Abstract
Dissemination of bacterial clones carrying plasmid-mediated resistance genes is a major factor contributing to the increasing prevalence of antibiotic resistance. Understanding the evolution of successful clones and the association to mobile resistance elements are therefore crucial. In this study, we determined the sequence of a 145 kb IncC multi-drug resistance plasmid (pK71-77-1-NDM), harbouring resistance genes to last-resort antibiotics including carbapenems. We show that the plasmid is able to transfer into a range of genetically diverse clinical Escherichia coli strains and that the fitness cost imposed on the host is often low. Moreover, the plasmid is stably maintained under non-selective conditions across different genetic backgrounds. However, we also observed a lower conjugation frequency and higher fitness cost in the E. coli sequence type (ST) 73 background, which could partially explain why this clone is associated with a lower level of antibiotic resistance than other E. coli clones. This is supported by a bioinformatical analysis showing that the ST73 background harbours plasmids less frequently than the other studied E. coli STs. Studying the evolution of antibiotic resistance in a clinical context and in diverse genetic backgrounds improves our understanding of the variability in plasmid-host associations.
Subject terms: Evolution, Microbiology
Introduction
The development of antibiotic resistance in bacterial populations is an inevitable evolutionary consequence of the selective pressure exerted by the use and misuse of antibiotics. Two main routes allow bacterial evolution towards antibiotic resistance: mutations in the chromosome or acquisition of mobile genetic elements (MGEs) harbouring resistance-determinants.
Plasmids are extrachromosomal, independently replicating, most often circular and transferable DNA molecules that constitute the most prominent group of MGEs facilitating horizontal spread of antibiotic resistance1. Transferable plasmids harbouring resistance-determinants are widespread among clinically relevant Gram-negative pathogens like Escherichia coli2. This includes plasmids encoding extended-spectrum β-lactamases (ESBLs) or carbapenemases conferring resistance to β-lactams such as cephalosporins and carbapenems3. In E. coli, a wide diversity of plasmid associated ESBL and carbapenemase genes have been identified including blaCTX-M, blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48-like4. Interestingly, the molecular epidemiology of E. coli supports that certain resistance genes are clearly linked to specific dominant E. coli clones and plasmid backbones like the association of blaCTX-M-15 with IncF family plasmids and sequence type (ST) 1312. It has been suggested that E. coli ST131 may dominate as a successful multi-drug resistant clone due to the ability to offset the fitness cost of plasmid acquisition and maintenance via compensatory mutations in gene regulatory regions5,6. In contrast, other resistance genes like blaNDM show a more broad diversity, both with respect to host genetic backgrounds and plasmid backbones2,7. In addition to successful multi-drug resistant E. coli clones, other clones like ST73 have been shown to be equally successful, but generally susceptible to antibiotics8,9.
The success of a plasmid, and consequently a plasmid associated resistance gene, is constrained by several factors like conjugation rate, incompatibility with other plasmids in the same cell, stability and fitness cost10–14. The majority of studies investigating these mechanisms and plasmid-host interactions are performed using laboratory-adapted strains, environmental bacteria and/or plasmids with limited clinical relevance. A scarcity of studies has focused on clinically relevant pathogenic bacteria using plasmids with resistance-determinants observed in the clinical setting15,16. With respect to carbapenemase-encoding plasmids, it has been shown that a clinical blaNDM-1-encoding plasmid was stably maintained in laboratory strains of E. coli and Klebsiella pneumoniae, but imposed a significant fitness cost17. In contrast, we have previously shown that acquisition of clinical plasmids carrying blaKPC-2 or blaVIM-1 genes by plasmid-naïve clinical E. coli strains of different genetic backgrounds resulted in low to moderate reductions in fitness cost (1.1-3.6%) and that the fitness cost and plasmid stability were both plasmid and host dependent18. This relatively low impact on host fitness exerted by clinical carbapenemase-encoding plasmids has also been shown for a blaKPC-2-positive plasmid in K. pneumoniae19.
