Abstract
Background
The accurate identification of the exon/intron boundaries is critical for the correct annotation of genes with multiple exons. Donor and acceptor splice sites (SS) demarcate these boundaries. Therefore, deriving accurate computational models to predict the SS are useful for functional annotation of genes and genomes, and for finding alternative SS associated with different diseases. Although various models have been proposed for the in silico prediction of SS, improving their accuracy is required for reliable annotation. Moreover, models are often derived and tested using the same genome, providing no evidence of broad application, i.e. to other poorly studied genomes.
Results
With this in mind, we developed the Splice2Deep models for SS detection. Each model is an ensemble of deep convolutional neural networks. We evaluated the performance of the models based on the ability to detect SS in Homo sapiens, Oryza sativa japonica, Arabidopsis thaliana, Drosophila melanogaster, and Caenorhabditis elegans. Results demonstrate that the models efficiently detect SS in other organisms not considered during the training of the models. Compared to the state-of-the-art tools, Splice2Deep models achieved significantly reduced average error rates of 41.97% and 28.51% for acceptor and donor SS, respectively. Moreover, the Splice2Deep cross-organism validation demonstrates that models correctly identify conserved genomic elements enabling annotation of SS in new genomes by choosing the taxonomically closest model.
Conclusions
The results of our study demonstrated that Splice2Deep both achieved a considerably reduced error rate compared to other state-of-the-art models and the ability to accurately recognize SS in other organisms for which the model was not trained, enabling annotation of poorly studied or newly sequenced genomes. Splice2Deep models are implemented in Python using Keras API; the models and the data are available at https://github.com/SomayahAlbaradei/Splice_Deep.git.
Abbreviations: Acc, accuracy; AcSS, acceptor splice site; AUC, area under curve; CNN, convolutional neural network; CONV, convolutional layers; DNA, deoxyribonucleic acid; DL, deep learning; DoSS, donor splice site; DT, decision trees; FC, fully connected layer; ML, machine learning; NB, naive Bayes; NN, neural network; POOL, pooling layer; ReLU, rectified linear unit layer; RF, random forest; RNA, ribonucleic acid; Sn, sensitivity; SS, splice site; Sp, specificity; SVM, support vector machine
Keywords: Splice sites, Splicing, Deep-learning, Prediction, Bioinformatics
1. Background
Variability in intron-exon boundaries enables alternative splicing. Alternative splicing affords eukaryotic organisms the ability to generate various transcripts and encode multiple proteins from the same gene locus. During the transcription process, introns are spliced out, and different combinations of exons join together to form mature RNA. When mature RNA contains information for protein synthesis, we denote it as a messenger RNA (mRNA). Thus, the accurate annotation of the exon and intron boundaries, referred to as splice sites (SS), is critical for the correct annotation of genes contained in a genomic DNA sequence. The SS at the exon/intron boundary corresponds to the donor site (DoSS), while the SS at the intron/exon boundary is the acceptor site (AcSS). The DoSS and AcSS are characterized in most cases by canonical GT and AG dinucleotides in DNA (Burset et al., 2000), respectively. In silico detection of SS facilitates delineating the internal structure of multi-exon genes, functional annotation of new genomes, and finding alternative SS linked to aberrant organism phenotype and diseases (Leegwater et al., 2016; Roshon et al., 2003; Morrison et al., 2013; Kurmangaliyev and Gelfand, 2008; Iso-Touru et al., 2019; Buckley et al., 2009; Sahakyan and Balasubramanian, 2016).
Current computational approaches for SS detection implement different strategies for feature extraction and deploy various machine learning (ML) methods to build classifiers. Although some approaches implemented methods based on probabilistic models (Pertea et al., 2001; Zhang et al., 2010), the majority of the implemented methods are based on ML techniques. Feature extraction and feature selection are among the main challenges for deriving robust ML models for SS prediction.
Several feature extraction techniques were proposed to characterize SS. For example, as a pre-processing step, Markov models are used to extract nucleotide dependencies and sequence compositional features surrounding SS in terms of probabilistic parameters that were used as features to characterize the SS (Baten et al., 2006; Goel et al., 2015). Information related to mono/di/trinucleotide distribution surrounding the SS in DNA sequences is also used as features to characterize the sequences surrounding the SS (Wei et al., 2013; Huang et al., 2006), as well as the chemical properties of each nucleotide, and positional and density information (Bari et al., 2014). A combination of features based on dinucleotide association score, nucleotide dependency, and positions was also used to describe SS (Meher et al., 2016a).
Notably, some of the extracted features may be irrelevant or redundant in the SS prediction task and could result in an unnecessarily more complex ML model. Therefore, feature selection is a common step that follows feature extraction (Bengio et al., 2012; Bins, 2002; Magana-Mora and Bajic, 2017; Alshahrani et al., 2017). For example, feature selection based on the F-score was used to identify informative features for improving model performance (Wei et al., 2013).
