Abstract
The gut microbiota is emerging as a promising target for the management or prevention of inflammatory and metabolic disorders in humans. Many of the current research efforts are focused on the identification of specific microbial signatures, more particularly for those associated with obesity, type 2 diabetes, and cardiovascular diseases. Some studies have described that the gut microbiota of obese animals and humans exhibits a higher Firmicutes/Bacteroidetes ratio compared with normal-weight individuals, proposing this ratio as an eventual biomarker. Accordingly, the Firmicutes/Bacteroidetes ratio is frequently cited in the scientific literature as a hallmark of obesity. The aim of the present review was to discuss the validity of this potential marker, based on the great amount of contradictory results reported in the literature. Such discrepancies might be explained by the existence of interpretative bias generated by methodological differences in sample processing and DNA sequence analysis, or by the generally poor characterization of the recruited subjects and, more particularly, the lack of consideration of lifestyle-associated factors known to affect microbiota composition and/or diversity. For these reasons, it is currently difficult to associate the Firmicutes/Bacteroidetes ratio with a determined health status and more specifically to consider it as a hallmark of obesity.
Keywords: Microbiota, gut, obesity, dysbiosis, Firmicutes, Bacteroidetes, marker
1. Introduction
The gut microbiota is a complex community of microorganisms inhabiting the gastrointestinal tract that have established a close symbiotic relationship with their human host. It plays a crucial role in health maintenance, allowing the metabolism of indigestible dietary components and the synthesis of some vitamins, preventing pathogen colonization, and contributing to the maturation and education of the immune system [1]. The human gut microbiota is mostly composed by two dominant bacterial phyla, Firmicutes and Bacteroidetes that represent more than 90% of the total community, and by other subdominant phyla including Proteobacteria, Actinobacteria, and Verrucomicrobia [2]. This composition remains relatively unaffected by acute perturbations, as its plasticity allows it to rapidly return to its initial composition [3]. However, it is continuously exposed to various stress factors associated with modern lifestyles including, among others, the consumption of chlorinated water and that of food additives and contaminants such as heavy metals, pesticides, antibiotics, organic pollutants, and mycotoxins. These factors could chronically modify its composition (dysbiosis), selecting more virulent microorganisms leading to deleterious effects on the host health [3]. Gut dysbiosis is also associated with various pathologic conditions affecting the gastrointestinal tract (diarrhea, irritable bowel syndrome) [4], the immune system (allergy, multiple sclerosis, type 1 diabetes, inflammatory bowel diseases, rheumatoid arthritis) [4,5,6], the central nervous system (Alzheimer and Parkinson diseases, autism) [6,7], as well as the energy metabolism of the host (obesity, type 2 diabetes, atherosclerosis) [8], even if it is not yet clear whether these alterations are cause or consequence of these disorders. More particularly, the relationship between the two dominant phyla, expressed as the Firmicutes/Bacteroidetes ratio, has been associated with several pathological conditions. Accordingly, the gut microbiota is emerging as a promising target for the nutritional or therapeutic prevention and management of these diseases.
Characterizing the bacterial populations involved in the dysbiosis is therefore important, as this might be helpful to adopt alternative strategies for management of diseases. For example, the increase of potentially pathogenic species could be treated through targeted antimicrobial therapies while the disappearance of beneficial commensals could be addressed by the administration of specific probiotics such as Lactobacillus rhamnosus GG, L. reuteri DSM 17938, L. plantarum DSM 9843, and Bifidobacterium lactis Bb-12 [9].
The general purpose of this review is to discuss the relevance of the Firmicutes/Bacteroidetes ratio as marker of obesity. First, we will describe the evidence suggesting an association between the Firmicutes/Bacteroidetes ratio and the obesity, or rejecting this relationship. Next, we will expose the possible reasons for these contradictions, in particular differences in the methods of analysis of the microbiota, the control of the interfering factors (diet, antibiotics, etc.), and the possible biases in the recruitment process of the subjects. Ultimately, we re-analyzed the 16S rRNA gene sequence data from nine published studies to allow direct comparisons among their Firmicutes/Bacteroidetes ratio. Thus, we will show that obesity is associated with multiple taxonomic signatures, consistent with the high heterogeneity of the gut microbiome observed in the healthy population.
2. The Obesity and Its Relationship With an Increased Firmicutes/Bacteroidetes Ratio
Obesity is a complex, multifactorial, disease due to various factors including the host genetic background, decreased physical activity, and excess food intake. In the last few decades, the gut microbiota has been proposed as an additional factor favoring fat storage, weight gain, and insulin resistance [10]. Indeed, the gut microbiota is involved in energy homeostasis by extracting energy from foodstuffs through fermentation processes and formation of short chain fatty acid (SCFAs) [11,12]. It also increases villous vascularization, leading to improved nutrient absorption [13], and decreased AMPK levels and ß-oxidation in the muscular tissue. In addition, the microbiota modulates inhibits the release of fasting induced adipose factor (Fiaf), an inhibitor of lipoprotein lipase (LPL) activity, resulting in the subsequent storage of triglycerides in the adipose tissue and liver [13]. Finally, it influences the development of metabolic endotoxemia and low-grade inflammation [14]. Furthermore, the obese phenotype in mice was shown to be transmissible by transplanting the gut microbiota of conventional obese mice [12,15] to normal-weight germ-free animals.
Subsequently, the research efforts were focused on the identification of bacterial taxa involved in the development of obesity. Alterations affecting the dominant phyla Firmicutes and Bacteroidetes were first described in obese animals and subjects who exhibited increased abundances of Firmicutes at the expense of Bacteroidetes [16]. When these subjects were submitted to a calorie-restricted diet for one year, they showed an increase of their Bacteroidetes abundance and the normalization of their Firmicutes/Bacteroidetes ratio, in parallel with weight loss. These studies were supported by studies in animals fed high-fat or high-fiber diets showing higher Firmicutes and Bacteroidetes abundances, respectively [17,18]. Similar findings were reported in children living in rural African areas, who consumed a traditional diet rich in fiber and showed higher proportions of Bacteroidetes and lower of Firmicutes, compared to children from western countries whose diet included large amounts of protein, fat, sugar, and starch [19]. Based on these results and others obtained from obese animals and humans [13,20,21,22,23,24,25], it has been proposed that the Firmicutes were more effective in extracting energy from food than Bacteroidetes, thus promoting a more efficient absorption of calories and the subsequent weight gain [25]. This might be related to the observations of Turnbaugh et al. [21] in twins discordant for obesity; the microbiome of the obese twin was enriched in genes coding for nutrient transporters while that from the lean twin was enriched in genes coding for enzymes associated with carbohydrate metabolism [15]. These data suggest that the alterations in the bacterial composition/diversity are generally associated with changes in the metabolic profile of the microbiota that also influence host health. Accordingly, in the last decade, the Firmicutes/Bacteroidetes ratio has been frequently considered as a possible hallmark for obesity [26,27].
3. Controversies over the Altered Firmicutes/Bacteroidetes Ratio in Obesity
However, in opposition to these results, a number of studies did not observe any modifications of this parameter or even reported decreased Firmicutes/Bacteroidetes ratio in obese animals and humans [11,28,29,30,31,32]. The fact that, in most of the studies, the obese patients showed lesser bacterial diversity than the lean subjects, suggests the existence of other compositional changes at family, genus, or species level, which might be more relevant than the Firmicutes/Bacteroidetes ratio [33].
