Abstract
Each year, almost 4.1 million people are diagnosed with gastrointestinal (GI) cancers. Due to late detection of this disease, the mortality is high, causing approximately 3 million cancer-related deaths annually, worldwide. Although the incidence and survival differs according to organ site, earlier detection and improved prognostication have the potential to reduce overall mortality burden from these cancers. Epigenetic changes, including aberrant promoter DNA methylation, are common events in both cancer initiation and progression. Furthermore, such changes may be identified non-invasively with the use of PCR based methods, in bodily fluids of cancer patients. These features make aberrant DNA methylation a promising substrate for the development of disease biomarkers for early detection, prognosis and for predicting response to therapy. In this article, we will provide an update and current clinical perspectives for DNA methylation alterations in patients with colorectal, gastric, pancreatic, liver and esophageal cancers, and discuss their potential role as cancer biomarkers.
Keywords: Biomarkers, Detection, DNA methylation, Gastrointestinal cancers, Prognosis
1. Introduction
Gastrointestinal (GI) cancers include malignancies arising in the esophagus, stomach, liver and bile ducts, gallbladder, pancreas, the small intestine, colon and rectum. Together they account for approximately 4.1 million new cases and 3 million deaths, annually worldwide [1]. Colorectal and gastric cancer are the two most common GI cancers worldwide, affecting 1.4 million and 952 000 people each year, respectively. Each cancer type leads to around 700 000 deaths per year. Liver cancer is also quite common with an incidence of 782 000 new cases, and the very high mortality rate causes ~746 000 deaths annually. Cancers of the pancreas (338 000) and esophagus (456 000) are relatively less common, but the poor overall survival (OS) in these two malignancies still make these cancer types as some of the leading causes of cancer-related deaths (330 000 and 400 000, respectively) [1]. Most patients with early-stage GI cancers are asymptomatic, leading to an increased risk for their diagnosis at advanced stages, where the treatment options are limited and patient outcomes are often poor. This clinical challenge underscores the need for identification and development of robust molecular markers that can facilitate reduced mortality rates, either through earlier detection and/or through better prediction of tumor response to specific therapies.
Epigenetic changes are common in all types of cancers, including GI cancers, and contribute to both cancer initiation and progression. ‘Epigenetics’ is defined as heritable changes in gene expression that do not cause permanent alteration of the underlying DNA sequences, and include e.g. DNA methylation, histone modifications and non-coding RNAs. The most commonly studied epigenetic alteration in cancer, including in GI cancer, is aberrant DNA methylation. DNA methylation is predominantly found at the 5′-position of cytosine residues (5mC or 5-methyl cytosine) followed by a guanine dinucleotide sequences (CpG). Some regions in the genome are characterized by a particularly high CpG content termed ‘CpG islands’, and are present in approximately 60% of human gene promoters [2]. Normally these CpG islands are unmethylated in normal cells, and allow active transcription of the gene involved. However, in cancer cells, these islands are frequently targeted for hypermethylation–an alteration that causes transcriptional repression of the associated gene, including tumor suppressors. Gene silencing by promoter hypermethylation occurs in the majority of cancer types, and occurs more frequently compared to genetic mutations. Since aberrant promoter DNA methylation may additionally be detected in various bodily fluids, including bile, feces, and blood, such markers are gaining a lot of attention as potential noninvasive cancer biomarkers for the early detection, prognosis and for predicting the treatment outcomes. Furthermore, DNA methylation is a highly stable mark that can easily be detected by PCR-based technologies, making it suitable for clinical use.
This review focuses on DNA methylation changes in GI cancers, including colorectal, gastric, esophageal, pancreatic, and liver cancers and their potential value as cancer biomarkers. Other epigenetic aberrations, including miRNAs, are outside the scope of this text and reviewed elsewhere [3–7].
2. Aberrant DNA methylation in colorectal cancer
2.1. Diagnostic biomarkers
Colorectal cancer develops in a step-wise manner through clear premalignant precursor adenomas by gaining increasingly more dysplastic features and eventually progressing to malignant carcinomas [8]. Colonoscopy, considered as the gold standard for screening patients with colorectal cancer, has the potential to detect and remove these precursor lesions. The method is, however, invasive, expensive and further hampered by low compliance rates [9]. Fecal occult blood test (FOBT) and fecal immunochemical test (FIT), the most commonly used screening tests in Europe and other western countries have low sensitivity and specificity [10], mandating the need for improved biomarkers for detection of early stage colorectal cancers. In the colon, epigenetic alterations have been observed also in aberrant crypt foci [11] and in the pre-malignant adenomas [12], highlighting the clinical potential of exploiting aberrant promoter methylation for the early detection of colorectal adenomas and cancers. In this context, several blood and fecal-based DNA methylation biomarkers with clinical potential have been reported for colorectal cancer (reviewed in [13–16]).
2.1.1. Blood
Non-invasive tests, including blood-based assays, have the potential to improve overall patient compliance compared to colonoscopy. Indeed, this has also been shown for methylation of SEPT9 [17], which is included in the FDA approved Epi proColon plasma test (Epigenomics AG, Berlin, Germany). The sensitivities for detecting cancer and advanced adenomas have been reported between 48 and 72%, and 11–22%, respectively, with specificities ranging from 80 to 95% [18,19]. Since positive test results have been observed in patients with other malignant and non-malignant conditions, clinical parameters should be evaluated along with the test results and suspected neoplasms should be confirmed by colonoscopy [20]. Although single markers have shown promise for cancer detection, biomarker panels might be more reliable considering tumor heterogeneity and technical challenges. In a recent prospective study the methylation of BCAT1 and IKZF1 was analyzed in plasma samples from > 2000 people, including 129 with cancer [21]. The two-marker blood test had a sensitivity and specificity of 66% and 94%, respectively. The sensitivity for advanced adenomas was, however, only 6%. The methylation frequency of these markers was reduced after surgery in several patients, motivating ClinicalGenomics to develop the Colvera™ test for monitoring tumor recurrence [21,22]. Two additional biomarker panels with high sensitivity and specificity for colorectal cancer have been detected [23,24]. The first panel (CNRIP1, FBN1, INA, MAL, SNCA, and SPG20) showed a combined sensitivity of 94% for colorectal cancers and 93% for adenomas, with an area under the ROC curve (AUC) of 0.98 and 0.97, respectively [23]. The second panel (CDO1, DCLK1, SFRP1, ZNF331 and ZSCAN18) displayed a sensitivity of 95% for colorectal cancers and a specificity of 98% with an AUC of 0.98 [24]. Preliminary data showed that a selection of these markers performed well also in plasma and fecal samples, with a combined AUC 0.82 and 0.87, respectively [25]. The high AUC (0.93) for advanced adenomas in plasma samples underscore the promise of these markers for early detection of colorectal cancer.
2.1.2. Feces
Colonocytes from tumors are constantly shed into the lumen, and identification of aberrant methylation in fecal samples represents a good source for cancer specific detection of colorectal cancer. Aberrant methylation of VIM was included in the first commercial fecal test for colorectal cancer detection (ColoSure, LabCorp) [26]. The sensitivities for cancers and adenomas have been reported between 33 and 81% and 15–45%, respectively, with specificities ranging from 82 to 100% [26,27]. The FDA approved Cologuard (Exact Sciences) evaluates methylation of BMP3 and NDRG4, mutant KRAS, β-actin and hemoglobin. In a large population based study (n ~ 10 000), the fecal test reached a sensitivity of 92% for cancers and 42% for advanced precancerous lesions with a specificity of 87% [28]. Importantly, the sensitivity for advanced precancerous lesions was higher for Cologuard compared to FIT alone [28]. An observational cross-sectional cohort study was recently initiated in the Netherlands, aiming at including 4000 individuals for evaluating Cologuard and FIT as alternative screening methods for colorectal cancer [29].
2.2. Prognostic and predictive biomarkers
The tumor-node-metastasis (TNM) classification system is used for colorectal cancer staging, to predict prognosis and for deciding treatment. Patients within the same TNM stage do however, show high variability when it comes to both survival and response to therapy. Proper selection of patients within stages that may benefit from chemotherapy and/or targeted therapy may increase survival and treatment efficacy, and simultaneously reduce the toxicity and cost of such treatments. Abnormal CpG island methylation is a source for prognostic and predictive information. Examples of such markers are listed in Table 1 and reviewed in [13].
Table 1.
Prognostic and predictive DNA methylation biomarkers for colorectal cancer.
| Gene /panel | Material | Cancer samples | Stage | Effect of methylation | Survival / Response | Method | Ref |
|---|---|---|---|---|---|---|---|
| APC1A, CDKN2A, RASSF1A | Tissue | 111 | I-IV | Poor survival | *RR = 2.20, P = .037 | Pyrosequencing | [174] |
| AGBL4, FLI1, TWIST1 | Plasma | 353 | I-IV | Poor DFS | HR= 2.00, P = .001 | MALDI-TOF | [175] |
| *HR = 0.495, P = .495 | |||||||
| CDKN2A | Tissue | 902 | I-IV | Poor OS | HR = 1.36, P = .004 | qMSP | [176] |
| *HR = 1.03, P = .78 | |||||||
| CDKN2A | Tissue | 111 | I-IV | Poor survival | P = .036 | Pyrosequencing | [174] |
| *P = .065 | |||||||
| CDKN2A, KRAS mut | Tissue | 86 | I-III | Poor OS | *HR = 3.0, P = .036 | MSP | [177] |
| CDKN2A, NEUROG | Tissue | 497 (FOLFOX) | II-III | Worse OS | *HR = 2.89, P = .002 | qMSP | [178] |
| CDX2 | Tissue | 72 | I-IV | Poor OS | *HR = 3.02, P = .030 | MSP | [179] |
| CIMP | Tissue | 1035 | I-IV | Poor OS | *HR = 1.89, P < .001 | qMSP | [39] |
| Poor TTR | *HR = 1.86, P < .001 | ||||||
| CIMP | Tissue | 206 | I-IV | Poor OS | P = .029 | qMSP | [33] |
| *CIMP not included | |||||||
| CIMP | Tissue | 734 | I-IV | Poor OS | P = .009, *P = .451 | qMSP | [34] |
| Poor DFS | P = .008, *P = .337 | ||||||
| CIMP | Tissue | 190 (cohort 1) | I-IV | Poor CSS | *HR = 1.84, P > .05 | qMSP | [35] |
| 574 (cohort 2) | *HR = 1.10, P > .05 | ||||||
| CIMP | Tissue | 272 (MSS) | I-IV | Poor relative survival | *HR = 2.90, P = .001 | MSP | [36] |
| CIMP | Tissue | 649 | I-IV | Improved CSS | *HR = 0.44, P < .05 | qMSP | [32] |
| CIMP | Tissue | 24 (CIMP+) | II-III | 5-FU improved survival | P = .022 | qMSP | [40] |
| 100 (CIMP−) | 5-FU no survival benefit | P = .988 | |||||
| CIMP | Tissue | 67 (CIMP+) | III | 5-FU improved survival | P = .002 | MSP | [41] |
| 140 (CIMP−) | 5-FU no survival benefit | P = .60 | |||||
| CIMP | Tissue | 615 | III | Poor OS | HR = 1.36, P = .044 | qMSP | [42] |
| *P = .031 | |||||||
| CIMP | Tissue | 145 (CIMP+) | III | IFL (vs 5-FU/LV) improved OS | HR = 0.62, P = .07 | qMSP | [42] |
| 470 (CIMP−) | IFL (vs FU/LV) worse OS | HR = 1.38, P = .049 | |||||
| CIMP | Tissue | 12 (CIMP+) | II-III | 5-FU worse DFS | P = .023 | MSP | [46] |
| 38 (CIMP−) | 5-FU no survival benefit | P = .146 | |||||
| CIMP/MLH1 | Tissue | 115 | II | Poor DFS | *HR = 3.16, P = .011 | qMSP | [180] |
| Poor OS | *HR = 4.70, P = .002 | ||||||
| CREB1 | Tissue | 33 (rectum) | II-III | Predictive of lower response to radiation | P = .031(histology) | qMSP | [181] |
| DIRAS1 | Tissue | 146 | I-IV | Poor OS | P = .012 | MSP | [182] |
| FOXE1 | Tissue | 396 | I-IV | Poor OS | P = .008 | Illumina HM450 | [183] |
| HLTF | Serum | 103 | IV | Poor OS | *HR = 1.8, P = .04 | qMSP | [49] |
| HLTF | Serum | 77 | I-IV | Poor OS | RR = 3.0, P = .008 | qMSP | [184] |
| HOPX | Tissue | 170 | III | Poor DSS | *HR = 1.40, P = .035 | qMSP | [185] |
| HPP1 | Plasma | 467 | IV | Poor OS | HR = 1.86, P < .05 | qMSP | [48] |
| HPP1 | Plasma (after therapy initiated) | 467 | IV | Predictive of non-responders to bevacizumab comb therapy | AUC = 0.77, NPV = 97.7 (RECIST) | qMSP | [48] |
| HPP1 | Serum | 103 | IV | Poor OS | *HR = 1.6, P < .05 | qMSP | [49] |
| HPP1 | Serum | 77 | I-IV | Poor OS | RR = 5.1, P = .001 | qMSP | [184] |
| HPP1, HLTF | Serum | 77 | I-IV | Poor OS | *RR = 3.4, P = .007 | qMSP | [184] |
| IGFBP3 | Tissue | 34 (training) | II | Improved RFS | *HR = 6.46, P = .012 | qMSP | [186] |
| 81 (validation) | *HR = 2.40, P = .029 | ||||||
| IGFBP3 | Tissue | 425 | II-III | Improved DFS | *HR = 0.49, P < .01 | Pyrosequencing | [47] |
| IGFBP3 | Tissue | 89 (methylated) | II-III | Chemotherapy no survival benefit | P = .20 | Pyrosequencing | [47] |