To further investigate the impact of plasmid acquisition, we studied the conjugation frequency, fitness cost and stability of a ~145 kb IncC blaNDM-1-encoding clinical plasmid in a genetically diverse collection of clinical E. coli strains.
Results and discussion
Characterization of the blaNDM-1 plasmid and donor strain
PacBio sequencing of the blaNDM-1-carrying E. coli donor strain K71-77 resulted in a circularized chromosome (4,934,660 bp) and two circularized plasmids, pK71-77-1-NDM (145,272 bp) and pK71-77-2 (117,597 bp). The genomic data confirmed that K71-77 belongs to phylogroup A and ST41020, which is considered an emerging international high-risk clone21. The isolate carried fimH24 encoding type I fimbriae and chromosomal mutations in gyrA (S83L and D87N), parC (S80I) and parE (S458A) conferring fluoroquinolone resistance common to ST41021. No acquired resistance genes were identified on the chromosome.
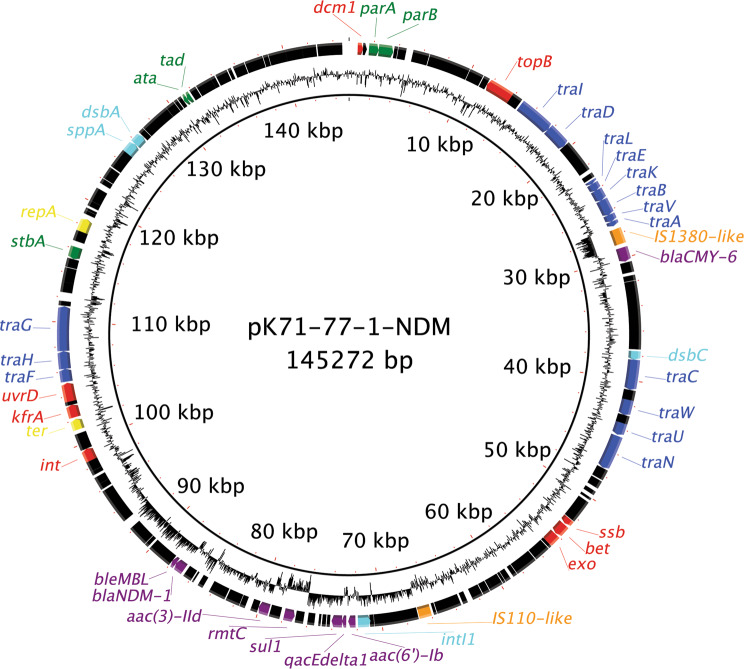
The blaNDM-1-carrying plasmid (pK71-77-1-NDM) is an IncC type 1 plasmid (also termed IncA/C2) based on the R1 and R2 regions and i1 and i2 segments22 as well as cgST1.2 according to the cgPMLST IncA/C scheme23,24. In addition to blaNDM-1, pK71-77-1-NDM carried the plasmid-mediated AmpC gene blaCMY-6, the 16S rRNA methylase gene rmtC, two variants of aminoglycoside acetyltransferase genes aac(6’)-Ib and aac(3)-II, the sulfonamide resistance gene sul1 and the bleomycin resistance gene bleMBL (Fig. 1). pK71-77-1-NDM displayed ~99% identity over its entire sequence with other IncC blaNDM-encoding plasmids like pNDM-EcoGN568 (GenBank acc. no. KJ802404), pNDM102337 (GenBank acc. no. JF714412), pNDM10505 (GenBank acc. no. JF503991) and pNDM-PstGN576 (GenBank acc. no. KJ802405). The second plasmid, pK71-77-2 is an IncF (IncFIA, IncFIB, IncFII) plasmid of replicon sequence type F36:A4:B1, encoding resistance to aminoglycosides (aac(6′)-Ib-cr), β-lactams (blaOXA-1) and chloramphenicol (catB4).
Figure 1.
Genetic map of pK71-77-1-NDM. Specific CDSs are colour coded as follows: DNA metabolism, red; plasmid maintenance, green; conjugative transfer, blue; mobile element, orange; other, turquoise; antimicrobial resistance, purple; replication, yellow. The map was constructed using the BLAST Ring Image Generator45.
