Abstract
Objective:
The aim of the study was to explore specific microRNAs (miRs) in rectal cancer that would predict response to radiation and identify target pathways that may be exploited for neoadjuvant therapies.
Summary Background Data:
Chemoradiotherapy (CRT) response is a predictor of survival in rectal cancer. Studies have demonstrated changes in RNA expression correlate with chemoradiation sensitivity across cancers.
Methods:
Forty-five rectal cancer patients, partial responders (PR = 18), nonresponders (NR = 13), and complete responders (CR = 14) to CRT, as defined by a tumor regression score, were examined. miRs differentially expressed, using NanoString microArray profiling, were validated with qPCR. We quantified 1 miR and its downstream targets in patient samples. Chemosensitivity was measured in HCT-116, a human colorectal carcinoma cell line, using inhibitors of SHP2 and RAF.
Results:
miR-451a, 502–5p, 223–3p, and 1246werethe most upregulated miRs (>1.5-fold change) in a NanoString profiling miR panel. qPCR revealed a decrease in expression of miR-451a in NRs. EMSY and CAB39, both downstream targets of miR-451a and involved in carcinogenesis (shown in TCGA) were increased in NRs (qPCR). Both targets are associated with worse survival in colorectal cancer. Inhibition of miR-451a in HCT-116 cells significantly decreased cell proliferation with treatment of SHP2 and RAF inhibitors. Conclusions: An integrated analysis of rectal cancer miRs may yield biomarkers of radioresistance and offer treatment targets for resensitization.
Keywords: miRNA, radiation therapy, rectal cancer
In rectal cancer, the response to preoperative chemoradiotherapy (CRT) is an important predictor of local tumor control and patient survival.1 Advancements in rectal cancer therapy have established the role of radiation therapy (RT) as an optimal treatment in the neoadjuvant setting.2–6,7 Concurrent implementation of chemotherapy further improves rate of local recurrence.8 Patients who have received neoadjuvant therapy and obtained a complete clinical response may be followed by a “watchful waiting” nonoperative strategy.9 This trial is based on approximately 30% of patients obtaining a complete pathological response following CRT. 8,10 In this exciting shift in rectal cancer surgery paradigm, no validated biomarkers are available to predict an individual’s response to CRT. More significantly, neoadjuvant therapies that may reset radiation sensitivity have not been identified.
Recent studies have examined microRNAs (miRs) to better understand the molecular underpinnings of the pathogenesis, advancement, and outcomes associated with colorectal cancer (CRC).11 miRs are small noncoding RNA molecules that bind the messenger RNA, mostly at the 3’-untranslated region (3’-UTR), and either decrease the expression or stop translation of mRNA. miRs are significant in multiple cancers, including colorectal cancer.12,13,14 miRs have been associated with chemoradiation resistance in CRC,15 esophageal carcinoma,16,17 small cell lung carcinoma,18 and gliomas.18 No study has identified a miR profile in rectal cancer to elicit predictors of CRT response.
We therefore sought to identify a miR signature in samples of rectal cancer patients stratified by radiation response. We hypothesized specific miRs would be differentially expressed between nonand partial responders. We aimed to validate this profile in a subset of non-, partial, and complete responders. We also identified downstream genes of miRs upregulated in partial responders, and exploited these target genes with potential neoadjuvant therapies.
METHODS
Patient Selection
Patients were identified with a rectal cancer diagnosis from the years 2000 to 2016 in the Oregon Colorectal Cancer Registry (OCCR).19 The registry is maintained by Salem Hospital and the Oregon Health & Science University, and catalogs patients treated for colorectal cancer at both institutions. Investigational Research Board (IRB) approval was granted for the study, and a written informed consent was obtained from all participating patients. Pretreatment formalin-fixed paraffin-embedded (FFPE) specimens, posttreatment surgical FFPE specimens, and plasma are stored onsite. Patients were categorized as either non-, partial, or complete pathological responders, based on a pathological tumor regression score. Nonresponders had >50% tumor, partial responders had <50%, and complete responders were identified with either no or small singular cancer cells remaining on pathologic review.20
Tissue Collection
Pre- and posttreatment specimens were prepared on 5-μm-thick FFPE slides, which were independently assessed by 2 pathologists. Pretreatment specimens were obtained at time of diagnosis. Posttreatment specimens were obtained, following standard CRTand surgical resection. Pretreatment specimens were used for miR profiling, and posttreatment surgical FFPE specimens were used for confirmatory qPCR analysis. In patients with complete response, the tumor bed was identified by scar tissue, which was used for analysis. Macrodissection of slides was used. Slides containing >20% necrosis, or <50% tumor, were not used for study. Plasma samples were collected at the time of surgery and stored at −80°C.
