Abstract
Parasitic weeds represent a major threat to agricultural production across the world. Little is known about which host genetic pathways determine compatibility for any host–parasitic plant interaction. We developed a quantitative assay to characterize the growth of the parasitic weed Phelipanche aegyptiaca on 46 mutant lines of the host plant Arabidopsis thaliana to identify host genes that are essential for susceptibility to the parasite. A. thaliana host plants with mutations in genes involved in jasmonic acid biosynthesis/signaling or the negative regulation of plant immunity were less susceptible to P. aegyptiaca parasitization. In contrast, A. thaliana plants with a mutant allele of the putative immunity hub gene Pfd6 were more susceptible to parasitization. Additionally, quantitative PCR revealed that P. aegyptiaca parasitization leads to transcriptional reprograming of several hormone signaling pathways. While most tested A. thaliana lines were fully susceptible to P. aegyptiaca parasitization, this work revealed several host genes essential for full susceptibility or resistance to parasitism. Altering these pathways may be a viable approach for limiting host plant susceptibility to parasitism.
Keywords: Phelipanche aegyptiaca, Parasitic plants, Parasite resistance, Parasite susceptibility, Arabidopsis thaliana, Plant immunity
Introduction
The parasitic weed Phelipanche aegyptiaca (syn. Orobanche aegyptiaca) is an obligate holoparasite, lacking the capacity for photosynthesis and fully dependent on parasitization of a host plant for nutrients and completion of its lifecycle. P. aegyptiaca, which is commonly known as Egyptian broomrape, is a major biotic constraint to crop production throughout much Eastern Europe, Asia and Northern Africa (Parker, 2013). P. aegyptiaca is able to parasitize a remarkably broad host range of dicotyledonous plants including crop and non-crop species. The crop-containing plant families for which P. aegyptiaca is a major threat are Solanaceae, Fabaceae, Apiaceae and Cucurbitaceae (Parker, 2013). Additionally, P. aegyptiaca can successfully parasitize the model host plant Arabidopsis thaliana which makes it a promising model parasitic plant (Goldwasser, Plakhine & Yoder, 2000; Westwood, 2000), despite the significant challenges of working with P. aegyptiaca in a laboratory setting.
Phelipanche aegyptiaca is a member of the Orobanchaceae family, which includes the overwhelming majority of agriculturally relevant parasitic weeds. Other members of the family that are major pests in agricultural production systems include Striga hermonthica (purple witchweed), a widespread and devastating parasite of cereal crops throughout Africa (Spallek, Mutuku & Shirasu, 2013), Orobanche cumana (sunflower broomrape), one of the primary biotic constraints to sunflower production in Europe and Asia (Molinero-Ruiz et al., 2015) and Orobanche cernua (nodding broomrape), a costly weed of tomato and other solanaceous crops throughout Africa, Asia and Europe (Parker, 2013). Although much has been written about the ability of parasitic weeds to germinate in response to host-specific chemical cues (Zwanenburg, Pospíšil & Ćavar Zeljković, 2016), develop haustoria to invade host plant tissues (Yoshida et al., 2016), and extract nutrient through vascular connections (Irving & Cameron, 2009), relatively little is known about the host plant immune responses and parasitic plant virulence mechanisms throughout these stages of plant–parasite interactions (See Kaiser et al. (2015) and Clarke et al. (2019) for review).
Because of the close physical association and shared angiosperm characteristics between parasitic plants and their hosts, mechanical and chemical controls are largely ineffectual in the control of parasitic weeds. Development of genetically resistant host crops is the most promising strategy for managing parasitic weeds (Rubiales, Rojas-Molina & Sillero, 2016). Unfortunately, to date only a few resistance (R) genes or resistance-associated quantitative trait loci have been identified for parasitic weeds. A classic nucleotide-binding-site, Leucine-rich-repeat (LRR) R gene that confers resistance to S. gesnerioides was cloned from cowpea (Li & Timko, 2009). A pattern recognition receptor (PRR) in tomato that confers resistance to the stem parasite Cuscuta reflexa was identified in tomato (Hegenauer et al., 2016). Recently, another LRR receptor-like kinase was identified as responsible for sunflower resistance to O. cumana (Duriez et al., 2019). Several other resistance loci have been deployed in the management of O. cumana on sunflower, but the parasite rapidly overcomes such resistance (Molinero-Ruiz et al., 2015).
An alternative genetic strategy for the control of parasitic weeds is the alteration of host plant genes that are essential for parasite attachment or development, so called susceptibility genes (Van Schie & Takken, 2014). For example, genes involved in the biosynthesis of parasite germination stimulants or transport of critical nutrients to the parasite are potential susceptibility genes. Identifying the genetic pathways that underpin host compatibility for parasite attachment and development is an essential first step toward finding susceptibility genes. To that end, we developed an assay to quantify the ability of P. aegyptiaca to successfully attach and develop on A. thaliana roots, and quantified susceptibility to parasite attachment and development for 46 mutant lines of A. thaliana. The primary goal in this work was to test the impact on a parasitic plant of host plant hormone signaling and immunity-related genes that have been either demonstrated or hypothesized to be involved in plant susceptibility to other pathogen classes.
Materials and Methods
Plant growth and quantifying parasite attachment and development
The parasite attachment assay was conducted using a randomized incomplete block design due to large number of tested genotypes. For each experimental block, four mutant plant lines were randomly selected from the pool of all mutant lines in the collection. Approximately 25 A. thaliana seeds of each of the four mutant lines plus the wild type background ecotype were stratified in water at 4 °C for 2 days in then planted in Sunshine #1 potting mix (Sungro, Agawam, MA, USA). Plants were grown in a Conviron ATC40 growth chamber at 20 °C, 12-h light cycle and light intensity of 90 µmol m2s−1 for 10 days. Eight Polyethylene (PE) bags with glass fiber grade A (GFA) paper (Whatman, Maidstone, UK) backings were made as previously described (Westwood, 2000) with dimensions of 26 cm × 9 cm (Fig. 1A). The 10-day-old A. thaliana seedlings were gently removed from the soil and the roots were washed with water until free of soil. The plants were transplanted such that the roots were positioned between the PE bag and the GFA paper and the hypocotyl extended from the top of the bag (Fig. 1A). One plant of each of the four mutant lines and one plant of the wild type background ecotype were randomly distributed into each of the PE bags and placed under a laboratory growth light with a 12-h light cycle for 12 days. P. aegyptiaca seeds were sterilized following previously described protocols (Westwood, 2000) and placed on wet GFA paper in a sealed Petri plate (Fisher Scientific, Waltham, MA, USA) for six days to condition the seeds (Westwood, 2000). The strigolactone analog rac-GR24 was then applied to the conditioned P. aegyptiaca seeds at a concentration of 2 mg/L to stimulate germination. The Petri dishes were resealed and stored in the dark for an additional 24 h. Conditioned and stimulated P. aegyptiaca seeds were inoculated onto the roots of the A. thaliana plants in the PE bags (12 days after transplanting from soil) using a fine-tipped paintbrush. By placing germinated P. aegyptiaca seeds immediately adjacent to developed roots, the impact of differential root development among the various tested Arabidopsis mutants was limited. The seeds were aligned to be within 0.3 mm of roots that had grown since the transplanting (lighter colored roots) and approximately 35 seeds were placed along the roots of each plant in each bag.
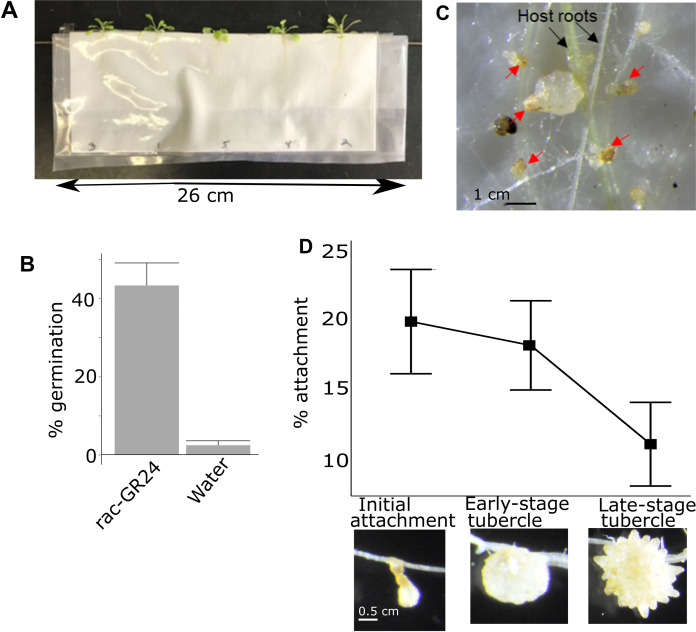
Figure 1. Pipeline for quantifying attachment and tubercle development rate of P. aegyptiaca on A. thaliana.
(A) Example of A. thaliana lines growing in PE bags before inoculation with P. aegyptiaca. Plants were randomly distributed. (B) Typical germination rates of P. aegyptiaca seedlings following conditioning and treatment with germination stimulants n = 3 plates for each germination stimulant with approximately 100 total seeds per plate. (C) Example of germinated and ungerminated P. aegyptiaca radicles (red arrows) on A. thaliana roots. Germinated seedlings are counted to quantify the attachment and tubercle development rates. (D) Rate of attachment and tubercle development of P. aegyptiaca on wildtype (Col) A. thaliana roots for the three parasite development stages considered in this work—initial haustorial connection and attachment, early-stage tubercle, and late-stage tubercle n = 8. The attachment rate is the number of seeds that reached that stage of development divided by the total number of germinated seeds.
