Abstract
Introduction:
Centronuclear myopathies (CNMs) are a subtype of congenital myopathies (CMs) characterized by muscle weakness, predominant type 1 fibers, and increased central nuclei. SPEG (striated preferentially expressed protein kinase) mutations have recently been identified in seven CM patients (six with CNMs). We report two additional patients with SPEG mutations expanding the phenotype and evaluate genotype-phenotype correlations associated with SPEG mutations.
Methods/Results:
Using whole exome/genome sequencing in CM families, we identified novel recessive SPEG mutations in two patients. Patient 1, with severe muscle weakness requiring respiratory support, dilated cardiomyopathy, ophthalmoplegia, and findings of non-specific CM on muscle biopsy carried a homozygous SPEG mutation (p.Val3062del). Patient 2, with milder muscle weakness, ophthalmoplegia, and CNM carried compound heterozygous mutations (p.Leu728Argfs*82) and (p.Val2997Glyfs*52).
Discussion:
The two patients add insight into genotype-phenotype correlations of SPEG-associated CMs. Clinicians should consider evaluating a CM patient for SPEG mutations even in the absence of CNM features.
Keywords: Centronuclear myopathies (CNMs), Striated preferentially expressed protein kinase (SPEG), Congenital myopathies (CMs), Next generation sequencing (NGS), Cardiomyopathy, Myotubularin (MTM1)
Introduction
Congenital myopathies (CMs) are a group of muscle diseases that commonly present at birth or during infancy with muscle weakness and hypotonia. The clinical presentation ranges from mild hypotonia causing delays in achieving motor skills to severe muscle weakness causing death from respiratory involvement1. Centronuclear myopathies (CNMs) are a subtype characterized by increased central nuclei within myofibers, and often associated with disruption of excitation-contraction coupling1,2. Approximately 60–80% of CNMs are caused by dominant DNM2 mutations, dominant and recessive RYR1 and CACNA1S mutations, recessive BIN1 mutations, and X-linked recessive MTM1 mutations3–8. Recently, recessive SPEG mutations have been identified in six CNM patients and one patient with non-CNM CM9–12. Here, we report two additional unrelated patients with CMs caused by recessive SPEG mutations, compare the clinical findings of all nine patients, and discuss genotype-phenotype correlations thereby improving the understanding of SPEG-related CM.
Methods
Patient Recruitment and Genetic Analysis
For patient 1, a CGH array was initially performed and then whole exome sequencing (WES) was performed in a diagnostic setting with a parent–offspring trio approach as previously described13. For Patient 2, the patient and her family were enrolled in an IRB-approved study (NINDS Protocol 12-N-0095). WES was initially performed through the NIH Intramural Sequencing Center using the Nimblegen SeqCap EZ Exome+UTR Library and Illumina HiSeq, and variants were analyzed using Varsifter14. Whole genome sequencing (WGS) was then performed by the Genomics Platform at the Broad Institute using Illumina HiSeq X Ten v2 chemistry, and variants were analyzed using Variant Effect Predictor.
Histopathology Studies
The muscle biopsy samples were frozen and processed using standard histological techniques15.
Results
Clinical Description
Patient 1 was the first child of healthy consanguineous parents, with normal intellect and no family history of neuromuscular disease. He has been reported in a large series of cardiomyopathy patients with minimal clinical information13. The pregnancy was reportedly uncomplicated, and he was delivered by vacuum extraction at 37 weeks gestation. At birth, he presented with severe hypotonia and left-sided inguinal hernia. At age 4, he developed progressive proximal muscle weakness and was noted to have marked atrophy of his lower leg muscles, pes planovalgus, and a high-arched palate. His history was significant for recurrent abdominal pain and diarrhea, recurrent otitis media, frequent upper airway infections, recurrent pneumonias, and multiple bone fractures (distal ulna, medial condyle, distal tibia, all after trauma). His serum creatine kinase level ranged from 9–60 U/l (normal <171 U/l)16. At age 6, an electrocardiogram (EKG) showed biventricular hypertrophy and an echocardiogram demonstrated severe left ventricular dilation with poor muscle contractility. He was started on digoxin, captopril and diuretics, tube feeding, and nocturnal noninvasive ventilation. At age 7, a gastrostomy tube was inserted. At age 12, ophthalmoplegia and mild lumbar torsion-scoliosis was diagnosed. His dilated cardiomyopathy was progressive; his shortening fraction decreased from 20% at age 10 to 9% at age 16 with severe mitral valve insufficiency. Despite maximum support, he died at age 17 due to cardiopulmonary insufficiency.