Different ML techniques that use such “manually-crafted” features as inputs are then applied to perform the SS prediction task; examples are, support vector machine (SVM) (Baten et al., 2006; Goel et al., 2015; Wei et al., 2013; Huang et al., 2006; Bari et al., 2014; Chen and Lin, 2006; Dror et al., 2004; Li et al., 2017; Meher et al., 2016b; Sonnenburg et al., 2007), random forest (RF) (Meher et al., 2016a), decision trees (DT) (Lopes et al., 2007), and naïve Bayes (NB) (Kamath et al., 2014) models. Despite the great success of these ML techniques, the extraction and selection of features from raw data are not straightforward. The main problem remains in difficulty associated with extracting sufficiently discriminative information from raw data. As a consequence, recent approaches used deep learning (DL) techniques to avoid the limitations associated with the “manually-crafted” features. The use of DL models has achieved exceptional results in different genomic-related studies (Zeng et al., 2016; Nguyen et al., 2016; Leung et al., 2014; Alipanahi et al., 2015; Quang and Xie, 2016; Sønderby et al., 2015; Lee and Yoon, 2015; Albalawi et al., 2019; Kalkatawi et al., 2019; Magana-Mora et al., 2017). Concerning SS, DL models have been used for automatic feature extraction and detection of SS in RNA (Zhang et al., 2016) and DNA (Zuallaert et al., 2018; Du et al., 2018). DeepSS (Du et al., 2018) is a recent DL model trained and tested on Homo sapiens and Caenorhabditis elegans datasets to predict SS. It consists of two stacked convolution-pooling layers followed by two fully-connected layers, where the last layer performs a SoftMax function that outputs a prediction score. DeepSS outperformed different previously proposed state-of-the-art models (Goel et al., 2015; Meher et al., 2016b; Meher et al., 2014). Another recent DL model, Splicerover (Zuallaert et al., 2018), was trained and tested on H. sapiens and Arabidopsis thaliana datasets for SS prediction. This model implements several alternating convolutional pooling layers followed by a fully-connected layer with a SoftMax function that outputs a prediction score. Splicerover also outperformed several state-of-the-art models (Bari et al., 2014; Sonnenburg et al., 2007; Lee and Yoon, 2015; Degroeve et al., 2004).
Although considerable improvements have been achieved using DL models, there is a need to further improve SS characterization to increase the accuracy of the models and improve models that detect splice junctions (Jaganathan et al., 2019; Zhang et al., 2018). The DL models that predict SS were also developed based on a limited number of selected organisms. In this study, we derived independent models for five different organisms, namely, H. sapiens, A. thaliana, Oryza sativa japonica, Drosophila melanogaster, and C. elegans. We then performed cross-organism validation to assess the capability of the models to accurately predict SS in other organisms, not explicitly used to train the models. Therefore, the models can be used to annotate SS in new genomes by choosing the SS detection model trained on the most closely related organism. Finally, it is essential to mention that our model, Splice2Deep, aims to recognize the SS in the primary DNA genomic sequence that would be transcribed into donor/acceptor sites in the corresponding RNA, although we may not always make that distinction explicitly in the text.
2. Results
The main contribution of this study is the development of Splice2Deep that consists of an ensemble of DL models for the improved and generic detection of SS. To evaluate the performance of Splice2Deep, we derived separate models. We computed the statistical measures shown in Table 1 for the five considered organisms, namely: H. sapiens, A. thaliana, O. sativa japonica, D. melanogaster, and C. elegans. Moreover, we also report the results using the area under the receiver operating characteristic curve (AUC). Table 2 shows the performance obtained for the five different considered organisms using 60%, 15%, and 25% of the data as training, validation, and testing, respectively.
Table 1.
Statistical measures used to assess the performance of the models.
| Measure | Equation |
|---|---|
| Accuracy (Acc) | |
| Specificity (Sp) | |
| Sensitivity (Sn) | |
| F1 Score (F1) | |
| Error rate | 1 − Accuracy |
Table 2.
Performance metrics for the detection of donor and acceptor SS by Splice2Deep on five organisms.
| Organism | Acc | Sp | Sn | F1 | AUC | |
|---|---|---|---|---|---|---|
| AcSS | Homo sapiens | 96.91 | 97.80 | 95.61 | 96.91 | 98.69 |
| Arabidopsis thaliana | 95.21 | 94.86 | 95.53 | 95.22 | 98.31 | |
| Oryza sativa japonica | 93.89 | 93.62 | 94.16 | 93.92 | 97.52 | |
| Drosophila melanogaster | 94.07 | 95.04 | 94.09 | 94.07 | 98.16 | |
| Caenorhabditis elegans | 98.08 | 97.78 | 98.38 | 98.09 | 99.49 | |
| DoSS | Homo sapiens | 97.38 | 98.83 | 95.93 | 96.38 | 99.10 |
| Arabidopsis thaliana | 95.59 | 95.67 | 95.50 | 95.58 | 98.69 | |
| Oryza sativa japonica | 94.33 | 94.41 | 94.25 | 94.33 | 98.30 | |
| Drosophila melanogaster | 90.52 | 93.71 | 90.46 | 91.52 | 96.56 | |
| Caenorhabditis elegans | 97.68 | 97.74 | 97.63 | 97.69 | 99.48 |
2.1. Comparing Splice2Deep to other SS detection models
To gain more insight into the performance and value of Splice2Deep, we compared its accuracy to that of other available tools, namely: GeneSplicer (Pertea et al., 2001), SplicePredictor (Brendel et al., 2004), DeepSS (Du et al., 2018), and Splicerover (Zuallaert et al., 2018). Table 3 shows the accuracy obtained from the tools using the same testing data used to evaluate Deep2Splice. Some of the considered organisms were not used to train the existing tools, and a method was not provided to re-train the model. These cases are indicated using N/A (see Table 3). We also provide the results for additional statistical measures, such as Sp and Sn (see Additional file 1: Table S1).
Table 3.
Comparing the SS prediction accuracy of Splice2Deep and state-of-the-art tools using five well-studied organisms. Results in bold represent the best performing model. N/A indicates that the tool has not and cannot be trained for that specific organism.
| Organism | Gene-Splicer | Splice-Predictor | DeepSS | Splicerover | Splice2Deep | |
|---|---|---|---|---|---|---|
| AcSS | Homo sapiens | 83.31 | 88.01 | 94.85 | 95.35 | 96.91 |
| Arabidopsis thaliana | 87.76 | 92.13 | N/A | 94.35 | 95.21 | |
| Oryza sativa japonica | 84.21 | 89.42 | N/A | N/A | 93.89 | |
| Drosophila melanogaster | 88.66 | 88.69 | N/A | N/A | 94.07 | |
| Caenorhabditis elegans | N/A | N/A | 93.32 | N/A | 98.08 | |
| DoSS | Homo sapiens | 79.48 | 88.2 | 94.76 | 96.18 | 97.38 |
| Arabidopsis thaliana | 90.85 | 92.49 | N/A | 94.25 | 95.59 | |
| Oryza sativa japonica | 86.17 | 87.5 | N/A | N/A | 94.33 | |
| Drosophila melanogaster | 90.19 | 88.79 | N/A | N/A | 90.52 | |
| Caenorhabditis elegans | N/A | N/A | 94.01 | N/A | 97.68 |
Table 4 provides a comparison of the error rate, defined as error rate = 1 − Acc, and the relative improvement of the error rate compared to the best performing tool for each organism. The Splice2Deep model consistently reduced the relative error rate compared to the other state-of-the-art models.