The attachment rate for each individual A. thaliana plant was quantified 15 days after inoculation. Every P. aegyptiaca seed was examined under a dissecting microscope (Zeiss Stemi SV11) at 12×g magnification and classified as either non-germinated, germinated but not attached, or attached. Not attached and attached germinated seeds were distinguished by probing with a 0.3 mm wide dissecting probe to test for adherence of the parasite radicle to the A. thaliana roots. The attached P. aegyptiaca plants were further classified as either early-stage attachment/haustoria connection, early-stage tubercle, or late-stage tubercle which correspond to the stages 3, 4.1 and 4.2 in the Parasitic Plant Genome Project datasets (Westwood et al., 2012; Yang et al., 2014) (Fig. 1D). The 0.3 mm dissecting probe was used to distinguish the three attached classifications as follows: an attached radicle thinner than 0.3 mm was categorized as an attached radicle/initial attachment (stage 3). A tubercle formed that is thicker than 0.3 mm but has no secondary roots was categorized as an early-stage tubercle (stage 4.1) and coincides with the completion of the vascular connection/feeding bridge between the parasite and the host plant. A tubercle with secondary roots longer than 0.3 mm was categorized as a late-stage tubercle (stage 4.2). The rate of attachment for each of the three stages was determined by calculating the ratio of all of the P. aegyptiaca seeds that had reached at least the designated stage (i.e., the rate of initial attachment included the count of late-stage tubercles plus early-stage tubercles) divided by the total number of germinated P. aegyptiaca seeds (i.e., the total number of all stages plus those seeds that germinated but did not attach to hosts).
Statistical analysis of attachment rates
We used a generalized linear mixed model (SAS/STAT(R) 9.2 User’s Guide SE, 2016) to analyze different rates of parasitization at each of the three developmental stages. This model is based on the binomial distribution with the logit link. The SAS script and example files are uploaded to the Ag Data Commons (https://data.nal.usda.gov/dataset/data-multiple-immune-pathways-control-susceptibility-arabidopsis-thaliana-parasitic-weed-phelipanche-aegyptiaca). The one fixed effect of interest was treatment of the plants. The treatments are the different host plant genetic pathways which were compared to the respective wildtype in our model (Col-0, Ler-0, or Ws-2). Both bag and position were set as fixed effects to account for any systematic variability within both bag and position. The experiment was specified as a random block, which allows us to determine significance of the fixed effects above and beyond experiment to experiment variability. This also accounts for the fact that results within an experiment may be more related than results across experiments. Our response variable was the ratio of attachment stage total to overall germinated seed total (described above). We compared 95% and 99% confidence intervals on the odds ratio estimates at each of the three stages to determine which treatments are statistically significantly different from the wildtype. An odds ratio greater than one indicates that P. aegyptiaca is more successful on the mutant line relative to the wildtype. An odds ratio less than one indicates that P. aegyptiaca is less successful on the mutant line than on the wildtype.
Type III tests of fixed effects indicated that there was no significant variation dependent on the bag. However, there was a significant effect dependent on the position of the plant within the bag for early and late stage tubercle attachment rates (Table S1). Tukey–Kramer analysis of the least square means of the differences of position with adjustment for multiple comparisons showed that significant differences (p < 0.05) always included plants on the edge of the bag (positions 1 and 5). We hypothesize that these differences were due to the edge of the GFA paper drying out more unevenly than the center of the GFA paper. Therefore, it is essential to randomize the position of the plants within each bag, as was done here.
Time-lapse photography
A PE bag containing A. thaliana eco. Col-0 was inoculated with stimulated P. aegyptiaca seeds as described above. The bags were mounted on a metal hanger inside a 11.3-L Rubbermaid container that had a 70 mm diameter hole cut in the side for insertion of a macro camera lens and the top portion was covered with aluminum foil with a narrow opening so that the A. thaliana shoots could be exposed to light (12-h light-dark cycles) while the parasites were in darkness. The camera was a Sigma SD14 digital camera with a 70 mm Sigma DG macro lens. The camera flash provided the light source for the photography. The A. thaliana was watered through a tube that fed directly into the bag, so that the plant did not have to be disturbed during the time-lapse photography. For recording aboveground plant growth, a tomato (Solanum lycopersicum) was used as host because Arabidopsis does not support robust aboveground growth in our experimental setup. A Nikon D5100 camera with a Nikon DX AF-S Nikkor lens was used. Pictures were taken at 9-min intervals for 37 days over the course of the above and below ground segments. Sigma Photo Pro software (Sigma Corporation of America, Ronkonkoma, NY, USA) was used to convert the images from raw X3F format into JPEG. Adobe Premiere Pro CC 2017 was then used to assemble the images and for video editing to make the time-lapse video at 24 fps.
Quantitative RT-PCR
Three biological replicates of approximately 40 0.5 cm root sections were harvested from both inoculated and time-matched mock-inoculated A. thaliana eco. Col-0 plants at early attachment stage, early vascularization/early tubercle stage, and late tubercle stage (six total experimental conditions with three biological replicates and three technical replicates each). RNA was extracted using a Qiagen RNeasy kit. cDNA was synthesized using Superscript cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s recommended protocol. qRT-PCRs were performed by StepOnePlus Real-Time PCR System (Invitrogen, Carlsbad, CA, USA) with Power SYBR green master mix (Invitrogen, Carlsbad, CA, USA). The reaction condition was 2 min at 50 °C and 10 min at 95 °C followed by 40 cycles at 95 °C for 15 s, at 56 °C for 15 s and 72 °C for 15 s. Dissociation curves were evaluated to confirm the specificity. Relative quantification was calculated by 2−∆∆Ct method using qBase+ software. Three housekeeping genes (SAND, UBQ10 and UFP) were used for the normalization. geNorm analysis revealed that geNorm M-value of all three genes (SAND, UBQ10 and UFP) were 0.762, 0.709 and 0.619, relatively (qBASE+, Biogazelle). The recommended cutoff value is one or less. Calibrated, normalized relative quantification values for each gene were compared between the mock-inoculated and Phelipanche-inoculated plants for each parasite developmental stage using a two-tailed, two-sample unequal variance t-test. The primers used are described in Table S2. All primers were previously designed in other studies: (Jacobs et al., 2011) (VSP2, MYB51, EXPPT1, BOI, SID2, WRKY53), (Paponov et al., 2008) (IAA13, IAA2, ACS6), (Nguyen et al., 2016) (ARR10, UBQ10), (Šašek et al., 2014) (SAND), (Tran, Chen & Wang, 2017) (UFP), (Pegadaraju et al., 2007) (PAD4), (Zhang et al., 2014) (PR1). All primers were validated to be specific to A. thaliana DNA and to not amplify any product from cDNA prepared from early tubercle stage P. aegyptiaca tissue.
Results
A quantitative screen for host plant resistance and susceptibility to parasitic weeds
We developed a quantitative assay to measure the susceptibility of A. thaliana–P. aegyptiaca attachment and tubercle development based on previously described techniques (Westwood, 2000). A. thaliana plants grown in PE bags with GFA backing (Fig. 1A) allow for host roots to be directly inoculated with P. aegyptiaca. All P. aegyptiaca seeds were stimulated with the strigolactone analog rac-GR24 (Yoneyama et al., 2010), which is an effective germination stimulant for P. aegyptiaca (Fig. 1B). P. aegyptiaca attachment and development can be monitored in this growth system. Our analysis of time-lapse video of P. aegyptiaca (Video S1—available at https://figshare.com/articles/PhelipancheParasitization_mov/11894388) growth highlighted the acceleration in parasite growth following development of the floral meristem. Therefore, the challenge for our assay of parasite growth on mutants was to devise a method that could provide sensitive, reproducible measures of parasite success during a developmental stage characterized by relatively small changes in growth. The number of P. aegyptiaca seedlings at various growth stages can be counted under a dissecting microscope (Fig. 1C). Graphical representation of the typical attachment and tubercle development rates from inoculation of P. aegyptiaca onto wildtype A. thaliana is shown in Fig. 1D. All data from all parasite attachment experiments are in Table S3. Because of the substantial number of comparisons made in this analysis (three different stages for each of 46 mutant lines compared to wildtype), we primarily focus on differences that met the more stringent 99% confidence limit cutoff (see “Methods”) to minimize false positives. Based on this cutoff, only between 2 and 9 mutant lines showed altered susceptibility to P. aegyptiaca parasitization at any specific stage compared to wildtype A. thaliana (Table S4).
Host genetic pathways essential for susceptibility to P. aegyptiaca
Phelipanche aegyptiaca is a successful pathogen of A. thaliana (Goldwasser, Plakhine & Yoder, 2000); therefore, our assay of attachment and development rates is more suited to identify mutations that attenuate A. thaliana susceptibility to P. aegyptiaca parasitization than mutations that increase susceptibility. The 46 tested mutant lines include numerous disruptions in key signaling pathways (Table 1). It is important to note that many of these genes are involved in several different genetic pathways due to the substantial crosstalk among many of the signaling pathways. Examples include pad4 and eds1 which are involved in salicylic acid (SA) signaling, but also critical for several R gene-mediated responses (Cui et al., 2017; Rustérucci et al., 2001) and the substantial crosstalk between jasmonic acid (JA), SA and ethylene signaling (Koornneef & Pieterse, 2008).
