Abstract
Background
The basic/helix-loop-helix (bHLH) transcription factor family exists in all three eukaryotic kingdoms as important participants in biological growth and development. To date, the comprehensive genomic and functional analyses of bHLH genes has not been reported in cucumber (Cucumis sativus L.).
Results
Here, a total of 142 bHLH genes were identified and classified into 32 subfamilies according to the conserved motifs, phylogenetic analysis and gene structures in cucumber. The sequences of CsbHLH proteins were highly conserved based on the results of multiple sequence alignment analyses. The chromosomal distribution, synteny analysis, and gene duplications of these 142 CsbHLHs were further analysed. Many elements related to stress responsiveness and plant hormones were present in the promoter regions of CsbHLH genes based on a cis-element analysis. By comparing the phylogeny of cucumber and Arabidopsis bHLH proteins, we found that cucumber bHLH proteins were clustered into different functional clades of Arabidopsis bHLH proteins. The expression analysis of selected CsbHLHs under abiotic stresses (NaCl, ABA and low-temperature treatments) identified five CsbHLH genes that could simultaneously respond to the three abiotic stresses. Tissue-specific expression profiles of these five genes were also analysed. In addition, 35S:CsbHLH041 enhanced the tolerance to salt and ABA in transgenic Arabidopsis and in cucumber seedlings, suggesting CsbHLH041 is an important regulator in response to abiotic stresses. Lastly, the functional interoperability network among the CsbHLH proteins was analysed.
Conclusion
This study provided a good foundation for further research into the functions and regulatory mechanisms of CsbHLH proteins and identified candidate genes for stress resistance in cucumber.
Keywords: Abiotic stresses, bHLH family, Cucumber, CsbHLH041, Expression patterns, Regulatory networks
Background
Basic helix-loop-helix (bHLH) transcription factors form one of the largest families of TFs and exist widely in all three eukaryotic kingdoms [1, 2]. The bHLH TFs are named for their own structural characteristics [3], which are mostly composed of conserved 60 amino acid residues. According to the different functions, they can be divided into two parts: the basic region and the HLH region [4]. The basic region is distributed at the N-terminus of the bHLH conserved domain and contains approximately 15 to 20 residues, which are related to DNA binding [5, 6]. The HLH domain is distributed at the C-terminus of the gene sequence, composing of two amphipathic α-helices mainly constituting of hydrophobic residues linked by a loop region of variable sequence and length. The HLH domain is an essential structure for the formation of homologous or heterologous dimers in bHLH TFs [6, 7].
According to the evolutionary origin, sequence similarity, DNA binding patterns, and functional types, in animals, bHLH transcription factors are mainly divided into six categories, A-F, containing 45 subgroups [8, 9]. In plants, the bHLH gene family has been divided into 15–26 groups [10], and even up to 32 groups when atypical bHLH proteins are included [2]. In Arabidopsis, 167 bHLH proteins are divided into 21 subfamilies [2, 11]; the 165 bHLH family members in rice are classified into 22 subfamilies [12]; and the 159 bHLH proteins are divided into 21 subfamilies in tomato [13]. Currently, increasing numbers of bHLH proteins have been found in plants, and their functional research is gradually increasing.
In plants, the bHLH genes are involved in processes such as metabolic regulation, plant growth and development, and response to environmental signals. The first member of the bHLH family discovered was the maize R gene, which was shown to play a key role in anthocyanin synthesis [14]. Subsequently, an increasing number of bHLHs have been shown to be involved in a wider range of physiological pathways. For example, Phytochrome Interacting Factors (PIFs) have been reported to respond to light signals [15]; overexpression of PRE1 activates gibberellin-dependent responses in Arabidopsis thaliana [16]; AtGL3, AtEGL3 and AtTT8 have been demonstrated to be involved in anthocyanin and PA biosynthesis [17, 18]; while AtGL3, AtEGL3 and AtMYC1 also regulate trichome formation and root hair patterning [19]. In addition, some bHLH TFs are also considered to be able to respond to a variety of abiotic stresses and improve plant stress tolerance, including tolerance to drought tolerance, salt and cold. In wheat, overexpression of bHLH39 increases tolerance to salt stress [20]. The bHLH TFs often function by forming homodimers or heterodimers with other proteins. For example, MYC3 and MYC4 transcription factors all can interact with multiple JAZ proteins (such as JAZ1, JAZ4, and JAZ9) to jointly regulate the JA signalling pathway [21]. The MYB-bHLH-WD40 complexes are involved in different processes, such as the biosynthesis of anthocyanins and PAs, leaf trichome formation and root hair patterning [22]. In summary, bHLH in plants can form homologous or heterologous complexes with bHLH proteins or other proteins to extend their biological functions.
Cucumber (Cucumis sativus L.) is an economically important crop cultivated worldwide [23]. The functions of the AtbHLH family have been widely studied in Arabidopsis thaliana [2]. However, genome-wide information on members of the CsbHLH family has not been reported. In this study, we identified and characterized 142 bHLH family genes in cucumber. They were classified into 32 subgroups and could be distributed over seven chromosomes. Their gene structures, conserved motifs, synteny analysis, gene duplications and cis-elements in promoters also have been investigated. In addition, the expression levels of some CsbHLH genes were measured by qRT-PCR to study their responses to low temperature (4 °C), salt (NaCl) and ABA stress, for which all tested genes were stress-responsive. The protein interaction network among the CsbHLH proteins was predicted, which could help to understand the possible functional mechanism of CsbHLH proteins. Furthermore, overexpression of CsbHLH041 showed increased salt resistance and ABA resistance compared with controls in cucumber and Arabidopsis. We hope that this work will provide useful resources for further studies on the functions and regulatory mechanisms of a potentially important CsbHLH protein, which plays a crucial role in the regulation of abiotic stress responses in cucumber.
Results
Identification and analysis of cucumber bHLH genes
To identify CsbHLH family genes in cucumber, we used the BlastP programme to search against the cucumber genome database by using 166 Arabidopsis bHLH proteins [2, 10] and the consensus protein sequences of the bHLH domain, with Hidden Markov Model (HMM) profile (PF00010) as queries. We obtained 164 putative members of the CsbHLH family. To confirm the reliability of the bHLH genes in the cucumber genome, we used Pfam (http://pfam.janelia.org/) and SMART (http://smart.embl-heidelberg.de/) [24] to search for the presence of the bHLH domain in the amino acid sequences of the 164 proteins. Only 142 proteins had the corresponding conserved bHLH domain, which were named CsbHLH1 to CsbHLH142 according to their sequence similarity and phylogenies with individual AtbHLH proteins. Finally, the specific information for the 142 typical bHLH genes, including the gene ID, amino acids length, chromosomal locations, and gene length were present in Table 1. The lengths of the CsbHLH protein sequences varied from 84 residues (CsaV3_1G005290) to 960 residues (CsaV3_1G043790), and the isoelectric points (pI) varied from 4.57 (CsaV3_2G030090) to 11.79 (CsaV3_6G028530).