Table 4.
Relative error rates associated with SS detection when using Splice2Deep and the best performing SS prediction tools.
| Splice site | Organism | Best performing model | Error rate of the best performing model (%) | Error rate of Deep2Splice (%) | Relative error rate reduction (%) |
|---|---|---|---|---|---|
| AcSS | Homo sapiens | Splicerover | 4.65 | 3.09 | 33.55 |
| Arabidopsis thaliana | Splicerover | 5.65 | 4.79 | 15.22 | |
| Oryza Sativa japonica | SplicePredictor | 10.58 | 6.11 | 42.25 | |
| Drosophila melanogaster | SplicePredictor | 11.31 | 5.93 | 47.57 | |
| Caenorhabditis elegans | DeepSS | 6.68 | 1.92 | 71.26 | |
| Average | 41.97 | ||||
| DoSS | Homo sapiens | Splicerover | 3.82 | 2.62 | 45.80 |
| Arabidopsis thaliana | Splicerover | 5.75 | 4.41 | 23.30 | |
| Oryza Sativa japonica | SplicePredictor | 12.50 | 5.67 | 54.64 | |
| Drosophila melanogaster | SplicePredictor | 9.81 | 9.48 | 3.36 | |
| Caenorhabditis elegans | DeepSS | 5.95 | 5.03 | 15.46 | |
| Average | 28.51 | ||||
3. Discussion
To make Splice2Deep generic, we trained the incorporated models on five well-studied organisms, and performed cross-organism model validation. For each of the five considered organisms, we developed two independent DL models: one to predict AcSS and the other to predict DoSS. Independent DL models were used to characterize the local upstream flanking region, local downstream flanking region, and the region surrounding the SS to derive an ensemble of DL models. In other words, we extracted features from two local windows and one window surrounding the SS to improve the performance of the DL models. We also exploit the fact that exons (if they contain a portion of the protein-coding sequence) typically exhibit a periodicity of three, where this appeared upstream of DoSS and downstream of AcSS. From the results shown in Table 3, Deep2Splice consistently outperformed the state-of-the-art models for the five different considered organisms and was able to reduce the relative error rate by 41.97% and 28.51% compared to the best performing model for AcSS and DoSS, respectively (Table 4). Thus, Deep2Splice providing more accurate SS prediction should enable better characterization of the internal structure of multi-exon genes and should be useful for finding alternative SS linked to aberrant organism phenotype and diseases. However, the real advantage of Deep2Splice is its proven ability for SS cross-organism prediction, which demonstrates it could be used to annotate organisms beyond the well-studied ones.
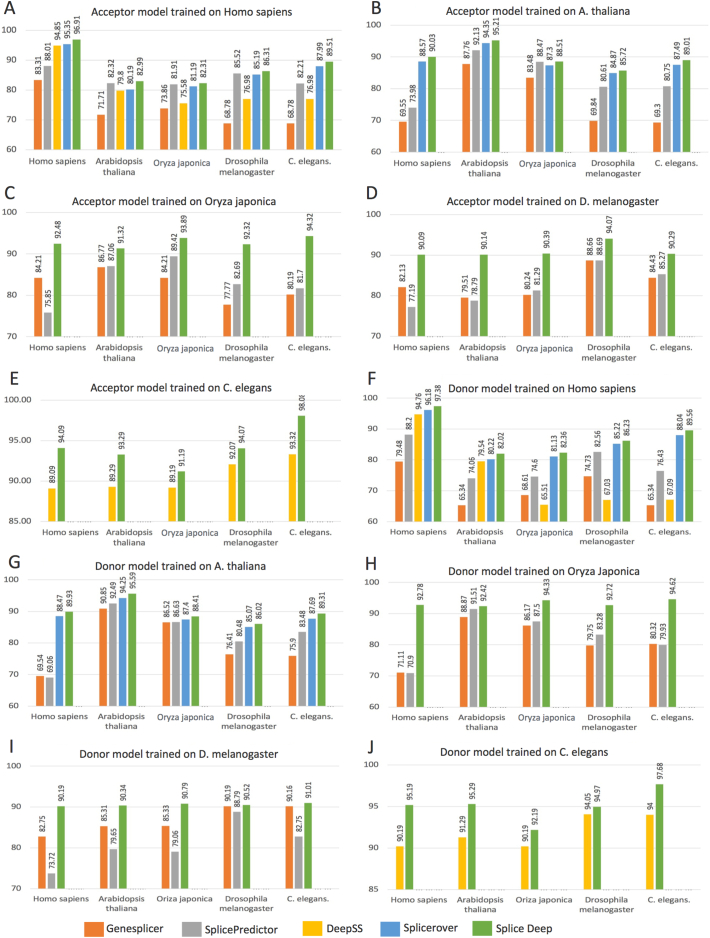
Evaluating the performance of the models for the annotation of poorly studied organisms is not possible as we do not have any reference to compare or compute any statistical measure of performance. On the other hand, by performing cross-organism model testing, i.e., testing the model with data from an organism for which it was not trained, we may approximately estimate the generalization capabilities of the model for annotating diverse organisms. For example, a cross-organism validation consists of testing a model derived from A. thaliana on the other four genomes for which it was not trained (H. sapiens, C. elegans, O. sativa japonica, and D. melanogaster) and perform the same test with other tools that are applicable (i.e., that were trained on A. thaliana as well). We performed this cross-organism validation for each of the five derived models for each organism. Fig. 1 shows the accuracy results for the cross-organism model validation, and Additional file 2: Table S2, Additional file 3: Table S3, Additional file 4: Table S4, Additional file 5: Table S5, Additional file 6: Table S6: Tables S2–S6 show the results for the other considered performance measures. Interestingly, in some cases, Splice2Deep trained on organism X and tested on organism Y achieved better results than other tools trained on organism Y and tested on the same organism Y. For instance, Splice2Deep model trained on C. elegans achieved an accuracy of 94.07% when tested on D. melanogaster (Fig. 1E), compared to 88.69% achieved by SplicePredictor model trained and tested on D. melanogaster (Fig. 1D). Similarly, Splice2Deep model trained on C. elegans achieved an accuracy of 91.19% when tested on O. sativa japonica (Fig. 1E), compared to 89.42% achieved by SplicePredictor model trained and tested on O. sativa japonica (Fig. 1C). These results demonstrate that Splice2Deep is more adapt to annotate the poorly studied or newly sequenced genomes than the other models.
