Abstract
Mouse Ptchd3 (patched domain containing 3) was previously identified as a male germ-cell specific gene. The protein product of this gene has been found on the surface of mouse, rat and human sperm. Since Ptchd3 contains a conserved patched domain, we hypothesize that it functions as a membrane receptor for the hedgehog ligand. Herein, we used a Ptchd3 knockout mouse model to study its function in mouse development and spermatogenesis. We found that Ptchd3 knockout mice were born and lived normally. The fertility and sperm production of knockout males were not changed. Moreover, our data indicated that the expression levels of several hedgehog signaling genes were not affected in mutant testis. Taken together, these findings demonstrate that Ptchd3 is a non-essential gene in mouse development and spermatogenesis.
Keywords: Ptchd3, Hedgehog signaling, Spermatogenesis, Knockout, Male germ cells
Abbreviations: Ptchd3, patched domain containing 3; Hh, hedgehog; Dhh, desert hedgehog
1. Introduction
Evolutionarily conserved Hedgehog (Hh) signaling plays important roles in animal development as well as in various human cancers (Riobo and Manning 2007), through driving cell proliferation, promoting cell survival, and directing cell differentiation. Hh function is mediated through its membrane receptor Patched.
Desert hedgehog (Dhh), which is specifically expressed in testicular Sertoli cells, has been shown to play an essential role in spermatogenesis (Bitgood et al. 1996; Clark et al. 2000). Male mice with a Dhh null mutation are viable but sterile. Spermatogenesis in Dhh knockout males is blocked at the pachytene primary spermatocyte stage and, consequently, no mature sperm are produced. However, the molecular mechanism of Dhh function during spermatogenesis remains elusive.
We have previously classified a male germ cell-specific gene Ptchd3 (patched domain containing 3) (Fan et al. 2007), which is conserved among many organisms, including human, mouse and zebrafish. The Ptchd3 mRNA is detected in primary spermatocytes (leptotene, zygotene and pachytene) and secondary spermatocytes (round spermatids). The protein product of this gene has been found on the surface of mouse, rat and human sperm. Ptchd3 contains a conserved Patched domain and is predicted to have Hh receptor activity. Therefore, we hypothesize that male germ cell-specific Ptchd3 is an Hh receptor relaying the Dhh signal from Sertoli cells to developing germ cells and is required for spermatogenesis.
Herein, we test our hypothesis by generating and analyzing a Ptchd3 knockout mouse model. Our data demonstrate that Ptchd3 is not essential for mouse development, spermatogenesis or fertility.
2. Materials and methods
2.1. Chemicals
All chemicals were purchased from Sigma Aldrich (St. Louis, MO, USA) unless stated elsewhere.
2.2. Mice
The Ptchd3 knockout chimeric male mice were generated at the University of California- Davis under the project CSD 24758 of Knockout Mouse Project (https://www.komp.org/geneinfo.php?geneid=75664). The chimeric mice (from the BL3085–6 ES cell line), which contained a targeting allele of Ptchd3tm1a(KOMP)Wtsi, were transferred to Marshall University animal facility and crossed with C57BL/6 female mice to produce F1 Ptchd3+/− heterozygous mice. The F1 mice were inter-crossed to obtain F2 Ptchd3 homozygous knockout and wild-type littermate mice.
Mouse genotyping was performed by polymerase chain reaction (PCR) on tail genomic DNA. The primer pairs used to detect the knockout amplicon (389 bp) were Ptchd3-loxF (5’-GAGATGGCGCAACGCAATTAATG-3′) and Ptchd3-R (5′- CAACTGTATCCCTCAAGAAACAAGCC-3′). The other set of primers, Ptchd3-F (5′- GCATGGCTGACTCACTTTCTTGACC-3′) and Ptchd3-ttR (5′-.
GGGTTATATTTTGGGATTGCTGGCCC-3′) were used to detect the wild-type amplicon (543 bp).
The animal care and experiments described within were reviewed and approved by the Institutional Animal Care and Use Committee of Marshall University, and were performed in accordance with the Guiding Principles for Care and Use of Laboratory Animals.
