Abstract
There is considerable interest in defining metabolic reprogramming in human diseases, which is recognized as a hallmark of human cancer. Although radiotracers have a long history in specific metabolic studies, stable isotope-enriched precursors coupled with modern high resolution mass spectrometry and NMR spectroscopy have enabled systematic mapping of metabolic networks and fluxes in cells, tissues and living organisms including humans. These analytical platforms are high in information content, are complementary and cross-validating in terms of compound identification, quantification, and isotope labeling pattern analysis of a large number of metabolites simultaneously. Furthermore, new developments in chemoselective derivatization and in vivo spectroscopy enable tracking of labile/low abundance metabolites and metabolic kinetics in real-time. Here we review developments in Stable Isotope Resolved Metabolomics (SIRM) and recent applications in cancer metabolism using a wide variety of stable isotope tracers that probe both broad and specific aspects of cancer metabolism required for proliferation and survival.
Keywords: SIRM, cancer metabolism, NMR, mass spectrometry, model systems
1. Introduction
Isotope tracing approaches in metabolic studies have a long history, where a biological system is given an isotopically enriched precursor such that its metabolic transformations can be tracked by the tracer atom. Due to the much better availability as well as ease and high sensitivity of detection, radioisotopes were much preferred over stable isotopes in early tracer studies and have been instrumental in deciphering both central (e.g. the Krebs cycle) and secondary metabolic pathways [5]. However, stable isotope tracers are non-hazardous, more amenable to human studies, and can be analyzed by both NMR and mass spectrometry (MS). Radioisotopes are NMR-silent except for 3H, which compromises the structural elucidation of the transformed products. Recent recognition that our understanding of cell and tissue metabolism in various human disease is grossly inadequate has driven greatly improved analytical techniques (especially NMR and MS) concomitant with the dramatically increased availability of various stable isotope-enriched precursors. Both advances have led to a resurgence of interest in utilizing stable isotope tracer approaches in metabolic studies. When coupling stable isotope tracers with advanced metabolomic technologies, such as Stable Isotope-Resolved Metabolomics (SIRM) [6–8], metabolic networks can now be systematically and rigorously tracked using the combined structural capabilities of NMR and MS. Figure 1 shows the pipeline of SIRM studies in different biological systems, which can be readily incorporated into a systems biochemistry workflow. Since 2000, more than 1300 articles on cancer metabolism with stable isotope tracing have been published using these search terms via Clarivate Web of Science.
Figure 1. SIRM Pipeline.
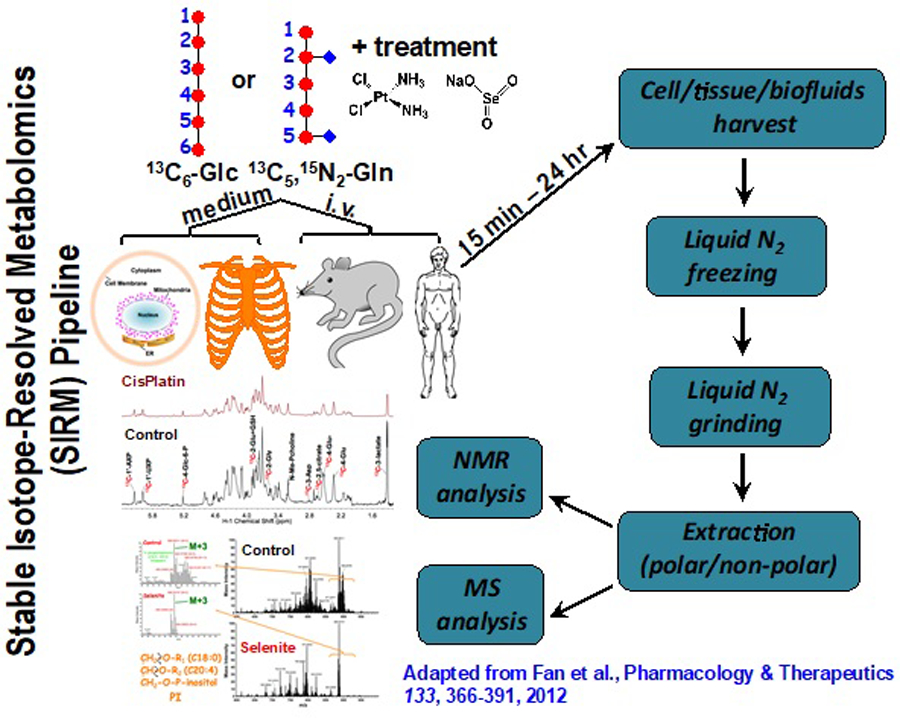
Stable isotope-enriched precursors are introduced to the biological system for a defined period, followed by harvesting the tissue, biofluid or medium, metabolite extraction and analysis by MS and NMR. The isotopomers and isotopologue distributions are then incorporated into metabolic network maps and flux models. Such results may also be combined with genomics, proteomics and in vivo NMR information acquired on the same samples.
In this review, we focus on recent applications of SIRM in cancer metabolism. Earlier work [9–19] and MS and NMR methodologies [20–27] have been extensively reviewed elsewhere.
2. Analytical Tools for SIRM: NMR and Mass spectrometry
The additional neutron(s) in the nucleus of a rare stable isotope enables simple detection by both NMR and mass spectrometry. For example, of the natural stable isotopes of carbon, nitrogen, and hydrogen, the rare (1.1 % abundance) 13C atom is ~1 Da heavier than the abundant 12C atom (98.9%), and similarly for 15N (0.35%) versus 14N and 2H (0.02%) versus 1H. However, two or more different tracer atoms such as 13C, 15N, and 2H cannot be distinguished in the same samples with typical resolution-MS. The advances in ultra high-resolution Fourier transform mass spectrometry (UHR-FTMS) with mass-to-charge ratio (m/z) resolution to the fifth decimal place for most metabolites now make it feasible to distinguish molecules with multiple stable isotope atoms of multiple elements. This is because the additional neutron mass in for example, 13C is different from that in 15N or 2H, so that their isotopologues (identical compounds differing by the number of tracer atoms [28]) are distinguishable by UHR-FTMS (Figure 2). This makes multiplexed Stable Isotope Resolved Metabolomics (mSIRM) experiments with the use of multiple tracer atoms readily feasible [29]
Figure 2. UHR-FTMS distinguishes isotopes of identical nominal mass.

Panel A indicates the m/z difference between 2H and 13C or 13C and 15N in the mamu range as well as the expected resolving power of a commercial FTMS instrument. Panel B shows the resolution of 2H, 13C, and 15N in ATP in a crude cancer cell extract (adapted from [3]). LW is the width at half height in milli atomic mass units (mamu).
Stable isotopes such as 13C can also be detected directly by NMR, whereas 12C and radioactive 14C are invisible to NMR. More importantly, 15N and 13C can be observed indirectly by the attached proton, thereby affording much higher sensitivity than direct detection [30]. As these nuclei resonate at widely different radiofrequencies (i.e. chemical shifts), mSIRM can also be achieved.
The power of NMR and MS techniques in metabolic studies lies in their versatility, complementarity and rigor in determining isotopically enriched metabolite structures and concentrations, particularly in terms of the number (isotopologues) and positions (positional isotopomers [28]) of the tracer atoms present. The two techniques are cross-validating as they analyze metabolite structures and quantify them based on entirely different physical principles. They are also complementary as NMR can more readily provide information on positional isotopomers than MS and vice versa for information on isotopologues of given metabolites. Combining both types of information with the appropriate experimental design more robustly reveal the paths of metabolic transformations and detailed metabolic fluxes (e.g. via time course and/or flux modeling analysis). This can all be achieved in unfractionated extracts of cells, tissues, or organs, and sometimes in whole organisms.
Furthermore, an added, unique advantage of NMR is its ability to observe metabolic transformations and their rates (fluxes) in live tissues or organisms in situ. Traditionally, in vivo NMR mainly used 31P NMR to determine the levels of ATP, inorganic phosphate, sugar phosphates and phosphocreatine as well as ATP turnover rates and intracellular pH [31, 32]. In recent years, stable isotope tracers have been used in both animal systems and humans [33–37] especially in combination with the new hyperpolarization methods (e.g. dynamic nuclear polarization or DNP) for short segments of metabolic pathways [38, 39].
3. Stable isotope tracers for metabolic studies
The most commonly used stable isotope in metabolic studies – such as in cancer metabolism research – is 13C. There are numerous 13C-enriched precursors commercially available, including several isotopomers of D-glucose (e.g. [U-13C], [13C-1,2], [13C-1,6], [13C-3,4]) for probing different routes of glucose metabolism, 13C-octanoate [40, 41], -palmitate and -acetate for probing fatty acid metabolism and fates of intracellular 13C-acetyl CoA [42, 43], 13C-fructose, and 13C-amino acids. It is also common to use 15N-enriched amino acids either as 15N only or double element labeled versions such as [U-13C,15N]-glutamine [44] for multiplexed SIRM studies. Such a wide variety of stable isotope-labeled sources opens a rich avenue of experimental designs for interrogating the vast and intricate metabolic networks in mammalian systems. Table 1 summarizes some commonly used, commercially available stable isotope tracers and example metabolic pathways they probe.
Table 1.
Common stable-isotope enriched compounds and their uses
| Tracer | Pathways sampled | references |
|---|---|---|
| [U-13C]-glucose | glycolysis, Krebs cycle, PPP, glycogen synthesis, hexosamine biosynthesis pathway, serine-glycine-one carbon metabolism, nucleotide and lipid synthesis | [10, 45] |
| [13C-1,2]-glucose | Non-oxidative versus oxidative branches of the PPP | [46, 47] |
| [13C-3,4]-glucose | anaplerosis via pyruvate carboxylation | [48, 49] |
| [13C-1]-pyruvate | Lactic fermentation | [50] |
| [U-13C]-lactate | Krebs cycle, gluconeogenesis | [51–54] |
| [U-13C,15N]-glutamine | glutaminolysis, Krebs cycle, gluconeogenesis, transamination, nucleotide and lipid synthesis | [55–59] |
| [U-13C]-palmitate [U-13C]-octanoate [U-13C]-oleate |
beta-oxidation, fatty acid synthesis, fatty acid uptake | [41, 60–62] [63] |
| [U-13C]-serine | Serine and one-carbon metabolism; purine biosynthesis; lipid synthesis (PS, sphingolipids) | [64] [65] |
| 2H3-serine | Serine and one-carbon metabolism; purine biosynthesis; lipid synthesis (PS, sphingolipids) | [29] |
| [U-13C]-glycine | Purine biosynthesis; glutathione biosynthesis | [65, 66] |
| 13C branched chain amino acids | Amino acid metabolism | [67–69] |
| 15N arginine, citrulline | Arginine/NO metabolism | [70] |
| [U-13C]-glycerol | Lipid synthesis, gluconeogenesis-PPP interactions | [41, 71, 72] |
| 2H2O | Lipid synthesis in vivo | [73, 74] |
See also Cambridge Isotope Laboratories (CIL) note: MET_RSCH_CANCER (2/13/18)
4. Selective detection of a class of stable isotope-enriched metabolites
Phosphorus is an important biological element, but the major stable isotope 31P is close to 100% abundant. 31P is a spin ½ nucleus with a gyromagnetic ratio 40% as large as that of the proton. Therefore it is a good NMR nucleus, and can be used to filter or edit a complex mixture for the detection of phosphorylated compounds, which may also be enriched with 13C or 15N. The simplest editing experiment is the 1H{31P} HSQC experiment which detects only metabolites that contain the three bond 31P-O-CH coupling. These include all phosphorylated metabolites such as nucleotides, phosphosugars and intermediates of the pentose phosphate pathway [75]. This experiment greatly simplifies the 1H NMR spectrum of a complex unfractionated biological extract, A wide variety of edited NMR experiments can be used to select for different biologically important nuclei particularly 13C and 15N. However, it may be difficult to assign metabolites reliably based on this experiment alone. HSQC-TOCSY experiments relay magnetization to additional protons to enable mapping of more of the covalent network in phosphorylated metabolites [22, 75]. If the protons are attached to 13C, then their 13C satellites will be observable and quantifiable in the HSQC-TOCSY spectrum, and thus provide information on the site-specific enrichment in phosphorylated metabolites [20]. Such 13C and/or 15N isotopomer analysis of metabolites via spectral editing decreases spectral crowding that can overwhelm low abundant metabolites, and such selection is difficult to accomplish with MS analysis. An alternative approach to achieve selective detection of metabolites is the use of chemoselective derivatization as described below.