Fig. 1.
Accuracy results obtained from the cross-organism model validation. A–E) Cross-organism validation results for the prediction of AcSS, F–J) cross-organism validation results for the prediction of DoSS.
4. Conclusions
Interest in the prediction of genomic signals goes far beyond model organisms. Therefore, a critical problem associated with many genomic signal prediction tools is that predicting SS in genomes that were not used to train the models may not be sufficiently accurate. The lack of evidence for the broader application of such models limits its use despite the conceptual promise such models hold. In this study, we addressed this problem of practical usability by firstly deriving more accurate prediction models using DL; and secondly, by developing five different models broadly tested in a cross-organism manner. The results showed that our models were able to capture conserved splicing mechanisms. Splice2Deep exhibits an average error rate reduction of 41.97% for AcSS and 28.51% for DoSS relative to the current state-of-the-art models. Moreover, Splice2Deep allows users to select the taxonomically closest organism of interest, making our models both accurate as well as useful. One possible avenue for further improvement of the usability of Splice2Deep would be to add additional organism models, which could be a subject of future work.
5. Methods
5.1. Datasets
In this study, we extracted genomic DNA sequences surrounding the SS from five different organisms (H. sapiens, A. thaliana, O. sativa japonica, D. melanogaster, and C. elegans), using their respective gene annotation available at Ensembl (Zerbino et al., 2018). Table 5 describes the used annotation for each organism and the number of positive (true) SS samples. For each annotated AcSS and DoSS, bedtools (Quinlan and Hall, 2010) were used to extract the surrounding DNA sequences of the considered SS. We extracted from both upstream and downstream flanking segments of SS 300 nucleotides. This resulted in a sequence of 602 nucleotides (300-SS-300). Sequences with false SS (i.e., those dinucleotides that are the same as SS but are not involved in the splice site machinery) were extracted. In accordance to previous studies for the detection of genomic signals (Albalawi et al., 2019; Kalkatawi et al., 2019; Magana-Mora et al., 2017; Xie et al., 2013; Ashoor et al., 2011; Magana-Mora et al., 2013; Kalkatawi et al., 2012; Kalkatawi et al., 2013), we selected the number of false SS to match the number of the true SS as determined by the available genome annotation (i.e., equal to the number of positive samples). False SS (negative) samples were mined from the chromosomes whose average GC content is most close to the average GC content of the considered genome. Therefore, false SS were thus obtained from chromosomes 21, 2, 1, 2L, and I from H. sapiens, A. thaliana, O. sativa japonica, D. melanogaster, and C. elegans, respectively.
Table 5.
Annotation for each organism and the number of positive and negative SS samples.
| Organism | Number of sequences | Assembly & Genebuild reference | |
|---|---|---|---|
| DoSS | Homo sapiens | 250,400 (true) 250,400 (false) |
GRCh38.p12 (Zerbino et al., 2018) |
| Arabidopsis thaliana | 110,299 (true) 110,299 (false) |
TAIR10 (Cheng et al., 2016) | |
| Oryza sativa japonica | 103,426 (true) 103,426 (false) |
IRGSP-1.0 (Sakai et al., 2013) | |
| Drosophila melanogaster | 30,118 (true) 30,118 (false) |
BDGP6.22 (Thurmond et al., 2019) | |
| Caenorhabditis elegans | 77,387 (true) 77,387 (false) |
WBcel235 (Lee et al., 2017) | |
| AcSS | Homo sapiens | 248,150 (true) 248,150 (false) |
GRCh38.p12 (Zerbino et al., 2018) |
| Arabidopsis thaliana | 112,318 (true) 112,318 (false) |
TAIR10 (Cheng et al., 2016) | |
| Oryza sativa japonica | 104,028 (true) 104,028 (false) |
IRGSP-1.0 (Sakai et al., 2013) | |
| Drosophila melanogaster | 28,703 (true) 28,703 (false) |
BDGP6.22 (Thurmond et al., 2019) | |
| Caenorhabditis elegans | 77,763 (true) 77,763 (false) |
WBcel235 (Lee et al., 2017) |
5.2. The Splice2Deep method
In what follows, we explain the most important facets of developing a robust DL model. We explain the strategy of selecting the appropriate data representation, determining the configuration of the DL model, as well as search strategy within the large hyperparameter space aiming to find the optimized set of model parameters.
