Abstract
The performance of ruminant livestock has been shown to benefit from the enhanced nutritive value and herbage yield associated with clover incorporation in the grazing sward. However, little research to date has been conducted investigating the effects of mixed swards containing white clover on the composition of the rumen microbiome. In this study, the rumen microbial composition of late lactation dairy cows grazing perennial ryegrass only (PRG; n = 20) or perennial ryegrass and white clover (WCPRG; n = 19) swards, was characterised using 16S rRNA amplicon sequencing. PERMANOVA analysis indicated diet significantly altered the composition of the rumen microbiome (P = 0.024). Subtle shifts in the relative abundance of 14 bacterial genera were apparent between diets, including an increased relative abundance of Lachnospira (0.04 vs. 0.23%) and Pseudobutyrivibrio (1.38 vs. 0.81%) in the WCPRG and PRG groups, respectively. The composition of the archaeal community was altered between dietary groups, with a minor increase in the relative abundance of Methanosphaera in the WCPRG observed. Results from this study highlight the potential for sward type to influence the composition of the rumen microbial community.
Subject terms: Applied microbiology, Microbiome
Introduction
Forage is recognised as the cheapest source of nutrition for ruminant livestock1 and predominates as the main source of feed in temperate climatic regions such as Ireland2. The addition of white clover (Trifolium repens) in the grazing sward has been reported to increase herbage yield3–5 and enhance animal performance in both dairy and sheep systems6–10. However the positive response to milk production associated with clover inclusion at pasture can be varied11,12. Legumes tend to have increased protein content and a reduced proportion of fibre in comparison to grasses13. Indeed, it is these attributes that are common to legumes, which has resulted in the majority of authors concluding improvements to animal performance to arise from increased feed intakes (due to reduced NDF) and overall increase to the sward nutritive value (as a result of increased CP) associated with clover inclusion in pastoral systems4,7,10.
A reduction to the fibrous proportion of diets is associated with increased feed consumption in ruminants14. The decreased fibre proportion in legumes compared to grasses13 most likely results from a combination of the increased pectin15,16 and reduced hemicellulose content in clover17. Differences in the carbohydrate composition between both forages results in a contrasted microbial degradation when ingested. The degradation of the pectin component of the cell wall in legumes18 results in a more rapid rate of degradability in the rumen19 most likely explaining the associated increase in feed intake when clover is fed20, due to a swift reduction in particle size.
The regulatory capacity of diet on the composition and functionality of the rumen microbiome has long been recognised resulting from the effects of a varied supply of growth substrates for differing microbes21–23. Subsequently, differences in the composition of the rumen microbial communities are known to influence animal performance24–26. Therefore, all dietary strategies under investigation and/or currently utilised on farm, should consider animal performance in tandem with rumen microbial composition and function.
To the best of our knowledge, only two studies have evaluated the effects of grazing swards containing white clover on the composition of the rumen microbiome using next generation sequencing techniques27,28. Both studies reported minor or no effects on the rumen microbial composition, although the potential for white clover inclusion in the sward to cause changes in the rumen microbiota has been suggested27.
Monocultures of white clover were investigated by Bowen et al.27, which would not regularly be grazed at farm level. As a result, the effect of white clover feeding, under practical on farm conditions, on the rumen microbiome, has only been conducted in one study and requires further validation. In addition, the grazing of clover swards has been reported to have a variable effect on methane production12,20,29,30. However, no simultaneous data on the composition of the rumen microbiome and methane production of ruminants grazing clover swards is available.
Therefore the aim of this study was to investigate if the inclusion of white clover in the grazing sward (WCPRG) would alter the composition and predicted functionality of the rumen bacteria and archaea communities of dairy cows, in comparison to monocultures of perennial ryegrass (PRG). Rumen microbial samples were obtained from a previous experiment investigating the effects of sward type on animal performance and enteric methane production12. An additional aspect of this study was to determine if minor differences in methane yield associated with sward type, would be reflective of alterations to the rumen archaeal population.
Results
Animal performance and herbage analysis has previously been reported by Enriquez-Hidalgo et al.12.
DNA Extraction and Sequencing Run Performance
No evidence of DNA degradation associated with the long term storage of samples was present with agarose gels revealing bands predominantly smeared between 12 kb and 1 kb positions for all samples. This range of smearing is to be expected due to DNA shearing associated with the bead beating method utilised31.
A total of 13,513,572 reads were obtained from sequencing on the Illumina MiSeq with an average 168,307 ± 36,207 reads per sample. Following quality filtering, merging and removal of chimeric sequences, there was a total of 5,543,164 reads with 138,579 ± 32,416 reads per sample.
A correlation of (rs = 0.976) was observed between the theoretical composition of the ZymoBIOMICSTM DS standard and that of our generated DS library. Run performance was deemed to have been performed to an optimal standard based on the correlation between our internal positive control and the theoretical composition supplied by ZymoBIOMICSTM.
Microbial Community Structure and Composition
No effect of sward type was observed on alpha diversity in the rumen microbial community structure, with no difference detected in Shannon or Simpson metrics (P = 0.23 and P = 0.34), respectively. Due to poor species classification, all ASVs within each sample were classified to the genus level. This resulted in a total of 304 taxa being identified at the genus level across all samples.
Across both diets, bacteria and archaea had an average relative abundance of 98.32 and 1.68%, respectively. At the phylum level, Bacteroidetes and Firmicutes dominated, with Prevotellaceae, Lachnospiraceae and Ruminococcaceae residing as the most abundant family of taxa across all samples. Finally, Prevotella 1 (40.58%) was the most abundant taxa identified in all samples when classified at the genus level. Other abundant genera of microbes observed across all samples included Christensenellaceae R-7 group (5.59%), Rikenellaceae RC9 gut group (4.81%), Fibrobacter (4.69%) and Ruminococcaceae NK4A214 group (3.93%).