Regarding this point, the hypothesis of the metabolic endotoxemia proposes that the increased adiposity and development of systemic inflammation could be due to the chronic exposure to lipopolysaccharide (LPS), a pro-inflammatory molecule derived from Gram-negative bacteria, which would pass from the gut lumen into the bloodstream [14]. This hypothesis does not fit with the decreased abundance of Bacteroidetes reported in obesity, since this phylum is the main group of Gram-negative bacteria in the gut microbiota [28]. Such discrepancy could be explained by the fact that the endotoxic activity of the LPS from the bacteria belonging to the Bacteroidetes phylum is considered as lower than that from other Gram-negative bacteria like those belonging to the Proteobacteria phylum. Interestingly, increases of Proteobacteria have also been observed in obese subjects or animals, and the administration of Enterobacter, a member of the phylum Proteobacteria, in germ-free mice results in the development of obesity and insulin resistance in these animals [34,35,36].
On the other hand, the increased Firmicutes/Bacteroidetes ratio did not correlate with the production of SCFAs observed in obese individuals. Indeed, MacFarlane et al. reported that Bacteroidetes mainly produce acetate and propionate, whereas Firmicutes produce more butyrate [36]. Butyrate is considered a health-promoting molecule due to its capacity to [37] increase insulin sensitivity [38], exert anti-inflammatory activities [39], regulate energy metabolism, and increase leptin gene expression [40]. Propionate, in the colon, stimulates GLP-1 and PYY release by L-entero-endocrine cells, resulting in the inhibition of appetite [41]. It may also reach the portal circulation, being mainly captured by the liver where it participates in hepatic gluconeogenesis and reduces the expression of enzymes involved in the de novo synthesis of fatty acids and cholesterol [42]. Acetate is also absorbed and reaches the systemic circulation and peripheral organs including adipose tissue, muscle, and brain. In the liver, contrary to propionate, it stimulates the hepatic synthesis of lipids [43], contributing to dyslipidemia. In the brain, it activates the parasympathetic nervous system, promoting the secretion of insulin and ghrelin, by the pancreas and the gastric mucosa, respectively [44]. These events result in increased fat storage and appetite that contribute to obesity. Based on these results, acetate is generally considered as more obesogenic. The increased Firmicutes/Bacteroidetes ratio in obese individuals would mean a higher butyrate and lower propionate and acetate production in these subjects, a finding that is partially contradictory with the respective anti-obesogenic and obesogenic effects of these SCFAs. An explanation is that the butyrate-producing bacteria decrease in the obese individuals and are progressively replaced by other bacteria belonging to the same phylum, resulting in lower production of butyrate in the colonic lumen. For example, increased abundances of Staphylococcus spp. and Lactobacillus reuteri (both from the phylum Firmicutes) have been reported in obese people, and positively correlated with energy intake and plasma >C-reactive protein (CRP), respectively [23,45]. On the contrary, the decreased abundance of the butyrate-producing Faecalibacterium prausnitzii (Firmicutes phylum) correlated negatively with the intensity of low-grade inflammation in obese subjects and patients with type 2 diabetes patients [46,47]. Obesity status was also associated with lower abundance of A. muciniphila (Verrucomicrobia phylum), a mucin degrading bacterium contributing to the stabilization of the gut barrier function, the secretion of antibacterial peptides, and control of inflammation [48,49].
On the other hand, the heterogeneity of the results in humans, regarding the Firmicutes/Bacteroidetes ratio, could be due to the insufficient number of subjects included in most of the studies, making their statistical power insufficient to detect small variations. Trying to clarify this issue, several meta-analyses pooling the high-throughput sequencing amplicons obtained from different studies in obese patients have been performed [50,51,52]. With this aim, all the sequences were downloaded and processed using a unique pipeline. They were screened de novo to remove the chimera sequences and assigned to operational taxonomic units (OTUs). Accordingly, all eventual bias associated with methodological differences (described below) were prevented and data of individual studies were compared or grouped for overall analysis. After clustering the sequences according to the subject’s Body Mass Index (BMI), no differences in the abundance of Firmicutes and Bacteroidetes or the Firmicutes/Bacteroidetes ratio were detected between obese and normal-weight individuals. Only a small reduction of diversity was detected in the microbiota from the obese subjects. The authors projected that it was necessary to recruit approximately 160,000, 6300, 1600, and 700 individuals per group to detect a 1%, 5%, 10%, and 15% difference in the Firmicutes/Bacteroidetes ratio, respectively [50], i.e., a far greater than the number of subjects recruited in most of these studies. In summary, these data indicate that most studies lack the power to detect modest differences between healthy and obese subjects, suggesting that the Firmicutes/Bacteroidetes ratio is not a robust marker of microbiome dysbiosis associated with obesity.
4. Origin of Disparities in Obesity-Associated Gut Microbiome Research. The Methodological Discrepancies between Studies
The discrepant results previously reported may also be explained by differences in sample processing and data analysis including the method of DNA extraction, the selection of the amplified 16S rRNA region (choice of the primers), the sequencing method, and the bioinformatic analysis (taxonomy database and taxonomy assignment algorithm used) [51,53,54]. It is therefore challenging to eliminate the bias introduced by primer design, library preparation, DNA isolation methods, and PCR amplification artifacts, that can result in the over- or under-representation of individual taxa within complex communities [51,53,54]. In addition, sample storage can also influence the identification of bacterial communities. While refrigeration at 4 °C did not affect significantly the fecal microbial composition and its diversity, compared to control samples stored at −80 °C, the use of preservative buffers (RNAlater, OMNIgene.GUT, Tris-EDTA) seemed to alter the microbiota profile [55]. This observation is important since rapid freezing to −80 °C, commonly considered as the best practice, is not always practicable in studies involving sample collection at the domicile of the subjects [55]. Conversely, DNA extraction protocols based on the use of Phenol:Chloroform:Isoamyl alcohol are more efficient in extracting DNA from Gram-positive bacteria [53]. In another study, bacterial DNA from fecal samples obtained through the Human Microbiome Project was extracted using the same protocol and subsequently amplified with different primers targeting the V1-3/V2 or the V3-5/V4 regions of the rRNA. Compared to the V3-5/V4 region, the analysis of the V1-3/V2 region shows an enrichment in both Erysipelotrichi and Verrucomicrobia populations and a depletion of Actinobacteria and Gamma Proteobacteria [53]. Primer mismatches can represent another problem since they produce selective amplification and then prevent the correct assessment of the bacterial diversity [56]. Sequences not correctly matched by the primers are not correctly amplified, leading to a lesser representation of the corresponding microorganisms or even to their non-identification when the number of amplified sequences is below the detection limit [57]. Consequently, an evaluation of the bacterial coverage is necessary to correctly interpret the data obtained from the samples by next-generation sequencing (NGS) [57]. However, studies using the same set of primers have also reported contradictory results regarding gut microbiota composition [21,58,59], suggesting that primer-related bias is not the main reason for the discrepancies observed among studies [51].