Table 1.
bHLH genes in Cucumber
| CsbHLH | Gene ID | Location | Gene length | Amino acid length | pI |
|---|---|---|---|---|---|
| 001 | CsaV3_1G005290 | Chr1:3503806–3,504,411 | 1909 | 84 | 5.04 |
| 002 | CsaV3_1G011300 | Chr1:6972358–6,976,069 | 4167 | 236 | 5.78 |
| 003 | CsaV3_3G022420 | Chr3:19737551–19,739,874 | 958 | 502 | 5.7 |
| 004 | CsaV3_3G049150 | Chr3:40071324–40,074,834 | 1400 | 689 | 5.11 |
| 005 | CsaV3_3G001710 | Chr3:1295970–1,297,898 | 5601 | 643 | 6.21 |
| 006 | CsaV3_3G000850 | Chr3:656062–658,628 | 6057 | 448 | 8.65 |
| 007 | CsaV3_1G039580 | Chr1:24945063–24,953,384 | 1684 | 319 | 5.78 |
| 008 | CsaV3_2G007370 | Chr2:3725743–3,731,272 | 605 | 707 | 6.09 |
| 009 | CsaV3_4G032110 | Chr4:22635255–22,641,052 | 3486 | 551 | 6.25 |
| 010 | CsaV3_1G043790 | Chr1:28804330–28,811,620 | 6167 | 960 | 6.4 |
| 011 | CsaV3_2G015700 | Chr2:13017831–13,023,291 | 1802 | 336 | 5.64 |
| 012 | CsaV3_6G000530 | Chr6:351129–356,108 | 2050 | 645 | 5.51 |
| 013 | CsaV3_3G007980 | Chr3:6919411–6,921,670 | 2306 | 650 | 5.83 |
| 014 | CsaV3_2G010120 | Chr2:6877325–6,878,870 | 2524 | 323 | 6.02 |
| 015 | CsaV3_7G025510 | Chr7:14980031–14,984,292 | 3589 | 533 | 6.06 |
| 016 | CsaV3_6G009090 | Chr6:7311297–7,315,687 | 3711 | 486 | 6.35 |
| 017 | CsaV3_2G028950 | Chr2:18953613–18,956,324 | 3528 | 348 | 8.3 |
| 018 | CsaV3_7G007460 | Chr7:4647975–4,650,800 | 3316 | 335 | 6.64 |
| 019 | CsaV3_6G044570 | Chr6:26373287–26,375,739 | 3723 | 330 | 5.3 |
| 020 | CsaV3_6G044560 | Chr6:26366139–26,368,813 | 11,538 | 309 | 5.81 |
| 021 | CsaV3_5G026500 | Chr5:21650877–21,655,135 | 2678 | 624 | 5.04 |
| 022 | CsaV3_6G044730 | Chr6:26485325–26,487,050 | 1415 | 342 | 4.86 |
| 023 | CsaV3_2G030090 | Chr2:19685080–19,689,447 | 4059 | 363 | 4.57 |
| 024 | CsaV3_6G043370 | Chr6:25541506–25,544,586 | 8321 | 416 | 5.2 |
| 025 | CsaV3_6G044580 | Chr6:26382822–26,384,588 | 3478 | 276 | 6.05 |
| 026 | CsaV3_2G035250 | Chr2:23586273–23,591,605 | 3855 | 239 | 6.77 |
| 027 | CsaV3_2G008770 | Chr2:5179234–5,186,154 | 7290 | 246 | 4.92 |
| 028 | CsaV3_6G008940 | Chr6:7177946–7,179,613 | 5695 | 432 | 5.42 |
| 029 | CsaV3_6G014370 | Chr6:10430376–10,432,021 | 6113 | 308 | 4.98 |
| 030 | CsaV3_5G033960 | Chr5:27092009–27,094,880 | 4184 | 372 | 5.77 |
| 031 | CsaV3_6G036080 | Chr6:20032486–20,036,831 | 1493 | 242 | 9.03 |
| 032 | CsaV3_1G033410 | Chr1:20481133–20,483,811 | 5529 | 256 | 9.24 |
| 033 | CsaV3_1G009900 | Chr1:6174783–6,178,372 | 6920 | 551 | 5.6 |
| 034 | CsaV3_2G001440 | Chr2:370393–376,506 | 1545 | 543 | 8.37 |
| 035 | CsaV3_1G031920 | Chr1:18957508–18,969,046 | 2275 | 242 | 5.03 |
| 036 | CsaV3_7G004510 | Chr7:3234760–3,236,331 | 1320 | 211 | 6.77 |
| 037 | CsaV3_4G034440 | Chr4:24394485–24,395,550 | 3489 | 239 | 7.1 |
| 038 | CsaV3_4G029740 | Chr4:19342473–19,344,831 | 5460 | 253 | 6.17 |
| 039 | CsaV3_3G000950 | Chr3:733695–738,681 | 1011 | 773 | 5.11 |
| 040 | CsaV3_2G026610 | Chr2:18201502–18,202,748 | 2627 | 240 | 7.26 |
| 041 | CsaV3_1G040580 | Chr1:25826012–25,829,490 | 1816 | 492 | 7.03 |
| 042 | CsaV3_6G037080 | Chr6:20849605–20,855,801 | 4137 | 651 | 5.91 |
| 043 | CsaV3_6G041730 | Chr6:24304724–24,306,996 | 1246 | 235 | 6.42 |
| 044 | CsaV3_1G003910 | Chr1:2423148–2,424,832 | 2711 | 266 | 6.87 |
| 045 | CsaV3_3G013690 | Chr3:10293079–10,294,740 | 2245 | 191 | 8.64 |
| 046 | CsaV3_1G006280 | Chr1:4002735–4,008,902 | 4367 | 566 | 9.01 |
| 047 | CsaV3_1G002260 | Chr1:1450554–1,451,954 | 1411 | 261 | 6.16 |
| 048 | CsaV3_3G039100 | Chr3:32125102–32,130,846 | 2115 | 370 | 5.74 |
| 049 | CsaV3_5G033600 | Chr5:26846039–26,850,063 | 5332 | 571 | 6.09 |
| 050 | CsaV3_1G006650 | Chr1:4277323–4,279,125 | 2566 | 279 | 6.1 |
| 051 | CsaV3_6G001900 | Chr6:1303975–1,307,218 | 4986 | 248 | 9.26 |
| 052 | CsaV3_1G037610 | Chr1:23567571–23,568,986 | 1928 | 309 | 4.78 |
| 053 | CsaV3_6G037070 | Chr6:20836169–20,845,986 | 5647 | 263 | 6.08 |
| 054 | CsaV3_2G026190 | Chr2:17969523–17,971,339 | 1844 | 330 | 5.15 |
| 055 | CsaV3_3G034600 | Chr3:29292016–29,296,383 | 2259 | 695 | 5.24 |
| 056 | CsaV3_3G044120 | Chr3:35995420–35,997,451 | 4852 | 272 | 5.09 |
| 057 | CsaV3_2G005070 | Chr2:2751170–2,752,663 | 3663 | 318 | 5.74 |
| 058 | CsaV3_2G014750 | Chr2:12321166–12,324,655 | 1661 | 343 | 6.2 |
| 059 | CsaV3_7G027630 | Chr7:17195587–17,199,883 | 1509 | 317 | 6 |
| 060 | CsaV3_2G025890 | Chr2:17778099–17,780,726 | 3295 | 342 | 6.26 |
| 061 | CsaV3_2G030500 | Chr2:20027274–20,029,389 | 2181 | 372 | 4.71 |
| 062 | CsaV3_3G015900 | Chr3:11810548–11,813,843 | 2822 | 547 | 6.88 |
| 063 | CsaV3_7G000080 | Chr7:185656–189,146 | 2627 | 457 | 5.85 |
| 064 | CsaV3_1G000190 | Chr1:132006–133,915 | 2323 | 276 | 7.03 |
| 065 | CsaV3_3G028610 | Chr3:25161463–25,169,489 | 3059 | 540 | 5.7 |
| 066 | CsaV3_2G003660 | Chr2:1833037–1,837,221 | 6474 | 422 | 6.1 |
| 067 | CsaV3_4G002800 | Chr4:1745825–1,749,669 | 8026 | 168 | 7.65 |
| 068 | CsaV3_1G045830 | Chr1:31650489–31,656,184 | 4367 | 395 | 6.39 |
| 069 | CsaV3_4G026430 | Chr4:15704947–15,706,451 | 3065 | 196 | 6.51 |
| 070 | CsaV3_3G007090 | Chr3:6381666–6,383,510 | 5744 | 359 | 5.94 |
| 071 | CsaV3_3G049050 | Chr3:40003473–40,010,806 | 1802 | 322 | 7.73 |
| 072 | CsaV3_1G005810 | Chr1:3727778–3,731,264 | 2031 | 443 | 9.02 |
| 073 | CsaV3_2G026540 | Chr2:18154564–18,158,701 | 3855 | 380 | 5.01 |
| 074 | CsaV3_4G035310 | Chr4:24884098–24,888,333 | 2900 | 403 | 5.7 |
| 075 | CsaV3_3G020750 | Chr3:16961726–16,964,548 | 7333 | 248 | 7.09 |
| 076 | CsaV3_5G018750 | Chr5:14293739–14,297,926 | 3510 | 534 | 5.2 |
| 077 | CsaV3_4G034660 | Chr4:24537693–24,539,435 | 1700 | 409 | 6.38 |
| 078 | CsaV3_7G026520 | Chr7:16047841–16,051,016 | 3844 | 490 | 6.04 |
| 079 | CsaV3_1G001960 | Chr1:1286661–1,290,828 | 5186 | 196 | 7.64 |
| 080 | CsaV3_5G040480 | Chr5:31836009–31,840,665 | 2228 | 245 | 5.67 |
| 081 | CsaV3_6G002130 | Chr6:1472037–1,473,566 | 1504 | 161 | 5.12 |
| 082 | CsaV3_1G028780 | Chr1:15708212–15,711,935 | 2358 | 423 | 6.34 |
| 083 | CsaV3_5G024030 | Chr5:18711066–18,713,891 | 3741 | 285 | 6.21 |
| 084 | CsaV3_4G000380 | Chr4:228607–230,307 | 5797 | 342 | 4.89 |
| 085 | CsaV3_4G034980 | Chr4:24696914–24,698,535 | 1065 | 245 | 6.18 |
| 086 | CsaV3_3G014190 | Chr3:10642450–10,643,959 | 1742 | 203 | 7.81 |
| 087 | CsaV3_7G035000 | Chr7:22142359–22,144,409 | 1621 | 394 | 6.54 |
| 088 | CsaV3_2G016810 | Chr2:14091802–14,092,813 | 4235 | 216 | 9.77 |
| 089 | CsaV3_6G012850 | Chr6:8973920–8,976,038 | 2826 | 262 | 9.23 |
| 090 | CsaV3_5G031540 | Chr5:25725825–25,732,273 | 1729 | 679 | 7.3 |
| 091 | CsaV3_2G011050 | Chr2:8296805–8,299,080 | 1004 | 496 | 5.5 |
| 092 | CsaV3_2G013060 | Chr2:10626443–10,627,763 | 5030 | 280 | 9.14 |
| 093 | CsaV3_3G048260 | Chr3:39394424–39,397,324 | 4187 | 326 | 4.94 |
| 094 | CsaV3_1G039160 | Chr1:24662874–24,666,933 | 2825 | 359 | 5.94 |
| 095 | CsaV3_7G008580 | Chr7:5322491–5,324,405 | 2588 | 254 | 7.22 |
| 096 | CsaV3_2G029940 | Chr2:19575197–19,577,442 | 4258 | 308 | 6.01 |
| 097 | CsaV3_3G011010 | Chr3:8707121–8,711,973 | 6448 | 382 | 5.08 |
| 098 | CsaV3_3G021970 | Chr3:19073471–19,076,098 | 2460 | 377 | 5.08 |
| 099 | CsaV3_6G028530 | Chr6:16812784–16,815,830 | 2030 | 207 | 11.79 |
