Abstract
Many Shiga toxin-producing Escherichia coli (STEC) strains, including the serogroups of O157 and most of the top six non-O157 serotypes, are frequently associated with foodborne outbreaks. Therefore, they have been extensively studied using next-generation sequencing technology. However, related information regarding STEC O45 strains is scarce. In this study, three environmental E. coli O45:H16 strains (RM11911, RM13745, and RM13752) and one clinical E. coli O45:H2 strain (SJ7) were sequenced and used to characterize virulence factors using two reference E. coli O45:H2 strains of clinical origin. Subsequently, whole-genome-based phylogenetic analysis was conducted for the six STEC O45 strains and nine other reference STEC genomes, in order to evaluate their evolutionary relationship. The results show that one locus of enterocyte effacement pathogenicity island was found in all three STEC O45:H2 strains, but not in the STEC O45:H16 strains. Additionally, E. coli O45:H2 strains were evolutionarily close to E. coli O103:H2 strains, sharing high homology in terms of virulence factors, such as Stx prophages, but were distinct from E. coli O45:H16 strains. The findings show that E. coli O45:H2 may be as virulent as E. coli O103:H2, which is frequently associated with severe illness and can provide genomic evidence to facilitate STEC surveillance.
Keywords: Shiga-toxin producing Escherichia coli O45, comparative genomics, locus of enterocyte effacement, Stx prophages
1. Introduction
Shiga toxin-producing Escherichia coli (STEC), as one of the major foodborne pathogens, can produce Shiga toxins and cause severe human disease, such as diarrhea, hemorrhagic colitis, and hemolytic–uremic syndrome (HUS) [1,2,3]. STEC strains have been characterized by serotyping based on the O-antigen, determined by the polysaccharide portion of the cell wall lipopolysaccharide (LPS) and the H-antigen related to the flagella protein [4]. E. coli O157 and the “top six” non-O157 serogroups, including O26, O45, O103, O111, O121, and O145, are the primary STEC pathotypes frequently associated with foodborne outbreaks around the world [5,6]. Among the non-O157 STEC serotypes, STEC O45 has been identified as a cause of sporadic cases of bloody diarrhea [1]. In 2005, the first STEC O45 outbreak occurred in New York City, in which a total of 52 inmates were sick with diarrhea or bloody diarrhea, likely resulting from exposure to an ill food worker [7,8]. Subsequently, two E. coli O45:H2-associated outbreaks were reported to have caused 18 illnesses in the United States, and contaminated smoked game meat and goats were implicated as sources of contamination in these outbreaks [9].
The extensive research into STEC has utilized the advantages conferred by whole-genome sequencing technology and genomic analyses in the characterization of genetic information and the virulence factors associated with the diversity and evolution of STEC pathogens [10,11,12]. Bielaszewska et al. found that the E. coli O104:H4 strain associated with a large outbreak in Germany in 2011 was a clone of the E. coli O104:H4 (HUSEC041) strain isolated from the German outbreak in 2001 [13]. Another study reported that two E. coli O145:H28 outbreak strains (RM13714 and RM13716), linked to a U.S. lettuce-associated outbreak and a Belgium ice cream-associated outbreak, respectively, shared a common lineage with five different STEC O157:H7 strains, including the E. coli Sakai strain implicated in a Japanese outbreak [14,15]. Additionally, Ju et al. (2012) conducted phylogenetic analysis on bacterial whole-genome sequencing of STEC O26, O111, and O103 strains, and found a close phylogenetic relationship between E. coli O26:H11 and E. coli O111:H11 strains [16].
Other studies have demonstrated that the pathogenicity evolution of most STEC strains was highly associated with the acquisition of mobile virulence elements, such as Stx prophages, virulence plasmids, and pathogenetic genomic islands [13,14,17]. Specifically, the primary virulence factors of STEC pathogens, stx genes, were located on the Stx prophage sequences and could be transferred to other susceptible strains by phage transduction [18,19]. For example, two HUS-associated STEC O111:H− strains (95JB1 and 95NR1) shared high genomic homology; however, the strain 95NR1 contained two additional Stx2 prophages that contributed to a higher HUS morbidity than 95JB1 [20]. In addition, the locus of enterocyte effacement (LEE) pathogenicity islands containing espB, espD, espA, tir, and eae, were virulence factors of STEC strains in charge of the production of proteins associated with intimate adherence to intestinal epithelial cells and the formation of attaching-and-effacing (A/E) lesions [19,21,22]. Several studies have indicated that LEE pathogenicity islands could be horizontally transferred between two different E. coli strains through specific insert loci, including selC, pheU, and pheV in the bacterial chromosome [23,24]. Moreover, several LEE-positive STEC strains, including those of the O26, O103, O111, O145, and O157 serogroups, have been frequently isolated from HUS patients [25]. Therefore, these findings reveal that the aforementioned virulence genes are essential for the pathogenicity evolution of STEC pathogens and are highly associated with the development of severe human illness, such as HUS [14,26].
Due to the close association with foodborne outbreaks and human illness, the U.S. Department of Agriculture (USDA) Food Safety and Inspection Service (FSIS) has declared E. coli O157 and the “top six” non-O157 serogroups of STEC, including O26, O45, O103, O111, O121, and O145, to have zero tolerance in raw ground beef and tenderized beef [27]. Many studies regarding the genomic characterization and evolutionary relatedness of these STEC strains have been conducted to minimize the spread of the pathogens and to enhance STEC surveillance related to foodborne outbreaks [28,29,30]. However, similar information about STEC O45 is lacking. Therefore, the objectives of this study were to characterize four STEC O45 strains isolated from environmental and clinical samples using next-generation sequencing technology, and to evaluate the pathogenicity of these STEC O45 strains in comparison to other clinical and outbreak-associated STEC strains.
2. Materials and Methods
2.1. Bacterial Strains
Three environmental E. coli O45:H16 strains (RM11911, RM13745, and RM13752) and one clinical E. coli O45:H2 strain (SJ7) were subject to whole-genome sequencing in the present study. The E. coli O45:H2 strain SJ7 was obtained from the Centers for Disease Control and Prevention (CDC), and was originally derived from a patient’s stool. The E. coli O45:H16 strain RM11911 was isolated from a standing water sample, and two E. coli O45:H16 strains (RM13745 and RM13752) were isolated from cattle feces in California. All strains were stored in the culture collections of the Produce Safety and Microbiology Research Unit in the USDA, Agricultural Research Service (ARS), Western Regional Research Center (WRRC). All strains were stored at −80 °C and grown in 10 mL of tryptic soy broth (TSB, Difco, Becton Dickinson, Sparks, MD) in a sterile 15-mL conical tube at 37 °C with shaking at 175 rpm before use.