Another important factor is the platform used for sequencing the 16S rRNA amplicons. Several platforms (Ion Torrent PGM, Illumina MiSeq, Illumina HiSeq, and Roche GS FLX+) are currently available, which use different sequencing chemistry and can also introduce internal biases, for example by influencing the detection and abundance of microorganisms with low or high genomic GC content [60]. In addition, an eventual impact of the adapters and barcodes added to the sequencing primers, that are specific for each platform, cannot be discarded [60].
To determine how the sequencing platforms can affect the results of microbiota composition, chicken cecum microbiota was analyzed through three different sequencing platforms (Illumina MiSeq, Ion Torrent PGM, and Roche 454 GS FLX) using the same set of primers (8F and 338R) [60]. The authors observed differences in the relative abundance of specific genera according to the platform used, confirming previous studies [61,62,63]. Despite the phylogenic differences induced by the different performance of the three sequencing platforms, the authors observed that the sample discrimination according to the treatment persisted, whatever the platform used, suggesting that the biological conclusions remained valid [60]. Several pipelines are currently available for removing the chimeric sequences generated by the sequencing platforms. However, they can provide discrepant results. For example, in a study comparing three analysis pipelines (QIIME, Mothur, and MG-RAST) to characterize the microbiota from preterm infants by NGS sequencing [64], the authors observed significant differences in the effective number of detected genera according to the program used. In addition, differences were also observed in the proportion of some phyla. Proteobacteria was detected in lower abundance by MG-RAST while Mothur detected Actinobacteria at a slightly higher abundance [64]. Therefore, the study of complex microbial communities requires the selection of the most accurate and appropriate methodological approaches to generate and analyze sequencing of 16S rRNA gene. These biases are unavoidable and can impact the overall results of a study.
Technological aspects, therefore, can strongly impact the identification of bacterial taxa, overshadowing biological differences in the samples, especially when the sample size is small [51]. This problem is illustrated by the results of a meta-analysis using amplicons from high-throughput sequencing studies in obese subjects and showing that the gut microbiota composition clustered by study rather than by subject’s BMI, suggesting that the per-study effect was greater than the biological effect [51].
5. Origin of Disparities in Obesity-Associated Gut Microbiome Research. The Selection of Subjects Included in Studies
Comparisons of the results between studies are generally complicated by the inadequate characterization of the studied populations and the fact that many lifestyle-associated factors, not considered in the studies, contribute towards shaping the gut microbiota composition. Therefore, the discrepant results observed between studies evaluating the Firmicutes/Bacteroidetes ratio in obese and lean subjects could be explained by the fact that these co-variables have not been adequately considered. Table 1 describes the different parameters related to the subject that were considered in the main studies from the last decade. It can be observed that some factors such as the intake of antibiotics or dietary factors such as probiotics, prebiotics, and symbiotics are not reported, and that the qualitative and quantitative description of the diet as well as the intensity of physical activity are largely uncontrolled. Information about the intake of macronutrients, micronutrients, and non-nutrients such as dietary fibers and phytochemicals is important, considering that part of them may reach the colon and impact the microbiota. In addition, it is not clearly established whether the changes observed in the gut microbiota of the obese subjects are due to variations in their dietary intake or to obesity per se [65]. This phenomenon is also well described for non-digestible carbohydrates and also for the dietary proteins that may reach the colon and influence bacterial composition and microbial production of SCFAs and other potentially toxic, metabolites (H2S, p-cresol, phenol, NH3, etc.). Non-absorbed dietary iron also (including iron supplements) affects the microbiota, as well as dietary phenolic compounds that may act as prebiotics. The impact of dietary lipids is poorly known; animals fed diets with high fat (supra-physiologic) contents exhibit gut dysbiosis but it is unclear whether this also occurs in humans [66]. Interestingly, Hildebrandt et al. showed that rats fed a high-fat diet displayed gut dysbiosis, independently of their obese phenotype [18]. Food additives such as low-caloric sweeteners and emulsifiers have been shown to alter microbiota, increasing its potential virulence and favoring metabolic disorders in animals and humans [67,68]. Finally, a number of pollutants including heavy metals, persistent organic pollutants, and pesticides, which are currently increasing in the human environment, also alter the gut microbial ecosystem [69]. Accordingly, the identification and characterization of these confounding factors is, therefore, particularly important when comparing lean and obese populations, which clearly differ by their lifestyles. Not taking them into account might generate bias and lead to result misinterpretations.
Table 1.
Consideration of factors affecting the microbiota and that should be controlled, in studies analyzing microbiota in obese and normal-weight adults. These factors include the intake of antibiotics and that of prebiotics/probiotics before/during the study, the characterization of the diet consumed by the subjects, the intensity of their physical activity, and the presence of interfering pathologies that appear as exclusion criteria. Only studies analyzing the microbiota through molecular methods (qPCR or sequencing) were selected. Results corresponding to the relative abundance of Firmicutes and Bacteroidetes and the Firmicutes/Bacteroidetes ratio are shown when available.
