Abstract
With the advent of checkpoint inhibition, immunotherapy has revolutionized the clinical management of several cancers, but has demonstrated limited efficacy in mammary carcinoma. Transcriptomic profiling of cancer samples defined distinct immunophenotypic categories characterized by different prognostic and predictive connotations. In breast cancer, genomic alterations leading to the dysregulation of mitogen-activated protein kinase (MAPK) pathways have been linked to an immune-silent phenotype associated with poor outcome and treatment resistance. These aberrations include mutations of MAP3K1 and MAP2K4, amplification of KRAS, BRAF, and RAF1, and truncations of NF1. Anticancer therapies targeting MAPK signaling by BRAF and MEK inhibitors have demonstrated clear immunologic effects. These off-target properties could be exploited to convert the immune-silent tumor phenotype into an immune-active one. Preclinical evidence supports that MAPK-pathway inhibition can dramatically increase the efficacy of immunotherapy. In this review, we provide a detailed overview of the immunomodulatory impact of MAPK-pathway blockade through BRAF and MEK inhibitions. While BRAF inhibition might be relevant in melanoma only, MEK inhibition is potentially applicable to a wide range of tumors. Context-dependent similarities and differences of MAPK modulation will be dissected, in light of the complexity of the MAPK pathways. Therapeutic strategies combining the favorable effects of MAPK-oriented interventions on the tumor microenvironment while maintaining T-cell function will be presented. Finally, we will discuss recent studies highlighting the rationale for the implementation of MAPK-interference approaches in combination with checkpoint inhibitors and immune agonists in breast cancer.
Keywords: breast cancer immunotherapy, immune phenotypes, MAP2K4 and MAP3K1 mutations, MAPK modulation, MEK inhibition, transcriptomics
Introduction
The advent of checkpoint inhibitors (e.g. PD-1/PD-L1 and CTLA4 antagonists) has revolutionized the field of cancer immunology. In several tumor types, this therapeutic approach has shown to induce a durable response in a considerable proportion of patients, with a dramatic impact on overall survival [1–10]. In breast cancer, the highest response rate is observed in triple-negative (TN) and PD-L1-positive, and/or displaying a higher level of tumor-infiltrating lymphocytes (TILs) [11]. The overall response rate in this subset of patients was ∼20% (range 13–44%), according to data from three early-phase clinical trials [6,11]. As the majority of patients do not respond to treatment, the understanding of mechanisms associated with immunologic resistance is critical to developing more effective immunotherapeutic approaches.
Transcriptome profiling of tumor lesions has identified hierarchically relevant molecular pathways linked to tumor elimination [12–15]. Overlapping signatures have been observed in tumors characterized by favorable prognosis [16–18], responsiveness to immunotherapeutic approaches [e.g. vaccination and interleukin (IL)-2] [19,20], adoptive-cell therapy (ACT) [21], and checkpoint inhibition [22–25]), or in post-treatment lesions that will subsequently undergo regression [13,20,24,26,27]. These pathways are also activated during other forms of immune-mediated rejection such as allograft rejections or autoimmunity [28,29]. We refer to them as the Immunologic Constant of Rejection (ICR) [12,14–17,30–34]. More specifically, the ICR is summarized by a gene expression signature including genes involved in Th-1 signaling (IFNG, TXB21, CD8B, IL12B, STAT1, and IRF1), T-helper 1 (Th)-1 chemoattraction sustained by M1 polarization (such as the CXCR3 and CCR5 ligands CXCL9, CXCL10, and CCL5) and cytotoxic functions (GNLY, PRF, GZMA, GZMB, and GZMH) [31]. In tumor samples, the expression of such pro-inflammatory transcripts is associated with the counter-activation of suppressive mechanisms, as signified by the expression of IDO1, CTLA4, and CD274 (or PD-L1), PDCD1 (or PD1), and FOXP3 [30,31]. Based on the level expression of the ICR transcripts, two divergent immune phenotypes can be envisioned [16,30]. The immune-active phenotype is characterized by the co-ordinated activation of the ICR modules and is associated with responsiveness to immunotherapy, chemotherapy (at least in breast cancer [16,35]), and a more favorable prognosis. Conversely, the lack of the expression of ICR transcripts typifies the immune-silent cancer, which is associated with poor prognosis and resistance to treatment. It is likely that the tumor genetic program, the genetic makeup of the host, and environmental factors such as the gut microbiota can contribute to the development of these divergent phenotypes [30,36,37]. Among these variables, genomic alterations of tumor cells that are potentially implicated in the modulation of an effective antitumor immune response have been increasingly scrutinized as potential therapeutic targets.
Studies mostly conducted in melanoma have identified cancer cell-intrinsic features associated with the immune-silent phenotype in humans and/or lack of responsiveness to immunotherapy. These traits include the genomic activation of the WNT/β-catenin [38] and PI3K–AKT pathways [39], and the expression of transcripts encoding for proteins with mechanical barrier functions such as filaggrin and desmosomal proteins [40,41].
It has been recently observed that distinct dysregulation of the mitogen-activated protein kinase (MAPK) pathways sustained by genomic alterations are associated with lack of spontaneous antitumor response in mammary carcinoma [31,42].
Moreover, MAPK modulation is emerging as a promising strategy to counteract primary immune resistance and to enhance the efficacy of immunotherapy across tumor types. Here, we analyze the studies that have linked MAPK dysregulation with a differential antitumor immune response and provide an overview of the immune-regulatory role of the MAPK pathways. Finally, we discuss the immunologic effects induced by MAPK inhibitors and their potential implications for breast cancer treatment in combination with checkpoint inhibitors and immune agonists.
Genetic dysregulations of the MAPK pathways and immune phenotypes
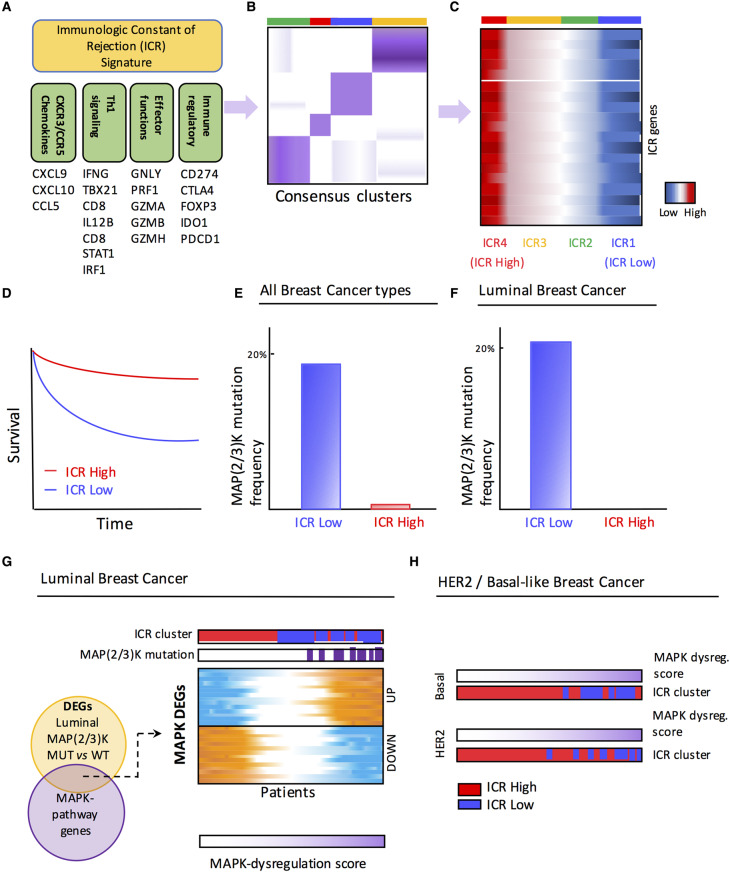
To elucidate the relationship between tumor genetic programs and immune responsiveness in breast cancer, we analyzed copy number variation, somatic mutations, and transcriptomic profile from more than 1000 samples submitted to the TCGA (The Cancer Genome Atlas) consortium and an additional meta-cohort consisting of ∼2000 gene expression profiles [43,44]. In this analysis, partially summarized in Figure 1, we demonstrated that ICR genes can segregate breast cancer in distinct phenotypes (Figure 1A–C), reflecting different immunologic orientation. The two opposite immune phenotypes are referred to here as ICR High and ICR Low. The co-ordinated activation of the ICR pathways, observed in the ICR High tumors, was associated with prolonged survival (Figure 1D) [31]. This observation was confirmed in the validation meta-cohort [31,43,44]. Only a small proportion of luminal samples (15%) displayed the Th-1/ICR High phenotype in contrast with 32% of basal-like and 20% of HER2-enriched tumors. Analysis of differentially mutated genes between ICR Low and ICR High groups revealed a 10-fold enrichment of MAP3K1 or MAP2K4 mutations in the immune-silent ICR Low tumors (Figure 1E). In fact, MAP2K4 and MAP3K1 were the only genes displaying a higher mutation rate in this immune phenotype. This association remained statistically highly significant when the associative analysis was restricted to luminal samples (Figure 1F). By comparing luminal samples harboring the MAP2K4 or MAP3K1 mutations [MAP(2/3)K-mut], we defined an MAPK-dysregulation signature consisting of 40 genes differentially expressed in luminal MAP(2/3)K-mut vs. wild type and belonging to the MAPK pathways (Figure 1G). The MAPK-dysregulation score, which was based on the expression of these 40 genes, could stratify not only the luminal, but also the other molecular subtypes (i.e. basal-like and HER2) according to their immunophenotypic features. The ICR1 Low tumors had the highest level of the MAPK-dysregulation score (Figure 1H). Superimposable ICR stratification was obtained by applying the MAPK-dysregulation score to the validation cohort. Increased expression of downstream molecules of the ERK and JNK cascades was observed in the ICR Low group (Figure 2). Although these mutations have been predicted to be deleterious [45], the expression levels of MAP3K1 and MAP2K4 were increased in mutated samples, which might indicate a compensatory mechanism for the loss-of-function mutations. [31,44]. These findings suggest the existence of a causal nexus between MAPK pathway dysregulation and the breast cancer immune phenotype [31] (Figure 1B,C). Therefore, dysregulation of the MAPK pathways, either sustained by MAP3K1 or MAP2K4 mutations or alternative mechanisms, could be implicated in the development of the unfavorable cancer immune phenotype (Figure 2). Moreover, Loi et al. [42], by analyzing the residual disease of TN breast cancer patients treated with neoadjuvant chemotherapy, observed a significant correlation between the low density of TILs and high levels of ERK–MAPK transcriptomic and genetic alterations, such as amplifications in KRAS, BRAF, and RAF1, and truncation in NF1. This observation suggests that dysregulation of the ERK–MAPK pathway by breast cancer cells may facilitate immune-evasion mechanisms [42].