| 100 | CsaV3_6G037460 | Chr6:21181685–21,184,516 | 4024 | 299 | 5.66 |
| 101 | CsaV3_4G029750 | Chr4:19361035–19,364,776 | 2871 | 211 | 9.21 |
| 102 | CsaV3_6G033930 | Chr6:18737868–18,742,274 | 1923 | 333 | 5.71 |
| 103 | CsaV3_6G036240 | Chr6:20141242–20,142,683 | 4656 | 97 | 9.18 |
| 104 | CsaV3_3G012210 | Chr3:9421374–9,425,037 | 4979 | 227 | 5.7 |
| 105 | CsaV3_3G022870 | Chr3:20405212–20,408,271 | 3243 | 236 | 6.13 |
| 106 | CsaV3_3G042970 | Chr3:34884792–34,886,594 | 1529 | 244 | 8.4 |
| 107 | CsaV3_6G018830 | Chr6:13512926–13,515,085 | 1667 | 253 | 7.06 |
| 108 | CsaV3_2G030310 | Chr2:19895721–19,897,132 | 4390 | 240 | 9.2 |
| 109 | CsaV3_1G002670 | Chr1:1668618–1,674,219 | 2118 | 359 | 8.65 |
| 110 | CsaV3_3G045440 | Chr3:37108680–37,112,535 | 1645 | 430 | 6.69 |
| 111 | CsaV3_5G012430 | Chr5:7900420–7,905,450 | 2159 | 451 | 6.33 |
| 112 | CsaV3_1G011460 | Chr1:7106592–7,110,120 | 3046 | 360 | 4.75 |
| 113 | CsaV3_7G008090 | Chr7:5057906–5,059,973 | 4406 | 255 | 6.11 |
| 114 | CsaV3_5G026380 | Chr5:21538651–21,541,239 | 4345 | 173 | 9.15 |
| 115 | CsaV3_UNG229040 | scaffold115:93241–95,547 | 1441 | 274 | 9.42 |
| 116 | CsaV3_3G027730 | Chr3:24067204–24,073,678 | 9817 | 529 | 5.88 |
| 117 | CsaV3_7G003870 | Chr7:2853486–2,854,424 | 6196 | 313 | 5.32 |
| 118 | CsaV3_3G016560 | Chr3:12365460–12,367,641 | 2831 | 253 | 8.6 |
| 119 | CsaV3_1G012350 | Chr1:7668571–7,671,887 | 2272 | 205 | 6.16 |
| 120 | CsaV3_5G003430 | Chr5:2204438–2,205,442 | 3080 | 256 | 7.75 |
| 121 | CsaV3_6G047120 | Chr6:27815147–27,818,761 | 2674 | 337 | 6.13 |
| 122 | CsaV3_1G042640 | Chr1:27559193–27,563,048 | 2452 | 438 | 7.72 |
| 123 | CsaV3_3G005540 | Chr3:4716554–4,722,201 | 1766 | 439 | 6.41 |
| 124 | CsaV3_6G046660 | Chr6:27537459–27,541,954 | 1725 | 357 | 8.44 |
| 125 | CsaV3_5G003420 | Chr5:2191536–2,193,265 | 1497 | 261 | 5.59 |
| 126 | CsaV3_5G003410 | Chr5:2180337–2,183,163 | 4495 | 252 | 7.01 |
| 127 | CsaV3_6G049510 | Chr6:28902722–28,905,982 | 3614 | 405 | 5.35 |
| 128 | CsaV3_4G003860 | Chr4:2368080–2,373,266 | 3260 | 357 | 8.51 |
| 129 | CsaV3_7G031270 | Chr7:19779388–19,783,470 | 749 | 420 | 8.28 |
| 130 | CsaV3_3G039080 | Chr3:32107556–32,110,621 | 3490 | 367 | 9.16 |
| 131 | CsaV3_6G045070 | Chr6:26672336–26,673,833 | 938 | 229 | 10.26 |
| 132 | CsaV3_6G051560 | Chr6:29996501–29,997,250 | 1571 | 250 | 5.66 |
| 133 | CsaV3_7G027460 | Chr7:17033739–17,042,484 | 2825 | 692 | 5.66 |
| 134 | CsaV3_5G037950 | Chr5:30085680–30,087,603 | 1391 | 92 | 9.09 |
| 135 | CsaV3_1G002240 | Chr1:1440343–1,441,301 | 2067 | 93 | 9.09 |
| 136 | CsaV3_5G032530 | Chr5:26313766–26,315,796 | 1914 | 96 | 9.17 |
| 137 | CsaV3_1G009880 | Chr1:6153304–6,155,828 | 4261 | 373 | 6.61 |
| 138 | CsaV3_7G033460 | Chr7:21082838–21,087,117 | 3175 | 298 | 6.78 |
| 139 | CsaV3_7G007860 | Chr7:4913547–4,914,938 | 8745 | 211 | 6.35 |
| 140 | CsaV3_4G010010 | Chr4:7769516–7,771,744 | 4296 | 333 | 5.11 |
| 141 | CsaV3_1G003270 | Chr1:2026829–2,032,886 | 4082 | 619 | 9.19 |
| 142 | CsaV3_5G031750 | Chr5:25868912–25,871,372 | 4279 | 365 | 5.84 |
Phylogenetic analysis, gene structure and conserved motif analysis of CsbHLH gene family
To confirm the structural characteristics of CsbHLH proteins, we performed multi-sequence alignment (MSA) analysis on 142 CsbHLH proteins. All 142 CsbHLH proteins contained the characteristic regions of bHLH: two helix regions, one loop region and one basic region (Fig. 1). Additionally, the conserved amino acids with a sequence identity greater than 50% in bHLH domains, were present as light blue or purple colour (Fig. 1a). Sequence logos were produced using the 142 CsbHLH homologous domain amino acid sequences (Fig. 1b). The CsbHLH proteins in cucumber contained 17 conserved amino acids of bHLH domain, which were present in the bHLH gene family of Arabidopsis and Moso bamboo [2, 25]. As shown in Fig. 1b, we could clearly observe that key amino acid residues Arg-10, Arg-11, Leu-21 and Leu-53 were highly conserved (92, 87, 96, and 90%, respectively) in the 142 CsbHLH proteins. Subsequently, a phylogenetic tree was constructed on the 142 CsbHLH proteins, which were divided into 32 subgroups (C1-C32) based on the clades over 50% bootstrap support (Fig. 2a).
Fig. 1.

Conserved amino acids and multiple sequence alignment schematic diagrams of the CsbHLHs bHLH domains. (a) Multiple sequence alignments of CsbHLH proteins. The CsbHLH conserved sequences were marked with a purple background for an amino acid identity greater than 75% and a light blue background for an amino acid identity greater than 50%. The bHLH domains were labelled. (b) Sequence logo of CsbHLH domains. The overall height of each stack represented the conservation of the sequence at that position
Fig. 2.
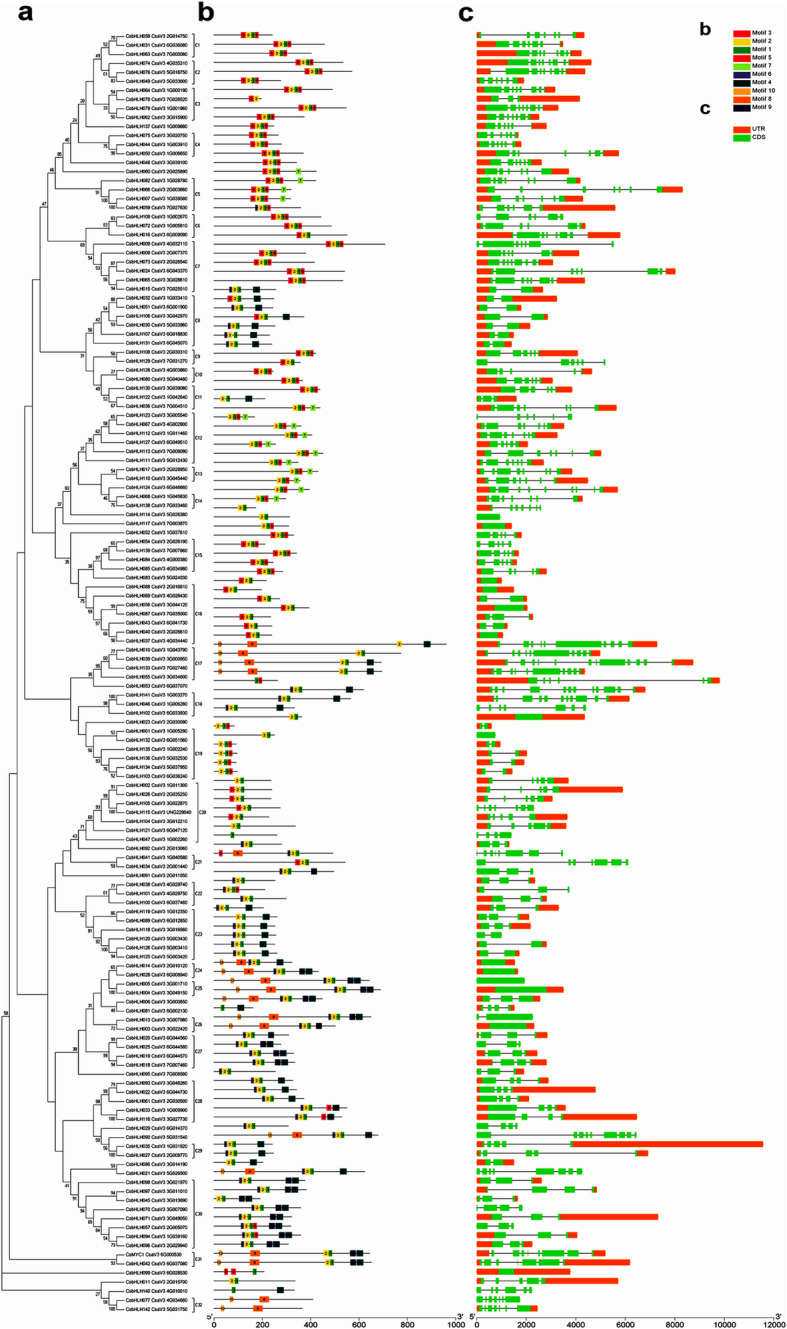
Phylogenetic relationships, gene structure and conserved protein motifs in bHLH genes from cucumber. (a) The phylogenetic tree was constructed based on the full-length protein sequences of 142 CsbHLH proteins using MEGA 7.0 software. The tree showed the 32 phylogenetic subgroups (C1-C32) with high bootstrap value. (b) Conserved motifs in CsbHLH proteins. The motifs, numbers 1–10, were displayed in different coloured boxes. The sequence logos and E values for each motif were shown in Fig. S1. (c) Exon-intron structure of CsbHLH genes. Exons and introns were indicated by green boxes and single lines, respectively. Blue boxes represented upstream or downstream. The length of each gene was listed in Table 1
We then performed gene structure analysis of CsbHLH gene to support the phylogenetic analysis, which showed that CsbHLHs in the same subgroups presented similar numbers of exons and introns, and regardless of intron sizes, the CsbHLH genes in the same subgroups had similar intron-exon gene structures (Fig. 2c).
To further investigate the specific motifs of CsbHLH proteins in the same subgroup, we used the MEME tool to identify 10 conserved motifs. The different numbers of conserved motifs were present in 142 CsbHLH proteins (Fig. 2b). Moreover, a similar motif existed in CsbHLH proteins of the same subgroup. For instance, all proteins of subgroup 23 contained motifs 1, 2, 4 and 6, and motif 5 was identified in most CsbHLH proteins. We also found that certain motifs were absent in certain subgroups. For example, motif 4 was absent in all proteins of the 1, 2 and 3 subgroups (Fig. 2b).
In general, the results of conserved motif and gene structure analyses further confirmed the results of the phylogenetic analysis, indicating that proteins within the same subgroup may have similar functions.
Synteny analysis of bHLH genes in cucumber, Arabidopsis and tomato
Through the analysis of the genome distribution of CsbHLH genes, we found the 142 CsbHLHs (except CsaV3_UNG229040) all could be located on chromosomes 1–7 (Fig. 3a; Table 1; Fig. S2). According to the description reported by [26] to determine the duplication of CsbHLH genes, we analysed the syntenic regions. The cucumber genome contained 231 segmental duplication blocks and 1468 tandem duplication gene pairs. We obtained five tandem duplication gene pairs (CsbHLH019 / CsbHLH020; CsbHLH019 / CsbHLH025; CsbHLH120 / CsbHLH125; CsbHLH125 / CsbHLH126; CsbHLH038 / CsbHLH101) and seven segmental duplication gene pairs (CsbHLH112 / CsbHLH127; CsbHLH040 / CsbHLH037; CsbHLH054 / CsbHLH085; CsbHLH060 / CsbHLH074; CsbHLH001 / CsbHLH135; CsbHLH141 / CsbHLH046; CsbHLH050 / CsbHLH044) in cucumber CsbHLH family (Fig. 3a; Table S1).