Table 1. The 46 mutant lines considered in this study.
| Gene | Involved in1 | Background | Citation | ABRC # |
|---|---|---|---|---|
| 35s:ERF1-2 | Ethylene response/ JA signaling | Col | Lorenzo et al. (2003) | CS6143 |
| 35s:PMR4 | Penetration resistance | Col | Ellinger et al. (2013) | n/a |
| aba1 | ABA signaling | Col | Alonso et al. (2003) | SALK_027326C |
| arr1-2/arr10-1/arr11-1 | Cytokinin signaling | Ws-2 | Mason et al. (2005) | CS6993 |
| aux1-7 | Auxin distribution | Col | Pickett, Wilson & Estelle (1990) | CS3074 |
| aux1-7; ein2 | Auxin distribution/ethylene perception | Col | CS8843 | |
| axr1-3 | Auxin, cytokinin, JA, ethylene signaling | Col | Lincoln, Britton & Estelle (1990) | CS3075 |
| bak1-4 | Master regulator of pattern-triggered immunity | Col | Alonso et al. (2003); Kemmerling et al. (2007) | SALK_116202C |
| bkk1 | Master regulator of pattern-triggered immunity | Col | Alonso et al. (2003); He et al. (2007) | SALK_057955C |
| cpr5 | Negative regulator of systemic acquired resistance and programed cell death | Col/No-0 | Boch et al. (1998) | CS3770 |
| csn5 | Core targeted immunity hub | Col | Alonso et al. (2003); Mukhtar et al. (2011) | SALK_027705 |
| dcl1-7 | RNAi | Ler | Robinson-Beers, Pruitt & Gasser (1992) | CS3089 |
| dde2-2 | JA biosynthesis | Col | Von Malek et al. (2002) | CS65993 |
| dde2-2/ein2-1/pad4-1/sid2-2 | JA biosynthesis, ethylene signaling, SA signaling | Col | Tsuda et al. (2009) | CS66007 |
| dde2-2/pad4-1 | JA biosynthesis, SA signaling | Col | Tsuda et al. (2009) | CS65998 |
| dde2-2/sid2-2 | JA biosynthesis, SA signaling | Col | Tsuda et al. (2009) | CS65999 |
| edr1-1 | Ethylene-dependent stress responses | Col | Frye & Innes (1998) | CS67959 |
| eds1-1 | Negative regulator R gene-mediated resistance | Ws-2 | Parker et al. (1996) | n/a |
| ein2-1 | Ethylene signaling | Col | Alonso et al. (1999) | n/a |
| gai1 | Giberellic acid signaling | Ler | Koorneef et al. (1985) | CS63 |
| gai-t6 rga-t2 rgl1-1 rgl2-1 | DELLA genes, giberellic acid signaling | Col | Navarro et al. (2008) | n/a |
| jar1-1 | JA biosynthesis | Col | Staswick, Su & Howell (1992) | n/a |
| jar1-1/axr1-3 | JA signaling, auxin signaling | Col | Tiryaki & Staswick (2002) | CS67934 |
| jar1-1/mlo2-11 | JA signaling, penetration resistance | Col | Consonni et al. (2006) | CS9723 |
| jaz3/jaz4-1/jaz9-1 | JA signaling (repressor) | Col | n/a | |
| Jaz3 (T3) | JA signaling (overexpression Jaz3) | Col | n/a | |
| jin1-1 | Jasmonic acid signaling | Col | Berger, Bell & Mullet (1996) | n/a |
| lsd1-2 | Negative regulation of cell death and disease resistance | Col | Kaminaka et al. (2006) | CS68738 |
| lsu2-1 | Core targeted immunity hub | Col | Alonso et al. (2003); Mukhtar et al. (2011) | SALK_031648c |
| lsu2-2 | Core targeted immunity hub | Col | Alonso et al. (2003); Mukhtar et al. (2011) | SALK_126244c |
| mkk1/mkk2 | Map kinase signaling | Col | Qiu et al. (2008) | n/a |
| mlo2-5; pen2-1 | Penetration resistance | Col | Consonni et al. (2006) | CS9717 |
| mpk4 | Map kinase signaling | Col | Qiu et al. (2008) | n/a |
| NahG | SA antagonist | Col | Lawton (1995) | n/a |
| ndr1-1 | R gene-mediated resistance, systemic acquired resistance | Col | Century, Holub & Staskawicz (1995) | CS6358 |
| pad4-1 | SA and SA-independent defense responses | Col | Glazebrook, Rogers & Ausubel (1996) | CS3806 |
| pad4-1/ndr1-1 | SA signaling, systemic acquired resistance, R gene-mediated resistance | Col | n/a | |
| pen2-3 | Penetration resistance | Col | Lipka et al. (2005) | CS66946 |
| pepr1/pepr2 | Perception of DAMPs | Col | Krol et al. (2010) | n/a |
| pfd6-1 | Core targeted immunity hub | Col | Alonso et al. (2003); Mukhtar et al. (2011) | n/a |
| rar1 | Master regulator of R gene-mediated immunity | Col | Muskett et al. (2002) | n/a |
| rdr1-1/rdr2-1/rdr6-15 | RNAi (known problems with pleiotropy) | Col | Garcia-Ruiz et al. (2010) | CS66485 |
| rdr6-11 | RNAi | Col | Peragine et al. (2004) | CS24285 |
| rpm1 | R gene-mediate immunity | Col | Grant et al. (1995) | n/a |
| sgt1 | Master regulator of R gene-mediate immunity | Col | Austin et al. (2002) | n/a |
| sid2-2 | SA biosynthesis | Col | Nawrath & Métraux (1999) | n/a |
| tir1 | Auxin perception | Col | Ruegger et al. (1998) | CS3798 |
Note:
An incomplete representation of the pathways in which the gene is involved.
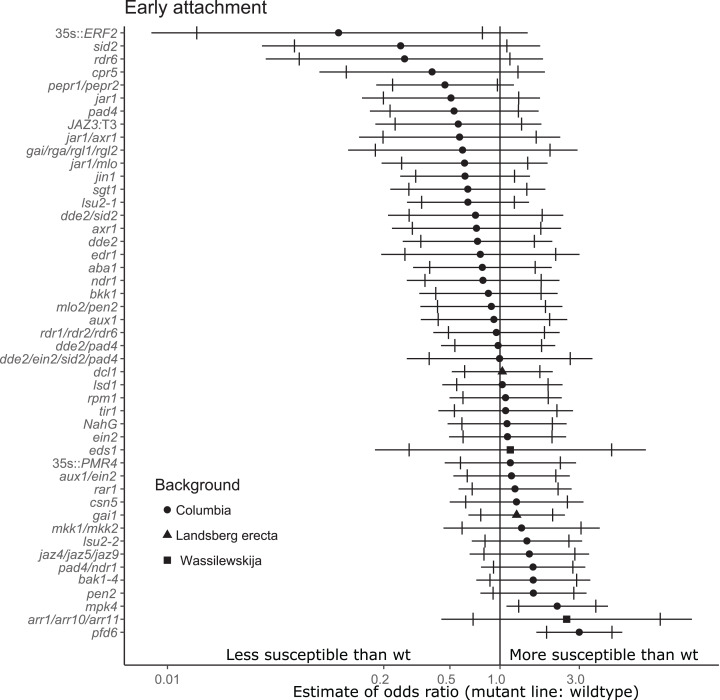
For the stage of initial parasitization/haustoria development, no mutant lines were less susceptible than wildtype A. thaliana at the 99% confidence level (Fig. 2). This result demonstrates that none of the tested host endogenous signaling pathways are essential for P. aegyptiaca to form haustoria and attach to host roots. However, plants overexpressing Ethylene Response Factor 2 (ERF2), a transcription factor upregulated in the presence of ethylene that activates numerous immune responses (Catinot, Huang & Zimmerli, 2015), were significantly less susceptible than wildtype to P. aegyptiaca initial attachment at the 95% confidence level, suggesting that elevated host ethylene levels may attenuate initial attachment. The attachment rates were more variable within and among experiments for the early attachment stage compared to the two later stages measuring tubercle growth (Table S3).
Figure 2. Mutations in multiple immunity-related genes significantly affect the susceptibility of A. thaliana to initial parasitization by P. aegyptiaca.
Marker points indicate the estimated odds ratio of the rate of early attachment of P. aegyptiaca on the mutant line relative to wildtype A. thaliana. An odds ratio greater than one indicates that P. aegyptiaca is more successful on the mutant line than wildtype. An odds ratio less than one indicates that P. aegyptiaca is less successful on the mutant line than wildtype. The shapes of the marker points depict the ecotype background for each mutant line. The capped error bars represent the 95% confidence interval and the uncapped error bars (overlapping but extending) represent the 99% confidence interval. A confidence interval that does not cross the vertical line at attachment rate = 1 (the normalized attachment rate to wildtype Arabidopsis) is considered statistically different at the indicated confidence level. Data represent at least 14 replicates from at least two independent experiments for each mutant line.