Figure 1. MAPK dysregulation and immune phenotypes in breast cancer.
Overview of the analytic pipeline that identified MAP2K4/MAP3K1 [MAP(2/3)K]-driven alteration as genetic determinants of breast cancer immune phenotypes. (A) The ICR signature includes genes underlying Th-1 polarization, related chemokines (CXCR3 and CCR5 ligands), genes associated with the activation of cytotoxic effector functions, and counter-activating immune-regulatory genes. (B) Consensus clustering based on expression of the ICR signature generates four groups of breast tumors with distinct immune phenotypes. (C) Schematic representation of the heat map of ICR genes (vertical axis) with tumors (horizontal axis) sorted by ICR cluster. ICR4 tumors are associated with high expression of ICR genes (ICR High), while ICR1 tumors display low ICR gene expression (ICR Low). (D) Patients with ICR High tumors show a significantly improved survival compared with patients with ICR Low tumors. (E and F) Frequency of MAP2K4 or MAP3K1 [MAP(2/3)K] mutations in ICR Low breast tumors and ICR High tumors, in all breast cancer types (E) and in luminal breast tumors (F). (G) Genes differentially expressed (DEGs; N = 40) between MAP(2/3)K-mutated luminal breast cancer and MAP(2/3)K wild-type (WT) belonging to MAPK signaling (MAPK DEGs) were used to generate an MAPK-dysregulation score. (H) MAP2/3K dysregulation score can segregate the ICR High and ICR Low patients in HER2-enriched and basal-like breast cancer as well. Figure was readapted from Hendrickx et al. [31].
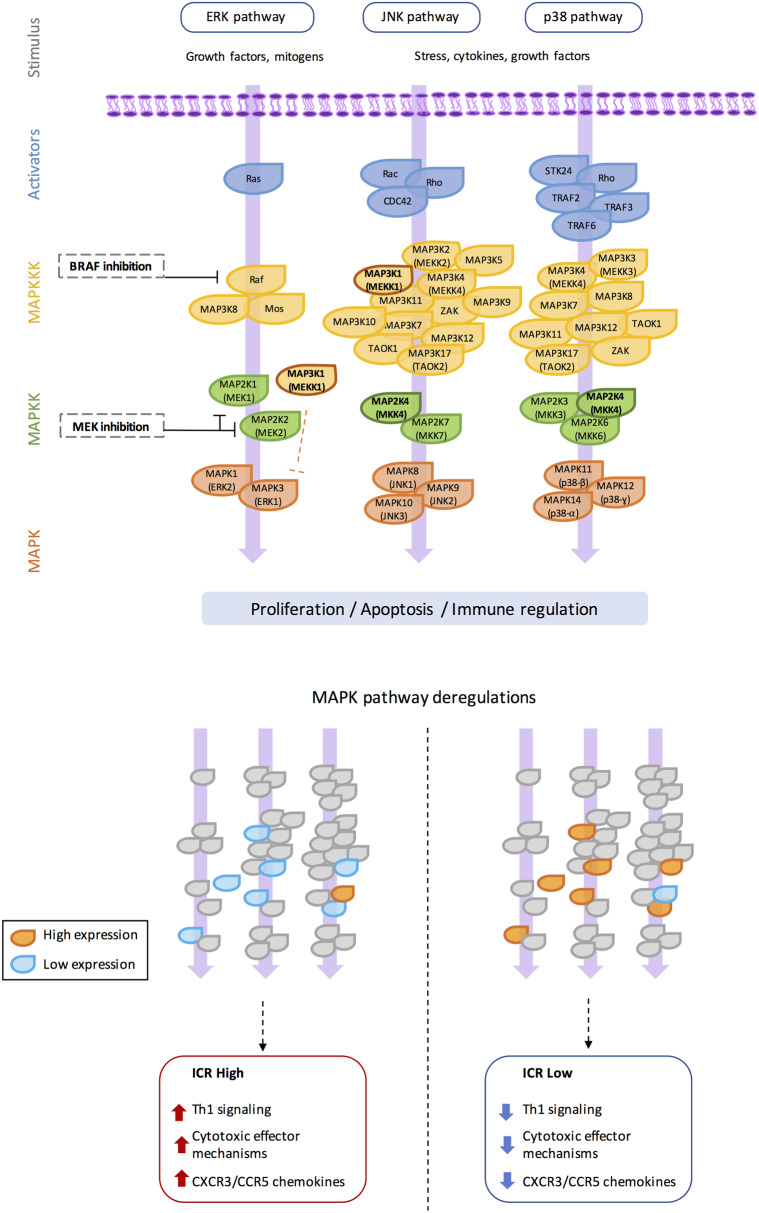
Figure 2. The MAPK signaling network and perturbations associated with immune phenotype.
The MAPK signaling network is composed of three main cross-talking cascades (i.e. ERK, JNK, and p38). The two MAPK dispositions based on MAPK-dysregulated genes associated with the opposite immune phenotypes are represented in this simplified version of the MAPK pathways.
MAPK pathways and immune response
The MAPK network is an established key regulator in cancer [46]. MAPK signaling can be exerted through three primary cross-signaling pathways: ERK, p38, and JNK, each comprising several levels of kinases (Figure 2). MAPK signaling can be activated by internal and external stimuli, which results in either a tumor suppressive or oncogenic event. The differential outcome is driven by the complexity of the signaling pathways, along with spatial and temporal regulation. This, in part, explains why different cancer types feature different responses to MAPK signaling interventions, thereby guiding different treatment approaches according to tumor types [47,48]. In addition, genomic alterations of any of the upstream or downstream molecules of the MAPK cascades can alter the pathway activity and therapeutic response.
While MAPK pathways are classified according to their downstream kinases, they are tightly interlocked as they share several intermediate mediators. As a consequence, treatment-induced interference with one of the MAPK pathways might deregulate the other ones. Even more, the effects of MAPK pathways are signaling- and cell-specific, thus increasing the complexity of predicting the net biological effect of targeted MAPK inhibition. For example, MAP3K1, which mediates both ERK–MAPK, and JNK–MAPK activity, has more than 25 published binding partners [49] including kinases, scaffolds, and adapters, but also nuclear and transcription factors, and microfilament, tight junction, and GTPase-activation proteins. Furthermore, the MAP3K1 protein contains 2 functional domains; a kinase domain and a plant homeodomain which is an E3 ubiquitin ligase. As a result, MAP3K1 has been reported to exhibit contradictory functions, either activating of inactivating ERK– and JNK–MAPK pathways. The full-length MAP3K1 induces activation of MAP2K4/7-JNK and MAP2K1/2-ERK pathways, promoting cell survival while caspase cleavage, which generates the soluble active kinase domain, triggers apoptosis [48]. MAP3K1 also catalyzes the poly-ubiquitylation of c-Jun and ERK1/2, resulting in their degradation by proteasomes, leading to their degradation and inhibition of the ERK–MAPK pathway [48]. Mutations of MAP3K1 have been found to reduce tumor cell migration using wound-healing assays as a result of lower adhesive properties [50,51]. It is unknown whether reduced tumor cell migration can influence the antitumor immune response by altering the permeability of the tumor microenvironment and/or immune cell infiltration.
Even though the MAPK signaling has been extensively studied in the context of their oncogenic function and proliferative stimulus, these complex systems also regulate several functions of the innate and adaptive immunity. The understanding of the immune-regulatory functions exerted by MAPK pathways is critical to implementing integrative immunomodulatory strategies targeting these kinases. Below, we summarize some relevant aspects of the MAPK pathways’ immunomodulation.