Systematic and reliable compound identification in crude extracts already requires significant effort, even with the combination of multidimensional NMR, tandem high-resolution MS, and extensive compound libraries. The problems are greatly exacerbated in SIRM studies where the multiple isotopomers and isotopologues of the many labeled metabolites must be identified and quantified. In fact, each isotopologue and isotopomer of a biochemical is essentially a different metabolite because they have different metabolic provenance, which are clues that their function and fate may be different. That is, the metabolic biosynthetic history is “encoded” in the NMR and MS spectra of each enriched biochemical.
This spectral encoding precludes identification by simple NMR and MS spectral matching. Moreover the presence of labeled metabolites causes greater spectral crowding, and also reduces the detectability of given metabolites by NMR and MS as each metabolite signal is split into multiple signals.
Chemoselective derivatization is a means of reducing spectral crowding while enhancing detection both by NMR (via spectral editing) and by MS. Several chemoselective probes have been reported, including cholamine for carboxylate groups [76], N-(2-15N-aminooxyethyl)-N,N-dimethyl-1-dodecylammonium (QDA) for aldehydes and ketones [1, 77, 78], N-(2-15N-iodoacetamido)-N,N-dimethyl-1-dodecylammonium (QDE) for sulfhydryl [4] and ethylchoroformate (ECF) for amino, hydroxyl, and carboxyl groups [29]. Each chemoselective derivatizing agent places a spectrally discernable tag on one or a few particular functional groups, which not only help metabolite identification but also quantification by NMR and MS. For example, the QDA or QDE probe contains a permanent positive charge, which facilitates ionization and thus detectability in MS (Figure 3). In addition, stable isotope-enriched versions of QDA or QDE (e.g. 13CD3-QDA) can be prepared and reacted with metabolites as 1:1 mixture with the unlabeled probe. This greatly facilitates the identification and quantification of carbonyl or thiol containing metabolites simply by detecting a pair of adducts with an m/z difference of 4.02188 using UHR-FTMS (Figure 3).
Figure 3. Chemoselective probe design for facilitating MS-based structural analysis and quantification.
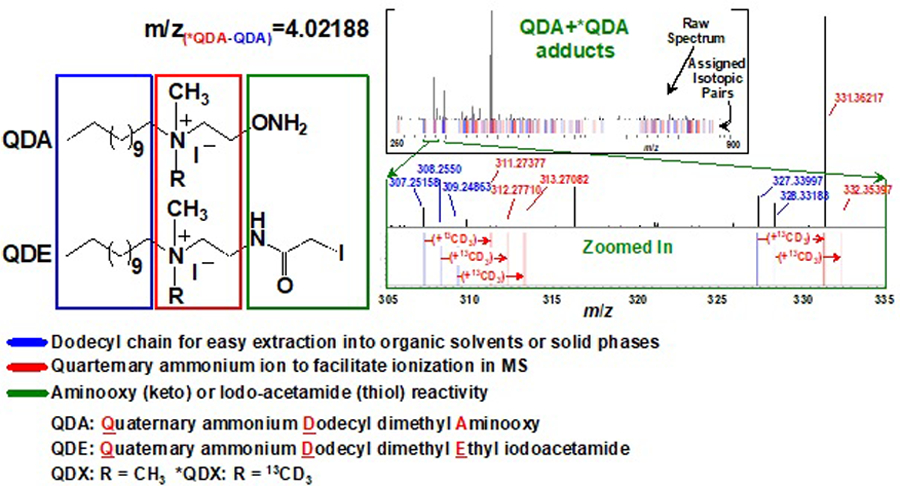
The probe design includes the carbonyl-selective aminooxy for QDA [1] or thiol-selective iodoacetamide groups for QDE [4], plus two common structural motifs of a dodecyl chain for extractability into organic phases and a quaternary ion for ready ionization in MS. Each probe can also be prepared as a pair with R equal to CH3 and 13CD3 (*QDA or *QDE). The resulting adducts of metabolites with a 1:1 mixture of QDA and *QDA can be readily discerned by an m/z difference of 4.02188 by UHR-FTMS, as illustrated by a “bar-code”-like profile of the adduct pairs in the spectrum on the top right (adapted from [1]).
Furthermore, this QDA probe can be modified to bear a 15N-aminooxy group, which reacts with carbonyl metabolites such as pyruvate to yield a ketoxime adduct with 3-bond coupling of 15N to the adjacent protons (Figure 4A). This coupling enables 15N-edited HSQC detection of the adduct in both 1D and 2D experiments, which would otherwise be impractical to discern in unedited 1H NMR analysis (Figure 4B–C).
Figure 4. Chemoselective derivatization enables NMR detection of carbonyl metabolites.
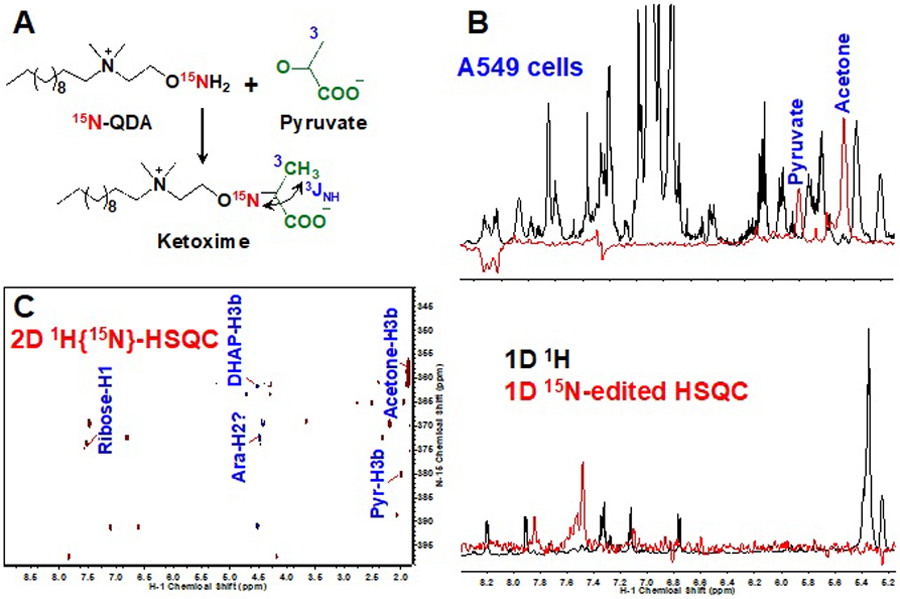
The QDA probe from Figure 3 was modified to contain a 15N aminooxy group, which forms a ketoxime adduct upon reaction with carbonyl metabolites such as pyruvate (A). The resulting three-bond coupling between 15N and neighboring protons of carbonyl metabolites enables their detection in crude A549 cell extracts by 1D (B) and 2D (C) 15N-edited HSQC experiments. Also shown in B is the comparison of unedited 1H and 15N-edited HSQC spectra with clear detection of carbonyl metabolites in the latter but not in the former
Thus, chemoselective derivatization can be coupled with stable isotope enrichment and spectral editing to facilitate the analysis of low abundance metabolites whose NMR or MS signals are typically swamped by those of more abundant metabolites. It is also important to note that the derivative formation helps stabilize labile (e.g. 4-hydroxynonenal) and volatile metabolites (e.g. acetone) for NMR and MS analysis.
5. Applications in Cancer Metabolism
As noted above, metabolic reprogramming is now recognized as a hallmark of human cancer [79] and cancer metabolism studies have most frequently taken advantage of the stable isotope tracer approach. The transformation of a normal cell to a highly proliferative cancer state is associated with numerous metabolic perturbations, which are often interactively linked to changes in the transcriptional program driven by alterations of critical oncoproteins and/or tumor suppressors. During growth, the increasingly hostile tumor microenvironment can further promote metabolic reprogramming to support tumor progression [80–82]. Most solid tumors up-regulate glycolysis and convert a high fraction of the glucose consumed to lactate (i.e. the Warburg effect), which is excreted along with a proton to acidify the extracellular space. In order to support cell growth, survival, and eventual metastasis, other aspects of the metabolism must also be up-regulated, especially the synthesis of precursors for nucleic acids, lipids, complex carbohydrates and proteins, e.g. serine and glycine (for purine biosynthesis, one-carbon metabolism), aspartate (for pyrimidine biosynthesis), citrate (for lipid biosynthesis), UDP-glucose (for glycogen synthesis), non essential amino acids such as alanine, glutamate, proline for protein synthesis [83], and glutamate (for antioxidant glutathione synthesis). It is also becoming clear that the metabolism of essential amino acids, particularly the branched chain amino acids Ile, Leu and Val are important in the development of some cancers [84].
Enhanced lactic fermentation is readily determined using 13C-glucose, by tracking the 13C fate in cell culture and organ systems from the uptake of 13C-glucose to the excretion of 13C-lactate in terms of both levels and fractional enrichment as well as the fraction of glucose conversion to lactate. The excretion of 13C-Ala and 13C-Glu (due to exchange with extracellular cystine) are often observed in the 13C-glucose tracer studies of cancer cells and tissues, which informs the status of Ala metabolism via the alanine aminotransferase (ALT) activity and the demand for glutathione (cysteine being a precursor). The fate of individual 13C atoms of glucose into intracellular metabolites can also be traced, to map both central catabolic and anabolic pathways that are keys to supporting cancer cell proliferation and survival. These include, but not limited to, the pentose phosphate pathway (PPP), the Krebs cycle, Ser-Gly-one carbon metabolism, and the synthesis of glutathione, non-essential amino acids, purine/pyrimidine nucleotides, nucleotide sugars (e.g. UDP N-acetylglucosamine or UDP-GlcNAc), glycogen, and lipids. Moreover, the incorporation of newly synthesized (i.e. labeled) non-essential amino acids into proteins and nucleotides into RNA can be determined following proper hydrolysis of the macromolecules [29, 85]; such “metabolomics of biomacromolecules” is in its infancy, but these large sinks and sources of metabolites are critical to detect and quantify.
The PPP is important both for generating cytoplasmic NADPH (required for anabolic and detoxification metabolism) and ribose for nucleotide synthesis. Thus, proliferating cancer cells have a strong requirement for the PPP activity. However, it is often unclear the extent of which the PPP contributes to NADPH production via the oxidative branch in cancer cells. It is feasible to discriminate the activity of the oxidative from non-oxidative branches using specific 13C isotopomers of glucose, such as [13C-1,2]-glucose analyzed via MS analysis [86] or a mixture of [13C-1]- and [13C-2]-glucose by NMR analysis [41].