Plasmid conjugative transfer
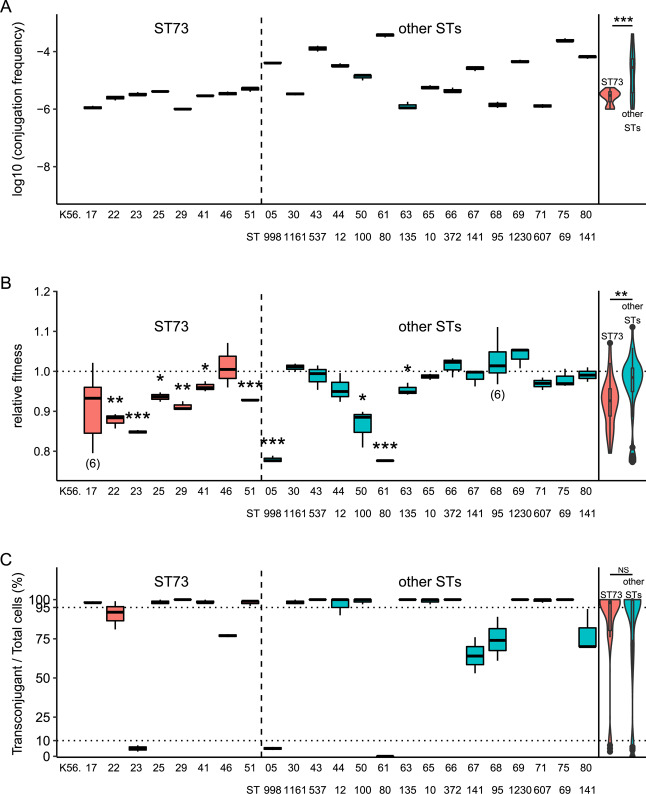
We mated the E. coli K71-77 isolate carrying pK71-77-1-NDM, in liquid for 3 hours (1:1 donor:recipient ratio), with 23 different uropathogenic recipient strains belonging to 15 different STs (Table 1). The recipient strains were selected from the ECO-SENS collection found to be devoid of phenotypic resistance to 24 antibiotics and plasmid-naïve based on plasmid-replicon typing and S1 nuclease pulsed-field gel electrophoresis25. This enabled us to introduce rifampicin resistance as a selection marker in the conjugation experiment and to avoid interactions with other plasmids. Plasmid pK71-77-1-NDM was successfully transferred by conjugation to all recipient strains, but with varying frequencies, ranging from 1 × 10−6 to 4.1 × 10−4 transconjugants per donor (Fig. 2A). In general, strains belonging to ST73 acquired the plasmid less efficiently than other STs (Wilcoxon test, p < 0.001; Fig. 2A). This finding corroborates the potential of IncC plasmids to spread into different genetic backgrounds and play a significant role in the dissemination of antibiotic resistance22. However, the results indicate that there are differences in the conjugation frequencies dependent on the genetic background as also shown for other resistance plasmids18,26. Thingholm et al. also showed that the transfer frequency of a plasmid carrying blaCTX-M-15 to ST73 was lower than for other genetic backgrounds26.
Table 1.
E. coli strains used in the study20,25,36. K71-77 harbours plasmids pK71-77-1-NDM and pK71-77-2. All K56- strains are spontaneous rifR mutants of the isolate in25.
| Strain | Sequence type | Phylogroup |
|---|---|---|
| K71-77 | ST410 | A |
| K56-5 | ST998 | B2 |
| K56-17 | ST73 | B2 |
| K56-22 | ST73 | B2 |
| K56-23 | ST73 | B2 |
| K56-25 | ST73 | B2 |
| K56-29 | ST73 | B2 |
| K56-30 | ST1161 | B2 |
| K56-41 | ST73 | B2 |
| K56-43 | ST537 | B2 |
| K56-44 | ST12 | B2 |
| K56-46 | ST73 | B2 |
| K56-50 | ST100 | A |
| K56-51 | ST73 | B2 |
| K56-61 | ST80 | B2 |
| K56-63 | ST135 | B2 |
| K56-65 | ST10 | A |
| K56-66 | ST372 | B2 |
| K56-67 | ST141 | B2 |
| K56-68 | ST95 | B2 |
| K56-69 | ST1230 | A |
| K56-71 | ST607 | A |
| K56-75 | ST69 | D |
| K56-80 | ST141 | B2 |
Figure 2.