RNA Isolation
miRs were isolated from FFPE patient slides, using an RNeasy FFPE Kit (Qiagen catalog # 73504). miRs were isolated from patient plasma, using mirRNeasy Plasma/Serum Kit (Qiagen cat#217184). cDNA synthesis was performed, using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Cat#4368814). For miR cDNA synthesis, specific miR primers were used, following the Taqman Small RNA Assay Protocol. qPCR was performed with the Taqman Gene Expression Assays (Applied Biosystems), using the Applied Biosystems ViiA7 qRT-PCR instrument. miR values were normalized to qPCR reactions performed in quadruplicate. miR values were normalized to the Lifetech probes and primers (housekeeping control RNU48 or GAPDH, Life Technologies), using the delta-delta CT method.21
MicroRNA Profiling
miRs isolated from 12 pretreatment FFPE specimens of 6 non-and6 partial responders were compared using the NanoString Technology platform;3 μL of RNA was added to mi RNA tag ligation reaction with reporter and capture probes. Raw counts for each assay were collected using NanoString’s data analysis application nSolver. Normalization of the data was carried out using the NanoString nSolver analysis application, and each assay was normalized based on the mean counts of the 100 most-abundant miRNA. A total of 798 human miRs were profiled. Upregulated and downregulated miRs with differences >1.5-fold between groups were identified. Among these 14 candidate miRs, those significantly altered in CRC were the focus of further investigation.
Downstream Target Analysis
Using the Cancer Genome Atlas (TCGA) data, downstream targets of miR-451a in CRC tumors were assessed.22 SurvExpress, an online biomarker validation for cancer gene expression tool, was used to better understand the target expression profiles.23
In Vitro Models
HCT-116 cells (American Type Culture Collection) were cultured in McCoy’s media supplemented with 10% FBS and antibiotics. Cells were tested and found negative for mycoplasma contamination before use in the assays described. Cells were reverse-transfected with miR-451a-5p mimics or inhibitors with their respective controls (purchased from Life Technologies), using Lipofectamine RNAiMAX (Invitrogen) per manufacturer’s instructions. We report highly efficient transfection (>90%) of mimics and inhibitors in cell culture, as measured by fluorescent miR control. Cells were irradiated in a Shepherd137 cesium irradiator at a rate of 1.34 cGy min. HCT-116 cells were initially transfected in a 6-well plate. Cells were then transferred to a 96-well plate 16hours post-transfection (2000cells/well). In the chemosensitivity model, HCT-116 cells were treated with SHP2 inhibitor (Cayman Chemical, cat#20000) (3.5μM), and RAF inhibitor (selective paradox breaker; SelleckChem, cat#S7964) (20μM) at 24hours post transfection. Cell proliferation was analyzed at 48hours postirradiation, using Cell Titer-Glo, per the manufacturer’s instructions.
Statistical Analysis
Clinicopathological data were collected, and response groups were compared, using χ2 tests and the Freeman–Halton extension of Fisher exact test. NanoString profiling was performed by a NanoString proof of concept study. Deidentified specimens were sent to NanoString, and data were returned normalized per standard protocol. Differences in gene expression determined by qPCR among pathological non-, partial, and complete responders groups were evaluated with Mann-Whitney tests. Correlations were calculated between miR-451a and downstream genes stratified by response to CRT. Differences in serum treatment groups were evaluated with exact F tests from a multivariate ANOVA. A significant difference was declared if the P value was ≤0.05. Marginal significance was declared if the P value was >0.05, but ≤0.10. All analyses were completed with JMP 13 and SAS 9.4.
RESULTS
We examined 45 patients who underwent treatment for rectal cancer from 2000 to 2016 captured in the OCCR with available tissue for exploratory analysis. There were a total of 13 nonresponders, 18 partial responders, and 14 complete responders. For 3 patients, demographic data were not available. The level of response was significantly different across pathological T-stage and marginally significant for M-stage patients (Table 1).
TABLE 1.
Study population demographics
| Variables | Total % (n = 45) | Response |
p | ||
|---|---|---|---|---|---|
| None (n = 13) | Partial (n = 18) | Complete (n = 14) | |||
| Sex | 0.518* | ||||
| Male | 23 (51%) | 5 | 11 | 7 | |
| Female | 19 (42%) | 6 | 7 | 6 | |
| Missing | 3 (7%) | ||||
| Age | 0.8525† | ||||
| <50 | 16 (36%) | 5 | 7 | 4 | |
| >50 | 26 (58%) | 6 | 11 | 9 | |
| Missing | 3 (7%) | ||||
| Tumor difference | 0.7553† | ||||
| Well | 3 (7%) | 2 | 1 | 0 | |
| Moderate | 26 (58%) | 7 | 10 | 9 | |
| Poor | 5 (11%) | 2 | 2 | 1 | |
| Dysplasia | 1 (2%) | 0 | 1 | 0 | |
| Missing | 10 (22%) | ||||
| pT stage | <0.0001† | ||||
| T0 | 13 (29%) | 0 | 0 | 13 | |
| T1 | 4 (9%) | 0 | 4 | 0 | |
| T2 | 6 (13%) | 2 | 4 | 0 | |
| T3 | 16 (36%) | 8 | 8 | 0 | |
| T4 | 3 (7%) | 1 | 2 | 0 | |
| Missing | 3 (7%) | ||||
| pN stage | 0.1031† | ||||
| N0 | 28 (62%) | 5 | 11 | 12 | |
| N1 | 8 (18%) | 4 | 3 | 1 | |
| N2 | 6 (13% | 2 | 4 | 0 | |
| Missing | 3 (7%) | ||||
| pM stage | 0.0550† | ||||
| M0 | 38 (84%) | 8 | 17 | 13 | |
| M1 | 4 (9%) | 3 | 1 | 0 | |
| Missing | 3 (7%) | ||||
| Alive | 34 (76%) | 9 | 13 | 12 | 0.3761† |
| Not alive | 8 (18%) | 2 | 5 | 1 | |
| Missing | 3 (7%) | ||||
χ2 test.