Fig. 3.
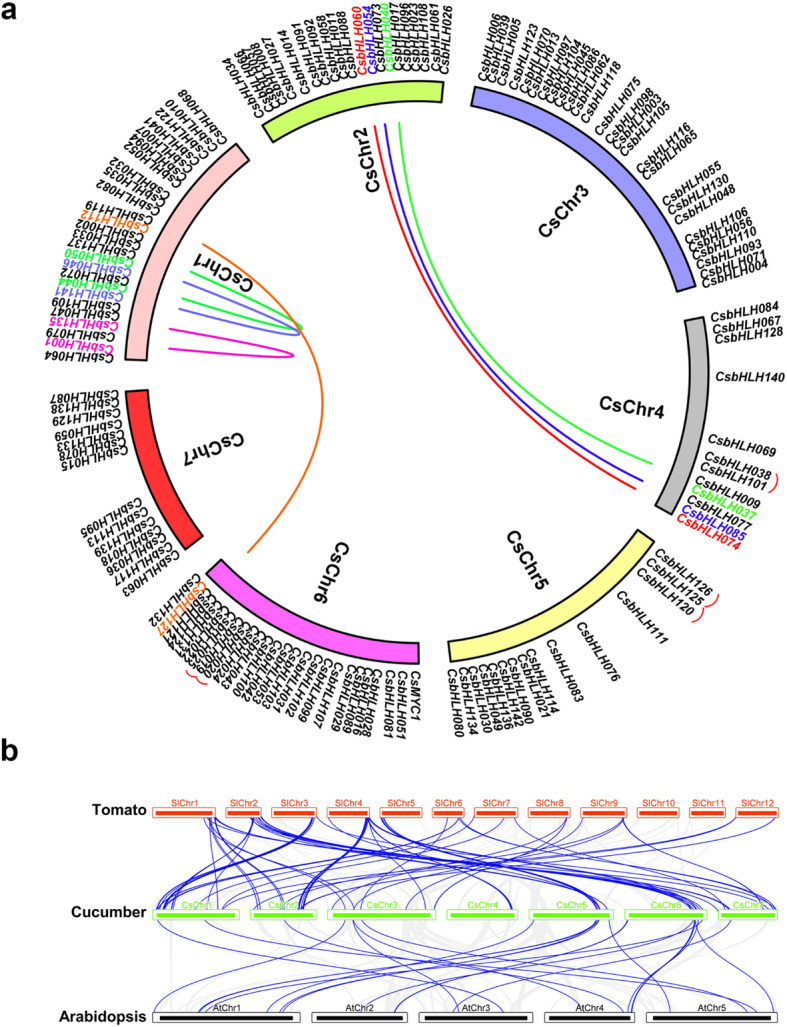
Gene duplication and synteny analysis of CsbHLH genes. (a) Schematic representations of the chromosomal distribution and interchromosomal relationships of CsbHLH genes. Different line colours represented different segmental duplicated CsbHLH gene pairs, among which the two genes of the same segmental duplicated gene pair were labelled in the same colour. The red lines in the outer ring indicated tandem duplication gene pairs. (b) Synteny analysis of bHLH genes between cucumber and Arabidopsis and tomato. Blue lines indicated collinear blocks of the bHLH gene within the cucumber and Arabidopsis and tomato genomes
In order to further illuminate the phylogenetic mechanisms of CsbHLH family, we constructed a comparison of the syntenic map of cucumber related to tomato and Arabidopsis, respectively (Fig. 3b). We found that CsbHLH024, CsbHLH040 and CsbHLH054 genes were associated with more than two syntenic gene pairs between cucumber and tomato. Moreover, for instance, CsbHLH020 and CsbHLH049 genes were also corresponded to two syntenic gene pairs between cucumber and Arabidopsis, indicating that these bHLH genes may play a key role in evolution. In addition, we found certain collinear pairs were present between cucumber and both Arabidopsis and tomato (such as CsbHLH132, CsbHLH135 and CsbHLH136) (Fig. 3b; Table S2), illustrating that before the ancestral divergence, these orthologous pairs might have already present. Meanwhile, some CsbHLH genes were not associated with syntenic gene pairs in Arabidopsis or tomato, indicating that they might have been peculiar to cucumber during the course of evolution.
Cis-elements in the promoters of CsbHLH genes in cucumber
According to the studies reported by [27], many bHLH genes may be able to respond to a variety of abiotic stresses. We isolated the 2-kb promoter regions of the CsbHLH genes to identify the potential cis-elements (Table S3), in which a number of CsbHLH genes particularly presented elements associated with plant hormones (such as auxin, abscisic acid and gibberellic acid) and stress responsiveness (such as drought inducibility and low temperature). Moreover, the promoter regions of some CsbHLH genes contained an MYB binding site involved in flavonoid biosynthetic gene regulation, which might be involved in the synthesis of flavonoid in cucumber (Fig. S3; Table S3). In addition, the promoter regions of CsbHLH genes contained G-Box and Box-4 elements related to light responsiveness. The cis-regulatory elements in CsbHLH promoters included the plant light-responsive elements, plant growth- and development-responsive elements, and responding to diverse stresses (Table S3).
To further analyse whether there is co-expression of CsbHLH genes with the same cis-elements, we constructed a co-expression network of CsbHLH genes, based on the available RNA-seq data of 10 cucumber tissues regarding correlations between cucumber bHLH genes [26]. The co-expression network containing 23 CsbHLH genes (nodes) and 191 correlations (edges) showed that each of the CsbHLH genes had multiple co-expression genes with same cis-elements (Fig. S4; Table S3). The result indicated the co-expression of genes may be related to the same cis-elements in their promoter regions.
Function prediction of CsbHLHs based on phylogenetic analyses
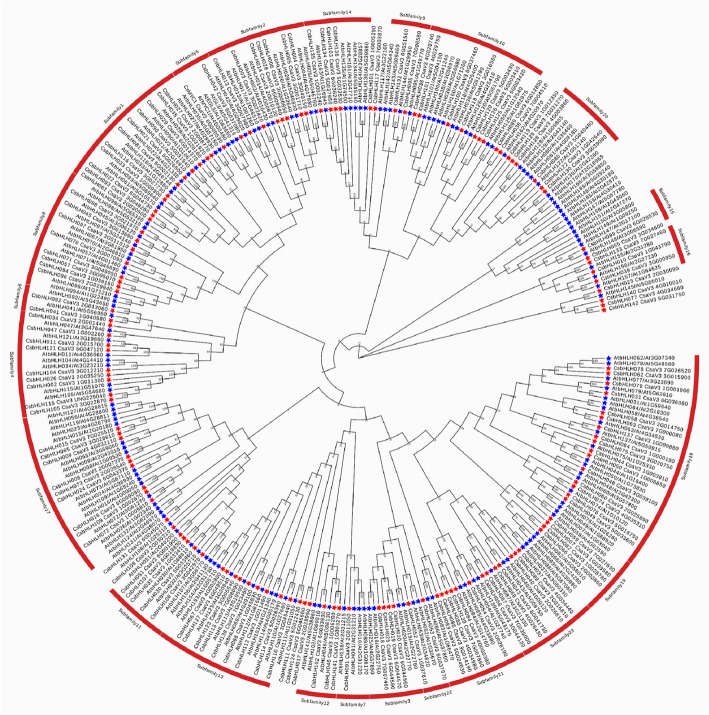
Previous studies have identified and verified the function of numerous bHLH proteins in Arabidopsis [28, 29]. However, the biological functions of CsbHLHs are known little in cucumber. In this study, we performed phylogenetic analyses of 166 AtbHLHs and 142 CsbHLHs proteins to identify the genetic relationship of the bHLH proteins in cucumber and Arabidopsis, so as to preliminarily explore the functions of CsbHLH proteins [2, 10] (Fig. 4).
Fig. 4.
Evolutionary tree analysis (circle tree) and subfamily classifications of bHLHs proteins in cucumber and Arabidopsis thaliana. The evolutionary tree was constructed using the Neighbour-Joining method with 1000 bootstrap replication. The evolutionary distances were computed using poisson correction. The analysis involved 142 cucumber bHLH protein sequences and 166 Arabidopsis thaliana bHLH protein. Red stars represented the CsbHLH proteins and blue represented the AtbHLH proteins
Finally, we divided the 308 bHLH proteins into 23 subfamilies, and predicted the functions of CsbHLHs according to their verified functional homologs in the same subfamily (Table S4). As shown in Table S4, most of the proteins of subfamilies 1, 2, 4, 10, 13, 14 and 18 responded to different biotic and abiotic stresses [30, 31], such as drought [32], cold [33] and salt [34]. Some of the proteins in subfamilies 4 and 10 might be involved in iron regulation, regulating the iron homeostasis [35]. The proteins of subfamilies 19 and 23 have been identified to regulate flower development [36], and the members of subfamilies 3, 8, 9, 16 and 21 might participate in the development of multiple plant organs [37–39]. There were PIFs in subfamily 17, related to light signal transduction and protect the normal growth and development of plants [15]. The members of subfamily 5 regulate the flavonoid biosynthesis and cell differentiation of root epidermis [22]. The detailed possible functions of CsbHLHs are listed in Table S4.
In general, although the evolutionary relationships could not be clearly deciphered for the functions of all genes, the analysis was meaningful and necessary.