2.3. Testis histology
These experiments were carried out as reported in our previous studies (Fan et al. 2006a). Briefly, testes were removed from four mice in both the wild-type and Ptchd3 homozygous knockout groups. The testes were immediately immersed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. After overnight fixation at 4 °C, the testes were dehydrated in alcohol and embedded in paraffin. The testis sections (6 μm in thickness), were stained with hematoxylin and eosin. Images of the sections were captured with a Leica DMI 4000B microscope with a CCD digital camera (Buffalo Grove, IL, USA).
2.4. Sperm count and morphological analysis
The cauda epididymis was removed from 10-weeks-old Ptchd3 knockout and wild-type littermate mice in the M199*2 medium [M199 (Invitrogen, Carlsbad, CA, USA), 3.5 mM sodium pyruvate, 1000 i.u. penicillin-streptomycin, 3.0% bovine serum albumin]. The cauda was incised several times and incubated at 37 °C, 5% CO2 for 15 min to allow sperm to release from the epididymis. Sperm were collected after a nylon-mesh filtration and counted with a hemocytometer. Sperm morphology was analyzed with a Leica DMI 4000B microscope equipped with a CCD digital camera (Buffalo Grove, IL, USA).
2.5. In vivo fertility assay
Each adult male mouse (12-weeks-old) was mated with two adult C57BL/6 females (10-weeks-old) for two weeks. The number of offspring from each pregnant female was counted after birth.
2.6. Reverse transcription-polymerase chain reaction (RT-PCR)
Testis total RNAs were isolated as reported previously (Fan et al. 2006b). RT-PCR was conducted by the random primer method as described in the RT-for-PCR kit (Clontech). The primers for β-actin were (5’-GTGGGCCGCTCTAGGCACCAA-3′ and 5’-CTCTTTGATGTC ACGCACGATTTC-3′). The primers FP1 (5′-CACCCAGCTCATCTACTTAGC-3′, located on exon 1) and RP2 (5′-GAGCAGGGTTGTTCCTGTATAG-3′, located on exon 4) were used to produce an amplicon of 700 bp to identify the long isoform Ptchd3b (Fan et al. 2007). The primers for Ptch1 were forward 5’-TATGCTCGCTCTGGAGCACA-3′ and reverse 5’-TCTGTGGCTTCCACAATCAC-3′. The primers for Ptch2 were forward 5′- CAATGATGACTGTGGAGCTC-3′ and reverse 5′- AGAACCAGCAAGCATGAGCA-3′. The primers for Smoothened were forward 5′- ACCTCAATGAACCCTCAGCT-3′ and reverse 5′- CTCAGCCTCCATTAGGTTAG-3′. The primers for Gli1 were forward 5′- TCGGAAGTCCTATTCACGCCTTGA-3′ and reverse 5′- CCATGCACTGTCTTCACGTGTTTG-3′. The primers for Wsb2 were forward 5’ TAAGCACGGTAAGCAGATCCAGGT-3′ and reverse 5′- CCAGATCCTGAGCAGCCTGTCATC-3′. PCR parameters: 94 °C for 2 min, 1 cycle; 94 °C for 20 s, 58 °C for 20 s, 72 °C for 90 s, 30 cycles; followed by a 6-min extension at 72 °C. All PCR products were analyzed on 1% agarose gel.
2.7. DNA sequencing and bioinformatics
The purified Ptchd3 RT-PCR products were sent to the genomic core facility at Marshall University for conventional sequencing. The DNA sequence, predicted protein sequence and predicted protein conserved domain were analyzed by CLC Main Workbench 6.0 (CLC bio).
2.8. Statistical analysis
Data were presented as mean ± standard deviation. Student's t-test was performed to evaluate pairwise differences (p < .05 was considered significant).