5.2.1. Representation of data
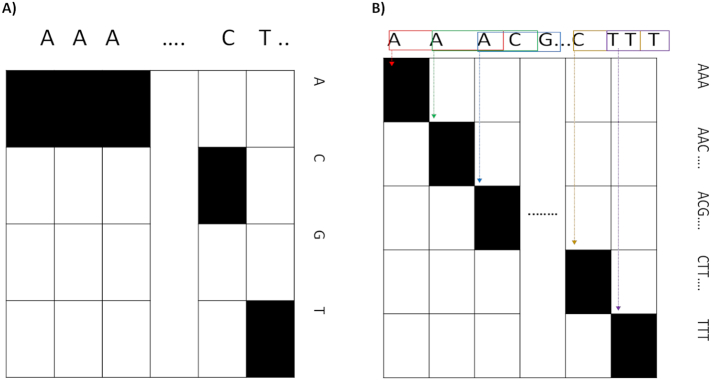
In order to develop a reliable model, we first represent each sequence window with appropriate embedding based on whether this window is found within an exon or intron region. Therefore, we assumed that the upstream region of a DoSS frequently contains coding sequences, while its downstream region consists of non-coding sequences. Conversely, the upstream region of an AcSS consists of non-coding sequences, while its downstream region frequently contains coding sequences. Based on this consideration, in the case of a candidate DoSS, we embedded the upstream window sequence in a two-dimensional (2D) space with trinucleotides, while embedding the downstream window sequence (a non-coding region) in a 2D space with single nucleotides. For AcSS, we embedded the upstream window sequence (a non-coding region) in a 2D space with single nucleotides, while embedding the downstream window sequence in a 2D space with trinucleotides. Finally, we embedded the surrounding window sequence in a 2D space with single nucleotides. This embedding process is shown in Fig. 2A. The 2D space embedding for mononucleotide is a 4 × L matrix, where L is the length of the window. Each nucleotide is encoded as 4 × 1 vector e.g., A [1,0,0,0], C [0,1,0,0], G [0,0,1,0], and T [0,0,0,1]. The 2D space embedding for trinucleotide is a matrix with 64 × (L − 2), and each trinucleotide is represented as 64 × 1 vector, e.g., AAA [1,0, …,0], AAC [0,1, …,0], …, and TTT [0,0, …,1]. Because we cannot be certain that reading frames are not interrupted by intron/exon boundaries (Tomita et al., 1996), the trinucleotides are formed in an overlapping manner, where we scan the whole sequence in a window and shift only one nucleotide each time until L − k + 1 (similar to overlapped k-mers where k = 3). For example, the AACGTT sequence would result in AAC, ACG, CGT, and GTT trinucleotides.
Fig. 2.
Data representation. A) Mononucleotide embedding with length (4 × L), and B) trinucleotide embedding with length (64 × L).
5.2.2. Deep learning model and parameter tuning
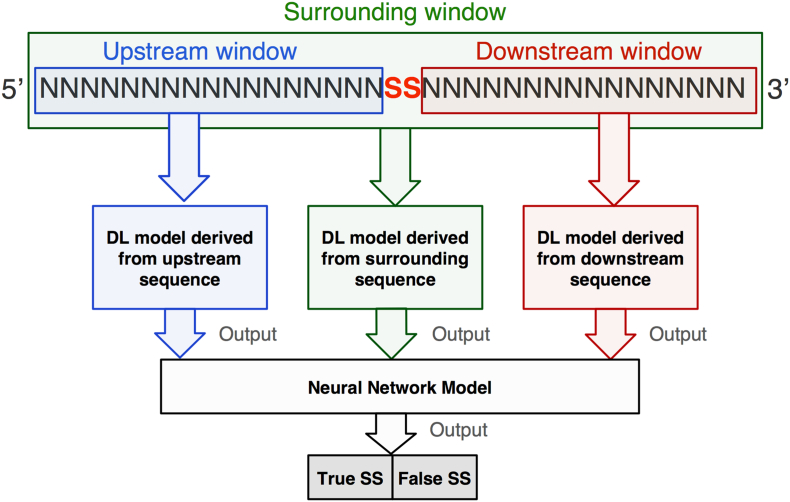
The Splice2Deep model receives as input a DNA sequence as raw data and performs feature extraction and feature selection using DL from the flanking regions described above. That is, given a sequence S, our model computes a score f(S) based on a deep convolutional neural network (CNN), which consists of stacking a sequence of layers: 1) sequence encoding, 2) convolutional layers (CONV), 3) rectified linear unit layer (ReLU), 4) pooling layer (POOL), 5) fully connected layer (FC), and 6) SoftMax layer. The outputs from the CNN models are then used as inputs to an artificial neural network (NN) binary classifier to predict whether the given sequence represents a true or false SS. The structure of Splice2Deep method is shown in Fig. 3.
Fig. 3.
Splice2Deep model overview. Local and surrounding windows. ‘SS’ refers to splice site and ‘N’ to nucleotides.
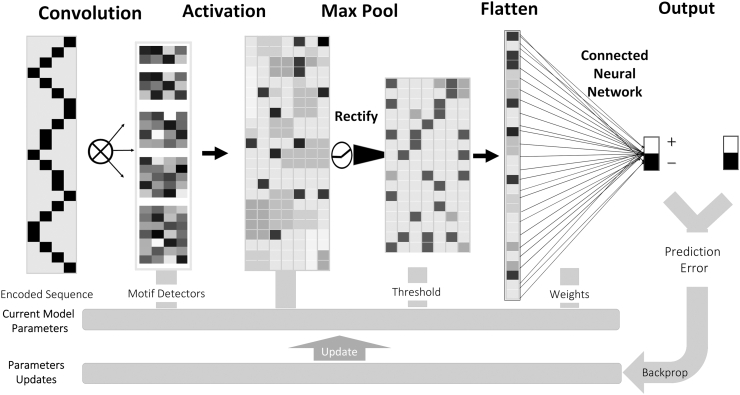
The CONV layer is the core building block of a CNN. Each of the DL models consists of k CONV layers (motif detectors) that capture motifs important for SS detection. The size of each filter depends on the encoded input. In each CONV layer, we scan the embedded sequence with the assigned filter, a stride of one and zero paddings to extract features. The CONV layers provide inputs to a nonlinear layer that contains a rectified linear unit (ReLU) as activation function (Glorot et al., 2011), defined as f(x) = max (0, x), where x is the output of the CONV layer. The POOL layer is used to select the best features obtained from the previous layer. Then a dropout layer added to each POOL layer to avoid overfitting by randomly removing some neurons during the training of the DL model. The outputs from all CONV layers are concatenated and flattened as a vector to make it suitable for an FC layer that uses a conventional NN with 500 neurons. The output classification layer that has two output neurons with SoftMax activation functions, receives inputs from the FC layer.