2.2. Whole-Genome Sequencing, Assembly, and Annotation of STEC O45 Strains
Genomic DNA was extracted from the cultures grown to the mid-exponential-phase in 10 mL TSB using a Quick-DNA Miniprep Plus Kit (Zymo Research, Irvine, CA), according to the manufacturer’s instructions. The DNA library was constructed using a SMARTbell Express Template Prep kit 2.0 (PacBio, Menlo Park, CA) and subsequently sequenced on a Pacific Biosciences Sequel II with v1 reagents (PacBio, Menlo Park, CA) by SNPsaurus (Eugene, OR) [31]. An average of 251,060 single-end reads per bacterial strain were generated and subjected to de novo assembly using Flye 2.4.1 with default parameters. The resulting assemblies of each strain were generated with more than 100× coverage. The genome completeness of four strains was analyzed by BUSCO 3.0.2, and each assembled genome reached a score of above 98% completeness, which was identified as a complete genome [32]. The assembled contigs were also confirmed as bacterial chromosomes or plasmids by the use of BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Nucleotides) [33]. The resulting genome sequences were subject to annotation using the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/) and deposited in GenBank. The circular maps were generated using the CGview server [34]. The accession numbers of the chromosome and plasmids for E. coli O45:H2 (SJ7) and three E. coli O45:H16 strains (RM11911, RM13745, and RM13752) are included in Table 1.
Table 1.
Genomic features of the chromosomes and plasmids for six Shiga toxin-producing Escherichia coli (STEC) O45 strains analyzed in this study, including three environmental and three clinical strains.
| Feature | E. coli O45:H16 | E. coli O45:H2 | ||||
|---|---|---|---|---|---|---|
| RM11911 | RM13745 α | RM13752 | SJ7 α | FWSEC0003 α | 2011C-4251 | |
| Chromosome | ||||||
| Size (kbp) | 5310 | 5264 | 5264 | 5444 | 5532 | 5440 |
| %GC | 51 | 51 | 51 | 50.7 | 50.7 | 50.7 |
| No. CDSs | 5194 | 5132 | 5131 | 5278 | 5735 | 5765 |
| No. rRNA | 22 | 22 | 22 | 22 | 22 | 22 |
| No. tRNA | 91 | 90 | 90 | 96 | 103 | 96 |
| No. Prophages | 14 | 12 | 12 | 17 | 18 | 17 |
| No. Genomic islands (GI) | 44 | 40 | 44 | 48 | 55 | 51 |
| Accession number * | CP044313 | CP044312 | CP044311 | CP044315 | reference | reference |
| Plasmid | ||||||
| Size (kbp) | 175 | 70/45 | 99 | 74/53 | 95/52 | 68 |
| %GC | 47.6 | 48.7/48.8 | 47.2 | 49.1/49.2 | 47.2/48.8 | 49.2 |
| Accession number * | CP051656 | CP051654, CP051655 | CP051653 | CP051657, CP051658 | reference | reference |
* The accession numbers are provided for the strains, including three environmental E. coli O45:H16 strains and one clinical E. coli O45:H2 strain (SJ7), which were sequenced in this study with the given accession numbers. Additionally, “reference” indicates the chromosome and plasmid of two clinical strains (FWSEC0003 and 2011C-4251) obtained from the National Center for Biotechnology Information (NCBI) with the accession numbers indicated in Table S1. α The strains contained two different plasmids.
2.3. Determination of Virulence and Antibiotic Resistance Genes
Two reference E. coli O45:H2 strains (FWSEC0003 and 2011C-4251) were obtained from the NCBI database to facilitate the genomic analyses of virulence and antibiotic resistance genes of STEC O45 strains. E. coli clinical O45:H2 (FWSEC0003) was previously isolated from human feces in Canada [35], and the E. coli O45:H2 strain (2011C-4251) was isolated from samples of human feces obtained in the United States [36]. The virulence gene profiles of six strains (four experimental strains and two reference strains) were obtained using VirulenceFinder 2.0 [37]. The prediction of antibiotic resistance genes in each bacterial chromosome was conducted using ResFinder 3.2 [38] and Abricate 0.5 (https://github.com/tseemann/abricate). The setting with a minimum of both 95% nucleotide sequence identity and coverage was used to screen for these genes.
2.4. Identification of the Mobile Genetic Elements: Prophages and Genomic Islands
The identification of prophage and prophage-like sequences from each bacterial genome was conducted using the PHASTER web server [39]. Genomic islands were analyzed using the IslandViewer4 web server [40]. The default parameters were used for all the software.
2.5. Whole-Genome-Based Phylogenetic Analysis
Six STEC O45 strains, including four strains sequenced in this study and two reference strains from the NCBI database, as well as nine different STEC strains of clinical and outbreak origin (Table S1) were subject to phylogenetic analysis by whole-genome multilocus typing (wg-MLST). Specifically, a total of 14,837 loci and 2513 core loci located on the entire genomes of 15 E. coli strains were analyzed and phylogenetic networks were constructed via a complete linkage method using BioNumerics 7.6 (Applied Maths, Kortrijk, Belgium). The GenBank accession numbers of 11 reference bacterial genomes obtained from the public database are indicated in Table S1.
2.6. Comparative Genomics of E. coli O45:H2 and E. coli O103:H2
Based on the results of wg-MLST, two reference E. coli O103:H2 strains (12009 and 2015C-3163) of clinical origin were subject to comparative genomics analysis with STEC O45:H2 on the virulence genes and mobile genetic elements to evaluate their genetic relatedness using the BLAST Ring Image Generator (BRIG) with a minimum nucleotide sequence identity of 50% [41]. The crucial virulence factors of STEC pathogens, including Stx prophages and LEE pathogenicity islands, were extracted from each genome and subsequently aligned using the MAFFT algorithm in Geneious (Version 11.1.5, Biomatters, New Zealand). The phylogenetic trees of Stx prophages were constructed using Mega-X (Version 10.0.5) with the maximum likelihood algorithm and visualized using the Interactive Tree Of Life (ITOL) webserver [42]. The aligned sequences and the consensus identity of the LEE pathogenicity islands were visualized using Geneious (Version 11.1.5, Biomatters, New Zealand) [43,44].
3. Results
3.1. Genomic Features of Three E. coli O45:H16 Environmental Strains
The chromosomes of the three E. coli O45:H16 strains RM11911, RM13745, and RM13752 had genome sizes of 5,310,338, 5,264,698, and 5,264,517 bp, containing a total of 5194, 5264, and 5264 regions of coding DNA sequence (CDS), respectively; 91% of which were annotated with known functions (Table 1). RM11911, RM13745, and RM13752 strains also contained 22 rRNA and 90–91 tRNAs in each chromosome. Regarding the prediction of numerous mobile elements, RM11911 harbored 14 prophages, including a 65,626 bp Stx prophage carrying an stx1a gene, whereas both RM13745 and RM13752 contained 12 prophages, including a 65,626 bp Stx1a prophage located at the same insertion site of each bacterial genome. Three E. coli O45:H16 strains also contained two additional virulence factors, iroN and lpfA genes, which were associated with the utilization of the siderophore enterobactin and bacterial adhesion and colonization in the intestine, respectively. Additionally, the antibiotic resistance gene mdfA was detected in each chromosome of E. coli O45:H16, which was attributed to a broad spectrum of drug-resistant features of these strains.
The RM11911 strain contained one large circular plasmid, pRM11911, which was 175,089 bp in length, with an average G+C content of 47.6%. The plasmid pRM11911 also harbored several virulence factors, including the hyl operon (hylC, hylA, hylB, and hylD) encoding hemolysin, afa family (afaA-VIII, afaB-VIII, afaC-VIII, afaD-VIII, afaE-VIII, and afaF-VII) encoding adhesins, cdt family (cdtA, cdtB, and cdtC) encoding cytolethal distending toxin B, espP encoding extracellular serine protease, cnf1 encoding cytotoxic necrotizing factor 1, and iha encoding the IrgA homolog adhesin (Table 2). The RM13745 strain harbors two plasmids: pRM13745-1 (45,063 bp) and pRM13745-2 (70,650 bp). The virulence gene-screening results showed that espP and afa families were present in pRM13745-1, while iha and hyl operons were present in pRM13745-2. RM13752 had one plasmid (pRM13752), with a genome size of 99,009 bp that contained the virulence genes, including hyl operon and cnf1. No antibiotic resistance genes were found in any of these plasmids.