Fisher–Freeman–Halton test (extension of Fisher exact test for a 3xk table).
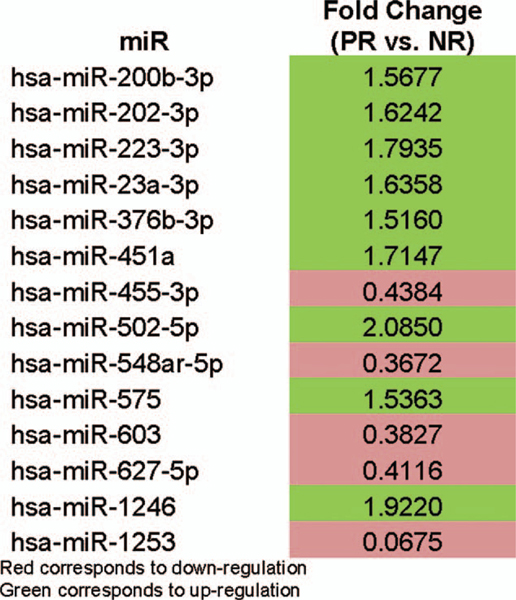
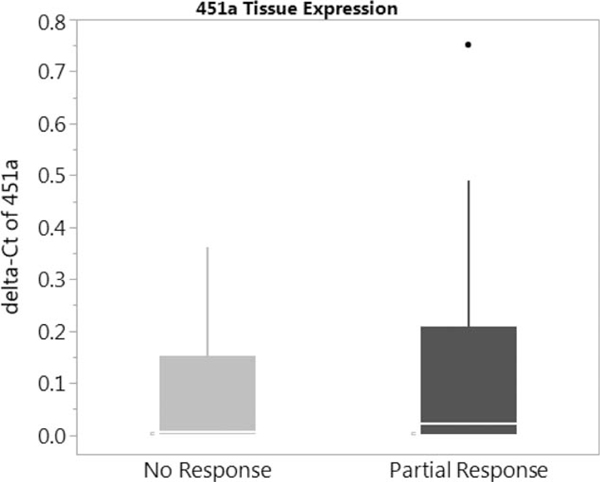
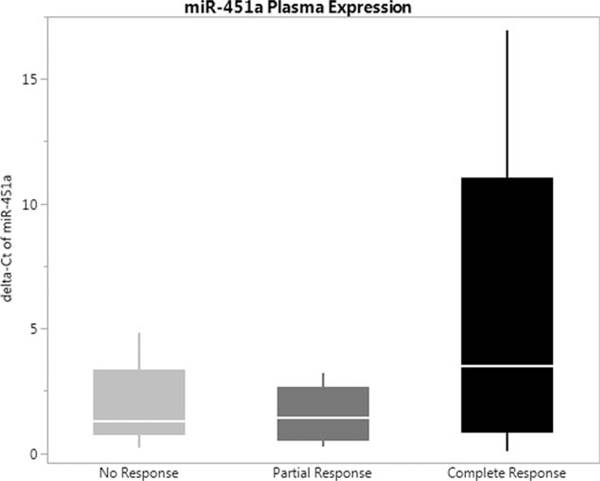
Using NanoString profiling, we identified 4 upregulated miRs, miR-451a, 502–5p, 223–3p, and 1246 (>1.5-fold change) (Fig. 1). Prior data demonstrated significant influence of miR-451a in colorectal cancer in vitro and in vivo models24 (see Supplementary Figures 1, 2, 3, http://links.lww.com/SLA/B286) miR-451a was validated in our cohort out of the four upregulated mIRs shown above. Exploration of posttreatment tissue was completed, using qPCR. There was no significant difference between non- or partial responders; there was, however, a slight increase in expression of miR-451a in the partial responders (Fig. 2). Patient-matched posttreatment plasma was also assessed for miR-451a, and expression was higher in the complete responders versus non- and partial responders (Fig. 3).
FIGURE 1.
NanoString miR panel generated from 6 non- and 6 partial responders pretreatment.
FIGURE 2.
miR-451a expression profile in posttreatment tissue of non- and partial responders.
FIGURE 3.
rmR-451a plasma concentration profile in posttreatment non-, partial, and complete responders.