The ERK–MAPK pathway
The classic ERK kinases have been identified as the downstream effectors of the RAS oncogenes. The ERK–MAPK pathway mainly exerts proliferative effects, although it is now clear that dysregulation of this pathway is associated with other aspects of the tumor phenotype [46]. The activation of the ERK–MAPK pathway favors differentiation into the CD4 lineage [52] and is critical for CD4 T cells’ Th-2 polarization as it is required for IL-4 receptor function [53]. In addition, ERK kinases trigger the production of IL-10 and negatively regulate the production of IL-12 by macrophages and dendritic cells, with a consequent induction of Th cell differentiation into a Th2 type [54]. ERK1 is dispensable for CD8 T-cell activation, whereas ERK2 is required for the proliferation of CD8 T cells activated in the absence of a co-stimulatory signal [55].
Additionally, this pathway induces the production of TNF, and inhibits the secretion of INF-β and iNOS [54,56–58].
The p38–MAPK pathway
The p38–MAPK pathway plays a pleiotropic role in cell survival, as it can either sustain proliferation or induce apoptosis in a cell type-specific manner depending on the type of stimulus [59]. This pathway is critical for the production of inflammatory cytokines. The p38–MAPK pathway exerts positive regulation on Th-1 differentiation, and it is selectively activated in Th-1, but not in Th-2, cells [60–62]. In both CD4 and CD8 T cells, p38 positively regulates production of IFN-γ [63,64]. In dendritic cells, activation of this pathway induces production of IL-12, which is crucial for Th-1 polarization [65].
The JNK–MAPK pathway
Similarly to the p38–MAPK pathway, the JNK–MAPK pathway can induce pro- or anti-apoptotic effects dependent on the stimulus, tissue type, and strength of the signal [66]. The JNK–MAPK pathway exerts a pro-inflammatory role in macrophages inducing M1 differentiation [54,67]. Studies in JNK1 and JNK2 knockout mice demonstrated that JNK reduce the proliferative response of activated Th cells, but facilitate the polarization into a Th-1 phenotype [68,69]. In fact, genetic loss of MAP3K2(MEKK2) abrogates IL-2 production [70]. Moreover, the ablation of MKP5 results in a selective increase in JNK–MAPK activity in CD4 T cells, which exhibit hypoproliferation upon T-cell receptor (TCR)-mediated activation [71]. Conversely, suppression of JNK–MAPK activity in CD8 T cells through JNK1 knockout induces hypoproliferation and reduced production of IL-2 [72,73]. It has been shown that MAP3K1 plays a negative regulatory role in the expansion of CD8 T cells in hematopoietic MAP3K1-deficient mice challenged with a viral infection [74]. B-cell proliferation and germinal center formation have also been shown to be regulated by MAPK signaling [75].
Immunomodulatory effects of MAPK-pathway blockade induced by BRAF and MEK inhibitors alone or in combination with immunotherapy
BRAF inhibition
With the clinical implementation of BRAF and MEK inhibitors in metastatic melanoma, several observations regarding the immune-regulatory mechanisms of this pharmacological intervention emerged. BRAF mutations are reported in ∼50–60% of melanoma patients [76–78]. About 90% of BRAF mutations consist of a single nucleotide mutation in the 600 codon (V600). Among them, ∼90% of mutations are represented by the valine to glutamic acid substitution (V600E), which is associated with a 400-fold increased activity of the protein, resulting in hyperactivation of the ERK–MAPK pathways [77,79–82]. Because of this oncogenic addiction, inhibition of the ERK–MAPK pathway through specific BRAF inhibitors targeting the V600E mutation (i.e. vemurafenib and dabrafenib) induces strong antiproliferative effects. BRAF inhibition alone, however, induces only transient remission, with most of patients relapsing a few months after treatment because of acquired secondary resistance often due to reactivation of the ERK–MAPK pathway [83–85]. Moreover, BRAF inhibition mediates paradoxically activation of the MAPK pathways in cells with wild-type BRAF and strong upstream signaling in the MAPK pathways, resulting in a higher rate of adverse events, including skin-related toxicity, basal cell carcinomas, and keratoacanthomas [86,87]. Both phenomena can be counteracted by targeting the ERK–MAPK pathway downstream from BRAF through MEK (MAP2K1/MAP2K2) inhibition. Data from three phase 3 clinical trials have demonstrated that combined inhibition of MEK and mutant BRAF kinases results in increased tumor response, prevents or delays MAPK-driven acquired resistance, and decreases the frequency and severity of some of adverse events [88–90].
The effects of the inhibition of the ERK–MAPK by targeting mutant BRAF, which might be relevant to melanoma only, or by targeting MEK, which is potentially applicable to a wider range of tumors, share some similarities but are also characterized by a different immunologic impact (Figure 3). In general, these two approaches induce similar immunologic changes in tumor cells (e.g. increase in the expression of MHCs and tumor-associated antigens, and decrease in the production of immunosuppressive cytokines), but different modulation of T-cell functions. While BRAF inhibition does not negatively affect T-cell functions, as it targets the mutated form of BRAF, MEK inhibition can inhibit T-cell activation and proliferation. These aspects are discussed in more detail below.
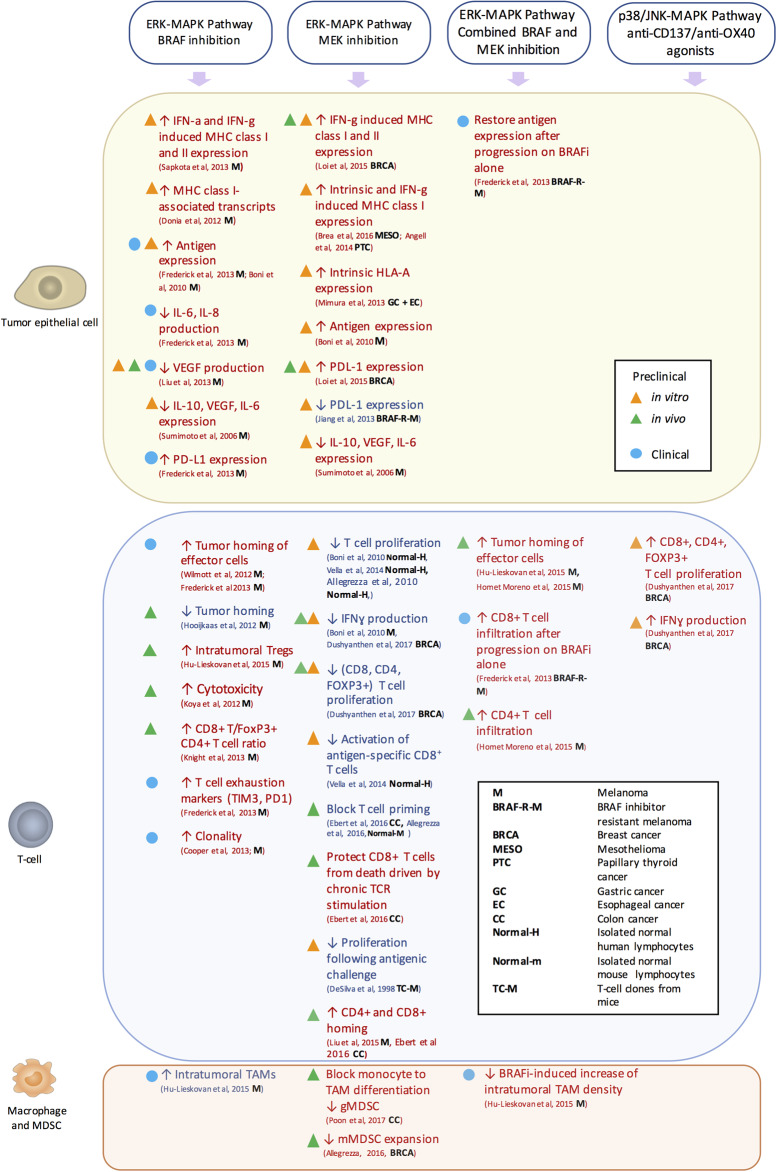
Figure 3. Immunologic impact of MAPK-targeted interventions per cell population.
The immunologic effects of MAPK-pathway interventions are represented. Preclinical in vitro (orange triangle), in vivo (green triangle), and clinical (blue circle) evidence of the direct and indirect effects of these interventions are summarized per cell population that comprises the tumor microenvironment. The investigated tumor type is indicated for each of these studies by abbreviation (bold) as defined in the figure. Effects that favor or counteract tumor rejection are showed in red and blue, respectively.
A seminal study on BRAF V600E melanoma cell lines has shown that MEK inhibition and RNA interference for BRAF V600E can reduce the production of immunosuppressive cytokines such as VEGF, IL-10, and IL-6 [91]. However, inhibition of the ERK–MAPK pathway with MEK or BRAF inhibitors in V600E-mutated melanoma cell lines and tumor digests results in increased levels of melanocyte differentiation antigens, followed by enhanced recognition by T cells [92]. Consistently, the evaluation of human melanoma biopsies before and after treatment with the BRAF inhibitor demonstrated a down-regulation of VEGF [93]. In a clinical context, treatment with a BRAF inhibitor was associated with increased expression of melanoma antigens [94], decrease in immunosuppressive cytokines (IL-6 and IL-8) [94], and increased tumor homing [94,95] and clonality of effector CD8 T cells [96]. On the other hand, TIL activation was associated with concomitant expression of exhaustion markers such as PD-1 and TIM3 [94]. Moreover, a better response was observed in patients who had a high proportion of pre-existing dominant T-cell clones [96]. Conversely, decreased density of TILs after BRAF inhibition was found in an inducible BRAF-mutated melanoma mouse model [97]. Furthermore, some in vitro studies have shown that BRAF inhibitors enhance the induction of MHC I/II molecules by IFN-γ and IFN-α and the up-regulation of MHC-associated transcripts [98,99], while others did not [92,100,101]. However, no change of HLA I expression was observed in tumor biopsies after BRAF treatment [94].