Although accelerated lactic fermentation is a common adaptation in cancer cells, there are numerous other adaptations that generate metabolic energy and anabolic precursors that are equally important. It was long conjectured that mitochondria are non-functional in cancers. However, tracer studies with 13C-glucose, 13C-Gln and 13C-fatty acids clearly demonstrated that cancer cells in general oxidize a wide range of substrates in the mitochondria, even under hypoxia. Indeed, many established cancer cells have a high requirement for glutamine which also serves as the nitrogen donor in important amidotransferase reactions such as those in nucleotide [57] and UDP-GlcNAc biosynthesis, and as a source of Glu which is the most common co-substrate for aminotransferases. We have shown in SIRM studies using NMR and MS that many aspects of mitochondrial metabolism including anaplerotic pyruvate carboxylation is activated in vivo [87, 88] and ex vivo in human lung cancer tissues [82, 87]
Mitochondrial metabolism, the PPP, and glycolysis together are essential to fueling nucleotide biosynthesis, which is fundamental to cell proliferation [89]. As indicated above, the ribose subunit of the free nucleotides is directly derived from glucose (or glycogen) as shown by tracer studies with 13C-glucose [85]. In contrast, the nucleobases derive from multiple sources. Uracil (and subsequently cytosine) derives primarily from aspartate, which is mainly synthesized de novo in cells and tissues as the extracellular concentration of Asp is very low and it is not efficiently transported. Asp can be synthesized via transamination of oxaloacetate with Glu in both the cytoplasm (via GOT1) and the mitochondria (via GOT2). Using [U-13C,15N]-Gln as the tracer, the elevated presence of the m+5 (i.e.13C4,15N1) isotopologue of Asp [25] is evidence for the activation of the aminotransferases GOT1/2 [90]. This transamination is also part of the aspartate/malate shuttle system for transferring reducing equivalents from the cytoplasm to the mitochondria for oxidation via the electron transport chain. Under respiration deficiency, pyruvate may act as an alternative electron acceptor and utilize pyruvate carboxylase to maintain Asp synthesis [91–93]. Indeed, we determined that pyruvate carboxylase is activated in vivo in human lung cancer by employing 13C6-glucose infusion into patients coupled with SIRM analysis [87]. We also determined that Gln is a better substrate for Asp and uracil ring synthesis than glucose in cancer cells by comparing 13C incorporation from 13C6-glucose and [U-13C,15N]-Gln into the two products [85]. Interestingly, Asn is not a precursor of Asp in mammalian cells owing to the lack of asparaginase, but it can rescue cell growth and survival under low Gln availability [94].
Glycine-derived one-carbon metabolism plays a central role in cell cycle control during cell growth and replication [95]. Purine synthesis requires glycine as a direct donor of two carbons and one nitrogen. Glycine is also a source of the one-carbon unit in formyl tetrahydrofolate (THF) that supplies two additional purine ring carbons. Although glycine is freely available in the serum, it is also synthesized in cells from serine, which in turn is made from the glycolytic intermediate 3-phosphoglycerate. This pathway is up-regulated in some human cancers [95, 96] and as much as 8–9% of the glucose utilization may be shunted to serine synthesis [96]. In addition, purine synthesis requires Gln and we have shown that the three nitrogen atoms in the ring of purine nucleotides are derived from Gln by using [U-13C,15N] Gln as a tracer. Two nitrogen atoms derive from the amidotransferase reactions and the third is from Asp, which should have received its amino nitrogen via transamination from Gln-derived Glu [25]. Other than fueling nucleotide biosynthesis, one-carbon metabolites such as 5-Methyl THF is the source of methyl groups for replenishing S-adenosylmethionine (SAM) from S-adenosylhomocysteine (SAH). SAM is the universal methyl donor for a wide range of methyl transferases involved in epigenetic methylation of histones as well as cytosine in DNA [97].
Although many cancers are driven by alterations in oncoproteins and tumor suppressors, which often lead to reprogramming of cell metabolism, there are also cases where loss or gain of function directly in enzymes is key to carcinogenesis. For example, loss-of-function alterations in fumarate hydratase (FH) and succinate dehydrogenase (SDH) are important to the development of several aggressive familial cancers of the kidney [98–101]. Such loss of enzyme activity results in the accumulation of high levels of fumarate or succinate, respectively, which are derived predominantly from glutamine based on 13C tracer studies in cell cultures [56, 102] (cf. Figure 5). Cells expressing these inactive enzymes display substantial reductive carboxylation via a reversal of the isocitrate dehydrogenase (IDH) reaction to support lipogenesis for growth. The accumulated fumarate and succinate are potent inhibitors of the α–ketoglutarate-dependent dioxygenases, including prolyl hydroxylases (e.g. those that hydroxylate hypoxia-inducible factor Hif1α for marking for proteasomal degradation) and the epigenetic modulating enzymes Ten-eleven translocation enzyme (TET) (Fig. 5) and jumonji-domain histone K/R demethylases.
Figure 5. Metabolism and epigenetic regulation.
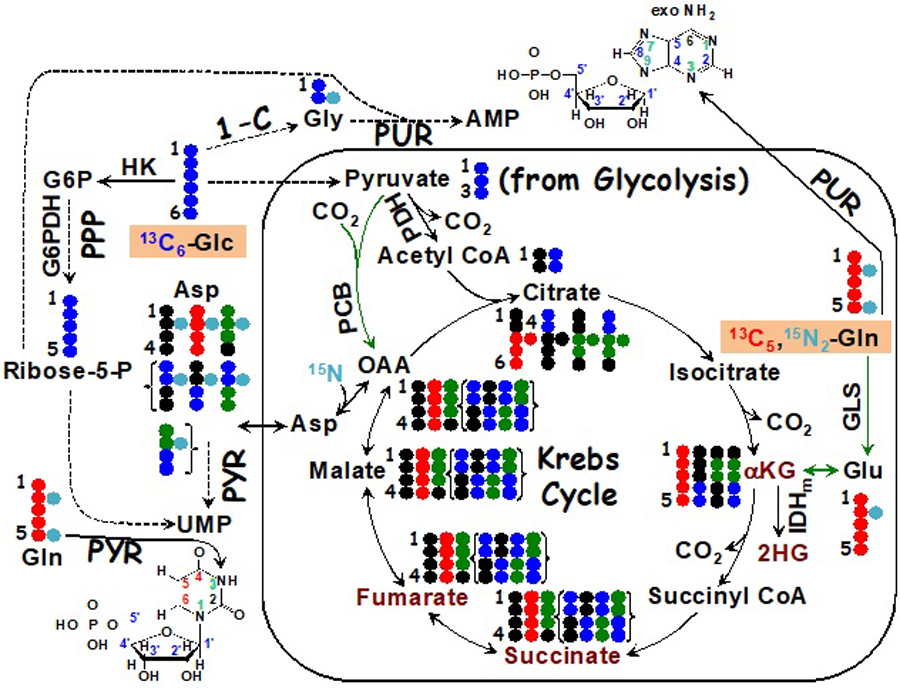
The diagram depicts 13C tracing from 13C6-glucose ( ) or 13C/15N tracing from 13C5,15N2 glutamine (
) or 13C/15N tracing from 13C5,15N2 glutamine ( ) to the synthesis of nucleotides and effectors (dark red text) involved in epigenetic modifications, including αketoglutarate (αKG), 2-hydroxyglutarate (2HG), succinate, and fumarate. IDH1/2 mutants produce the histone demethylation inhibitor 2HG from αKG while inactivating SDH and FH mutations generate high levels of succinate and fumarate, which are inhibitors of αKG-dependent dioxygenases including TET involved in demethylation of methylcytosine in DNA. Green arrows denote anaplerotic inputs into the Krebs cycle mediated by pyruvate carboxylase (PCB) (
) to the synthesis of nucleotides and effectors (dark red text) involved in epigenetic modifications, including αketoglutarate (αKG), 2-hydroxyglutarate (2HG), succinate, and fumarate. IDH1/2 mutants produce the histone demethylation inhibitor 2HG from αKG while inactivating SDH and FH mutations generate high levels of succinate and fumarate, which are inhibitors of αKG-dependent dioxygenases including TET involved in demethylation of methylcytosine in DNA. Green arrows denote anaplerotic inputs into the Krebs cycle mediated by pyruvate carboxylase (PCB) ( ) and glutaminase (GLS) reactions. { } encloses scrambled 13C labeling patterns of succinate, fumarate, malate, and Asp after one turn of the Krebs cycle. ●: 12C; OAA: oxaloacetate; HK: hexokinase; G6PDH: glucose-6-phosphate dehydrogenase; PDH: pyruvate dehydrogenase; IDHm: isocitrate dehydrogenase mutant; PUR: purine synthesis; PYR: pyrimidine synthesis; PPP: pentose phosphate pathway; 1-C: one-carbon pathway; exo: exocyclic.
) and glutaminase (GLS) reactions. { } encloses scrambled 13C labeling patterns of succinate, fumarate, malate, and Asp after one turn of the Krebs cycle. ●: 12C; OAA: oxaloacetate; HK: hexokinase; G6PDH: glucose-6-phosphate dehydrogenase; PDH: pyruvate dehydrogenase; IDHm: isocitrate dehydrogenase mutant; PUR: purine synthesis; PYR: pyrimidine synthesis; PPP: pentose phosphate pathway; 1-C: one-carbon pathway; exo: exocyclic.
In addition to loss of function enzyme variants, there are rare gain-of-function mutations in the genes encoding IDH1 and 2, which are NADPH-dependent cytoplasmic and mitochondrial isoforms, respectively. These mutations alter the enzyme activity from oxidative decarboxylation of isocitrate to α–ketoglutarate (αKG) to reduction of αKG to 2-hydroxyglutarate (2HG) with NADPH [103, 104]. 2HG is now known as an oncometabolite that accumulates to high levels in certain gliomas [105] and in acute myeloid leukemia (AML) [106]. This has the combined effect of depleting NADPH (which is needed both for anabolic purposes and for detoxifying excess H2O2 production) and αKG (which inhibits the αKG-dependent dioxygenases [107, 108]) as well as inhibiting histone demethylation to block cell differentiation [109].
NADPH is a key coenzyme and the NADPH/NADP+ ratio is tightly regulated in cells to support anabolic reactions and anti-oxidative defense. The possible sources of the hydride have been elegantly determined using different deuterated substrates that result in the production of NADPD [110, 111]. When combined with genetic modification of specific enzymes, tracing deuterium (D or 2H) can also provide information about compartmentation of different NADPH-producing activity such as PPP and reactions catalyzed by malic enzymes and IDHs [110], which prompts a need for reassessing NADPH production in cancer cells.
6. Future directions
In most cases, metabolic reprogramming in human cancer involves quantitative (i.e. altered production of existing metabolites) rather than qualitative changes (formation of new metabolites due to e.g. gain-of-function mutations such as in IDH [112]). Regardless of the nature of the reprogrammed events, stable isotope tracing has opened many new avenues for resolving and quantifying metabolic fates in atomic detail that are crucial to deciphering reprogrammed metabolic networks required for supporting cancer cell proliferation, survival and metastasis. Tracer data are also essential for rigorous metabolic flux analysis (e.g. [113, 114]), which can inform the changes in the kinetics of individual enzymes and/or transporters in given metabolic networks in-cell, in-tissue, and in vivo, in response to disease development or therapeutic interventions. This information is valuable in its own right but is even more powerful when linked to functional genomic data for elucidating altered metabolic regulatory network [115, 116].
6.1. Cancer metabolomic studies in better preclinical models
The majority of the stable isotope tracer studies have been performed in 2D cell cultures, which cannot recapitulate proper cell-cell and cell-extracellular matrix interactions. These microenvironment factors can have a significant impact on cell behavior and functions including gene expression profiles and drug responses [117–121]. This is a key reason for the U.S. National Cancer Institute to abandon the NCI-60 cell panel for drug screening purposes. The development of 3D cell cultures including spheroids of single cell types and organoids of multiple cell types [118, 122] can circumvent these drawbacks of 2D cell cultures. Multiple 3D cell culturing techniques are now available including scaffold (e.g. Matrigel, hydrogel) and scaffold-free (hanging drop, micropatterned/U-shape cell repellent plates) methods [118, 122, 123]. They variously suffer from technical challenges such as long spheroid formation times with variable efficiency and/or difficult handling for high throughput assays [120]. For adaptation to SIRM studies, there are added challenges from matrix contamination (e.g. from Matrigel) that interferes with metabolite qunatification and/or limitations in scaling-up.