Dietary Effect on Rumen Microbial Community
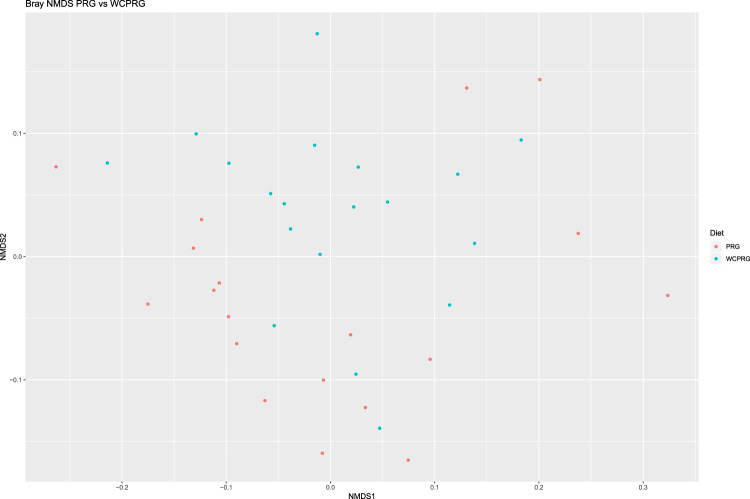
A NMDS plot of overall microbial community sample data, generated using Bray-Curtis dissimilarity analysis (Fig. 1), displayed a moderate degree of clustering of samples based on diet with respect to the microbial communities. Community composition was deemed to have been altered by diet based on the results of the PERMANOVA analysis (P = 0.024).
Figure 1.
Bray-Curtis NMDS plot highlighting differences in the bacterial and archaeal community composition between treatments. Different colour dots represent samples obtained from animals grazing different swards. Red = Perennial ryegrass (PRG), Blue = Perennial ryegrass and white clover (WCPRG).
Cyanobacteria were the only phyla with a relative abundance that was significantly altered by diet, with an increased abundance noted in the PRG group (Table 1). Dietary composition resulted in a significant increase in the abundance of Veillonellaceae in the PRG group (Table 2). Additionally, a minor but significant, rise in the abundance of Desulfovibrionaceae was observed in the PRG group.
Table 1.
Mean relative abundance (%) of the ten most abundant phylum in each dietary group.
| Kingdom | Phylum | WCPRG | PRG | P-value | adj P- value |
|---|---|---|---|---|---|
| Bacteria | Bacteroidetes | 48.63% | 47.53% | 0.58 | 0.73 |
| Bacteria | Firmicutes | 31.97% | 31.41% | 0.64 | 0.77 |
| Bacteria | Kiritimatiellaeota | 5.25% | 6.66% | 0.09 | 0.24 |
| Bacteria | Fibrobacteres | 3.74% | 3.42% | 0.22 | 0.43 |
| Bacteria | Tenericutes | 2.66% | 2.24% | 0.10 | 0.26 |
| Bacteria | Cyanobacteria | 1.27% | 2.10% | 0.00 | <0.05 |
| Bacteria | Spirochaetes | 1.50% | 1.51% | 0.77 | 0.88 |
| Archaea | Euryarchaeota | 1.29% | 1.58% | 0.03 | 0.15 |
| Bacteria | Patescibacteria | 1.33% | 1.11% | 0.02 | 0.15 |
| Bacteria | Proteobacteria | 1.17% | 1.17% | 0.89 | 0.96 |
PRG = Perennial ryegrass only; WCPRG = Perennial ryegrass and white clover sward;
P- values were generated using the Wilcoxon rank sum test, with correction for FDR. Adjusted p values deemed significant when adj p-value (P < 0.05) and trends (P < 0.10).
Table 2.
Mean relative abundance (%) of the ten most abundant microbes, classified to the taxonomic level of family, in each dietary group.
| Kingdom | Phylum | Class | Order | Family | WCPRG | PRG | P-value | adj P- value |
|---|---|---|---|---|---|---|---|---|
| Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | 43.90% | 43.20% | 0.86 | 0.94 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 13.00% | 12.13% | 0.32 | 0.51 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | 11.23% | 10.46% | 0.25 | 0.51 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Christensenellaceae | 5.03% | 5.24% | 0.62 | 0.71 |
| Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | 4.04% | 4.33% | 0.19 | 0.49 |
| Bacteria | Fibrobacteres | Fibrobacteria | Fibrobacterales | Fibrobacteraceae | 4.18% | 3.93% | 0.43 | 0.60 |
| Bacteria | Firmicutes | Negativicutes | Selenomonadales | Veillonellaceae | 2.40% | 3.39% | <0.001 | 0.02 |
| Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | F082 | 2.50% | 2.35% | 0.37 | 0.53 |
| Bacteria | Firmicutes | Negativicutes | Selenomonadales | Acidaminococcaceae | 1.69% | 2.26% | 0.01 | 0.18 |
| Bacteria | Spirochaetes | Spirochaetia | Spirochaetales | Spirochaetaceae | 1.63% | 1.69% | 0.93 | 0.97 |
PRG = Perennial ryegrass only; WCPRG = Perennial ryegrass and white clover sward;
p- values were generated using the Wilcoxon rank sum test, with correction for FDR. Adjusted p values deemed significant when adj p-value (P < 0.05) and trends (P < 0.10).
When classified at the genus level and corrected for FDR, the relative abundance of 15 taxa, with an average abundance of > 0.1% in at least one dietary group were found to be significantly, or tentatively, affected by diet (Table 3). Most genera influenced by diet belonged to the phylum Firmicutes (n = 11). A total of 4 genera belonging to the family Lachnospiraceae had relative abundances that differed between the groups. An increase in the relative abundance of, Pseudobutyrivibrio and Lachnoclostridium 10 was evident in the PRG animals whereas there was a rise in the relative abundance of Lachnospira and Lachnoclostridium 1 in the WCPRG group. Within Veillonellaceae, the abundance of Selenomonas 1 was decreased in the animals grazing the WCPRG swards. In the family Ruminococcaceae, a near significant increase in the abundance of Ruminococcaceae UCG-014 occurred in the WCPRG.
Table 3.