Expression analysis of CsbHLH genes under different stress conditions and in different tissues
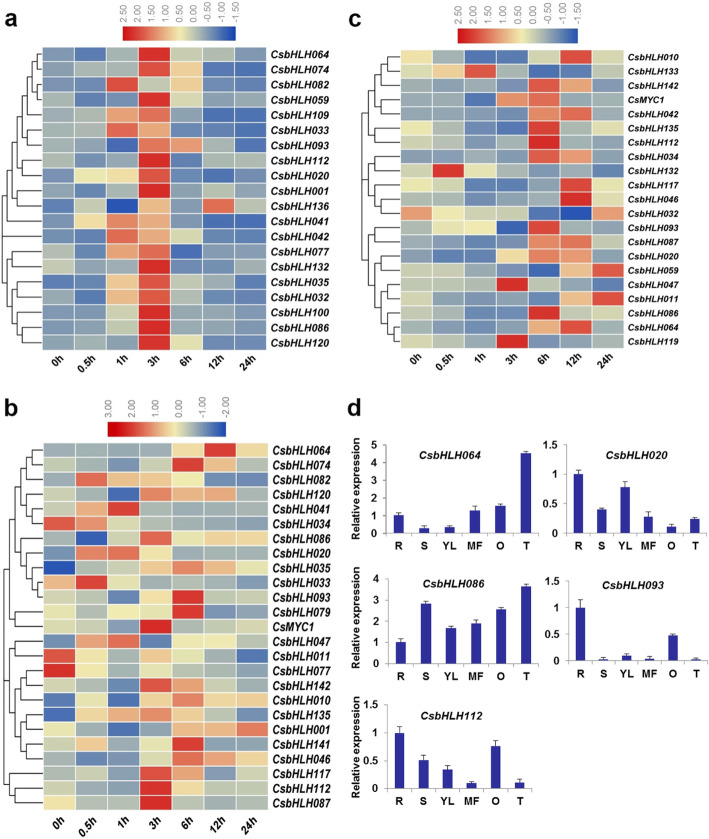
To identify which CsbHLH genes play important roles in abiotic stress responses, we carefully screened 21, 20 and 25 bHLH genes based on the cis-acting elements containing low temperature, defense and stress responsive and abscisic acid (ABA) elements in the promoters of bHLH genes, respectively, and detected their transcriptional changes with treatments of low temperature (4 °C), salt (NaCl) and ABA, respectively. As expected, all the CsbHLH genes screened responded to stress treatments under the respective stress conditions (Fig. 5). For example, the expression levels of the 20 CsbHLHs were all positive in response to salt stress, and many of them were upregulated after one hour of salt treatment and achieved the highest expression level 3 h later, and then gradually declined. The expressions of CsbHLH033, CsbHLH041 and CsbHLH082 were the highest after NaCl treatment for just 1 h, but the expressions levels of CsbHLH136 reached its maximum after 12 h. CsbHLH041 was the most susceptible to salt stress (increased by approximately 37-fold) (Fig. 5a). Under ABA treatment, the transcriptional levels of CsbHLH020, CsbHLH041 and CsbHLH064 were more than 10-fold higher than those of untreated level (CsbHLH020: the highest nearly 61-fold; CsbHLH041: the highest nearly 55-fold; CsbHLH064: the highest nearly 19-fold). In contrast, the expression levels of four of the CsbHLHs genes were significantly down-regulated under ABA treatment (CsbHLH011, CsbHLH033, CsbHLH034 and CsbHLH077), as could be seen in Fig. 5b. The expression levels of 20 of the 21 CsbHLH were up-regulated at some time points after the 4 °C treatment, while only CsbHLH032 was decreased (Fig. 5c). We found the CsbHLH020, CsbHLH064, CsbHLH086, CsbHLH093 and CsbHLH112 genes could simultaneously respond to the three abiotic stresses (Fig. 5).
Fig. 5.
Relative expression analysis of the CsbHLH genes under different stress conditions and different tissues. Expression patterns of CsbHLH genes under NaCl (100 mM) treatment (a), ABA (100 μM) treatment (b) and low temperature (4 °C) treatment (c). (d) Tissue-specific expression profiles of five cucumber bHLH genes. Total RNA was isolated from roots (R), stems (S), young leaves (YL), male flowers (MF), ovary (O) and tendrils (T), respectively. The cucumber β-actin gene was performed as an internal control, and three independent samples were used for these experiments. Error bars indicated standard errors (SE)
The expression patterns of genes under different conditions are often related to their functions. Therefore, we used qRT-PCR to detect the expression patterns for CsbHLH020, CsbHLH064, CsbHLH086, CsbHLH093 and CsbHLH112 abiotic stress-responsive CsbHLHs in different tissues. The expression patterns of the five CsbHLH genes showed different tissue specificities (Fig. 5d). For instance, CsbHLH093 and CsbHLH112 had higher expression levels in ovaries and roots, but lower expression levels in tendrils and male flowers (Fig. 5d). On the contrary, both CsbHLH064 and CsbHLH086 were highly expressed in tendrils and male flowers. The expression levels of CsbHLH020 in young leaves and roots were higher than that in other tissues (Fig. 5d). These results suggested that CsbHLH genes might play key roles in plant developmental and physiological processes.
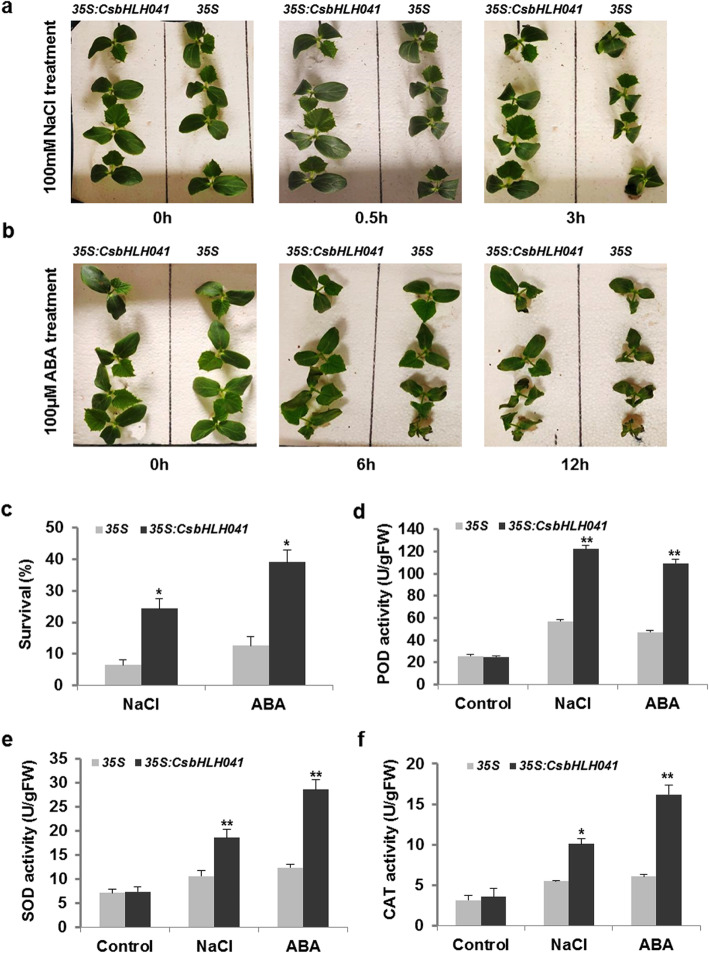
CsbHLH041 enhanced tolerance to NaCl and ABA in transgenic Arabidopsis and cucumber
CsbHLH041 expression was significantly induced by salt and ABA in cucumber (Fig. 5a-b). Therefore, we used Agrobacterium-mediated transient transformation of cucumber cotyledons to clarify CsbHLH041 tolerance to salt and ABA. After 0.5 h of 100 mM NaCl treatment, serious wilting occurred in the seedlings overexpressing 35S empty vector compared with over-expression CsbHLH041, and the wilting difference was more obvious after 3 h of NaCl treatment (Fig. 6a). After 12 h, the survival rate of the transgenic seedlings (24%) was markedly higher than that of the 35S empty vector seedlings (6%), showing that over-expression of CsbHLH041 resulted in significant salt resistance (Fig. 6c). After 6 h of ABA treatment, the transgenic seedlings were more vigorous than 35S empty vector seedlings (Fig. 6b). With the extension of ABA treatment time, the 35S cucumber seedlings showed visible symptoms of ABA-induced damage, such as drying, wilting, and even death, with survival of only 12%. While some CsbHLH041 transgenic plants remained green with expanded cotyledons, and the survival rate was up to approximately 40% (Fig. 6b-c).
Fig. 6.
Overexpression of CsbHLH041 increased salt and ABA tolerance in cucumber seedlings. Phenotypes of 35S empty vector and 35S:CsbHLH041 cucumber seedlings treated with 100 mM NaCl (a) and 100 μM ABA (b) at different time periods during hydroponic growth. (c) Survival of 35S empty vector and 35S:CsbHLH041 cucumber seedlings after 12 h of salt and ABA treatments. Comparison of the antioxidant enzyme activity between 35S empty vector and 35S:CsbHLH041 cucumber seedlings under salt and ABA treatment: (d) peroxidase (POD) activity, (e) superoxide dismutase (SOD) activity, (f) catalase (CAT) activity. The bars showed the SE. * and ** indicate significant differences at P < 0.05 and P < 0.01, respectively
To clarify the possible factors underlying the enhanced NaCl and ABA resistance, we examined the enzymatic activities in the ROS clearance system under NaCl and ABA treatments, respectively. Without the NaCl or ABA treatment, the enzymatic activities of POD, SOD and CAT in 35S and 35S:CsbHLH041 transgenic seedlings were no significant difference (Fig. 6d-f). Nevertheless, both NaCl treatment and ABA treatment could significantly activate more enzymatic scavenging activities in the CsbHLH041 transgenic plants than in the 35S empty vector seedlings (Fig. 6d-f).