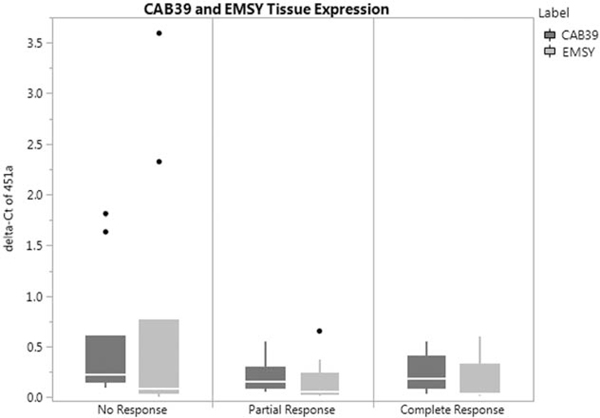
Our prior in vitro and in vivo models demonstrated miR-451a directly binds and downregulates CAB39 and EMSY24 (see Supplementary Figure 4, http://links.lww.com/SLA/B286). These 2 proteins are expressed in 14% and 6% of CRC, which was associated with worse survival.22 The expression profile of CAB39 and EMSYin our patient cohort demonstrated a nonsignificant increase in the non-responders when compared with partial and complete responders (Fig. 4). Results of tissue and plasma expression of miR-451a and genes are summarized in Table 2.
FIGURE 4.
Downstream target protein expression profile in posttreatment non-, partial, and complete responders.
TABLE 2.
PCR quantification of posttreatment tissue and plasma miR-451a, CAB39, and EMSY
| miR | Total % (n = 45) | Response |
p | ||
|---|---|---|---|---|---|
| None (n = 13) | Partial (n = 18) | Complete (n = 14) | |||
| miR-451a | |||||
| Tissue (mean delta-CT, SD) | 43 | 0.0747 (0.1163) | 0.1275 (.2122) | 0.1748 (.3818) | 0.2111* |
| Plasma (mean delta-CT, SD) CAB39 | 17 | 1.8878 (1.7501) | 1.5567 (1.1662) | 5.6908 (6.4993) | 0.6437* |
| Tissue (mean delta-CT, SD) EMSY | 35 | 0.5109 (0.6180) | 0.2082 (.1524) | 0.2360 (.1836) | 0.4820* |
| Tissue (mean delta-CT, SD) | 37 | 0.6875 (1.185) | 0.1326 (.1804) | 0.1541 (.2134) | 0.5275* |
Mann-Whitney test.
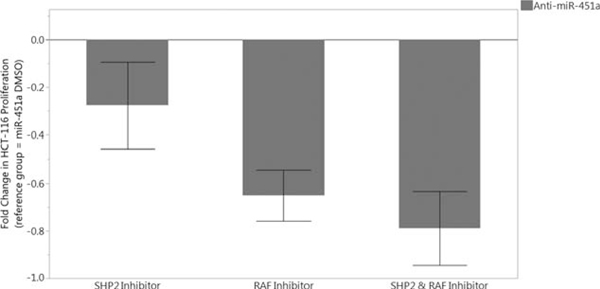
CAB39 and EMSY proteins are correlated with poorer overall survival.22 Patients with increased CAB39 and EMSY are shown to have upregulated in a number of signaling pathways, including RAF kinases and SHP2 enzymes.22 We have demonstrated miR-451a directly binds and downregulates CAB39 and EMSY24 (see Supplementary Figure 4, http://links.lww.com/SLA/B286). We inhibited RAF and SHP2 in HCT-116 cells reverse-transfected with anti-miR451a. We found cells treated with SHP2, and RAF inhibitors demonstrated a significant decrease in cell proliferation. Specifically, the combined treatment of 20 μM RAF inhibitor and 3.5 μM SHP2 inhibitor was the most effective at preventing cell proliferation (P < 0.0001) (Fig. 5).
FIGURE 5.
Resetting radiosensitivity in HCT-116 cell lines with treatment of SHP2 and RAF inhibitors.
DISCUSSION
Our findings suggest miR-451a is a potential predictor of radiosensitivity in rectal cancer. More importantly, we have identified downstream genes, CAB39 and EMSY, controlled by miR451a, that may be targetable by SHP2 and RAF inhibitors. Rectal cancer patients with low miR-451a expression, who will potentially have a limited response to CRT, may therefore benefit from the addition of SHP2 and RAF inhibitors. Modulation of CRT response willfacilitatelessinvasivesurgicaltreatment,suchaslocalexcision, or a decision to omit surgery altogether.1,25,26 Rectal cancer surgery is not a benign procedure, and has complications including, but not limited to, fecal incontinence, permanent stoma, and risk of anastomotic leak. The addition of radiation-sensitizing drugs to neoadjuvant radiation therapy would, therefore, benefit rectal cancer patients.
Multiple studies have validated miR-451a associations with dysregulation of human malignancies, including lung, gastric, breast, gliomas, and leukemia.27 Studies have reported miR-451a involvement in colorectal cancer tumorigenesis. In patient tissue samples, miR-451a expression was associated with decreased cancer cell proliferation and increased sensitivity to radiation.14 In that study, authors proposed the mechanism of action of miR-451a was on macrophage migration inhibitory factor (MIF), a proinflammatory cytokine involved in innate immunity. Others corroborated these findings by demonstrating that expression of miR-451a in colono-spheres decreased tumorigencity via a similar pathway.15 Therefore, miR-451a may act as a tumor suppressor or radiosensitizer, which is supported by our findings. We have shown miR-451a expression improves radiation treatment response within the in vivo and in vitro models.24 We are the first to report on its predictive value in the setting of radiation sensitivity in rectal cancer. We aimed to reproduce these findings in the plasma of our cohort, as prior reports have identified a liquid biomarker to detect colorectal adenomas.28 Our data suggest miR-451a plasma expression may also be a predictor, and is easier and more cost-effective than tissue analysis. If this finding is not reproduced in prospective studies, however, tissue obtained at the time of diagnostic biopsy can still be used to quantify miR-451a expression.