Treatment associated differences classified to genus level with mean relative abundance (%) for each group.
| Kingdom | Phylum | Class | Order | Family | Genus | WCPRG | PRG | P-value | adj P- value |
|---|---|---|---|---|---|---|---|---|---|
| Bacteria | Actinobacteria | Coriobacteriia | Coriobacteriales | Atopobiaceae | Olsenella | 0.21% | 0.15% | 0.0022 | 0.032 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnospira | 0.23% | 0.04% | <0.001 | <0.001 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Pseudobutyrivibrio | 0.81% | 1.38% | <0.001 | <0.001 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcus_2 | 0.16% | 0.09% | <0.001 | <0.01 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnoclostridium_1 | 0.13% | 0.07% | <0.001 | <0.01 |
| Bacteria | Firmicutes | Negativicutes | Selenomonadales | Veillonellaceae | Selenomonas_1 | 2.05% | 2.77% | <0.001 | <0.01 |
| Bacteria | Firmicutes | Erysipelotrichia | Erysipelotrichales | Erysipelotrichaceae | Kandleria | 0.35% | 0.16% | <0.001 | 0.015 |
| Bacteria | Firmicutes | Negativicutes | Selenomonadales | Veillonellaceae | Selenomonas | 0.05% | 0.15% | <0.001 | 0.015 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnoclostridium_10 | 0.13% | 0.19% | 0.0016 | 0.025 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcaceae_UCG-014 | 1.35% | 0.86% | 0.0051 | 0.06 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminiclostridium_6 | 0.10% | 0.07% | 0.006 | 0.067 |
| Bacteria | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Saccharofermentans | 1.58% | 1.24% | 0.007 | 0.072 |
| Bacteria | Patescibacteria | Saccharimonadia | Saccharimonadales | Saccharimonadaceae | Candidatus_Saccharimonas | 0.71% | 0.51% | 0.0034 | 0.045 |
| Bacteria | Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio | 0.07% | 0.12% | 0.0016 | 0.025 |
| Archaea | Euryarchaeota | Methanobacteria | Methanobacteriales | Methanobacteriaceae | Methanobrevibacter | 1.13% | 1.54% | 0.0046 | 0.058 |
PRG = Perennial ryegrass only; WCPRG = Perennial ryegrass and white clover sward.
P- values were generated using the Wilcoxon rank sum test, with correction for FDR. Adjusted p values deemed significant when adj p-value (P < 0.05) and trends (P < 0.10).
Only microbes with a relative abundance of > 0.1%, in one and/or both groups reported.
Diet also had a minor effect on the relative abundance of lowly abundant bacteria including an increase to the relative abundance of genera Ruminococcus 2, Candidatus Saccharimonas, Kandleria and Olsenella, in WCPRG cows and Desulfovibrio and Selenomonas in the PRG group. In addition, the relative abundance of Methanobrevibacter was increased in the PRG animals when abundance was calculated as a proportion of all 16S rRNA sequences.
Effect of Sward Type on Rumen Archaeal Community
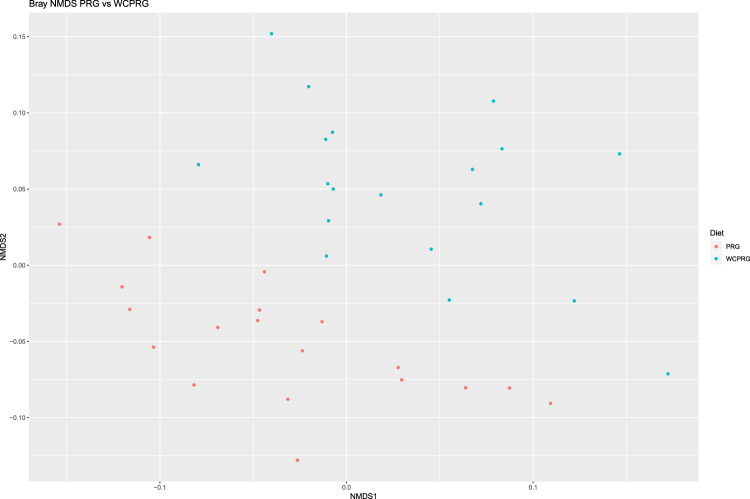
Overall, the archaeal relative abundance accounted for a small proportion (1.68%) of all sequences generated. Within both dietary groups, Methanobacteriaceae dominated at the family level with the genera Methanobrevibacter accounting for 79.1% of archaea across all samples. Calculating the abundance of methanogens relative to archaeal sequences only, resulted in a degree of sample clustering based on diet (Fig. 2). In addition, an increase in the relative abundance of Methanobrevibacter (81.98 vs. 76.21%; adj P < 0.001) within the PRG diet with a subtle shift towards an increase in the abundance of Methanosphaera (10.19 vs. 15.40%; adj P < 0.001) in the WCPRG group, was observed. No dietary effect was observed on the remaining genera of methanogens examined in the current study.
Figure 2.
Bray-Curtis NMDS plot highlighting differences in the archaeal community composition between treatments. Different colour dots represent samples obtained from animals grazing different swards. Red = Perennial ryegrass (PRG), Blue = Perennial ryegrass and white clover (WCPRG).
Predicted Metabolic Pathway Analysis
Two metabolic pathways, with a relative read abundance greater than 0.01%, were found to be influenced by diet. Metabolic pathways associated with other glycan degradation (adj P < 0.001; Fig. 3) and membrane and intracellular structural molecules (adj P < 0.01; Fig. 4) were predicted to have a higher expression in rumen samples collected from WCPRG cows. Diet did not have an impact on pathways predicted to be connected to methane metabolism.
Figure 3.
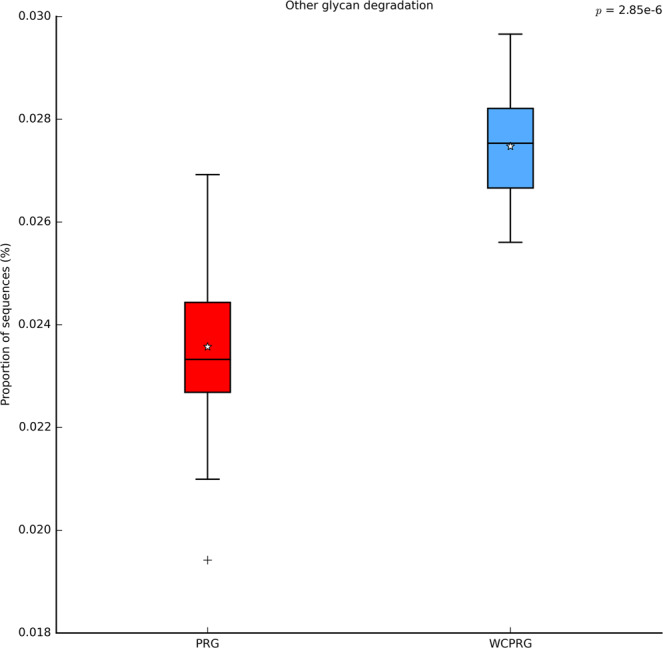
Relative read abundance for pathways predicted to be associated with other glycan degradation. PRG = Perennial ryegrass, WCPRG = Perennial ryegrass and white clover.