To improve generalization performance on the testing set, we applied an early stopping technique, which monitors the validation error rate and stops the training if the error is not decreasing for five consecutive epochs. This DL model structure allows our network to learn richer features as the sequence progresses through the network. The structure of CNN is depicted in Fig. 4. One critical step in developing DL models is the hyperparameter selection. For this, we considered a grid search technique to select the best combination of different parameters. We used Keras (Chollet, 2015) library in python to construct our DL model. The best performing parameters based on the validation set were used to derive the final DL model. Table 6 shows the search space for the CNN models with the best performing parameters highlighted in bold. The outputs of each of the three CNN models (resulting from using the downstream, upstream, and surrounding regions) are used as features for the NN model that serves as the final binary classifier for candidate SS (Fig. 3). We used Python MLPclassifier from Scikit-learn (Pedregosa et al., 2011) to derive the NN model. The resulting NN has an input layer with three inputs (CNN outputs), a hidden layer with two nodes and a single output node. We used a grid search technique to select the best parameter combinations for the NN model. Table 6 shows the search space for the NN with the best performing parameters in bold.
Fig. 4.
Splice2Deep learning model. It takes DNA sequence as input embedded in 2D (either 4 × L or 64 × L), apply k motif detectors (filters), max pooling, flatten, fully connected layer using SoftMax to output scores.
Table 6.
Grid search space for the tuning of the CNN and NN hyperparameters.
| CNN model hyperparameters | Search space | |
|---|---|---|
| Activation function | [tanh, relu] | |
| Number of neurons on FC layer | [128, 250, 512, 1024] | |
| Initialization mode | [uniform, glorot_normal] | |
| Batch size | [16, 32, 64, 128] | |
| Dropout rate | [0.01, 0.1, 0.2, 0.3, 0.4, 0.5] | |
| Optimizer | [SGD, Adam, Nadam] | |
| 4 × L embedding | Number of filters | [16, 32, 64] |
| Filter length | [1, 2, 3, 4, 5, 6, 7, 8, 9] | |
| Filter width | [4] | |
| 64 × L embedding | Number of filters | [16, 32, 64] |
| Filter length | [3, 5, 10, 21, 31, 41, 50, 61] | |
| Filter width | [64] | |
| NN model hyperparameters | Search space |
|---|---|
| Size of nodes | [1,2,3,4,5] |
| Activation function | [tanh, relu] |
| Solver | [‘lbfgs’, ‘sgd’, ‘adam’] |
The following are the supplementary data related to this article.
Sensitivity (Sn) and specificity (Sp) comparison of Splice2Deep and the state-of-the- art tools for the prediction of SS on the five selected organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Homo sapiens model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Arabidopsis thaliana model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Oryza sativa japonica model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Drosophila melanogaster model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Caenorhabditis elegans model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
The datasets generated and/or analyzed during the current study are publicly available online at https://github.com/SomayahAlbaradei/Splice_Deep.git.
Funding
VBB has been supported by the King Abdullah University of Science and Technology (KAUST) Base Research Fund (BAS/1/1606-01-01); ME has been supported by KAUST Office of Sponsored Research (OSR) Award no. FCC/1/1976-17-01. TG has also been supported by the King Abdullah University of Science and Technology (KAUST) Base Research Fund (BAS/1/1059-01-01).
Authors' contributions
The study is conceptualized by VBB and design made by VBB and SA. SA developed the models. SA, AMM, BJ, MT, ME, TG, and VBB analyzed the data. AMM performed data extraction and data curation. SA and AMM wrote the manuscript (with support from BRJ). SA, AMM, BRJ, MU, MT, ME, and VBB contributed to the discussion. ME and VBB reviewed/edited the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Acknowledgements
We are grateful to Mohammad Shoaib Amini for consultation with data extraction. This research made use of the resources of the GPU clusters at King Abdullah University of Science & Technology (KAUST) in Thuwal, Saudi Arabia.
Contributor Information
Magbubah Essack, Email: magbubah.essack@kaust.edu.sa.
Boris R. Jankovic, Email: boris.jankovic@kaust.edu.sa.