To understand the action of miR-451a at a proteomic level, we interrogated proteins involved in CRC and tissue radiation sensitivity. Using a bioinformatics approach, we explored miRWalk, an atlas of miR targets.29 EMSY and CAB39 were 2 proteins involved in CRC and radiation sensitivity. CAB39, also known as calcium-binding protein 39, is involved in STK11 activity and localization, thereby influencing the PI3K/AKT signaling pathway, which is involved in cell survival.30 It has been correlated to miR-451a in human glioma.30 EMSY is also closely involved with cancer development. In vitro models demonstrate its inhibitor effects on BRCA2, which is lost when mutations in BRCA2 are present.31 Within the TCGA, CRC patients with CAB39 and EMSY mutations had a worse survival, albeit they represent a small minority of CRC patients.22 This was similarly reflected in SurvExpress.23 CAB39 and EMSY are therefore both likely negative prognosticators in rectal cancer. This information was reflected in our patient population, specifically in individuals who were nonresponders. There was a higher expression of CAB39 and EMSY miRs in the nonresponders when compared with the partial and complete responders. This was supported in the tissue array data, where CAB39 and EMSY were upregulated in CRC tissue, but not normal tissue specimens.
Based on our findings, CAB39 and EMSY, negative prognosticators of rectal cancer, are likely associated with the upregulation of SHP2 and RAF. SHP2 and RAF are targetable enzymes involved in tumorigenesis pathways. SHP2 is a nonreceptor protein tyrosine phosphatase, encoded by PTPN11.32 It is intricately involved in the RAS/ERK signaling pathway.33,34 Activating mutations of SHP2 are found in multiple cancer subtypes, including lung and breast cancer.35,36 SHP2 inhibitors suppress tumor cell growth by targeting enzymes upstream from BRAF, which acts to sensitive concurrent treatment with vemurafenib.37,38 Although this compound is still in study, there exists a selective and orally bioavailable SHP2 inhibitor that may be used in a select patient cohort.39,40 BRAF is a wellknown enzyme in the RAF family, mutated in CRC MSI-H patients. It is currently the focus of numerous studies using FDA-approved BRAF inhibitors. BRAF mutations result in downstream activation of MEK and ERK, which stimulates growth-promoting gene products.41,42 Unfortunately, CRC studies of RAF inhibitors reported incidences of tumor resistance, likely secondary to the redundant networks of the MAP kinase pathway.42 Recent work on a pan-RAF inhibitors has demonstrated improved antitumor activity in CRC BRAF-mutated human models, providing additional hope for this therapeutic intervention.43 Our data suggest that in individuals with decreased or inhibited miR-451a expression, treatment with SHP2 and RAF inhibitors would decrease cell survival. Therefore, in individuals with low miR-451a who are likely poor RT responders, the addition of SHP2 or RAF inhibitors to the neoadjuvant treatment profile may reset the radiosensitivity of the tumor and improve RT response.
Our research is limited. Owing to the retrospective nature of this study, we were not able to obtain all pre- and posttreatment specimens for analysis. Pretreatment tissue would have ideally been used for validation of the miR profile. In addition, our miR profile is not complete, as the complete responders would have provided more insightful data. Although we attempted profiling compete responders, sample hybridization and quality control were unsatisfactory. We examined 1 miR in our generated profile, based on our biostatistical exploration of the TCGA. We plan to explore additional miRs in our profile. The majority of our experiments were completed using colorectal cell lines, particularly HCT-116. We attempted replication of gain and loss experiments in rectal cancer lines (SW837 and SW1463), which both failed to proliferate after transfection. We are currently expanding our research to include patient tissue-derived tumor cell and organoid cultures.
Future work will address these issues with prospective tissue and plasma sample collection to correlate miR-451a, CAB39, and EMSY in preoperative tissue biopsy, postoperative tissue samples, and normal colon. Multi-institutional collaborations are being established to increase our tissue and plasma repository. We are the first to demonstrate a trend of miR-451a expression in tissue and plasma as a predictor of radiation sensitivity. We have proposed mechanisms by which radiation response can be reset. Additional studies will clarify the role of SHP2 and RAF inhibitors on CAB39 and EMSY pathways. Clarifying a subset of patients with sensitivities to SHP2 and RAF inhibitors in a phase I trial should be anticipated, with eventual clinical application to improve CRT response in rectal cancer.
Supplementary Material
DISCUSSANTS.
Dr. Susan Galandiuk (Louisville, KY):
Dr. Tsikitis and colleagues have presented an interesting study that evaluates microRNA expression in both tissue and plasma of rectal cancer patients who have undergone chemoradiation and attained either a complete, partial, or no response following this treatment. This is an important field of investigation because of the increasing utilization of this watch-and-wait approach to treating patients with apparent complete response with observation alone without surgery. It is a dilemma for both the patient and the surgeon not knowing for certain if a complete response has occurred.