| 157 (unmethylated) | Chemotherapy survival benefit | P = .040 | |||||
| KISS1 | Tissue | 100 (validation 1) | I-III | Poor DSS and OS | P = .034 (DSS), P = .015 (OS) | MSP, BGS | [187] |
| 190 (validation 2) | P = .030 (DSS) | ||||||
| MGMT | Plasma | 49 (TMZ) | IV | Improved PFS | P = .008 | MethylBEAMing | [53] |
| MGMT | Tissue | 68 | IV | Improved response to dacarbazine | DCR = 44% (methylated), DCR = 6% (unmethylated), P = .012 | MSP | [188] |
| MGMT | Tissue | 61 (training) | IV | Improved response to alkylating agents | PPV = 0.67, NPV = 0.89 (RECIST) | MethylBEAMing | [53] |
| 21 (validation) | PPV = 0.5, NPV = 0.67 (RECIST) | ||||||
| MGMT | Tissue | 111 | I-IV | Improved survival | *RR = .36, P = .023 | Pyrosequencing | [174] |
| MLH1 | Tissue | 195 | I-IV | Improved OS | *HR = .56, P = .01 | MS-MLPA | [189] |
| PCDH10, SPARC, UCHL1 | Tissue | 71 (5-FU/LV) | II | Poor DFS and OS | P = .069 (DFS), P = .139 (OS) | qMSP | [190] |
| 72 (surveillance) | Improved DFS and OS | P = .031 (DFS), P = .003 (OS) | |||||
| RARb2 | Tissue | 73 | I-IV | Poor OS | P = .026 | MSP | [191] |
| RET | Tissue | 233 (series 1) | II | Poor OS | *HR = 2.46, P < .05 | MSP, pyrosequencing | [192] |
| 231 (series 2) | *HR = 1.13, P > .05 | ||||||
| 294 (series 3) | *HR = 1.91, P < .05 | ||||||
| SEPT9 | Serum (after 1y follow-up) | 137 | II-III | Poor CSS | *HR = 2.69, P < .05 | qMSP | [193] |
| SFRP2 | Tissue | 77 | I-IV | Poor OS | *HR = 5.15, P = .041 | MSP | [194] |
| SHISA3 | Tissue | 120 (OS) | I-IV | Poor OS | *HR = 2.9, P = .002 | Pyrosequencing | [195] |
| 83 (DFS) | II-III | Poor DFS | *HR = 4.0, P = .003 | ||||
| SLFN11 | Tissue | 128 | T1-T4/N0-N2 | Poor OS | *HR = 0.44, P = .022 | MSP | [196] |
| Poor RFS | *HR = 0.44, P = .023 | ||||||
| SYK | Tissue | 139 | I-IV | Poor OS | *HR = 1.77, P = .001 | MSP | [197] |
| SYNPO2 | Tissue | 31 (training) | I-IV | Poor DSS and OS | P = 0.132 (DSS) | MSP, BGS | [198] |
| 100 (validation 1) | II | P = .046 (DSS), P = .031 (OS) | |||||
| 48 (validation 2) | II-III | P = .012 (DSS), P = .009 (OS) | |||||
| TAC1 | Serum (after 6 months follow-up) | 144 | II-III | Poor CSS | *HR = 4.12, P ≤ .001 | qMSP | [193] |
| Poor DFS | *HR = 5.72, P < .001 | ||||||
| TFAP2E | Tissue | 74 (cohort I) | IV | Lower response to 5-FU | P < .001 (RECIST) | qMSP | [51] |
| 36 (cohort II) | IV | P < .001 (RECIST) | |||||
| 42 (cohort III) | P < .001 (histology) | ||||||
| 68 (cohort IV) | P < .001 (histology) | ||||||
| TFAP2E | Tissue | 193 | I-III | Improved OS | *HR = 2.24, P = .025 | MS-HRM | [199] |
| Improved RFS | *HR = 2.44, P = .025 | ||||||
| TFAP2E | Tissue | 154 (chemotherapy) | II-III | Improved OS | *HR = 2.55, P = .017 | MS-HRM | [199] |
| Improved RFS | *HR = 2.36, P = .032 | ||||||
| WNT5A | Tissue | 126 (treated with 5-FU) | I-IV | Improved PFS | P < .005 | MSP | [52] |
| WRN | Tissue | 88 (treated with irinotecan) | NA | Improved OS | P < .001 | MSP | [200] |
Abbreviations: BGS, bisulfite genomic sequencing; COBRA, combined bisulfite restriction analysis; DCR, disease control rate (= partial response+ stable disease); DFS, disease free survival; DSS, disease specific survival; HR, hazard ratio; MSP, methylation specific PCR; MSS, microsatellite stable; NA, not available; NPV. Negative predictive value; OS, overall survival; PFS, progression free survival; PPV, positive predictive value; qMSP, quantitative methylation specific PCR; RECIST, response evaluation criteria in solid tumors; RFS, recurrence/relapse-free survival; RR, relative risk; TTR, time to recurrence.
Multivariate.
A subset of colorectal cancers (15–20%) develop through the CpG island methylator phenotype (CIMP) pathway and are characterized by particularly high levels of aberrantly methylated genes [30,31]. Although it has been suggested that CIMP confers improved survival [32], most studies report that CIMP is associated with a worse prognosis [33–36], including two recent meta-analyses [37,38]. Additionally, a recent study, analyzing more than 1000 tumor samples, found that CIMP was associated with inferior survival, both across all patients and specifically within patients with MSS and MSS BRAF mutated tumors. The finding remained significant in multivariate analyses adjusted for stage [39]. Furthermore, CIMP has been suggested to have predictive value, although opposing findings have been reported. Some studies have shown that patients with CIMP positive colorectal cancers display better survival when treated with 5-FU based adjuvant chemotherapy in addition to surgery [40,41], or that these patients may have a survival benefit when treated with irinotecan in addition to 5-FU [42]. However, others have reported that CIMP is not predictive of response towards adjuvant chemotherapy, or that chemotherapy does not increase survival in patients with CIMP tumors [43–45]. It has furthermore been reported that patients with CIMP positive tumors experienced a worse survival when treated with chemotherapy compared to surgery alone [46].
As illustrated in Table 1, some genes with prognostic potential have also been suggested to possess predictive value. For instance, Perez-Carbonell et al. reported that patients with IGFBP methylation had an improved disease free survival (DFS), and further that stage II and III patients with high levels of IGFBP3 methylation did not seem to benefit from adjuvant 5-FU-based chemotherapy [47]. Methylation of HPP1 in blood has also, in addition to being associated with worse survival, been suggested as a potential marker for the identification of patients who have likely not benefitted from a combination chemotherapy with bevacizumab [48,49]. Furthermore, patients with MSI tumors, normally caused by epigenetic silencing of MLH1, have been shown to have either no benefit or to have increased mortality when treated with 5-FU [50]. Similarly, Ebert et al. observed a strong association between TFAP2E methylation and lack of response towards 5-FU based chemotherapy, by analyzing four independent patient cohorts with primary rectal cancer and metastatic colorectal cancers [51]. Methylation may also predict improved response towards chemotherapy. For instance, a significantly higher proportion of patients with WNT5A methylation responded to 5-FU-based chemotherapy compared to patients with an unmethylated promoter [52]. Also MGMT methylation has been reported to associate with better response and improved median progression free survival (PFS) in metastatic colorectal cancer patients treated with alkylating agents [53].
2.3. Summary and perspectives
A decrease in colorectal cancer death rates has been observed in western countries the last years [54,55], largely explained by improvements in early detection. However, current screening strategies are not optimal considering cost, invasiveness and compliance, and less invasive screening methods are warranted. Non-invasive DNA methylation biomarkers have shown great promise for colorectal cancer detection, including the two FDA approved tests Epi proColon (blood) and Cologuard (feces). Both tests do, however, have room for improvements when it comes to sensitivity and specificity. Several other DNA methylation biomarkers have also been suggested, including biomarkers panels, but validation in independent and large population based series are needed.
When it comes to prognostication and prediction of treatment response, reliable DNA methylation biomarkers for colorectal cancer are still lacking. Several independent studies have, however, shown that colorectal cancer patients with CIMP have an inferior survival. Once a consensus marker panel to identify CIMP is defined and routinely analyzed with a quantitative method, it is likely that CIMP can provide valuable information about prognosis and potentially guide treatment.
3. Aberrant DNA methylation in gastric cancer
3.1. Diagnostic biomarkers
The incidence of gastric cancer has declined in most western countries, but remains one of the most common causes of cancer-related deaths in Asia [1]. The most important approach to reduce the mortality in these patients is by early detection and curative resection of malignant lesions. In Asian countries, including Korea and Japan where the incidence of gastric cancer is particularly high, screening programs, including endoscopy and barium-meal have been implemented [56]. These methods are however, not optimal considering availability, cost, and participation rate [56].
Infection with Heliobacter pylori (H.pylori) and Epstein-Barr virus (EBV) are considered as major risk factors for gastric cancer. Both infections are associated with increased levels of promoter DNA methylation [57–59]. As is the case in colorectal cancer, promoter DNA methylation of several tumor suppressor genes have been identified in premalignant stages of gastric carcinomas [60,61], highlighting their suitability as biomarkers for early cancer detection.
3.1.1. Blood
A number of hypermethylated genes with potential for early detection have been identified in blood specimens from gastric cancer patients (reviewed in [5,62,63]). In a study from 2002, Lee et al. reported frequent methylation of CDH1 (57%), DAPK (48%), GSTP1 (15%), CDKN2A (p15, 56%), and CDKN2A (p16, 52%) in sera from 54 cancer patients, but in none of the 30 age-matched serum samples from individuals without cancer [64]. Combining the markers further increased the sensitivity (83%) without compromising the specificity (100%) [64]. Since this initial study, methylation of CDH1 [65,66] and CDKN2A [65–67] have been confirmed as potential biomarkers for gastric cancer detection in blood samples. Another example is RNF180, with a sensitivity of 76% (150/198) and specificity of 100% (23/23) in tissue samples [68]. Further analyses in 32 plasma samples confirmed a high sensitivity (56–63%) and specificity (91–100%) [68]. Bernal et al. identified frequent promoter methylation (95%) of RPRM in plasma samples from 43 gastric cancer patients, but rarely in the 31 controls (10%), suggesting RPRM also as a potential biomarker for early detection of gastric cancer [69].
3.1.2. Gastric washes/gastric juice and feces
Gastric washes represent an alternative source for detecting aberrant DNA methylation is gastric cancer. By analyzing the methylation levels of six genes (ADAM23, GDNF, MINT25, MLF1, PRDM5, RORA) in gastric washes from 20 cancer patients and 48 controls Watanabe and colleagues found that a combination of the markers MINT25, PRDM5 and GDNF achieved a high sensitivity (95%) and specificity (92%). Among individual genes, MINT25 displayed the highest sensitivity (90%) and specificity (96%) [70]. Recently, it was reported that BARHL2 methylation in gastric washes had promising potential for detection of gastric cancer with significantly higher methylation frequency in the 128 analyzed cancer samples compared to the 32 control samples [71]. Additionally, the methylation levels detected in the cancer patients decreased significantly after endoscopic resection, suggesting that BARHL2 methylation could be also useful for monitoring tumor recurrence. Furthermore, promoter methylation of BARHL2 in exosomal DNA from gastric juice could discriminate between cancer (n = 20) and controls (n = 10) with a sensitivity and specificity of 90% and 100%, respectively [71].
Finally, RASSF2 and SFRP2 promoter methylation has been found in 57% of fecal samples from gastric cancer patients (n = 21), with 89% specificity [72]. The same genes were also positive for colorectal cancer (75%), indicating that the assay could be useful for detection of both cancer types.
3.2. Prognostic and predictive biomarkers
The TNM staging system is used to predict prognosis and to decide treatment for gastric cancer patients. Similar to colorectal cancer, this pathological system is suboptimal and better markers for predicting prognosis and response to specific drugs are needed.
Promoter hypermethylation of several genes have been reported to be associated with worse prognosis in gastric cancer (Table 2 and reviewed in [63,73,74]). For instance, BCL6 B methylation was analyzed in 309 patients with gastric cancer from two different cohorts, and in 20 normal tissue specimens [75]. Combining the two cohorts, 55% of the cancer patients displayed BCL6 B promoter methylation. No methylation was detected in the normal samples, confirming its tumor specificity. It was further shown that BCL6 B methylation was an independent predictor of worse OS [75]. Methylation of either BNIP3 or DAPK was shown to confer a significantly poorer OS and PFS in a study including 80 tissue samples from gastric cancer patients. Methylation was additionally predictive of lower response to 5-FU based chemotherapy [76]. Kato et al. noted that methylation of DAPK together with TMS1was associated with poor prognosis, and also lower response towards 5-FU among patients with recurrent disease [77]. Recently, it was reported that methylation of NDRG4 was predictive for poor prognosis among Chinese gastric cancer patients. Validation in the TCGA data which included 357 gastric patients, did however, imply an improved prognosis for patients with NDRG4 promoter methylation, suggesting that different ethnicities, therapies, and methylation detection methods could influence both the methylation frequencies and survival [78]. Additional examples of methylated genes that confer improved prognosis include BMP4 methylation, which also caused sensitization to cisplatin [79], IGF2 methylation [80], and methylation of MLH1 [81].