Table 2.
The number of various virulence genes present in the chromosomes and plasmids of six STEC O45 strains analyzed in this study.
| Virulence Genes | E. coli O45:H16 | E. coli O45:H2 | ||||
|---|---|---|---|---|---|---|
| RM11911 | RM13745 | RM13752 | SJ7 | FWSEC0003 | 2011C-4251 | |
| Chromosome | ||||||
| stx 1a | 1α | 1 | 1 | 1 | 1 | 1 |
| stx 2a | - | - | - | - | - | 1 |
| gad | 2 | 2 | 2 | 2 | 2 | 2 |
| iss | 2 | 2 | 2 | 1 | 2 | 1 |
| iha | - | - | - | - | - | - |
| iroN | 1 | 1 | 1 | - | - | - |
| lpfA | 1 | 1 | 1 | - | - | - |
| cif | - | - | - | 1 | 1 | 1 |
| eae | - | - | - | 1 | 1 | 1 |
| efa1 | - | - | - | 1 | 1 | 1 |
| espA | - | - | - | 1 | 1 | 1 |
| espB | - | - | - | 1 | 1 | 1 |
| espF | - | - | - | 1 | 1 | 1 |
| espI | - | - | - | 1 | 1 | 1 |
| espJ | - | - | - | - | - | - |
| nleA | - | - | - | 1 | 1 | 1 |
| nleB | - | - | - | 3 | 3 | 3 |
| nleC | - | - | - | 1 | 1 | - |
| tir | - | - | - | 1 | 1 | 1 |
| tccP | - | - | - | - | - | - |
| Plasmid | ||||||
| hyl | 1 | 1 | 1 | 1 | 1 | 1 |
| afa | 1 | - | - | - | - | - |
| cdt | 1 | - | - | - | - | - |
| esp | 1 | 1 | - | - | - | - |
| cnf1 | 1 | - | 1 | - | - | - |
| iha | 1 | 1 | - | - | - | - |
| ehxA | - | - | - | 1 | - | 1 |
| etpD | - | - | - | 1 | 1 | 1 |
| stcE | - | - | - | 1 | 1 | 1 |
| toxB | - | - | - | - | - | - |
| katP | - | - | - | - | - | - |
α The numerical value indicates the number of genes present, and “-” means that no virulence gene was detected.
3.2. Genomic Features of a Clinical E. coli O45:H2 Strain
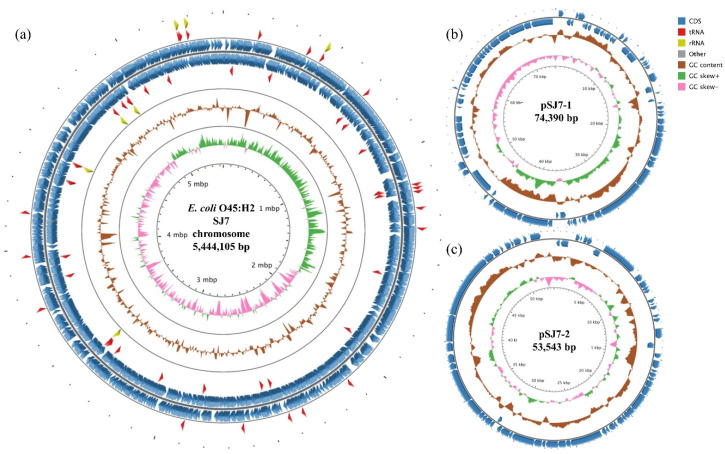
The E. coli O45:H2 strain SJ7, isolated from a patient stool, contained a 5,444,105 bp chromosome which was approximately 130–180 kb smaller than the genomes of the three E. coli O45:H16 strains (Table 1). The chromosome of SJ7 consisted of 5278 CDSs, 22 rRNA, and 96 tRNA (Figure 1a). Ninety-one percent of CDSs were annotated with known functions. A total of 17 prophages, including one Stx1a prophage which was 65,813 bp in length, were predicted in the bacterial chromosome. Most of all, strain SJ7 contained a LEE pathogenicity island integrated at the pheV tRNA locus. Several non-LEE-encoded type III translocated virulence genes, including nelA, nelB, nelC, espI, and cif, were also present in the chromosome. No antibiotic resistance genes were found in the genome of SJ7.
Figure 1.
Circular genome maps of the chromosome (a), plasmid pSJ7-1 (b), and plasmid pSJ7-2 (c) for the clinical STEC O45:H2 strain SJ7 generated using CGview server. For the map of chromosome (a), the rings from the inside out represent the GC skew (green and pink), GC content (brown), coding DNA sequences (CDSs) (blue), tRNA (red), and rRNA (yellow). For the maps of two plasmids (pSJ7-1 and pSJ7-2), the same color codes are used, and no tRNA and rRNA were detected. The annotated functions of the CDSs are not indicated.
SJ7 had two plasmids: pSJ7-1 and pSJ7-2 (Figure 1b,1c, Table 2). Plasmid pSJ7-1 had a genome of 74,390 bp with an average GC content of 49.1% and three virulence genes, including stcE (encoding metalloprotease), ehxA (encoding enterhemolysin), and etpD (encoding type II secretion protein). The second-largest plasmid, pSJ7-2, was 53,543 bp in length with 49.2% GC content, and contained no virulence genes. Neither plasmids harbored antibiotic resistance genes.
The two other clinical E. coli O45:H2 strains (FWSEC0003 and 2011C-4251) were obtained from the NCBI database to facilitate the genomic characterization of STEC O45:H2 strains (Table 1). Similar to the strain SJ7, FWSEC0003 and 2011C-4251 had a genome size of 5,532,455 and 5,440,026 bp, respectively, with an average GC content of 50.7% and a similar number of CDSs (>5700). FWSEC0003 contained an Stx1a prophage (56,560 bp), whereas 2011C-4251 had two Stx prophages consisting of a 64,500 bp Stx1a prophage and a 62,499 bp Stx2a prophage. Furthermore, both FWSEC0003 and 2011C-4251 strains encoded the LEE pathogenicity island, and several non-LEE-encoded type III translocated virulence factors, including nelA, nelB, espI, and cif (Table 2).
2011C-4251 had a plasmid (68,062 bp) which contained the virulence genes ehxA, etpD, and stcE, similar to SJ7. FWSEC0003 had two plasmids (95,228 and 52,940 bp), and the virulence genes etpD and stcE were found in the smaller plasmid. Furthermore, no antibiotic resistance genes were found in the plasmids of these two strains (Table 2).