BRAF inhibition in combination with immunotherapy
In view of the favorable immunomodulatory effects of BRAF inhibition observed in some studies, this treatment has been combined with immunotherapeutic approaches to enhance their efficacy. Indeed, in xenograft mouse models, BRAF inhibition decreased VEGF production by mutated melanoma cells and augmented tumor infiltration of adoptively transferred T cells in mice, resulting in enhanced efficacy of ACT [93,101]. Moreover, BRAF inhibition increased in vivo cytotoxic activity and intratumoral cytokine secretion of adoptively transferred lymphocytes in a syngeneic fully immunocompetent mouse model, augmenting the efficacy of ACT [101]. This phenomenon was attributed to a paradoxical activation of the ERK–MAPK pathway in wild-type T cells [101]. However, improved therapeutic antitumor activity was observed for BRAF inhibition combined with a CD137 agonist but not with anti-CTLA4, anti-PD-1, and anti-Tim3 antibodies in melanoma mouse models [100]. Despite the potential benefit of combinatorial approaches, a clinical study investigating concomitant administration of the BRAF inhibitor vemurafenib and anti-CTLA-4 in metastatic melanoma was stopped due to unacceptable hepatotoxicity [102], probably sustained by paradoxical activation of the ERK–MAPK pathway in liver-resident (wild-type) cells [87]. The effects of MAPK-immunotherapy combinations are summarized in Table 1.
Table 1. In vivo preclinical studies combining MAPK inhibition with immunotherapeutic approaches.
| Reference | Setting | Tested combinations | Efficacy comparison | Effect on tumor microenvironment |
|---|---|---|---|---|
| Liu et al. [93] | BRAFV600E melanoma | BRAFi + ACT | BRAFi + ACT > either therapy alone | BRAFi increased tumor infiltration of adoptively transferred T cells by inhibiting VEGF production in tumor cells. |
| Koya et al. [101] | BRAFV600E melanoma | BRAFi + ACT | BRAFi + ACT > either therapy alone | BRAFi did not alter expansion, distribution, or tumor accumulation of adoptively transferred T cells; BRAFi paradoxically increased MAPK signaling, in vivo cytotoxic activity, and intratumoral cytokine secretion by adoptively transferred T cells. |
| Knight et al. [100] | BRAFV600E melanoma | BRAFi + anti-CD137/anti-CTLA-4/anti-PD-1/anti-Tim3 | BRAFi + anti-CD137 > either therapy alone BRAFi + anti-CTLA4/anti-PD-1/anti-Tim3 = either therapy alone |
Combined antitumoral activity was observed between BRAFi and anti-CD137. |
| Hooijkaas et al. [97] | BRAFV600E melanoma | BRAFi + anti-CTLA4 | BRAFi + anti-CTLA4 > anti-CTLA4 BRAFi + anti-CTLA4 = BRAFi |
BRAFi led to a decreased frequency of tumor-resident T cells, NK cells, MDSCs, and macrophages, which could not be restored by the addition of anti-CTLA4. |
| Homet Moreno et al. [116] | BRAFV600E melanoma | BRAFi + MEKi + anti-PD-1 BRAFi + MEKi + anti-CD137 + anti-OX40 + anti-PD-L1 |
BRAFi + MEKi + anti-PD-1 >
BRAFi + MEKi + anti-PD-L1 combined with either anti-CD137, anti-OX40, or both > BRAFi + MEKi + anti-CD137 + OX40 |
Combined BRAFi and MEKi increased CD8+ and CD4+ T-cell, and TAM infiltration. (BRAFi + MEKi vs. no therapy) (BRAFi + MEKi + anti-PD-1 vs. anti-PD-1 monotherapy). |
| Hu-Lieskovan et al. [114] | BRAFV600E melanoma | BRAFi + MEKi + ACT BRAFi + MEKi + anti-PD-1 |
BRAFi + MEKi + ACT >
BRAFi + MEKi + anti-PD-1 >
|
BRAFi + MEKi treatment increased (both endogenous and adoptively transferred) effector T-cell homing. BRAFi + MEKi increased tumor antigen expression both in control tumors and ACT-treated tumors. BRAFi + MEKi up-regulated MHC molecules in ACT-treated tumor, but not in control tumors. |
| Liu et al. [111] | Colon carcinoma | MEKi + anti-PD-1/anti-PD-L1/anti-CTLA4 | MEKi + anti-PD-1 = MEKi + anti-PD-L1 = MEKi + anti-CTLA4 > either therapy alone | MEKi combined with anti-PD-1 increased tumor-infiltrating CD8+ T cells. |
| Ebert et al. [112] | Colon carcinoma | MEKi + anti-PD-L1 | MEKi + anti-PD-L1 > either therapy alone | Combination MEKi + anti-PD-L1 provoked changes that largely mirrored MEKi treatment only, based on expression of 94 immune-related genes. |
| Poon et al. [106] | Colon carcinoma | MEKi + anti-CTLA4 | MEKi + anti-CTLA4 > either therapy alone | Combination of anti-CTLA-4 with MEKi annihilated Cox-2 and Arg1 up-regulation induced by CTLA-4 treatment. |
| Loi et al. [42] | Breast cancer | MEKi + anti-PD-L1 | MEKi + anti-PD-L1 > either therapy alone | Significant synergy between MEKi and anti-PD-L1 (in the LACZ model). Increased expression of antigen presentation and processing genes when MEKi was combined with anti-PD-L1. |
| Dushyanthen et al. [115] | Breast cancer | MEKi + anti-CD137 MEKi + anti-OX40 MEKi + anti-CD137 + anti-PD-1 MEKi + anti-OX40 + anti-PD-1 |
MEKi + anti-CD137 > either therapy alone MEKi + anti-OX40 > either therapy alone MEKi + anti-CD137 + anti-PD-1 >
|
Anti-CD137 and anti-OX40 T-cell agonist antibodies prevent MEKi-induced decrease in CD8+, CD4+, and FOXP3+ T-cell proliferation and MEKi-induced reduction in IFN-γ production by CD8+ T cells. |
MEK inhibition
Similarly to BRAF inhibition, MEK inhibition can favorably modify the tumor microenvironment. Most of the studies have shown that MEK inhibition can increase the expression of intrinsic and IFN-γ-induced HLA/MHC I/II in cancer cell lines, including melanoma, mesothelioma, prostate, gastric, and esophageal cancer cell lines [103–105]. Similarly to BRAF inhibition, treatment of mutant melanoma cell lines with MEK inhibition enhances the expression of melanoma-differentiation antigens [92] and decreases the production of IL-10, IL-6, and VEGF [91]. Moreover, in a syngeneic colon carcinoma model, MEK inhibition was found to prevent monocyte to tumor-associated macrophage (TAM) differentiation, substantiated by the increase in the intratumoral frequency of intermediary differentiated monocytes expressing MHC II, and the decrease in intratumoral granulocytic myeloid-derived suppressive cells (gMDSCs) [106].
Conversely, because of the critical role of the ERK–MAPK pathway in T-cell function, MEK inhibition can theoretically dampen T-cell antitumor activity. Studies performed in the late 1990s have shown that MEK inhibition blocks T-cell proliferation in response to antigenic stimulation, but it does not affect IL-2-mediated proliferation [107], in agreement with ERK1/2 knockout experiments [55]. However, Boni et al. [92] observed that MEK inhibition decreases production of IFN-γ by T cells and inhibits T-cell proliferation after stimulation with IL-2 and OKT3, while Vella et al. [108] showed that MEK inhibition suppresses in vitro T-lymphocyte proliferation, cytokine production, and antigen-specific expansion. Similarly, Allegrezza et al. [109] observed that MEK inhibition equally inhibits proliferation of naive, memory, and effector memory T cells, and profoundly blocks T-cell priming. These effects could be reverted by IL-15 administration [109]. These immunosuppressive mechanisms, observed in vitro, are perhaps only transient and did not fully translate in vivo. In fact, in a KRAS-inducible breast cancer model, MEK inhibition prevented tumor progression in vivo through an immune-mediated mechanism, rather than by direct inhibition of tumor-cell proliferation. This mechanism, which required CD8 T cells, was mediated by the blockade of monocytic myeloid-derived suppressive cell (mMDSC) expansion [110]. Moreover, MEK inhibition increases the number of CD4 TILs and did not negatively affect the density of CD8 TILs in a fully competent colon carcinoma syngeneic mouse model [111]. These findings were mechanistically explained by another study using the same model. Ebert et al. [112] demonstrated that MEK inhibition profoundly blocks naive CD8 T-cell priming. At the same time, this pharmacological inhibition increases the number of antigen-specific effector CD8 T cells in the tumor and protects CD8 TILs from death driven by chronic TCR stimulation [112].