| Population | Anthropometric and Biochemical Factors | Factors Influencing the Gut Microbiota | Gut Microbiota Analysis (Ob vs. Nw) | Ref | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | Recruited Subjects/Sample Size | Age (Years) 1 | BMI (kg/m2) 1 | Biochemical Parameters | Antibiotic 2 | Diet | Intake of Pre-/Probiotic | Physical Activity | Exclusion Factors | Method | F 3 | B 3 | F/B 4 | |
| Brazil |
|
|
|
FPG, FPI, TC, HDL LDL, TG | ≤3 months | Yes | ND | ND | Diagnosed diseases, Pregnancy, Lactation | qPCR | ND | ND | ND | [70] |
| Canada |
|
|
|
ND | Last 5 years | Yes | ND | ND | All drugs influencing gastrointestinal functions, Inflammatory bowel diseases, Malabsorption, Gastrointestinal infection, Short bowel syndrome, Illness or surgery requiring hospitalization | Sequencing | ↑ | = | ↑ | [71] |
| China |
|
|
|
ND | ≤2 weeks | ND | ND | ND | Diabetes, Diarrhea, Constipation, Long-term medication | Sequencing | = | = | ND | [72] |
| Germany |
|
|
|
ND | ≤6 months | ND | ND | ND | ND | qPCR | ↓ | ↑ | ↓ | [30] |
| France |
|
|
|
ND | ≤1 month | ND | ND | ND | Gastric bypass, Colon cancer, Inflammatory bowel diseases, Diarrhea | qPCR | = | = | ND | [73] |
| France |
|
|
|
ND | ND | ND | Yes (only Probiotic) | ND | ND | qPCR | = | ↓ | ↑ | [24] |
| India |
|
|
|
ND | ≤3 months | ND | Yes | ND | ND | Sequencing | = | = | = | [31] |
| Japan |
|
|
|
TC, TG, HDL, ALT, AST, HbA1c | Yes | ND | ND | Yes | Chronic bowel or liver diseases, Colorectal cancer, Chemotherapy or radiotherapy, Immunosuppressants | T-RFLP and Sequencing | = | ↑ | ↓ | [74] |
| Thailand |
|
|
|
FPG, Lipids, TG, TC, HDL, LDL | ≤2 weeks | ND | ND | ND | Chronic inflammatory diseases, Diarrhea | qPCR | ↓ | ↓ | ND | [75] |
| UK |
|
|
|
ND | During the course of the study | ND | ND | ND | Metabolic syndrome Gastrointestinal problems | FISH | ND | = | ND | [29] |
| Ukraine |
|
|
|
ND | ND | ND | ND | Yes | Oncology diseases, Endocrinology diseases, Anorexia, Psychiatric disorders, Chronic diseases | qPCR | ↑ | ↓ | ↑ | [76] |
| USA |
|
|
|
ND | ≤3 months | ND | Yes | ND | ND | Sequencing | ND | = | ND | [28] |
| USA |
|
|
|
Glc, FPG, Ins, FPI | ≤3 months | Yes | Yes (only Probiotic) | ND | Smokers, Gastrointestinal diseases, Antacids and laxatives, Transit time | Sequencing | = | = | = | [11] |
| USA |
|
|
|
ND | ≤6 months | Yes | ND | ND | Detailed medical and lifestyle questionnaire (not detailed) | Sequencing | = | ↓ | ND | [21] |
| USA |
|
|
|
ND | ND | ND | ND | ND | ND | Sequencing | ↑ | ↓ | ↑ | [16] |
| USA |
|
|
|
ND | Yes | Yes | Yes | ND | ND | Sequencing | ND | ND | ND | [77] |
1 Values expressed as mean ± SEM or [range]; 2 Time prior to the study; 3 Relative abundance of Firmicutes (F) and Bacteroidetes (B); Proportion; 4 Firmicutes/Bacteroidetes ratio; * BMI-for-age percentile; ** ranges; SEM. standard error of the mean; BMI. body mass index; Ob. obese; Ow. overweight; Nw. Normal-weight; ND. not-determined; Sequencing. 16S rDNA pyrosequencing; T-RFLP. Terminal restriction fragment length polymorphism; FISH. Fluorescence in situ hybridization; ↑. Significantly increased; ↓. Significantly decreased; =. Not significantly different; GLC. glucose; FPG. Fasting plasma glucose; INS. Insulin; FPI. Fasting plasma insulin; FC. Fecal calprotectin; ALT. Alanine aminotransferase; AST. Aspartate aminotransferase; HDL. high-density lipoprotein cholesterol; LDL. Low-density lipoprotein; Cholesterol; TG. Triglycerides; TC. Total cholesterol; HbA1c. glycated haemoglobin (A1c).
Physical activity is another important confounding variable; it determines intestinal motility and the transit time of the chime through the different segments of the gastrointestinal tract, which, in turn, affects the composition of the microbiota [78]. Physical activity has been correlated with the Firmicutes/Bacteroidetes ratio in animals [79] and humans, independently of the diet [80]. In addition, the frequency and intensity of physical activity also determine the proportion of fat and lean masses, eventually making the BMI stratification irrelevant. Thus, a sedentary subject may be normal-weight (i.e., with a BMI < 24.9) but have an unbalanced fat mass while an athlete may have a BMI corresponding to that of an overweight or obese subject, but with low fat mass and high lean mass [81]. The impact of these individual differences on the microbiota composition remains unknown. It might be therefore useful to consider parameters other than BMI to determine whether the individual is healthy or not. In fact, the richness of human gut microbiota was shown to be inversely correlated with metabolic markers such as overall adiposity, insulin resistance, dyslipidemia, and low-grade inflammation [59,82]. Interestingly, the classification of lean and obese subjects according to their cardiometabolic health status showed better dissimilarities in the microbial community [83].
Within a determined range of BMI, the marker bacterial taxa associated with obesity and cardiometabolic disorders were exacerbated in unhealthy individuals [83]. As previously stated, such parameters can be altered in lean subjects while they may be in the normal range for subjects with high BMIs, limiting the interpretation of the observed changes in the composition of the microbiota. Therefore, the individuals recruited in the studies need to be better characterized to control the confounding factors and avoid erroneous association between obesity and microbiota.
6. The Obesity Might be Associated with Multiple Taxonomic Signatures?
The pattern of the gut microbiota associated with obesity is not homogeneous throughout the world and differs according to the geographical location of the studies [84,85,86,87]. Microbes in the gastrointestinal tract are under selective pressure largely due to lifestyles that change according to the socioeconomic status prevailing in the different continents and countries. These geographic differences include the access to specific food materials, differences in the genetic background of the inhabitants including the presence of single nucleotide polymorphisms, and differences in host energy requirements due to the climatic conditions [19,88,89]. For example, the Inuits live in a cold climate and mostly have access to a diet rich in animal proteins and fats which, in turn, determines a specific microbiota composition by selecting bacteria capable of metabolizing these macronutrients, independent of their BMI [90]. A meta-analysis revealed that the Firmicutes/Bacteroidetes ratio was higher in the populations living at high latitudes, suggesting that extraction of energy from food by the gut microbiota could be greater in these regions [88]. This finding is supported by a study carried out in mice maintained in a cold environment that also showed a higher Firmicutes/Bacteroidetes ratio in these animals, similarly to that observed in obese or animals fed a high-fat diet [91]. Notably, this study shows that the gut microbiota contributes to the adaptation to cold exposure through its ability to harvest energy.