Figure 4.
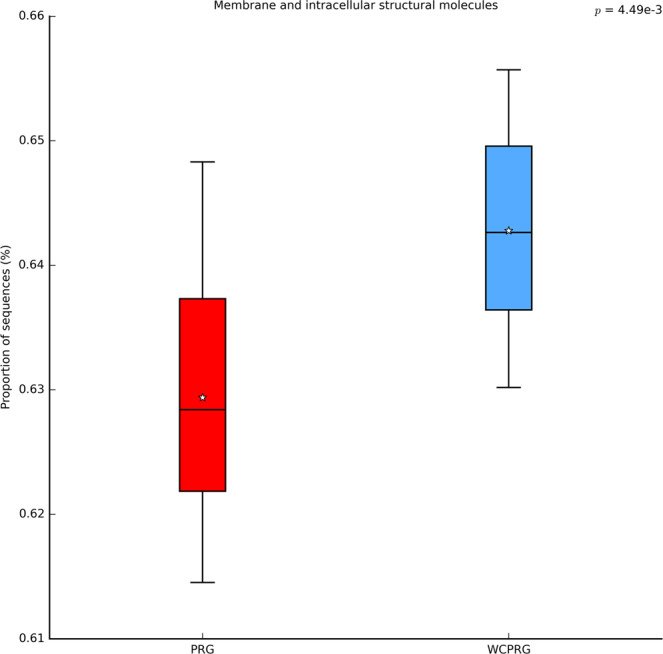
Relative read abundance for pathways predicted to be associated with membrane and intracellular structural molecules. PRG = Perennial ryegrass, WCPRG = Perennial ryegrass and white clover.
Discussion
The addition of white clover to the grazing sward has been shown to boost herbage yield and reduce the dependence on synthetic fertiliser3–5. Equally, animal performance has been shown to potentially benefit from an enhanced dietary nutritive value and digestibility with the incorporation of white clover into the grazing platform6–10, albeit response can be varied11,12.
The grazing of clover monocultures has previously been suggested to illicit stresses, such as low pH and/or bloat, on the rumen in conjunction with an elevated relative abundance of genera belonging to the phylum Proteobacteria27. However, data generated from this study would suggest the grazing of swards with 24% clover content to be free of imbalances to the rumen microbiota as the relative abundance of Proteobacteria between diets was consistent.
White clover has previously been shown to have higher pectin content in comparison to perennial ryegrass15,16. Therefore, elevated expression of pathways predicted to be associated with other glycan degradation in the WCPRG may be indicative of an increased supply of pectin in the diet. Members of the genera Lachnospira, such as L. multiparus, are predominantly pectin degrading bacteria32. Therefore, the observed greater abundance of this genus in the WCPRG cows is most likely due to higher pectin content in white clover when compared to grasses. This is similar to the findings of previous studies focused on legume forages27,33.
The ability to degrade pectin has been identified in other ruminal bacteria such as Prevotella and Butyrivibrio34. The genus Prevotella has been identified as a core rumen microbial resident21. In addition, the genus has been labelled a catabolic generalist35 due the ability of individual member species to degrade a variety of substrates such as starch, hemicellulose, xylan and pectin36,37. As a result, the dominance of Prevotella within both dietary groups is most likely reflective of the numerous growth substrates utilised by members of the genera. Butyrivibrio also has the ability to degrade a similar cohort of plant structural carbohydrates38, most likely leading to the similar relative abundance of the bacteria across both dietary groups.
The genomes of Lachnospira, Prevotella and Butyrivibrio have recently been reported to encode for pectin methyl esterases (PME)39, which are responsible for the conversion of pectin to methanol40. The abundance of methylotrophic methanogens such as Methanosphaera, is suggested to be limited by the availability of methanol in the rumen40. Therefore, the minor shift towards an increased abundance of Methanosphaera relative to the overall archaeal community, in the WCPRG dietary group, may be due to an enhanced supply of methyl compounds arising from the fermentation of pectin by the previously mentioned bacteria.
White clover is more readily fermented in the rumen in comparison to grasses41 and has been shown to be both associated with reduced ruminal retention time42 and increased feed intake4,20,29. Such characteristics of clover degradation may be a combination of the reduced fibre content in clover14 and cellular fracture, rather than indentation, of clover plant cells during the fermentation process18.
The model proposed by Janssen43 suggests that a faster rumen passage rate is likely to negatively impact microbial growth rates when substrates for growth are reduced. Recent work by others has identified Pseudobutyrivibrio to be a secondary coloniser of ingested fibre44,45, and therefore most likely unsuited to conditions associated with increased ruminal passage rate. In addition members of the genera Pseudobutyrivibrio are known fermenters of hemicellulose38. While in depth chemical analysis was not performed on the swards under investigation in this study12, perennial ryegrass has been shown to have a greater proportion of hemicellulose in comparison to white clover16,17. Therefore, the increase in the relative abundance of Pseudobutyrivibrio observed in the grass only group may have potentially benefited from an increased availability of hemicellulose. Also it is possible that clover may have some ‘as of yet’ unknown inhibitory effect on the microbe as a reduction in the abundance of the genus in vitro has previously been shown when incubated on red clover in comparison to PRG46.
Key characteristics of mixed clover and grass swards are indicative of their potential as a possible mitigation strategy for pastoral based systems. For example, a decreased rumen retention time has previously been associated with reductions in methane yield47,48 while reducing the proportion of fibre in the diet is also related to lower methane output49. We observed a minor reduction in the relative abundance of Methanobrevibacter in the clover sward when microbial abundance was calculated relative to all 16S rRNA sequences. However, as qPCR was not conducted as part of this study it is difficult to determine if the actual abundance of Methanobrevibacter was reduced. Equally, while the utilisation of 16S rRNA methods allowed for the simultaneous study of bacterial and archaeal populations in this study, the mcrA gene has proved a reliable biomarker for investigations of methanogenesis50. As a result, comparing the abundance of methanogens relative to bacteria on the bases of 16S rRNA sequencing, when studying methanogenesis, has limitations, including a lack of data on methanogen activity51. However, differences in the abundance of individual members of the methanogen population are known to be associated with methane production52. With this said when the abundance of methanogens was calculated relative to the archaeal sequences only, a subtle shift towards an increased abundance Methanosphaera was noted in WCPRG diet. In addition to this, a clear separation in the archaeal community structure between both dietary groups was apparent.