(A) Conjugation frequency of pK71-77-1-NDM from E. coli K71-77 into rifR K56 ECO-SENS strains. Vertical axis indicates the log10 of the conjugation frequency. (B) Relative fitness of rifR K56 ECO-SENS strains carrying pK71-77-1-NDM. Vertical axis indicates the fitness of plasmid-carrying strains relatively to their isogenic plasmid-free counterpart. (C) Stability of plasmid pK71-77-1-NDM in rifR K56 ECO-SENS strains. Vertical axis indicates the percentage of plasmid-carrying cells in the population. For all plots the horizontal axes indicate the rifR K56 strains’ suffix, and ST for non-ST73 strains; left panel: strains belonging to ST73, middle panel: strains belonging to other STs, right panel: overall comparison between ST73 and other STs. NS, ** and *** denote respectively: non-significant (p value > 0.05), p value < 0.01 and p value <0.001 (Wilcoxon test). The graphic illustrations were performed in R44.
Fitness effect and stability of pK71-77-1-NDM
To investigate if genetic background impacts the plasmid’s stability and fitness effect, we selected one transconjugant per strain and verified the presence of pK71-77-1-NDM by PCR (IncA/C2 replicon and blaNDM-1) and antibiotic resistance profile. Absence of the additional plasmid pK71-77-2 was confirmed by PCR for the IncFIA replicon. Host background was confirmed by RAPD-PCR-profiling and Sanger sequencing of the fumC allele.
We calculated exponential growth rates of the plasmid-free and plasmid-carrying strains by measuring the optical densities of monocultures as a function of time. The ratio between the growth rates of isogenic strains (differing only in the presence of pK71-77-1-NDM) served as a proxy to determine the fitness effect of plasmid carriage for each strain background. Plasmid pK71-77-1-NDM imposed detectable fitness costs varying between 2.5% to 22.7% in 10 of the 23 strains (T test, p < 0.05; Fig. 2B), mainly imposed on strains belonging to ST73 (Wilcoxon test, p = 0.002; Fig. 2B). Our data here indicate that the effect on fitness is strongly dependent on the recipient background, providing further support to previous reports18,27–29. The fitness cost of plasmids has been attributed to multiple factors14 and the mechanistic basis for this difference in plasmid cost is unclear. However, it is well known that acquisition of plasmids impacts the expression of chromosomal genes and consequently physiological processes that could cause a fitness effect19,30. One could therefore speculate that fitness differences upon plasmid acquisition could be due to variability in the interactions of plasmid-encoded proteins on the cellular networks in the recipients, which are likely to be distinct based on the diversity in genetic backgrounds. A detailed analysis would be required to elucidate the factors implicated in the variability of fitness effects between the different genetic backgrounds. The lack of initial fitness cost of pK71-77-1-NDM in more than half of the strains is worrying as this indicates potential for successful plasmid maintenance in different genetic backgrounds and in non-selective environments without the need for compensation. Others and we have shown that clinical plasmids frequently impose a limited fitness cost, including plasmids harbouring blaNDM18,31–33.
We further propagated the plasmid-carrying strains for ≈300 generations in the absence of selection to assess plasmid stability. Most strains maintained pK71-77-1-NDM stably in the population (Fig. 2C). For 14 strains, the plasmid was retained in more than 95% of the cells, while for three strains less than 10% of the cells carried pK71-77-1-NDM after 300 generations. We observed no difference in plasmid stability between strains belonging to ST73 and the other genetic backgrounds (Wilcoxon test, p > 0.05; Fig. 2C). Overall, this indicates that pK71-77-1-NDM can be stably maintained in variable genetic backgrounds under non-selective conditions further corroborating the epidemic potential of the plasmid. Like other IncC plasmids, pK71-77-1-NDM harbours a putative toxin-antitoxin system that could contribute to the maintenance of the plasmid23,34.