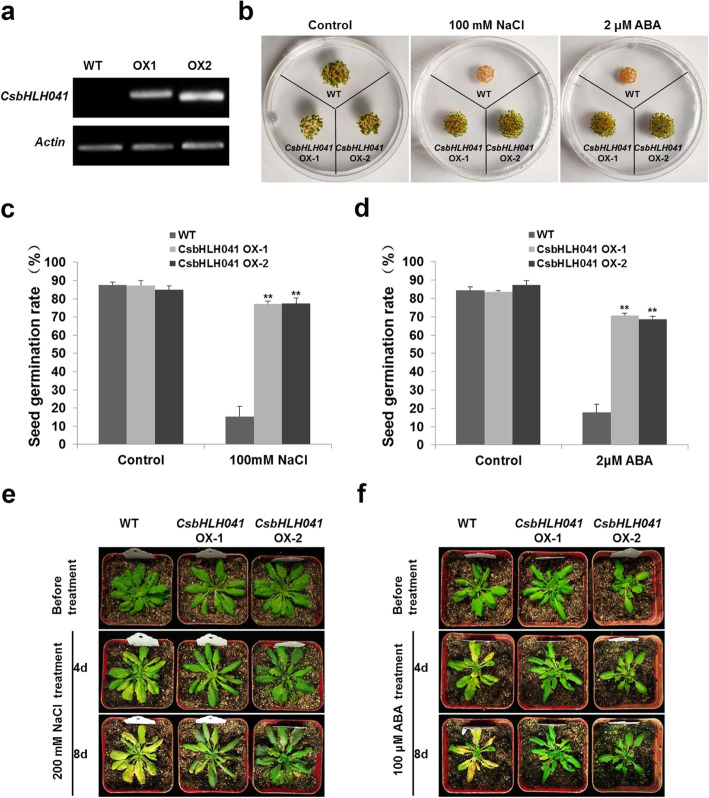
To further explore the function of CsbHLH041 resistance to abiotic stress in plants, transgenic Arabidopsis plants overexpressing CsbHLH041 driven by the CaMV35S promoter were generated. Two independent homozygous lines with relatively high expression levels, CsbHLH041 OX1 and CsbHLH041 OX2, were selected for the analysis (Fig. 7a). The salt and ABA tolerance of CsbHLH041 transgenic plants were assessed. There were no differences in seed germination between WT and CsbHLH041 transgenic Arabidopsis on 1/2 MS (Control) (Fig. 7b). However, the germination ratio of transgenic plants seeds was markedly higher than WT seeds in 1/2 MS medium containing 100 mM NaCl or 2 μM ABA (Fig. 7b-d). Subsequently, the 3-week-old seedlings of CsbHLH041 transgenic lines and wild-type (WT) plants were treated with 200 mM NaCl and 100 μM ABA, respectively. The leaves of WT plants turned severely yellow after 4 days of 200 mM NaCl or 100 μM ABA treatment, while CsbHLH041 transgenic lines were still growing with green leaves (Fig. 7e-f). After 8 days, the difference in NaCl or ABA resistance between WT plants and CsbHLH041 transgenic lines was more obvious, which suggested that CsbHLH041 transgenic plants were more tolerant to salt and ABA stresses than WT.
Fig. 7.
CsbHLH041 transgenic Arabidopsis showed enhanced salt and ABA tolerance. (a) Relative expression of CsbHLH041 in Col-0 (WT) and two T3 generation transgenic lines by semi-quantitative PCR. The actin8 gene was used as an internal control. The original, uncropped gel image was provided as Additional file 9. (b) Germination of WT seeds of Col-0 and CsbHLH041 transgenic lines OX-1, OX-2 on 1/2 MS supplemented with 100 mM NaCl and 2 μM ABA after 7 days of cultivation at 22 °C. (c) and (d) Seed germination rate for the corresponding (b), respectively. Three biological replications were performed. Asterisks indicated a significant difference **p < 0.01 compared with the corresponding controls. The growth of Col-0 (WT) and CsbHLH041 transgenic lines after 200 mM NaCl (e) and 100 μM ABA (f) treatments
The protein interaction network predictions for CsbHLH orthologs in Arabidopsis that were crucial for the abiotic stress response
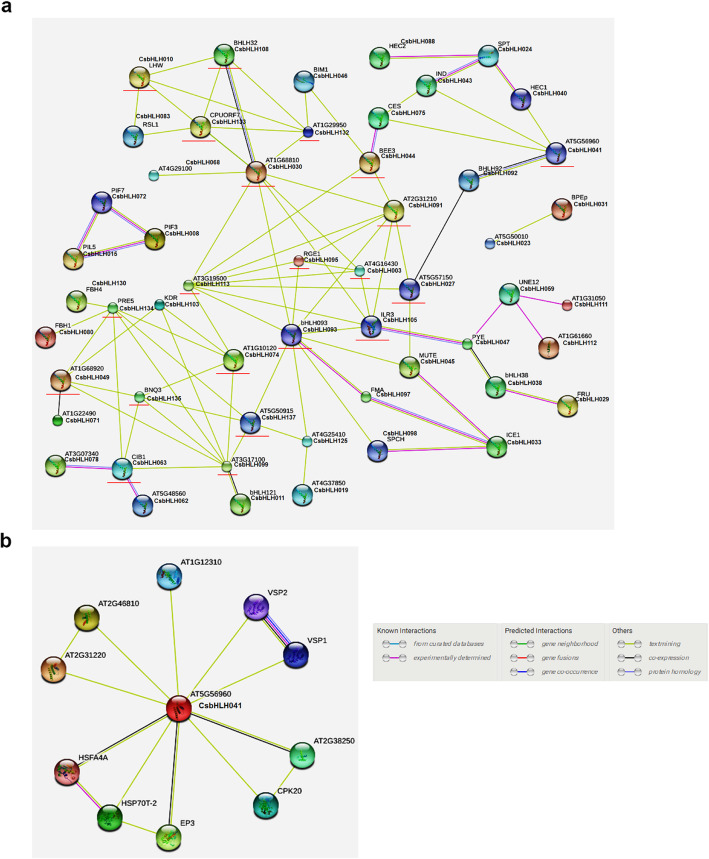
Network interaction analysis has been demonstrated to be an effective method to analyse the gene function [40]. We used the software STRING 10 to predict the protein interaction network among the 142 CsbHLH proteins (Fig. 8a). Numerous CsbHLH transcription factors interacted with multiple CsbHLHs, consistent with previous reports demonstrating that the binding activity of specific DNA sequences depends on the homodimers or heterodimers formed by the interactions of bHLH proteins [2]. As shown in Fig. 8a, there were 21 proteins that had correlation with more than four other bHLH proteins, which may make them play important roles in regulating plant stress responses and growth, and detailed informations about these orthologs were showed in Table S6.
Fig. 8.
Protein interaction network for CsbHLHs based on CsbHLH orthologs in Arabidopsis. Protein interaction network predictions of CsbHLHs (a) and CsbHLH041 (b). Red lines indicated proteins that were predicted to interact with more than four other bHLH proteins. CsbHLH proteins were shown next to Arabidopsis orthologs
In our study, CsbHLH041 responded significantly to salt and ABA treatments, and CsbHLH041 could enhance tolerance to NaCl and ABA in transgenic Arabidopsis and cucumber (Fig. 5a-b; Fig. 6; Fig. 7). The function of bHLH proteins are mainly realized through the formation of heterodimers or homodimers with other transcription factors, which are essential for their binding to downstream target genes [2]. AT5G56960, the CsbHLH041 homologous gene, was at the centre of the protein association network, indicating that it played main roles in regulating different functional proteins (Fig. 8b; Table S6). For example, EP3 might play a role in both normal plant growth and disease resistance [41]. VSP1 and VSP2 are anti-insect proteins and respond to methyl jasmonate and wounding, in which their defense function were correlated with its acid phosphatase activity [42, 43]. The predicted gene association network provides useful resources for subsequent research.
Discussion
Characterization of the cucumber bHLH family
The basic helix-loop-helix (bHLH) transcription factor family is the second largest family in eukaryotes [10, 44] and extensive studies of bHLH families have been identified in various plants [2]. For example, 166 bHLH genes have been identified in Arabidopsis [2, 10], 115 bHLH genes in Nelumbo nucifera [45], 188 bHLH genes in apple [40], 167 bHLH genes in rice [12] and 159 bHLH genes in tomato [13]. The bHLH TFs have been involved in multiple biological processes in plants, especially in regulating defense against biotic and abiotic stresses [46]. However, we know very little about bHLHs in cucumber. In our study, 142 bHLH cucumber genes were identified and characterized. According to phylogenetic analyses, the 142 CsbHLHs were divided into 32 subgroups (Fig. 2a), and multiple sequence analysis indicated that the conserved bHLH domains existed in all 142 CsbHLH proteins (Fig. 1). For instance, the two amino acid residues Leu-21 and Leu-53 were relatively conserved in the helical region that are essential for the formation of dimers. Moreover, the conservative sequence analyses indicated that almost all 142 CsbHLH proteins had the conserved 1 and 2 motifs. The analyses of gene structure and the motif further supported the phylogenetic relationship for the 142 CsbHLH genes (Fig. 2b-c). To sum up, these results showed that all 142 CsbHLHs had the characteristics of the bHLH family, confirming the reliability of the bHLH genes discovered in cucumber.
Phylogenetic analysis and evolution of cucumber bHLH genes
In the model plant Arabidopsis, the bHLH gene family has been systematically analysed [2, 11]. To explore the evolutionary relationships between 142 CsbHLH proteins in cucumber and 166 AtbHLH proteins in Arabidopsis, a phylogenetic tree was constructed based on the protein of 308 bHLHs, which clustered into 23 subfamilies (Fig. 4). There are differences in anatomy and physiology between cucumber and Arabidopsis, so some clades may have different modes of expansion in the bHLH family of cucumber and Arabidopsis. As shown in Fig. 4 and Table S4, not all bHLH members in cucumber were included in these 23 subfamilies, which suggested that there were differences between cucumber and Arabidopsis during the process of evolution.
Studies had shown that gene duplication events played a crucial role in the rapid expansion and evolution of gene families [26]. In the cucumber genome, we identified 231 segmental duplication events and 1468 tandem duplication gene pairs (Table S1). Seven segmental duplication events and five tandem duplication gene pairs were found in the CsbHLH family (Fig. 3a). In general, the gene functions of a clade are highly conserved among different plant species, but it is not absolute. Therefore, it is of great significance to accurately identify the true orthologs between plant species based on synteny analysis. The results showed that the cucumber genome had extensive synteny with the Arabidopsis and tomato genomes, and 944 and 983 syntenic blocks between the cucumber and Arabidopsis and tomato genome were identified, respectively (Table S5). Many CsbHLH genes showed a linear relationship with the tomato and Arabidopsis genes, respectively (Fig. 3b; Table S2).
Previous studies have shown that orthologous genes are usually distributed in the same clade, and have similar functions. In our study, many CsbHLH proteins were grouped into some functional clades of Arabidopsis, providing valuable information for studying the functions of CsbHLHs. CsMYC1 and CsbHLH042 were grouped into subfamily5 along with AtGL3, AtEGL3, AtMYC1 and AtTT8, and were highly homologous to these proteins. In Arabidopsis, AtGL3, AtEGL3 and AtTT8 have been demonstrated to be key regulators of anthocyanin and PA biosynthesis [22]. Moreover, AtGL3, AtEGL3 and AtMYC1 were shown to regulate trichome formation and root hair patterning [19, 47]. Therefore, it is possible that CsMYC1 and CsbHLH042 may control trichome formation and PA biosynthesis in cucumber.