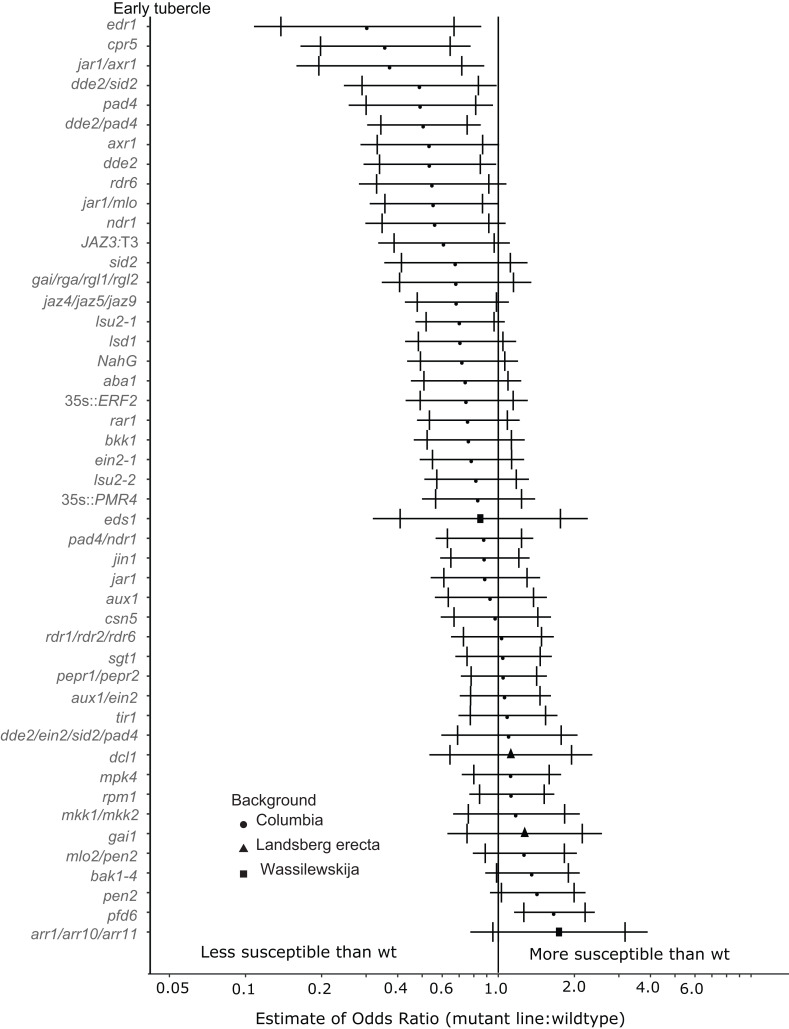
Eight A. thaliana mutant lines were attenuated in the ability to support P. aegyptiaca early-stage tubercle formation at the 99% confidence limit (Fig. 3). Five of the less susceptible mutants involve knockouts in jasmonic acid biosynthesis or signaling (dde2, dde2/pad4, dde2/sid2, jar1/mlo, jar1/axr1) suggesting that P. aegyptiaca requires functional host JA signaling and biosynthesis to form successful tubercles. However, neither the jar1 nor jin1 single mutant is less susceptible to P. aegyptiaca tubercle formation. pad4 plants were also less susceptible to P. aegyptiaca tubercle development. PAD4 is involved in both SA signaling and TIR R-gene signaling. Because other mutants defective in SA accumulation (sid2 and NahG) and R-gene-mediated responses (sgt1, rar1 and rpm1) were not significantly less susceptible to parasite attachment, the reason pad4 plants are less susceptible at the early tubercle stage remains uncertain. Lastly, the mutants cpr5 and edr1 were substantially less susceptible to early-stage tubercle formation. Both of these mutants have been previously shown to constitutively express multiple plant defense pathways and be more resistant to several microbial biotrophic plant pathogens (Bowling et al., 1997; Christiansen et al., 2011; Tang, Christiansen & Innes, 2005; Yoshida et al., 2002).
Figure 3. Mutations in multiple immunity-related genes significantly affect the susceptibility of A. thaliana to early tubercle development by P. aegyptiaca.
Marker points indicate the estimated odds ratio of the rate of early attachment of P. aegyptiaca on the mutant line relative to wildtype A. thaliana. An odds ratio greater than one indicates that P. aegyptiaca is more successful on the mutant line than wildtype. An odds ratio less than one indicates that P. aegyptiaca is less successful on the mutant line than wildtype. The shapes of the marker points depict the ecotype background for each mutant line. The capped error bars represent the 95% confidence interval and the uncapped error bars (overlapping but extending) represent the 99% confidence interval. A confidence interval that does not cross the vertical line at attachment rate = 1 (the normalized attachment rate to wildtype Arabidopsis) is considered statistically different at the indicated confidence level. Data represent at least 14 replicates from at least two independent experiments for each mutant line.
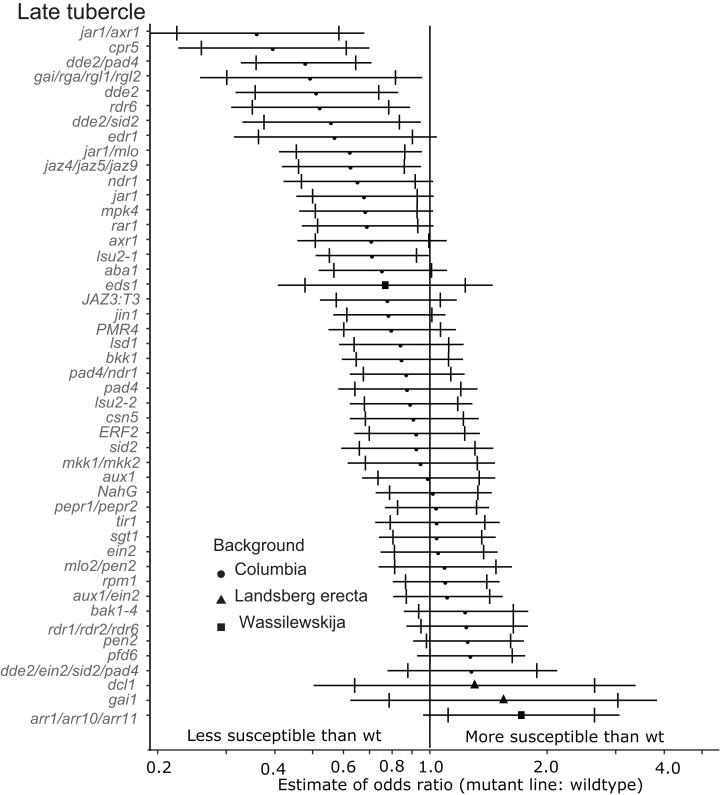
Six of the mutants (jar1/axr1, jar1/mlo, cpr5, dde2/pad4, dde2 and dde2/sid2) identified as being less susceptible to early-stage tubercles also supported significantly fewer late-stage P. aegyptiaca tubercles at the 99% confidence limit (Fig. 4). Additionally, the jaz4/5/9 mutant, which is defective in JA responses because the JAZ4, JAZ5 and JAZ9 proteins are all transcriptional regulators triggered by jasmonate (Pauwels & Goossens, 2011; Yan et al., 2014), also supported fewer late-stage tubercles than WT A. thaliana. The JA biosynthesis mutant jar1 supported fewer late stage tubercles at the 95% confidence level giving further support to the hypothesis that several genes involved in host JA signaling and biosynthesis are essential for P. aegyptiaca parasitism.
Figure 4. Mutations in multiple immunity-related genes significantly affect the susceptibility of A. thaliana to supporting late tubercle development by P. aegyptiaca.
Marker points indicate the estimated odds ratio of the rate of early attachment of P. aegyptiaca on the mutant line relative to wildtype A. thaliana. An odds ratio greater than one indicates that P. aegyptiaca is more successful on the mutant line than wildtype. An odds ratio less than one indicates that P. aegyptiaca is less successful on the mutant line than wildtype. The shapes of the marker points depict the ecotype background for each mutant line. The capped error bars represent the 95% confidence interval and the uncapped error bars (overlapping but extending) represent the 99% confidence interval. A confidence interval that does not cross the vertical line at attachment rate = 1 (the normalized attachment rate to wildtype Arabidopsis) is considered statistically different at the indicated confidence level. Data represent at least 14 replicates from at least two independent experiments for each mutant line.
In addition to the JA signaling and biosynthesis mutant lines, the DELLA quad mutant (gai/rga/rgl1/rgl2) and rdr6 mutant lines supported fewer late-stage tubercles at the 99% confidence level. The DELLA genes (GAI, RGR and RGLs) are negative growth regulators controlled by Gibberellic acid (GA), and this quad mutant is more resistant to the biotrophic pathogen Pseudomonas syringae and more susceptible to the necrotrophic pathogen Alterneria brassisicola (Navarro et al., 2008).
Host genetic pathways essential for resistance to P. aegyptiaca
Two tested mutants, mpk4 and pfd6-1, were more susceptible to initial P. aegyptiaca parasitization (Fig. 2). PFD6 is a subunit of the prefoldin complex, and pfd6 mutants are defective in microtubule function and formation (Gu et al., 2008) and is a putative hub of the A. thaliana immune system (Mukhtar et al., 2011). pfd6-1 mutants were also significantly more susceptible to supporting early-stage tubercle development by P. aegyptiaca (Fig. 3). We, therefore, hypothesize that PFD6 is an important component of the host plant immune response against plant parasitization.
None of the tested mutant lines supported significantly more late-stage P. aegyptiaca tubercles compared to wildtype A. thaliana at the 99% confidence level (Fig. 4). However, pfd6 did support significantly more late-stage tubers at the 95% confidence level. Additionally, the triple mutant arr1/arr10/arr11, which is deficient in cytokinin signaling (Mason et al., 2005), also supported more late-stage P. aegyptiaca tubercles at the 95% confidence level.