Patient 2 is a 6.5-year-old female. She was born at term via Cesarean section and presented with a weak cry, respiratory distress, hypotonia, and reduced deep tendon reflexes. She had bilateral vocal cord paralysis, and a gastrostomy tube was placed at age 4 weeks due to swallowing concerns. She attained head control at 4 months, rolled over at 6–9 months, got into a sitting position at 9–12 months, crawled at 18 months, pulled to stand at 18–20 months, and walked at 2 years. EKG at 3 years and 10 months revealed sinus tachycardia; an echocardiogram was normal. Her serum creatine kinase level was within normal limits at 89 IU/l. At age 4, she had mild lower facial weakness, axial hypotonia and proximal muscle weakness (MRC 3–4/5 range) with subgravity neck flexion. She has nearly complete ophthalmoplegia, bilateral ptosis, and intermittent strabismus. She has a high-arched palate and nasal speech. She has a weak cough and has had recurrent respiratory infections. She has difficulty feeding by mouth and receives all nutrition via a gastrostomy tube. Although she has a history of delayed motor milestones, she continues to demonstrate improvements. At age 4.5, she was still unable to run and jump. A nerve conduction study at age 5 showed a reduced compound muscle action potential (CMAP) amplitude of 2.3 mV (normal > 3.0 mV) of the ulnar motor nerve recorded at the abductor digiti minimi muscle.
Muscle Biopsy Findings
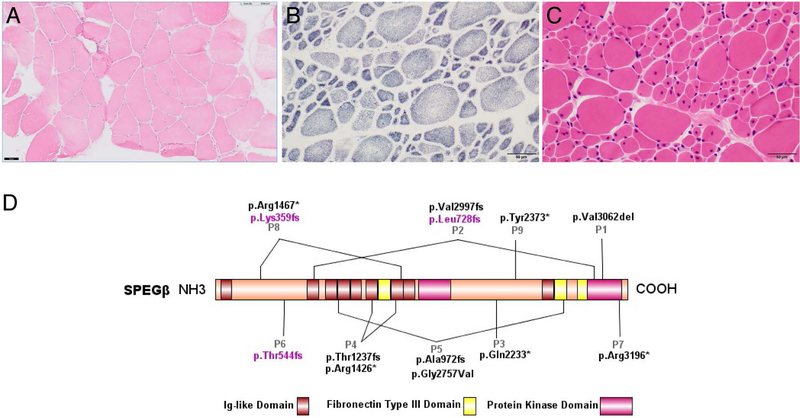
Patient 1 had a quadriceps muscle biopsy at age 9, which is consistent with non-CNM CM and shows a mild increase in fiber size variability, several atrophic fibers, and only a few internal/central nuclei (<20% of fibers) (Figure 1a). No clear fiber size hypertrophy is noted. Patient 2 had a quadriceps muscle biopsy at age 3, which is consistent with CNM and shows good fiber type differentiation without clear fiber type predominance, hypotrophic Type 1 fibers and hypertrophic Type 2 fibers, and many central nuclei (~50% of fibers and 60% of Type 1 fibers) (Figure 1b–c). Electron microscopy for Patient 2 shows a few myofibers with unstructured cores.
Figure 1:
Histological examination of patients’ muscle biopsies and SPEG schematic. (A) Hematoxylin & eosin (H&E) staining of Patient 1’s muscle biopsy specimen, performed at 9 years of age. The muscle biopsy shows a mild increase in fiber size variability, several atrophic fibers, and only a few internal/central nuclei, consistent with non-CNM CM. (B) SDH and (C) H&E staining of Patient 2’s muscle biopsy, performed at 3 years of age. The muscle biopsy reveals marked variability in fiber size with hypotrophic type 1 fibers and hypertrophic type II fibers with many central nuclei, consistent with CNM. Scale bar 50μm for all images. (D) Schematic of SPEGβ domain organization with positions of identified mutations generated by IBS (Illustrator for Biological Sequences). Mutations affecting both SPEGα and SPEGβ are in black, while mutations affecting only SPEGβ are in pink.