Double MAPK blockade through BRAF and MEK inhibition in combination with immunotherapy
When combined with BRAF inhibition, MEK inhibition can potentially balance the overreacting effector cells to prevent exhaustion [113]. In fact, concurrent administration of BRAF and MEK inhibitors with ACT resulted in dramatic tumor response, increased T-cell infiltration, and effector functions in a syngeneic immunocompetent melanoma mouse model [114]. This was accompanied by increase in melanoma-associated antigens [114]. However, MEK inhibition was able to overcome the increase in intratumoral T-regulatory cells and macrophages observed after treatment with BRAF and ACT [114].
In a clinical context, the concomitant treatment with an MEK inhibitor was able to restore melanoma-associated antigen expression and CD8 T-cell infiltration in a patient experiencing progression following BRAF inhibition [94].
MEK inhibition in combination with immunotherapy and implications for breast cancer treatment
In view of the negative correlation between MAPK dysregulation and density of TILs in patients with TN breast cancer treated with neoadjuvant chemotherapy, Loi et al. [42] assessed the immune-regulatory impact of MEK inhibition in this setting. They showed that MEK inhibition increases the IFN-γ-inducible expression of MHC I/II and PD-L1 in TN breast cancer cell lines. These observations were validated in vivo by analyzing MHCI/II and PD-L1 expression of tumor cells from lesions generated by orthotopic injection of TN cell lines into syngeneic mice treated with MEK inhibition [42]. Moreover, combined MEK inhibition and anti-PD-L1 in orthotopic TN and HER2 breast cancer models resulted in enhanced tumor control, associated with an increase in the intratumoral expression of immune-related transcripts [42].
Consistently, in colon carcinoma syngeneic mouse models, the combination of MEK inhibition with either anti-PD-1 [111,112], anti-PD-L1 [111], or anti-CTLA4 [106,111] results in a synergistic effect. In a subsequent study on TN mouse models, the investigators showed that MEK inhibition can have a detrimental effect on T-cell homing, proliferation, and effector functions [115]. However, T-cell functions were restored by the administration of anti-137 and anti-OX40 agonists [115]. Similarly, in mutant melanoma models, the addition of either OX-40 or 137 agonists enhanced the efficacy of the triple combination of BRAF inhibitor, MEK inhibitor, and PD1 blockade [116]. Furthermore, the investigators convincingly demonstrated that the rescue of immune-effector functions was due to the redirection of the signaling from the classical ERK–MAPK pathway to the alternative p-38–MAPK/JNK–MAPK pathways mediated by OX-40 or CD137 agonists [115].
Altogether these data provide a solid rationale for combining MEK inhibition with immune agonist and/or checkpoint inhibitors in TN breast cancer. A clinical trial exploring the combination of the anti-PD1 pembrolizumab and the MEK inhibitor binimetinib in TN breast cancer (NCT02322814) is ongoing. An additional ongoing trial is evaluating the combination of anti-PD-L1 avelumab together with the CD-137 agonist utomilumumab across several tumor types, including TN breast cancer (NCT02554812).
In view of the association between the degree of intratumoral immune response and the MAPK dysregulation in luminal and HER2 tumors, we propose that combined targeted-immunotherapeutic strategies should also be evaluated in these subtypes (Figure 4). Moreover, the effect of MEK inhibition on cancer cells harboring MAP2K4 or MAP3K1 mutations needs to be elucidated.
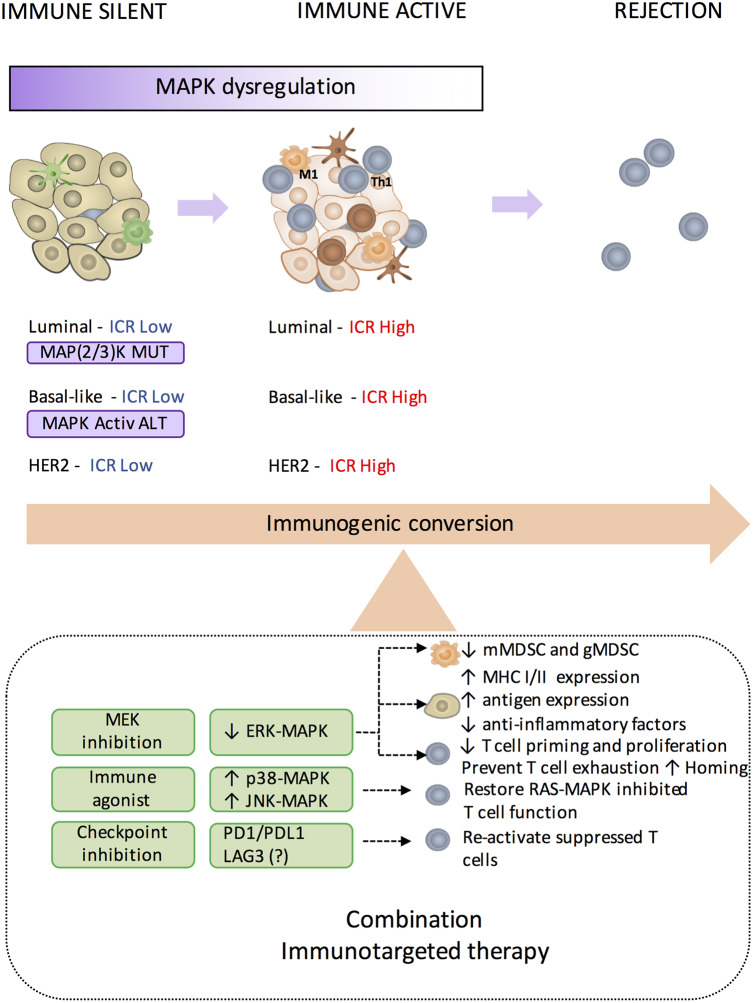
Figure 4. The immunogenic conversion.
Immune-silent breast cancer characterized by low TIL density or low expression of genes included in the immunologic constant of rejection (ICR Low) frequently displays dysregulation of MAPK pathways, either caused by genomic alterations (in purple) [i.e. MAP2K4 or MAP3K1 mutations in luminal breast cancer; Hendrickx et al. [31]] and MAPK-activating alterations (e.g. amplifications in KRAS, BRAF, and RAF1, and truncations in NF1) in basal-like tumors (Loi et al. [42]), or by alternative mechanisms. To convert these tumors into an immune-active phenotype characterized by high expression of ICR genes (ICR High) and then trigger tumor rejection, combination therapy consisting of MEK inhibition (MEKi), immune checkpoint inhibition (e.g. PD1 blockade; Dushyanthen et al. [115]), and perhaps other checkpoint inhibitions and immune agonist antibodies (Dushyanthen et al. [115]) is suggested. We refer to this as the ‘immunogenic conversion’.
In conclusion, we believe that the immunogenic conversion of the immune-silent tumors through the modulation of oncogenic pathways conducive to immunosuppression could dramatically expand the number of patients who will benefit from immunotherapeutic approaches.
Summary
Genomic alterations causing dysregulations of MAPK pathways are associated with an immune-silent phenotype of breast cancer and could represent a mechanism of immune evasion.
Immunologic effects of MAPK-pathway blockade on tumor cells promote the development of an immune-active tumor phenotype, providing a rationale to combine MAPK-pathway inhibitors with immunotherapy.
As MEK inhibition has a negative impact on T cells, strategies to rescue T-cell function, for example, by immune agonist antibodies are essential to achieve maximal therapeutic efficacy.
Reprogramming the immune phenotype from an immune-silent to an immune-active phenotype by targeting oncogenic pathways could extend the efficacy of immunotherapeutic approaches to patients who were previously considered immunotherapy-resistant.
Abbreviations
- ACT
adoptive-cell therapy
- CTLA4
cytotoxic T-lymphocyte-associated protein 4
- DEG
differentially expressed genes
- ERK
extracellular signal-regulated kinase
- gMDSC
granulocytic myeloid-derived suppressive cell
- HLA
human leukocyte antigen
- ICR
Immunologic Constant of Rejection
- IFNG
interferon gamma
- IL
interleukin
- IRF1
interferon regulatory factor 1
- JNK
c-Jun N-terminal kinase
- MAP (2/3)K-mut
MAP2K4 or MAP3K1 mutations
- MAPK
mitogen-activated protein kinase
- MEK
mitogen-activated protein kinase kinase
- MHC
major histocompatibility complex
- mMDSC
monocytic myeloid-derived suppressive cell
- PD-(L)1
programmed cell death protein (ligand) 1
- STAT1
signal transducer and activator of transcription 1
- TAM
tumor-associated macrophages
- TCR
T-cell receptor
- Th
T helper
- TIL
tumor-infiltrating lymphocyte
- TN
triple negative
- T-reg
T-regulatory cell
- VEGF
vascular endothelial growth factor
Author Contribution
All the authors critically revised the draft and contributed to the writing of the final manuscript.