Table 2.
Prognostic and predictive DNA methylation biomarkers for gastric cancer.
| Gene /panel | Material | Cancer samples | Stage | Effect of methylation | Survival/Response | Method | Ref |
|---|---|---|---|---|---|---|---|
| APC | Serum | 79 | I-IIIAB | Poor OS | *HR = 4.6, P = .046 | MSP | [201] |
| APC, CDH1 | Serum | 58 | I-IV | Poor survival | P = .0006 | qMSP | [202] |
| ASC/TMS1 | Tissue | 200 | T1-T4/N0-N3 | Poor OS | *HR = 0.536, P = .001 | MSP | [203] |
| BCL6B | Tissue | 208 (cohort 1) | I-IV | Poor OS | *RR = 2.14, P = .001 | COBRA, BGS | [75] |
| 101 (cohort 2) | *RR = 1.85, P = .02 | ||||||
| BNIP | Tissue | 80 | IV or recurrence | Poor OS | P = .031 | MSP | [76] |
| BNIP, DAPK | Tissue | 80 | IV or recurrence | Lower response to 5-FU | P = .003 (RECIST) | MSP | [76] |
| BNIP, DAPK | Tissue | 80 | IV or recurrence | Poor OS | P = .001 | MSP | [76] |
| Poor PFS | P = .002 | ||||||
| CACNA2D3 | Tissue | 53 | Advanceda | Poor survival | *OR = 4.38, P = .002 | MSP, BGS | [204] |
| CDH1 | PPW | 92 | I-IV | Poor DFS | RR = 333, P < .001 | qMSP | [82] |
| CDH1 | Serum | 97 | I-IV | Poor OS | P < .05 | MSP | [83] |
| CDH1 | Tissue | 73 | II-III | Poor DFS | *RR = 2.28, P < .001 | MSP | [84] |
| Poor OS | *RR = 1.94, P = .004 | ||||||
| CDKN2A | Tissue | 119 | I-IV | Poor survival | P < .0001 | MSP | [87] |
| CDKN2A | Tissue | 38 (chemotherapy) | I-IV | Improved DFS | *RR = 0.093, P = .043 | MSP | [88] |
| CDO1 | Peritoneal washes | 102 | I-IV | Poor OS | P = .0004 | qMSP | [205] |
| Poor RFS | P = .005 | ||||||
| CHFR | Tissue | 12 | Advanceda or recurrence | Improved response to TXL or TXT | 86% PR or NC (CHFR+), 20% PR or NC (CHFR−) | MSP | [206] |
| CIMP | Tissue | 68 | I-IV | Improved OS | P = .069 | COBRA | [81] |
| CIMP/MLH1 | Tissue | 68 | I-IV | Improved OS | *P = .031 | COBRA | [81] |
| DAPK | Tissue | 81 | I-IV | Poor OS | P = .045 | MSP | [77] |
| DAPK | Tissue | 80 | IV or recurrence | Lower response to 5-FU | P = .012 (RECIST) | MSP | [76] |
| DAPK | Tissue | 80 | IV or recurrence | Poor PFS | P = .007 | MSP | [76] |
| DAPK and TMS1 | Tissue | 81 | I-IV | Poor OS | P < .001 | MSP | [77] |
| Poor RFS | P < .0001 | ||||||
| DAPK and TMS1 | Tissue | 43 (5-FU) | IV or recurrence | Poor OS | P = .083 | MSP | [77] |
| Poor TTR | P = .008 | ||||||
| DAPK1 | Tissue | 141 | I-IV | Poor OS | P = .04 | qMSP | [207] |
| *OR = 1.13, P > .05 | |||||||
| DKK2 | Tissue | 92 | I-IV | *HR = 2.05, P = .041 | Pyrosequencing | [208] | |
| DLEC1 | Tissue | 148 | I-IV | Poor RFS | *HR = 2.43, P = .025 | qMSP | [209] |
| FLNc | Tissue | 119 | I-IV | Poor survival | P = .03 | MSP | [87] |
| GFRA3 | Tissue | 24 | I-IV | Poor OS | *P = .01 | Illumina HM27K | [210] |
| IGF2 | Leukocytes | 299 | I-IV | Improved OS | *HR = 0.42, P = .001 | qMSP | [80] |
| MGMT | Tissue | 79 | I-IV | Poor DFS | P < .02 | MSP | [211] |
| MGMT | Tissue | 119 | I-IV | Poor survival | P = .02 | MSP | [87] |
| MINT2 | Serum | 92 | I-IV | Poor DFS | *RR = 4.11, P < .05 | qMSP | [212] |
| PPLF | *RR = 3.26, P < .05 | ||||||
| MLH1 | Tissue | 68 | I-IV | Improved OS | P = .026 | COBRA | [81] |
| NDRG4 | Tissue | 110 | I-IV | Poor OS | *HR = 1.887, P = .020 | qMSP, pyrosequencing | [78] |
| PAX5 | Tissue | 460 | T1-T4/N0-N3 | Poor OS | *HR = 1.39, P = .005 | BGS | [213] |
| PAX5 | Tissue | 187 | I-IV | Poor OS | *RR = 2.10, P = .01 | MSP, BGS | [214] |
| PAX6 | Tissue | 141 | I-IV | Poor OS | P < .001 | qMSP | [207] |
| *OR = 1.51, P > .05 | |||||||
| PCDH10 | Tissue | 104 | I-IV | Poor OS | *RR = 1.73, P = .039 | COBRA | [61] |
| PCDH10 | Tissue | 471 | T1-T4/N0-N3 | Poor OS | *HR = 1.41, P = .011 | BGS | [215] |
| RARb | Tissue | 141 | I-IV | Poor OS | P < .001 | qMSP | [207] |
| *OR = 2.04, P > .05 | |||||||
| RASSF10 | Serum | 82 | I-IV | Poor OS | *OR = 13.96, P < .001 | BGS | [216] |
| Poor DFS | *OR = 13.11, P < .001 | ||||||
| RASSF1A | Tissue | 141 | I-IV | Poor OS | P = .004 | qMSP | [207] |
| *OR = 1.72, P > .05 | |||||||
| RASSF2A | Tissue | 119 | I-IV | Poor survival | P = .01 | MSP | [87] |
| RKIP | Tissue | 76 | I-IV | Poor survival | *OR = 0.22, P = .042 | MSP | [217] |
| RNF180 | Plasma | 32 | I-IV | Poor CSS | *RR = 2.13, P = .02 | qMSP | [68] |
| RPRM | Tissue | 49 (5-FU + cisplatin) | IV or recurrence | Poor DSS | P = .05 | qMSP | [86] |
| RPRM | Tissue | 68 | Advanceda | Poor DSS | *HR = 2.148, P = .026 | qMSP | [86] |
| SPARC | Tissue | 185 | I-IV | Poor OS | *RR = 2.75, P < .001 | MSP | [218] |
| TIMP3 | PPW | 92 | I-IV | Poor DFS | *HR = 1.72, P < .001 | qMSP | [219] |
| TIMP3 | Serum | 92 | I-IV | Poor DFS | *HR = 1.55, P = .03 | qMSP | [219] |
| XAF1 | Serum | 202 | I-IV | Poor DFS | *HR = 5.71, P < .001 | qMSP | [85] |
Abbreviations: BGS, bisulfite genomic sequencing; COBRA, combined bisulfite restriction analysis; DSS, disease specific survival; HR, hazard ratio; MSP, methylation specific PCR; NC, no change; OR, odds ratio; OS, overall survival; PPLF, preoperative peritoneal lavage fluid; PPW, preoperative peritoneal washes; PR; partial response; qMSP, quantitative methylation specific PCR; RECIST, response evaluation criteria in solid tumors; RFS, relapse/recurrence free survival; RR, relative risk; TTP, time to progression; TXT, Taxotere; TXL, Taxol.
Invading muscuaris propria.
Multivariate.
Several of the markers reported as potential markers for detection of gastric cancer have also been suggested to possess prognostic or predictive value. Methylation of CDH1, both in preoperative peritoneal washes [82], serum [83], and tissue samples [84] has for instance been reported to be significantly associated with worse survival. In addition to showing that XAF1 was frequently methylated in a cancer-specific manner, in the serum from gastric cancer patients, Ling et al. reported that patients with XAF1 promoter methylation had significantly poorer survival compared to those with an unmethylated promoter. XAF1 methylation in sera was additionally associated with tumor recurrence [85]. Methylation of RPRM was reported to predict both poor survival among patients with advanced gastric cancer, and poor response towards 5-FU and cisplatin [86]. Also CDKN2A has been suggested both as a marker for detection, and as a potential marker for poor prognosis. Shi et al. analyzed 119 tumor samples for promoter methylation of e.g. CDKN2A, and found that methylation of this gene, in addition to MGMT, RASSF2 and FLNc, was associated with poor survival, both across all samples and when only tumors without residual disease were included [87]. Methylation of CDKN2A has further been reported to be significantly associated with improved survival in patients treated with 5-FU [88].
CIMP has also been identified in gastric cancer [89]. In contrast to colorectal cancer, most studies suggest a lower mortality for patients with CIMP positive gastric cancer [81,90], although an unfavorable prognosis has also been reported [91,92]. A meta-analysis by Zong et al., which included 12 studies and 1000 patients, concluded that CIMP was not a prognostic marker for gastric cancer [93]. Most other studies included much fewer samples than this, making it difficult to infer true clinical potential of these biomarkers.
3.3. Summary and perspectives
The majority of the patients with gastric cancer have regional or distant metastases at the time of diagnosis [54]. The failure to detect cancer at a stage where clinical intervention is effective obviously contributes to the high gastric cancer mortality. Although screening programs have been implemented in some high-risk countries, the available methods are not optimal. Several DNA methylation biomarkers with high sensitivity and specificity for non- or minimally invasive detection of gastric cancer have been identified. However, the number of samples analyzed for these markers is a limiting factor, and validation of candidate markers in larger and independent samples series is needed. This is also the case for prognostic and predictive candidate DNA methylation biomarkers.
4. Aberrant DNA methylation in esophageal cancer
4.1. Diagnostic biomarkers
Esophageal cancer is a highly fatal malignancy. There are two main classes of histopathologically distinct esophageal cancer, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). ESCC accounts for about 90% of cases worldwide [94] and is especially prevalent in Eastern Asia and Southern and Eastern Africa, whereas EAC, associated with an increasing incidence, is more commonly seen in Western countries [95].
4.1.1. Esophageal adenocarcinoma
EAC is usually diagnosed at a late stage, with few treatment options, explaining the underlying reason for high mortality rates associated with this disease. The cancer can develop from the premalignant Barrett’s esophagus [96], and risk stratification of EAC in individuals with Barrett’s esophagus is an area of active research. CDKN2A was one of the first genes to be identified as methylated during the neoplastic progression in Barrett’s esophagus [97,98]. By analyzing multiple clinical specimens from a handful of patients, Eads and colleagues demonstrated that hypermethylation of CDKN2A, along with promoter methylation of APC and ESR1 was present in large contiguous fields [99]. These researches further found significantly higher incidences of hypermethylation in tissues from patients with dysplasia or cancer, compared to patients with no further progression of their disease, confirming that abnormal DNA methylation represents a clinical tool in stratifying Barrett’s esophagus patients at risk of developing cancer [100].
From targeted analyses, several additional genes have been suggested as biomarkers for predicting disease progression in Barrett’s esophagus patients [101–105]. An eight-gene biomarker panel, including CDKN2A, RUNX3, HPP1, NELL1, TAC1, SST, AKAP12, and CDH13, was validated in a multicenter, double-blind, tissue-based study. The panel predicted approximately half of the Barrett’s esophagus-associated EACs analyzed, with a high specificity [106]. A recent 27 K methylation array based study identified an alternative panel of four DNA methylation biomarkers: SLC22A18, PIGR, GJA12 and RIN2. The markers were validated in a retrospective cohort and subsequently in a multicenter study. The biomarker panel achieved an area under the ROC curve of 0.988 to distinguish Barrett’s esophagus and EAC patients, and was able to stratify patients from the prospective cohort into distinct risk groups [107].
4.1.2. Esophageal squamous cell carcinoma
An integrated genomic characterization of ESCC and EAC from the TCGA research network recently demonstrated that these two subtypes could be differentiated not only by histology but also by their molecular features, including the DNA methylation levels [108]. Interestingly, ESCC seem to resemble squamous carcinomas of other organs more than they do with EAC.
CDKN2A was one of the first genes to be identified as methylated also in ESCC [109]. Analyzing CDKN2A and 13 additional gene promoters, Guo and colleagues could illustrate accumulation of promoter methylation through the histological progression from squamous dysplasia to ESCC [110], underscoring the potential of using DNA methylation biomarkers for early detection of ESCC. In line with this, Ishii et al. confirmed, from analyses of 14 gene promoters, that ESCC patients had accumulated low levels of DNA methylation already in the non-neoplastic esophageal epithelium. The frequency of methylation increased through intraepithelial neoplasia to advanced ESCC; hence most likely contributing to the pathogenesis of this malignancy [111]. The genes with the highest methylation frequencies in ESCC cases were MGMT (80%) and SFRP1 (64%) [111]. DNA methylation in ESCC has not been as extensively studied as is the case in EAC; however RARB2 (up to 70%) [112] and FHIT (up to 69%) [113] are examples of genes reported to be frequently methylated. Interestingly, studies that have analyzed both EAC and ESCC seem to find significantly lower methylation frequencies of specific genes in the latter group ((RPRM) [114], APC [115]).