Theoretically, hydrogenotrophic methanogenesis has a greater net energy availability (−131 ΔG°′ (kJ/mol CH4))53 in comparison to methylotrophic methanogenesis (−113 to −112.5 ΔG°′ (kJ/mol CH4))54. However, as alluded to by Kelly et al.40, the lower H2 requirement of methylotrophic methanogenesis results in increased free energy availability in comparison to the CO2 reduction pathway. Previously, we referenced methanol to be the limiting factor for growth of Methanosphaera rather than H2. As a result, it could be possible that the increased supply of methanol, coupled with both the suggested negative effect of feed intake on members of Methanobrevibacter55 and more energetically favourable methanogenesis pathway, sustained a competitive advantage for Methanosphaera in the WCPRG group. While differences in the proportions of members of the archaeal community between both diets have been previously associated with reduced methane yield56,57, community shifts were minor which is reflective of the similar level of daily methane output and slight reduction in methane yield associated with the WCPRG diet.
Our research has shown the inclusion of white clover to the grazing sward to be accompanied by subtle changes to the composition of the rumen microbiome. Results from this study highlight the effect of different sward types on the composition of the rumen. It is assumed that more abundant microbes in a normal functioning rumen prosper due to their fermentation pathways providing an advantage, over less abundant microbial inhabitants, to utilise available substrates43. Considering this, it is probable that variation in the substrates supplied by the different swards and/or variation in ruminal retention time altered conditions in the rumen between the two groups. However, the subtle changes in nutrient flow to the rumen, do not appear to have negative consequences on the microbial composition indicative of ruminal stress, such as low pH or bloat, when grazing of swards with a 24% clover. To achieve benefits to animal performance, a 30% white clover inclusion rate is deemed necessary58. Therefore, future research may look to investigate if white clover, at an inclusion rate of 30% or greater, has similar impacts on the rumen microbiome.
Methods
Animal handling and sampling procedures carried out in this experiment were recommended for licencing by two qualified signatories at University College Dublin and approved for licencing by the Irish Department of Health and Children in accordance with the legislative requirements under section 8 of the Cruelty to Animals Act, 1876.
The experiment described here is based on an earlier study which examined the effect of sward composition on milk production and enteric methane emissions of dairy cows grazing either perennial ryegrass dominant pasture with or without the inclusion of white clover. A more thorough description of the animal model, methane measurement methodology and sward characteristics is reported in the original study12.
Animal and Grazing Model, DMI and Enteric Methane Emissions Estimates
Briefly, a total of 40 spring calving late lactation dairy cows, balanced by breed (Holstein Friesian, Norwegian Red and Norwegian Red × Holstein Friesian) were selected in April 2011 from the main herd at the Dairygold Research Farm (Teagasc, Animal and Grassland Research and Innovation Centre, Moorepark, Fermoy, Co. Cork, Ireland). Cows were randomly assigned to two herds and rotationally grazed swards containing either perennial ryegrass (Lolium perenne) only (PRG; n = 20) or perennial ryegrass and white clover (Trifolium repens) (WCPRG; n = 20). A postgrazing sward height (PostGSH) of 4 cm above ground height was targeted to allow individual animals a daily herbage allowance (HA) of 16 kg of DM. Additionally, cows were allocated a supplementation of 1 kg of concentrate/cow per day (CP = 154.1, NDF = 40.9, and ash = 102.8 g/kg of DM). Concentrate supplementation was withdrawn three days prior to the measurement of dry matter intake (DMI), followed by an increase in HA to 17 kg of DM/cow per day.
The PRG swards contained a 50:50 mixture of tetraploid (AstonEnergy) and diploid (Tyrella) PRG varieties sown at 37 kg/ha. The WCPRG sward included the same relative PRG combination utilised in the PRG sward (37 kg/ha) in addition to a medium leaf white clover mixture of 50:50 (Chieftain and Crusader) sown at a rate of 5 kg/ha.
Sample Collection and DNA Extraction
During the methane measurement period, approximately 50 ml of rumen digesta was sampled from each animal using transoesophageal rumen sampling (FLORA rumen scoop; Guelph, Ontario, Canada). After collection, individual rumen samples were immediately squeezed through three layers of synthetic cheesecloth and snap frozen followed by storage at −20 °C and by subsequent long-term storage at −80 °C until DNA was extracted. Further details in relation to the processing of samples at collection are reported in Enriquez-Hidalgo et al.59.
One sample was mislaid resulting in a total number of 39 samples (PRG n = 20; WCPRG n = 19) for microbial DNA extraction. Using the repeated bead beating and column purification method31, DNA was extracted from approximately 250 mg of the frozen rumen sample. DNA quality was assessed on agarose gels (see Supplementary Fig. S1) with the concentration of extracted DNA quantified on the Nanodrop 1000 spectrophotometer.
Library Preparation and Sequencing
A total of 39 amplicon libraries were generated from 25 ng of individual rumen microbial DNA from a total of 39 animals by performing two rounds of PCR amplification as outlined in the Illumina Miseq 16S Sample Preparation Guide with minor modifications to cycle length, as outlined by McGovern et al.60. An additional library was generated as an internal positive control using the ZymoBIOMICSTM Microbial Community Standard DNA (DS) (Zymo Research Corp., Irvine, CA, United States) to assess library preparation and sequencing performance. The first round of PCR amplification, targeting the V4 hypervariable region of the 16S rRNA gene, was performed using the 515 F/806 R primers61, designed with Nextera over hang adapters, and 2x KAPA Hifi HotStart ReadyMix DNA polymerase (Roche Diagnositics, West Sussex, UK). Cycle conditions were as follows: 95 °C for 3 minutes, 20 cycles at 95 °C for 30 seconds, 55 °C for 30 seconds, 72 °C for 30 seconds and then 72 °C for 5 minutes.
Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen, Manchester, UK). Following purification, amplicons were subject to a second round of PCR to permit attachment of dual indices and Illumina sequencing adapters using the Nextera XT indexing kit (Illumina, San Diego, CA, USA). Cycle conditions for the second round of PCR were 95 °C for 3 minutes, 8 cycles at 95 °C for 30 seconds, 55 °C for 30 seconds, 72 °C for 30 seconds and then 72 °C for 5 minutes followed by an additional PCR purification with the QIAquick PCR Purification Kit (Qiagen, Manchester, UK). Confirmation of amplicon generation was conducted visually on a 2% agarose gel. Amplicons were pooled together in equal concentration and subject to gel purification using the QIAquick Gel Extraction Kit (Qiagen, Manchester, UK) to remove adapter primers and further purified to remove any residues of agarose using the QIAquick PCR purification kit (Qiagen, Manchester, UK).
Pooled sample purity and quantity was analysed on the Nanodrop 1000 with further validation on the Qubit fluorometer and using the KAPA SYBR FAST universal kit with Illumina Primer Premix (Roche Diagnositics, West Sussex, UK). Following this, the library pool was diluted and denatured as per the Illumina Miseq 16S Sample Preparation Guide with sequencing conducted on the Illumina MiSeq using the 500 cycle version 2 MiSeq reagent kit (Illumina, San Diego, CA, USA).
Sequencing Analysis
All 16S rRNA gene amplicons were processed in R (version 3.4.2) using DADA2 (version 1.9.0) (https://benjjneb.github.io/dada2/tutorial.html) and submitted to the pipeline as described62 with minor modifications. Quality checks of both forward and reverse reads were initiated based on a visualisation of the average Q score for each sample with the aim to ensure mean Q scores of > 30 were upheld for forward and reverse reads. To achieve this, forward reads were trimmed to a length of 240 bp and reverse reads trimmed to 200 bp. The removal of primer sequences was conducted using the trimLeft function. Identical sequences were combined using the dereplication function followed by the merging of forward and reverse reads. Following this an amplicon sequence variant (ASV) table was constructed after which chimeric sequences were removed and taxonomy assigned to sequences variants using the SILVA database (version 132). Sample metadata, sequence taxonomy, and ASVs were combined into a phyloseq object using phyloseq (version 1.22.3)63 for further analysis. Predictions of metabolic pathways for each sample, based on the generated ASVs, were conducted using CowPI64.
Statistical Analysis
Analysis of animal performance data has been previously reported by Enriquez-Hidalgo et al.12. The generated ASV table and sequence taxonomies were analysed in R (version 3.4.2). Of the 39 samples sequenced, one sample was excluded from the analysis due to having a substantially lower sequencing depth compared to all other samples leaving a total of 38 successfully sequenced samples evenly split between the dietary groups (n = 19 per group). Taxa with a read count of less than two were removed for statistical analysis. The relative abundance of taxa was calculated for each sample at the genus level, as a percentage of total read count, in phyloseq. Two non-metric multidimensional scaling (NMDS) plots, based on Bray-Curtis dissimilarity were generated to visualise differences in the overall microbial communities (Fig. 1) and archaeal community (Fig. 2) between the dietary treatments. A PERMANOVA test, based on 9,999 permutations and a significance level of P < 0.05, was carried using the R package vegan (version 2.5.4)65 implemented through microbiome (version 1.0.2)66 to investigate differences in the overall community structure amongst samples, based on treatment, at the level of genus. The Wilcoxon rank sum test, with Benjamini Hochberg (BH) correction for false discovery rate (FDR) was implemented for the identification of significant treatment differences in the overall relative abundance of taxa and the archaeal only proportion, based on adjusted P-values (adj P) of <0.05. Only taxa with a relative abundance of >0.1% in at least one of the dietary groups, was considered in treatment comparisons. The dietary grouping of animals was the only descriptive variable included in the comparison of microbial communities between treatments, due to the balanced statistical design of the original study12 and dietary focus of the current study. A Student’s T-Test was used to compare diversity metrics microbial community (species level) structure comparisons. Statistical analysis of predicted pathways was carried out using STAMP (version 2.1.3)67. Comparison of predicted pathways was conducted using the proportion of reads that were annotated to each individual metabolic pathway as a percentage of the total reads (read relative abundance). Statistical analysis of read relative abundance was conducted using a Student’s T-Test, with BH correction for FDR (adj P < 0.05) to determine differences based on dietary group. Spearman’s rank correlation coefficient was used to determine the correlation of the theoretical composition of the ZymoBIOMICSTM DS standard and that of over generated DS library.
Supplementary information
Acknowledgements
The molecular rumen microbiome work was funded by the FACCE ERA GAS ‘RumenPredict’ grant 16/RD/ERAGAS/1RUMENPREDICT-ROI2017. The animal experiment received funding from the Irish Dairy Levy Trust and from the European Community’s Seventh Framework Programme (FP7/2007–2013) under the grant agreement no. FP7-244983 (MULTISWARD). P.S is funded by a Walsh Fellowship award (RMIS 0364).
Author contributions
Conceived and designed molecular experiments: S.W., D.K and A.K. Conceived and designed animal experiments: D.H. and D.E.H. Performed the experiments: S.W., A.K., D.K., M.M., D.E.H., D.H. and P.S. Analysed data: P.S., M.M., Contributed to reagents/materials/analysis tools: S.W., D.K., M.M., D.H., D.E.H, P.S. Result interpretation and wrote paper: P.S., D.E.H., D.H., A.K., D.K. and S.W. All authors agree to be accountable for all aspects of the work.
Data availability
16S rRNA gene amplicon sequence data for this experiment is accessible at the National Centre for Biotechnology Information Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) accession number PRJNA550258.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-66028-3.