A recently developed matrix-free 3D culture method, magnetic 3D bioprinting (M3DB) can overcome these difficulties. This technique utilizes nanoparticles composed of gold, iron oxide, and poly-L-lysine to magnetize cells, followed by spheroid or organoid assembly within minutes to hours under mild magnetic forces in 6- to 384-well cell repellent plates [120].
We have employed this system in a SIRM study of a patient-derived organoid (PDO) culture subjected to [13C6]-Glc ± anti-cancer selenite treatments in a 96-well plate. As shown in Figure 6, [13C6]-Glc transformations into the metabolites of glycolysis, PPP, Krebs cycle, gluconeogenesis (GNG), and purine/pyrimidine/glutathione biosynthesis pathways were readily observed, in addition to multiple other pathways (e.g. UDPGlcNAc synthesis) of importance to cancer cell growth and development (not shown). It is also clear that this PDO was insensitive to SeO3 treatment based on the metabolic responses, unlike A549 cells in 2D cultures with extensive growth and metabolic changes after only 24 h of 6.25 µM SeO3 treatment [115, 124]. This study demonstrates the feasibility of performing SIRM studies in high-throughput formats on cancer cell organoids or spheroids. Future developments in this area such as SIRM coupled with 3D co-cultures of different cell types [125, 126] and microfluidics-based 3D cultures [123] will help transform our understanding on the influence of tumor microenvironment on cancer and stromal cell metabolism.
Figure 6. SIRM study on patient-derived organoids (PDO) in response to anti-cancer selenite.
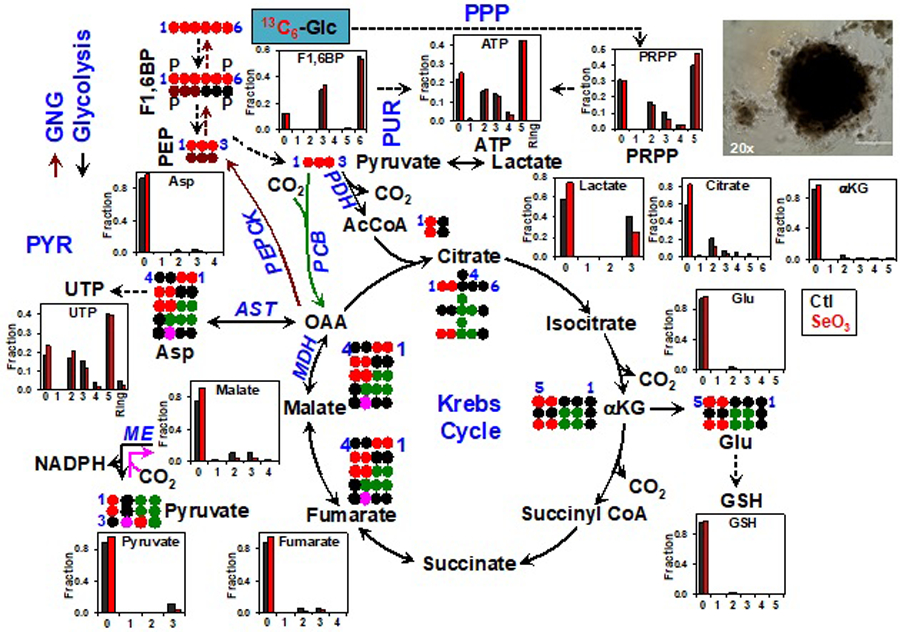
PDO was isolated from a non-small cell lung cancer patient (UK022) tumor xenograft in Matrigel and subsequently grown as M3DB cultures (example 20x image shown) in [13C6]-Glc+DMEM medium ± 10 µM selenite (SeO3) for 48 hr. Atom-resolved tracing from [13C6]-Glc to glycolytic, Krebs cycle, and gluconeogenic (GNG) metabolites is shown along with the 13C labeling patterns of these metabolites in response to SeO3. Also shown are the SeO3-induced changes in 13C labeling patterns of the products of PPP, purine (PUR)/pyrimidine (PYR) synthesis, and glutathione (GSH) synthesis. The numbers in X-axis represent the number of 13C atoms (isotopologues) in each metabolite while “Ring” in ATP and UTP indicates the sum of all 13C-nucleobase isotopologues.  denote GNG and anaplerotic inputs into the Krebs cycle mediated by PCB, respectively. ●: 12C;
denote GNG and anaplerotic inputs into the Krebs cycle mediated by PCB, respectively. ●: 12C;  : PDH-, PCB-, or malic enzyme (ME) initiated Krebs cycle reactions, respectively;
: PDH-, PCB-, or malic enzyme (ME) initiated Krebs cycle reactions, respectively;  : GNG; F1,6BP: fructose-1,6-bisphosphate; PEP: phosphoenolpyruvate; PRPP: phosphoribosylpyrophosphate; PEPCK: PEP carboxykinase; MDH: malate dehydrogenase; AST: aspartate amino transferase. All other symbols and abbreviations are as in Fig. 5.
: GNG; F1,6BP: fructose-1,6-bisphosphate; PEP: phosphoenolpyruvate; PRPP: phosphoribosylpyrophosphate; PEPCK: PEP carboxykinase; MDH: malate dehydrogenase; AST: aspartate amino transferase. All other symbols and abbreviations are as in Fig. 5.
Another common preclinical model for human cancer is tumor xenografts in mice including those of established cancer cells or preferably patient-derived tumor xenografts (PDTX). Stable isotope tracer studies have been performed in vivo on mouse xenograft models via intravenous (i.v.) injection of tracers as a bolus [127] and/or continuous infusion [128]. These tracer introduction methods involve physical constraints and/or the use of anesthetics and minor surgery, which traumatize, to variable extents, the animals and thereby alter their individual baseline metabolism. In addition, the duration of the tracer infusion is typically minutes to hours, which limits the observed pathways to those with relatively rapid turnover such as glycolysis, PPP, and the Krebs cycle. Pathways that involve de novo synthesis of lipids, proteins, and nucleotides require much longer periods of tracer administration, which is problematic with the i.v. method. We have recently introduced stable isotope tracer delivery via a liquid diet, which is non-invasive with no restriction on the tracer duration [2]. Using this method, we observed de novo protein synthesis in different mouse organs from 18 h of [13C6]-Glc feeding (Figure 7). When applied to PDTX mouse models, we noted distinct glucose metabolism between primary tumor and its distant lymph node metastases [2]. Future applications of such tracer delivery method can greatly facilitate metabolic network analysis including flux modeling in vivo in animal models of human cancer.
Figure 7. ECF-UHR-FTMS analysis indicates differential [13C6]-glucose incorporation into proteins in 5 mouse organs.
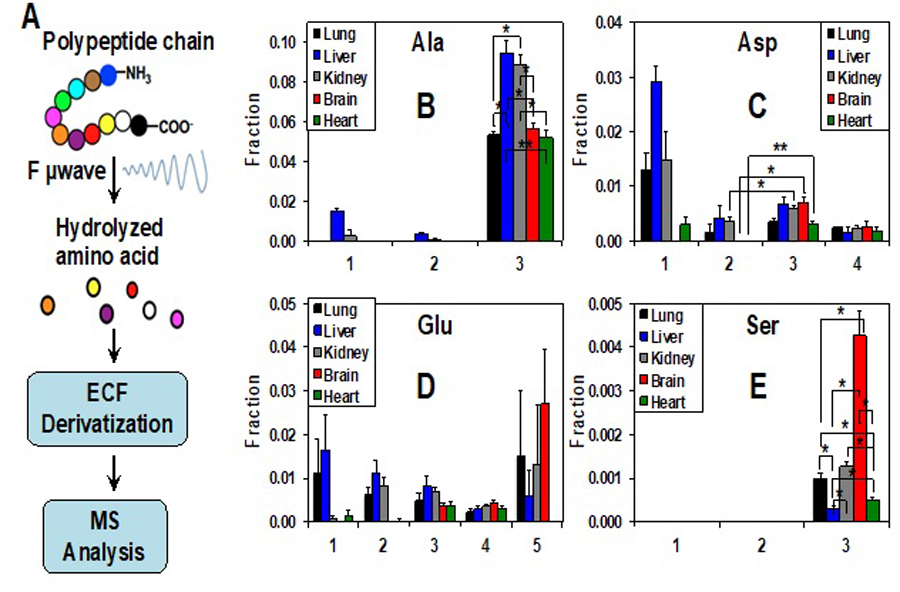
Extracted tissue proteins were hydrolyzed in 6N HCl by focused-beam microwave (F µwave) digestion. The free amino acids liberated were derivatized with ethyl chloroformate (ECF) before analysis by UHR-FTMS (A). Fractional enrichment distribution of 13C isotopologues of the proteinaceous amino acids Ala, Asp, Glu, and Ser is shown in B to E, respectively. The x-axis represents the number of 13C atoms present in each isotopologue. Values shown were mean ± SEM (n=3). * and ** in B, E denote false discovery q values for liver or kidney versus lung, brain, and heart; 0.01<q<0.05 and 0.001<q<<0.01, respectively. * in C denotes p values (<0.05–0.01) for 13C3- (3) versus 13C2-Asp (2) for kidney, brain, and heart (reprinted from Fig. S6, ref [2]. under a Creative Commons Attribution 4.0 International License http://creativecommons.org/licenses/by/4.0/).
6.2. High throughput multi-dimensional NMR
To greatly expand 1D NMR resolution, to gain much more structural insights, and to enhance isotopomer distribution analysis, further development in multidimensional NMR is needed, particularly in terms of the speed of data acquisition. There have been numerous developments in this area, notably non-uniform sampling [129, 130] and projection-reconstruction [131] [132] among others [133–135]. There are also hardware approaches, including the use of multiple receivers for the simultaneous acquisition of 2 or more 2D NMR data [136, 137]. The number of increments needed for multidimensional NMR can also be reduced using selective excitation methods, which could be especially useful for sugar phosphates where the spectral dispersion is low. The combination of sparse sampling in 2 dimensions [138], multiple receivers coupled with relaxation enhancement could decrease the total experimental time by more than an order of magnitude, which should enable fast-throughput high-dimensional SIRM NMR analysis in the future.
6.3. Improved quantification and isotopomer analysis by MS
Despite the very high sensitivity, mass accuracy, and resolution of UHR-FTMS, identifying and resolving different isotopologues of numerous metabolites in crude extracts remains a technical challenge. UHR-FTMS models capable of mSIRM currently range in performance from >350,000 resolution at m/z=400 (R400) upwards to a few million, and systematic analysis of labeled metabolites certainly benefits from further improvement of the UHR-FTMS hardware both in terms of sensitivity and m/z resolution (e.g. R400 up to 106 for Thermo’s Orbitrap Fusion™ Lumos™ and R400 >106 Bruker’s SolariX XR FTMS). It is also advantageous to improve on the fragmentation methods for UHR-FTMS so that more complex, substructure-specific MS/MS data can be acquired for metabolite identification and labeled position (isotopomer) analysis. For example, the recent development in UVPD (UV photodissociation) [139] technique for UHR-FTMS enables fragmentation at fatty acyl double bonds, yielding at once double-bond positions and narrowing of stable isotope locations. Both are traditionally tediously difficult tasks and will be a valuable addition to lipid pathway analysis in SIRM studies. Moreover, the increasing choices of stable isotope tracer standards will greatly facilitate robust MS quantification of unlabeled and labeled metabolites alike. This is synergistic with the high throughput direct-introduction UHR-FTMS methods that are undergoing both active improvements in nanoelectrospray, DESI, DART and others, and emergent techniques such laser-thermal desorption and “paperspray” among others [140]. All these (and other) developments in MS greatly extend the choice of experimental designs and tracer standards for standard addition-based quantification of labeled metabolites such as the use of deuterated amino acid standards for quantifying 13C and/or 15N labeled amino acids in crude extracts, anchored by UHR-FTMS [27, 29].