Genetic Results
Copy number variant analysis for Patient 1 using array-CGH identified a deletion of chromosome 4q35.2 (190,462,807–191,041,681; 579 kb), a deletion of chromosome 7q11.22 (66,692,376–68,103,955; 1,412 Mb), and copy neutral homozygosity of 6 regions >10 Mb, confirming consanguinity. Both deletions did not correlate to a phenotype and were identified in his father; the first includes BC087857 and the second does not include any genes. Sanger sequencing of FKRP, SEPN1, and RYR1 was unrevealing. Trio WES identified a homozygous mutation in exon 38 of SPEG, c.9185_9187delTGG (p.(Val3062del)). The amino acid at this position is highly conserved and located in the protein kinase domain, which is critical for SPEG function. This variant was heterozygous in the parents and unaffected sister.
WES analysis for Patient 2 initially identified a maternally inherited c.2183delT (p.(Leu728Argfs*82)) mutation in exon 10 of SPEG. Due to regions of low coverage, WGS was then performed, and identified the same maternally inherited mutation in compound heterozygosity with a paternally inherited 25 base pair insertion in exon 38, c.8962_8963insCGGGGCGAACGTTCGTGGCCAAGAT (p.(Val2997Glyfs*52)). These variants result in frameshifts and thus are classified as loss-of-function. The variants identified in both patients were predicted deleterious by MutationTaster and absent from ExAC, gnomAD, and 1000 Genomes databases.
Discussion
We report two additional patients with SPEG-associated CMs, Patient 1 with muscle pathology consistent with non-specific CM and Patient 2 with muscle pathology consistent with CNM (P1 and P2 in Figure 1d). The clinical and molecular findings of all nine patients reported so far including ours are summarized in Table 1 and pathological findings are described in Supplementary Table 1.
Table 1:
Clinical and Molecular Findings in Individuals Carrying SPEG Mutations
| Patient/Sex | P1/M (this study) | P2/F (this study) | P3/F9 | P4/F9 | P5/M9 | P6/M10 | P7/M10 | P8/M12 | P9/M11 |
|---|---|---|---|---|---|---|---|---|---|
| Age (when reported) | Died at 17 years | 6.5 years | Died at 3 weeks | 6 years | 1.5 years | 3 years | 7 years | 10 years | Died at 19 weeks |
| SPEG exons | Exon 38 | Exon 10 and 38 | Exon 30 | Exons 18 and 13 | Exons 10 and 35 | Exon 4 | Exon 40 | Exon 4 and 20 | Exon 30 |
| Allele 1 (maternal) | c.9185_9187delTGG; p.Val3062del | c.2183delT; p.Leu728fs | c.6697C>T; p.Gln2233* | c.4276C>T; p.Arg1426* | c.2915_2916delCCinsA; p.Ala972fs | c.1627–1628insA; p.Thr544fs | c.9586C>T; p.Arg3196* | c.1071_1074dup; p.Lys359fs | c.7119C>A p.Tyr2373* |
| Allele 2 (Paternal) | same as above | c.8962_8963ins25; p.Val2997fs | same as above | c.3709_3715+29del36; p.Thr1237fs | c.8270G>T; p.Gly2757Val | same as above | same as above | c.4399C>T; p.Arg1467* | same as above |
| Family History | Consanguineous parents, one healthy sister | No known consanguinity | Consanguineous parents, two sisters died early | No known consanguinity | No known consanguinity, sibling died early | parents from village in Turkey | Likely consanguineous | Non-consanguineous | Consanguineous parents |
| Birth History | Full term, severely hypotonic | Full-term, hypotonic | Full-term, breech delivery, severely hypotonic | Severely hypotonic | Born at 36 weeks of gestation, severely hypotonic | Full-term, hypotonic | Full-term, poor fetal movements | Uneventful pregnancy, hypotonic | Uneventful pregnancy, severely hypotonic |
| Neurological Findings | symmetric atrophy of lower extremities, wheel chair bound at 17 years | normal early motor milestones, walked at 2 years, unable to run or jump | Died of severe muscle weakness | Sit unsupported at 2.5 years, unable to walk unsupported | Head control at 16 months, sit unsupported at 18 months | Head control - 6 months, sit unsupported - 12 months, unable to walk | Head control at 18 months, sitting at 30 months, walking- 4 years | sit −11 months, walk - 30 months, short distances | Contracture of right ankle and lacked deep tendon reflex, antigravity movement at 1 week |