References
- 1.Finneran E, et al. Stochastic simulation of the cost of home-produced feeds for ruminant livestock systems. J. Agri. Sci. 2012;150:123–139. [Google Scholar]
- 2.O’Brien D, Moran B, Shalloo L. A national methodology to quantify the diet of grazing dairy cows. J. Dairy Sci. 2018;101:8595–8604. doi: 10.3168/jds.2017-13604. [DOI] [PubMed] [Google Scholar]
- 3.Enriquez-Hidalgo D, Gilliland TJ, Egan M, Hennessy D. Production and quality benefits of white clover inclusion into ryegrass swards at different nitrogen fertilizer rates. J. Agri.Sci. 2018;156:378–386. [Google Scholar]
- 4.Guy C, et al. Comparison of perennial ryegrass, Lolium perenne L., ploidy and white clover, Trifolium repens L., inclusion for herbage production, utilization and nutritive value. Grass For. Sci. 2018;73:865–877. [Google Scholar]
- 5.Guy C, et al. White clover incorporation at high nitrogen application levels: results from a 3-year study. Anim.Prod. Sci. 2020;60:187–191. [Google Scholar]
- 6.Dineen M, Delaby L, Gilliland T, Mccarthy B. Meta-analysis of the effect of white clover inclusion in perennial ryegrass swards on milk production. J. Dairy Sci. 2018;101:1804–1816. doi: 10.3168/jds.2017-12586. [DOI] [PubMed] [Google Scholar]
- 7.Egan M, Lynch MB, Hennessy D. Including white clover in nitrogen fertilized perennial ryegrass swards: effects on dry matter intake and milk production of spring calving dairy cows. J. Agric. Sci. 2017;155:657–668. [Google Scholar]
- 8.Grace C, et al. Grazing multispecies swards improves ewe and lamb performance. Animal. 2018;13:1–9. doi: 10.1017/S1751731118003245. [DOI] [PubMed] [Google Scholar]
- 9.Egan M, Galvin N, Hennessy D. Incorporating white clover (Trifolium repens L.) into perennial ryegrass (Lolium perenne L.) swards receiving varying levels of nitrogen fertilizer: Effects on milk and herbage production. J. Dairy Sci. 2018;101:3412–3427. doi: 10.3168/jds.2017-13233. [DOI] [PubMed] [Google Scholar]
- 10.McClearn B, et al. The effect of perennial ryegrass ploidy and white clover inclusion on milk production of dairy cows. Anim. Prod. Sci. 2020;60:143–147. [Google Scholar]
- 11.Humphreys J, Casey IA, Laidlaw AS. Comparison of milk production from clover-based and fertilizer-N-based grassland on a clay-loam soil under moist temperate climatic conditions. Irish. J. Agr. Food. Res. 2009;48:189–207. [Google Scholar]
- 12.Enriquez-Hidalgo D, Gilliland T, Deighton MH, O’Donovan M, Hennessy D. Milk production and enteric methane emissions by dairy cows grazing fertilized perennial ryegrass pasture with or without inclusion of white clover. J. Dairy Sci. 2014;97:1400–1412. doi: 10.3168/jds.2013-7034. [DOI] [PubMed] [Google Scholar]
- 13.Lee MA. A global comparison of the nutritive values of forage plants grown in contrasting environments. J. Plant Res. 2018;131:641–654. doi: 10.1007/s10265-018-1024-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen MS. Effects of diet on short-term regulation of feed intake by lactating dairy cattle. J. Dairy Sci. 2000;83:1598–1624. doi: 10.3168/jds.S0022-0302(00)75030-2. [DOI] [PubMed] [Google Scholar]
- 15.Thomson D. 1984. The nutritive value of white clover. British Grassland Society Occasional Symposium. Forage Legumes. 1984;16:78–92. [Google Scholar]
- 16.Ulyatt MJ. Studies on the causes of the differences in pasture quality between perennial ryegrass, short-rotation ryegrass, and white clover. New Zeal. J. Agr. Res. 1971;14:352–367. [Google Scholar]
- 17.Fraser TJ, Rowarth JS. Legumes, herbs or grass for lamb performance? Proc N Z Grassl Assoc. 1996;58:49–52. [Google Scholar]
- 18.Cheng KJ, Fay JP, Howarth RE, Costerton JW. Sequence of events in the digestion of fresh legume leaves by rumen bacteria. Appl. Environ. Microbiol. 1980;40:613–625. doi: 10.1128/aem.40.3.613-625.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chesson A, Monro JA. Legume pectic substances and their degradation in the ovine rumen. J. Sci. Food Agri. 1982;33:852–859. [Google Scholar]
- 20.Niderkorn V, et al. Associative effects between fresh perennial ryegrass and white clover on dynamics of intake and digestion in sheep. Grass For. Sci. 2017;72:691–699. [Google Scholar]
- 21.Hungate, R. E. Chapter X - Variations in the Rumen.The Rumen and its Microbes. Academic Press (1966).
- 22.Henderson G, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellison MJ, et al. Diet and feed efficiency status affect rumen microbial profiles of sheep. Small Ruminant Res. 2017;156:12–19. [Google Scholar]
- 24.Carberry CA, Kenny DA, Han S, McCabe MS, Waters SM. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 2012;78:4949–4958. doi: 10.1128/AEM.07759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shabat SK, et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016;10:2958–2972. doi: 10.1038/ismej.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGovern E, et al. 16S rRNA sequencing reveals relationship between potent cellulolytic genera and feed efficiency in the rumen of bulls. Front. Microbiol. 2018;9:1842. doi: 10.3389/fmicb.2018.01842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowen JM, McCabe MS, Lister SJ, Cormican P, Dewhurst RJ. Evaluation of Microbial Communities Associated With the Liquid and Solid Phases of the Rumen of Cattle Offered a Diet of Perennial Ryegrass or White Clover. Front. Microbiol. 2018;9:2389. doi: 10.3389/fmicb.2018.02389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Callaghan TF, et al. 2018. Pasture feeding changes the bovine rumen and milk metabolome. Metabolites. 2018;8:27. doi: 10.3390/metabo8020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J, Woodward S, Waghorn G, Clark D. Methane emissions by dairy cows fed increasing proportions of white clover (Trifolium repens) in pasture. Proc. N. Z. Grassland Ass. 2004;66:151–155. [Google Scholar]
- 30.Hammond KJ, et al. Effects of feeding fresh white clover (Trifolium repens) or perennial ryegrass (Lolium perenne) on enteric methane emissions from sheep. Anim.Feed Sci. Tech. 2011;166:398–404. [Google Scholar]
- 31.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. 2004;36:808–12. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 32.Stewart C. S., Flint H. J., Bryant M. P. The rumen bacteria in The Rumen Microbial Ecosystem (ed. Hobson P. N. and Stewart C. S.) 10–72 (Springer, 1997).