3.3. Phylogenetic Analysis of 15 STEC Strains Using Whole-Genome Multilocus Typing
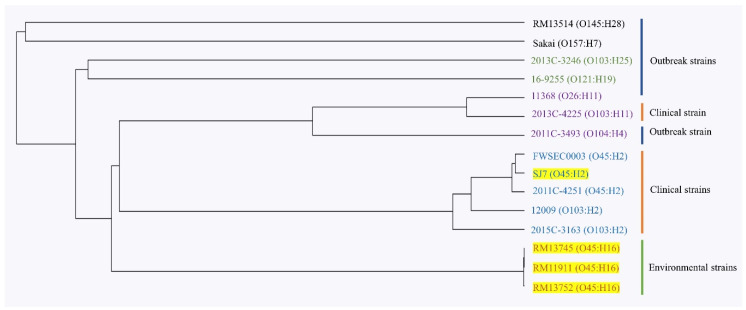
The evolutionary relatedness of 15 STEC strains, including four E. coli O45 strains (RM11911, RM13745, RM13752, and SJ7) sequenced in this study, two reference E. coli O45 strains (FWSEC0003 and 2011C-4251), and nine reference STEC strains with different serotypes, was investigated through whole-genome multilocus typing analysis. The results show that these STEC strains grouped into five clusters in the phylogenetic tree (Figure 2). Two outbreak strains, E. coli O145:H28 (RM13514) and E. coli O157:H7 (Sakai), grouped into cluster 1, while a clinical E. coli O103:H25 (2013C-3264) and an outbreak-associated E. coli O121:H19 (16-9255) strain were located in cluster 2. Additionally, cluster 3 contained a clinical E. coli O103:H11 strain (2013C-4225) and two outbreak strains of E. coli O26:H11 (11368) and E. coli O104:H4 (2011C-3493). Cluster 4 was composed of three E. coli O45:H2 strains (SJ7, FWSEC0003, and 2011C-4251) and two clinical E. coli O103:H2 strains (12009 and 2015C-3163), whereas three environmental E. coli O45:H16 strains (RM11911, RM13745, and RM13752) grouped into cluster 5. Additionally, the results showed that the three clinical E. coli O45:H2 strains shared a common evolutionary lineage with two E. coli O103:H2 strains (cluster 4), but differed from the environmental E. coli O45:H16 strain (cluster 5) and other STEC strains of clinical and outbreak origin (Figure 3). The findings indicate significant genomic differences between the E. coli O45:H16 and E. coli O45:H2 strains.
Figure 2.
Whole-genome multilocus typing-based phylogenetic analysis of three environmental E. coli O45:H16 strains (RM13745, RM11911, and RM13752) and one clinical E. coli O45:H2 strain (SJ7) with eleven reference strains. The phylogenetic tree indicates five distinct clusters, with the strain names shown in black color for cluster 1, green color for cluster 2, purple color for cluster 3, blue cluster for cluster 4, and orange color for cluster 5. The strains highlighted in yellow were sequenced this study. The sources of isolation (outbreak, clinical, and environmental) for these 15 strains are indicated right next to the strain names.
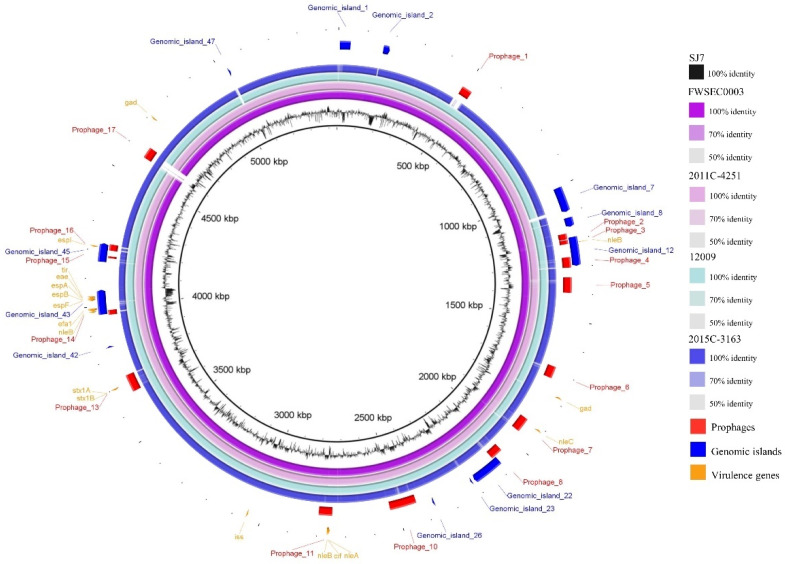
Figure 3.
Comparative analysis of the complete chromosome of the STEC O45:H2 strain SJ7 with two STEC O45:H2 and two STEC O103:H2 reference strains. The BLAST comparison of STEC chromosomes and the chromosome of E. coli O45:H2 strain SJ7 was generated using the BLAST Ring Image Generator (BRIG) with a minimum nucleotide sequence identity of 50%. The bacterial chromosomes from inside out represent E. coli O45:H2 SJ7 (black), E. coli O45:H2 FWEC0003 (dark purple), E. coli O45:H2 2011C-4251 (light purple), E. coli O103:H2 12009 (light blue), and E. coli O103:H2 2015C-3163 (dark blue). Color codes for different degrees of nucleotide sequence homologies based on comparison to SJ7 are listed. The prophages (red), genomic islands (navy), and virulence genes (yellow) annotated in the SJ7 chromosome are present at the outermost three rings.
3.4. Comparative Genomics of E. coli O45:H2 and E. coli O103:H2 Strains
Due to the close genetic relatedness, the comparative genomics analysis was further performed on the backbones and mobile genetic elements of the three E. coli O45:H2 and two E. coli O103:H2 strains to investigate their evolutionary relationship. The whole-genome sequences of two E. coli O45:H2 (FWSEC0003 and 2011C-4251) reference strains and two E. coli O103:H2 (12009 and 2015C-3163) reference strains were compared against the E. coli O45:H2 strain (SJ7) using BLASTn, and the results show that the genomic backbones of these strains shared more than 95% identity covering over 95% of the SJ7 chromosome (Figure 3). Furthermore, non-homologous regions were identified among the genomes of E. coli O45:H2 and E. coli O103:H2 strains, particularly in the sequences of their mobile genetic elements. In the three E. coli O45:H2 chromosomes, prophage sequences were the primary non-homologous regions. For example, a 42,590-bp prophage (prophage_17) present in the genome of SJ7 was absent in both genomes of FWSEC0003 and 2011C-4251. Additionally, both FWSEC0003 and 2011C-4251 contained a prophage sequence sharing a low nucleotide sequence similarity to prophage_7 located in similar regions in the chromosome of SJ7. Another prophage sequence (prophage_6) in SJ7 shared low homology to the counterpart prophage sequence in 2011C-4251. On the other hand, genomic heterology of the mobile genetic elements was also observed for the E. coli O45:H2 strain (SJ7) and two E. coli. O103:H2 (12009 and 2015C-3163) strains. The prophage_1 and prophage_17 sequences found in SJ7 were absent in similar regions in the chromosomes of 2015C-3163 and 12009, respectively. Additionally, there was a region of low homology located at the sequence of genomic island_7 in SJ7 compared to the counterpart in both E. coli O103:H2 strains. Minor variations in specific regions, such as genomic island_1, prophage_7, and prophage_10, in E. coli O45:H2 (SJ7) were observed in comparison to both E. coli O103:H2 strains. These results indicate that the presence of different mobile genetic elements, and prophages in particular, was primarily attributed to the heterology of the closely related E. coli O45:H2 and E. coli O103:H2 strains.