| Eye Findings | Ophthalmoplegia | Ophthalmoplegia, bilateral ptosis | No known evaluation | Ophthalmoplegia | None | ophthalmoplegia, mild ptosis | None | None | None |
| Respiratory Issues | non-invasive ventilation during night, recurrent pneumonia | Weak cough | Insufficient respiratory efforts | Tracheostomy, mechanical ventilation dependent | brief NICU stay for respiratory issues, no assisted ventilation | NICU for apnea, no intubations, recurrent lung infections | non-invasive ventilation during first 48 hours of life | None | Intubation required immediately after birth, weaned at 10 weeks for palliative care |
| Feeding Issues | Gastrostomy tube from age 6 | gastrostomy tube | Gastrostomy tube early in life | Gastrostomy tube early in life | NG feeding | None | NG feeding until day 13 | Gastrostomy tube from age 9 | Gastrostomy tube |
| Cardiac Issues | Dilated cardiomyopathy at age 7, severe mitral valve insufficiency | No cardiomyopathy at 3 years 10 months, sinus tachycardia | No cardiac evaluation | Dilated cardiomyopathy | Dilated cardiomyopathy, mitral valve insufficiency | None | Dilated cardiomyopathy mild mitral insufficiency | Reduced myocardial contraction, no ventricular dilation at 5 year | Enlarged atria, abnormal trabeculation of left ventricle |
| Skeletal Issues | Torsion scoliosis | Ulnar fracture at age 4, condyle fracture at age 5, tibia fracture at age 11 (all after trauma) | Not applicable | None | None | Pectus excavatum and mild scoliosis | None | Scoliosis developed at age 4 | Not applicable |
SPEG is alternatively spliced into 4 tissue-specific isoforms: APEG (aortic preferentially expressed gene), BPEG (brain preferentially expressed gene), and SPEGα and SPEGβ (expressed in skeletal and cardiac muscle)17. SPEGβ is the longer isoform with SPEGα missing amino acids 1–8549. Clinical data from Patients 6 and 8 suggests that SPEGα may partially rescue mutations affecting only SPEGβ, possibly preserving cardiac function10,12. This appears to be the case for Patient 2, who carries one variant sparing SPEGα, and has not yet developed signs of cardiac dysfunction. In contrast, Patient 1 carried a homozygous mutation affecting SPEGα and SPEGβ, and developed dilated cardiomyopathy, also seen in Patients 3, 4, 5 and 7 carrying mutations affecting both isoforms9,10. Interestingly, Patient 9, who also carries a mutation affecting both isoforms, developed non-compaction cardiomyopathy11.
Skeletal muscle dysfunction seems more severe in patients with mutations affecting both isoforms, as seen in Patients 1, 3, and 9 dying early, and Patient 4 needing constant mechanical ventilation9,11. The other two patients with both isoforms affected are Patients 5 and 79,10. In Patient 5, the disease was relatively mild, likely due to one variant being missense while all SPEG variants described so far have been loss-of-function, suggesting haploinsufficiency9. In Patient 7, the milder phenotype may be due to the mutation being very close to the C-terminus, thereby escaping nonsense mediated decay and potentially having less effect on protein function10. Overall, these findings suggest SPEGα has a critical role in skeletal and cardiac function and the disease is more severe when both isoforms are affected. Future studies should investigate the role of SPEGα in compensating for mutant SPEGβ. The clinical features of all patients have phenotypic similarities, most notably the presence of respiratory problems (Patients 1–7, and 9), eye involvement (Patients 1,2, 4, and 6), and scoliosis (Patients 1, 6, and 8)9–12.