Ultrahigh sensitivity also means that there is an increased tendency to carry out what is really trace analysis at femtomole or lower quantities in the sample, for those low abundance but highly potent metabolites. Even moderate abundance metabolites become trace level in small samples, such as single cells [141, 142]. For example, in a cell of volume 1–2 pL, a fairly typical volume for an epithelial cancer cell, the amount of substrate present at a cellular concentration of 1 μM is 1–2 amol, which with an ionization efficiency of 1% in electrospray after chromatographic separation implies only a few thousand ions reach the detector.
6.4. Informatics needs for pathway mapping and analysis
Once the labeling patterns of metabolites are determined in tracer studies, they are valuable for rigorous reconstruction of metabolic networks, including compartmentalized events across cells and tissues [26, 55, 143, 144]. [2, 45, 66, 145–150] Although multiple databases and tools are available for pathway mapping, such as KEGG [151, 152], HMDB [153], Metacyc, Recon3D and others [154–160], automated reconstruction of atom-resolved pathways based on tracer data remains challenging [159]. There is also a general lack of delineation of pathway compartmentation, particularly in terms of tissue specificity in these databases. Databases of compartment-delineated, atom-resolved metabolic networks and tracer-friendly pathway reconstruction tools are urgently needed. Progress is being made on atom-resolved metabolic atlases and reconstructions [158, 160–163], but they are not yet ready for general use.
Accurate reconstruction of metabolic networks at the atomic level is the foundation for metabolic flux analysis, including Flux Balance Analysis (FBA) and Kinetic Modeling Analysis (KMA). The FBA approaches use reaction stoichiometries and steady state assumptions [164] such that detailed kinetic parameters of the enzymes/transporters participating in the networks are not required. The resulting flux models do not provide individual enzyme/transporter kinetics, nor can the model predict flux changes in response to perturbations of participant proteins. The KMA approaches require a full set of enzyme/transporter kinetic parameters to solve sets of differential equations either at steady state, or under non-steady state conditions [165–173]. The models established provide individual enzymes/transporter kinetics and can predict how metabolic networks respond to changes in protein components. Currently, parameterization of these sets of equations requires extensive expert input and is time-consuming. Future informatics development in assisting the parameterization effort, e.g. narrowing the initial parameter space, is of particular importance to encourage non-expert in engaging the kinetic flux modeling approaches. The power of these approaches lies not only in quantitative understanding of reprogrammed metabolism in cancer or other human diseases but also in facilitating system level integration of reprogrammed metabolism with gene expression and proteomics changes. Such integration is fundamental to deciphering the regulatory metabolic networks and in turn gaining mechanistic insights into disease progression and therapeutic efficacy.
By overcoming the multitude of educational and technical challenges that currently limit the experimental, analytical, and informatics tools already in place and under development, we fully expect that the application of stable isotope tracers to metabolic research in cancers or other diseases will greatly expand and flourish in the future.
Highlights.
Stable isotope resolved metabolomics maps and quantifies metabolic networks
Stable isotope tracing tracks known and novel reprogrammed metabolism
NMR and mass spectrometry complement, cross-validate, and maximize isotope distribution analysis in metabolites
7. Acknowledgments
This work was supported in part by grants: 1P01CA163223–01A1 (to ANL and TWMF), 1U24DK097215–01A1 (to RMH, TWMF, and ANL).
Abbreviations:
- 1D, 2D, 3D
one-, two-, three-dimensional
- ECF
ethyl chloroformate
- HSQC
heteronuclear single quantum coherence
- mamu
milli atomic mass units
- PPP
Pentose Phosphate Pathways
- QDA
N-(2-15N-aminooxyethyl)-N,N-dimethyl-1-dodecylammonium
- QDE
N-(2-15N-iodoacetamido)-N,N-dimethyl-1-dodecylammonium
- TOCSY
Total Correlation Spectroscopy
- SIRM
Stable Isotope Resolved Metabolomics
- UHR-FT-MS
ultrahigh resolution Fourier transform mass spectrometry
Footnotes
Some of this text was published as an application note by Cambridge Isotopes Laboratories [MET_RSCH_CANCER (2/13/18)].
8. References
- [1].Mattingly SJ, Xu T, Nantz MH, Higashi RM, Fan TWM, A carbonyl capture approach for profiling oxidized metabolites in cell extracts, Metabolomics, 8 (2012) 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sun RC, Fan TW-M, Deng P, Higashi RM, Lane AN, Scott TL, Sun Q, Warmoes MO, Yang Y, Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing, Nature Communications, 8 (2017) 1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lorkiewicz P, Higashi RM, Lane AN, Fan TW, High information throughput analysis of nucleotides and their isotopically enriched isotopologues by direct-infusion FTICR-MS, Metabolomics : Official journal of the Metabolomic Society, 8 (2012) 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gori SS, Lorkiewicz PK, Ehringer DS, Belshoff AC, Higashi RM, Fan TW-M, Nantz MH, Profiling Thiol Metabolites and Quantification of Cellular Glutathione using FT-ICR-MS Spectrometry, Analytical & Bioanalytical Chemistry, 406 (2014) 4371–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].deHevesy G, A Scientific Career, Pergamon Press, New York, 1962. [Google Scholar]
- [6].Lane AN, Fan TW-M, Bousamra M II, Higashi RM, Yan J, Miller DM, Stable Isotope-Resolved Metabolomics (SIRM) in Cancer Reseach with Clinical Applications of Non-Small Cell Lung Cancer., Omics, 15 (2011) 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM, Altered Regulation of Metabolic Pathways in Human Lung Cancer Discerned by 13C Stable Isotope-Resolved Metabolomics (SIRM)) Molecular Cancer 8 (2009) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bruntz RC, Higashi RM, Lane AN, Fan TW-M, Exploring Cancer Metabolism using Stable Isotope Resolved Metabolomics (SIRM), J. Biol. Chem, 292 (2017) 11601–11609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fan TWM, Lane AN, Higashi RM, The promise of metabolomics in cancer molecular therapeutics, Current Opinion in Molecular Therapeutics, 6 (2004) 584–592. [PubMed] [Google Scholar]
- [10].Lane AN, Fan TW, Higashi RM, Isotopomer-based metabolomic analysis by NMR and mass spectrometry., Biophysical Tools for Biologists, 84 (2008) 541–588. [DOI] [PubMed] [Google Scholar]
- [11].Harrigan GG, Brackett DJ, Boros LG, Medicinal chemistry, metabolic profiling and drug target discovery: A role for metabolic profiling in reverse pharmacology and chemical genetics, Mini-Rev. Med. Chem, 5 (2005) 13–20. [DOI] [PubMed] [Google Scholar]
- [12].Cascante M, Selivanov V, Ramos-Montoya A, Application of Tracer-Based Metabolomics and Flux Analysis in Targteted Cancer Drug Design, in: Fan TW-M, Lane AN, Higashi RM (Ed.) The Handbook of Metabolomics, Springer, New York, 2012, pp. 299–320. [Google Scholar]
- [13].Koppenol WH, Bounds PL, Dang CV, Otto Warburg’s contributions to current concepts of cancer metabolism, Nature Reviews Cancer, 11 (2011) 325–337. [DOI] [PubMed] [Google Scholar]
- [14].Dang CV, Hamaker M, Sun P, Le A, Gao P, Therapeutic targeting of cancer cell metabolism, Journal of Molecular Medicine-Jmm, 89 (2011) 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shaw RJ, Cantley LC, Decoding key nodes in the metabolism of cancer cells: sugar & spice and all things nice, F1000 Biology Reports, 4 (2012) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tennant DA, Duran RV, Gottlieb E, Targeting metabolic transformation for cancer therapy, Nature Rev. Cancer, 10 (2010) 267–277. [DOI] [PubMed] [Google Scholar]
- [17].Chokkathukalam A, Kim D-H, Barrett MP, Breitling R, Creek DJ, Stable isotope-labeling studies in metabolomics: new insights into structure and dynamics of metabolic networks, Bioanalysis, 6 (2014) 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Boroughs LK, DeBerardinis RJ, Metabolic pathways promoting cancer cell survival and growth, Nature Cell Biology, 17 (2015) 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kaushik AK, DeBerardinis RJ, Applications of metabolomics to study cancer metabolism, BBA Reviews on Cancer, 1870 (2018) 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fan TW, Lane AN, Structure-based profiling of Metabolites and Isotopomers by NMR., Progress in NMR Spectroscopy, 52 (2008) 69–117 [Google Scholar]
- [21].Lane AN, Fan TW, Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using 1H TOCSY, Metabolomics, 3 (2007) 79–86. [Google Scholar]
- [22].Fan TW-M, Lane AN, NMR-based Stable Isotope Resolved Metabolomics in Systems Biochemistry., J. Biomolec. NMR 49 (2011) 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Higashi RM, Structural Mass Spectrometry for Metabolomics in: Fan TW, Higashi RM, Lane AN (Ed.) Handbook of Metabolomics Methods, Humana Press, New York, 2011. [Google Scholar]
- [24].Fan TW-M, Lorkiewicz P, Sellers K, Moseley HNB, Higashi RM, Lane AN, Stable isotope-resolved metabolomics and applications to drug development, Pharmacology and Therapeutics, 133 (2012) 366–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lane AN, Fan TW-M, NMR-based Stable Isotope Resolved Metabolomics in systems biochemistry, Arch. Biochem. Biophys, 628 (2017) 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fan TW-M, Lane AN, Applications of NMR to Systems Biochemistry, Prog. NMR Spectrosc, 92 (2016) 18–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Higashi RM, Fan TW-M, Lorkiewicz PK, Moseley HNB, Lane AN, Stable Isotope Labeled Tracers for Metabolic Pathway Elucidation by GC-MS and FT-MS, in: Raftery D (Ed.) Mass Spectrometry Methods in Metabolomics, Humana Press; USA: 2014, pp. 147–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].McNaught AD, Wilkinson A, IUPAC. Compendium of Chemical Terminology, 2nd ed. (the “Gold Book”). Blackwell Scientific Publications, Oxford, 1997. [Google Scholar]
- [29].Yang Y, Fan WW-M, Lane AN, Higashi RM, Chloroformate Derivatization for Tracing the Fate of Amino Acids in Cells by Multiple Stable Isotope Resolved Metabolomics (mSIRM), Anal. Chim. Acta 976 (2017) 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bodenhausen G, Ruben DJ, Natural Abundance N-15 Nmr by Enhanced Heteronuclear Spectroscopy, Chemical Physics Letters, 69 (1980) 185–189. [Google Scholar]
- [31].Gadian DG, NMR and its applications to living systems, 2nd ed., Oxford University Press, Oxford, U.K., 1995. [Google Scholar]
- [32].Befroy DE, Rothman DL, Petersen KF, Shulman GI, P-31-Magnetization Transfer Magnetic Resonance Spectroscopy Measurements of In Vivo Metabolism, Diabetes, 61 (2012) 2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thelwall PE, Yemin AY, Gillian TL, Simpson NE, Kasibhatla MS, Rabbani ZN, Macdonald JM, Blackband SJ, Gamcsik MP, Noninvasive in vivo detection of glutathione metabolism in tumors, Cancer Research, 65 (2005) 10149–10153. [DOI] [PubMed] [Google Scholar]