3.5. Virulence Factors Located on Mobile Genetic Elements
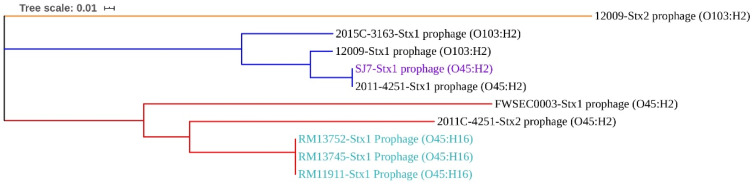
The virulence factors of E. coli O45:H2 and E. coli O103:H2 were analyzed, and the results show that all virulence genes of each strain, except for gad, were located on different prophages and genomic islands (Figure 3). Stx prophages and LEE pathogenicity islands were among the most important virulence factors contributing to the pathogenicity of these pathogenic strains. The Stx prophages in the chromosomes of E. coli O45:H2 and E. coli O103:H2 were predicted and compared to investigate their genetic similarity (Figure 4). The phylogenetic analysis of Stx prophages showed that the Stx2a prophage in E. coli O103:H2 (12009) was classified as being alone in cluster 1. The Stx1a prophages from two E. coli O45:H2 strains (SJ7 and 2011C-4251) and two E. coli O103:H2 strains (12009 and 2015C-3163) were grouped into cluster 2. The Stx1a prophage and Stx2a prophage from E. coli O45:H2 (FWSEC0003) and E. coli O45:H2 (2011C-4251), respectively, were classified into cluster 3. Based on the phylogenetic results, the high sequence homology between the Stx1a prophage of E. coli O45:H2 strain (SJ7) and the two E. coli O103:H2 strains likely indicates that E. coli O45:H2 might exhibit a similar Stx prophage-associated pathogenicity to E. coli O103:H2.
Figure 4.
Maximum likelihood phylogenetic analysis based on the MAFFT algorithm of the Stx prophages from the genomes of six STEC O45 strains and two STEC O103:H2 strains. The phylogenetic tree indicates three distinct clusters among the Stx prophages, in orange for cluster 1, navy for cluster 2, and red for cluster 3. Stx prophages highlighted in purple and light blue indicates they were obtained from the bacterial strains sequenced in this study. All Stx prophage sequences in the bacterial chromosome were predicted using the PHASTER web server.
Additionally, one of the essential virulence factors of STEC strains, the LEE pathogenicity island, was also present among the genomes of these E. coli O45:H2 and E. coli O103:H2 strains (Figure 5). LEE pathogenicity islands had an average sequence size ranging from 61,863 to 88,869 bp and were located at the pheU or pheV tRNA locus. All LEE pathogenicity islands contained the core virulence genes (espF, espB, espA, eae, and tir) in the highly conserved region (Figure 5). All strains, except for E. coli O103:H2 (2015C-3163), contained the LEE pathogenicity islands harboring extra virulence genes, nleB and efa1, which were located in the non-homologous region of the LEE pathogenicity island. The comparative genomics of LEE pathogenicity islands from the selected strains also showed that the E. coli O103:H2 strain (12009), associated with a diarrhea patient from a previous foodborne infection in Japan, and the three clinical E. coli O45:H2 strains (SJ7, FWSEC00033, and 2011C-4251) shared a high nucleotide sequence identity, demonstrating that these E. coli O45:H2 strains could harbor similar human pathogenesis, causing diarrhea.
Figure 5.
Nucleotide sequence alignments of the locus of enterocyte effacement (LEE) pathogenicity islands from the genomes of three clinical STEC O45:H2 strains (SJ7 (sequenced in this study), FWSEC003, and 2011C-4251), and two clinical STEC O103:H2 strains (12009 and 2015C-3163) using the MAFFT algorithm. The consensus identity of the alignment of five LEE pathogenicity islands indicates the mean pairwise nucleotide sequence identified for all pairs in the column: green = 100% identity; green-brown = <100% but >30% identity; and red = <30% identity. Virulence genes within the sequence of each LEE pathogenicity island (black) are annotated and colored (yellow).
4. Discussion
Due to the advancement of next-generation sequencing technology, the high accuracy of complete bacterial genome information has dramatically facilitated the genomic characterization of STEC pathogens and investigation of the evolutionary relationship between different pathogenic E. coli strains. A number of studies have utilized whole-genome sequencing technology to predict the parallel evolution of distinct STEC strains, including the O26, O111, O103, O145, and O157 serogroups [14,45]. Their findings have not only provided genomic evidence related to the clinical impact of these strains, but also facilitated the surveillance of these STEC serotypes. However, similar studies, including on published complete genomes of STEC O45 strains, are lacking. Currently, only two complete genome sequences of STEC O45:H2 strains (FWSEC0003 and 2011C-4251) are available in the public database. Therefore, in this study, four STEC O45 strains, from environmental and clinical samples, were sequenced and subjected to comprehensive genomic characterization, particularly for crucial virulence factors, and a comparison with other reference bacterial genomes was conducted to understand whether or not STEC O45 poses a similar food safety threat as the other serogroups.
4.1. STEC O45:H16 Strains Are Genomically Conserved
In this study, the results showed that the genome features of three E. coli O45:H16 strains (RM11911, RM13745, and RM13752) contained similar virulence factors, most of which were located on similar prophage sequences. All three E. coli O45:H16 strains contained the same Stx1a prophage (65,626 bp) with 100% nucleotide sequence homology (Figure 4). A previous study in our lab also demonstrated that Stx1a prophages from STEC O45 genomes were more conservative than those detected from other serotypes of STEC genomes [46]. Additionally, these three STEC O45:H16 strains contained a virulence factor—the iss gene—contributing to the serum resistance of E. coli strains [47,48]. A previous study revealed that the tested E. coli strains contained three iss alleles, which were likely disseminated among E. coli bacteria through horizontal gene transfer [49]. In this study, each of the three O45:H16 strains contained two different iss alleles, which were located on the Stx1a prophage and a suspected 50,703-bp prophage sequence, respectively. These findings show that the three environmental E. coli O45:H16 strains in this study were genomically conserved, with the virulence factors being carried by certain prophages.
4.2. STEC O45:H2 Evolved from a Common Ancestor with STEC O103:H2
The current results of wg-MLST analysis showed that three STEC O45:H2 strains (SJ7, FWSEC0003, 2011C-4251) and two STEC O103:H2 strains (12009 and 2015C-3163) have a close evolutionary relationship (Figure 2). Moreover, the comparative genomics of E. coli O45:H2 and E. coli O103:H2 revealed that the genomic backbone of these strains share 99% homology. Previous studies have demonstrated that a large percentage of E. coli O103:H2 strains have a high level of virulence in human infection and are commonly associated with diarrhea and HUS [50,51]. In particular, E. coli O45:H2 strains (SJ7, FWSEC0003, 2011C-4251) were shown to contain the crucial virulence factors, eae and tir, that are found in most STEC O103:H2 strains and are involved in the formation of A/E lesions [52]. Additionally, E. coli O45:H2 and E. coli O103:H2 used in this study were found to contain non-LEE-encoded type III translocated virulence genes, which are related to bacterial colonization and the development of HUS [53,54].