7. The Heterogeneity of the Gut Microbiome in the Populations
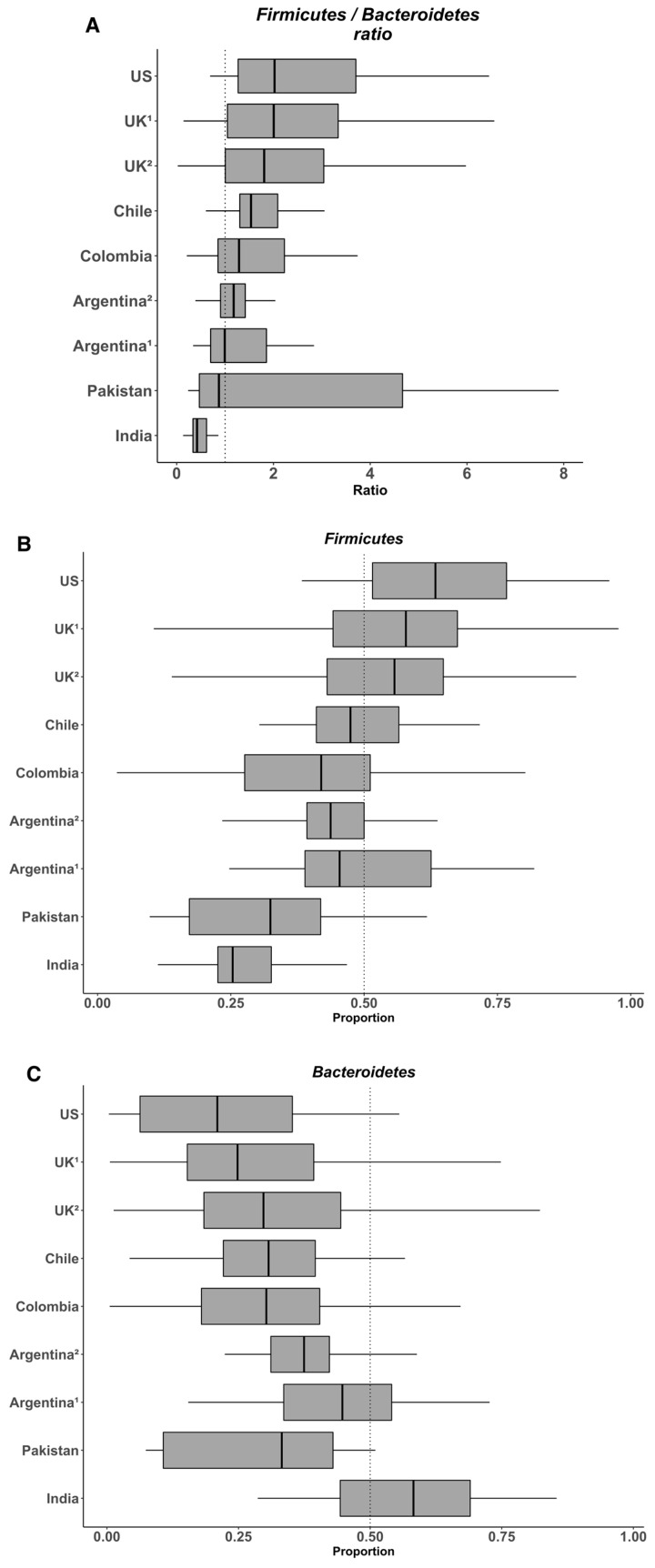
To assess the relevance of the Firmicutes/Bacteroidetes ratio as a taxonomic signature of obesity, we used data from microbiota composition from nine published studies carried out in seven countries (USA, United Kingdom, India, Pakistan, Chile, Argentina, and Colombia) and including 728 healthy subjects. High-throughput 16S rRNA gene sequence data from previous studies, corresponding to V3–V4 or V4 hypervariable regions and generated by the Illumina MiSeq Platform, were collected (Table 2). To allow direct comparisons among sequences from different studies, all the reads were filtered using the DADA2 pipeline, then aligned and trimmed to the same length (80 bp) using Mothur, followed by the taxonomic identification using the DADA2 pipeline based on the identification of Exact Sequence Variants [92]. We considered that a sequence length of 80 bp was adequate for analyzing the microbial communities at the Phylum level (less than 0.02% of the reads were not assigned). Using a unique pipeline and reads generated by a same sequencing platform, we expected to eliminate all bias generated by sequencing and bioinformatic tools, as commented above. Subsequently, we analyzed the relative abundance of Firmicutes and Bacteroidetes and their ratio. Overall, the data indicate that the abundance of Firmicutes in the gut microbiota of healthy individuals varies between 11% to 95% and that of Bacteroidetes between 0.6% to 86.6% (Figure 1). Considering this variability, it seems difficult to observe significant changes for these two phyla in obese people. In their meta-analysis, Finucane et al. observed that the variations for both Firmicutes and Bacteroidetes abundance were much larger among studies than between lean and obese individuals within any study [52]. In agreement with these findings, we recently observed a high variability of both Firmicutes and Bacteroidetes, between 25%–67% and 4%–64% respectively, in the fecal microbiota of young healthy Chilean volunteers, despite rigorous inclusion criteria including the control of anthropometrical and biochemical markers, biomarkers of systemic and colonic inflammation (plasma IL-6 and high sensitivity C-reactive protein and fecal calprotectin, respectively), and dietary intake [93]. Again, the heterogeneity of the diet is probably the main factor explaining such variations in the healthy population and this might eventually make difficult the identification of specific microbial signatures. For example, on the one hand, Wu et al. showed in 100 healthy individuals with known dietary habits that the microbiota from those consuming protein and fat-based diets were enriched with Bacteroides whereas that from those consuming carbohydrate-based diets were enriched with Prevotella [59], results from which these authors formulated the concept of “enterotype”. On the other hand, Balamurugan et al. compared the gut microbiota from a tribal population (Malayalis) living in the northern part of Tamil Nadu (India) that consumed a restricted diet due to cultural and religious beliefs, with that from healthy villagers from the same region as controls. Both populations exhibited a high abundance of Firmicutes (85.9% and 63.5%, for the Malaiyalis and controls, respectively) and low abundance of Bacteroidetes (2.65% and 0.45%, respectively), resulting in very high Firmicutes/Bacteroidetes ratio (34.0 and 92.9, respectively), though the individuals from both populations were lean [94]. Although the Malaiyalis population had a restricted, homogenous diet, a high variation in the proportions of both Firmicutes and Bacteroidetes was observed, confirming that factors other than diet influence this ratio.
Table 2.
Description of studies considered in this study for evaluating the variability of the Firmicutes and Bacteroidetes.
| Country | Accession Number | Effective & | Age (Years) | BMI (kg/m2) | Sequencing Platform | HypervariaBle Region | Ref |
|---|---|---|---|---|---|---|---|
| USA | PRJNA290926 | 68 | 53.1 ± 10.8 | 22.0 ± 1.9 | MiSeq Illumina | V4 region | [95] |
| UK 1 | PRJEB6702 | 230 | 61.2 ± 10.1 | 22.4 ± 1.8 | MiSeq Illumina | V4 region | [96] |
| UK 2 | PRJEB6705 | 189 | 60.0 ± 9.5 | 22.3 ± 1.8 | MiSeq Illumina | V4 region | [96] |
| Pakistan | PRJNA554535 | 20 | 37.7 ± 12.1 | 22.08 ± 3.1 | MiSeq Illumina | V3–V4 region | [97] |
| India | PRJEB28290 | 80 | Range 18–55 * | 23.9 ± 3.2 * | MiSeq Illumina | V3–V4 region | [98] |
| Colombia | PRJEB33360 | 83 | 52.1 ± 18.6 | 25.1 ± 3.9 | MiSeq Illumina | V3–V4 region | [99] |
| Chile | PRJEB16755 | 32 | 25.0 ± 3.9 | 22.5 ± 1.6 | MiSeq Illumina | V3–V4 region | [93] |
| Argentina 1 | PRJNA503303 | 28 | 35.2 ± 8.3 * | 23.9 ± 3.4 * | MiSeq Illumina | V3–V4 region | [100] |
| Argentina 2 | Personal data ** | 28 | 40.2 ± 4.4 | 22.6 ± 2.0 | MiSeq Illumina | V4 region | This study |
& Effective obtained after the bioinformatic processing; * Data obtained from publishing data (not-recalculated due to the lack of individual data); ** Data submitted for publication, provided by Susan Pesoa, co-author of this work; UK 1, UK 2, Argentina 1 and Argentina 2 are studies reported in the Figure 1.
Figure 1.
Variability in the Firmicutes/Bacteroidetes ratio (A) and the relative abundances of Firmicutes (B) and Bacteroidetes (C) in the gut microbiota from several healthy populations. Box plots were constructed using R. In the box and whisker plots, the line shows the median; the box, the interquartile range; and the whiskers, the highest and lowest values.