In summary, this study expands the genetic heterogeneity of SPEG-associated CMs and further elucidates genotype-phenotype correlations to help guide appropriate clinical screening and management. The phenotype of SPEG-associated CM is varied and expanding, including ophthalmoplegia, and diagnostic markers that were initially considered, such as the presence of centralized nuclei on muscle biopsy and dilated cardiomyopathy, do not capture all cases. Thus, it is important for clinicians to consider evaluating a patient with congenital myopathy for SPEG mutations using WES even in the absence of type 1 fiber predominance, central nuclei, or cardiomyopathy.
Supplementary Material
Acknowledgements
The authors thank the families for their participation in the study, Daniel Ezzo for help with data analysis, Dr. Anne Rutkowski and CureCMD for help with patient recruitment, and Gilberto (Mike) Averion and Christopher Mendoza for clinical support. For Patient 2, initial whole exome sequencing was funded by the Clinical Center Genomics Opportunity, which is sponsored by the National Human Genome Research Institute, the NIH Deputy Director for Intramural Research, and the NIH Clinical Center. Whole genome sequencing for the same patient was performed at the Broad Center for Mendelian Genomics (CMG) (UM1 HG008900), funded by the National Human Genome Research Institute with supplemental funding provided by the National Heart, Lung, and Blood Institute under the Trans-Omics for Precision Medicine program and the National Eye Institute. CGB was supported by NIH Intramural Research Program funding from the National Institute of Neurological Disorders and Stroke. AMD was supported by the National Institute of General Medical Sciences (T32GM007753). PBA was supported by NIH/NIAMS 1R01AR068429-01.
Abbreviations
- CGH
comparative genomic hybridization
- CMs
congenital myopathies
- CMG
Center for Mendelian Genomics
- CNMs
centronuclear myopathies (CNMs)
- EKG
electrocardiography
- SPEG
Striated preferentially expressed gene
- WES
whole exome sequencing
- WGS
whole genome sequencing
Footnotes
Ethical Publication Statement: We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Disclosure of Conflicts of Interest: None of the authors has any conflict of interest to disclose.
References:
- 1.Nance JR, Dowling JJ, Gibbs EM, Bonnemann CG. Congenital myopathies: an update. Curr Neurol Neurosci Rep 2012;12(2):165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierson CR, Tomczak K, Agrawal P, Moghadaszadeh B, Beggs AH. X-linked myotubular and centronuclear myopathies. J Neuropathol Exp Neurol 2005;64(7):555–564. [DOI] [PubMed] [Google Scholar]
- 3.Bevilacqua JA, Monnier N, Bitoun M, Eymard B, Ferreiro A, Monges S, Lubieniecki F, Taratuto AL, Laquerriere A, Claeys KG, Marty I, Fardeau M, Guicheney P, Lunardi J, Romero NB. Recessive RYR1 mutations cause unusual congenital myopathy with prominent nuclear internalization and large areas of myofibrillar disorganization. Neuropathol Appl Neurobiol 2011;37(3):271–284. [DOI] [PubMed] [Google Scholar]
- 4.Bitoun M, Maugenre S, Jeannet PY, Lacene E, Ferrer X, Laforet P, Martin JJ, Laporte J, Lochmuller H, Beggs AH, Fardeau M, Eymard B, Romero NB, Guicheney P. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet 2005;37(11):1207–1209. [DOI] [PubMed] [Google Scholar]
- 5.Ceyhan-Birsoy O, Agrawal PB, Hidalgo C, Schmitz-Abe K, DeChene ET, Swanson LC, Soemedi R, Vasli N, Iannaccone ST, Shieh PB, Shur N, Dennison JM, Lawlor MW, Laporte J, Markianos K, Fairbrother WG, Granzier H, Beggs AH. Recessive truncating titin gene, TTN, mutations presenting as centronuclear myopathy. Neurology 2013;81(14):1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laporte J, Hu LJ, Kretz C, Mandel JL, Kioschis P, Coy JF, Klauck SM, Poustka A, Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet 1996;13(2):175–182. [DOI] [PubMed] [Google Scholar]