- [34].Thelwall P, Simpson N, Rabbani Z, Clark M, Pourdeyhimi R, Macdonald J, Blackband S, Gamcsik M, In vivo MR studies of glycine and glutathione metabolism in a rat mammary tumor., NMR Biomed, 25 (2011) 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wolak J, Rahimi-Keshari K, Jeffries RE, Joy MP, Todd A, Pediatikakis P, Deawar BJ, Winnike JH, Favorov O, Elston TC, Grace LM, Kurhanewicz J, Vigneron D, Holmuhamedov E, Macdonald JM, Noninvasive fluxomics in mammals by nuclear magnetic resonance spectroscopy, in: T.W.-M. F, Lane AN, Higashi RM (Ed.) The Handbook of Metabolomics, Springer, New York, 2012, pp. 321–392. [Google Scholar]
- [36].Mason GF, Petersen KF, de Graaf RA, Shulman GI, Rothman DL, Measurements of the anaplerotic rate in the human cerebral cortex using C-13 magnetic resonance spectroscopy and [1-C-13] and [2-C-13] glucose, Journal of Neurochemistry, 100 (2007) 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Patel AB, de Graaf RA, Mason GF, Rothman DL, Shulman RG, Behar KL, The contribution of GABA to glutamate/glutamine cycling and energy metabolism in the rat cortex in vivo, Proceedings of the National Academy of Sciences of the United States of America, 102 (2005) 5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brindle KM, Imaging Metabolism with Hyperpolarized C-13-Labeled Cell Substrates, Journal of the American Chemical Society, 137 (2015) 6418–6427. [DOI] [PubMed] [Google Scholar]
- [39].Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PEZ, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, Reed G, Carvajal L, Small EJ, Munster P, Weinberg VK, Ardenkjaer-Larsen JH, Chen AP, Hurd RE, Odegardstuen L-I, Robb FJ, Tropp J, Murray JA, Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized 1-C-13 Pyruvate, Science Translational Medicine, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Winnike JH, Pediaditakis P, Wolak JE, McClelland RW, Watkins PB, Macdonald JM, Stable isotope resolved metabolomics of primary human hepatocytes reveals a stressed phenotype, Metabolomics, 8 (2011) 34–49. [Google Scholar]
- [41].Lane AN, Tan J, Wang Y, Yan J, Higashi RM, Fan TW-M, Probing the metabolic phenotype of breast cancer cells by multiple tracer stable isotope resolved metabolomics, Metabolic Engineering, 43 (2017) 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kamphorst JJ, Chung MK, Fan J, R. JD., Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate, Cancer & Metabolism, 2 (2014) 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pichumani K, Mashimo T, Vemireddy V, Ijare OB, Mickey B, Malloy CR, Marin-Valencia I, Baskin DS, Bachoo RM, Maher EA, Measurement of 13 C turnover into glutamate and glutamine pools in brain tumor patients, FEBS Lett, 591 (2016) 3548–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bruntz R, Higashi RM, Lane AN, Fan TW-M, Exploring Cancer Metabolism using Stable Isotope Resolved Metabolomics (SIRM), J. Biol. Chem, 292 (2017) 11601–11609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sellers K, Fox MP, Bousamra M, Slone S, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, Lane AN, Fan TW-M, Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation, J. Clin. Invest, 125 (2015) 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lee W-NP, Boros LG, Puigjaner J, Bassilian S, Lim S, Cascante M, Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2–13C2]glucose, Am J Physiol Endocrinol Metab, 274 (1998) E843–851. [DOI] [PubMed] [Google Scholar]
- [47].Vizan P, Boros LG, Figueras A, Capella G, Mangues R, Bassilian S, Lim S, Lee W-NP, Cascante M, K-ras Codon-Specific Mutations Produce Distinctive Metabolic Phenotypes in Human Fibroblasts, Cancer Res, 65 (2005) 5512–5515. [DOI] [PubMed] [Google Scholar]
- [48].Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, DeBerardinis RJ, Pyruvate carboxylase is required for glutamine-independent growth of tumor cells, Proceedings of the National Academy of Sciences of the United States of America, 108 (2011) 8674–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Crown SB, Ahn WS, Antoniewicz MR, Rational design of (1)(3)C-labeling experiments for metabolic flux analysis in mammalian cells, BMC Syst Biol 6(2012) 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wilson DM, Keshari KR, Larson PE, Chen AP, Hu S, Van Criekinge M, Bok R, Nelson SJ, Macdonald JM, Vigneron DB, Kurhanewicz J, Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo, J Magn Reson, 205 (2010) 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fan TW, Yuan P, Lane AN, Higashi RM, Wang Y, Hamidi AB, Zhou R, Guitart X, Chen G, Manji HK, Kaddurah-Daouk R, Stable isotope-resolved metabolomic analysis of lithium effects on glial-neuronal metabolism and interactions, Metabolomics, 6 (2010) 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Leithner K, Hrzenjak A, Trotzmuller M, Moustafa T, Kofeler HC, Wohlkoenig C, Stacher E, Lindenmann J, Harris AL, Olschewski A, Olschewski H, PCK2 activation mediates an adaptive response to glucose depletion in lung cancer, Oncogene, 34 (2015) 1044–1050 [DOI] [PubMed] [Google Scholar]
- [53].Peltz M, He TT, Adams G.A.t., Chao RY, Meyer DM, Jessen ME, Characterizing lung metabolism with carbon-13 magnetic resonance spectroscopy in a small-animal model: evidence of gluconeogenesis during hypothermic storage, Transplantation, 80 (2005) 417–420. [DOI] [PubMed] [Google Scholar]
- [54].Katz J, Wals P, Lee WN, Isotopomer studies of gluconeogenesis and the Krebs cycle with 13C-labeled lactate, J Biol Chem, 268 (1993) 25509–25521. [PubMed] [Google Scholar]
- [55].Le A, Lane AN, Hamaker M, Bose S, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJC, Lorkiewicz PK, Higashi RM, Fan TW-M, Dang CV, Myc induction of hypoxic glutamine metabolism and a glucose-independent TCA cycle in human B lymphocytes Cell Metabolism, 15 (2012) 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mullen AR, Wheaton WW, Jin ES, Chen P-H, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ, Reductive carboxylation supports growth in tumour cells with defective mitochondria, Nature, 481 (2011) 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yuneva MO, Fan TW-M, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Matés JM, Alonso FJ, Wang C, Seo Y, Chen X, Bishop JM, The Metabolic Profile of Tumors Depends on Both the Responsible Genetic Lesion and Tissue Type, Cell Metabolism, 15 (2012) 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB, Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis, Proceedings of the National Academy of Sciences of the United States of America, 104 (2007) 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Vercoutere B, Durozard D, Baverel G, Martin G, Complexity of glutamine metabolism in kidney tubules from fed and fasted rats, Biochem. J, 378 (2004) 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Duan XY, Kasumov T, Kovacs W, David F, Kelleher JK, Krisans S, Brunengraber H, Acetyl-CoA generated in peroxisomes of CHO and HepG2 cells is preferentially incorporated into sterols versus fatty acids: studies with U-13C12 dodecanedioate, Faseb Journal, 20 (2006) A1467–A1467. [Google Scholar]
- [61].Winnike JH, Pediaditakis P, Wolak JE, McClelland RW, Watkins PB, Macdonald JM, Stable isotope resolved metabolomics of primary human hepatocytes reveals a stressed phenotype, Metabolomics, 8 (2012) 34–49. [Google Scholar]
- [62].Magkos F, Mittendorfer B, Stable isotope-labeled tracers for the investigation of fatty acid and triglyceride metabolism in humans in vivo, Clin Lipidol, 4 (2009) 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Li J, Song J, Zaytseva YY, Liu Y, Rychahou P, Jiang K, Starr ME, Kim JT, Harris JW, Yiannikouris FB, Katz WS, Nilsson PM, Orho-Melander M, Chen J, Zhu H, Fahrenholz T, Higashi RM, Gao T, Morris AJ, Cassis LA, Fan TWM, Weiss HL, Dobner PR, Melander O, Jia J, E. BM., An obligatory role for neurotensin in high-fat-diet-induced obesity, Nature, 19 (2016) 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cowin GJ, Willgoss DA, Bartley J, Endre ZH, Serine isotopomer analysis by 13C-NMR defines glycine-serine interconversion in situ in the renal proximal tubule, Biochim Biophys Acta 1310 (1996) 32–40. [DOI] [PubMed] [Google Scholar]
- [65].Labuschagne C, van den Broek N, Mackay G, Vousden K, Maddocks OD, Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells., Cell Rep, 7 (2014) 1248–1258. [DOI] [PubMed] [Google Scholar]
- [66].Thelwall PE, Simpson NE, Rabbani ZN, Clark MD, Pourdeyhimi R, Macdonald JM, Blackband SJ, Gamcsik MP, In vivo MR studies of glycine and glutathione metabolism in a rat mammary tumor, NMR Biomed 25 (2012) 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li T, Zhang Z, Kolwicz SC, Abell L, Roe ND, Kim M, Zhou B, Cao Y, Ritterhoff J, Gu HW, Raftery D, Sun HP, Tian R, Defective Branched-Chain Amino Acid Catabolism Disrupts Glucose Metabolism and Sensitizes the Heart to Ischemia-Reperfusion Injury, Cell Metabolism, 25 (2017) 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Green CR, Wallace M, Divakaruni AS, Phillips SA, Murphy AN, Ciaraldi TP, Metallo CM, Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis, Nature Chemical Biology, 12 (2016) 15-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, Muir A, Chin CR, Freinkman E, Jacks T, Wolpin BM, Vitkup D, Vander Heiden MG, Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers, Science, 353 (2016) 1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Engelen MPKJ, Safar AM, Bartter T, Koeman F, Deutz NEP, Reduced arginine availability and nitric oxide synthesis in cancer is related to impaired endogenous arginine synthesis, Clinical Science, 130 (2016) 1185–1195. [DOI] [PubMed] [Google Scholar]
- [71].Kurland IJ, Alcivar A, Bassilian S, Lee WN, Loss of [13C]glycerol carbon via the pentose cycle. Implications for gluconeogenesis measurement by mass isotopomer distribution analysis, J Biol Chem 275 (2000) 36787–36793. [DOI] [PubMed] [Google Scholar]
- [72].Qi J, Lang W, Geisler JG, Wang P, Petrounia I, Mai S, Smith C, Askari H, Struble GT, Williams R, Bhanot S, Monia BP, Bayoumy S, Grant E, Caldwell GW, Todd MJ, Liang Y, Gaul MD, Demarest KT, Connelly MA, The use of stable isotope-labeled glycerol and oleic acid to differentiate the hepatic functions of DGAT1 and −2. J Lipid Res 53 (2012) 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lee WNP, Bassilian S, Guo ZK, Schoeller D, Edmond J, Bergner EA, Byerley LO, Measurement of Fractional Lipid-Synthesis Using Deuterated Water ((H2o)-H-2) and Mass Isotopomer Analysis, American Journal of Physiology, 266 (1994) E372–E383. [DOI] [PubMed] [Google Scholar]
- [74].Diraison F, Yankah V, Letexier D, Dusserre E, Jones P, Beylot M, Differences in the regulation of adipose tissue and liver lipogenesis by carbohydrates in humans, Journal of Lipid Research, 44 (2003) 846–853. [DOI] [PubMed] [Google Scholar]
- [75].Gradwell MJ, Fan TWM, Lane AN, Analysis of phosphorylated metabolites in crayfish extracts by two-dimensional H-1-P-31 NMR heteronuclear total correlation spectroscopy (heteroTOCSY), Analytical Biochemistry, 263 (1998) 139–149. [DOI] [PubMed] [Google Scholar]