Additionally, the wg-MLST result in this study showed that E. coli O145:H28 (RM13514) and E. coli O157:H7 (Sakai) shared a close phylogenetic relationship, which is in agreement with the findings of a previous study demonstrating that STEC O145:H28 and O157:H7 strains evolved from a common evolutionary lineage [55]. The author also found that these two STEC serotypes could be further classified into sublineages based on the presence of different virulence factors, such as Stx prophages and the large virulence plasmids. However, in this study, the most highly heterogeneous regions between E. coli O45:H2 and E. coli O103:H2 strains fell on the sequences corresponding to prophages and genomic islands which were not associated with virulence factors (Figure 3). These findings likely suggest that due to a high homology of the virulence factors, E. coli O45:H2 may possess a similar pathogenicity to E. coli O103:H2 and could potentially cause severe human diseases such as HUS. Thus, the critical findings of the close genetic relatedness between the clinical STEC O103:H2 and STEC O45:H2 strains could facilitate future development of the antimicrobial strategies for the control of STEC O45 strains because some antimicrobial agents, such as weak acids, or lytic bacteriophages, effective in mitigating STEC O103 could also be tested to control STEC O45 strains [56,57]. Other natural antimicrobials that are capable of displaying a wide spectrum of antimicrobial activity and overcoming the emergence of antibiotic resistance in bacterial pathogens could also be considered [58].
4.3. Mobile Elements Play a Key Role in Driving the Virulence Evolution of STEC
Mobile genetic elements play a key role in shaping the bacterial genome and virulence evolution of STEC [59,60,61]. The current results also show that most virulence factors were located on mobile genetic elements, including plasmids, prophages, and genomic islands. In this study, the four STEC O45 strains contained different plasmids. Even the three E. coli O45:H16 environmental strains, which shared high nucleotide sequence similarity of the bacterial chromosomes, had plasmids with different genome sizes and contained various virulence genes, including afa, cdt, esp, cnf1, and iha. Several studies have indicated that the plasmids of LEE-negative STEC strains, which were found to contain encoded virulence factors, can also cause severe human diseases such as HUS, an ability which is frequently observed for LEE-positive STEC pathogens [62,63,64,65]. Additionally, a previous study conducted by Michelacci et al. reported that one LEE-negative STEC strain, isolated from an HUS patient, contained a plasmid harboring several virulence factors, including ehxA, sta1, and a novel variant of the faeG, in particular [64]. Notably, the faeG, encoding the production of ETEC F4 fimbriae, is commonly found in the pathogenic E. coli strains related to causing swine diseases, but not in those that could affect human health. Therefore, the presence of faeG in the plasmid of this human clinical strain provided genetic evidence for virulence gene transfer among plasmids of different STEC strains, which contributed to the pathogenicity evolution of the STEC strains.
Prophages represent an essential member of mobile genetic elements and are highly distributed in the genomes of different E. coli strains [66]. Many studies have indicated that stx, encoded in Stx prophage sequence, is the main virulence factor of STEC [46,67,68,69]. The results of this study showed that three E. coli O45:H16 strains carried a similar Stx prophage, whereas the phylogenetically related E. coli O45:H2 and E. coli O103:H2 strains carried distinct Stx prophages. The current results were supported by the finding of our previous study that the distribution of different Stx prophages is highly associated with specific STEC serotypes. Some serotypes of STEC strains, such as O103, were able to accept a wide range of different Stx prophages, but other serotypes, like O121, were only susceptible to infection with genetically conserved Stx prophages [46]. Therefore, the diversity of Stx prophages from E. coli O45:H2 (SJ7) and E. coli O103:H2 strains used in this study likely implies that these strains are susceptible to accepting exogenous genes, rather than E. coli O45:H16 strains. Additionally, other prophages predicted from all of the STEC O45 and O103 strains in this study also contained several virulence factors, including non-LEE encoded type III translocated effectors (nleA, nleB, nleC, and cif), serine protease autotransporters (espI), and increased serum survival gene (iss). Similar results were also found in a previous study, in which the genes encoding type III secretion in E. coli were located on a vast prophage region, which acted as a crucible for the evolution of pathogenicity in numerous E. coli species [70]. Interestingly, the results of this study show that the prophages carrying non-LEE encoded type III translocated effectors were only identified in LEE-positive strains, including E. coli O45:H2 and E. coli O103:H2 strains, but were absent in the E. coli O45:H16 LEE-negative strains. This phenomenon will need to be investigated in future studies.
5. Conclusions
This is the first study to report the genomic characterization of STEC O45 strains of different origins. The whole-genome-based phylogenetic analysis revealed that the environmental E. coli O45:H16 strains were phylogenetically distinct from the clinical E. coli O45:H2 strains, whereas both clinical E. coli O45:H2 and E. coli O103:H2 shared a common evolutionary ancestor. Furthermore, most of the crucial virulence factors from E. coli O45:H2 and E. coli O103:H2 shared high nucleotide sequence similarity and were located on mobile genetic elements, such as prophages, genomic islands, or plasmids, which is strongly associated with the pathogenicity evolution of these STEC strains. The findings of this study provide better insights into the genomic characterization, evolutionary relatedness, and virulence evolution of STEC O45 strains and indicate that E. coli O45:H2 strains still pose potential threats to public health and should, therefore, be included in epidemiological surveillance.
Acknowledgments
The authors would like to thank Alexandra Salvador for her assistance in the strain preparation and the extraction of plasmids.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/5/782/s1: Table S1: Information on reference Shiga toxin-producing E. coli genomes obtained from NCBI in this study.
Author Contributions
Y.Z. was responsible for strain preparation, data analysis, and manuscript preparation. Y.-T.L. was responsible for manuscript preparation. X.S. assisted the experiment design. V.C.H.W. conceived and supervised the study, aided in experiment design, and reviewed and edited the manuscript. All authors have read and agreed to the manuscript.
Funding
This research was funded by the USDA-ARS CRIS projects 2030-42000-050-00D.
Conflicts of Interest
The submitted work was not carried out in the presence of any personal, professional, or financial relationships that could potentially be construed as a conflict of interest.
References
- 1.Brooks J.T., Sowers E.G., Wells J.G., Greene K.D., Griffin P.M., Hoekstra R.M., Strockbine N.A. Non-O157 Shiga toxin–producing Escherichia coli infections in the United States, 1983–2002. J. Infect. Dis. 2005;192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 2.Tarr P.I., Gordon C.A., Chandler W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 3.Paton J.C., Paton A.W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 1998;11:450–479. doi: 10.1128/CMR.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gyles C.L. Shiga toxin-producing Escherichia coli: An overview1. J. Anim. Sci. 2007;85:E45–E62. doi: 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 5.Gyles CDC National Shiga toxin-producing Escherichia coli (STEC) surveillance. [(accessed on 18 April 2020)]; Available online: https://www.cdc.gov/nationalsurveillance/ecoli-surveillance.html.