- 33.Bryant MP, et al. Predominant bacteria in the rumen of cattle on bloat-provoking ladino clover pasture. J. Dairy Sci. 1960;43:1435–1444. [Google Scholar]
- 34.Marounek M, Dušková D. Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Letters in App. Microbiol. 1999;29:429–433. [Google Scholar]
- 35.Emerson EL, Weimer PJ. Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 2017;101:4269–4278. doi: 10.1007/s00253-017-8150-7. [DOI] [PubMed] [Google Scholar]
- 36.Matsui H, et al. Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr. Microbiol. 2000;41:45–49. doi: 10.1007/s002840010089. [DOI] [PubMed] [Google Scholar]
- 37.Rubino F, et al. Divergent functional isoforms drive niche specialisation for nutrient acquisition anduse in rumen microbiome. ISME J. 2017;11:932–944. doi: 10.1038/ismej.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause DO, et al. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol. Rev. 2003;27:663–693. doi: 10.1016/S0168-6445(03)00072-X. [DOI] [PubMed] [Google Scholar]
- 39.Rode LM, Genthner BS, Bryant MP. Syntrophic association by cocultures of the methanol-and CO2-H2-utilizing species Eubacterium limosum and pectin-fermenting Lachnospira multiparus during growth in a pectin medium. Appl. Environ. Microbiol. 1981;42:20–22. doi: 10.1128/aem.42.1.20-22.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly WJ, et al. Occurrence and expression of genes encoding methyl-compound production in rumen bacteria. Animal Microbiome. 2019;1:15. doi: 10.1186/s42523-019-0016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minson, D.J. & Wilson, J.R. Prediction of intake as an element of forage quality in Forage quality, evaluation, and utilization (ed. Fahey G. C.) 533–563 (American Society of Agronomy, 1994).
- 42.Dewhurst RJ, Fisher WJ, Tweed JKS, Wilkins RJ. Comparison of Grass and Legume Silages for Milk Production. 1. Production Responses with Different Levels of Concentrate. J. Dairy Sci. 2003;86:2598–2611. doi: 10.3168/jds.S0022-0302(03)73855-7. [DOI] [PubMed] [Google Scholar]
- 43.Janssen PH. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed. Sci. Technol. 2010;160:1–22. [Google Scholar]
- 44.Huws SA, et al. Temporal dynamics of the metabolically active rumen bacteria colonizing fresh perennial ryegrass. FEMS Microbiol. Ecol. 2016;92:fiv137. doi: 10.1093/femsec/fiv137. [DOI] [PubMed] [Google Scholar]
- 45.Mayorga OL, et al. Temporal metagenomic and metabolomic characterization of fresh perennial ryegrass degradation by rumen bacteria. Front. Microbiol. 2016;7:1854. doi: 10.3389/fmicb.2016.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elliott CL, et al. Using ‘Omic Approaches to Compare Temporal Bacterial Colonization of Lolium perenne, Lotus corniculatus, and Trifolium pratense in the Rumen. Front. Microbiol. 2018;9:2184–2184. doi: 10.3389/fmicb.2018.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinares-Patiño C, Ulyatt J, Lassey M, N. Barry K. T. & W. Holmes, C. Rumen function and digestion parameters associated with differences between sheep in methane emissions when fed chaffed lucerne hay. J. Agric. Sci. 2003;140:205–214. [Google Scholar]
- 48.Goopy JP, et al. Low-methane yield sheep have smaller rumens and shorter rumen retention time. Br. J. Nutr. 2014;111:578–585. doi: 10.1017/S0007114513002936. [DOI] [PubMed] [Google Scholar]
- 49.Moss, A.R., Jouany, J.P. & Newbold, J. Methane production by ruminants: its contribution to global warming. Ann. Zootech. 49, 231–253, EDP Sciences (2000).
- 50.Wilkins D, Lu XY, Shen Z, Chen J, Lee PK. Pyrosequencing of mcrA and archaeal 16S rRNA genes reveals diversity and substrate preferences of methanogen communities in anaerobic digesters. Appl. Environ. Microbiol. 2015;8:604–613. doi: 10.1128/AEM.02566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGovern E, et al. Plane of nutrition affects the phylogenetic diversity and relative abundance of transcriptionally active methanogens in the bovine rumen. Sci. Reports. 2017;7:1–10. doi: 10.1038/s41598-017-13013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tapio I, Snelling TJ, Strozzi F, Wallace RJ. The ruminal microbiome associated with methane emissions from ruminant livestock. J. Anim. Sci. Biotechnol. 2017;8:7. doi: 10.1186/s40104-017-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N Y. Acad. Sci. 2008;1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- 55.McCabe, M.S. et al. Illumina MiSeq phylogenetic amplicon sequencing shows a large reduction of an uncharacterised Succinivibrionaceae and an increase of the Methanobrevibacter gottschalkii clade in feed restricted cattle. PloS One, 10 (2015). [DOI] [PMC free article] [PubMed]
- 56.Shi W, et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 2014;24:1517–1525. doi: 10.1101/gr.168245.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramayo‐Caldas Y, et al. Identification of rumen microbial biomarkers linked to methane emission in Holstein dairy cows. J. Anim. Breed. Genet. 2020;137:49–59. doi: 10.1111/jbg.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Annicchiarico P, Tomasoni C. Optimizing legume content and forage yield of mown white clover–Italian ryegrass mixtures through nitrogen fertilization and grass row spacing. Grass Forage Sci. 2010;65:220–226. [Google Scholar]
- 59.Enriquez-Hidalgo D, et al. Effect of rotationally grazing perennial ryegrass white clover or perennial ryegrass only swards on dairy cow feeding behaviour, rumen characteristics and sward depletion patterns. Livest. Sci. 2014;169:48–62. [Google Scholar]
- 60.Mcgovern E, Waters SM, Blackshields G, Mccabe MS. Evaluating Established Methods for Rumen 16S rRNA Amplicon Sequencing With Mock Microbial Populations. Front. Microbiol. 2018;9:1365–1365. doi: 10.3389/fmicb.2018.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 2011;108:4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMurdie, P. J. & Holmes, H. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 8, (2013). [DOI] [PMC free article] [PubMed]
- 64.Wilkinson, T.J. et al. CowPI: a rumen microbiome focussed version of the PICRUSt functional inference software. Front. Microbiol. 9, (2018). [DOI] [PMC free article] [PubMed]
- 65.Oksanen, J. et al. vegan: Community Ecology Package. R package.version 2.5-4. (2019). https://CRAN.R-project.org/package=vegan (2019).
- 66.Lahti, L. et al. Tools for microbiome analysis in R. Version 1.9.19. URL: http://microbiome.github.com/microbiome (2017).
- 67.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene amplicon sequence data for this experiment is accessible at the National Centre for Biotechnology Information Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) accession number PRJNA550258.