Finally, using a bead-array analysis of more than 800 cancer-related genes Lima et al. identified 37 CpG sites that were differentially methylated between ESCCs and surrounding tissues. Of these, TFF1 was methylated both in ESCC and surrounding tissue in contrast to healthy esophageal mucosa, suggesting that it could be an early event and potential biomarker for ESCC [116].
4.1.3. Blood
Although several DNA methylation biomarkers have been suggested for esophageal cancer, only few of them have been analyzed in a noninvasive manner in blood. Methylation of APC has been found in the plasma from 25% (13/52) of the EAC patients and in 6% (2/32) of ESCC patients [115]. Furthermore, CDKN2A methylation has been found in 23% (7/31) of ESCC patients harboring a methylated primary tumor [117]. Among four additional genes analyzed by Li and coworkers in serum from 45 ESCC patients and 15 controls, CDH1 achieved the highest AUC value of 0.822 followed by DAPK (0.800), RASSF1A (0.778), and RARB (0.567) [118]. Zhai and coworkers used methylation arrays to identify hundreds of differentially methylated loci between cell-free circulating DNA from a limited number of EAC and Barrett’s esophagus patients. The results have so far not been validated by an independent method or in larger clinical series [119].
4.2. Prognostic and predictive biomarkers
DNA methylation biomarkers with prognostic value for esophageal cancer are generally scarce and often analyzed in small samples series (n < 100; reviewed in [100]). Suggested biomarkers are APC [115,120], TAC1 [121], and NELL1 [122]. In EAC, promoter methylation of multiple genes has additionally been suggested as a predictor of poor prognosis [123]. In ESCCs, FHIT has been analyzed in a larger cohort (n = 257), and aberrant promoter methylation was significantly associated with a poor prognosis for early stage (I &II) cancers (multivariate HR = 5.81, P = 0.009) [124]. Finally, in a recent study CDO1 methylation was suggested as an independent prognostic factor for ESCC patients (multivariate HR = 2.00, P = 0.03) based on analyses of 169 cancer samples [125].
4.3. Summary and perspectives
Because of late detection, only 11–18% of the patients with esophagus cancer survive 5 years [126]. DNA methylation biomarkers present in premalignant or early stage cancer could contribute to earlier detection thereby reducing the mortality. However, at present, only a small number of the identified biomarker candidates for esophagus cancer have been analyzed in non-invasive material, and the few studies that have been performed are limited by few samples. The same goes for prognostic and predictive biomarkers. As such, before epigenetic biomarkers become clinically useful in esophagus cancer, more and larger studies are needed, preferentially with the use of quantitative methods.
5. Aberrant DNA methylation in pancreatic cancer
5.1. Diagnostic biomarkers
Pancreatic ductal adenocarcinomas (PDACs), the most prevalent subtype of pancreatic cancer, evolves through non-invasive precursor lesions, most commonly pancreatic intraepithelial neoplasias (PanINs) [127]. These lesions typically grow slowly, and may take between 10 and 15 years to develop into malignant lesions that can further metastasize [128], providing an ideal opportunity for the early detection and curative treatment of affected patients. The microscopic PanINs are usually not visible by pancreatic imaging, and other markers, such as CA19–9, is inadequate as its level may also be elevated in benign diseases [127,129]. This highlights the need for biomarkers that can both reliably identify the high-grade PanINs (with high probability of malignancy) and facilitate further differentiation between malignant and benign disease. Interestingly, several epigenetically silenced genes have been detected in the non-invasive PanINs, including NTPX2, SARP2, RPRM, and LHX1 [130].
5.1.1. Pancreatic juice
Pancreatic juice contains exfoliated cells from all parts of the pancreas, and represents a good source for detecting aberrations in the pancreatic ductal epithelium. Indeed, several genes commonly methylated in PDACs are also detectable in pancreatic juice of patients with invasive pancreatic cancer. Methylation of ≥2 of the fives genes CCND2, TFPI2, PENK, NPTX2, and FOXE1 has been shown to distinguish between patients with cancer (n = 11) and individuals without neoplasia (n = 64), including chronic pancreatitis, with high sensitivity (82%) and specificity (100%) [131]. In another study, by using reduced representation bisulfite sequencing (RRBS) several differentially methylated regions (DMRs) between cancer and benign and normal control samples were identified [132]. These regions were both technically (by methylation specific PCR or MSP) and biologically validated, before tested by quantitative MSP (qMSP) assays in a pilot study of 102 pancreatic juice samples from 61 patients with cancer, 22 with chronic pancreatitis and 19 with normal pancreas. The AUC values for methylation of CD1D, KCNK12, CLEC11A, NDRG4, IKZF1, and PKRCB for detection of cancer compared with normal pancreas and chronic pancreatitis ranged from 0.83–0.92 and 0.73–0.92, respectively. The most promising gene, CD1D, could discriminate between pancreatic cancer and normal tissues with a75% sensitivity and 95% specificity. Compared to patients with chronic pancreatitis, the specificity was 91% [132]. Several additional aberrantly methylated genes have been identified in pancreatic juice from cancer patients, including NPTX2 (67%), SARP2 (46%), and CLDN5 (42%) [133], PENK (67%) and CDKN2A (11%) [134], and APC (71%) [135].
5.1.2. Blood
Genes aberrantly methylated in cell free DNA in plasma represent another potential source for non-invasive pancreatic cancer detection. By analyzing patients with PDAC (n = 95), and non-malignant controls (chronic pancreatitis, n = 97, and healthy controls, n = 27), a diagnostic prediction model was generated, including age > 65, and promoter methylation of BMP3, RASSF1A, BNC1, MESTv2, TFPI2, APC, SFRP1 and SFRP2. The model displayed an AUC of 0.86, with a sensitivity of 76% and a specificity of 83% for pancreatic cancer [136]. By analyzing 42 serum samples from patients with pancreatic cancer and 26 samples from healthy controls, methylation of BNC1 and ADAMS1 could detect cancer with a sensitivity of 79% and 48%, respectively [137]. The specificity was also high; 89% for BNC1 and 92% for ADAMTS1. Combining the two markers resulted in a sensitivity of 81% and a specificity of 85%. Interestingly, the genes were frequently methylated in tissue specimens from PanINs (ADAMST1: 25%, BNC1: 70%) and stage I invasive cancers (ADAMST1: 63%, BNC1: 97%), high-lightening their potential as early detection markers. Furthermore, the combination of ADAMTS1 and BNC1 methylation showed superior sensitivity compared with CA19–9 [137]. Analyzing > 300 blood samples from cancer patients and 300 control samples in a test and validation series by Illumina arrays, Pedersen et al. identified several differently methylated sites between cancer and normal samples, which potentially could aid in the detection of pancreatic cancer [138].
5.1.3. Brush samples
Finally, aberrant methylation for pancreatic cancer detection has also been observed in brush samples. Parsi et al. analyzed the methylation levels of TFPI2, NPTX2 and CCND2 in biliary brush samples from 41 patients with PDAC, 10 patients with biliary tract cancer, and from 66 individuals with benign biliary tract diseases. At least one methylation positive gene was detected in 73% of patients with pancreatic cancer, in 80% of patients with biliary tract cancer, and in 14% of patients with a benign stricture, concluding that the detection of aberrantly methylated genes in endoscopic brush samples are promising tools to differentiate benign and malignant biliary strictures [139].
5.2. Prognostic and predictive biomarkers
Since most pancreatic cancer patients present with advanced disease (> 80%), where the prognosis is poor regardless of the treatment, less focus has been put on trying to identify biomarkers with prognostic and predictive potential. Still, some methylation candidates have been proposed. In a study analyzing promoter methylation in whole-blood samples from 30 pancreatic cancer patients, methylation of the two genes TNFRSF10C and ACIN1 was found to be significantly associated with shorter OS (P = 0.023 and P = 0.012, respectively) [140]. By analyzing 87 primary cancers, Sato et al. observed that patients with RPRM methylation had a significantly shorter OS compared to those without methylation (univariate HR = 2.2, P = 0.009). In multivariate analyses, adjusted for tumor size and differentiation, patients with RPRM methylation still tended to have poorer prognosis, although the results were not statistically significant (HR = 1.8, P = 0.07) [141]. It has been further shown that patients with hypermethylated MUC1 and MUC4 had significantly improved survival compared to patients with hypomethylated promoters [142]. Finally, by analyzing 11 samples with RRBS Thomson and colleagues identified increased methylation in the promoter region of three genes (FAM150A, ONECUT1, and RASSF10), which strongly correlated with worse survival [143].
About 75% of patients treated with gemcitabine, the first- line treatment for pancreatic cancer patients, demonstrate no response to such treatments. Tan et al. generated 30 pancreatic cancer xenografts from surgically resected primary carcinomas and treated them with gemcitabine. Methylation analyses (the Golden Gate methylation cancer panel I) identified two genes, GSTM1 and ONECU, that were differentially methylated between the 10 xenografts that responded and the 20 that were non-responsive to treatment. The finding was validated in 12 pancreatic cancer cell lines treated with gemcitabine, suggesting that hypermethylation of the two genes may sensitize patients to this type of drug treatment [144].
5.3. Summary and perspectives
Pancreatic cancer is considered a largely incurable disease, with a 5-year survival rate of 9% [54]. The poor prognosis is mainly caused by cancer detection after metastases, where effective treatment options are lacking. Identifying robust biomarkers for earlier detection thus have a large potential impact if successful, as earlier detection could enable surgical resection of the tumor and thereby reduce mortality from pancreatic cancer. Although some epigenetic biomarkers have been reported for pancreatic cancer detection, including CD1D, their accuracy is not optimal, underscoring that more research is needed. This is also the case for potentially prognostic and predictive DNA methylation biomarkers in pancreatic cancer. Such markers are therefore not anticipated to reach the clinic in the near future.
6. Aberrant DNA methylation in liver cancer
6.1. Diagnostic biomarkers
Hepatocellular carcinoma (HCC) and cholangiocarcinoma (CCA) are the two most frequent primary liver cancers, accounting for > 80% of the cases. Most liver cancers (~85%) occur in Asia, and can largely be explained by the continent’s high rate of chronic infections with hepatitis B- (HBV) and C- viruses (HCV), which are known risk factors for HCC and CCA [145–147]. For CCA, primary sclerosing cholangitis (PSC) is a common predisposing condition, with a lifetime risk of CCA between 5 and 10% [148]. Distinguishing between benign biliary strictures and malignant changes are difficult in PSC patients, and in up to 37% of cases, CCA is not detected until laparotomy is performed in connection with intended liver transplantation or at autopsy [149]. CA19–9 is a commonly used biomarker for CCA, but is elevated in > 30% of patients with PSC [148]. Serological tests, including α-fetoprotein (AFP), are commonly used diagnostic biomarkers for HCC, but may show positive test results in other malignancies and in healthy pregnant women [150]. For both HCC and CCA, the diagnosis is challenging due to the non-specific clinical presentation, resulting in late detection [147]. With more sensitive and specific biomarkers increased number of patients could potentially qualify for curative treatment, including surgery [151].
6.1.1. Brush, bile and blood samples
6.1.1.1. Cholangiocarcinoma (CCA)
Aberrant DNA methylation has been observed both in patients with PSC and in CCA precursor lesions (biliary intraepithelial neoplasia; BiIN), emphasizing the suitability of such markers for early detection of CCA [152]. As previously mentioned (section 5.1), the methylation levels of TFPI2, NPTX2 and CCND2 could detect biliary tract cancers (n = 10) from biliary brush samples with a sensitivity of 80% and a specificity of 86% compared to patients with benign liver conditions [139]. The usefulness of brush samples to differentiate between benign and malignant biliary strictures has further been confirmed by Andresen and colleagues. In an initial study analyzing tissue specimens from 39 CCA patients and 54 samples from individuals with non-malignant liver disease, a biomarker panel comprising CDO1, DCLK1, SFRP1 and ZSCAN18 was identified with high sensitivity (87%) and specificity (100%) for detection of CCA [153]. These genes, in addition to several other markers, were further validated in biliary brush samples comprising a test series (15 CCAs, 20 PSC controls) and a validation series (34 CCAs and 34 PSC controls) [151]. Across both series, a four marker panel (CDO1, CNRIP1, SEPT9, and VIM) could detect CCA with a sensitivity of 85% and a specificity of 98%, with an AUC of 0.944. Importantly, the biomarker panel outperformed standard brush cytology, underscoring the potential to improve detection of CCA, especially among patients with PSC [151].
Bile may also be suitable for CCA detection. Shin et al. reported that methylation of the marker panel CCND2, CDH13, GRIN2B, RUNX3, and TWIST1 achieved 70% sensitivity in a training set (n = 20), 74% sensitivity in the first validation set (n = 33), and 83% sensitivity in the second validation set (n = 24), with 100% specificity for all sets (total n = 48) [154]. Finally, promoter hypermethylation of SHOX2 and SEPT9 was shown to display a sensitivity of 45% and a specificity of 99% when analyzed in plasma from 20 CCA patients and 100 controls [155].