Parasitization by P. aegyptiaca alters the transcription of several immunity-related genes
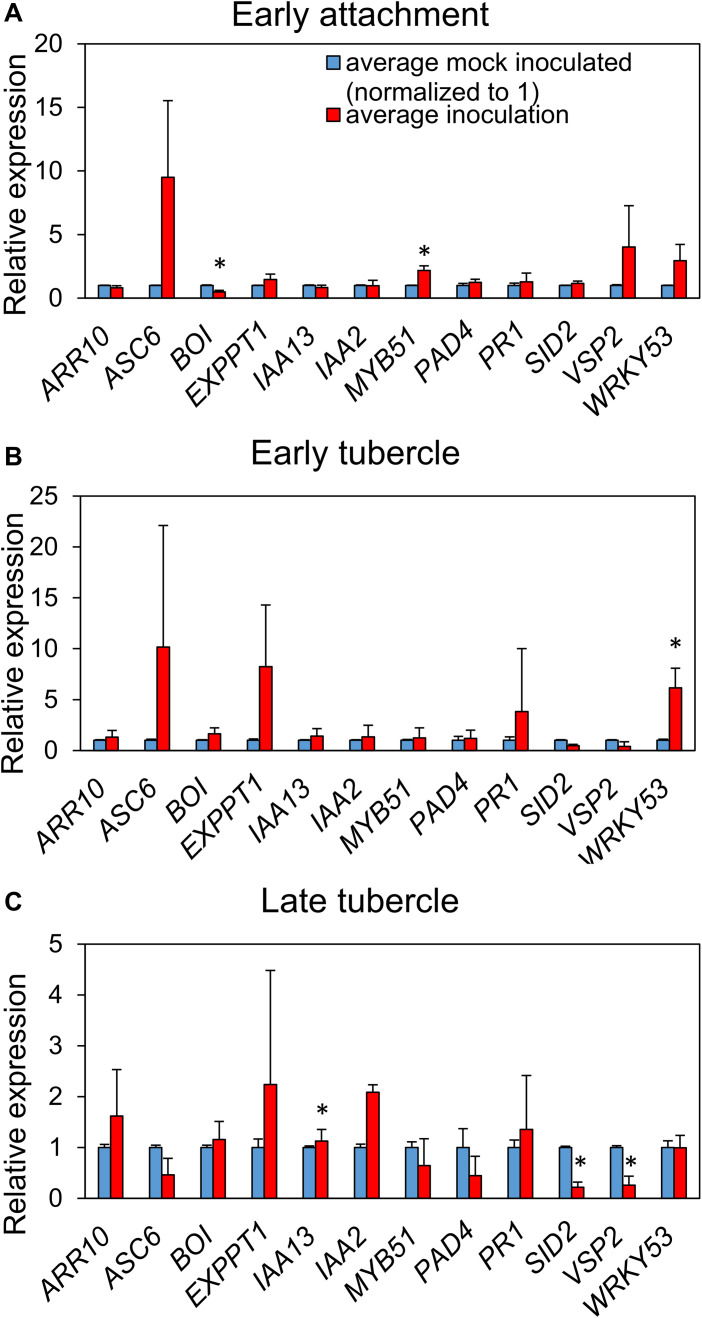
We monitored transcript levels of 12 defense-and hormone signaling-associated genes of A. thaliana (Table S2) following P. aegyptiaca parasitization. Significant upregulation of the glucosinolate biosynthetic gene MYB51 and downregulation of the negative cell death regulator BOI were observed during initial haustorial attachment (Fig. 5). During early stage tubercle development, the pattern-trigged immunity (PTI) marker gene WRKY 53 is significantly upregulated. During late-stage tubercle development, only smaller magnitude changes in gene expression were observed, most notably significant down-regulation in the JA marker gene VSP2 and the SA marker gene SID2. Whether these changes are more directly associated with host defense responses or parasite virulence strategies remains unknown.
Figure 5. P. aegyptiaca parasitization alters the transcription of marker genes for multiple hormone-signaling pathways.
Relative expression of twelve genes of interest were determined in the roots of both inoculated and mock-inoculated A. thaliana eco. Col-0 plants across the three studied stages: early attachment (A), early-stage tubercle development (B) and late-stage tubercle development (C). Data bars depict the expression of the 12 indicated genes relative to three stably-expressed housekeeping genes (SAND, UBQ10 and UFP) from three biological and five technical replicates. Error bars represent the standard error. Data were normalized to set the relative expression of each gene in the mock-inoculated condition to one. Asterisks indicate statistically different means compared to mock-inoculated plants at the same growth stage based on paired t-test (p < 0.05).
Discussion
The importance of several genes for A. thaliana resistance and susceptibility to P. aegyptiaca was revealed by our novel quantitative screen for plant parasitism. Given the artificial nature of the PE bag growth system, these observations are a starting point for elucidating the host molecular pathways that dictate whether a host is resistant or susceptible to attack from a parasitic plant. Notably, the mpk4, gai1 and cpr5 plants exhibited marked reduced growth compared to wildtype in the PE bags. Additionally, substantial crosstalk among many of the studied pathways (Koornneef & Pieterse, 2008), complicates our ability to definitively list which host pathways are critical for supporting plant parasitism. Nevertheless, this data provides the foundation for understanding the host genetics of susceptibility to plant parasitism. We discuss the host pathways that appear to be the most important below.
JA/SA signaling
Several host genes involved in JA biosynthesis and signaling are (e.g., dde2, jar1, jaz4/5/9) are required for full host susceptibility to development of parasite tubercles (Figs. 3 and 4). Several of these mutants also supported fewer early-stage haustorial attachments (Fig. 2), but not significantly as determined by our stringent cutoff (Table S4). Therefore, we conclude that aberrations in host endogenous JA signaling can compromise compatibility to P. aegyptiaca parasitization. Further supporting this hypothesis, JA signaling genes were previously identified as upregulated in response to compatible parasitization of Lotus japonicus by P. aegyptiaca (Hiraoka, Ueda & Sugimoto, 2008), A. thaliana by O. ramosa (Dos Santos et al., 2003), sorghum by Striga (Hiraoka & Sugimoto, 2008) and Medicago truncatula by Orobanche crenata (Die et al., 2007). The common interpretation of these studies is that hosts induce JA-related defense responses to deter the parasite, but all of these hosts are susceptible to the respective parasites. Exogenous jasmonate treatment was previously shown to limit P. aegyptiaca parasitism (Bar-Nun & Mayer, 2008), suggesting that general perturbations in JA signaling may attenuate the compatibility of host plants to root parasitization.
Jasmonic acid signaling plays a critical role in numerous components of plant response to pathogen attack, including antagonism to SA signaling (Gutjahr & Paszkowski, 2009; Kazan & Manners, 2008). A reasonable hypothesis is that the JA mutants are less susceptible to parasitism because of elevated SA levels. High levels of SA are often associated with reduced virulence of biotrophic plant pathogens (Thomma et al., 2001). Application of exogenous SA was previously shown to reduce parasitism of clover by the closely related parasite Orobanche minor (Kusumoto et al., 2007). However, several SA-deficient mutants (e.g., sid2 and NahG) were not more susceptible to parasite attachment and development. Additionally, a few mutants with known defects in SA accumulation and/or signaling (ndr1 and pad4) were actually less susceptible to early-and late-stage tubercle development at the 95% confidence interval. Taken together, these results suggest that perturbations to functional crosstalk between SA and JA signaling limit the ability of P. aegyptiaca to parasitize A. thaliana. Therefore, we hypothesize that wildtype-functional SA and JA signaling is required by the parasite, potentially because these pathways are precisely targeted and manipulated during parasitism to support the attachment and development of the parasite. This hypothesis is further supported by the observation that the DELLA quad mutant plants are significantly less susceptible to late-stage tubercle development. The DELLA quad mutant was previously shown to be much more susceptible to biotrophs, likely due to aberrations in SA–JA crosstalk (Navarro et al., 2008). The DELLA genes are repressed by GA and parasitized tissue likely contains low levels of GA based on the expression of the marker gene EXP-PT1 (Fig. 5).
Putative hubs and regulators of plant immunity
pfd6 plants were significantly more susceptible to P. aegyptiaca parasitization than wildtype host plants (Figs. 2–4). Even though our assay is biased toward identifying mutant host plants that are less susceptible to P. aegyptiaca parasitization, the increased susceptibility of pfd6 was one of the strongest and most consistent phenotypes identified. PFD6 is a putative immunity hub protein (Mukhtar et al., 2011) involved in microtubule dynamics (Gu et al., 2008). It is possible that microtubules are being deployed to prevent parasitization, and disruption of microtubule formation is sufficient to support increased parasitization. Several plant pathogens target microtubules as part of their attack (Hardham, 2013; Lee et al., 2012). Overexpression of PFD6 or genes involved in similar functions may be sufficient to increase the resistance of host plants against parasitic plants.
Obligate holoparasites, such as P. aegyptiaca, are analogous to biotrophic microbial plant pathogens because they rely on living host tissue for the completion of their lifecycle. Multiple mutants that have hyper-activated immunity and are resistant to several biotrophic plant pathogens (cpr5 and edr1) were substantially more resistant to P. aegyptiaca as well. This finding suggests that increased activation of known immune pathways that function in the control of biotrophic microbial plant pathogens is sufficient, in some instances, to also limit the severity of infestation by parasitic plants.
Auxin/cytokinin
The arr1/10/11 triple mutant supported more development of late-stage P. aegyptiaca tubercles than wildtype A. thaliana (Fig. 4) but only at the 95% confidence level. This finding suggests that host plant insensitivity to cytokinins leads to increased parasite attachment. The related parasite Phtheirospermum japonicum was recently shown to translocate cytokinins into host tissue to alter host development (Spallek et al., 2017). The reduced parasite attachment rates to axr1 plants (Figs. 3 and 4) potentially contradict the hypothesis that altered cytokinin signaling enhances host susceptibility to P. aegyptiaca. Though initially identified as an essential component of Arabidopsis response to auxin treatment, AXR1 is also involved in sensitivity to ethylene (Timpte et al., 1995), cytokinin (Li, Kurepa & Smalle, 2013) and methyl-jasmonate (Tiryaki & Staswick, 2002). axr1 mutant plants are expected to have lower sensitivity to cytokinins due to increased stability of the ARR5 protein, a negative regulator of cytokinin responses (Li, Kurepa & Smalle, 2013). We hypothesize that the reduced susceptibility to parasitization of the axr1 A. thaliana genotype is due to reduced sensitivity to ethylene (see below) as opposed to reduced sensitivity to cytokinins. Alternatively, reduced sensitivity to auxin signaling may explain the reduced susceptibility of axr1 mutants to P. aegyptiaca. Local auxin biosynthesis and response gene activation were recently implicated as essential for haustorial formation by the P. japonicum (Ishida et al., 2016). However, the auxin signaling mutants tir1 and aux1 were not differentially susceptible to formation of P. aegyptiaca tubercles.