References
- Albalawi F., Chahid A., Guo X., Albaradei S., Magana-Mora A., Jankovic B.R., Uludag M., Neste C.V., Essack M., Laleg-Kirati T.-M. Hybrid model for efficient prediction of Poly (A) signals in human genomic DNA. Methods. 2019;166:31–39. doi: 10.1016/j.ymeth.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Alipanahi B., Delong A., Weirauch M.T., Frey B.J. Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015;33(8):831. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- Alshahrani M., Soufan O., Magana-Mora A., Bajic V.B. DANNP: an efficient artificial neural network pruning tool. PeerJ Computer Science. 2017;3 [Google Scholar]
- Ashoor H., Magana-Mora A., Jankovic B.R., Kamau A., Awara K., Chowdary R., Archer J.A.C., Bajic V.B. Recognition of translation initiation sites in Arabidopsis thaliana. In: Lecca P., Tulpan D., Rajaraman K., editors. Systemic Approaches in Bioinformatics and Computational Systems Biology: Recent Advances. IGI Global; 2011. pp. 105–116. [Google Scholar]
- Bari A., Reaz M.R., Jeong B.-S. Effective DNA encoding for splice site prediction using SVM. MATCH Commun Math Comput Chem. 2014;71:241–258. [Google Scholar]
- Baten A.K., Chang B.C., Halgamuge S.K., Li J. BMC Bioinformatics: 2006. BioMed Central; 2006. Splice site identification using probabilistic parameters and SVM classification; p. S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengio Y., Courville A.C., Vincent P. Unsupervised feature learning and deep learning: a review and new perspectives. CoRR, abs/12065538. 2012;2012:1. [Google Scholar]
- Bins F.J. 2002. Feature Selection From Huge Feature Sets in the Context of Computer Vision. [Google Scholar]
- Brendel V., Xing L., Zhu W. Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics. 2004;20(7):1157–1169. doi: 10.1093/bioinformatics/bth058. [DOI] [PubMed] [Google Scholar]
- Buckley K.M., Florea L.D., Smith L.C. A method for identifying alternative or cryptic donor splice sites within gene and mRNA sequences. Comparisons among sequences from vertebrates, echinoderms and other groups. MC Genomics. 2009;10:318. doi: 10.1186/1471-2164-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M., Seledtsov I., Solovyev V. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28(21):4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-W., Lin C.-J. Feature Extraction. Springer; 2006. Combining SVMs with various feature selection strategies; pp. 315–324. [Google Scholar]
- Cheng C.-Y., Krishnakumar V., Chan A., Schobel Seth, Town C.D. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2016;89(4):789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- Chollet F. GitHub Repository. 2015. Keras. [Google Scholar]
- Degroeve S., Saeys Y., De Baets B., Rouzé P., Van De Peer Y. SpliceMachine: predicting splice sites from high-dimensional local context representations. Bioinformatics. 2004;21(8):1332–1338. doi: 10.1093/bioinformatics/bti166. [DOI] [PubMed] [Google Scholar]
- Dror G., Sorek R., Shamir R. Accurate identification of alternatively spliced exons using support vector machine. Bioinformatics. 2004;21(7):897–901. doi: 10.1093/bioinformatics/bti132. [DOI] [PubMed] [Google Scholar]
- Du X., Yao Y., Diao Y., Zhu H., Zhang Y., Li S. DeepSS: exploring splice site motif through convolutional neural network directly from DNA sequence. IEEE Access. 2018;6:32958–32978. [Google Scholar]
- Glorot X., Bordes A., Bengio Y. Deep sparse rectifier neural networks. In: Geoffrey G., David D., Miroslav D., editors. Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics. Vol. 15. Proceedings of Machine Learning Research; PMLR: 2011. pp. 315–323. [Google Scholar]
- Goel N., Singh S., Aseri T.C. An improved method for splice site prediction in DNA sequences using support vector machines. Procedia Computer Science. 2015;57:358–367. [Google Scholar]
- Huang J., Li T., Chen K., Wu J. An approach of encoding for prediction of splice sites using SVM. Biochimie. 2006;88(7):923–929. doi: 10.1016/j.biochi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Iso-Touru T., Wurmser C., Venhoranta H., Hiltpold M., Savolainen T., Sironen A., Fischer K., Flisikowski K., Fries R., Vicente-Carrillo A. A splice donor variant in CCDC189 is associated with asthenospermia in Nordic Red dairy cattle. BMC Genomics. 2019;20(286) doi: 10.1186/s12864-019-5628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganathan K., Panagiotopoulou S.K., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B. Predicting splicing from primary sequence with deep learning. Cell. 2019;176(3):535–548. doi: 10.1016/j.cell.2018.12.015. (e524) [DOI] [PubMed] [Google Scholar]
- Kalkatawi M., Rangkuti F., Schramm M., Jankovic B.R., Kamau A., Chowdary R., Archer J.A., Bajic V. Dragon PolyA Spotter: predictor of poly(A) motifs within human genomic DNA sequences. Bioinformatics. 2012;28(1):127–129. doi: 10.1093/bioinformatics/btr602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkatawi M., Rangkuti F., Schramm M., Jankovic B.R., Kamau A., Chowdary R., Archer J.A., Bajic V. Dragon PolyA Spotter: predictor of poly(A) motifs within human genomic DNA sequences. Bioinformatics. 2013;29(11):1484. doi: 10.1093/bioinformatics/btt161. [DOI] [PubMed] [Google Scholar]
- Kalkatawi M., Magana-Mora A., Jankovic B., Bajic V.B. DeepGSR: an optimized deep-learning structure for the recognition of genomic signals and regions. Bioinformatics. 2019;35(7):1125–1132. doi: 10.1093/bioinformatics/bty752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath U., De Jong K., Shehu A. Effective automated feature construction and selection for classification of biological sequences. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0099982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmangaliyev Y.Z., Gelfand M.S. Computational analysis of splicing errors and mutations in human transcripts. BMC Genomics. 2008;9:13. doi: 10.1186/1471-2164-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Yoon S. International Conference on Machine Learning: 2015. 2015. Boosted categorical restricted Boltzmann machine for computational prediction of splice junctions; pp. 2483–2492. [Google Scholar]
- Lee R.Y.N., Howe K.L., Harris T.W., Arnaboldi V., Cain S., Chan J., Chen W.J., Davis P., Gao S., Grove C. WormBase 2017: molting into a new stage. Nucleic Acids Res. 2017;46(4):D869–D874. doi: 10.1093/nar/gkx998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater P.A., Vos-Loohuis M., Ducro B.J., Boegheim I.J., van Steenbeek F.G., Nijman I.J., Monroe G.R., Bastiaansen J.W.M., Dibbits B.W., van de Goor L.H. Dwarfism with joint laxity in Friesian horses is associated with a splice site mutation in B4GALT7. BMC Genomics. 2016;(839):17. doi: 10.1186/s12864-016-3186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung M.K., Xiong H.Y., Lee L.J., Frey B.J. Deep learning of the tissue-regulated splicing code. Bioinformatics. 2014;30(12):i121–i129. doi: 10.1093/bioinformatics/btu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li J., Huo L., Li W., Du X. Proceedings of the International Conference on Bioinformatics and Computational Intelligence. Vol. 2017. ACM; 2017. Prediction of splice site using support vector machine with feature selection; pp. 1–5. [Google Scholar]