6.1.1.2. Hepatocelluar carcinoma (HCC)
As early as 1999, it was shown that promoter methylation of CDKN2A was detected in plasma/serum of HCC patients (n = 22, 59% sensitivity) with a 100% specificity, including 38 patients with chronic hepatitis/cirrhosis and 10 healthy controls [156]. In addition to CDKN2A, several genes, mainly in tissue specimens but also in blood samples, were found to be frequently methylated in HCC (reviewed in [150,157–160]). A recent metaanalysis highlighted six genes (CDH1, CDKN2A, GSTP1, RASSF1A, RUNX3 and WIF1) which, across more than three studies, were significantly hypermethylated in serum from HCC compared to normal samples [161]. In addition, a marker panel consisting of APC, GSTP, RASSF1A and SFRP1 has been evaluated in plasma samples from 72 HCC patients, 37 patients with benign liver disease and 41 samples from normal controls. The panel achieved a sensitivity of 93% and a specificity of 82%, with an AUC of 0.933. Compared to benign controls, the sensitivity, specificity and AUC were 85%, 82% and 0.877, respectively [162]. Finally, using a genome wide approach in 62 HCC patients, several DMRs regions were identified between tumor and adjacent non-tumor tissues. For a subset of the markers, pyrosequencing of 38 plasma samples was performed resulting in a combined sensitivity of 87% for HCC detection [163].
6.2. Prognostic and predictive biomarkers
The survival of patients with liver cancer is generally poor due to late disease detection where effective treatment options are lacking. As such, incidence often mirrors death. Better ways of predicting prognosis and response to e.g. liver transplantation and surgery could allocate resources to those that most likely would benefit from such treatments.
6.2.1. CCA
Based on 36 methylation probes (Illumina HM450) a methylation signature was generated that could predict poor survival among patients with CCA both in a training set (n = 221; multivariate HR = 13.35, P < 0.001) and in a validation set (n = 83, P = 0.01) [164]. Furthermore, the signature could accurately predict tumor recurrence in the training set (multivariate HR = 5.8, P < 0.001). By analyzing 79 patients with CCA, Lee and colleagues found that methylation of APC, CDKN2A and TIMP3 was significantly associated with worse OS in univariate analyses [165]. Methylation of DAPK was in another study shown to be an interdependent predictor of poor survival while analyzing 37 patients with biliary tract carcinomas (multivariate HR = 8.71, P = 0.024) [166].
6.2.2. HCC
In line with CCA, genome-wide methylation studies have been used to identify subgroups of HCC patients with poor prognosis [167,168]. Single genes have also been proposed to have prognostic value, including methylation of RASSF1 in plasma (n = 72), which was shown to be an independent prognostic factor for OS (multivariate HR = 3.26, P = 0.003) [162]. Similarly, hypomethylation of S100A8 was significantly associated with both worse OS (multivariate HR = 1.71, P < 0.05) and PFS (multivariate HR = 1.77, P < 0.05), analyzing 52 HCC tissue samples [169]. Finally, SOCS3 methylation was recently reported to predict treatment response and prognosis in 246 HCC patients receiving transarterial chemoembolization (TACE) treatment (multivariate HR = 3.44, P < 0.001) [170].
6.3. Summary and perspectives
Most HCC and CCAs are detected at a stage where curative options are limited and the prognosis is poor. Identifying accurate biomarkers for early detection, especially among patients that are at high risk of developing these cancers, still remains an unmet clinical need. Several interesting epigenetic biomarkers have been suggested for liver cancer detection, including a biomarker panel consisting of CDO1, CNRIP, SEPT9 and VIM for detection of CCA in biliary brush samples. These biomarkers do however, require validation, preferentially in blood. Although bile and biliary brush samples are interesting sources for detection of epigenetic biomarkers, the potential risk of pancreatitis during collection of such material underscore the need for non-invasive analyses.
Only a small percentage of patients with HCC and CCA are fit for surgery or liver transplantation. Biomarkers to better identify patients most likely to benefit from such treatment are naturally attractive. Although some epigenetic biomarkers for this purpose have been suggested, they are far from entering the clinic.
7. Conclusions and perspectives
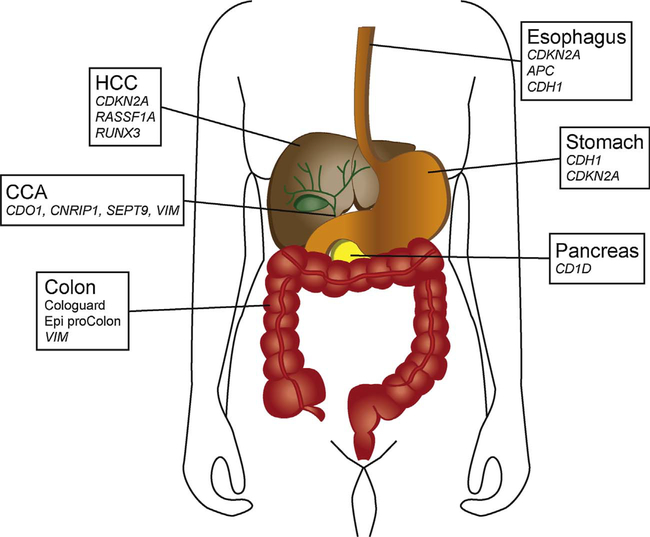
Epigenetic changes are common in gastrointestinal cancers, and are frequently found in pre-malignant conditions and early stage cancers in different biological fluids, rendering such aberration quite promising for early cancer detection. Fig. 1 summarizes some of these promising DNA methylation biomarkers. Of note, few epigenetic biomarkers have so far been implemented in the clinic. Suboptimal methods and design of biomarker discovery studies, selective and incomplete reporting combined with small samples sizes and lack of independent validation, have been highlighted as some of the explanations for the discrepancy between the number of reported promising biomarkers and those that are in fact implemented in the clinic [171]. Regarding validation of potential biomarkers, the use of sensitive technologies, proper unbiased optimization and standardization of commonly used methods is critically important [172]. In light of this, and based on the limited sample series analyzed so far and the general lack of independent validation, non-invasive biomarkers for most of the GI cancer types are far from being adopted for clinical applications. The exception is colorectal cancer, where the two DNA methylation-based tests Epi ProColon and Cologuard have been approved by the FDA for non-invasive cancer detection. The use of non-invasive material, especially blood, for detection is attractive from a compliance perspective. Blood is furthermore easily accessible, and may give a better picture of the tumor heterogeneity compared to a single biopsy [173], which is especially important considering prognostic and predictive biomarkers. Such tests, however, do have the potential of detecting any lesion, irrespective of location, which may represent a pitfall for a diagnostic test. As evident from Fig. 1, several genes are hypermethylated in more than one GI cancer type, including the Epi ProColon SEPT9, which has also been reported by Epigenomics themselves [20]. For improved cancer-type specificity of a diagnostic test, adding tissue specific markers is one of the options. Another alternative is site-specific sampling, such as feces for colon, gastric washes for gastric, bile for bile duct, and pancreatic juice for pancreatic cancer detection. Several promising biomarkers have been identified using this approach, including Cologuard for detection of colorectal cancer.
Fig. 1.
Potential non-or minimally-invasive DNA methylation biomarkers for the detection of different gastrointestinal cancers. FDA approved markers are marked in bold, validated markers or markers that are reported to be frequently methylated in larger non-invasive patients series are in normal typing. Smaller sample series have been analyzed for CCA, esophageal-, HCC and pancreatic- cancer compared to colorectal and gastric cancer. Abbreviations: CCA, cholangiocarcinoma; HCC, hepatocellular carcinoma.
Currently, no epigenetic biomarker has been approved for prognostic or for predicting response to treatment in any of the GI cancers. However, with increasing application of genome-wide methylation analyses, identification of additional methylation markers are anticipated. Such changes may in the future play a major role in GI cancer detection, for determining prognosis and for predicting response to specific treatments. Proper study-design, quantitative methods and large samples series with independent validation are, however, important factors to succeed in the discovery and development of clinically relevant biomarkers.
Acknowledgements
This work was supported by grants from the South-Eastern Norway Regional Health Authority (to G.E. Lind), the Research Council of Norway (project number 239961, to G.E. Lind), and the Centre of Excellence funding scheme (project number 179571). This work was also supported by the CA72851, CA181572, CA184792, CA187956 and CA202797 grants from the National Cancer Institute, National Institute of Health; RP140784 from the Cancer Prevention Research Institute of Texas; grants from the Sammons Cancer Center and Baylor Foundation, as well as funds from the Baylor Scott & White Research Institute, Dallas, TX, USA awarded to A.G.
Abbreviations
- AUC
area under the ROC curve
- CCA
cholangiocarcinoma
- EAC
esophageal adenocarcinoma
- ESCC
Esophageal squamous cell carcinoma
- FDA
US Food and Drug Administration
- FIT
fecal immunochemical test
- FOBT
fecal occult blood test
- GI
gastrointestinal
- HCC
hepatocellular carcinoma
- MSP
methylation specific PCR
- OS
overall survival
- PDAC
pancreatic ductal adenocarcinoma
- PFS
progression free survival
- PSC
primary sclerosing cholangitis
- qMSP
quantitative methylation specific PCR
- ROC
receiver operating characteristics curve
Footnotes
Conflict of interest
None.
References
- [1].Ferlay J, Soerjomataram I, Ervik M, et al. , GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11, International Agency for Research on Cancer, Lyon, France, 2013. Internet, Available from: http://globocan.iarc.fr, Accessed on 07/06/2017. [Google Scholar]
- [2].Portela A, Esteller M, Epigenetic modifications and human disease, Nat. Biotechnol. 28 (2010) 1057–1068. [DOI] [PubMed] [Google Scholar]
- [3].Ishiguro H, Kimura M, Takeyama H, Role of microRNAs in gastric cancer, World J. Gastroenterol. 20 (2014) 5694–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tsai MM, Wang CS, Tsai CY, et al. , Potential diagnostic, prognostic and therapeutic targets of MicroRNAs in human gastric cancer, Int. J. Mol. Sci. 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Toiyama Y, Okugawa Y, Goel A, DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer, Biochem. Biophys. Res. Commun. 455 (2014) 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shigeyasu K, Toden S, Zumwalt TJ, et al. , Emerging role of MicroRNAs as liquid biopsy biomarkers in gastrointestinal cancers, Clin. Cancer Res. 23 (2017) 2391–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Link A, Goel A, MicroRNA in gastrointestinal cancer: a step closer to reality, Adv. Clin. Chem. 62 (2013) 221–268. [DOI] [PubMed] [Google Scholar]
- [8].Fearon ER, Vogelstein B, A genetic model for colorectal tumorigenesis, Cell 61 (1990) 759–767. [DOI] [PubMed] [Google Scholar]
- [9].Taylor DP, Cannon-Albright LA, Sweeney C, et al. , Comparison of compliance for colorectal cancer screening and surveillance by colonoscopy based on risk, Genet. Med. 13 (2011) 737–743. [DOI] [PubMed] [Google Scholar]
- [10].Schreuders EH, Ruco A, Rabeneck L, et al. , Colorectal cancer screening: a global overview of existing programmes, Gut 64 (2015) 1637–1649. [DOI] [PubMed] [Google Scholar]
- [11].Hanley MP, Hahn MA, Li AX, et al. , Genome-wide DNA methylation profiling reveals cancer-associated changes within early colonic neoplasia, Oncogene 36 (2017) 5035–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ahlquist T, Lind GE, Costa VL, et al. , Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers, Mol. Cancer 7 (2008) 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Okugawa Y, Grady WM, Goel A, Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers, Gastroenterology 149 (2015) 1204–1225 e1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hashimoto Y, Zumwalt TJ, Goel A, DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer, Epigenomics 8 (2016) 685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Goel A, Boland CR, Epigenetics of colorectal cancer, Gastroenterology 143 (2012) 1442–1460 e1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim MS, Lee J, Sidransky D, DNA methylation markers in colorectal cancer, Cancer Metastasis Rev. 29 (2010) 181–206. [DOI] [PubMed] [Google Scholar]
- [17].Adler A, Geiger S, Keil A, et al. , Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany, BMC Gastroenterol. 14 (2014) 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Church TR, Wandell M, Lofton-Day C, et al. , Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer, Gut 63 (2014) 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Potter NT, Hurban P, White MN, et al. , Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma, Clin. Chem. 60 (2014) 1183–1191. [DOI] [PubMed] [Google Scholar]
- [20].Inc EIFU Epi ProColon 2.0 CE- Rev 7, Instructions For Use, (2016) (In).