- [76].Tayyari F, Gowda G, Gu H, Raftery D, 15N-cholamine--a smart isotope tag for combining NMR- and MS-based metabolite profiling, Anal Chem, 85 (2013) 8715–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lane AN, Arumugam S, Lorkiewicz PK, Higashi RM, Laulhe S, Nantz MH, Moseley HN, Fan TW, Chemoselective detection and discrimination of carbonyl-containing compounds in metabolite mixtures by 1H-detected 15N nuclear magnetic resonance, Magnetic resonance in chemistry : MRC, 53 (2015) 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Deng P, Higashi RM, Lane AN, Bruntz RC, Sun RC, Ramakrishnam Raju MV, Nantz MH, Qi Z, Fan TW, Quantitative profiling of carbonyl metabolites directly in crude biological extracts using chemoselective tagging and nanoESI-FTMS, Analyst, 143 (2017) 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hanahan D, Weinberg RA, Hallmarks of Cancer: The Next Generation, Cell, 144 (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [80].Liu W, Glunde K, Bhujwalla ZM, Raman V, Sharma A, Phang JM, Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments, Cancer Research, 72 (2012) 3677–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ho P-C, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui Y-C, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, Kaech SM, Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses, Cell, 162 (2015) 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Fan TW, Warmoes MO, Sun Q, Song H, Turchan-Cholewo J, Martin JT, Mahan A, Higashi RM, Lane AN, Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator beta-glucan in a two-case ex vivo non-small-cell lung cancer study, Cold Spring Harb Mol Case Stud, 2 (2016) a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liu W, Le A, Lane AN, Fan TW-M, Dang CV, Phang JM, The reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses to c-MYC., Proc. Natl. Acad. Sci. USA, 109 (2012) 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, Tayyari F, McSkimming D, Kannan N, Tojo A, Edison AS, Ito T, Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia, Nature, 545 (2017) 500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fan TW-M, Tan JL, McKinney MM, Lane AN, Stable Isotope Resolved Metabolomics Analysis of Ribonucleotide and RNA Metabolism in Human Lung Cancer Cells., Metabolomics, 8 (2012) 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Boren J, Cascante M, Marin S, Comin-Anduix B, Centelles JJ, Lim S, Bassilian S, Ahmed S, Lee WNP, Boros LG, Gleevec (ST1571) influences metabolic enzyme activities and glucose carbon flow toward nucleic acid and fatty acid synthesis in myeloid tumor cells, Journal of Biological Chemistry, 276 (2001) 37747–37753. [DOI] [PubMed] [Google Scholar]
- [87].Sellers K, Fox MP, Bousamra M II, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, Lane AN, Fan TWM, Pyruvate carboxylase is critical for non–small-cell lung cancer proliferation, The Journal of Clinical Investigation, 125 (2015) 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM, Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM), Molecular cancer, 8 (2009) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lane AN, Fan TW-M, Regulation of mammalian nucleotide metabolism and biosynthesis, Nucleic Acids Res, 43 (2015) 2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Thornburg JM, Nelson KK, Lane AN, Arumugam S, Simmons A, Eaton JW, Telang S, Chesney J, Targeting Aspartate Aminotransferase in Breast Cancer., Breast Cancer Research, 10 (2008) R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sullivan LB., Gui DY., Hosios AM., Bush LN., Freinkman E., V.H. MG., Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells Cell, 162 (2015) 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, David MDM Sabatini, An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis, Cell, 162 (2015) 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cardaci S, Zheng Liang, MacKay Gillian, van den Broek Niels J.F., MacKenzie Elaine D., Nixon Colin, Stevenson David, Tumanov Sergey, Bulusu Vinay, Kamphorst Jurre J., Vazquez Alexei, Fleming Stewart, Schiavi Francesca, Kalna Gabriela, Blyth Karen, Strathdee Douglas, Gottlieb E, Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis, Nat. Cell Biol, 17 (2015) 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Pavlova NN, Hui S, Ghergurovich JM, Fan J, Intlekofer AM, White RM, Rabinowitz JD, Thompson CB, Z. J., As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid, Cell Metab, 27 (2018) 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tedeschi PM, Markert EK, Gounder M, Lin H, Dvorzhinski D, Dolfi SC, Chan LL, Qiu J, DiPaola RS, Hirshfield KM, Boros LG, Bertino JR, Oltvai ZN, Vazquez A, Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells, Cell Death Dis, 4 (2013) e877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM, Functional genomics reveal that the serine synthesis pathway is essential in breast cancer, Nature, 476 (2011) 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hardwick J, Lane AN, Brown T, Epigenetic modifications of cytosine: biophysical properties, regulation and function in mammalian DNA Bioessays, 40 (2018) 1700199. [DOI] [PubMed] [Google Scholar]
- [98].Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, Stewart L, Duray P, Tourre O, Sharma N, Choyke P, Stratton P, Merino M, Walther MM, Linehan WM, Schmidt LS, Zbar B, Mutations in the Fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America, American Journal of Human Genetics, 73 (2003) 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wei MH, Toure O, Glenn GM, Pithukpakorn M, Neckers L, Stolle C, Choyke P, Grubb R, Middelton L, Turner ML, Walther MM, Merino MJ, Zbar B, Linehan WM, Toro JR, Novel mutations in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer, Journal of Medical Genetics, 43 (2006) 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Linehan WM, Srinivasan R, Schmidt LS, The genetic basis of kidney cancer: a metabolic disease., Nature Reviews Urology, 7 (2010) 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ricketts CJ, Shuch B, Vocke CD, Metwalli AR, Bratslavsky G, Middelton L, Yang YF, Wei MH, Pautler SE, Peterson J, Stolle CA, Zbar B, Merino MJ, Schmidt LS, Pinto PA, Srinivasan R, Pacak K, Linehan WM, Succinate Dehydrogenase Kidney Cancer: An Aggressive Example of the Warburg Effect in Cancer, Journal of Urology, 188 (2012) 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Saxena N, Maio N, Crooks DR, Ricketts CJ, Yang Y, Wei M-H, Fan TW-M, Lane AN, Sourbier C, Rouault TA, Linehan WM, SDHB-Deficient Cancers: The Role of Mutations That Impair Iron Sulfur Cluster Delivery, JNCI, djv287 (2016). [DOI] [PMC free article] [PubMed]
- [103].Dang L, Jin S, Su SM, IDH mutations in glioma and acute myeloid leukemia, Trends in Molecular Medicine, 16 (2010) 387–397. [DOI] [PubMed] [Google Scholar]
- [104].Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Heiden MGV, Su SM, Cancer-associated IDH1 mutations produce 2-hydroxyglutarate, Nature, 462 (2009) 739–U752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM, Cancer-associated IDH1 mutations produce 2-hydroxyglutarate Nature, 465 (2010) 966-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW, Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations, Journal of Experimental Medicine, 207 (2010) 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, X. Y., Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases, cancer Cell, 19 (2011) 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB, IDH mutation impairs histone demethylation and results in a block to cell differentiation, Nature, 483 (2012) 474–U130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O/’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB, IDH mutation impairs histone demethylation and results in a block to cell differentiation, Nature, advance online publication (2012). [DOI] [PMC free article] [PubMed]
- [110].Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM, Tracing Compartmentalized NADPH Metabolism in the Cytosol and Mitochondria of Mammalian Cells, Molecular Cell, 55 (2014) 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Fan J, Ye JB, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD, Quantitative flux analysis reveals folate-dependent NADPH production, Nature, 510 (2014) 298-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM, Cancer-associated IDH1 mutations produce 2-hydroxyglutarate Nature, 465 (2010) 966–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Selivanov VA, Marin S, Lee PW, Cascante M, Software for dynamic analysis of tracer-based metabolomic data: estimation of metabolic fluxes and their statistical analysis, Bioinformatics, 22 (2006) 2806–2812. [DOI] [PubMed] [Google Scholar]
- [114].Young JD, INCA: a computational platform for isotopically non-stationary metabolic flux analysis, Bioinformatics, 30 (2014) 1333–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Fan T, Bandura L, Higashi R, Lane A, Metabolomics-edited transcriptomics analysis of Se anticancer action in human lung cancer cells, Metabolomics, 1 (2005) 325–339 [Google Scholar]
- [116].Fan TW, Lane AN, Applications of NMR spectroscopy to systems biochemistry, Prog Nucl Magn Reson Spectrosc, 92–93 (2016) 18–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ravi M, Paramesh V, Kaviya SR, Anuradha E, Paul Solomon FDP, 3D Cell Culture Systems: Advantages and Applications, J. Cell Physiol, 230 (2014) 16–26. [DOI] [PubMed] [Google Scholar]
- [118].Sant S, Johnston PA, The production of 3D tumor spheroids for cancer drug discovery, Drug Discov Today Technol, 23 (2017) 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Ekert JE, Johnson K, Strake B, Pardinas J, Jarantow S, Perkinson R, Colter DC, Three-dimensional lung tumor microenvironment modulates therapeutic compound responsiveness in vitro--implication for drug development, PLoS One, 9 (2014) e92248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Tseng H, Gage JA, Shen T, Haisler WL, Neeley SK, Shiao S, Chen J, Desai PK, Liao A, Hebel C, Raphael RM, Becker JL, Souza GR, A spheroid toxicity assay using magnetic 3D bioprinting and real-time mobile device-based imaging, Scientific reports, 5 (2015) 13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lane AN, Higashi RM, Fan TWM, Preclinical models for interrogating drug action in human cancers using Stable Isotope Resolved Metabolomics (SIRM), Metabolomics, 12 (2016) 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Thakuri PS, Liu C, Luker GD, Tavana H, Biomaterials-Based Approaches to Tumor Spheroid and Organoid Modeling, Adv Healthc Materials, 7 (2018) 1700980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Rodrigues T, Kundu B, Silva-Correia J, Kundu SC, Oliveira JM, Reis RL, Correlo VM, Emerging tumor spheroids technologies for 3D in vitro cancer modeling, Pharmacol Ther, 184 (2018) 201–211 [DOI] [PubMed] [Google Scholar]
- [124].Fan TWM, Higashi RM, Lane AN, Integrating metabolomics and transcriptomics for probing Se anticancer mechanisms, Drug Metabolism Reviews, 38 (2006) 707–732. [DOI] [PubMed] [Google Scholar]
- [125].Jaganathan H, Gage J, Leonard F, Srinivasan S, Souza GR, Dave B, Godin B, Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation, Scientific reports, 4 (2014) 6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Tseng H, Gage JA, Raphael RM, Moore RH, Killian TC, Grande-Allen KJ, Souza GR, Assembly of a three-dimensional multitype bronchiole coculture model using magnetic levitation, Tissue Eng Part C Methods, 19 (2013) 665–675. [DOI] [PubMed] [Google Scholar]
