead WW. Clinical implications and challenges of artificial intelligence and deep learning. JAMA. 2018;320:1107–1108. doi: 10.1001/jama.2018.11029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The slide images and the corresponding cancer information were uploaded from the Genomic Data Commons portal (https://portal.gdc.cancer.gov/) and were in whole or in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). These data were publicly available without restriction, authentication, or authorization. The datasets for the independent cohorts generated and/or analyzed during the current study are available from the corresponding author (X.J.C.) upon reasonable request and through collaborative investigations.
The codes that were used to train and validate the deep-learning model in the manuscript are available at https://github.com/drmaxchen-gbc/HCC-deep-learning. It also used other open-source codes (inception V3), which were available at https://github.com/openslide/openslide-python.