- [21].Pedersen SK, Symonds EL, Baker RT, et al. , Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia, BMC Cancer 15 (2015) 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Young GP, Pedersen SK, Mansfield S, et al. , A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer, Cancer Med 5 (2016) 2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lind GE, Danielsen SA, Ahlquist T, et al. , Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas, Mol. Cancer 10 (2011) 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vedeld HM, Andresen K, Eilertsen IA, et al. , The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers, Int. J. Cancer 136 (2015) 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Speight G, Workman N, Agashe P, et al. , The translation of epigenetic colorectal cancer tissue based diagnostic biomarkers into both faecal and blood based diagnostic biomarkers.[abstract], Washington, DC. Philadelphia (PA): AACR, Proceedings of the 104th Annual Meeting of the American Association for Cancer Research; 2013 Apr 6–10, 73 2013, p. 3511, , 10.1158/1538-7445 (Cancer Res 2013, Suppl. 8, Abstract nr 3511). [DOI] [Google Scholar]
- [26].Ned RM, Melillo S, Marrone M, Fecal DNA testing for Colorectal Cancer Screening: the ColoSure test, PLoS Curr 3 (2011) RRN1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Amiot A, Mansour H, Baumgaertner I, et al. , The detection of the methylated Wif-1 gene is more accurate than a fecal occult blood test for colorectal cancer screening, PLoS One 9 (2014) e99233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. , Multitarget Stool DNA Testing for Colorectal-Cancer Screening, N. Engl. J. Med. 371 (2014) 187–188. [DOI] [PubMed] [Google Scholar]
- [29].van Lanschot MC, Carvalho B, Coupe VM, et al. , Molecular stool testing as an alternative for surveillance colonoscopy: a cross-sectional cohort study, BMC Cancer 17 (2017) 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Toyota M, Ahuja N, Ohe-Toyota M, et al. , CpG island methylator phenotype in colorectal cancer, Proc. Natl. Acad. Sci. U.S.A 96 (1999) 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Weisenberger DJ, Siegmund KD, Campan M, et al. , CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer, Nat. Genet. 38 (2006) 787–793. [DOI] [PubMed] [Google Scholar]
- [32].Ogino S, Nosho K, Kirkner GJ, et al. , CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer, Gut 58 (2009) 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim JH, Shin SH, Kwon HJ, et al. , Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers, Virchows Arch. 455 (2009) 485–494. [DOI] [PubMed] [Google Scholar]
- [34].Bae JM, Kim JH, Cho NY, et al. , Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location, Br. J. Cancer 109 (2013) 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dahlin AM, Palmqvist R, Henriksson ML, et al. , The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status, Clin. Cancer Res. 16 (2010) 1845–1855. [DOI] [PubMed] [Google Scholar]
- [36].Barault L, Charon-Barra C, Jooste V, et al. , Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases, Cancer Res. 68 (2008) 8541–8546. [DOI] [PubMed] [Google Scholar]
- [37].Juo YY, Johnston FM, Zhang DY, et al. , Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis, Ann. Oncol. 25 (2014) 2314–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zong L, Abe M, Ji J, et al. , Tracking the correlation between CpG island methylator phenotype and other molecular features and clinicopathological features in human colorectal cancers: a systematic review and meta-Analysis, Clin. Transl. Gastroenterol. 7 (2016) e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vedeld HM, Merok M, Jeanmougin M, et al. , CpG island methylator phenotype identifies high risk patients among microsatellite stable BRAF mutated colorectal cancers, Int. J. Cancer 141 (2017) 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Min BH, Bae JM, Lee EJ, et al. , The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy, BMC Cancer 11 (2011) 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Van Rijnsoever M, Elsaleh H, Joseph D, et al. , CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer, Clin. Cancer Res. 9 (2003) 2898–2903. [PubMed] [Google Scholar]
- [42].Shiovitz S, Bertagnolli MM, Renfro LA, et al. , CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer, Gastroenterology 147 (2014) 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ward RL, Cheong K, Ku SL, et al. , Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability, J. Clin. Oncol. 21 (2003) 3729–3736. [DOI] [PubMed] [Google Scholar]
- [44].Cohen SA, Wu C, Yu M, et al. , Evaluation of CpG island methylator phenotype as a biomarker in colorectal cancer treated with adjuvant oxaliplatin, Clin. Colorectal Cancer 15 (2016) 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jover R, Nguyen TP, Perez-Carbonell L, et al. , 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer, Gastroenterology 140 (2011) 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang Y, Long Y, Xu Y, et al. , Prognostic and predictive value of CpG island methylator phenotype in patients with locally advanced nonmetastatic sporadic colorectal cancer, Gastroenterol. Res. Pract. 2014 (2014) 436985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Perez-Carbonell L, Balaguer F, Toiyama Y, et al. , IGFBP3 methylation is a novel diagnostic and predictive biomarker in colorectal cancer, PLoS One 9 (2014) e104285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Herbst A, Vdovin N, Gacesa S, et al. , Methylated free-circulating HPP1 DNA is an early response marker in patients with metastatic colorectal cancer, Int. J. Cancer 140 (2017) 2134–2144. [DOI] [PubMed] [Google Scholar]
- [49].Philipp AB, Stieber P, Nagel D, et al. , Prognostic role of methylated free circulating DNA in colorectal cancer, Int. J. Cancer 131 (2012) 2308–2319. [DOI] [PubMed] [Google Scholar]
- [50].Ribic CM, Sargent DJ, Moore MJ, et al. , Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer, N. Engl. J. Med. 349 (2003) 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ebert MP, Tanzer M, Balluff B, et al. , TFAP2E-DKK4 and chemoresistance in colorectal cancer, N. Engl. J. Med. 366 (2012) 44–53. [DOI] [PubMed] [Google Scholar]
- [52].Jiang G, Lin J, Wang W, et al. , WNT5A promoter methylation is associated with better responses and longer progression-Free survival in colorectal cancer patients treated with 5-Fluorouracil-Based chemotherapy, Genet. Test Mol. Biomarkers 21 (2017) 74–79. [DOI] [PubMed] [Google Scholar]
- [53].Barault L, Amatu A, Bleeker FE, et al. , Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer, Ann. Oncol. 26 (2015) 1994–1999. [DOI] [PubMed] [Google Scholar]
- [54].Howlader NNA, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (Eds.), SEER Cancer Statistics Review, 1975–2014, National Cancer Institute, Bethesda, MD, 2017(based on November 2016 SEER data submission, posted to the SEER web site, April 2017. In), https://seer.cancer.gov/csr/1975_2014/. [Google Scholar]
- [55].Malvezzi M, Carioli G, Bertuccio P, et al. , European cancer mortality predictions for the year 2017, with focus on lung cancer, Ann. Oncol. 28 (2017) 1117–1123. [DOI] [PubMed] [Google Scholar]
- [56].Leung WK, Wu M-s Kakugawa Y, et al. , Screening for gastric cancer in Asia: current evidence and practice, Lancet Oncol. 9 (2008) 279–287. [DOI] [PubMed] [Google Scholar]
- [57].Maekita T, Nakazawa K, Mihara M, et al. , High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk, Clin. Cancer Res. 12 (2006) 989–995. [DOI] [PubMed] [Google Scholar]
- [58].Graham DY, Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits, Gastroenterology 148 (2015) 719–731 (e713). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Comprehensive molecular characterization of gastric adenocarcinoma, Nature 513 (2014) 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kang GH, Shim YH, Jung HY, et al. , CpG island methylation in premalignant stages of gastric carcinoma, Cancer Res. 61 (2001) 2847–2851. [PubMed] [Google Scholar]
- [61].Yu J, Cheng YY, Tao Q, et al. , Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer, Gastroenterology 651 (2009) e641. [DOI] [PubMed] [Google Scholar]
- [62].Qu Y, Dang S, Hou P, Gene methylation in gastric cancer, Clin. Chim. Acta 424 (2013) 53–65. [DOI] [PubMed] [Google Scholar]
- [63].Sapari NS, Loh M, Vaithilingam A, Soong R, Clinical potential of DNA methylation in gastric cancer: a meta-analysis, PLoS One 7 (2012) e36275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee TL, Leung WK, Chan MW, et al. , Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma, Clin. Cancer Res. 8 (2002) 1761–1766. [PubMed] [Google Scholar]
- [65].Koike H, Ichikawa D, Ikoma H, et al. , Comparison of methylation-specific polymerase chain reaction (MSP) with reverse transcriptase-polymerase chain reaction (RT-PCR) in peripheral blood of gastric cancer patients, J. Surg. Oncol. 87 (2004) 182–186. [DOI] [PubMed] [Google Scholar]
- [66].Ichikawa D, Koike H, Ikoma H, et al. , Detection of aberrant methylation as a tumor marker in serum of patients with gastric cancer, Anticancer Res. 24 (2004) 2477–2481. [PubMed] [Google Scholar]
- [67].Abbaszadegan MR, Moaven O, Sima HR, et al. , p16 promoter hypermethylation: a useful serum marker for early detection of gastric cancer, World J. Gastroenterol. 14 (2008) 2055–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cheung KF, Lam CN, Wu K, et al. , Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer, Cancer 118 (2012) 947–959. [DOI] [PubMed] [Google Scholar]
- [69].Bernal C, Aguayo F, Villarroel C, et al. , Reprimo as a potential biomarker for early detection in gastric cancer, Clin. Cancer Res. 14 (2008) 6264–6269. [DOI] [PubMed] [Google Scholar]
- [70].Watanabe Y, Kim HS, Castoro RJ, et al. , Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes, Gastroenterology 136 (2009) 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yamamoto H, Watanabe Y, Oikawa R, et al. , BARHL2 methylation using gastric wash DNA or gastric juice exosomal DNA is a useful marker for early detection of gastric cancer in an H. pylori-Independent manner, Clin. Transl. Gastroenterol. 7 (2016) e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Nagasaka T, Tanaka N, Cullings HM, et al. , Analysis of fecal DNA methylation to detect gastrointestinal neoplasia, J. Natl. Cancer Inst. 101 (2009) 1244–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Otani K, Li X, Arakawa T, et al. , Epigenetic-mediated tumor suppressor genes as diagnostic or prognostic biomarkers in gastric cancer, Expert Rev. Mol. Diagn. 13 (2013) 445–455. [DOI] [PubMed] [Google Scholar]
- [74].Gao Y, Zhang K, Xi H, et al. , Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis, Oncotarget 8 (2017) 6330–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xu L, Li X, Chu ES, et al. , Epigenetic inactivation of BCL6B, a novel functional tumour suppressor for gastric cancer, is associated with poor survival, Gut 61 (2012) 977–985. [DOI] [PubMed] [Google Scholar]
- [76].Sugita H, Iida S, Inokuchi M, et al. , Methylation of BNIP3 and DAPK indicates lower response to chemotherapy and poor prognosis in gastric cancer, Oncol. Rep. 25 (2011) 513–518. [DOI] [PubMed] [Google Scholar]
- [77].Kato K, Iida S, Uetake H, et al. , Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer, Int. J. Cancer 122 (2008) 603–608. [DOI] [PubMed] [Google Scholar]
- [78].Chen X, Yang Y, Liu J, et al. , NDRG4 hypermethylation is a potential biomarker for diagnosis and prognosis of gastric cancer in Chinese population, Oncotarget 8 (2017) 8105–8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ivanova T, Zouridis H, Wu Y, et al. , Integrated epigenomics identifies BMP4 as a modulator of cisplatin sensitivity in gastric cancer, Gut 62 (2013) 22–33. [DOI] [PubMed] [Google Scholar]
- [80].Yuasa Y, Nagasaki H, Oze I, et al. , Insulin-like growth factor 2 hypomethylation of blood leukocyte DNA is associated with gastric cancer risk, Int. J. Cancer 131 (2012) 2596–2603. [DOI] [PubMed] [Google Scholar]
- [81].Shigeyasu K, Nagasaka T, Mori Y, et al. , Clinical significance of MLH1 methylation and CpG island methylator phenotype as prognostic markers in patients with gastric cancer, PLoS One 10 (2015) e0130409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yu QM, Wang XB, Luo J, et al. , CDH1 methylation in preoperative peritoneal washes is an independent prognostic factor for gastric cancer, J. Surg. Oncol. 106 (2012) 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ikoma H, Ichikawa D, Koike H, et al. , Correlation between serum DNA methylation and prognosis in gastric cancer patients, Anticancer Res. 26 (2006) 2313–2316. [PubMed] [Google Scholar]
- [84].Graziano F, Arduini F, Ruzzo A, et al. , Prognostic analysis of E-cadherin gene promoter hypermethylation in patients with surgically resected, node-positive, diffuse gastric cancer, Clin. Cancer Res. 10 (2004) 2784–2789. [DOI] [PubMed] [Google Scholar]
- [85].Ling ZQ, Lv P, Lu XX, et al. , Circulating methylated XAF1 DNA indicates poor prognosis for gastric cancer, PLoS One 8 (2013) e67195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ooki A, Yamashita K, Yamaguchi K, et al. , DNA damage-inducible gene, reprimo functions as a tumor suppressor and is suppressed by promoter methylation in gastric cancer, Mol. Cancer Res. 11 (2013) 1362–1374. [DOI] [PubMed] [Google Scholar]
- [87].Shi J, Zhang G, Yao D, et al. , Prognostic significance of aberrant gene methylation in gastric cancer, Am. J. Cancer. Res. 2 (2012) 116–129. [PMC free article] [PubMed] [Google Scholar]