- 7.Nicot AS, Toussaint A, Tosch V, Kretz C, Wallgren-Pettersson C, Iwarsson E, Kingston H, Garnier JM, Biancalana V, Oldfors A, Mandel JL, Laporte J. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet 2007;39(9):1134–1139. [DOI] [PubMed] [Google Scholar]
- 8.Schartner V, Romero NB, Donkervoort S, Treves S, Munot P, Pierson TM, Dabaj I, Malfatti E, Zaharieva IT, Zorzato F, Abath Neto O, Brochier G, Lornage X, Eymard B, Taratuto AL, Bohm J, Gonorazky H, Ramos-Platt L, Feng L, Phadke R, Bharucha-Goebel DX, Sumner CJ, Bui MT, Lacene E, Beuvin M, Labasse C, Dondaine N, Schneider R, Thompson J, Boland A, Deleuze JF, Matthews E, Pakleza AN, Sewry CA, Biancalana V, Quijano-Roy S, Muntoni F, Fardeau M, Bonnemann CG, Laporte J. Dihydropyridine receptor (DHPR, CACNA1S) congenital myopathy. Acta Neuropathol 2017;133(4):517–533. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal PB, Pierson CR, Joshi M, Liu X, Ravenscroft G, Moghadaszadeh B, Talabere T, Viola M, Swanson LC, Haliloglu G, Talim B, Yau KS, Allcock RJ, Laing NG, Perrella MA, Beggs AH. SPEG Interacts with Myotubularin, and Its Deficiency Causes Centronuclear Myopathy with Dilated Cardiomyopathy. Am J Hum Genet 2014;95(2):218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Castiglioni C, Kacar Bayram A, Fattori F, Pekuz S, Araneda D, Per H, Erazo R, Gumus H, Zorludemir S, Becker K, Ortega X, Bevilacqua JA, Bertini E, Cirak S. Insights from genotype-phenotype correlations by novel SPEG mutations causing centronuclear myopathy. Neuromuscul Disord 2017;27(9):836–842. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Schanzer A, Kampschulte B, Daimaguler HS, Logeswaran T, Schlierbach H, Petzinger J, Ehrhardt H, Hahn A, Cirak S. A novel SPEG mutation causes non-compaction cardiomyopathy and neuropathy in a floppy infant with centronuclear myopathy. Acta Neuropathol Commun 2018;6(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lornage X, Sabouraud P, Lannes B, Gaillard D, Schneider R, Deleuze JF, Boland A, Thompson J, Bohm J, Biancalana V, Laporte J. Novel SPEG Mutations in Congenital Myopathy without Centralized Nuclei. J Neuromuscul Dis 2018;5(2):257–260. [DOI] [PubMed] [Google Scholar]
- 13.Herkert JC, Abbott KM, Birnie E, Meems-Veldhuis MT, Boven LG, Benjamins M, du Marchie Sarvaas GJ, Barge-Schaapveld D, van Tintelen JP, van der Zwaag PA, Vos YJ, Sinke RJ, van den Berg MP, van Langen IM, Jongbloed JDH. Toward an effective exome-based genetic testing strategy in pediatric dilated cardiomyopathy. Genet Med 2018. [DOI] [PubMed] [Google Scholar]
- 14.Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics 2012;28(4):599–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubowitz V, Sewry CA, Oldfors A. Muscle biopsy: A practical approach: Elsevier; 2013. [Google Scholar]
- 16.Schumann G, Bonora R, Ceriotti F, Clerc-Renaud P, Ferrero CA, Ferard G, Franck PF, Gella FJ, Hoelzel W, Jorgensen PJ, Kanno T, Kessne A, Klauker R, Kristiansen N, Lessinger JM, Linsinger TP, Misaki H, Panteghini M, Pauwels J, Schimmel HG, Vialle A, Weidemann G, Siekmann L. IFCC primary reference procedures for the measurement of catalytic activity concentrations of enzymes at 37 degrees C. Part 2. Reference procedure for the measurement of catalytic concentration of creatine kinase. Clin Chem Lab Med 2002;40(6):635–642. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh CM, Fukumoto S, Layne MD, Maemura K, Charles H, Patel A, Perrella MA, Lee ME. Striated muscle preferentially expressed genes alpha and beta are two serine/threonine protein kinases derived from the same gene as the aortic preferentially expressed gene-1. J Biol Chem 2000;275(47):36966–36973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.