Ethylene signaling
The ethylene signaling mutant lines, including ein2, did not support differential rates of P. aegyptiaca attachment and development compared to wildtype A. thaliana. Nevertheless, several observations suggest that ethylene signaling is an important part of host plant responses to P. aegyptiaca parasitism. First, host plants overexpressing Erf2, were substantially less susceptible to initial haustorial attachment by P. aegyptiaca (Fig. 2). Second, the dde2 mutant line (JA biosynthesis) exhibited substantially reduced susceptibility to P. aegyptiaca tubercle development, but the quad signaling mutant line (Tsuda et al., 2009) (dde2/pad4/sid2/ein2) was not less susceptible to parasitization. This result suggests that loss of function in SA signaling or ethylene signaling is sufficient to rescue the reduced susceptibility associated with defective host plant JA biosynthesis. Wildtype level P. aegyptiaca attachment rates were not rescued when infecting the double mutants dde2/pad4 and dde2/sid2 suggesting that the rescued attachment rate is due to the ein2 mutation in the dde2 background. We were not able to directly test this hypothesis because dde2/ein2 double mutant plants do not grow in the PE bag system for unknown reasons. Third, the resistant-to-parasitism mutant edr1 has perturbed ethylene signaling, specifically in the crosstalk of SA and ethylene signaling (Tang, Christiansen & Innes, 2005). However, the edr1 phenotype may be more predominant due to EDR1’s role as a negative regulator of plant immunity (see above). Given the high degree of crosstalk between JA and SA signaling with ethylene signaling (Koornneef & Pieterse, 2008), it will be challenging to determine the extent to which both of these host hormone-signaling pathways are independently required for successful P. aegyptiaca parasitization.
RNAi
RDR6 is involved in RNAi signaling and rdr6 mutant plants are less susceptible to late-stage P. aegyptiaca tubercle formation, suggesting that P. aegyptiaca may exploit host-derived RNAi pathways during parasitization. The distantly related parasitic plant Cuscuta campestris, for example, secretes sRNA molecules into host plant tissue to silence plant-immunity and auxin-signaling gene pathways (Shahid et al., 2018). However, other mutants deficient in RNAi signaling (dcl1and rdr1/2/6) are not less susceptible to P. aegyptiaca. Indeed, both dcl1 and rdr1/2/6 are among the most susceptible mutants tested in terms of late-stage tubercle development (Fig. 4). DCL1 is essential for most miRNA biogenesis (Bologna & Voinnet, 2014) and plays a role in the negative regulation in an RDR6-involved RNA-silencing pathway (Qu, Ye & Morris, 2008). It is possible that P. aegyptiaca exploits this RNA-silencing pathway during parasitization, hence dcl1 (negative regulator) mutants are highly susceptible and rdr6 mutants are significantly less susceptible to tubercle development. However, the differential susceptibility of the rdr6 and the rdr1/2/6 lines cannot be explained at this time. A more thorough examination is necessary to determine the role of RNAi in Arabidopsis–P. aegyptiaca interactions.
Pathways that appear not to be involved in susceptibility/resistance to P. aegyptiaca parasitization
It is important to note that the majority of tested mutant A. thaliana genotypes were not significantly different from wildtype in their susceptibility to P. aegyptiaca parasitization (Table S4). This result suggests that the ability of P. aegyptiaca to parasitize host plant tissue is robust, which is also supported by the relatively broad host range of P. aegyptiaca (Parker, 2013). Surprisingly, every tested A. thaliana mutant line was able to support some level of parasite attachment. In contrast, a recent similar screen of the obligate stem parasite Cuscuta reflexa on multiple tomato introgression lines revealed multiple genotypes where the host-parasite interaction was incompatible (Krause et al., 2018). We consider the lack of increased susceptibility of bak1-4 plants to be the most surprising result. We originally hypothesized that this mutant was the best candidate for a genotype more susceptible to parasitization because BAK1 is critical for many components of PTI (Chinchilla et al., 2009), which is a critical part of the plant immune response against many other classes of plant pests (Jones & Dangl, 2006) including the parasitic plant C. reflexa (Hegenauer et al., 2016). Future studies with the bak1-5 allele that specifically blocks innate immunity without impairing brassinosteroid signaling (Schwessinger et al., 2011) may be informative. mpk4 plants, which are deficient in map kinase signaling—another critical component of PTI (Rasmussen et al., 2012), were more susceptible to early attachment than wildtype (Fig. 2). But the increased susceptibility was not persistent through early and late stage P. aegyptiaca tubercle development. It is possible that P. aegyptiaca is already highly adept at subverting A. thaliana PTI. However, we show that the PTI marker gene WRKY53 is significantly upregulated in response to tubercle development (Fig. 5). Several of the borderline increased susceptibility genotypes, such as bak1-4, should be tested in the PE bag system using a related parasite such as O. minor that is largely incompatible on A. thaliana (Goldwasser, Plakhine & Yoder, 2000). Starting with an incompatible interaction will more readily enable discovery of host genes that underpin resistance to parasitism than the assay described here performed with the compatible P. aegyptiaca parasite.
Conclusion
This work demonstrates that the parasitization potential of P. aegyptiaca on A. thaliana is remarkably robust and not fully dependent on any of the genetic pathways tested here. However, P. aegyptiaca parasitization is at least partially limited in a small selection of A. thaliana mutant lines. The most robust phenotype revealed from this study is that multiple genes involved in JA signaling and biosynthesis are critical for full host plant susceptibility to P. aegyptiaca. Additionally, this work revealed that the putative immunity hub protein PFD6 is a critical component of the plant immune response that limits the severity of P. aegyptiaca parasitization. Further investigation into the identified genes that affect the degree of host plant susceptibility to parasitization will further elucidate the molecular mechanisms of plant parasitism. Alteration of these genetic pathways has the potential to help control parasitic weed infestation through either enhanced expression of resistance-associated genes or reduced expression of susceptibility-associated genes. Several of the susceptibility associated genes identified in this work cannot be knocked out without substantial negative impacts on the plants such as male sterility (e.g., dde2) or growth retardation (e.g., cpr5). This work serves as a starting point for understanding which genetic pathways are essential for plant susceptibility. Additionally, the viability of overexpressing PFD6 or other identified resistance-associated genes to control parasitic plant infection needs to be determined in future work.
Supplemental Information
Acknowledgments
We sincerely thank the Arabidopsis Biological Resource Center, John McDowell, Boris Vinatzer, Jeff Dangl, Ryan Anderson, Peter Morris, Nick Harberd, Georg Felix, Birgit Kemmerling, and Christian Voigt for providing seeds.
Funding Statement
This project was supported by the National Institute of Food and Agricultural through postdoctoral fellowship award 2015-67012-22821 to Christopher Clarke and award 135997 to James Westwood. Additional support was through the US National Science Foundation (IOS-1238057) to James Westwood. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Christopher R. Clarke, Email: christopher.clarke@usda.gov.
James Westwood, Email: westwood@vt.edu.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Christopher R. Clarke conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
So-Yon Park performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Robert Tuosto performed the experiments, prepared figures and/or tables, and approved the final draft.
Xiaoyan Jia performed the experiments, prepared figures and/or tables, and approved the final draft.
Amanda Yoder analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Jennifer Van Mullekom analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
James Westwood conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Data is available at the U.S. Department of Agriculture: Clarke, Christopher R.; Van Mullekom, Jennifer H.; Westwood, James H. (2019). Data from: Multiple immune pathways control susceptibility of Arabidopsis thaliana to the parasitic weed Phelipanche aegyptiaca. Ag Data Commons. DOI 10.15482/USDA.ADC/1503694. Accessed 13 May 2020.