- 6.Karmali M.A. Emerging public health challenges of Shiga toxin–producing Escherichia coli related to changes in the pathogen, the population, and the environment. Clin. Infect. Dis. 2016;64:371–376. doi: 10.1093/cid/ciw708. [DOI] [PubMed] [Google Scholar]
- 7.Schaffzin J.K., Coronado F., Dumas N.B., Root T.P., Halse T.A., Schoonmaker-Bopp D.J., Lurie M.M., Nicholas D., Gerzonich B., Johnson G.S., et al. Public health approach to detection of non-O157 Shiga toxin-producing Escherichia coli: Summary of two outbreaks and laboratory procedures. Epidemiol. Infect. 2011;140:283–289. doi: 10.1017/S0950268811000719. [DOI] [PubMed] [Google Scholar]
- 8.CDC Importance of culture confirmation of shiga toxin-producing Escherichia coli infection as illustrated by outbreaks of gastroenteritis--New York and North Carolina, 2005. MMWR. Morb. Mortal. Wkly. Rep. 2006;55:1042–1045. [PubMed] [Google Scholar]
- 9.Luna-Gierke R.E., Griffin P.M., Gould L.H., Herman K., Bopp C.A., Strockbine N., Mody R.K. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 2014;142:2270–2280. doi: 10.1017/S0950268813003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazen T.H., Michalski J., Luo Q., Shetty A.C., Daugherty S.C., Fleckenstein J.M., Rasko D.A. Comparative genomics and transcriptomics of Escherichia coli isolates carrying virulence factors of both enteropathogenic and enterotoxigenic E. coli. Sci. Rep. 2017;7:3513. doi: 10.1038/s41598-017-03489-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Font C.C., Lopez-Siles M., Ferrer-Guixeras M., Carulla L.N., Abella C., Garcia-Gil J., Martinez-Medina M. Comparative genomics reveals new single-nucleotide polymorphisms that can assist in identification of adherent-invasive Escherichia coli. Sci. Rep. 2018;8:2695. doi: 10.1038/s41598-018-20843-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien C.L., Bringer M.-A., Holt K., Gordon D., Dubois A.L., Barnich N., Darfeuille-Michaud A., Pavli P. Comparative genomics of Crohn’s disease-associated adherent-invasive Escherichia coli. Gut. 2016;66:1382–1389. doi: 10.1136/gutjnl-2015-311059. [DOI] [PubMed] [Google Scholar]
- 13.Bielaszewska M., Mellmann A., Zhang W., Köck R., Fruth A., Bauwens A., Peters G., Karch H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: A microbiological study. Lancet Infect. Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 14.Cooper K.K., Mandrell R.E., Louie J.W., Korlach J., Clark T., Parker C.T., Huynh S., Chain P., Ahmed S., Carter M.Q. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genom. 2014;15:17. doi: 10.1186/1471-2164-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michino H., Araki K., Minami S., Takaya S., Sakai N., Miyazaki M., Ono A., Yanagawa H. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 1999;150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 16.Ju W., Cao G., Rump L., Strain E., Luo Y., Timme R.E., Allard M., Zhao S., Brown E., Meng J. Phylogenetic analysis of non-O157 Shiga toxin-producing Escherichia coli strains by whole-genome sequencing. J. Clin. Microbiol. 2012;50:4123–4127. doi: 10.1128/JCM.02262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank C., Werber D., Cramer J.P., Askar M., Faber M., Der Heiden M.A., Bernard H., Fruth A., Prager R., Spode A., et al. Epidemic profile of Shiga-toxin–producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 18.Ogura Y., Mondal S.I., Islam R., Mako T., Arisawa K., Katsura K., Ooka T., Gotoh Y., Murase K., Ohnishi M., et al. The Shiga toxin 2 production level in enterohemorrhagic Escherichia coli O157:H7 is correlated with the subtypes of toxin-encoding phage. Sci. Rep. 2015;5:16663. doi: 10.1038/srep16663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan Y.S., Ng T.B. Shiga toxins: From structure and mechanism to applications. Appl. Microbiol. Biotechnol. 2015;100:1597–1610. doi: 10.1007/s00253-015-7236-3. [DOI] [PubMed] [Google Scholar]
- 20.McAllister L.J., Bent S., Petty N.K., Skippington E., Beatson S.A., Paton J.C., Paton A.W. Genomic Comparison of two O111:H− enterohemorrhagic Escherichia coli isolates from a historic hemolytic-uremic syndrome outbreak in Australia. Infect. Immun. 2016;84:775–781. doi: 10.1128/IAI.01229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2004;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ethelberg S., Olsen K.E.P., Scheutz F., Jensen C., Schiellerup P., Engberg J., Petersen A.M., Olesen B., Gerner-Smidt P., Mølbak K. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 2004;10:842–847. doi: 10.3201/eid1005.030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazen T.H., Sahl J.W., Fraser C.M., Donnenberg M., Scheutz F., Rasko D.A. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc. Natl. Acad. Sci. USA. 2013;110:12810–12815. doi: 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller D., Benz I., Liebchen A., Gallitz I., Karch H., Schmidt M.A. Comparative analysis of the locus of enterocyte effacement and its flanking regions. Infect. Immun. 2009;77:3501–3513. doi: 10.1128/IAI.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellmann A., Bielaszewska M., Köck R., Friedrich A.W., Fruth A., Middendorf B., Harmsen D., Schmidt M.A., Karch H. Analysis of collection of hemolytic uremic syndrome–associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 2008;14:1287–1290. doi: 10.3201/eid1408.071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heiman K.E., Mody R.K., Johnson S.D., Griffin P.M., Gould L.H. Escherichia coli O157 outbreaks in the United States, 2003–2012. Emerg. Infect. Dis. 2015;21:1293–1301. doi: 10.3201/eid2108.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delannoy S., Beutin L., Fach P. Discrimination of enterohemorrhagic Escherichia coli (EHEC) from Non-EHEC strains based on detection of various combinations of Type III effector genes. J. Clin. Microbiol. 2013;51:3257–3262. doi: 10.1128/JCM.01471-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katani R., Cote R., Kudva I.T., Debroy C., Arthur T.M., Kapur V. Comparative genomics of two super-shedder isolates of Escherichia coli O157:H7. PLoS ONE. 2017;12:e0182940. doi: 10.1371/journal.pone.0182940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma V.K., Akavaram S., Schaut R.G., Bayles D.O. Comparative genomics reveals structural and functional features specific to the genome of a foodborne Escherichia coli O157:H7. BMC Genom. 2019;20:196. doi: 10.1186/s12864-019-5568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorenz S.C., Gonzalez-Escalona N., Kotewicz M.L., Fischer M., Kase J.A. Genome sequencing and comparative genomics of enterohemorrhagic Escherichia coli O145:H25 and O145:H28 reveal distinct evolutionary paths and marked variations in traits associated with virulence & colonization. BMC Microbiol. 2017;17:183. doi: 10.1186/s12866-017-1094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Liao Y.-T., Salvador A., Sun X., Wu V.C.H. Investigating the whole-genome sequence of a new locus of enterocyte effacement-positive Shiga toxin-producing Escherichia coli O157:H7 strain isolated from river water. Microbiol. Resour. Announc. 2020;9 doi: 10.1128/MRA.00112-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 33.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 34.Grant J.R., Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181-4. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyson S., Peterson C.-L., Olson A., Tyler S., Knox N., Griffiths E., Dooley D., Hsiao W., Cabral J., Johnson R.P., et al. Eleven high-quality reference genome sequences and 360 draft assemblies of Shiga toxin-producing Escherichia coli isolates from human, food, animal, and environmental sources in Canada. Microbiol. Resour. Announc. 