Frequency of plasmid-carriage
ST73 is considered a major pandemic E. coli clone frequently shown to be the dominating clone causing urinary tract or bloodstream infections9,35. In a collection of pan-susceptible E. coli, ST73 was found to be the clone most frequently devoid of plasmids25. Since we observed lower conjugative transfer rates and higher plasmid fitness costs in ST73 compared to the other genetic backgrounds (Fig. 2A and B) we performed a bioinformatical approach to compare ST73 with the other STs we had tested experimentally in terms of plasmid content. We downloaded all the 14830 E. coli assembled genomes available in December 2018 from the NCBI GenBank (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/Escherichia_coli/latest_assembly_versions/). 2182 of these genomes were unambiguously classified in STs employed in our experiments, 305 of which belonged to ST73 (Table S2). Then, we used PlasmidFinder to identify the plasmid content of these genomes. We found that most genomes, 1836 out of 2182, harboured plasmids. 69% of ST73 genomes harboured plasmids, but this proportion was significantly higher, 87%, for genomes belonging to other STs (χ2 test, d.f. = 1, p = 6.3 × 10−15; Table 2). Our bioinformatic analyses of plasmid content in ST73 versus other STs are consistent with the experimental conjugation and relative fitness data suggesting that ST73, for some plasmids, appears to be a sub-optimal host.
Table 2.
Comparison of plasmid presence/absence between ST73 and the other STs studied. *Pearson’s χ2 test with Yates’ continuity correction.
| No. (%) of genomes | ST73 | Other STs | p-value* | |
|---|---|---|---|---|
| Plasmids | Presence | 210 (69) | 1626 (87) | 6.3 × 10−15 |
| Absence | 95 (31) | 251 (13) | ||
Conclusion
We show here that the conjugation frequency and fitness impact of a multi-drug resistance plasmid carrying the carbapenemase gene blaNDM-1 is dependent on the recipient host background. Moreover the lack of, or relatively low fitness cost imposed by pK71-77-1-NDM in diverse genetic backgrounds combined with stable maintenance in a non-selective environment shows the epidemic potential of the plasmid. The plasmid’s lower conjugation frequency and higher fitness cost in ST73 could partially explain why ST73 is less associated with antibiotic resistance than other successful epidemic clones. Our study also shows the importance of investigating the evolution of antibiotic resistance in clinical strains with diverse genetic backgrounds. One limitation of the study is the representation of genetic backgrounds relative to the overall E. coli population. Further work is required to investigate if there are other E. coli lineages that show similar properties as ST73. Investigations into the mechanistic aspects of these observations will be important for possible interventions to limit the spread of antimicrobial resistance15,16.
Methods
Strain collection and whole genome sequencing
Bacterial strains used in the study and relevant characteristics are summarized in Table 1. E. coli K71-77 previously shown to carry blaNDM-120,36 was sequenced using PacBio sequencing (Pacific Biosciences, Menlo Park, CA). Genomic DNA was isolated from an overnight culture using the GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO) and library preparations were performed according to the Pacific Biosciences 20 kb protocol with a final size selection of 9 kb using BluePippin (Sage Sciences, Beverly, MA, USA). Sequencing was performed using the Pacific Biosciences RSII sequencer, P6-C4 chemistry with 360 minutes movie time and one single-molecule real-time (SMRT) cell (Pacific Biosciences, Menlo Park, CA). The sequences were subsequently assembled using HGAP v.3 (Pacific Biosciences, SMRT Analysis Software v2.3.0) with default settings at The Norwegian Sequencing Centre (https://www.sequencing.uio.no/). The Minimus2 software of the Amos package was used to circularize contigs and the RS_Resequencing.1 software (Pacific Biosciences, SMRT Analysis Software v2.3.0) for correction of bases after circularization. Annotation of the sequences was performed using the NCBI Prokaryotic Genome Annotation Pipeline37.