References
- Alonso et al. (1999).Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284(5423):2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Alonso et al. (2003).Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Austin et al. (2002).Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE. Regulatory role of SGT1in early R gene-mediated plant defenses. Science. 2002;295(5562):2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- Bar-Nun & Mayer (2008).Bar-Nun N, Mayer AM. Methyl jasmonate and methyl salicylate, but not cis-jasmone, evoke defenses against infection of Arabidopsis thaliana by Orobanche aegyptiaca. Weed Biology and Management. 2008;8(2):91–96. doi: 10.1111/j.1445-6664.2008.00280.x. [DOI] [Google Scholar]
- Berger, Bell & Mullet (1996).Berger S, Bell E, Mullet JE. Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiology. 1996;111(2):525–531. doi: 10.1104/pp.111.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch et al. (1998).Boch J, Verbsky ML, Robertson TL, Larkin JC, Kunkel BN. Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Molecular Plant–Microbe Interactions. 1998;11(12):1196–1206. doi: 10.1094/MPMI.1998.11.12.1196. [DOI] [Google Scholar]
- Bologna & Voinnet (2014).Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annual Review of Plant Biology. 2014;65(1):473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- Bowling et al. (1997).Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9(9):1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catinot, Huang & Zimmerli (2015).Catinot J, Huang P-Y, Zimmerli L. Ethylene response factors in Arabidopsis immunity. Journal of Experimental Botany. 2015;67(5):1231–1241. doi: 10.1093/jxb/erv518. [DOI] [PubMed] [Google Scholar]
- Century, Holub & Staskawicz (1995).Century KS, Holub EB, Staskawicz BJ. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(14):6597–6601. doi: 10.1073/pnas.92.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla et al. (2009).Chinchilla D, Shan L, He P, De Vries S, Kemmerling B. One for all: the receptor-associated kinase BAK1. Trends in Plant Science. 2009;14(10):535–541. doi: 10.1016/j.tplants.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen et al. (2011).Christiansen KM, Gu Y, Rodibaugh N, Innes RW. Negative regulation of defence signalling pathways by the EDR1 protein kinase. Molecular Plant Pathology. 2011;12(8):746–758. doi: 10.1111/j.1364-3703.2011.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke et al. (2019).Clarke CR, Timko MP, Yoder JI, Axtell MJ, Westwood JH. Molecular dialog between parasitic plants and their hosts. Annual Review of Phytopathology. 2019;57(1):279–299. doi: 10.1146/annurev-phyto-082718-100043. [DOI] [PubMed] [Google Scholar]
- Consonni et al. (2006).Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, Somerville SC, Panstruga R. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nature Genetics. 2006;38(6):716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- Cui et al. (2017).Cui H, Gobbato E, Kracher B, Qiu J, Bautor J, Parker JE. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytologist. 2017;213(4):1802–1817. doi: 10.1111/nph.14302. [DOI] [PubMed] [Google Scholar]
- Die et al. (2007).Die JV, Dita MA, Krajinski F, González-Verdejo CI, Rubiales D, Moreno MT, Román B. Identification by suppression subtractive hybridization and expression analysis of Medicago truncatula putative defence genes in response to Orobanche crenata parasitization. Physiological and Molecular Plant Pathology. 2007;70(1–3):49–59. doi: 10.1016/j.pmpp.2007.06.001. [DOI] [Google Scholar]
- Dos Santos et al. (2003).Dos Santos CV, Letousey P, Delavault P, Thalouarn P. Defense gene expression analysis of Arabidopsis thaliana Parasitized by Orobanche ramosa. Phytopathology. 2003;93(4):451–457. doi: 10.1094/PHYTO.2003.93.4.451. [DOI] [PubMed] [Google Scholar]
- Duriez et al. (2019).Duriez P, Vautrin S, Auriac M-C, Bazerque J, Boniface M-C, Callot C, Carrère S, Cauet S, Chabaud M, Gentou F, Lopez-Sendon M, Paris C, Pegot-Espagnet P, Rousseaux J-C, Pérez-Vich B, Velasco L, Bergès H, Piquemal J, Muños S. A receptor-like kinase enhances sunflower resistance to Orobanche cumana. Nature Plants. 2019;5(12):1211–1215. doi: 10.1038/s41477-019-0556-z. [DOI] [PubMed] [Google Scholar]
- Ellinger et al. (2013).Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiology. 2013;161(3):1433–1444. doi: 10.1104/pp.112.211011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye & Innes (1998).Frye CA, Innes RW. An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell. 1998;10(6):947–956. doi: 10.1105/tpc.10.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz et al. (2010).Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010;22(2):481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, Rogers & Ausubel (1996).Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldwasser, Plakhine & Yoder (2000).Goldwasser Y, Plakhine D, Yoder JI. Arabidopsis thaliana susceptibility to Orobanche spp. Weed Science. 2000;48(3):342–346. doi: 10.1614/0043-1745(2000)048[0342:WBAE]2.0.CO;2. [DOI] [Google Scholar]
- Grant et al. (1995).Grant M, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R, Dangl J. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269(5225):843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Gu et al. (2008).Gu Y, Deng Z, Paredez AR, DeBolt S, Wang Z-Y, Somerville C. Prefoldin 6 is required for normal microtubule dynamics and organization in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(46):18064–18069. doi: 10.1073/pnas.0808652105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr & Paszkowski (2009).Gutjahr C, Paszkowski U. Weights in the balance: jasmonic acid and salicylic acid signaling in root-biotroph interactions. Molecular Plant–Microbe Interactions®. 2009;22(7):763–772. doi: 10.1094/MPMI-22-7-0763. [DOI] [PubMed] [Google Scholar]
- Hardham (2013).Hardham AR. Microtubules and biotic interactions. Plant Journal. 2013;75(2):278–289. doi: 10.1111/tpj.12171. [DOI] [PubMed] [Google Scholar]
- He et al. (2007).He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Current Biology. 2007;17(13):1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Hegenauer et al. (2016).Hegenauer V, Fürst U, Kaiser B, Smoker M, Zipfel C, Felix G, Stahl M, Albert M. Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science. 2016;353(6298):478–481. doi: 10.1126/science.aaf3919. [DOI] [PubMed] [Google Scholar]
- Hiraoka & Sugimoto (2008).Hiraoka Y, Sugimoto Y. Molecular responses of sorghum to purple witchweed (Striga hermonthica) parasitism. Weed Science. 2008;56(3):356–363, 358. doi: 10.1614/WS-07-136.1. [DOI] [Google Scholar]
- Hiraoka, Ueda & Sugimoto (2008).Hiraoka Y, Ueda H, Sugimoto Y. Molecular responses of Lotus japonicus to parasitism by the compatible species Orobanche aegyptiaca and the incompatible species Striga hermonthica. Journal of Experimental Botany. 2008;60(2):641–650. doi: 10.1093/jxb/ern316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving & Cameron (2009).Irving LJ, Cameron DD. Chapter 3 you are what you eat: interactions between root parasitic plants and their hosts. Advances in Botanical Research. 2009;50:87–138. [Google Scholar]
- Ishida et al. (2016).Ishida JK, Wakatake T, Yoshida S, Takebayashi Y, Kasahara H, Wafula E, De Pamphilis CW, Namba S, Shirasu K. Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell. 2016;28(8):1795–1814. doi: 10.1105/tpc.16.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs et al. (2011).Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, Kogel K-H, Schäfer P. Broad-spectrum suppression of innate immunity is required for colonization of arabidopsis roots by the fungus Piriformospora indica. Plant Physiology. 2011;156(2):726–740. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones & Dangl (2006).Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kaiser et al. (2015).Kaiser B, Vogg G, Fürst UB, Albert M. Parasitic plants of the genus Cuscuta and their interaction with susceptible and resistant host plants. Frontiers in Plant Science. 2015;6(227):45. doi: 10.3389/fpls.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaka et al. (2006).Kaminaka H, Näke C, Epple P, Dittgen J, Schütze K, Chaban C, Holt BF, Merkle T, Schäfer E, Harter K, Dangl JL. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO Journal. 2006;25(18):4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan & Manners (2008).Kazan K, Manners JM. Jasmonate signaling: toward an integrated view. Plant Physiology. 2008;146(4):1459–1468. doi: 10.1104/pp.107.115717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling et al. (2007).Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Müssig C, Thomma BPHJ, Albrecht C, De Vries SC, Hirt H, Nürnberger T. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Current Biology. 2007;17(13):1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Koorneef et al. (1985).Koorneef M, Elgersma A, Hanhart CJ, Van Loenen-Martinet EP, Van Rijn L, Zeevaart JAD. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiologia Plantarum. 1985;65(1):33–39. doi: 10.1111/j.1399-3054.1985.tb02355.x. [DOI] [Google Scholar]
- Koornneef & Pieterse (2008).Koornneef A, Pieterse CMJ. Cross talk in defense signaling. Plant Physiology. 2008;146(3):839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause et al. (2018).Krause K, Johnsen HR, Pielach A, Lund L, Fischer K, Rose JKC. Identification of tomato introgression lines with enhanced susceptibility or resistance to infection by parasitic giant dodder (Cuscuta reflexa) Physiologia Plantarum. 2018;162(2):205–218. doi: 10.1111/ppl.12660. [DOI] [PubMed] [Google Scholar]
- Krol et al. (2010).Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KAS, Becker D, Hedrich R. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. Journal of Biological Chemistry. 2010;285(18):13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto et al. (2007).Kusumoto D, Goldwasser Y, Xie X, Yoneyama K, Takeuchi Y, Yoneyama K. Resistance of red clover (Trifolium pratense) to the root parasitic plant Orobanche minor is activated by salicylate but not by jasmonate. Annals of botany. 2007;100(3):537–544. doi: 10.1093/aob/mcm148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton (1995).Lawton K. Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Molecular Plant–Microbe Interactions. 1995;8(6):863–870. doi: 10.1094/MPMI-8-0863. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2012).Lee AH-Y, Hurley B, Felsensteiner C, Yea C, Ckurshumova W, Bartetzko V, Wang PW, Quach V, Lewis JD, Liu YC, Börnke F, Angers S, Wilde A, Guttman DS, Desveaux D. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLOS Pathogens. 2012;8(2):e1002523. doi: 10.1371/journal.ppat.1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Kurepa & Smalle (2013).Li Y, Kurepa J, Smalle J. AXR1 promotes the Arabidopsis cytokinin response by facilitating ARR5 proteolysis. Plant Journal. 2013;74(1):13–24. doi: 10.1111/tpj.12098. [DOI] [PubMed] [Google Scholar]
- Li & Timko (2009).Li J, Timko MP. Gene-for-gene resistance in striga-cowpea associations. Science. 2009;325(5944):1094–1094. doi: 10.1126/science.1174754. [DOI] [PubMed] [Google Scholar]