8. Conclusions
In summary, the relative abundance of the Firmicutes and Bacteroidetes phyla is highly variable between subjects from a same population. This is probably due to many lifestyle-associated factors including diet, physical activity, food additives and contaminants, antibiotic consumption, physical activity, among others that influence the composition of the microbiota in the gastrointestinal tract. This could explain the contradictory results observed when comparing the microbiota between normal-weight and obese subjects, making it difficult to associate the Firmicutes/Bacteroidetes ratio with a determined health status. Though the gut microbiota could contribute to the development of obesity, the evidence suggesting an association between obesity and alterations of the Firmicutes/Bacteroidetes ratio is not convincing. In future studies, it is therefore necessary to improve the characterization of the subjects and to clearly identify the co-variables, which may affect microbiota composition and interfere with the interpretation of the results. In addition, the concept of a unique taxonomic signature associated with obesity appears compromised. Rather than investigate taxonomical marker of the obesity per se, obesity-associated gut microbiome studies should focus to identify taxonomic markers for stratifying patients into subgroups. Introducing microbiome patient stratification would improve the management of the obesity by personalizing treatment decisions through direct manipulation of patient microbiomes.
Acknowledgments
We thank Anne Lagomarcino for revising the English language of the manuscript.
Author Contributions
F.M. and M.G. wrote the manuscript. L.G. searched the studies and downloaded the sequencing data used for the evaluation of the Firmicutes/Bacteroidetes ratio. A.Z. and F.M. analyzed the sequencing data and edited the figures. S.P. provided sequencing data of the Argentina population. S.P., P.N., and R.B. reviewed the manuscript. All authors read and approved the final version of the manuscript.
Funding
This review was supported by the grant number 1191311 from the Chilean National Fund for Scientific and Technological Development program (FONDECYT).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar Reddy D. Role of the normal gut microbiota. World J. Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Candela M., Biagi E., Maccaferri S., Turroni S., Brigidi P. Intestinal microbiota is a plastic factor. Responding to environmental changes. Trends Microbiol. 2012;20:385–391. doi: 10.1016/j.tim.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Carding S., Verbeke K., Vipond D.T., Corfe B.M., Owen L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hills R.D., Pontefract B.A., Mishcon H.R., Black C.A., Sutton S.C., Theberge C.R. Gut Microbiome. Profound Implications for Diet and Disease. Nutrients. 2019;11:1613. doi: 10.3390/nu11071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bravo J.A., Julio-Pieper M., Forsythe P., Kunze W., Dinan T.G., Bienenstock J., Cryan J.F. Communication between gastrointestinal bacteria and the nervous system. Curr. Opin. Pharmacol. 2012;12:667–672. doi: 10.1016/j.coph.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Belkaid Y., Hand T. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157 doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yong V.B. The Role of the Microbiome in Human Health and Disease. An Introduction for Clinicians. BMJ. 2017;356 doi: 10.1136/bmj.j831. [DOI] [PubMed] [Google Scholar]
- 9.Mantegazza C., Molinari P., D’Auria E., Sonnino M., Morelli L., Zuccotti G.V. Probiotics and antibiotic-associated diarrhea in children. A review and new evidence on Lactobacillus rhamnosus GG during and after antibiotic treatment. Pharmacol. Res. 2018;128:63–72. doi: 10.1016/j.phrs.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Cardinelli C.S., Sala P.C., Alves C.C., Torrinhas R.S., Waitzberg D.L. Influence of Intestinal Microbiota on Body Weight Gain. A Narrative Review of the Literature. Obes. Surg. 2015;25:346–353. doi: 10.1007/s11695-014-1525-2. [DOI] [PubMed] [Google Scholar]
- 11.Jumpertz R., Le D.S., Turnbaugh P.J., Trinidad C., Bogardus C., Gordon J.I., Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. Anobesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 13.Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., Neyrinck A.M., Fava F., Tuohy K.M., Chabo C., et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 15.Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S., Sitaraman S.V., Knight R., Ley R.E., Gewirtz A.T. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 17.de Wit N., Derrien M., Bosch-Vermeulen H., Oosterink E., Keshtkar S., Duval C., de Vogel-van den Bosch J., Kleerebezem M., Müller M., van der Meer R. Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am. J. Physiol. Liver Physiol. 2012;303:G589–G599. doi: 10.1152/ajpgi.00488.2011. [DOI] [PubMed] [Google Scholar]
- 18.Hildebrandt M.A., Hoffmann C., Sherrill-Mix S.A., Keilbaugh S.A., Hamady M., Chen Y.-Y., Knight R., Ahima R.S., Bushman F., Wu G.D. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J.B., Massart S., Collini S., Pieraccini G., Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu P., Li M., Zhang J., Zhang T. Correlation of intestinal microbiota with overweight and obesity in Kazakh school children. BMC Microbiol. 2012;12:283. doi: 10.1186/1471-2180-12-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bervoets L., Van Hoorenbeeck K., Kortleven I., Van Noten C., Hens N., Vael C., Goossens H., Desager K.N., Vankerckhoven V. Differences in gut microbiota composition between obese and lean children. A cross-sectional study. Gut Pathog. 2013;5:10. doi: 10.1186/1757-4749-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armougom F., Henry M., Vialettes B., Raccah D., Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS ONE. 2009;4:e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krajmalnik-Brown R., Ilhan Z.-E., Kang D.-W., DiBaise J.K. Effects of Gut Microbes on Nutrient Absorption and Energy Regulation. Nutr. Clin. Pract. 2012;27:201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Bandt J.P., Waligora-Dupriet A.J., Butel M.J. Intestinal Microbiota in Inflammation and Insulin Resistance. Relevance to Humans. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14 doi: 10.1097/MCO.0b013e328347924a. [DOI] [PubMed] [Google Scholar]
- 27.Zou Y., Ju X., Chen W., Yuan J., Wang Z., Aluko R.E., He R. Rice Bran Attenuated Obesity via Alleviating Dyslipidemia, Browning of White Adipocytes and Modulating Gut Microbiota in High-Fat Diet-Induced Obese Mice. Food Funct. 2020;11 doi: 10.1039/C9FO01524H. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H., DiBaise J.K., Zuccolo A., Kudrna D., Braidotti M., Yu Y., Parameswaran P., Crowell M.D., Wing R., Rittmann B.E., et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duncan S.H., Lobley G.E., Holtrop G., Ince J., Johnstone A.M., Louis P., Flint H.J. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. (Lond.) 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- 30.Schwiertz A., Taras D., Schäfer K., Beijer S., Bos N.A., Donus C., Hardt P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 31.Patil D.P., Dhotre D.P., Chavan S.G., Sultan A., Jain D.S., Lanjekar V.B., Gangawani J., Shah P.S., Todkar J.S., Shah S., et al. Molecular analysis of gut microbiota in obesity among Indian individuals. J. Biosci. 2012;37:647–657. doi: 10.1007/s12038-012-9244-0. [DOI] [PubMed] [Google Scholar]