3. Results
3.1. Generation of Ptchd3 knockout mice
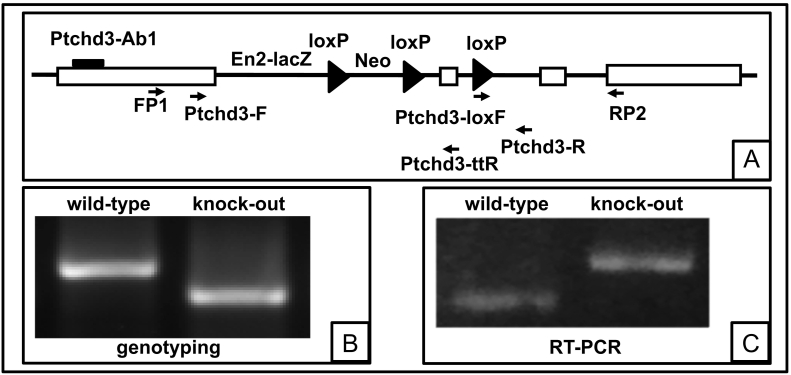
We previously reported that mouse Ptchd3 is a male germ-cell specific gene (Fan et al. 2007). To investigate whether Ptchd3 plays a role in mouse spermatogenesis and sperm function, we wanted to knock out this gene. The gene targeting vector (designated Ptchd3tm1a(KOMP)Wtsi) (the vector map is available at https://www.i-dcc.org/imits/targ_rep/alleles/9712/vector-image) was designed to create a mutant allele, which was initially a non-expressive form (conventional knockout first) but could be converted to a conditional allele (conditional knockout later) if lethal phenotype was observed (Fig. 1A). The crossing of Ptchd3 heterozygous mice produced Ptchd3 homozygous knockout mice with a normal Mendelian ratio (data not shown), indicating that Ptchd3 is not essential for embryonic development. Ptchd3 knockout mice grew and lived normally (data not shown), suggesting that Ptchd3 is not essential for overall health. Mouse genotyping was determined by PCR on tail genomic DNA (Fig. 1B). The size of wild-type and knock-out PCR product is 543 bp and 389 bp, respectively.
Fig. 1.
Generation of Ptchd3 knockout mice.
Panel A: A schematic diagram of the mouse Ptchd3 targeting allele Ptchd3tm1a(KOMP)Wtsi (KOMP project ID CSD24758). Exon 2 is flanked by the last two loxP sites. The positions of the PCR primers (Ptchd3-F/Ptchd3-ttR, Ptchd3-loxF/Ptchd3-R, and FP1/RP2) and the antigen (aa 131–150) recognized by the antibody (Ptchd3-Ab1) are illustrated.
Panel B: mouse genotyping by PCR on tail genomic DNA. Wild-type and knock-out mice were genotyped by the PCR primers Ptchd3-F/Ptchd3-ttR and Ptchd3-loxF/Ptchd3-R, respectively.
Panel C: RT-PCR analysis of Ptchd3 expression in wild-type and knock-out testes by the PCR primers FP1/RP2.
To confirm that the knockout mouse (which was identified by genotyping) indeed possessed mutated Ptchd3 gene, we carried out RT-PCR to examine the mRNA expression of Ptchd3. As shown in Fig. 1C, an expected PCR product of 700 bp was detected in the wild-type testis. In the knock-out testis, we could not detect the 700 bp wild-type PCR product. Instead, a mutated PCR product of ~800 bp was surprisingly found. To verify this result, the wild-type and mutated PCR products were purified and then subjected to conventional DNA sequencing. The sequencing data validated the 700 bp wild-type PCR product (data not shown). Meanwhile, the sequencing data also revealed that the ~800 bp mutated PCR product contained an insertion of 115 bp of mouse engrailed-2 (En2) gene, which was part of the original gene targeting vector (Fig. 1A and https://www.i-dcc.org/imits/targ_rep/alleles/9712/vector-image). The insertion was located between the exon 1 and exon 2 of Ptchd3 gene.