- Lincoln, Britton & Estelle (1990).Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2(11):1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka et al. (2005).Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, Llorente F, Molina A, Parker J, Somerville S, Schulze-Lefert P. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310(5751):1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- Lorenzo et al. (2003).Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15(1):165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason et al. (2005).Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17(11):3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero-Ruiz et al. (2015).Molinero-Ruiz L, Delavault P, Pérez-Vich B, Pacureanu-Joita M, Bulos M, Altieri E, Domínguez J. History of the race structure of Orobanche cumana and the breeding of sunflower for resistance to this parasitic weed: a review. Spanish Journal of Agricultural Research. 2015;13(4):e10R01. doi: 10.5424/sjar/2015134-8080. [DOI] [Google Scholar]
- Mukhtar et al. (2011).Mukhtar MS, Carvunis A-R, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, Pevzner SJ, Donovan SE, Ghamsari L, Santhanam B, Romero V, Poulin MM, Gebreab F, Gutierrez BJ, Tam S, Monachello D, Boxem M, Harbort CJ, McDonald N, Gai L, Chen H, He Y, Vandenhaute J, Roth FP, Hill DE, Ecker JR, Vidal M, Beynon J, Braun P, Dangl JL. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333(6042):596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett et al. (2002).Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sadanandom A, Shirasu K, Jones JDG, Parker JE. Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell. 2002;14(5):979–992. doi: 10.1105/tpc.001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro et al. (2008).Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Current Biology. 2008;18(9):650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- Nawrath & Métraux (1999).Nawrath C, Métraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen et al. (2016).Nguyen KH, Ha CV, Nishiyama R, Watanabe Y, Leyva-González MA, Fujita Y, Tran UT, Li W, Tanaka M, Seki M, Schaller GE, Herrera-Estrella L, Tran L-SP. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:3090–3095. doi: 10.1073/pnas.1600399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov et al. (2008).Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JAH, Palme K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Molecular Plant. 2008;1(2):321–337. doi: 10.1093/mp/ssm021. [DOI] [PubMed] [Google Scholar]
- Parker (2013).Parker C. The parasitic weeds of the Orobanchaceae. In: Joel DM, Gressel J, Musselman LJ, editors. Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Berlin: Springer; 2013. pp. 313–344. [Google Scholar]
- Parker et al. (1996).Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8(11):2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels & Goossens (2011).Pauwels L, Goossens A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23(9):3089–3100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju et al. (2007).Pegadaraju V, Louis J, Singh V, Reese JC, Bautor J, Feys BJ, Cook G, Parker JE, Shah J. Phloem-based resistance to green peach aphid is controlled by Arabidopsis PHYTOALEXIN DEFICIENT4 without its signaling partner ENHANCED DISEASE SUSCEPTIBILITY1. Plant Journal. 2007;52(2):332–341. doi: 10.1111/j.1365-313X.2007.03241.x. [DOI] [PubMed] [Google Scholar]
- Peragine et al. (2004).Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes & Development. 2004;18(19):2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, Wilson & Estelle (1990).Pickett FB, Wilson AK, Estelle M. The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiology. 1990;94(3):1462–1466. doi: 10.1104/pp.94.3.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu et al. (2008).Qiu J-L, Zhou L, Yun B-W, Nielsen HB, Fiil BK, Petersen K, MacKinlay J, Loake GJ, Mundy J, Morris PC. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiology. 2008;148(1):212–222. doi: 10.1104/pp.108.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Ye & Morris (2008).Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen et al. (2012).Rasmussen MW, Roux M, Petersen M, Mundy J. MAP kinase cascades in Arabidopsis innate immunity. Frontiers in Plant Science. 2012;3:169. doi: 10.3389/fpls.2012.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Beers, Pruitt & Gasser (1992).Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992;4(10):1237–1249. doi: 10.2307/3869410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubiales, Rojas-Molina & Sillero (2016).Rubiales D, Rojas-Molina MM, Sillero JC. Characterization of resistance mechanisms in faba bean (Vicia faba) against Broomrape Species (Orobanche and Phelipanche spp.) Frontiers in Plant Science. 2016;7(1409):1747. doi: 10.3389/fpls.2016.01747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger et al. (1998).Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes & Development. 1998;12(2):198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci et al. (2001).Rustérucci C, Aviv DH, Holt BF, III, Dangl JL, Parker JE. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell. 2001;13(10):2211–2224. doi: 10.1105/tpc.010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šašek et al. (2014).Šašek V, Janda M, Delage E, Puyaubert J, Guivarc’h A, López Maseda E, Dobrev PI, Caius J, Bóka K, Valentová O, Burketová L, Zachowski A, Ruelland E. Constitutive salicylic acid accumulation in pi4kIIIβ1β2 Arabidopsis plants stunts rosette but not root growth. New Phytologist. 2014;203(3):805–816. doi: 10.1111/nph.12822. [DOI] [PubMed] [Google Scholar]
- SAS/STAT(R) 9.2 User’s Guide SE (2016).SAS/STAT(R) 9.2 User’s Guide SE Overview: GLIMMIX Procedure. 2016. https://support.sas.com/documentation/cdl/en/statug/63033/HTML/default/viewer.htm#statug_glimmix_a0000001394.htm https://support.sas.com/documentation/cdl/en/statug/63033/HTML/default/viewer.htm#statug_glimmix_a0000001394.htm
- Schwessinger et al. (2011).Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLOS Genetics. 2011;7(4):e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid et al. (2018).Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, De Pamphilis CW, Westwood JH, Axtell MJ. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature. 2018;553(7686):82–85. doi: 10.1038/nature25027. [DOI] [PubMed] [Google Scholar]
- Spallek et al. (2017).Spallek T, Melnyk CW, Wakatake T, Zhang J, Sakamoto Y, Kiba T, Yoshida S, Matsunaga S, Sakakibara H, Shirasu K. Interspecies hormonal control of host root morphology by parasitic plants. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(20):5283–5288. doi: 10.1073/pnas.1619078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek, Mutuku & Shirasu (2013).Spallek T, Mutuku M, Shirasu K. The genus Striga: a witch profile. Molecular Plant Pathology. 2013;14(9):861–869. doi: 10.1111/mpp.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, Su & Howell (1992).Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(15):6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Christiansen & Innes (2005).Tang D, Christiansen KM, Innes RW. Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiology. 2005;138(2):1018–1026. doi: 10.1104/pp.105.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma et al. (2001).Thomma BPHJ, Penninckx IAMA, Cammue BPA, Broekaert WF. The complexity of disease signaling in Arabidopsis. Current Opinion in Immunology. 2001;13(1):63–68. doi: 10.1016/S0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- Timpte et al. (1995).Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant Journal. 1995;8(4):561–569. doi: 10.1046/j.1365-313X.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Tiryaki & Staswick (2002).Tiryaki I, Staswick PE. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiology. 2002;130(2):887–894. doi: 10.1104/pp.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, Chen & Wang (2017).Tran T, Chen S, Wang X. Root assays to study pattern-triggered immunity in plant-nematode interactions. European Journal of Plant Pathology. 2017;147(4):955–961. doi: 10.1007/s10658-016-1053-0. [DOI] [Google Scholar]
- Tsuda et al. (2009).Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLOS Genetics. 2009;5(12):e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schie & Takken (2014).Van Schie CCN, Takken FLW. Susceptibility genes 101: how to be a good host. Annual Review of Phytopathology. 2014;52(1):551–581. doi: 10.1146/annurev-phyto-102313-045854. [DOI] [PubMed] [Google Scholar]
- Von Malek et al. (2002).Von Malek B, Van der Graaff E, Schneitz K, Keller B. The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta. 2002;216(1):187–192. doi: 10.1007/s00425-002-0906-2. [DOI] [PubMed] [Google Scholar]
- Westwood (2000).Westwood JH. Characterization of the Orobanche–Arabidopsis system for studying parasite-host interactions. Weed Science. 2000;48(6):742–748. doi: 10.1614/0043-1745(2000)048[0742:COTOAS]2.0.CO;2. [DOI] [Google Scholar]
- Westwood et al. (2012).Westwood JH, De Pamphilis CW, Das M, Fernández-Aparicio M, Honaas LA, Timko MP, Wafula EK, Wickett NJ, Yoder JI. The parasitic plant genome project: new tools for understanding the biology of Orobanche and Striga. Weed Science. 2012;60(2):295–306. doi: 10.1614/WS-D-11-00113.1. [DOI] [Google Scholar]
- Yan et al. (2014).Yan H, Yoo M-J, Koh J, Liu L, Chen Y, Acikgoz D, Wang Q, Chen S. Molecular reprogramming of Arabidopsis in response to perturbation of jasmonate signaling. Journal of Proteome Research. 2014;13(12):5751–5766. doi: 10.1021/pr500739v. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2014).Yang Z, Wafula EK, Honaas LA, Zhang H, Das M, Fernandez-Aparicio M, Huang K, Bandaranayake PCG, Wu B, Der JP, Clarke CR, Ralph PE, Landherr L, Altman NS, Timko MP, Yoder JI, Westwood JH, De Pamphilis CW. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Molecular Biology and Evolution. 2014;32(3):767–790. doi: 10.1093/molbev/msu343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama et al. (2010).Yoneyama K, Awad AA, Xie X, Yoneyama K, Takeuchi Y. Strigolactones as germination stimulants for root parasitic plants. Plant and Cell Physiology. 2010;51(7):1095–1103. doi: 10.1093/pcp/pcq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida et al. (2016).Yoshida S, Cui S, Ichihashi Y, Shirasu K. The haustorium, a specialized invasive organ in parasitic plants. Annual Review of Plant Biology. 2016;67(1):643–667. doi: 10.1146/annurev-arplant-043015-111702. [DOI] [PubMed] [Google Scholar]
- Yoshida et al. (2002).Yoshida S, Ito M, Nishida I, Watanabe A. Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant Journal. 2002;29(4):427–437. doi: 10.1046/j.0960-7412.2001.01228.x. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang L, Du L, Shen C, Yang Y, Poovaiah BW. Regulation of plant immunity through ubiquitin-mediated modulation of Ca 2+ -calmodulin-AtSR1/CAMTA3 signaling. Plant Journal. 2014;78(2):269–281. doi: 10.1111/tpj.12473. [DOI] [PubMed] [Google Scholar]
- Zwanenburg, Pospíšil & Ćavar Zeljković (2016).Zwanenburg B, Pospíšil T, Ćavar Zeljković S. Strigolactones: new plant hormones in action. Planta. 2016;243(6):1311–1326. doi: 10.1007/s00425-015-2455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
Data is available at the U.S. Department of Agriculture: Clarke, Christopher R.; Van Mullekom, Jennifer H.; Westwood, James H. (2019). Data from: Multiple immune pathways control susceptibility of Arabidopsis thaliana to the parasitic weed Phelipanche aegyptiaca. Ag Data Commons. DOI 10.15482/USDA.ADC/1503694. Accessed 13 May 2020.