Similarly, in those patients with a partial response, it would be very desirable to have an intervention that would enable them to achieve a more complete response. Many studies are aimed at identification of biomarkers that would accurately identify individuals that are likely undergo a complete response or, as in this present article, identify ways of modifying this response. I think we will see many more such studies in the future.
I have several questions for the authors.
In the manuscript, the authors describe the Nanostring technology that they have used for microRNA screening. This was done on tissue samples; however, they do not accurately describe how this tissue was obtained. Was this done using laser capture microdissection or was this done using macro dissection?
During the screening process for using the 798 human microRNA, the authors generated a panel of 14 microRNA by comparing the partial responders to the nonresponders. Why were the complete responders not compared with the nonresponders?
Pretretreatment specimens were used for screening; however, posttreatment surgical FFPE specimens were used for confirmatory qPCR analysis. Why was the confirmatory analysis not performed on the same pretreatment specimens?
Among the differentially expressed microRNA, in the manuscript you describe that 9 of 14 were upregulated >1.5 fold when comparing partial to nonresponders, whereas 5 of 14 were downregulated. Why did you only focus on the upregulated microRNA? For example, one of the miRNA you describe, microRNA 1253, was downregulated almost 15-fold, whereas miR-451 that you focused on was only upregulated 1.7-fold. Also, when you discuss these microRNA, there is no mention as to what the significance of the fold change was.
Along these same lines, why did you only focus on 1 of the 9 upregulated microRNA? Some of the other upregulated microRNA had much higher fold changes than miR-451 that you described.
My last question focuses on the use of the HCT-116 cell line used for the in vitro model. This is a hereditary nonpolyposis colon cancer cell line that is microsatellite instable. Why did not you use a sporadic rectal cancer cell line?
Thank you for the opportunity to discuss this article.
Response from Dr. Vassiliki L. Tsikitis (Portland, OR):
Thank you, Dr. Galandiuk, for your excellent comments and discussion. To address your first question on how we obtained the tissue, we did microdissection. Our quality indicator was not to include slides that had >20% necrosis, we wanted to work on slides that have high percentage of tumor.
When it comes to the question of between complete, partial, and nonresponders, we did do this comparison analysis with the complete responders. I was very skeptical on analyzing the tissues of complete responders because we’re dealing with a scar/necrotic tissue and not certain on how to interpret those results. We did see however overexpression of the miR-451a. We were reassured on the interpretation of our results when we observed the over expression of the miR-451a in the plasma of complete responders as we demonstrated in our presentation.
2When it comes to your question on the pretreatment versus the posttreatment tissue validation analysis, we used the Nanostring technology and obtained our microarray signature from the pretreatment tissue; we used the the posttreatment tissue for the qPCR; it was a limitation of our study. We had pretreatment biopsies, and not enough tissue to continue with validation therefore we used surgical posttreatment tissue of the same patients to validate our microarray.
Excellent point about the miR signature, our goal is to analyze all those miRs. We concentrated on 451 because we have prior in vitro and in vivo analysis on 451 that has been extremely interesting and encouraging for us to proceed with functional gains and losses of that miR. And Kate Kelley, my wonderful resident who did an outstanding job during her 1 year in the laboratory, has concentrated on that 1 miR to study and understand its functional gains and losses. We will proceed with exploring the other miRs in our microarray signature.
Right on your point with the HCT-116 cell line, you are absolutely right. We did do a lot of our studies also in SW-480 and 620. We purchased a rectal cell line; however, it did not proliferate in cell culture; currently we have a couple of our patients’ tissues that we have grown cell cultures from and we are working on validating our presented study findings on those cultures.
I think that wraps up your questions. Thank you so very much.
Dr. David Allison (Toledo, OH):
This is a very nice study. I have 2 questions: first, were the microRNA patterns associated with sensitivity or resistance randomly acquired during the somatic mutations or other genetic changes associated with transformation and progression? Alternatively, were these microRNA patterns reflecting the genetic backgrounds of the patient’s normal cells? If the latter is true, is it possible that some patients’ tumors may be intrinsically sensitive to radiation and other patients’ tumors intrinsically resistant to radiation based on differences in the normal genomes? Second, very complex differences have been found between the gene expression patterns of normal and tumor cells growing in vivo and in vitro. Did you find similar and stable microRNA patterns for cells grown in vivo and in vitro, or did these patterns also vary with the growth stated? The answer to this question might give some insights into the basic mechanisms governing gene expression. I enjoyed your presentation.
Response from Dr. Vassiliki L. Tsikitis (Portland, OR):
We did see overexpression of 451 in vivo. We followed the in vivo results with xenografts study (submitted) and then proceeded with human tissue. Now, if we had explored all the miRs in our signature very likely some of the miRs may not have been validated with qPCR. We concentrated on the miR-451a because we did find promising results.
Dr. Allesandro Fichera (Seattle, WA):
I wanted to follow up on what Dr. Galandiuk just asked. We are looking at complete responders for nonoperative management in rectal cancer. We are also looking at eliminating radiation altogether and replacing it with definitive chemotherapy in the neoadjuvant setting. Have you looked at the mRNA signature in terms of chemosensitivity?