Funding
W.H. received funding from Qatar National Research Fund [JSREP07-012-3-005]. J.R.’s scholarship is supported by JSREP07-012-3-005.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Brahmer J.R., Tykodi S.S., Chow L.Q.M., Hwu W.-J., Topalian S.L., Hwu P. et al. (2012) Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emens L.A., Butterfield L.H., Hodi F.S., Marincola F.M. and Kaufman H.L. (2016) Cancer immunotherapy trials: leading a paradigm shift in drug development. J. Immunother. Cancer 4, 42 10.1186/s40425-016-0146-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heong V., Ngoi N. and Tan D.S.P. (2017) Update on immune checkpoint inhibitors in gynecological cancers. J. Gynecol. Oncol. 28, e20 10.3802/jgo.2017.28.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodi F.S., O'Day S.J., McDermott D.F., Weber R.W., Sosman J.A., Haanen J.B. et al. (2010) Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moehler M., Delic M., Goepfert K., Aust D., Grabsch H.I., Halama N. et al. (2016) Immunotherapy in gastrointestinal cancer: recent results, current studies and future perspectives. Eur. J. Cancer 59, 160–170 10.1016/j.ejca.2016.02.020 [DOI] [PubMed] [Google Scholar]
- 6.Nanda R., Chow L.Q.M., Dees E.C., Berger R., Gupta S., Geva R. et al. (2016) Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J. Clin. Oncol. 34, 2460–2467 10.1200/JCO.2015.64.8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postow M.A., Callahan M.K. and Wolchok J.D. (2015) Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 33, 1974–1982 10.1200/JCO.2014.59.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robert C., Thomas L., Bondarenko I., O'Day S., Weber J., Garbe C. et al. (2011) Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 364, 2517–2526 10.1056/NEJMoa1104621 [DOI] [PubMed] [Google Scholar]
- 9.Robert C., Long G.V., Brady B., Dutriaux C., Maio M., Mortier L. et al. (2015) Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- 10.Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F. et al. (2012) Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emens L.A. (2017) Breast cancer immunotherapy: facts and hopes. Clin. Cancer Res. 10.1158/1078-0432.CCR-16-3001. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedognetti D., Wang E., Sertoli M.R. and Marincola F.M. (2010) Gene-expression profiling in vaccine therapy and immunotherapy for cancer. Expert Rev. Vaccines 9, 555–565 10.1586/erv.10.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panelli M.C., Stashower M.E., Slade H.B., Smith K., Norwood C., Abati A. et al. (2007) Sequential gene profiling of basal cell carcinomas treated with imiquimod in a placebo-controlled study defines the requirements for tissue rejection. Genome Biol. 8, R8 10.1186/gb-2007-8-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang E., Worschech A. and Marincola F.M. (2008) The immunologic constant of rejection. Trends Immunol. 29, 256–262 10.1016/j.it.2008.03.002 [DOI] [PubMed] [Google Scholar]
- 15.Wang E., Bedognetti D. and Marincola F.M. (2013) Prediction of response to anticancer immunotherapy using gene signatures. J. Clin. Oncol. 31, 2369–2371 10.1200/JCO.2013.49.2157 [DOI] [PubMed] [Google Scholar]
- 16.Bedognetti D., Hendrickx W., Marincola F.M. and Miller L.D. (2015) Prognostic and predictive immune gene signatures in breast cancer. Curr. Opin. Oncol. 27, 433–444 10.1097/CCO.0000000000000234 [DOI] [PubMed] [Google Scholar]
- 17.Galon J., Angell H.K., Bedognetti D. and Marincola F.M. (2013) The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 39, 11–26 10.1016/j.immuni.2013.07.008 [DOI] [PubMed] [Google Scholar]
- 18.Miller L.D., Chou J.A., Black M.A., Print C., Chifman J., Alistar A. et al. (2016) Immunogenic subtypes of breast cancer delineated by gene classifiers of immune responsiveness. Cancer Immunol. Res. 4, 600–610 10.1158/2326-6066.CIR-15-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang E., Miller L.D., Ohnmacht G.A., Mocellin S., Perez-Diez A., Petersen D. et al. (2002) Prospective molecular profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 62, 3581–3586 PMID: [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss G.R., Grosh W.W., Chianese-Bullock K.A., Zhao Y., Liu H., Slingluff C.L. et al. (2011) Molecular insights on the peripheral and intratumoral effects of systemic high-dose rIL-2 (Aldesleukin) administration for the treatment of metastatic melanoma. Clin. Cancer Res. 17, 7440–7450 10.1158/1078-0432.CCR-11-1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedognetti D., Spivey T.L., Zhao Y., Uccellini L., Tomei S., Dudley M.E. et al. (2013) CXCR3/CCR5 pathways in metastatic melanoma patients treated with adoptive therapy and interleukin-2. Br. J. Cancer 109, 2412–2423 10.1038/bjc.2013.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R. et al. (2017) IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbst R.S., Soria J.-C., Kowanetz M., Fine G.D., Hamid O., Gordon M.S. et al. (2014) Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji R.-R., Chasalow S.D., Wang L., Hamid O., Schmidt H., Cogswell J. et al. (2011) An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol. Immunother. 61, 1019–1031 10.1007/s00262-011-1172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L. et al. (2015) Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen P.-L., Roh W., Reuben A., Cooper Z.A., Spencer C.N., Prieto P.A. et al. (2016) Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 10.1158/2159-8290.CD-15-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carretero R., Wang E., Rodriguez A.I., Reinboth J., Ascierto M.L., Engle A.M. et al. (2012) Regression of melanoma metastases after immunotherapy is associated with activation of antigen presentation and interferon-mediated rejection genes. Int. J. Cancer 131, 387–395 10.1002/ijc.26471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantovani A., Romero P., Palucka A.K. and Marincola F.M. (2008) Tumour immunity: effector response to tumour and role of the microenvironment. Lancet 371, 771–783 10.1016/S0140-6736(08)60241-X [DOI] [PubMed] [Google Scholar]
- 29.Spivey T.L., Uccellini L., Ascierto M.L., Zoppoli G., De Giorgi V., Delogu L.G. et al. (2011) Gene expression profiling in acute allograft rejection: challenging the immunologic constant of rejection hypothesis. J. Transl. Med. 9, 174 10.1186/1479-5876-9-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedognetti D., Hendrickx W., Ceccarelli M., Miller L.D. and Seliger B. (2016) Disentangling the relationship between tumor genetic programs and immune responsiveness. Curr. Opin. Immunol. 39, 150–158 10.1016/j.coi.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 31.Hendrickx W., Simeone I., Anjum S., Mokrab Y., Bertucci F., Finetti P. et al. (2017) Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. OncoImmunology 6, e1253654 10.1080/2162402X.2016.1253654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q., Tomei S., Ascierto M.L., De Giorgi V., Bedognetti D., Dai C. et al. (2014) Melanoma NOS1 expression promotes dysfunctional IFN signaling. J. Clin. Invest. 124, 2147–2159 10.1172/JCI69611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murtas D., Maric D., De Giorgi V., Reinboth J., Worschech A., Fetsch P. et al. (2013) IRF-1 responsiveness to IFN-γ predicts different cancer immune phenotypes. Br. J. Cancer 109, 76–82 10.1038/bjc.2013.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang E., Bedognetti D., Tomei S. and Marincola F.M. (2013) Common pathways to tumor rejection. Ann. N. Y. Acad. Sci. 1284, 75–79 10.1111/nyas.12063 [DOI] [PubMed] [Google Scholar]
- 35.Denkert C., von Minckwitz G., Brase J.C., Sinn B.V., Gade S., Kronenwett R. et al. (2015) Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 33, 983–991 10.1200/JCO.2014.58.1967 [DOI] [PubMed] [Google Scholar]
- 36.Perez-Chanona E. and Trinchieri G. (2016) The role of microbiota in cancer therapy. Curr. Opin. Immunol. 39, 75–81 10.1016/j.coi.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang E., Uccellini L. and Marincola F.M. (2012) A genetic inference on cancer immune responsiveness. Oncoimmunology 1, 520–525 10.4161/onci.19531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spranger S., Bao R. and Gajewski T.F. (2015) Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 39.Peng W., Chen J.Q., Liu C., Malu S., Creasy C., Tetzlaff M.T. et al. (2016) Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 6, 202–216 10.1158/2159-8290.CD-15-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ascierto M.L., Makohon-Moore A., Lipson E.J., Taube J.M., McMiller T.L., Berger A.E. et al. (2017) Transcriptional mechanisms of resistance to anti–PD-1 therapy. Clin. Cancer Res. 23, 3168–3180 10.1158/1078-0432.CCR-17-0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salerno E.P., Bedognetti D., Mauldin I.S., Deacon D.H., Shea S.M., Pinczewski J. et al. (2016) Human melanomas and ovarian cancers overexpressing mechanical barrier molecule genes lack immune signatures and have increased patient mortality risk. OncoImmunology 5, e1240857 10.1080/2162402X.2016.1240857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loi S., Dushyanthen S., Beavis P.A., Salgado R., Denkert C., Savas P. et al. (2015) RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin. Cancer Res. 22, 1499–1509 10.1158/1078-0432.CCR-15-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagalla S., Chou J.W., Willingham M.C., Ruiz J., Vaughn J.P., Dubey P. et al. (2013) Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 14, R34 10.1186/gb-2013-14-4-r34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roelands J., Decock J., Boughorbel S., Rinchai D., Maccalli C., Ceccarelli M. et al. (2017) A collection of annotated and harmonized human breast cancer transcriptome datasets, including immunologic classification. F1000Res. 6, 296 10.12688/f1000research.10960.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellis M.J., Ding L., Shen D., Luo J., Suman V.J., Wallis J.W. et al. (2012) Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 10.1038/nature11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhillon A.S., Hagan S., Rath O. and Kolch W. (2007) MAP kinase signalling pathways in cancer. Oncogene 26, 3279–3290 10.1038/sj.onc.1210421 [DOI] [PubMed] [Google Scholar]