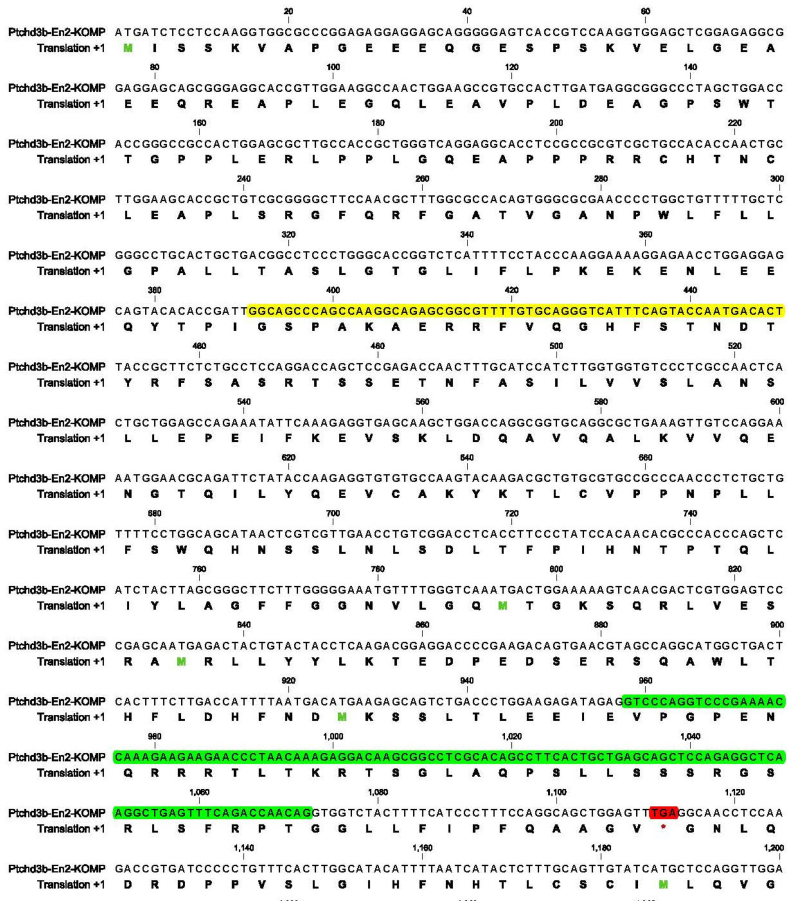
To find out what change of open reading frame might be caused by the 115 bp insertion, we first used bioinformatics tools to analyze the predicted protein sequence. As shown in Fig. 2, the 115 bp insertion (in green color) resulted in a partially mis-translated, truncated (stop codon in red color) protein of 370 amino acid residues. This mutant protein (named as Ptchd3b-En2-KOMP) does not contain a conserved Patched domain anymore and supposedly cannot function as a membrane receptor for the hedgehog ligand. As a comparison, wild-type Ptchd3b protein has 906 amino acid residues. The mutant protein Ptchd3b-En2-KOMP and the wild-type Ptchd3b protein possess the same first 320 amino acid residues. Thus, the 115 bp insertion in knockout mice caused loss of authentic Ptchd3 protein.
Fig. 2.
Predicted protein sequence of mutant Ptchd3. The 115 bp insertion (in green) affected authentic open reading frame and resulted in a partially mis-translated, truncated (stop codon in red color) protein of 370 amino acid residues. The antigen (aa 131–150) recognized by the antibody (Ptchd3-Ab1) is highlighted in yellow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We previously generated the rabbit polyclonal antibody Ptchd3-Ab1, which recognizes the amino acids 131–150 of Ptchd3 and can be used in sperm immunofluorescence assay (Fan et al. 2007). The mutant Ptchd3b-En2-KOMP protein is predicted to also contain the amino acids 131–150 of Ptchd3 (Fig. 2). Consistently, by using Ptchd3-Ab1, we were able to detect the same immunofluorescent signal in knockout sperm as in wild-type sperm (data not shown). Unfortunately, the attempt to apply Ptchd3-Ab1 in immunoblot assay was not successful.