- Lopes H.S., Erig Lima C.R., Murata N.J. A configware approach for high-speed parallel analysis of genomic data. Journal of Circuits, Systems, and Computers. 2007;16(4):527–540. [Google Scholar]
- Magana-Mora A., Bajic V.B. OmniGA: optimized omnivariate decision trees for generalizable classication models. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-04281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana-Mora A., Ashoor H., Jankovic B.R., Kamau A., Awara K., Chowdhary R., Archer J.A., Bajic V.B. Dragon TIS Spotter: an Arabidopsis-derived predictor of translation initiation sites in plants. Bioinformatics. 2013;29(1):117–118. doi: 10.1093/bioinformatics/bts638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana-Mora A., Kalkatawi M., Bajic V.B. Omni-PolyA: a method and tool for accurate recognition of Poly (A) signals in human genomic DNA. BMC Genomics. 2017;18(1) doi: 10.1186/s12864-017-4033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meher P.K., Sahu T.K., Rao A.R., Wahi S.D. A statistical approach for 5′ splice site prediction using short sequence motifs and without encoding sequence data. BMC bioinformatics. 2014;15(1):362. doi: 10.1186/s12859-014-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meher P.K., Sahu T.K., Rao A.R. Prediction of donor splice sites using random forest with a new sequence encoding approach. BioData mining. 2016;9(1):4. doi: 10.1186/s13040-016-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meher P.K., Sahu T.K., Rao A., Wahi S. Identification of donor splice sites using support vector machine: a computational approach based on positional, compositional and dependency features. Algorithms for molecular biology. 2016;11(1):16. doi: 10.1186/s13015-016-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison F.S., Locke J.M., Wood A.R., Tuke M., Pasko D., Murray A., Frayling T., Harries L.W. The splice site variant rs11078928 may be associated with a genotype-dependent alteration in expression of GSDMB transcripts. BMC Genomics. 2013;14:627. doi: 10.1186/1471-2164-14-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.G., Tran V.A., Ngo D.L., Phan D., Lumbanraja F.R., Faisal M.R., Abapihi B., Kubo M., Satou K. DNA sequence classification by convolutional neural network. J. Biomed. Sci. Eng. 2016;9(5):280. [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. The Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- Pertea M., Lin X., Salzberg S.L. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29(5):1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang D., Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016;44(11):e107. doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshon M., DeGregori J.V., Ruley H.E. Gene trap mutagenesis of hnRNP A2/B1: a cryptic 3′ splice site in the neomycin resistance gene allows continued expression of the disrupted cellular gene. BMC Genomics. 2003;4(2) doi: 10.1186/1471-2164-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A.B., Balasubramanian S. Long genes and genes with multiple splice variants are enriched in pathways linked to cancer and other multigenic diseases. BMC Genomics. 2016;17(225) doi: 10.1186/s12864-016-2582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Lee S., Tanaka T., Numa H., Kim J., Kawahara Y., Wakimoto H., Yang C.C., Iwamoto M., Abe T. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54(2) doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby S.K., Sønderby C.K., Nielsen H., Winther O. International Conference on Algorithms for Computational Biology. Vol. 2015. Springer; 2015. Convolutional LSTM networks for subcellular localization of proteins; pp. 68–80. [Google Scholar]
- Sonnenburg S., Schweikert G., Philips P., Behr J., Rätsch G. BMC Bioinformatics: 2007. BioMed Central; 2007. Accurate splice site prediction using support vector machines; p. S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J., Goodman J.L., Strelets V.B., Attrill H., Gramates L.S., Marygold S.J., Matthews B.B., Millburn M., Antonazzo G., Trovisco V. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47(D1):D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita M., Shimizu N., Brutlag D.L. Introns and reading frames: correlation between splicing sites and their codon positions. Mol. Biol. Evol. 1996;13(9):1219–1223. doi: 10.1093/oxfordjournals.molbev.a025687. [DOI] [PubMed] [Google Scholar]
- Wei D., Zhang H., Wei Y., Jiang Q. A novel splice site prediction method using support vector machine. Journal of Computational Information Systems. 2013;9(20):8053–8060. [Google Scholar]
- Xie B., Jankovic B.R., Bajic V.B., Song L., Gao X. Poly(A) motif prediction using spectral latent features from human DNA sequences. Bioinformatics. 2013;29(13):i316–i325. doi: 10.1093/bioinformatics/btt218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Edwards M.D., Liu G., Gifford D.K. Convolutional neural network architectures for predicting DNA–protein binding. Bioinformatics. 2016;32(12):i121–i127. doi: 10.1093/bioinformatics/btw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Giron C.G. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Peng Q., Zhang Q., Yan Y., Li K., Li J. Splice sites prediction of human genome using length-variable Markov model and feature selection. Expert Syst. Appl. 2010;37(4):2771–2782. [Google Scholar]
- Zhang Y., Liu X., MacLeod J.N., Liu J. 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM): 2016. IEEE; 2016. DeepSplice: deep classification of novel splice junctions revealed by RNA-seq; pp. 330–333. [Google Scholar]
- Zhang Y., Liu X., MacLeod J., Liu J. Discerning novel splice junctions derived from RNA-seq alignment: a deep learning approach. BMC Genomics. 2018;19(1):971. doi: 10.1186/s12864-018-5350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuallaert J., Godin F., Kim M., Soete A., Saeys Y., De Neve W., Hancock J. SpliceRover: interpretable convolutional neural networks for improved splice site prediction. Bioinformatics. 2018;34(24):4180–4188. doi: 10.1093/bioinformatics/bty497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity (Sn) and specificity (Sp) comparison of Splice2Deep and the state-of-the- art tools for the prediction of SS on the five selected organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Homo sapiens model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Arabidopsis thaliana model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Oryza sativa japonica model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Drosophila melanogaster model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Accuracy (Acc), sensitivity (Sn), and specificity (Sp) from the cross-organism validation obtained by using the Caenorhabditis elegans model and testing on the other organisms. N/A indicates that the tool is not available for that specific organism.
Data Availability Statement
The datasets generated and/or analyzed during the current study are publicly available online at https://github.com/SomayahAlbaradei/Splice_Deep.git.