E. coli K71-77 was used as donor strain in conjugation assays for the transfer of the blaNDM-1-encoding plasmid pK71-77-1-NDM. A sample of 23 previously characterized plasmid-naïve antibiotic susceptible clinical uropathogenic E. coli from the ECO-SENS collection were employed as recipient strains25. To facilitate selection during conjugation experiments rifampicin resistant (rifR) mutants of the recipient strains were generated by plating 100 µL of overnight cultures onto Lysogeny Broth (LB) agar supplemented with 100 mg/L rifampicin.
Plasmid conjugative transfer
E. coli K71-77 served as donor while the rifampicin-resistant ECO-SENS strains served as recipients for the conjugative transfer of pK71-77-1-NDM. Single overnight LB cultures of donor and recipient were diluted 100-fold in 10 mL of LB and incubated at 37 °C with shaking until exponential phase was reached (OD600 = 0.6). Donor and recipient strains (1 mL each) were mixed in a 1:1 ratio and incubated at 37 °C for 3 h. Overnight cultures were diluted and plated on LB while mating cultures were plated on LB supplemented with rifampicin (150 μg/mL) and ampicillin (100 μg/mL). Conjugation frequency was calculated as . Experiments were performed with three biological replicates.
The presence of pK71-77-1-NDM in transconjugants was assessed by PCR for plasmid replicon and carbapenemase genes (IncA/C2 and blaNDM-1) and antibiotic resistance profile using the EUCAST disc diffusion method38. Transconjugants were further PCR-screened for the IncFIA replicon to confirm that pK71-77-2, the other plasmid present in the donor strain, was not transferred to the transconjugants. Transconjugant identity was verified by RAPD-PCR-profiling39 and Sanger sequencing of the fumC allele40. Primers are listed in Table S1. For each isolate, one transconjugant was frozen and used for further experiments.
Plasmid stability
One transconjugant of each isolate was propagated by serial transfer for ≈300 generations, in three biological replicates. Serial transfers were performed in 1 mL of LB by diluting the previous culture 100-fold every 12 h. Plasmid presence was analysed at ≈300 generations by patching 100 colonies obtained from LB agar onto selective LB agar (ampicillin 100 μg/mL). After ≈300 generations, five plasmid-carrying clones per replicate were verified by PCR for plasmid replicon and carbapenemase genes and antibiotic resistance profile as described above. Additionally, RAPD-PCR-profiling was performed for one transconjugant per replicate.
Fitness assays
Relative fitness of unevolved transconjugants was determined for each isolate by measuring relative growth rates. Growth rates for plasmid-free and plasmid-carrying rifampicin-resistant ECO-SENS strains were assessed by measuring the optical density of monocultures at 600 nm every 10 min for a period of 24 h, at 37 °C on a VERSAmax microplate reader (Molecular Devices, LLC). For this, 250 μL of a 1:100 diluted overnight culture in LB were added to 96-well microtiter plates. Experiments were performed in three biological and three technical replicates. Growth rates were determined using GrowthRates software version 3.041.
Growth rates for each biological replicate resulted from the average of the three technical replicates. Fitness was calculated as: . We measured three additional biological replicates for strains exhibiting fitness values with >0.05 standard deviation. Values outside the interval [Q1 − .5 · (Q3 − Q1), Q3 + 1.5 · (Q3 − Q1)], where Q1 and Q3 are the lower and upper quartiles, were considered outliers and removed from the analysis. Shapiro-Wilk test was performed to assess normality of data.
Analyses of genomes
We downloaded the 14830 E. coli assembled genomes available at ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/Escherichia_coli/latest_assembly_versions/ in December 2018. Multilocus sequence typing (MLST) was determined with MLST version 2.0 (default settings, species = “ecoli”)42 for all genomes. Genomes identified as belonging to ST10, ST12, ST69, ST73, ST80, ST95, ST100, ST135, ST141, ST372, ST537, ST607, ST998, ST1161 or ST1230 were selected for further analyses (Table S2). Genomes which contained alleles with less than 100% identity or 100% coverage were discarded.