3.2. Normal spermatogenesis and fertility in Ptchd3 knockout mice
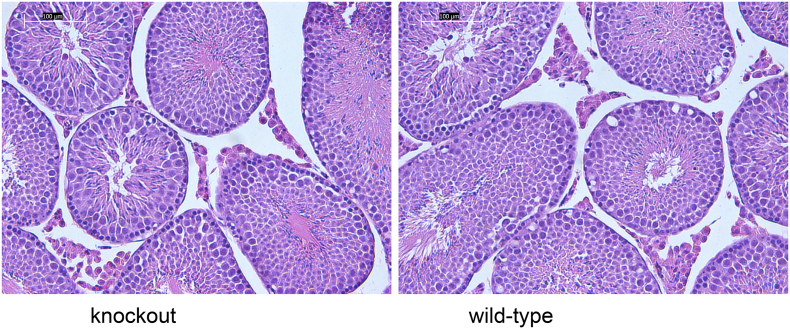
After successfully establishing a Ptchd3 knockout mouse model, we next investigated whether spermatogenesis and fertility were affected in mutant mice. We found that the morphology and weight of wild-type and knockout testes were not different (data not shown). Further testis histology analysis showed that mutant testes exhibited typical seminiferous tubules with different stages of spermatogenic cells (from spermatogonia to spermatozoa) (Fig. 3), suggesting that spermatogenesis is normal in Ptchd3 knockout testes.
Fig. 3.
Histological analysis of the testes. The testis sections (6 μm) of wild-type and Ptchd3 knockout mice were stained with hematoxylin and eosin (original magnification, x 20). The images were captured by a Leica DMI 4000B microscope. Normal spermatogenesis was seen in knockout testes.
We then examined cauda sperm and observed that, as compared with wild-type littermates, knockout mice produced sperm in the same number (Table 1, P = .27) and in the same morphology (data not shown). Finally, we conducted in vivo fertility assay. The result indicated that knockout males sired offspring as competently as wild-type littermates (Table 1, P = .31).
Table 1.
Cauda sperm number and in vivo fertility assay.
| Male genotype | Wild-type | Ptchd3 knockout |
|---|---|---|
| Cauda sperm number (X105) | 188 ± 22 (5)a | 179 ± 27 (5)a |
| Sired litter size | 5.3 ± 0.8 (6)b | 5.4 ± 0.9 (6)b |
Cauda sperm number and in vivo fertility assay was performed as described in the Materials and methods section. All mated females were pregnant. The number of tested males is indicated in parentheses. aP = 0.27. bP = 0.31.
Taken together, these results (Fig. 3 and Table 1) demonstrate that Ptchd3 is not essential for mouse spermatogenesis and male fertility, although we cannot rule out minor defects beyond our observation.
3.3. Normal expression of hedgehog signaling components in Ptchd3 knockout testis
We initially hypothesized that Ptchd3 might function as a membrane receptor for dessert hedgehog (Dhh), which is essential for mouse spermatogenesis (Bitgood et al. 1996). Hence, we wanted to determine if any biochemical changes in the hedgehog signaling pathway took place in Ptchd3 knockout testis. To address this, we performed RT-PCR to examine expression of several hedgehog signaling components. As shown in Fig. 4, the expression levels of membrane receptor Ptch1 and Ptch2, membrane signal transducer Smoothened, transcription factor Gli-1 and target gene Wsb2 (Sarraj et al. 2007) were not altered in Ptchd3 knockout testis, suggesting that Ptchd3 is not essential for hedgehog signaling in testis.
Fig. 4.
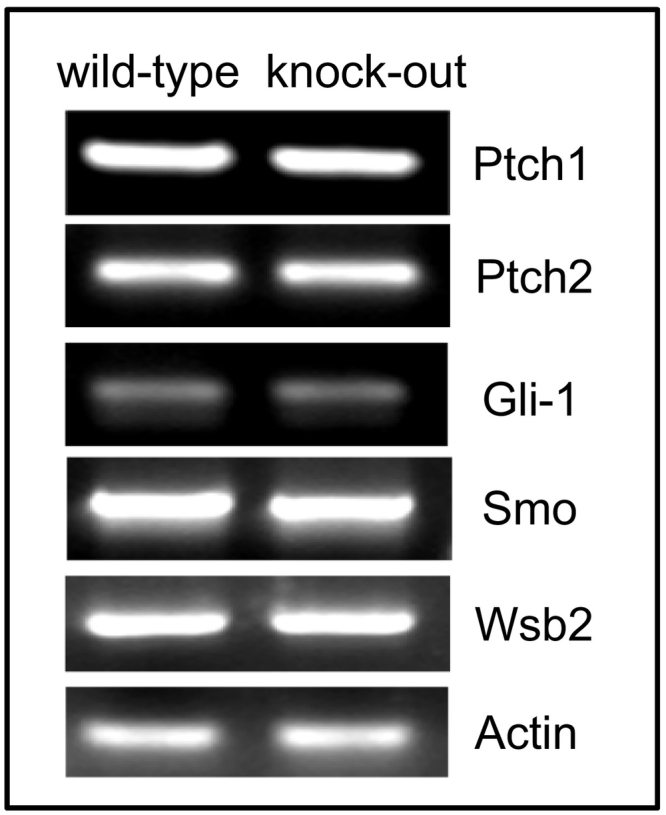
RT-PCR analysis of hedgehog signaling components in wild-type and Ptchd3 knockout testis. Beta-actin was used as the RT-PCR control. The experiment was repeated four times.