Cucumber bHLH genes may play important roles in abiotic stress tolerance
In the process of plant response to abiotic stress, bHLH TFs act as regulatory genes to regulate the expression changes of related stress genes, thus playing an important role in stress responses. Many studies have shown that bHLH TFs can respond to a range of stresses. For example, in addition to being involved in the morphogenesis of stomata, the TFs INDUCER OF CBF EXPESSION1 (ICE1) and ICE2 in Arabidopsis and their homologous genes in other species can play key roles in the response to low temperature stress [31, 46]. RERJ1 is upregulated in the event of physical damage and drought stress to plants [48]. All these examples indicate that bHLH TFs can play a certain role in response to abiotic stress. However, little is known about the functions of the bHLH gene family in cucumber. To better analyse the protein functions of the bHLH gene family in cucumber, we conducted a preliminary analysis of three aspects to reveal the functions of the CsbHLH gene family.
How cis-elements in the promoters of the bHLH genes respond to the environment will affect their roles in stimulating and regulating gene expression. Cis-element analyses indicated that there were a wide range of elements on the gene promoters of CsbHLH responding to different stresses, such as TCA-element, MBS and LTR (Fig. S3). MYB binding site involved in drought-inducibility existed in many CsbHLH gene promoters (Table S3), indicating that MYB TFs may regulate CsbHLHs expression in drought stress. The TC-rich and ABRE elements related to ABA-dependent or independent stress tolerance also appeared in some CsbHLH gene promoters [49]. In general, according to the cis-acting element contained on the promoters, these CsbHLH genes might play key roles responsing to various stresses in cucumber. In addition, the functions of 50 CsbHLHs were predicted, which were mainly related to stress responses and development processes (Table S4). For the third aspect, the regulatory networks for 142 CsbHLH genes were predicted, suggesting that a number of genes could respond to stimuli (Table S6). For example, bHLH093 and ICE1 were involved in the ABA signalling pathway, which were crucial for abiotic stress responses in plants [49, 50]. These results suggested that the bHLH gene family may also be involved in the response to stress, metabolic regulation, and plant development in cucumber, consistent with previous research [10, 12]. Subsequently, we analysed and screened CsbHLH genes that might respond to stress, as it is very important to improve stress tolerance of cucumber. According to cis-element analyses, the promoter regions of 60 CsbHLHs were rich in TC-rich cis-elements, suggesting that they may be involved in stress responses and defense (Fig. S3). Moreover, the promoters of 106 CsbHLHs contained the ABA-responsive element, responding to ABA stress and 41 CsbHLHs contained the LTR element, responding to cold stress. The phylogenetic analyses between Arabidopsis and cucumber further showed that 25 CsbHLHs might respond to abiotic stresses, such as ABA, salt, cold and drought (Table S4). Through comprehensive analysis, we carefully screened 21, 20 and 25 bHLH genes that were likely to respond to low temperature (4 °C), salt (NaCl) and ABA, respectively. The screened CsbHLH genes all responded to stress treatments under the respective stress conditions (Fig. 5). CsbHLH041 was induced by salt and ABA (Fig. 5a-b), and 35S:CsbHLH041 transgenic Arabidopsis thaliana and transient transformed cucumber cotyledons were shown to have enhanced tolerance to salt and ABA (Fig. 6; Fig. 7). In general, these results provided a good reference for further functional studies of CsbHLH gene family in cucumber.
Conclusions
Our study investigated the bHLH family genes in detail in cucumber. We also performed expression analyses of the selected genes under different stress treatments, and detailed functions of CsbHLH041 using the transgenic method. This work provides new insights into the functions and regulatory mechanisms of CsbHLH proteins in cucumber abiotic stress tolerance and growth and development.
Methods
Genome-wide identification of the CsbHLH genes in cucumber
To identify the CsbHLH gene family members from the entire cucumber genome database, 166 Arabidopsis bHLH proteins were used as query sequences and BlastP searches against the predicted cucumber proteins. In addition, the Hidden Markov Model (HMM) profile of the bHLH domain (PF00010) from the Pfam database (available online: http://pfam.janelia.org) was also applied as a query to search the bHLH genes. We further examined the bHLH domains of all candidate bHLH genes as described by [24].
Phylogenetic analysis and multiple sequence alignment
The sequence logos for bHLHs were obtained by submitting the multiple alignment sequences to the website (http://weblogo.berkeley.edu/logo.cgi) [51]. A phylogenetic tree was constructed with the aligned fully predicted protein sequences of 142 bHLH genes using MEGA7 (https://www.megasoftware.net/) [52]. The neighbour-joining (NJ) method was used with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed). The phylogenetic tree was visualized by plotting it using the EvolView tool (http://www.evolgenius.info). Classification of the CsbHLH genes was then performed according to their phylogenetic relationships with their corresponding Arabidopsis bHLH genes. Multiple sequence alignments were performed as described by [26].
Conserved motif and gene structure analysis
The 142 CsbHLH gene structures were analysed as described by [53]. Conserved motif structures in CsbHLHs were identified using MEME (http://meme-suite.org/index.html) [26].
Gene duplication and chromosomal distribution
The gene duplication events were assessed as described by [54]. According to the physical location information in the cucumber genome database, 142 CsbHLH genes were mapped to cucumber chromosomes as described by [26], and the syntenic analysis maps were completed using TBtools [26].
Analysis of the bHLH gene promoter in cucumber
We downloaded the entire cucumber genome sequence from the cucumber genome database (Chinese Long 9930) and extracted the 2-kb long sequences upstream of the transcription start site of these 142 CsbHLH genes. The cis-acting elements on the promoter regions of these genes were analysed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) software [55].
Plant materials and growth conditions
Cucumber (Cucumis sativus L. cv ‘Xintaimici’) seeds, provided by Professor Chenxing Cao (Shandong Agricultural University), were germinated on moist filter paper in an incubator at 28 °C for 1 day. The germinated seeds were sown into soil mixture in an ordinary illuminated incubator at Shandong Agricultural University. After 10 days, batches of 12 seedlings were transferred to a plastic tank filled with an aerated nutrient solution (pH 6.0–6.5) containing the following: Ca (NO3)2: 3.5 mM, KNO3: 7 mM, KH2PO4: 0.78 mM, MgSO4: 2 mM, H3BO3: 29.6 μM, MnSO4: 10 μM, Fe-EDTA: 50 μM, ZnSO4: 1.0 μM, H2MoO4: 0.05 μM and CuSO4: 0.95 μM. The experiment was carried out as previously described [56].
RNA extraction and qRT-PCR analysis
Total RNA was isolated from cucumber and Arabidopsis plants using an RNAprep pure Plant Kit (TianGen, Beijing, China), following the manufacturer’s instructions. Subsequently, reverse transcribed using the PrimeScript®1st Strand cDNA Synthesis Kit (Takara, Japan). The qRT-PCR reactions were performed using the UltraSYBR Mixture (with ROX I; Cwbiotech) with the iCycler iQ5 system (BioRad, CA, USA). The results were normalized to those of the cucumber ACTIN gene. Three biological replicates were used for each analysis. The primers used in this study are provided in Table S7.
Overexpression vector construction, Arabidopsis transformation and transient transformation in cucumber cotyledons
The full-length coding sequence of CsbHLH041 was recombined into the pCAMBIA1300 vector. The construct was transformed into Agrobacterium tumefaciens LBA4404, which was used for transformation of Arabidopsis plants and 8-d-old cucumber cotyledons [57]. The Arabidopsis seeds were Colombia (Col-0), which were bred in our laboratory. Homozygous T3 transgenic Arabidopsis lines were identified by hygromycin (300 mg/L) selection.
Abiotic stress tolerance assays and ABA sensitivity analysis
For Arabidopsis salt stress and ABA treatment, the seeds of CsbHLH041 T3-generation homozygous lines and Col-0 (WT) were sown in vermiculite soil in pots and cultured under normal conditions at 22 °C for 3 weeks. For salt treatment, the 3-week-old seedlings were watered with 200 mM NaCl solution every other day, and the growth of Col-0 (WT) and CsbHLH041 transgenic lines was observed every 4 days. For ABA treatments, the 3-week-old seedlings were watered with 100 μM ABA solution every other day, and phenotypes were evaluated every 4 days. To check the seed germination rate in response to salt stress and ABA treatment, the seeds of Col-0 (WT) and transgenic lines were surface sterilized and sown in 1/2 MS medium supplemented with 2 μM ABA or 100 mM NaCl, respectively, under normal conditions at 22 °C in a growth chamber. The germination rate was scored on the 7th day after culturing on the plates.
To determine the salt tolerance and ABA sensitivity in cotyledons of 8-d-old cucumber seedlings with transient infiltration of 35S and 35S:CsbHLH041, selected seedlings with equivalent growth were transferred to 6 L nutrient solution for hydroponic growth. Hoagland nutrient solution was used for culture, and the seedlings were grown hydroponically for 2 days before salt and ABA treatment. They were then treated with salt and ABA, and the final concentration in the medium was 100 mM and 100 μM, respectively. To ensure the reliability of the experiment, the cucumber seedlings with transient infiltration of 35S and 35S:CsbHLH041 were cultured in the same hydroponic box. The changes in transgenic and control seedlings were observed at different time periods.
Determination of physiological parameters
The cucumber cotyledons of 35S empty vector and 35S:CsbHLH041 seedlings were collected at different time points during salt and ABA stress treatment, then frozen in liquid nitrogen for subsequent experiments. The activity of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) were determined as previously described [58].
The functional annotations and protein association network predictions
We submitted the 142 CsbHLH protein sequences to the online server (version 10.0; http://string-db.org). For details was as described by [40].
Supplementary information
Additional file 1. Supplementary Figs. S1 to S4. (Fig. S1. Ten conserved motifs from 142 CsbHLH proteins; Fig. S2. Genome locations of the 142 CsbHLH genes on 7 chromosomes; Fig. S3. Cis-element analysis in the CsbHLH genes promoter regions; Fig. S4. Co-expression network of the CsbHLH genes).
Additional file 2: Table S1. Tandem duplication and Segmental duplication events.
Additional file 3: Table S2. Synteny analysis of bHLH genes in cucumber, Arabidpsis and tomato.
Additional file 4: Table S3.Cis-elements in the promoters of 142 CsbHLH genes.
Additional file 5: Table S4. Predicted functions of CsbHLHs with the function of their homologs verified in Arabidopsis by phylogenetic analysis.
Additional file 6: Table S5. Syntenic blocks between the cucumber and Arabidopsis and tomato genome.
Additional file 7: Table S6. String protein annotations.
Additional file 8: Table S7. Primers used for qRT-PCR.
Acknowledgments
We thank Dr. Chenxing Cao, College of Horticultural Science and Engineering, Shandong Agricultural University, for providing cucumber (Cucumis sativus L. cv ‘Xintaimici’) seeds.