- 32.Tims S., Derom C., Jonkers D.M., Vlietinck R., Saris W.H., Kleerebezem M., de Vos W.M., Zoetendal E.G. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7:707–717. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguirre M., Venema K. Does the Gut Microbiota Contribute to Obesity? Going beyond the Gut Feeling. Microorganisms. 2015;3:213–235. doi: 10.3390/microorganisms3020213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson C.L.J., Onnerfält J., Xu J., Molin G., Ahrné S., Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 2012;20:2257–2261. doi: 10.1038/oby.2012.110. [DOI] [PubMed] [Google Scholar]
- 35.Rizzatti G., Lopetuso L.R., Gibiino G., Binda C., Gasbarrini A. Proteobacteria. A Common Factor in Human Diseases. Biomed. Res. Int. 2017;2017:1–7. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fei N., Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Besten G., Den Eunen K., Van Groen A.K., Venema K., Reijngoud D., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid. Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Z., Yin J., Zhang J., Ward R.E., Martin R.J., Lefevre M., Cefalu W.T., Ye J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Säemann M.D., Böhmig G.A., Österreicher C.H., Burtscher H., Parolini O., Diakos C., Stöckl J., Hörl W.H., Zlabinger G.J. Anti-inflammatory effects of sodium butyrate on human monocytes. Potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14:2380–2382. doi: 10.1096/fj.00-0359fje. [DOI] [PubMed] [Google Scholar]
- 40.Soliman M.M., Ahmed M.M., Salah-Eldin A.-E., Abdel-Aal A.A.-A. Butyrate regulates leptin expression through different signaling pathways in adipocytes. J. Vet. Sci. 2011;12:319–323. doi: 10.4142/jvs.2011.12.4.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers E.S., Viardot A., Psichas A., Morrison D.J., Murphy K.G., Zac-Varghese S.E.K., MacDougall K., Preston T., Tedford C., Finlayson G.S., et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64:1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demigné C., Morand C., Levrat M.A., Besson C., Moundras C., Rémésy C. Effect of propionate on fatty acid and cholesterol synthesis and on acetate metabolism in isolated rat hepatocytes. Br. J. Nutr. 1995;74:209–219. doi: 10.1079/BJN19950124. [DOI] [PubMed] [Google Scholar]
- 43.Gao X., Lin S.-H., Ren F., Li J.-T., Chen J.-J., Yao C.-B., Yang H.-B., Jiang S.-X., Yan G.-Q., Wang D., et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun. 2016;7:11960. doi: 10.1038/ncomms11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry R.J., Peng L., Barry N.A., Cline G.W., Zhang D., Cardone R.L., Petersen K.F., Kibbey R.G., Goodman A.L., Shulman G.I. Acetate Mediates a Microbiome-Brain-β-Cell Axis to Promote Metabolic Syndrome. Nature. 2016;534 doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Million M., Angelakis E., Paul M., Armougom F., Leibovici L., Raoult D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb. Pathog. 2012;53:100–108. doi: 10.1016/j.micpath.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Balamurugan R., George G., Kabeerdoss J., Hepsiba J., Chandragunasekaran A.M.S., Ramakrishna B.S. Quantitative differences in intestinal Faecalibacterium prausnitzii in obese Indian children. Br. J. Nutr. 2010;103:335–338. doi: 10.1017/S0007114509992182. [DOI] [PubMed] [Google Scholar]
- 47.Furet J.-P., Kong L.-C., Tap J., Poitou C., Basdevant A., Bouillot J.-L., Mariat D., Corthier G., Dore J., Henegar C., et al. Differential Adaptation of Human Gut Microbiota to Bariatric Surgery-Induced Weight Loss. Links With Metabolic and Low-Grade Inflammation Markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dao M.C., Everard A., Aron-Wisnewsky J., Sokolovska N., Prifti E., Verger E.O., Kayser B.D., Levenez F., Chilloux J., Hoyles L., et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity. Relationship with gut microbiome richness and ecology. Gut. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 49.Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., Guiot Y., Derrien M., Muccioli G.G., Delzenne N.M., et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sze M.A., Schloss P.D. Looking for a Signal in the Noise. Revisiting Obesity and the Microbiome. MBio. 2016;7:e01018-16. doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walters W.A., Xu Z., Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finucane M.M., Sharpton T.J., Laurent T.J., Pollard K.S., Lim S., Vos T., Flaxman A., Danaei G., Shibuya K., Douketis J., et al. A Taxonomic Signature of Obesity in the Microbiome? Getting to the Guts of the Matter. PLoS ONE. 2014;9:e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozupone C.A., Stombaugh J., Gonzalez A., Ackermann G., Wendel D., Vazquez-Baeza Y., Jansson J.K., Gordon J.I., Knight R. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z., DeSantis T.Z., Andersen G.L., Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008;36:e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choo J.M., Leong L.E., Rogers G.B. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015;5:16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polz M.F., Cavanaugh C.M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 1998;64:3724–3730. doi: 10.1128/AEM.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao D.-P., Zhou Q., Chen C.-Y., Quan Z.-X. Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol. 2012;12:66. doi: 10.1186/1471-2180-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zupancic M.L., Cantarel B.L., Liu Z., Drabek E.F., Ryan K.A., Cirimotich S., Jones C., Knight R., Walters W.A., Knights D., et al. Analysis of the Gut Microbiota in the Old Order Amish and Its Relation to the Metabolic Syndrome. PLoS ONE. 2012;7:e43052. doi: 10.1371/journal.pone.0043052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.-Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allali I., Arnold J.W., Roach J., Cadenas M.B., Butz N., Hassan H.M., Koci M., Ballou A., Mendoza M., Ali R., et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017;17:194. doi: 10.1186/s12866-017-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kennedy K., Hall M.W., Lynch M.D.J., Moreno-Hagelsieb G., Neufeld J.D. Evaluating Bias of Illumina-Based Bacterial 16S rRNA Gene Profiles. Appl. Environ. Microbiol. 2014;80:5717–5722. doi: 10.1128/AEM.01451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones M.B., Highlander S.K., Anderson E.L., Li W., Dayrit M., Klitgord N., Fabani M.M., Seguritan V., Green J., Pride D.T., et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc. Natl. Acad. Sci. 2015;112:14024–14029. doi: 10.1073/pnas.1519288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schirmer M., Ijaz U.Z., D’Amore R., Hall N., Sloan W.T., Quince C. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 2015;43:e37. doi: 10.1093/nar/gku1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plummer E., Twin J., Bulach D.M., Garl S.M., Tabrizi S.N. A Comparison of Three Bioinformatics Pipelines for the Analysis of Preterm Gut Microbiota using 16S rRNA Gene Sequencing Data. J. Proteom. Bioinform. 2015;8 doi: 10.4172/jpb.1000381. [DOI] [Google Scholar]