4. Discussion
Our results demonstrate that mouse Ptchd3 is not an essential gene in embryonic development, postnatal life, testis development, spermatogenesis, and sperm physiology (morphology and fertility). These findings are somewhat surprising, since Ptchd3 gene is conserved in many organisms (Geer et al. 2010) and its protein is found on the mid-piece of mouse, rat and human sperm (Fan et al. 2007). However, our result indeed endorses a previous study on human PTCHD3 (Ghahramani Seno et al. 2011), which utilized population genomic screen to demonstrate that human PTCHD3 is a non-essential gene.
Based on protein domain structure, Ptchd3 belongs to the Patched (Ptch) family, which is the membrane receptor for hedgehog ligands (including sonic hedgehog, Indian hedgehog and dessert hedgehog) and has 6 members identified so far (including Ptch1, Ptch2, Ptchd1, Ptchd2, Ptchd3 and Ptchd4) (Geer et al. 2010). Hence, genetic redundancy may compensate the loss of one particular family member, such as Ptchd3 in this study. With this regard, Ptch1 and Ptch2 indeed have been shown to be expressed in developing germ cells in testis (Morales et al. 2009; Szczepny et al. 2006) and may functionally exchange with Ptchd3. Interestingly, previous studies also showed that Ptch2 null mice were fertile (Carpenter et al. 1998; Nieuwenhuis et al. 2006). Thus, it is possible that there are multiple Dhh receptors in male germ cells and losing anyone of them does not visibly compromise testis development, spermatogenesis and fertility.
Interestingly, genetic redundancy for the Ptch family members has been reported lately. Adolphe and colleagues studied null Ptch1, null Ptch2 and double mutant mice, in order to assess the function of the Ptch family members in epidermal development (Adolphe et al. 2014). They found that null Ptch1 alone produced some defects in epidermal development, but the cells were still able to develop eventually. However, the loss of both Ptch1 and Ptch2 inhibited the epidermal lineage specification and differentiation (Adolphe et al. 2014).
Thus far, Ptchd3 is the only Patched (Ptch) family member that has been shown on sperm (Fan et al. 2007). Since our data reveal that deletion of Ptchd3 does not affect sperm physiology, we predict that other Ptch family member(s) may be present on sperm. This interesting prediction needs to be addressed in the future. In addition, future studies are needed to determine whether Ptchd1, Ptchd2 and Ptchd4 are also expressed in male germ cells.
The gene targeting vector Ptchd3tm1a(KOMP)Wtsi was designed to create a Knockout First allele that was initially a non-expressive form, since of a splicing acceptor site at the end of mouse engrailed-2 (En2) gene and a poly(A) addition site at the end of lacZ gene (Fig. 1A and https://www.i-dcc.org/imits/targ_rep/alleles/9712/vector-image). However, we were able to detect the expression of a mutant mRNA in reverse-transcription polymerase chain reaction (Fig. 1B) and a mutant protein in sperm immunofluorescence assay (data not shown). The cause for this discrepancy is unknown.
Although the mutant protein Ptchd3b-En2-KOMP (370 amino acid residues) and the wild-type Ptchd3b protein (906 amino acid residues) possess the same first 320 amino acid residues, it is unlikely that the mutant protein can function as a Patched receptor, since the full Patched domain in the wild-type protein includes 127–906 amino acids. Nonetheless, our data cannot rule out the possibility that the mutant protein might perform an unknown, non-Patched related function.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We wish to thank Dr. Huahai Chen for initial maintenance of the Ptchd3 knockout mouse colony.