- 47.Pavese J.M., Ogden I.M., Voll E.A., Huang X., Xu L., Jovanovic B. et al. (2014) Mitogen-activated protein kinase kinase 4 (MAP2K4) promotes human prostate cancer metastasis. PLoS ONE 9, e102289 10.1371/journal.pone.0102289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham T.T., Angus S.P. and Johnson G.L. (2013) MAP3K1: genomic alterations in cancer and function in promoting cell survival or apoptosis. Genes Cancer 4, 419–426 10.1177/1947601913513950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suddason T. and Gallagher E. (2015) A RING to rule them all? Insights into the Map3k1 PHD motif provide a new mechanistic understanding into the diverse roles of Map3k1. Cell Death Differ. 22, 540–548 10.1038/cdd.2014.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuevas B.D., Abell A.N., Witowsky J.A., Yujiri T., Lassignal Johnson N., Kesavan K. et al. (2003) MEKK1 regulates calpain-dependent proteolysis of focal adhesion proteins for rear-end detachment of migrating fibroblasts. EMBO J. 22, 3346–3355 10.1093/emboj/cdg322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., Wang W., Hayashi Y., Jester J.V., Birk D.E., Gao M. et al. (2003) A role for MEK kinase 1 in TGF-β/activin-induced epithelium movement and embryonic eyelid closure. EMBO J. 22, 4443–4454 10.1093/emboj/cdg440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp L.L., Schwarz D.A., Bott C.M., Marshall C.J. and Hedrick S.M. (1997) The influence of the MAPK pathway on T cell lineage commitment. Immunity 7, 609–618 10.1016/S1074-7613(00)80382-9 [DOI] [PubMed] [Google Scholar]
- 53.Yamashita M., Kimura M., Kubo M., Shimizu C., Tada T., Perlmutter R.M. et al. (1999) T cell antigen receptor-mediated activation of the Ras/mitogen-activated protein kinase pathway controls interleukin 4 receptor function and type-2 helper T cell differentiation. Proc. Natl Acad. Sci. U.S.A. 96, 1024–1029 10.1073/pnas.96.3.1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arthur J.S.C. and Ley S.C. (2013) Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 13, 679–692 10.1038/nri3495 [DOI] [PubMed] [Google Scholar]
- 55.D'Souza W.N., Chang C.-F., Fischer A.M., Li M. and Hedrick S.M. (2008) The Erk2 MAPK regulates CD8 T cell proliferation and survival. J. Immunol. 181, 7617–7629 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dumitru C.D., Ceci J.D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J.-H. et al. (2000) TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103, 1071–1083 10.1016/S0092-8674(00)00210-5 [DOI] [PubMed] [Google Scholar]
- 57.Kaiser F., Cook D., Papoutsopoulou S., Rajsbaum R., Wu X., Yang H.-T. et al. (2009) TPL-2 negatively regulates interferon-β production in macrophages and myeloid dendritic cells. J. Exp. Med. 206, 1863–1871 10.1084/jem.20091059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mielke L.A., Elkins K.L., Wei L., Starr R., Tsichlis P.N., O'Shea J.J. et al. (2009) Tumor progression locus 2 (Map3k8) is critical for host defense against Listeria monocytogenes and IL-1β production. J. Immunol. 183, 7984–7993 10.4049/jimmunol.0901336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koul H.K., Pal M. and Koul S. (2013) Role of p38 MAP kinase signal transduction in solid tumors. Genes Cancer 4, 342–359 10.1177/1947601913507951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu H.-T., Yang D.D., Wysk M., Gatti E., Mellman I., Davis R.J. et al. (1999) Defective IL-12 production in mitogen-activated protein (MAP) kinase kinase 3-(Mkk3)-deficient mice. EMBO J. 18, 1845–1857 10.1093/emboj/18.7.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rincón M., Enslen H., Raingeaud J., Recht M., Zapton T., Su M.S. et al. (1998) Interferon-gamma expression by Th1 effector T cells mediated by the p38 MAP kinase signaling pathway. EMBO J. 17, 2817–2829 10.1093/emboj/17.10.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S. and Kaplan M.H. (2000) The p38 mitogen-activated protein kinase is required for IL-12-induced IFN-gamma expression. J. Immunol. 165, 1374–1380 PMID: [DOI] [PubMed] [Google Scholar]
- 63.Liu Y., Shepherd E.G. and Nelin L.D. (2007) MAPK phosphatases — regulating the immune response. Nat. Rev. Immunol. 7, 202–212 10.1038/nri2035 [DOI] [PubMed] [Google Scholar]
- 64.Merritt C., Enslen H., Diehl N., Conze D., Davis R.J. and Rincón M. (2000) Activation of p38 mitogen-activated protein kinase in vivo selectively induces apoptosis of CD8+ but not CD4+ T cells. Mol. Cell. Biol. 20, 936–946 10.1128/MCB.20.3.936-946.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson A.M., Mulcahy L.A., Porte J., Franks H.A., El Refaee M., Wang Q. et al. (2010) Role of mitogen-activated protein kinase and PI3K pathways in the regulation of IL-12-family cytokines in dendritic cells and the generation of T H-responses. Eur. Cytokine Netw. 21, 319–328 10.1684/ecn.2010.0219 [DOI] [PubMed] [Google Scholar]
- 66.Wagner E.F. and Nebreda Á.R. (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 9, 537–549 10.1038/nrc2694 [DOI] [PubMed] [Google Scholar]
- 67.Martinez F.O., Helming L. and Gordon S. (2009) Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27, 451–483 10.1146/annurev.immunol.021908.132532 [DOI] [PubMed] [Google Scholar]
- 68.Dong C. and Flavell R.A. (2000) Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res. 2, 179–188 PMCID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y.L. and Dong C. (2005) MAP kinases in immune responses. Cell Mol. Immunol. 2, 20–27 PMID: [PubMed] [Google Scholar]
- 70.Su B., Cheng J., Yang J. and Guo Z. (2001) MEKK2 is required for T-cell receptor signals in JNK activation and interleukin-2 gene expression. J. Biol. Chem. 276, 14784–14790 10.1074/jbc.M010134200 [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y., Blattman J.N., Kennedy N.J., Duong J., Nguyen T., Wang Y. et al. (2004) Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature 430, 793–797 10.1038/nature02764 [DOI] [PubMed] [Google Scholar]
- 72.Conze D., Krahl T., Kennedy N., Weiss L., Lumsden J., Hess P. et al. (2002) c-Jun NH2-terminal kinase (JNK)1 and JNK2 have distinct roles in CD8+ T cell activation. J. Exp. Med. 195, 811–823 10.1084/jem.20011508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sabapathy K., Kallunki T., David J.-P., Graef I., Karin M. and Wagner E.F. (2001) c-Jun NH2-terminal kinase (JNK)1 and JNK2 have similar and stage-dependent roles in regulating T cell apoptosis and proliferation. J. Exp. Med. 193, 317–328 10.1084/jem.193.3.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Labuda T., Christensen J.P., Rasmussen S., Bonnesen B., Karin M., Thomsen A.R. et al. (2006) MEK kinase 1 is a negative regulator of virus-specific CD8+ T cells. Eur. J. Immunol. 36, 2076–2084 10.1002/eji.200535163 [DOI] [PubMed] [Google Scholar]
- 75.Khiem D., Cyster J.G., Schwarz J.J. and Black B.L. (2008) A p38 MAPK-MEF2C pathway regulates B-cell proliferation. Proc. Natl Acad. Sci. 105, 17067–17072 10.1073/pnas.0804868105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J. et al. (2011) Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Colombino M., Lissia A., Capone M., De Giorgi V., Massi D., Stanganelli I. et al. (2013) Heterogeneous distribution of BRAF/NRAS mutations among Italian patients with advanced melanoma. J. Transl. Med. 11, 202 10.1186/1479-5876-11-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S. et al. (2002) Mutations of the BRAF gene in human cancer. Nature 417, 949–954 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- 79.Kumar R., Angelini S., Snellman E. and Hemminki K. (2004) BRAF mutations are common somatic events in melanocytic nevi. J. Invest. Dermatol. 122, 342–348 10.1046/j.0022-202X.2004.22225.x [DOI] [PubMed] [Google Scholar]
- 80.Pollock P.M., Harper U.L., Hansen K.S., Yudt L.M., Stark M., Robbins C.M. et al. (2003) High frequency of BRAF mutations in nevi. Nat. Genet. 33, 19–20 10.1038/ng1054 [DOI] [PubMed] [Google Scholar]