- [88].Mitsuno M, Kitajima Y, Ide T, et al. , Aberrant methylation of p16 predicts candidates for 5-fluorouracil-based adjuvant therapy in gastric cancer patients, J. Gastroenterol. 42 (2007) 866–873. [DOI] [PubMed] [Google Scholar]
- [89].Toyota M, Ahuja N, Suzuki H, et al. , Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype, Cancer Res. 59 (1999) 5438–5442. [PubMed] [Google Scholar]
- [90].An C, Choi IS, Yao JC, et al. , Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma, Clin. Cancer Res. 11 (2005) 656–663. [PubMed] [Google Scholar]
- [91].Park SY, Kook MC, Kim YW, et al. , CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features, Virchows Arch. 457 (2010) 415–422. [DOI] [PubMed] [Google Scholar]
- [92].Chen HY, Zhu BH, Zhang CH, et al. , High CpG island methylator phenotype is associated with lymph node metastasis and prognosis in gastric cancer, Cancer Sci. 103 (2012) 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zong L, Seto Y, CpG island methylator phenotype, Helicobacter pylori, Epstein-Barr virus, and microsatellite instability and prognosis in gastric cancer: a systematic review and meta-analysis, PLoS One 9 (2014) e86097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Pennathur A, Gibson MK, Jobe BA, Luketich JD, Oesophageal carcinoma, Lancet 381 (2013) 400–412. [DOI] [PubMed] [Google Scholar]
- [95].Thrift AP, The epidemic of oesophageal carcinoma: where are we now, Cancer Epidemiol. 41 (2016) 88–95. [DOI] [PubMed] [Google Scholar]
- [96].Barrett MT, Sanchez CA, Prevo LJ, et al. , Evolution of neoplastic cell lineages in Barrett oesophagus, Nat. Genet. 22 (1999) 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wong DJ, Barrett MT, Stoger R, et al. , p16INK4a promoter is hypermethylated at a high frequency in esophageal adenocarcinomas, Cancer Res. 57 (1997) 2619–2622. [PubMed] [Google Scholar]
- [98].Klump B, Hsieh CJ, Holzmann K, et al. , Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett’s esophagus, Gastroenterology 115 (1998) 1381–1386. [DOI] [PubMed] [Google Scholar]
- [99].Eads CA, Lord RV, Kurumboor SK, et al. , Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma, Cancer Res. 60 (2000) 5021–5026. [PubMed] [Google Scholar]
- [100].Eads CA, Lord RV, Wickramasinghe K, et al. , Epigenetic patterns in the progression of esophageal adenocarcinoma, Cancer Res. 61 (2001) 3410–3418. [PubMed] [Google Scholar]
- [101].Clement G, Braunschweig R, Pasquier N, et al. , Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett’s oesophagus patients at risk for malignant transformation, J. Pathol. 208 (2006) 100–107. [DOI] [PubMed] [Google Scholar]
- [102].Clement G, Braunschweig R, Pasquier N, et al. , Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett’s esophagus, Oncogene 25 (2006) 3084–3092. [DOI] [PubMed] [Google Scholar]
- [103].Zou H, Osborn NK, Harrington JJ, et al. , Frequent methylation of eyes absent 4 gene in Barrett’s esophagus and esophageal adenocarcinoma, Cancer Epidemiol. Biomarkers Prev. 14 (2005) 830–834. [DOI] [PubMed] [Google Scholar]
- [104].Schulmann K, Sterian A, Berki A, et al. , Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk, Oncogene 24 (2005) 4138–4148. [DOI] [PubMed] [Google Scholar]
- [105].Wang JS, Guo M, Montgomery EA, et al. , DNA promoter hypermethylation of p16 and APC predicts neoplastic progression in Barrett’s esophagus, Am. J. Gastroenterol. 104 (2009) 2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Jin Z, Cheng Y, Gu W, et al. , A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus, Cancer Res. 69 (2009) 4112–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Alvi MA, Liu X, O’Donovan M, et al. , DNA methylation as an adjunct to histopathology to detect prevalent, inconspicuous dysplasia and early-stage neoplasia in Barrett’s esophagus, Clin. Cancer Res. 19 (2013) 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Integrated genomic characterization of oesophageal carcinoma, Nature 541 (2017) 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Maesawa C, Tamura G, Nishizuka S, et al. , Inactivation of the CDKN2 gene by homozygous deletion and de novo methylation is associated with advanced stage esophageal squamous cell carcinoma, Cancer Res. 56 (1996) 3875–3878. [PubMed] [Google Scholar]
- [110].Guo M, Ren J, House MG, et al. , Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus, Clin. Cancer Res. 12 (2006) 4515–4522. [DOI] [PubMed] [Google Scholar]
- [111].Ishii T, Murakami J, Notohara K, et al. , Oesophageal squamous cell carcinoma may develop within a background of accumulating DNA methylation in normal and dysplastic mucosa, Gut 56 (2007) 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wang Y, Fang MZ, Liao J, et al. , Hypermethylation-associated inactivation of retinoic acid receptor beta in human esophageal squamous cell carcinoma, Clin. Cancer Res. 9 (2003) 5257–5263. [PubMed] [Google Scholar]
- [113].Noguchi T, Takeno S, Kimura Y, et al. , FHIT expression and hypermethylation in esophageal squamous cell carcinoma, Int. J. Mol. Med. 11 (2003) 441–447. [PubMed] [Google Scholar]
- [114].Hamilton JP, Sato F, Jin Z, et al. , Reprimo methylation is a potential biomarker of Barrett’s-Associated esophageal neoplastic progression, Clin. Cancer Res. 12 (2006) 6637–6642. [DOI] [PubMed] [Google Scholar]
- [115].Kawakami K, Brabender J, Lord RV, et al. , Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma, J. Natl. Cancer Inst. 92 (2000) 1805–1811. [DOI] [PubMed] [Google Scholar]
- [116].Lima SC, Hernandez-Vargas H, Simao T, et al. , Identification of a DNA methylome signature of esophageal squamous cell carcinoma and potential epigenetic biomarkers, Epigenetics 6 (2011) 1217–1227. [DOI] [PubMed] [Google Scholar]
- [117].Hibi K, Taguchi M, Nakayama H, et al. , Molecular detection of p16 promoter methylation in the serum of patients with esophageal squamous cell carcinoma, Clin. Cancer Res. 7 (2001) 3135–3138. [PubMed] [Google Scholar]
- [118].Li B, Wang B, Niu LJ, et al. , Hypermethylation of multiple tumor-related genes associated with DNMT3b up-regulation served as a biomarker for early diagnosis of esophageal squamous cell carcinoma, Epigenetics 6 (2011) 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Zhai R, Zhao Y, Su L, et al. , Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus, Neoplasia 14 (2012) 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zare M, Jazii FR, Alivand MR, et al. , Qualitative analysis of Adenomatous Polyposis Coli promoter: hypermethylation, engagement and effects on survival of patients with esophageal cancer in a high risk region of the world, a potential molecular marker, BMC Cancer 9 (2009) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Jin Z, Olaru A, Yang J, et al. , Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer, Clin. Cancer Res. 13 (2007) 6293–6300. [DOI] [PubMed] [Google Scholar]
- [122].Jin Z, Mori Y, Yang J, et al. , Hypermethylation of the nel-like 1 gene is a common and early event and is associated with poor prognosis in early-stage esophageal adenocarcinoma, Oncogene 26 (2007) 6332–6340. [DOI] [PubMed] [Google Scholar]
- [123].Brock MV, Gou M, Akiyama Y, et al. , Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma, Clin. Cancer Res. 9 (2003) 2912–2919. [PubMed] [Google Scholar]
- [124].Lee EJ, Lee BB, Kim JW, et al. , Aberrant methylation of Fragile Histidine Triad gene is associated with poor prognosis in early stage esophageal squamous cell carcinoma, Eur. J. Cancer 42 (2006) 972–980. [DOI] [PubMed] [Google Scholar]
- [125].Ushiku H, Yamashita K, Katoh H, et al. , Promoter DNA methylation of CDO1 gene and its clinical significance in esophageal squamous cell carcinoma, Dis. Esophagus 30 (2017) 1–9. [DOI] [PubMed] [Google Scholar]
- [126].Society AC, Global Cancer Facts & Figs, 2nd edition, American Cancer Society, Atlanta, 2011. [Google Scholar]
- [127].Vincent A, Herman J, Schulick R, et al. , Pancreatic cancer, Lancet 378 (2011) 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yachida S, Jones S, Bozic I, et al. , Distant metastasis occurs late during the genetic evolution of pancreatic cancer, Nature 467 (2010) 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hinton J, Callan R, Bodine C, et al. , Potential epigenetic biomarkers for the diagnosis and prognosis of pancreatic ductal adenocarcinomas, Expert Rev. Mol. Diagn. 13 (2013) 431–443. [DOI] [PubMed] [Google Scholar]
- [130].Sato N, Fukushima N, Hruban RH, Goggins M, CpG island methylation profile of pancreatic intraepithelial neoplasia, Mod. Pathol. 21 (2008) 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Matsubayashi H, Canto M, Sato N, et al. , DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease, Cancer Res. 66 (2006) 1208–1217. [DOI] [PubMed] [Google Scholar]
- [132].Kisiel JB, Raimondo M, Taylor WR, et al. , New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice, Clin. Cancer Res. 21 (2015) 4473–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Sato N, Fukushima N, Maitra A, et al. , Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays, Cancer Res. 63 (2003) 3735–3742. [PubMed] [Google Scholar]
- [134].Fukushima N, Walter KM, Uek T, et al. , Diagnosing pancreatic cancer using methylation specific PCR analysis of pancreatic juice, Cancer Biol. Ther. 2 (2003) 78–83. [DOI] [PubMed] [Google Scholar]
- [135].Ginesta MM, Diaz-Riascos ZV, Busquets J, et al. , APC promoter is frequently methylated in pancreatic juice of patients with pancreatic carcinomas or periampullary tumors, Oncol. Lett. 12 (2016) 2210–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Henriksen SD, Madsen PH, Larsen AC, et al. , Cell-free DNA promoter hypermethylation in plasma as a diagnostic marker for pancreatic adenocarcinoma, Clin. Epigenet. 8 (2016) 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Yi JM, Guzzetta AA, Bailey VJ, et al. , Novel methylation biomarker panel for the early detection of pancreatic cancer, Clin. Cancer Res. 19 (2013) 6544–6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Pedersen KS, Bamlet WR, Oberg AL, et al. , Leukocyte DNA methylation signature differentiates pancreatic cancer patients from healthy controls, PLoS One 6 (2011) e18223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Parsi MA, Li A, Li CP, Goggins M, DNA methylation alterations in endoscopic retrograde cholangiopancreatography brush samples of patients with suspected pancreaticobiliary disease, Clin. Gastroenterol. Hepatol. 6 (2008) 1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Dauksa A, Gulbinas A, Barauskas G, et al. , Whole blood DNA aberrant methylation in pancreatic adenocarcinoma shows association with the course of the disease: a pilot study, PLoS One 7 (2012) e37509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Sato N, Fukushima N, Matsubayashi H, et al. , Aberrant methylation of Reprimo correlates with genetic instability and predicts poor prognosis in pancreatic ductal adenocarcinoma, Cancer 107 (2006) 251–257. [DOI] [PubMed] [Google Scholar]
- [142].Yokoyama S, Higashi M, Kitamoto S, et al. , Aberrant methylation of MUC1 and MUC4 promoters are potential prognostic biomarkers for pancreatic ductal adenocarcinomas, Oncotarget 7 (2016) 42553–42565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Thompson MJ, Rubbi L, Dawson DW, et al. , Pancreatic cancer patient survival correlates with DNA methylation of pancreas development genes, PLoS One 10 (2015) e0128814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Tan AC, Jimeno A, Lin SH, et al. , Characterizing DNA methylation patterns in pancreatic cancer genome, Mol. Oncol. 3 (2009) 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Perz JF, Armstrong GL, Farrington LA, et al. , The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide, J. Hepatol. 45 (2006) 529–538. [DOI] [PubMed] [Google Scholar]
- [146].Shaib YH, El-Serag HB, Davila JA, et al. , Risk factors of intrahepatic cholangiocarcinoma in the United States: a case-control study, Gastroenterology 128 (2005) 620–626. [DOI] [PubMed] [Google Scholar]
- [147].Banales JM, Cardinale V, Carpino G, et al. , Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA), Nat. Rev. Gastroenterol. Hepatol. 13 (2016) 261–280. [DOI] [PubMed] [Google Scholar]
- [148].Razumilava N, Gores GJ, Cholangiocarcinoma, Lancet 383 (2014) 2168–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Boberg KM, Lind GE, Primary sclerosing cholangitis and malignancy, Best Pract. Res. Clin. Gastroenterol. 25 (2011) 753–764. [DOI] [PubMed] [Google Scholar]
- [150].Yin CQ, Yuan CH, Qu Z, et al. , Liquid biopsy of hepatocellular carcinoma: circulating tumor-Derived biomarkers, Dis. Markers 2016 (2016) 1427849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Andresen K, Boberg KM, Vedeld HM, et al. , Four DNA methylation biomarkers in biliary brush samples accurately identify the presence of cholangiocarcinoma, Hepatology 61 (2015) 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].O’Rourke CJ, Munoz-Garrido P, Aguayo EL, Andersen JB, Epigenome dysregulation in cholangiocarcinoma, Biochim. Biophys. Acta (2017). [DOI] [PubMed] [Google Scholar]
- [153].Andresen K, Boberg KM, Vedeld HM, et al. , Novel target genes and a valid biomarker panel identified for cholangiocarcinoma, Epigenetics 7 (2012) 1249–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Shin SH, Lee K, Kim BH, et al. , Bile-based detection of extrahepatic cholangiocarcinoma with quantitative DNA methylation markers and its high sensitivity, J. Mol. Diagn. 14 (2012) 256–263. [DOI] [PubMed] [Google Scholar]