We used PlasmidFinder version 2.0 (default settings, database = “Enterobacteriaceae”)43 to search for plasmids in the previously selected genomes. Statistical analyses and graphic illustrations were performed in R44.
Supplementary information
Acknowledgements
We thank Maria Chiara Di Luca for valuable and excellent technical assistance. This work was supported by the Northern Norway Regional Health Authority (Grant no. SFP1168-14) and a joint grant from Northern Norway Regional Health Authority/University Hospital North Norway and UiT The Arctic University of Norway (project A23270). The PacBio sequencing service was provided by the Norwegian Sequencing Centre (http://www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
Author contributions
Ø.S. and P.J.J. conceived the project; J.G. and J.K. performed experiments; J.G. and Ø.S. analysed the data; J.G., J.K. and Ø.S. wrote the initial manuscript; all authors reviewed the manuscript and approved the final version.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information File). PacBio sequences have been deposited in GenBank under BioProject PRJNA547487.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: João Alves Gama and Julia Kloos.
Supplementary information
is available for this paper at 10.1038/s41598-020-66239-8.
References
- 1.Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rozwandowicz M, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 4.Tangden T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J. Int. Med. 2015;277:501–512. doi: 10.1111/joim.12342. [DOI] [PubMed] [Google Scholar]
- 5.McNally A, et al. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. Plos Gen. 2016;12:e1006280. doi: 10.1371/journal.pgen.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNally A, et al. Diversification of colonization factors in a multidrug-resistant Escherichia coli lineage evolving under negative frequency-dependent selection. mBio. 2019;10:e00644–19. doi: 10.1128/mBio.00644-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu W, et al. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019;32:e00115–18. doi: 10.1128/CMR.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibreel TM, et al. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J. Antimicrob. Chemother. 2012;67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 9.Kallonen T, et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017;27:1437–1449. doi: 10.1101/gr.216606.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012;20:262–267. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Carroll AC, Wong A. Plasmid persistence: costs, benefits, and the plasmid paradox. Can. J, Microbiol. 2018;64:293–304. doi: 10.1139/cjm-2017-0609. [DOI] [PubMed] [Google Scholar]
- 12.Thomas, C.M. Plasmid incompatibility. In: Bell E. (eds) Molecular Life Sciences. Springer, New York, NY (2014).
- 13.Loftie-Eaton W, et al. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol. Biol. Evol. 2016;33:885–897. doi: 10.1093/molbev/msv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.San Millan, A. & MacLean, R.C. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5, MTBP-0016-2017 (2017). [DOI] [PMC free article] [PubMed]
- 15.San Millan A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 2018;26:978–985. doi: 10.1016/j.tim.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 16.MacLean RC, San Millan A. The evolution of antibiotic resistance. Science. 2019;365:1082–1083. doi: 10.1126/science.aax3879. [DOI] [PubMed] [Google Scholar]
- 17.Gottig S, Riedel-Christ S, Saleh A, Kempf VA, Hamprecht A. Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int. J. Antimicrob. Agents. 2016;47:430–435. doi: 10.1016/j.ijantimicag.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Di Luca MC, et al. Low biological cost of carbapenemase-encoding plasmids following transfer from Klebsiella pneumoniae to Escherichia coli. J. Antimicrob. Chemother. 2017;72:85–89. doi: 10.1093/jac/dkw350. [DOI] [PubMed] [Google Scholar]
- 19.Buckner MMC, et al. Clinically relevant plasmid-host interactions indicate that transcriptional and not genomic modifications ameliorate fitness costs of Klebsiella pneumoniae carbapenemase-carrying plasmids. mBio. 2018;9:e02303–17. doi: 10.1128/mBio.02303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuelsen Ø, et al. Identification of NDM-1-producing Enterobacteriaceae in Norway. J. Antimicrob. Chemother. 2011;66:670–672. doi: 10.1093/jac/dkq483. [DOI] [PubMed] [Google Scholar]