Response from Dr. Vassiliki L. Tsikitis (Portland, OR):
We do have samples of patients that have shown significant response just to neoadjuvant chemotherapy. We have not had the chance to do that yet on those samples, but definitely something to investigate.
Dr. David Shibata (Memphis, TN):
Congratulations on a great talk. It is very tough to work with the TCGA database. I congratulate you on doing that. It is quite a feat.
I am going to recapitulate a little bit what Dr. Galandiuk had mentioned about the use of posttreatment samples. I caution you in using some of that, particularly the tissues, because I think you have to look at your mouse models as well because you can have clonal selection as tumors respond to treatment, right? So the expression you see posttreatment may not reflect what you canuse as a biomarker or a treatment target pretreatment. I think that is very important.
The other question along those lines that I have for you relates to the fact that you saw a microRNA 451a increase in the serum of posttreatment patients. Now, you are presuming that this is coming from the tumor but you have complete responders. If you are measuring the expression levels in the tumor, but then you are saying that may be coming from another source, where is that source?
Response from Dr. Vassiliki L. Tsikitis (Portland, OR):
Very good point. On your first question about posttreatment, you are absolutely correct. I mean, to be able to say that I have a predictive marker to response to therapy, I need to have pretreatment tissue pretreatment plasma and that is what we are trying now to collect and study.
With your second point, you are correct. I understand this off the press and not presented today but we have examined plasma levels of miR-451a of normal non cancer individuals, and compared it with the cancer patients, ones that responded, partially responded, and not responded, and we see higher expression of the miR on the normal plasma. So it may be 451a is not just so much that it is modified by radiation, but maybe some of us individuals may have a higher of this level of the antioncomir that leads the tissue to greater response to radiation therapy. Again if that is true, then this miR could be a potential predictor to radiation treatment.
ACKNOWLEDGMENTS
The authors thank Mary Kwatkosky-Lawlor for her assistance with the review process and the preparation of the bibliography. We appreciate the assistance of Anand Lab members, Clayton Hudson, and Namita Chatterjee, PhD.
Funding: Funding was supported by an institutional Medical Research Foundation Grant of Oregon.
Footnotes
Disclosure: The authors report no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.annalsofsurgery.com).
REFERENCES
- 1.Maas M, Nelemans PJ, Valentini V, et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11:835–844. [DOI] [PubMed] [Google Scholar]
- 2.Gerard A, Buyse M, Nordlinger B, et al. Preoperative radiotherapy as adjuvant treatment in rectal cancer. Final results of a randomized study of the European Organization for Research and Treatment of Cancer (EORTC). Ann Surg. 1988;208:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahl O, Horn A, Morild I, et al. Low-dose preoperative radiation postpones recurrences in operable rectal cancer. Results of a randomized multicenter trial in western Norway. Cancer. 1990;66:2286–2294. [DOI] [PubMed] [Google Scholar]
- 4.Marsh PJ, James RD, Schofield PF. Adjuvant preoperative radiotherapy for locally advanced rectal carcinoma. Results of a prospective, randomized trial. Dis Colon Rectum. 1994;37:1205–1214. [DOI] [PubMed] [Google Scholar]
- 5.Medical Research Council Rectal Cancer Working Party. Randomised trial of surgery alone versus radiotherapy followed by surgery for potentially operable locally advanced rectal cancer. Lancet. 1996;348:1605–1610. [PubMed] [Google Scholar]
- 6.Improved survival with preoperative radiotherapy in resectable rectal cancer. Swedish Rectal Cancer Trial. N Engl J Med. 1997;336:980–987. [DOI] [PubMed] [Google Scholar]
- 7.Sauer R, Liersch T, Merkel S, et al. Preoperative versus postoperative chemo-radiotherapy for locally advanced rectal cancer: results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol. 2012;30:1926–1933. [DOI] [PubMed] [Google Scholar]
- 8.Bosset JF, Calais G, Mineur L, et al. Enhanced tumorocidal effect of chemotherapy with preoperative radiotherapy for rectal cancer: preliminary results—EORTC 22921. J Clin Oncol. 2005;23:5620–5627. [DOI] [PubMed] [Google Scholar]
- 9.Smith JD, Ruby JA, Goodman KA, et al. Nonoperative management of rectal cancer with complete clinical response after neoadjuvant therapy. Ann Surg. 2012;256:965–972. [DOI] [PubMed] [Google Scholar]
- 10.Martin ST, Heneghan HM, Winter DC. Systematic review and meta-analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. Br J Surg. 2012;99:918–928. [DOI] [PubMed] [Google Scholar]
- 11.Yan L, Zhao W, Yu H, et al. A comprehensive meta-analysis of MicroRNAs for predicting colorectal cancer. Medicine. 2016;95:e2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi S, Kopetz S, Davuluri R, et al. MicroRNAs, ultraconserved genes and colorectal cancers. Intl J Biochem Cell Biol. 2010;42:1291–1297. [DOI] [PubMed] [Google Scholar]
- 13.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandres E, Bitarte N, Arias F, et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–2290. [DOI] [PubMed] [Google Scholar]
- 15.Bitarte N, Bandres E, Boni V, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–1671. [DOI] [PubMed] [Google Scholar]
- 16.Wen J, Luo K, Liu H, et al. MiRNA expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neoadjuvant chemoradiotherapy. Ann Surg. 2016;263:942–948. [DOI] [PubMed] [Google Scholar]
- 17.Lynam-Lennon N, Heavey S, Sommerville G, et al. MicroRNA-17 is downregulated in esophageal adenocarcinoma cancer stem-like cells and promotes a radioresistant phenotype. Oncotarget. 2017;8:11400–11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Tong L, Meng H, et al. MiR-335 regulates the chemo-radioresistance of small cell lung cancer cells by targeting PARP-1. Gene. 2017;600:9–15. [DOI] [PubMed] [Google Scholar]
- 19.Gawlick U, Lu KC, Douthit MA, et al. Stage III & IV colon and rectal cancers share a similar genetic profile: a review of the Oregon Colorectal Cancer Registry. Am J Surg. 2013;205:608–612. discussion 612. [DOI] [PubMed] [Google Scholar]
- 20.Ryan R, Gibbons D, Hyland JM, et al. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathol. 2005;47:141–146. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparativeC(T) method. Nat Protoc 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 22.National Cancer Institute NHGRI. The Cancer Genome Atlas. The Cancer Genome Atlas 2015. Available at: https://cancergenome.nih.gov/. Accessed March 24, 2017.