- [155].Branchi V, Schaefer P, Semaan A, et al. , Promoter hypermethylation of SHOX2 and SEPT9 is a potential biomarker for minimally invasive diagnosis in adenocarcinomas of the biliary tract, Clin. Epigenet. 8 (2016) 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Wong IH, Lo YM, Zhang J, et al. , Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients, Cancer Res. 59 (1999) 71–73. [PubMed] [Google Scholar]
- [157].Wahid B, Ali A, Rafique S, Idrees M, New insights into the epigenetics of hepatocellular carcinoma, BioMed Res. Int. 2017 (2017) 1609575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Nishida N, Goel A, Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review, Curr. Genomics 12 (2011) 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Puszyk WM, Trinh TL, Chapple SJ, Liu C, Linking metabolism and epigenetic regulation in development of hepatocellular carcinoma, Lab. Invest. 93 (2013) 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Ozen C, Yildiz G, Dagcan AT, et al. , Genetics and epigenetics of liver cancer, New Biotechnol. 30 (2013) 381–384. [DOI] [PubMed] [Google Scholar]
- [161].Zhang C, Li J, Huang T, et al. , Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma, Oncotarget 7 (2016) 81255–81267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Huang ZH, Hu Y, Hua D, et al. , Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma, Exp. Mol. Pathol. 91 (2011) 702–707. [DOI] [PubMed] [Google Scholar]
- [163].Shen J, Wang S, Zhang YJ, et al. , Genome-wide DNA methylation profiles in hepatocellular carcinoma, Hepatology 55 (2012) 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Villanueva A, Portela A, Sayols S, et al. , DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma, Hepatology 61 (2015) 1945–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Lee S, Kim WH, Jung HY, et al. , Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma, Am. J. Pathol. 161 (2002) 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Tozawa T, Tamura G, Honda T, et al. , Promoter hypermethylation of DAP-kinase is associated with poor survival in primary biliary tract carcinoma patients, Cancer Sci. 95 (2004) 736–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Nishida N, Kudo M, Alteration of epigenetic profile in human hepatocellular carcinoma and its clinical implications, Liver Cancer 3 (2014) 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Mah WC, Thurnherr T, Chow PK, et al. , Methylation profiles reveal distinct subgroup of hepatocellular carcinoma patients with poor prognosis, PLoS One 9 (2014) e104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Liu K, Zhang Y, Zhang C, et al. , Methylation of S100A8 is a promising diagnosis and prognostic marker in hepatocellular carcinoma, Oncotarget 7 (2016) 56798–56810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Jiang BG, Wang N, Huang J, et al. , Tumor SOCS3 methylation status predicts the treatment response to TACE and prognosis in HCC patients, Oncotarget 8 (2017) 28621–28627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Ioannidis JPA, Bossuyt PMM, Waste, leaks, and failures in the biomarker pipeline, Clin. Chem. 63 (2017) 963–972. [DOI] [PubMed] [Google Scholar]
- [172].Lind GE, van Engeland M, Details matter: the role of genomic location and assay standardization in DNA methylation analyses, Epigenomics 9 (2017) 933–935. [DOI] [PubMed] [Google Scholar]
- [173].Warton K, Mahon KL, Samimi G, Methylated circulating tumor DNA in blood: power in cancer prognosis and response, Endocr. Relat. Cancer 23 (2016) R157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Nilsson TK, Lof-Ohlin ZM, Sun XF, DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients, Int. J. Oncol. 42 (2013) 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Lin PC, Lin JK, Lin CH, et al. , Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-Wide high-Resolution array, Ann. Surg. Oncol. 22 (Suppl. 3) (2015) S1419–1427. [DOI] [PubMed] [Google Scholar]
- [176].Shima K, Nosho K, Baba Y, et al. , Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: cohort study and literature review, Int. J. Cancer 128 (2011) 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Esteller M, Gonzalez S, Risques RA, et al. , K-ras and p16 aberrations confer poor prognosis in human colorectal cancer, J. Clin. Oncol. 19 (2001) 299–304. [DOI] [PubMed] [Google Scholar]
- [178].Lee DW, Han SW, Cha Y, et al. , Different prognostic effect of CpG island methylation according to sex in colorectal cancer patients treated with adjuvant FOLFOX, Clin. Epigenet. 7 (2015) 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Jiang G, Luo C, Sun M, et al. , Methylation of CDX2 as a predictor in poor clinical outcome of patients with colorectal cancer, Genet. Test Mol. Biomarkers 20 (2016) 710–714. [DOI] [PubMed] [Google Scholar]
- [180].Fu T, Liu Y, Li K, et al. , Tumors with unmethylated MLH1 and the CpG island methylator phenotype are associated with a poor prognosis in stage II colorectal cancer patients, Oncotarget 7 (2016) 86480–86489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Yokoi K, Yamashita K, Ishii S, et al. , Comprehensive molecular exploration identified promoter DNA methylation of the CRBP1 gene as a determinant of radiation sensitivity in rectal cancer, Br. J. Cancer 116 (2017) 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Zheng R, Gao D, He T, et al. , Methylation of DIRAS1 promotes colorectal cancer progression and may serve as a marker for poor prognosis, Clin. Epigenet. 9 (2017) 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Sugimachi K, Matsumura T, Shimamura T, et al. , Aberrant methylation of FOXE1 contributes to a poor prognosis for patients with colorectal cancer, Ann. Surg. Oncol. 23 (2016) 3948–3955. [DOI] [PubMed] [Google Scholar]
- [184].Wallner M, Herbst A, Behrens A, et al. , Methylation of serum DNA is an independent prognostic marker in colorectal cancer, Clin. Cancer Res. 12 (2006) 7347–7352. [DOI] [PubMed] [Google Scholar]
- [185].Katoh H, Yamashita K, Waraya M, et al. , Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer, Neoplasia 14 (2012) 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Fu T, Pappou EP, Guzzetta AA, et al. , IGFBP-3 gene methylation in primary tumor predicts recurrence of stage II colorectal cancers, Ann. Surg. 263 (2016) 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Moya P, Esteban S, Fernandez-Suarez A, et al. , KiSS-1 methylation and protein expression patterns contribute to diagnostic and prognostic assessments in tissue specimens for colorectal cancer, Tumour Biol. 34 (2013) 471–479. [DOI] [PubMed] [Google Scholar]
- [188].Amatu A, Sartore-Bianchi A, Moutinho C, et al. , Promoter CpG island hypermethylation of the DNA repair enzyme MGMT predicts clinical response to dacarbazine in a phase II study for metastatic colorectal cancer, Clin. Cancer Res. 19 (2013) 2265–2272. [DOI] [PubMed] [Google Scholar]
- [189].Jensen LH, Rasmussen AA, Byriel L, et al. , Regulation of MLH1 mRNA and protein expression by promoter methylation in primary colorectal cancer: a descriptive and prognostic cancer marker study, Cell Oncol. (Dordr.) 36 (2013) 411–419. [DOI] [PubMed] [Google Scholar]
- [190].Heitzer E, Artl M, Filipits M, et al. , Differential survival trends of stage II colorectal cancer patients relate to promoter methylation status of PCDH10, SPARC, and UCHL1, Mod. Pathol. 27 (2014) 906–915. [DOI] [PubMed] [Google Scholar]
- [191].Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, et al. , Hypermethylation of RARbeta2 correlates with high COX-2 expression and poor prognosis in patients with colorectal carcinoma, Tumour Biol. 31 (2010) 503–511. [DOI] [PubMed] [Google Scholar]
- [192].Draht MX, Smits KM, Tournier B, et al. , Promoter CpG island methylation of RET predicts poor prognosis in stage II colorectal cancer patients, Mol. Oncol. 8 (2014) 679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Tham C, Chew M, Soong R, et al. , Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer, Cancer 120 (2014) 3131–3141. [DOI] [PubMed] [Google Scholar]
- [194].Tang D, Liu J, Wang DR, et al. , Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer, Clin. Invest. Med. 34 (2011) E88–95. [DOI] [PubMed] [Google Scholar]
- [195].Tsai MH, Chen WC, Yu SL, et al. , DNA hypermethylation of SHISA3 in colorectal cancer: an independent predictor of poor prognosis, Ann. Surg. Oncol. 22 (Suppl. 3) (2015) S1481–1489. [DOI] [PubMed] [Google Scholar]
- [196].He T, Zhang M, Zheng R, et al. , Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer, Epigenomics 9 (2017) 849–862. [DOI] [PubMed] [Google Scholar]
- [197].Yang Z, Huo L, Chen H, et al. , Hypermethylation and prognostic implication of Syk gene in human colorectal cancer, Med. Oncol. 30 (2013) 586. [DOI] [PubMed] [Google Scholar]
- [198].Esteban S, Moya P, Fernandez-Suarez A, et al. , Diagnostic and prognostic utility of methylation and protein expression patterns of myopodin in colon cancer, Tumour Biol. 33 (2012) 337–346. [DOI] [PubMed] [Google Scholar]
- [199].Park SJ, Kim SM, Hong YS, et al. , TFAP2E methylation status and prognosis of patients with radically resected colorectal cancer, Oncology 88 (2015) 122–132. [DOI] [PubMed] [Google Scholar]
- [200].Agrelo R, Cheng WH, Setien F, et al. , Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer, Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 8822–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Balgkouranidou I, Matthaios D, Karayiannakis A, et al. , Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients, Mutat. Res. 778 (2015) 46–51. [DOI] [PubMed] [Google Scholar]
- [202].Leung WK, To KF, Chu ES, et al. , Potential diagnostic and prognostic values of detecting promoter hypermethylation in the serum of patients with gastric cancer, Br. J. Cancer 92 (2005) 2190–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Wu L, Zhang C, Wang X, et al. , Methylation of ASC/TMS1 promoter is associated with poor prognosis of patients with gastric cancer, Clin. Transl. Oncol. 18 (2016) 296–303. [DOI] [PubMed] [Google Scholar]
- [204].Wanajo A, Sasaki A, Nagasaki H, et al. , Methylation of the calcium channelrelated gene, CACNA2D3, is frequent and a poor prognostic factor in gastric cancer, Gastroenterology 135 (2008) 580–590. [DOI] [PubMed] [Google Scholar]
- [205].Ushiku H, Yamashita K, Ema A, et al. , DNA diagnosis of peritoneal fluid cytology test by CDO1 promoter DNA hypermethylation in gastric cancer, Gastric Cancer 20 (2017) 784–792. [DOI] [PubMed] [Google Scholar]
- [206].Koga Y, Kitajima Y, Miyoshi A, et al. , The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer, J. Gastroenterol. 41 (2006) 133–139. [DOI] [PubMed] [Google Scholar]
- [207].Yao D, Shi J, Shi B, et al. , Quantitative assessment of gene methylation and their impact on clinical outcome in gastric cancer, Clin. Chim. Acta 413 (2012) 787–794. [DOI] [PubMed] [Google Scholar]
- [208].Wang H, Duan XL, Qi XL, et al. , Concurrent hypermethylation of SFRP2 and DKK2 activates the wnt/beta-Catenin pathway and is associated with poor prognosis in patients with gastric cancer, Mol. Cells 40 (2017) 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Ye X, Feng G, Jiao N, et al. , Methylation of DLEC1 promoter is a predictor for recurrence in Chinese patients with gastric cancer, Dis. Markers 2014 (2014) 804023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Eftang LL, Klajic J, Kristensen VN, et al. , GFRA3 promoter methylation may be associated with decreased postoperative survival in gastric cancer, BMC Cancer 16 (2016) 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Park TJ, Han SU, Cho YK, et al. , Methylation of O(6)-methylguanine-DNA methyltransferase gene is associated significantly with K-ras mutation, lymph node invasion, tumor staging, and disease free survival in patients with gastric carcinoma, Cancer 92 (2001) 2760–2768. [DOI] [PubMed] [Google Scholar]
- [212].Han J, Lv P, Yu JL, et al. , Circulating methylated MINT2 promoter DNA is a potential poor prognostic factor in gastric cancer, Digestive Dis. Sci. 59 (2014) 1160–1168. [DOI] [PubMed] [Google Scholar]
- [213].Deng J, Liang H, Zhang R, et al. , Applicability of the methylated CpG sites of paired Box 5 Box 5 (PAX5) promoter for prediction the prognosis of gastric cancer, Oncotarget 5 (2014) 7420–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Li X, Cheung KF, Ma X, et al. , Epigenetic inactivation of paired box gene 5, a novel tumor suppressor gene, through direct upregulation of p53 is associated with prognosis in gastric cancer patients, Oncogene 31 (2012) 3419–3430. [DOI] [PubMed] [Google Scholar]
- [215].Hou YC, Deng JY, Zhang RP, et al. , Evaluating the clinical feasibility: the direct bisulfite genomic sequencing for examination of methylated status of protocadherin10 (PCDH10) promoter to predict the prognosis of gastric cancer, Cancer Biomarker 15 (2015) 567–573. [DOI] [PubMed] [Google Scholar]
- [216].Xue WJ, Feng Y, Wang F, et al. , The value of serum RASSF10 hypermethylation as a diagnostic and prognostic tool for gastric cancer, Tumour Biol. 37 (2016) 11249–11257. [DOI] [PubMed] [Google Scholar]
- [217].Guo W, Dong Z, Guo Y, et al. , Aberrant methylation and loss expression of RKIP is associated with tumor progression and poor prognosis in gastric cardia adenocarcinoma, Clin. Exp. Metastasis 30 (2013) 265–275. [DOI] [PubMed] [Google Scholar]
- [218].Chen ZY, Zhang JL, Yao HX, et al. , Aberrant methylation of the SPARC gene promoter and its clinical implication in gastric cancer, Sci. Rep. 4 (2014) 7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Yu JL, Lv P, Han J, et al. , Methylated TIMP-3 DNA in body fluids is an independent prognostic factor for gastric cancer, Arch. Pathol. Lab. Med. 138 (2014) 1466–1473. [DOI] [PubMed] [Google Scholar]