- 81.Tomei S., Wang E., Delogu L.G., Marincola F.M. and Bedognetti D. (2014) Non-BRAF-targeted therapy, immunotherapy, and combination therapy for melanoma. Expert Opin. Biol. Ther. 14, 663–686 10.1517/14712598.2014.890586 [DOI] [PubMed] [Google Scholar]
- 82.Tomei S., Bedognetti D., De Giorgi V., Sommariva M., Civini S., Reinboth J. et al. (2015) The immune-related role of BRAF in melanoma. Mol. Oncol. 9, 93–104 10.1016/j.molonc.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi H., Hugo W., Kong X., Hong A., Koya R.C., Moriceau G. et al. (2014) Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 4, 80–93 10.1158/2159-8290.CD-13-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trunzer K., Pavlick A.C., Schuchter L., Gonzalez R., McArthur G.A., Hutson T.E. et al. (2013) Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J. Clin. Oncol. 31, 1767–1774 10.1200/JCO.2012.44.7888 [DOI] [PubMed] [Google Scholar]
- 85.Van Allen E.M., Wagle N., Sucker A., Treacy D.J., Johannessen C.M., Goetz E.M. et al. (2014) The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 10.1158/2159-8290.CD-13-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oberholzer P.A., Kee D., Dziunycz P., Sucker A., Kamsukom N., Jones R. et al. (2012) RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J. Clin. Oncol. 30, 316–321 10.1200/JCO.2011.36.7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Su F., Viros A., Milagre C., Trunzer K., Bollag G., Spleiss O. et al. (2012) RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 366, 207–215 10.1056/NEJMoa1105358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Larkin J., Ascierto P.A., Dréno B., Atkinson V., Liszkay G., Maio M. et al. (2014) Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 371, 1867–1876 10.1056/NEJMoa1408868 [DOI] [PubMed] [Google Scholar]
- 89.Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J. et al. (2014) Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 371, 1877–1888 10.1056/NEJMoa1406037 [DOI] [PubMed] [Google Scholar]
- 90.Robert C., Karaszewska B., Schachter J., Rutkowski P., Mackiewicz A., Stroiakovski D. et al. (2015) Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 372, 30–39 10.1056/NEJMoa1412690 [DOI] [PubMed] [Google Scholar]
- 91.Sumimoto H., Imabayashi F., Iwata T. and Kawakami Y. (2006) The BRAF–MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J. Exp. Med. 203, 1651–1656 10.1084/jem.20051848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boni A., Cogdill A.P., Dang P., Udayakumar D., Njauw C.-N.J., Sloss C.M. et al. (2010) Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 70, 5213–5219 10.1158/0008-5472.CAN-10-0118 [DOI] [PubMed] [Google Scholar]
- 93.Liu C., Peng W., Xu C., Lou Y., Zhang M., Wargo J.A. et al. (2013) BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clin. Cancer Res. 19, 393–403 10.1158/1078-0432.CCR-12-1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frederick D.T., Piris A., Cogdill A.P., Cooper Z.A., Lezcano C., Ferrone C.R. et al. (2013) BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin. Cancer Res. 19, 1225–1231 10.1158/1078-0432.CCR-12-1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilmott J.S., Long G.V., Howle J.R., Haydu L.E., Sharma R.N., Thompson J.F. et al. (2012) Selective BRAF inhibitors induce marked T-cell infiltration into human metastatic melanoma. Clin. Cancer Res. 18, 1386–1394 10.1158/1078-0432.CCR-11-2479 [DOI] [PubMed] [Google Scholar]
- 96.Cooper Z.A., Frederick D.T., Juneja V.R., Sullivan R.J., Lawrence D.P., Piris A. et al. (2013) BRAF inhibition is associated with increased clonality in tumor-infiltrating lymphocytes. Oncoimmunology 2, e26615 10.4161/onci.26615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hooijkaas A., Gadiot J., Morrow M., Stewart R., Schumacher T. and Blank C.U. (2012) Selective BRAF inhibition decreases tumor-resident lymphocyte frequencies in a mouse model of human melanoma. Oncoimmunology 1, 609–617 10.4161/onci.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Donia M., Fagone P., Nicoletti F., Andersen R.S., Høgdall E., Straten P.t. et al. (2012) BRAF inhibition improves tumor recognition by the immune system. Oncoimmunology 1, 1476–1483 10.4161/onci.21940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sapkota B., Hill C.E. and Pollack B.P. (2013) Vemurafenib enhances MHC induction in BRAFV600E homozygous melanoma cells. Oncoimmunology 2, e22890 10.4161/onci.22890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Knight D.A., Ngiow S.F., Li M., Parmenter T., Mok S., Cass A. et al. (2013) Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. J. Clin. Invest. 123, 1371–1381 10.1172/JCI66236 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Koya R.C., Mok S., Otte N., Blacketor K.J., Comin-Anduix B., Tumeh P.C. et al. (2012) BRAF inhibitor vemurafenib improves the antitumor activity of adoptive cell immunotherapy. Cancer Res. 72, 3928–3937 10.1158/0008-5472.CAN-11-2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ribas A., Hodi F.S., Callahan M., Konto C. and Wolchok J. (2013) Hepatotoxicity with combination of vemurafenib and ipilimumab. N. Engl. J. Med. 368, 1365–1366 10.1056/NEJMc1302338 [DOI] [PubMed] [Google Scholar]
- 103.Angell T.E., Lechner M.G., Jang J.K., LoPresti J.S. and Epstein A.L. (2014) MHC class I loss is a frequent mechanism of immune escape in papillary thyroid cancer that is reversed by interferon and selumetinib treatment in vitro. Clin. Cancer Res. 20, 6034–6044 10.1158/1078-0432.CCR-14-0879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brea E.J., Oh C.Y., Manchado E., Budhu S., Gejman R.S., Mo G. et al. (2016) Kinase regulation of human MHC class I molecule expression on cancer cells. Cancer Immunol. Res. 4, 936–947 10.1158/2326-6066.CIR-16-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mimura K., Shiraishi K., Mueller A., Izawa S., Kua L.-F., So J. et al. (2013) The MAPK pathway is a predominant regulator of HLA-A expression in esophageal and gastric cancer. J. Immunol. 191, 6261–6272 10.4049/jimmunol.1301597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Poon E., Mullins S., Watkins A., Williams G.S., Koopmann J.-O., Di Genova G. et al. (2017) The MEK inhibitor selumetinib complements CTLA-4 blockade by reprogramming the tumor immune microenvironment. J. Immunother. Cancer 5, 63 10.1186/s40425-017-0268-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.DeSilva D.R., Jones E.A., Favata M.F., Jaffee B.D., Magolda R.L., Trzaskos J.M. et al. (1998) Inhibition of mitogen-activated protein kinase kinase blocks T cell proliferation but does not induce or prevent anergy. J. Immunol. 160, 4175–4181 PMID: [PubMed] [Google Scholar]
- 108.Vella L.J., Pasam A., Dimopoulos N., Andrews M., Knights A., Puaux A.-L. et al. (2014) MEK inhibition, alone or in combination with BRAF inhibition, affects multiple functions of isolated normal human lymphocytes and dendritic cells. Cancer Immunol. Res. 2, 351–360 10.1158/2326-6066.CIR-13-0181 [DOI] [PubMed] [Google Scholar]
- 109.Allegrezza M.J., Rutkowski M.R., Stephen T.L., Svoronos N., Tesone A.J., Perales-Puchalt A. et al. (2016) IL15 agonists overcome the immunosuppressive effects of MEK inhibitors. Cancer Res. 76, 2561–2572 10.1158/0008-5472.CAN-15-2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Allegrezza M.J., Rutkowski M.R., Stephen T.L., Svoronos N., Perales-Puchalt A., Nguyen J.M. et al. (2016) Trametinib drives T-cell–dependent control of KRAS-mutated tumors by inhibiting pathological myelopoiesis. Cancer Res. 76, 6253–6265 10.1158/0008-5472.CAN-16-1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu L., Mayes P.A., Eastman S., Shi H., Yadavilli S., Zhang T. et al. (2015) The BRAF and MEK inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin. Cancer Res. 21, 1639–1651 10.1158/1078-0432.CCR-14-2339 [DOI] [PubMed] [Google Scholar]
- 112.Ebert P.J.R., Cheung J., Yang Y., McNamara E., Hong R., Moskalenko M. et al. (2016) MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity 44, 609–621 10.1016/j.immuni.2016.01.024 [DOI] [PubMed] [Google Scholar]
- 113.Hu-Lieskovan S., Robert L., Homet Moreno B. and Ribas A. (2014) Combining targeted therapy with immunotherapy in BRAF-mutant melanoma: promise and challenges. J. Clin. Oncol. 32, 2248–2254 10.1200/JCO.2013.52.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hu-Lieskovan S., Mok S., Moreno B.H., Tsoi J., Robert L., Goedert L. et al. (2015) Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAFV600E melanoma. Sci. Transl Med. 7, 279ra41 10.1126/scitranslmed.aaa4691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dushyanthen S., Teo Z.L., Caramia F., Savas P., Mintoff C.P., Virassamy B. et al. (2017) Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer. Nat. Commun. 8, 606 10.1038/s41467-017-00728-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Homet Moreno B., Mok S., Comin-Anduix B., Hu-Lieskovan S. and Ribas A. (2016) Combined treatment with dabrafenib and trametinib with immune-stimulating antibodies for BRAF mutant melanoma. Oncoimmunology 5, e1052212 10.1080/2162402X.2015.1052212 [DOI] [PMC free article] [PubMed] [Google Scholar]