- 65.Conterno L., Fava F., Viola R., Tuohy K.M. Obesity and the gut microbiota. Does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr. 2011;6:241–260. doi: 10.1007/s12263-011-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morales P., Fujio S., Navarrete P., Ugalde J.A., Magne F., Carrasco-Pozo C., Tralma K., Quezada M., Hurtado C., Covarrubias N., et al. Impact of dietary lipids on colonic function and microbiota. An experimental approach involving orlistat-induced fat malabsorption in human volunteers. Clin. Transl. Gastroenterol. 2016;7 doi: 10.1038/ctg.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chassaing B., Koren O., Goodrich J.K., Poole A.C., Srinivasan S., Ley R.E., Gewirtz A.T. Dietary Emulsifiers Impact the Mouse Gut Microbiota Promoting Colitis and Metabolic Syndrome. Nature. 2015;519 doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suez J., Korem T., Zeevi D., Zilberman-Schapira G., Thaiss C.A., Maza O., Israeli D., Zmora N., Gilad S., Weinberger A., et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 69.Jin Y., Wu S., Zeng Z., Fu Z. Effects of environmental pollutants on gut microbiota. Environ. Pollut. 2017;222:1–9. doi: 10.1016/j.envpol.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 70.Teixeira T., Grześkowiak Ł.M., Salminen S., Laitinen K., Bressan J., Gouveia Peluzio M. do C. Faecal levels of Bifidobacterium and Clostridium coccoides but not plasma lipopolysaccharide are inversely related to insulin and HOMA index in women. Clin. Nutr. 2013;32:1017–1022. doi: 10.1016/j.clnu.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Rahat-Rozenbloom S., Fernandes J., Gloor G.B., Wolever T.M.S. Evidence for greater production of colonic short-chain fatty acids in overweight than lean humans. Int. J. Obes. 2014;38:1525–1531. doi: 10.1038/ijo.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao X., Zhang M., Xue J., Huang J., Zhuang R., Zhou X., Zhang H., Fu Q., Hao Y. Body Mass Index Differences in the Gut Microbiota Are Gender Specific. Front. Microbiol. 2018;9:1250. doi: 10.3389/fmicb.2018.01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Million M., Maraninchi M., Henry M., Armougom F., Richet H., Carrieri P., Valero R., Raccah D., Vialettes B., Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. (Lond.) 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Kasai C., Sugimoto K., Moritani I., Tanaka J., Oya Y., Inoue H., Tameda M., Shiraki K., Ito M., Takei Y., et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015 doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jinatham V., Kullawong N., Kespechara K., Gentekaki E., Popluechai S. Comparison of Gut Microbiota between Lean and Obese Adult Thai Individuals. Microbiol. Biotechnol. Lett. 2018;46:277–287. doi: 10.4014/mbl.1711.11003. [DOI] [Google Scholar]
- 76.Koliada A., Syzenko G., Moseiko V., Budovska L., Puchkov K., Perederiy V., Gavalko Y., Dorofeyev A., Romanenko M., Tkach S., et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017;17:120. doi: 10.1186/s12866-017-1027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davis S.C., Yadav J.S., Barrow S.D., Robertson B.K. Gut microbiome diversity influenced more by the Westernized dietary regime than the body mass index as assessed using effect size statistic. Microbiologyopen. 2017;6:e00476. doi: 10.1002/mbo3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu T.-W., Park Y.-M., Holscher H.D., Padilla J., Scroggins R.J., Welly R., Britton S.L., Koch L.G., Vieira-Potter V.J., Swanson K.S. Physical Activity Differentially Affects the Cecal Microbiota of Ovariectomized Female Rats Selectively Bred for High and Low Aerobic Capacity. PLoS ONE. 2015;10:e0136150. doi: 10.1371/journal.pone.0136150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Evans C.C., LePard K.J., Kwak J.W., Stancukas M.C., Laskowski S., Dougherty J., Moulton L., Glawe A., Wang Y., Leone V., et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS ONE. 2014;9:e92193. doi: 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clarke S.F., Murphy E.F., O’Sullivan O., Lucey A.J., Humphreys M., Hogan A., Hayes P., O’Reilly M., Jeffery I.B., Wood-Martin R., et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 81.Witt K.A., Bush E.A. College athletes with an elevated body mass index often have a high upper arm muscle area, but not elevated triceps and subscapular skinfolds. J. Am. Diet. Assoc. 2005;105:599–602. doi: 10.1016/j.jada.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 82.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J.-M., Kennedy S., et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 83.de la Cuesta-Zuluaga J., Corrales-Agudelo V., Carmona J.A., Abad J.M., Escobar J.S. Body size phenotypes comprehensively assess cardiometabolic risk and refine the association between obesity and gut microbiota. Int. J. Obes. (Lond.) 2017 doi: 10.1038/ijo.2017.281. [DOI] [PubMed] [Google Scholar]
- 84.Fallani M., Young D., Scott J., Norin E., Amarri S., Adam R., Aguilera M., Khanna S., Gil A., Edwards C.A., et al. Intestinal microbiota of 6-week-old infants across Europe. Geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 85.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Escobar J.S., Klotz B., Valdes B.E., Agudelo G.M. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014;14:311. doi: 10.1186/s12866-014-0311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Magne F., O’Ryan M.L., Vidal R., Farfan M. The human gut microbiome of Latin America populations. A landscape to be discovered. Curr. Opin. Infect. Dis. 2016 doi: 10.1097/QCO.0000000000000300. [DOI] [PubMed] [Google Scholar]
- 88.Suzuki T.A., Worobey M. Geographical variation of human gut microbial composition. Biol. Lett. 2014;10:20131037. doi: 10.1098/rsbl.2013.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dąbrowska K., Witkiewicz W. Correlations of Host Genetics and Gut Microbiome Composition. Front. Microbiol. 2016;7:1357. doi: 10.3389/fmicb.2016.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Girard C., Tromas N., Amyot M., Shapiro B.J. Gut Microbiome of the Canadian Arctic Inuit. mSphere. 2017;2:e00297-16. doi: 10.1128/mSphere.00297-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chevalier C., Stojanović O., Colin D.J., Suarez-Zamorano N., Tarallo V., Veyrat-Durebex C., Rigo D., Fabbiano S., Stevanović A., Hagemann S., et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell. 2015;163:1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 92.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2. High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fujio-Vejar S., Vasquez Y., Morales P., Magne F., Vera-Wolf P., Ugalde J.A., Navarrete P., Gotteland M. The Gut Microbiota of Healthy Chilean Subjects Reveals a High Abundance of the Phylum Verrucomicrobia. Front. Microbiol. 2017;8:1221. doi: 10.3389/fmicb.2017.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Balamurugan R., Sandya R., Pugazhendhi S., Ramakrishna B.S. Faecal microbiota of healthy adults in southern India. Comparison of a tribal and a rural population. Indian J. Med. Res. Res. 2017 doi: 10.4103/ijmr.IJMR_639_14. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baxter N.T., Ruffin M.T., Rogers M.A.M., Schloss P.D., Schloss P.D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8:37. doi: 10.1186/s13073-016-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T., et al. Human Genetics Shape the Gut Microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ahmad A., Yang W., Chen G., Shafiq M., Javed S., Ali Zaidi S.S., Shahid R., Liu C., Bokhari H. Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PLoS ONE. 2019;14:e0226372. doi: 10.1371/journal.pone.0226372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tandon D., Haque M.M., Saravanan R., Shaikh S., Sriram P., Dubey A.K., Mande S.S. A snapshot of gut microbiota of an adult urban population from Western region of India. PLoS ONE. 2018;13:e0195643. doi: 10.1371/journal.pone.0195643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Agudelo-Ochoa G.M., Valdés-Duque B.E., Giraldo-Giraldo N.A., Jaillier-Ramírez A.M., Giraldo-Villa A., Acevedo-Castaño I., Yepes-Molina M.A., Barbosa-Barbosa J., Benítez-Paéz A. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes. 2020:1–16. doi: 10.1080/19490976.2019.1707610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Belforte F.S., Fernandez N., Tonín Monzón F., Rosso A.D., Quesada S., Cimolai M.C., Millán A., Cerrone G.E., Frechtel G.D., Burcelin R., et al. Getting to Know the Gut Microbial Diversity of Metropolitan Buenos Aires Inhabitants. Front. Microbiol. 2019;10:965. doi: 10.3389/fmicb.2019.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]