Abbreviations
- bHLH
Basic Helix-Loop-Helix
- At
Arabidopsis thaliana
- Cs
Cucumis sativus L
- MS
Murashige and Skoog
- qRT-PCR
Quantitative reverse transcription-PCR
- CDS
Coding Sequence
- ABA
Abscisic acid
- pI
Isoelectric point
- WT
Wild type
Authors’ contributions
JL and ZR conceived and designed the experiments. JL, TW and JH performed the experiments. JL analyzed the data and wrote the manuscript. ZR revised the manuscript. All authors have read and approved this manuscript.
Funding
This work was supported by fundings from the National Natural Science Foundation of China (31672170 and 31872950), the Natural Science Foundation of Shandong Province (JQ201309), the Shandong “Double Tops” Program (SYL2017YSTD06) and the ‘Taishan Scholar’ Foundation of the People’s Government of Shandong Province (ts20130932). The funds played no role in study design, data analysis, and manuscript preparation.
Availability of data and materials
The data that support the results are included within the article and its additional files. Other relevant materials are available from the corresponding authors on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jialin Li, Email: 1149142195@qq.com.
Ting Wang, Email: 903067078@qq.com.
Jing Han, Email: 2359110855@qq.com.
Zhonghai Ren, Email: zhren@sdau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02440-1.
References
- 1.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 2.Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferré-D'Amaré AR, Prendergast GC, Ziff EB, Burley SK. Recognition by max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 4.Murre C, Mccaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 5.Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nair SK, Burley SK. Recognizing DNA in the library. Nature. 2000;404:715,717–715,718. doi: 10.1038/35008182. [DOI] [PubMed] [Google Scholar]
- 8.Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- 9.David C, Pierre K, Morgane TC, Gemma R, Valérie L, Elena S, Degnan BM, Michel V. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141(4):1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun H, Fan HJ, Ling HQ. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics. 2015;16:9. doi: 10.1186/s12864-014-1209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci U S A. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paik I, Kathare PK, Kim JI, Huq E. Expanding roles of PIFs in signal integration from multiple processes. Mol Plant. 2017;10(8):1035–1046. doi: 10.1016/j.molp.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, et al. Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47(5):591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- 17.Feyissa DN, Lovdal T, Olsen KM, Slimestad R, Lillo C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta. 2009;230(4):747–754. doi: 10.1007/s00425-009-0978-3. [DOI] [PubMed] [Google Scholar]
- 18.Appelhagen I, Jahns O, Bartelniewoehner L, Sagasser M, Weisshaar B, Stracke R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene. 2011;484(1–2):61–68. doi: 10.1016/j.gene.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10(2):63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Zhai Y, Zhang L, Xia C, Fu S, Zhao G, Jia J, et al. The wheat transcription factor, TabHLH39, improves tolerance to multiple abiotic stressors in transgenic plants. Biochem Biophys Res Commun. 2016;473(4):1321–1327. doi: 10.1016/j.bbrc.2016.04.071. [DOI] [PubMed] [Google Scholar]
- 21.Fernández-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S. Transcriptional control of flavonoid biosynthesis. Plant Signal Behav. 2014;9:e27522. doi: 10.4161/psb.27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41(12):1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 24.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37(Database issue):D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng X, Xiong R, Liu H, Wu M, Chen F, Hanwei Y, Xiang Y. Basic helix-loop-helix gene family: genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol Biochem. 2018;132:104–119. doi: 10.1016/j.plaphy.2018.08.036. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Pang B, Yan J, Wang T, Wang L, Chen C, et al. Comprehensive Analysis of Cucumber Gibberellin Oxidase Family Genes and Functional Characterization of CsGA20ox1 in Root Development in Arabidopsis. Int J Mol Sci. , 2018;19(10). [DOI] [PMC free article] [PubMed]
- 27.Babitha KC, Vemanna RS, Nataraja KN, Udayakumar M. Overexpression of EcbHLH57 Transcription Factor from Eleusine coracana L. in Tobacco Confers Tolerance to Salt, Oxidative and Drought Stress. PLoS One. 2015;10(9):e0137098. [DOI] [PMC free article] [PubMed]
- 28.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15(1):63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petridis A, Doll S, Nichelmann L, Bilger W, Mock HP. Arabidopsis thaliana G2-LIKE FLAVONOID REGULATOR and BRASSINOSTEROID ENHANCED EXPRESSION1 are low-temperature regulators of flavonoid accumulation. New Phytol. 2016;211(3):912–925. doi: 10.1111/nph.13986. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki-Sekimoto Y, Saito H, Masuda S, Shirasu K, Ohta H. Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal Behav. 2014;9(1):e27639. doi: 10.4161/psb.27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, et al. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013;9(7):e1003653. doi: 10.1371/journal.pgen.1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Hir R, Castelain M, Chakraborti D, Moritz T, Dinant S, Bellini C. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol Plant. 2017;160(3):312–327. doi: 10.1111/ppl.12549. [DOI] [PubMed] [Google Scholar]
- 33.Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17(8):1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmad A, Niwa Y, Goto S, Ogawa T, Shimizu M, Suzuki A, et al. bHLH106 integrates functions of multiple genes through their G-box to confer salt tolerance on Arabidopsis. PLoS One. 2015;10(5):e0126872. doi: 10.1371/journal.pone.0126872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurt F, Filiz E. Genome-wide and comparative analysis of bHLH38, bHLH39, bHLH100 and bHLH101 genes in Arabidopsis, tomato, rice, soybean and maize: insights into iron (Fe) homeostasis. Biometals. 2018;31(4):489–504. doi: 10.1007/s10534-018-0095-5. [DOI] [PubMed] [Google Scholar]
- 36.Sharma N, Xin R, Kim DH, Sung S, Lange T, Huq E. NO FLOWERING IN SHORT DAY (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development. 2016;143(4):682–690. doi: 10.1242/dev.128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, et al. Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 2009;151(3):1175–1185. doi: 10.1104/pp.109.143867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang KZ, Jiang M, Wang M, Xue S, Zhu LL, Wang HZ, et al. Phosphorylation of serine 186 of bHLH transcription factor SPEECHLESS promotes Stomatal development in Arabidopsis. Mol Plant. 2015;8(5):783–795. doi: 10.1016/j.molp.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Chakraborty M, Gangappa SN, Maurya JP, Sethi V, Srivastava AK, Singh A, et al. Functional interrelation of MYC2 and HY5 plays an important role in Arabidopsis seedling development. Plant J. 2019;99(6):1080–1097. doi: 10.1111/tpj.14381. [DOI] [PubMed] [Google Scholar]
- 40.Mao K, Dong Q, Li C, Liu C, Ma F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci. 2017;8:480. doi: 10.3389/fpls.2017.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passarinho PA, Van Hengel AJ, Fransz PF, de Vries SC. Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta. 2001;212(4):556–567. doi: 10.1007/s004250000464. [DOI] [PubMed] [Google Scholar]
- 42.Berger S, Bell E, Mullet JE. Two methyl Jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl Jasmonate and wounding. Plant Physiol. 1996;111:525–531. doi: 10.1104/pp.111.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi YH, Jing X, Lei J, Ahn JE, Koo YD, Yun DJ, et al. Stability of AtVSP in the insect digestive canal determines its defensive capability. J Insect Physiol. 2011;57(3):391–399. doi: 10.1016/j.jinsphys.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 44.Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao TY, Liu YY, Zhu HH, Zhang J, Yang JX, Fu Q, et al. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera. PeerJ. 2019;7:e7153. doi: 10.7717/peerj.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng XM, Zhao Q, Zhao LL, Qiao Y, Xie XB, Li HF, et al. The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 2012;12(1):22. doi: 10.1186/1471-2229-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao H, Wang X, Zhu D, Cui S, Li X, Cao Y, et al. A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J Biol Chem. 2012;287(17):14109–14121. doi: 10.1074/jbc.M111.280735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiribuchi K, Jikumaru Y, Kaku H, Minami E, Hasegawa M, Kodama O, et al. Yamane H. involvement of the basic helix-loop-helix transcription factor RERJ1 in wounding and drought stress responses in rice plants. Biosci Biotechnol Biochem. 2005;69:1042–1044. doi: 10.1271/bbb.69.1042. [DOI] [PubMed] [Google Scholar]
- 49.Chen L, Chen Y, Jiang J, Chen S, Chen F, Guan Z, Fang W. The constitutive expression of Chrysanthemum dichrum ICE1 in Chrysanthemum grandiflorum improves the level of low temperature, salinity and drought tolerance. Plant Cell Rep. 2012;31(9):1747–1758. doi: 10.1007/s00299-012-1288-y. [DOI] [PubMed] [Google Scholar]
- 50.An JP, Zhang XW, Bi SQ, You CX, Wang XF, Hao YJ. MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol 2019:222(2):735–751. [DOI] [PubMed]
- 51.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29:1023–1026. [PubMed] [Google Scholar]
- 54.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, De Peer YV, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q, Zhao P, Li J, Zhang C, Wang L, Ren Z. Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol Gen Genomics. 2014;289:103–124. doi: 10.1007/s00438-013-0789-x. [DOI] [PubMed] [Google Scholar]
- 57.Liu M, Zhang C, Duan L, Luan Q, Li J, Yang A, et al. CsMYB60 is a key regulator of flavonols and proanthocyanidans that determine the colour of fruit spines in cucumber. J Exp Bot. 2019;70(1):69–84. doi: 10.1093/jxb/ery336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Chen L, Shi Q, Ren Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020;291:110356. doi: 10.1016/j.plantsci.2019.110356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Figs. S1 to S4. (Fig. S1. Ten conserved motifs from 142 CsbHLH proteins; Fig. S2. Genome locations of the 142 CsbHLH genes on 7 chromosomes; Fig. S3. Cis-element analysis in the CsbHLH genes promoter regions; Fig. S4. Co-expression network of the CsbHLH genes).
Additional file 2: Table S1. Tandem duplication and Segmental duplication events.
Additional file 3: Table S2. Synteny analysis of bHLH genes in cucumber, Arabidpsis and tomato.
Additional file 4: Table S3.Cis-elements in the promoters of 142 CsbHLH genes.
Additional file 5: Table S4. Predicted functions of CsbHLHs with the function of their homologs verified in Arabidopsis by phylogenetic analysis.
Additional file 6: Table S5. Syntenic blocks between the cucumber and Arabidopsis and tomato genome.
Additional file 7: Table S6. String protein annotations.
Additional file 8: Table S7. Primers used for qRT-PCR.
Data Availability Statement
The data that support the results are included within the article and its additional files. Other relevant materials are available from the corresponding authors on reasonable request.