2019;8:e00625-19. doi: 10.1128/MRA.00625-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel P.N., Lindsey R.L., Garcia-Toledo L., Rowe L.A., Batra D., Whitley S.W., Drapeau D., Stoneburg D., Martin H., Juieng P., et al. High-quality whole-genome sequences for 77 Shiga toxin-producing Escherichia coli strains generated with PacBio sequencing. Genome Announc. 2018;6:e00391-18. doi: 10.1128/genomeA.00391-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joensen K., Scheutz F., Lund O., Hasman H., Kaas R.S., Nielsen E.M., Aarestrup F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16-21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertelli C., Laird M., Williams K.P., Lau B.Y., Hoad G., Winsor G.L., Brinkman F. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alikhan N.-F., Petty N.K., Ben Zakour N., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Boil. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Boil. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogura Y., Ooka T., Iguchi A., Toh H., Asadulghani M., Oshima K., Kodama T., Abe H., Nakayama K., Kurokawa K., et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. USA. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Liao Y.-T., Salvador A., Sun X., Wu V.C. Prediction, diversity, and genomic analysis of temperate phages induced from Shiga toxin-producing Escherichia coli strains. Front. Microbiol. 2020;10:3093. doi: 10.3389/fmicb.2019.03093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W.-Y., Li Y.-J., Fan C. Different loci and mRNA copy number of the increased serum survival gene of Escherichia coli. Can. J. Microbiol. 2018;64:147–154. doi: 10.1139/cjm-2017-0363. [DOI] [PubMed] [Google Scholar]
- 48.McPeake S., Smyth J., Ball H. Characterisation of avian pathogenic (APEC) associated with colisepticaemia compared to faecal isolates from healthy birds. Veter- Microbiol. 2005;110:245–253. doi: 10.1016/j.vetmic.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Johnson T., Wannemuehler Y.M., Nolan L.K. Evolution of the iss Gene in Escherichia coli. Appl. Environ. Microbiol. 2008;74:2360–2369. doi: 10.1128/AEM.02634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mariani-Kurkdjian P., Denamur E., Milon A., Picard B., Cave H., Lambert-Zechovsky N., Loirat C., Goullet P., Sansonetti P.J., Elion J. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J. Clin. Microbiol. 1993;31:296–301. doi: 10.1128/JCM.31.2.296-301.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarr P.I., Fouser L.S., Stapleton A., Wilson R.A., Kim H.H., Vary J.C., Clausen C.R. Hemolytic–uremic syndrome in a six-year-old girl after a urinary tract infection with Shiga-toxin–producing Escherichia coli O103:H2. N. Engl. J. Med. 1996;335:635–638. doi: 10.1056/NEJM199608293350905. [DOI] [PubMed] [Google Scholar]
- 52.Marchès O., Nougayrede J.-P., Boullier S., Mainil J., Charlier G., Raymond I., Pohl P., Boury M., Rycke J., Milon A., et al. Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 2000;68:2171–2182. doi: 10.1128/IAI.68.4.2171-2182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bugarel M., Martin A., Fach P., Beutin L. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: A basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 2011;11:142. doi: 10.1186/1471-2180-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelly M., Hart E., Mundy R., Marchès O., Wiles S., Badea L., Luck S., Tauschek M., Frankel G., Robins-Browne R., et al. Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect. Immun. 2006;74:2328–2337. doi: 10.1128/IAI.74.4.2328-2337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cooper K.K., Mandrell R.E., Louie J.W., Korlach J., Clark T., Parker C.T., Huynh S., Chain P.S.G., Ahmed S., Carter M.Q. Complete genome sequences of two Escherichia coli O145:H28 outbreak strains of food origin. Genome Announc. 2014;2:e00482-14. doi: 10.1128/genomeA.00482-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y.-T., Brooks J.C., Martin J.N., Echeverry A., Loneragan G.H., Brashears M. Antimicrobial interventions for O157:H7 and non-O157 Shiga toxin–producing Escherichia coli on beef subprimal and mechanically tenderized steaks. J. Food Prot. 2015;78:511–517. doi: 10.4315/0362-028X.JFP-14-178. [DOI] [PubMed] [Google Scholar]
- 57.Wang J.-Y., Niu Y.D., Chen J., Anany H., Ackermann H.-W., Johnson R.P., Ateba C.N., Stanford K., Bie X. Feces of feedlot cattle contain a diversity of bacteriophages that lyse non-O157 Shiga toxin-producing Escherichia coli. Can. J. Microbiol. 2015;61:467–475. doi: 10.1139/cjm-2015-0163. [DOI] [PubMed] [Google Scholar]
- 58.Gyawali R., Ibrahim S.A. Natural products as antimicrobial agents. Food Control. 2014;46:412–429. doi: 10.1016/j.foodcont.2014.05.047. [DOI] [Google Scholar]
- 59.Oliveira P.H., Touchon M., Cury J., Rocha E.P.C. The chromosomal organization of horizontal gene transfer in bacteria. Nat. Commun. 2017;8:841. doi: 10.1038/s41467-017-00808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Touchon M., De Sousa J.A.M., Rocha E.P.C. Embracing the enemy: The diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 2017;38:66–73. doi: 10.1016/j.mib.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 61.Ramisetty B.C.M., Santhosh R.S. Horizontal gene transfer of chromosomal type II toxin–antitoxin systems of Escherichia coli. FEMS Microbiol. Lett. 2015;363:238. doi: 10.1093/femsle/fnv238. [DOI] [PubMed] [Google Scholar]
- 62.Jarvis K.G., Girón J.A., Jerse A.E., McDaniel T.K., Donnenberg M., Kaper J. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haugum K., Johansen J., Gabrielsen C., Brandal L.T., Bergh K., Ussery D., Drabløs F., Afset J.E. Comparative genomics to delineate pathogenic potential in non-O157 Shiga toxin-producing Escherichia coli (STEC) from patients with and without haemolytic uremic syndrome (HUS) in Norway. PLoS ONE. 2014;9:e111788. doi: 10.1371/journal.pone.0111788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michelacci V., Maugliani A., Tozzoli R., Corteselli G., Chiani P., Minelli F., Gigliucci F., Arancia S., Conedera G., Targhetta C., et al. Characterization of a novel plasmid encoding F4-like fimbriae present in a Shiga-toxin producing enterotoxigenic Escherichia coli isolated during the investigation on a case of hemolytic-uremic syndrome. Int. J. Med. Microbiol. 2018;308:947–955. doi: 10.1016/j.ijmm.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Lorenz S.C., Monday S.R., Hoffmann M., Fischer M., Kase J.A. Plasmids from Shiga toxin-producing Escherichia coli strains with rare enterohemolysin gene (ehxA) subtypes reveal pathogenicity potential and display a novel evolutionary path. Appl. Environ. Microbiol. 2016;82:6367–6377. doi: 10.1128/AEM.01839-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kleinheinz K., Joensen K., Larsen M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allison H. Stx-phages: Drivers and mediators of the evolution of STEC and STEC-like pathogens. Futur. Microbiol. 2007;2:165–174. doi: 10.2217/17460913.2.2.165. [DOI] [PubMed] [Google Scholar]
- 68.Krüger A., Lucchesi P.M.A. Shiga toxins and Stx phages: Highly diverse entities. Microbiology. 2015;161:451–462. doi: 10.1099/mic.0.000003. [DOI] [PubMed] [Google Scholar]
- 69.Canchaya C., Fournous G., Brüssow H. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 2004;53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- 70.Tobe T., Beatson S.A., Taniguchi H., Abe H., Bailey C.M., Fivian A., Younis R., Matthews S., Marches O., Frankel G., et al. An extensive repetoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.