- 23.Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PloS One. 2013;8:e74250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruhl RKK, Espinosa-Diez C, Hudson C, Lanciault C, Tsikitis VL, Anand S. microRNA-451a regulates colorectal cancer radiosensitivity. bioRxiv 136234, 2017 Available at: 10.1101/136234. [DOI] [Google Scholar]
- 25.Borschitz T, Wachtlin D, Mohler M, et al. Neoadjuvant chemoradiation and local excision for T2–3 rectal cancer. Ann Surg Oncol. 2008;15:712–720. [DOI] [PubMed] [Google Scholar]
- 26.Habr-Gama A, Perez RO, Proscurshim I, et al. Patterns of failure and survival for nonoperative treatment of stage c0 distal rectal cancer following neoadjuvant chemoradiation therapy. J Gastrointest Surg. 2006;10:1319–1328. discussion 1328–1319. [DOI] [PubMed] [Google Scholar]
- 27.Pan X, Wang R, Wang ZX. The potential role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol Cancer Ther. 2013;12:1153–1162. [DOI] [PubMed] [Google Scholar]
- 28.Kanaan Z, Roberts H, Eichenberger MR, et al. A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258:400–408. [DOI] [PubMed] [Google Scholar]
- 29.miRWalk 2.0: a comprehensive atlas of predicted and validated miRNA-target interactions 2015; Available at: http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html. Accessed April 17, 2017, 2017.
- 30.Tian Y, Nan Y, Han L, et al. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol. 2012;40:1105– 1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes-Davies L, Huntsman D, Ruas M, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. [DOI] [PubMed] [Google Scholar]
- 32.Grossmann KS, Rosario M, Birchmeier C, et al. The tyrosine phosphataseShp2 in development and cancer. Adv Cancer Res. 2010;106:53–89. [DOI] [PubMed] [Google Scholar]
- 33.Chan RJ, Feng GS. PTPN11 is the first identified proto-oncogene that encodesa tyrosine phosphatase. Blood. 2007;109:862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matozaki T, Murata Y, Saito Y, et al. Protein tyrosine phosphatase SHP-2: aproto-oncogene product that promotes Ras activation. Cancer Sci. 2009;100:1786–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohi MG, Neel BG. The role of Shp2 (PTPN11) in cancer. Curr Opin Genet Dev. 2007;17:23–30. [DOI] [PubMed] [Google Scholar]
- 36.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nature reviews. Cancer. 2006;6:307–320. [DOI] [PubMed] [Google Scholar]
- 37.Prahallad A, Heynen GJ, Germano G, et al. PTPN11 is a central node inintrinsic and acquired resistance to targeted cancer drugs. Cell Rep. 2015;12:1978–1985. [DOI] [PubMed] [Google Scholar]
- 38.Schneeberger VE, Ren Y, Luetteke N, et al. Inhibition of Shp2 suppresses mutant EGFR-induced lung tumors in transgenic mouse model of lung adenocarcinoma. Oncotarget. 2015;6:6191–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen YN, LaMarche MJ, Chan HM, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. [DOI] [PubMed] [Google Scholar]
- 40.Garcia Fortanet J, Chen CH, Chen YN, et al. Allosteric Inhibition of SHP2: identification of a potent, selective, and orally efficacious phosphatase inhibitor. J Med Chem. 2016;59:7773–7782. [DOI] [PubMed] [Google Scholar]
- 41.Tie J, Desai J. Targeting BRAF mutant metastatic colorectal cancer: clinical implications and emerging therapeutic strategies. Target Oncol. 2015;10:179–188. [DOI] [PubMed] [Google Scholar]
- 42.Ahronian LG, Sennott EM, Van Allen EM, et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Disc. 2015;5:358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vakana E, Pratt S, Blosser W, et al. LY3009120, a panRAF inhibitor, has significant anti-tumor activity in BRAF and KRAS mutant preclinical models of colorectal cancer. Oncotarget. 2017;8:9251–9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.