- [127].Fan TW, Lane AN, Higashi RM, Yan J, Stable isotope resolved metabolomics of lung cancer in a SCID mouse model, Metabolomics, 7 (2011) 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, Cai L, Good L, Tu BP, Hatanpaa KJ, Mickey BE, Mates JM, Pascual JM, Maher EA, Malloy CR, Deberardinis RJ, Bachoo RM, Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo, Cell metabolism, 15 (2012) 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hoch JC, Maciejewski MW, Mobli M, Schuyler AD, Stern AS, Nonuniform Sampling and Maximum Entropy Reconstruction in Multidimensional NMR, Accounts of Chemical Research, 47 (2014) 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Mobli M, Maciejewski MW, Gryk MR, Hoch JC, Automatic maximum entropy spectral reconstruction in NMR, Journal of Biomolecular Nmr, 39 (2007) 133–139. [DOI] [PubMed] [Google Scholar]
- [131].Kupce E, Freeman R, Fast multi-dimensional Hadamard spectroscopy, Journal of Magnetic Resonance, 163 (2003) 56–63. [DOI] [PubMed] [Google Scholar]
- [132].Freeman R, Kupce E, New ways to record multidimensional NMR spectra, Current Analytical Chemistry, 2 (2006) 101–105. [Google Scholar]
- [133].Bingol K, Li D-W, Bruschweiler-Li L, Cabrera OA, Megraw T, Zhang F, Brueschweiler R, Unified and Isomer-Specific NMR Metabolomics Database for the Accurate Analysis of C-13-H-1 HSQC Spectra, Acs Chemical Biology, 10 (2015) 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wang C, He LD, Li DW, Bruschweiler-Li L, Marshall AG, Bruschweiler R, Accurate Identification of Unknown and Known Metabolic Mixture Components by Combining 3D NMR with Fourier Transform Ion Cyclotron Resonance Tandem Mass Spectrometry, Journal of Proteome Research, 16 (2017) 3774–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Hansen AL, Li DW, Wang C, Bruschweiler R, Absolute Minimal Sampling of Homonuclear 2D NMR TOCSY Spectra for High-Throughput Applications of Complex Mixtures, Angewandte Chemie-International Edition, 56 (2017) 8149–8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Kupce E, NMR with multiple receivers, Top Curr Chem., 335 (2013) 71–96. [DOI] [PubMed] [Google Scholar]
- [137].Pudakalakatti SM, Dubey A, Jaipuria G, Shubhashree U, Adiga SK, Moskau D, Atreya HS, A fast NMR method for resonance assignments: application to metabolomics, Journal of Biomolecular Nmr, 58 (2014) 165–173. [DOI] [PubMed] [Google Scholar]
- [138].Reardon PN, Marean-Reardon CL, Bukovec MA, Coggins BE, Isern NG, 3D TOCSY-HSQC NMR for Metabolic Flux Analysis Using Non-Uniform Sampling, Anal. Chem, 88 (2016) 2825–2831. [DOI] [PubMed] [Google Scholar]
- [139].Ryan E, Nguyen CQN, Shiea C, Reid GE, Detailed Structural Characterization of Sphingolipids via 193 nm Ultraviolet Photodissociation and Ultra High Resolution Tandem Mass Spectrometry, J Am Soc Mass Spectrom, 28 (2017) 1406–1419. [DOI] [PubMed] [Google Scholar]
- [140].Cooks RG, Manicke NE, Dill AL, Ifa DR, Eberlin LS, Costa AB, Wang H, Huang G, Ouyang Z, New ionization methods and miniature mass spectrometers for biomedicine: DESI imaging for cancer diagnostics and paper spray ionization for therapeutic drug monitoring, Faraday Discuss, 149 (2011) 247–267; discussion 333–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Javitt DC, Carter CS, Krystal JH, Kantrowitz JT, Girgis RR, Kegeles LS, Ragland JD, Maddock RJ, Lesh TA, Tanase C, Corlett PR, Rothman DL, M. G., Qiu M, Robinson J, Potter WZ, Carlson M, Wall MM, Choo TH, Grinband J, L. JA., Utility of Imaging-Based Biomarkers for Glutamate-Targeted Drug Development in Psychotic Disorders: A Randomized Clinical Trial., JAMA Psychiatry, 75 (2018) 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Qi M, Philip MC, Yang N, Sweedler JV, Single Cell Neurometabolomics, Acs Chemical Neuroscience, 9 (2018) 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Fan TW-M, Metabolomics-Edited Transcriptomics Analysis (Meta), in: McQueen CA (Ed.) Comprehensive Toxicology, Academic Press, Oxford, 2010, pp. 685–706. [Google Scholar]
- [144].Fan TW-M, Lane AN, Higashi RM, Stable Isotope Resolved Metabolomics Studies in ex vivo Tissue Slices, Bio-protocol, 6 (2016) e1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Malloy CR, Maher E, Marin-Valenica I, Mickey B, DeBerardinis RJ, Sherry AD, Carbon-13 Nuclear Magnetic Resonance for Analysis of Metabolic Pathways, in: Lutz N, Sweedler JV, Weevers RA (Eds.) Methodologies for Metabolomics: Experimental Strategies and Techniques, Cambridge University Press, Cambridge, 2013, pp. 415–445. [Google Scholar]
- [146].Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa KJ, Jindal A, Jeffrey FM, Choi CH, Madden C, Mathews D, Pascual JM, Mickey BE, Malloy CR, DeBerardinis RJ, Metabolism of U-13C glucose in human brain tumors in vivo, Nmr in Biomedicine, 25 (2012) 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Patel AB, De Graaf RA, Rothman DL, Behar KL, Mason GF, Evaluation of cerebral acetate transport and metabolic rates in the rat brain in vivo using H-1- C-13 -NMR, Journal of Cerebral Blood Flow and Metabolism, 30 (2010) 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Boumezbeur F, Petersen KF, Cline GW, Mason GF, Behar KL, Shulman GI, Rothman DL, The Contribution of Blood Lactate to Brain Energy Metabolism in Humans Measured by Dynamic C-13 Nuclear Magnetic Resonance Spectroscopy, Journal of Neuroscience, 30 (2010) 13983–13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Ljungqvist OH, Persson M, Ford GC, Nair KS, Functional heterogeneity of leucine pools in human skeletal muscle, American Journal of Physiology-Endocrinology and Metabolism, 273 (1997) E564–E570. [DOI] [PubMed] [Google Scholar]
- [150].Baumann PQ, Stirewalt WS, Orourke BD, Howard D, Nair KS, Precursor Pools Of Protein-Synthesis - A Stable-Isotope Study In A Swine Model, American Journal of Physiology, 267 (1994) E203–E209. [DOI] [PubMed] [Google Scholar]
- [151].Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K, KEGG: new perspectives on genomes, pathways, diseases and drugs, Nucleic Acids Research, 45 (2017) D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Kanehisa M, Enzyme Annotation and Metabolic Reconstruction Using KEGG, in: Kihara D (Ed.) Protein Function Prediction: Methods and Protocols 2017, pp. 135–145. [DOI] [PubMed]
- [153].Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, S. A., HMDB 3.0-The Human Metabolome Database in 2013, Nucleic Acids Research 41 (2013) D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD, The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases, Nucleic Acids Research, 44 (2016) D471–D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, Paley S, Subhraveti P, Karp PD, The MetaCyc database of metabolic pathways and enzymes, Nucleic Acids Research, 46 (2018) D633–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Hanson NW, Konwar KM, Hawley AK, Altman T, Karp PD, Hallam SJ, Metabolic pathways for the whole community, Bmc Genomics, 15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Karp PD, Caspi R, A survey of metabolic databases emphasizing the MetaCyc family, Archives of Toxicology, 85 (2011) 1015–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Swainston N, Smallbone K, Hefzi H, Dobson PD, Brewer J4, Hanscho M, Zielinski DC, Ang KS, Gardiner NJ, Gutierrez JM, Kyriakopoulos S, Lakshmanan M, Li S, Liu JK, Martínez VS, Orellana CA, Quek LE, Thomas A, Zanghellini J, Borth N, Lee DY, Nielsen LK, Kell DB, Lewis NE, M. P., Recon 2.2: from reconstruction to model of human metabolism, Metabolomics, 12 (2016) 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Gonzalez GAP, El Assal LRP, Noronha A, Thiele I, Haraldsdottir HS, Fleming RMT, Comparative evaluation of atom mapping algorithms for balanced metabolic reactions: application to Recon 3D, Journal of Cheminformatics, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Brunk E, Sahoo S, Zielinski D, Altunkaya A, Mih N, Prlic A, Sastry A, Gonzalez GAP, Danielsdottir AD, Noronha A, Aurich MK, Rose P, Fleming R, Draeger A, Burley SK, Nielsen J, Thiele I, Palsson B, Recon 3D: A resource enabling a three-dimensional view of human metabolism and disease, Abstracts of Papers of the American Chemical Society, 253 (2017). [Google Scholar]
- [161].Hadadi N, Hafner J, Shajkofci A, Zisaki A, Hatzimanikatis V, ATLAS of Biochemistry: A Repository of All Possible Biochemical Reactions for Synthetic Biology and Metabolic Engineering Studies, ACS Sythetic Biology, 5 (2016) 1155–1166. [DOI] [PubMed] [Google Scholar]
- [162].Bordbar A, Yurkovich TJT, Paglia G, Rolfsson O, Sigurjonsson OE, Palsson BO, Elucidating dynamic metabolic physiology through network integration of quantitative timecourse metabolomics, Scient. Rep, 7 (2017) 46249–46261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Shlomi T, Cabili MN, Herrgård MJ, Palsson BØ, R. E., Network-based prediction of human tissue-specific metabolism, Nat. Biotechnol, 26 (2008) 1003–1010. [DOI] [PubMed] [Google Scholar]
- [164].Orth JD, Thiele I, Palsson BO, What is flux balance analysis?, Nature Biotechnol, 28 (2010) 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Selivanov VA, Marin S, Lee PWN, Cascante M, Software for dynamic analysis of tracer-based metabolomic data: estimation of metabolic fluxes and their statistical analysis, Bioinformatics, 22 (2006) 2806–2812. [DOI] [PubMed] [Google Scholar]
- [166].Young JD, Walther JL, Antoniewicz MR, Yon H, Stephanopoulos G, An Elementary Metabolite Unit (EMU) based method of isotopically nonstationary flux analysis, Biotechnology and Bioengineering, 99 (2008) 686–699. [DOI] [PubMed] [Google Scholar]
- [167].Murphy TA, Dang CV, Young JD, Isotopically nonstationary 13C flux analysis of Myc-induced metabolic reprogramming in B-cells, Metabolic Engineering, 15 (2013) 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U, COPASI — a COmplex PAthway SImulator, Bioinformatics, 22 (2006) 3067–3074. [DOI] [PubMed] [Google Scholar]
- [169].Wiechert W, Noh K, Isotopically non-stationary metabolic flux analysis: complex yet highly informative, Current Opinion in Biotechnology, 24 (2013) 979–986. [DOI] [PubMed] [Google Scholar]
- [170].Qi Z, Voit EO, Identification of cancer mechanisms through computational systems modeling, Translational Cancer Research 3(2014) 233–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Shiraishi F, Yoshida E, Voit EO, An Efficient and Very Accurate Method for Calculating Steady-State Sensitivities in Metabolic Reaction Systems, Ieee-Acm Transactions on Computational Biology and Bioinformatics, 11 (2014) 1077–1086. [DOI] [PubMed] [Google Scholar]
- [172].Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, Frezza C, Ghesquiere B, Gottlieb E, Hiller K, Jones RG, Kamphorst JJ, Kibbey RG, Kimmelman AC, Locasale JW, Lunt SY, Maddocks OD, Malloy C, Metallo CM, Meuillet EJ, Munger J, Noh K, Rabinowitz JD, Ralser M, Sauer U, Stephanopoulos G, St-Pierre J, Tennant DA, Wittmann C, Vander Heiden MG, Vazquez A, Vousden K, Young JD, Zamboni N, Fendt S-M, A roadmap for interpreting (13)C metabolite labeling patterns from cells, Current opinion in biotechnology, 34 (2015) 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Zamboni N, Fendt S-M, Rühl M, Sauer U, 13C-based metabolic flux analysis, Nat. Protocols, 4 (2009) 878–892. [DOI] [PubMed] [Google Scholar]