Funding
This work was supported in part by the West Virginia state EPSCoR Research Challenge Fund, NASA West Virginia Space Grant Consortium, and by a grant from the National Institute of Child Health and Human Development (1R15HD062979).
Author contributions
G-Z. Zhu designed research; S.G., C.L., H.B., and G-Z. Zhu performed research; S.G., C.L., H.B., and G-Z. Zhu analyzed data; and S.G. and G-Z. Zhu wrote the paper.
References
- Adolphe C., Nieuwenhuis E., Villani R., Li Z., Kaur P., Hui C.C. Patched 1 and patched 2 redundancy has a key role in regulating epidermal differentiation. J Invest Dermatol. 2014;(7):1981–1990. doi: 10.1038/jid.2014.63. [DOI] [PubMed] [Google Scholar]
- Bitgood M.J., Shen L., McMahon A.P. Sertoli cell signaling by desert hedgehog regulates the male germline. Curr. Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- Carpenter D., Stone D.M., Brush J., Ryan A., Armanini M., Frantz G. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13630–13634. doi: 10.1073/pnas.95.23.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.M., Garland K.K., Russell L.D. Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biol. Reprod. 2000;63:1825–1838. doi: 10.1095/biolreprod63.6.1825. [DOI] [PubMed] [Google Scholar]
- Fan J., Akabane H., Graham S.N., Richardson L.L., Zhu G.Z. Sperm defects in mice lacking a functional Niemann-Pick C1 protein. Mol. Reprod. Dev. 2006;73:1284–1291. doi: 10.1002/mrd.20559. [DOI] [PubMed] [Google Scholar]
- Fan J., Graham M., Akabane H., Richardson L.L., Zhu G.Z. Identification of a novel male germ cell-specific gene TESF-1 in mice. Biochem. Biophys. Res. Commun. 2006;340:8–12. doi: 10.1016/j.bbrc.2005.11.152. [DOI] [PubMed] [Google Scholar]
- Fan J., Akabane H., Zheng X., Zhou X., Zhang L., Liu Q. Male germ cell-specific expression of a novel patched-domain containing gene Ptchd3. Biochem. Biophys. Res. Commun. 2007;363:757–761. doi: 10.1016/j.bbrc.2007.09.047. [DOI] [PubMed] [Google Scholar]
- Geer L.Y., Marchler-Bauer A., Geer R.C., Han L., He J., He S. The NCBI BioSystems database. Nucleic Acids Res. 2010:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahramani Seno M.M., Kwan B.Y., Lee-Ng K.K., Moessner R., Lionel A.C., Marshall C.R. Human PTCHD3 nulls: rare copy number and sequence variants suggest a non-essential gene. BMC Med Genet. 2011;12:45. doi: 10.1186/1471-2350-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales C.R., Fox A., El-Alfy M., Ni X., Argraves W.S. Expression of Patched-1 and smoothened in testicular meiotic and post-meiotic cells. Microsc. Res. Tech. 2009;72:809–815. doi: 10.1002/jemt.20733. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E., Motoyama J., Barnfield P.C., Yoshikawa Y., Zhang X., Mo R. Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia. Mol. Cell. Biol. 2006;26:6609–6622. doi: 10.1128/MCB.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo N.A., Manning D.R. Pathways of signal transduction employed by vertebrate hedgehogs. Biochem. J. 2007;403:369–379. doi: 10.1042/BJ20061723. [DOI] [PubMed] [Google Scholar]
- Sarraj M.A., McClive P.J., Szczepny A., Daggag H., Loveland K.L., Sinclair A.H. Expression of Wsb2 in the developing and adult mouse testis. Reproduction. 2007;133:753–761. doi: 10.1530/rep.1.01184. [DOI] [PubMed] [Google Scholar]
- Szczepny A., Hime G.R., Loveland K.L. Expression of hedgehog signalling components in adult mouse testis. Dev. Dyn. 2006;235:3063–3070. doi: 10.1002/dvdy.20931. [DOI] [PubMed] [Google Scholar]