- 21.Roer L, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. 2018;3:e00337–18. doi: 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambrose SJ, Harmer CJ, Hall RM. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid. 2018;99:40–55. doi: 10.1016/j.plasmid.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Hancock SJ, et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob. Agents Chemother. 2017;61:e01740–16. doi: 10.1128/AAC.01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bengtsson S, Naseer U, Sundsfjord A, Kahlmeter G, Sundqvist M. Sequence types and plasmid carriage of uropathogenic Escherichia coli devoid of phenotypically detectable resistance. J. Antimicrob. Chemother. 2012;67:69–73. doi: 10.1093/jac/dkr421. [DOI] [PubMed] [Google Scholar]
- 26.Thingholm KR, Hertz FB, Lobner-Olesen A, Frimodt-Moller N, Nielsen KL. Escherichia coli belonging to ST131 rarely transfers blaCTX-M-15 to fecal Escherichia coli. Infect. Drug Resist. 2019;12:2429–2435. doi: 10.2147/IDR.S208536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starikova I, et al. Fitness costs of various mobile genetic elements in Enterococcus faecium and Enterococcus faecalis. J. Antimicrob. Chemother. 2013;68:2755–2765. doi: 10.1093/jac/dkt270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kottara A, Hall JP, Harrison E, Brockhurst MA. Multi-host environments select for host-generalist conjugative plasmids. BMC Evol. Biol. 2016;16:70. doi: 10.1186/s12862-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kottara, A., Hall, J.P.J., Harrison, E. & Brockhurst, M.A. Variable plasmid fitness effects and mobile genetic element dynamics across Pseudomonas species. FEMS Microbiol. Ecol. 94, pii: 468909 (2018). [DOI] [PMC free article] [PubMed]
- 30.San Millan A, et al. Integrative analysis of fitness and metabolic effects of plasmids in Pseudomonas aeruginosa PAO1. ISME J. 2018;12:3014–3024. doi: 10.1038/s41396-018-0224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandegren L, Linkevicius M, Lytsy B, Melhus A, Andersson DI. Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 2012;67:74–83. doi: 10.1093/jac/dkr405. [DOI] [PubMed] [Google Scholar]
- 32.Wang R, et al. The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and blaNDM with low fitness cost. Int. J. Antimicrob. Agents. 2018;51:739–744. doi: 10.1016/j.ijantimicag.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Brolund, A., et al. Dynamics of resistance plasmids in extended-spectrum-β-lactamase-producing Enterobacteriaceae during postinfection colonization. Antimicrob. Agents Chemother. 63, pii: AAC.02201-18 (2019). [DOI] [PMC free article] [PubMed]
- 34.Harmer CJ, Hall RM. The A to Z of A/C plasmids. Plasmid. 2015;80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014;20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 36.Samuelsen, Ø., et al. Molecular and epidemiological characterization of carbapenemase-producing Enterobacteriaceae in Norway, 2007 to 2014. Plos one12, e0187832 (2017). [DOI] [PMC free article] [PubMed]
- 37.Tatusova T, et al. NCBI prokaryotic genome annotation pipeline. Nucleic acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matuschek E, Brown DF, Kahlmeter G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014;20:O255–266. doi: 10.1111/1469-0691.12373. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen KL, et al. Selection of unique Escherichia coli clones by random amplified polymorphic DNA (RAPD): Evaluation by whole genome sequencing. J. Microbiol. Methods. 2014;103:101–103. doi: 10.1016/j.mimet.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirth T, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall BG, Acar H, Nandipati A, Barlow M. Growth rates made easy. Mol. Biol. Evol. 2014;31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 42.Larsen MV, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Core Team., R: A language environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, Available online at, https://www.r-project.org/ (2018).
- 45.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information File). PacBio sequences have been deposited in GenBank under BioProject PRJNA547487.