Abstract
In this work, we report 2 cases of vancomycin-resistant Enterococcus faecium bacteremia with development of daptomycin resistance in 2 patients with acute myeloid leukemia and myelodysplastic syndrome. Mutations related to daptomycin-nonsusceptible phenotype in liaSR genes were found in all strains of the study, including those with a minimum inhibitory concentration <1 µg/mL collected before daptomycin therapy. Epidemiological investigation using core genome single nucleotide polymorphism and core genome multilocus sequence typing revealed clonality of all the isolates. In this study, we conclude that real-time genome sequencing of clinical isolates can provide rapid access to timely information on daptomycin-resistant genotypes that would help clinicians speed up and optimize the selection of the antibiotic for treatment.
Keywords: Daptomycin, Antibiotic Resistance, Bacteremia, Enterococcus faecium
Vancomycin-resistant Enterococcus faecium (VREfm) is a worldwide cause of nosocomial infection and has been declared a serious public health threat due to its ability to acquire and disseminate antibiotic resistance determinants [1]. Resistance to antibiotics prevalently used as primary treatment options for VREfm infections has already been reported [1, 2], dramatically reducing the therapeutic options available. Daptomycin (DAP) is a cyclic lipopeptide antibiotic that has been successfully used for the treatment of a variety of VREfm infections, being associated with better clinical outcomes than other commonly used antibiotics in the treatment of VREfm bacteremia [3]. Although the mechanisms of DAP resistance in VREfm are unclear, current knowledge suggests that the nonsusceptible phenotype for DAP is the result of a complex, multifactorial mechanism based on mutations in genes related to membrane homeostasis in the resistant strains [4].
For whole-genome sequencing, we additionally explored MinION by Oxford Nanopore Technologies, a portable and real-time sequencing platform that generates long-read sequences and has been successfully used in real-time genomic surveillance studies [5] and for the detection of antimicrobial resistance genes in different bacterial species [6].
Here, we present 2 cases of prolonged bacteremia with phenotypic development of DAP nonsusceptibility (DAP-NS) during treatment in 2 inpatients with acute myeloid leukemia (AML) and myelodysplastic syndrome. Whole-genome sequencing allowed us to detect the DAP-NS genotypes in all the strains analyzed, even in those with a minimum inhibitory concentration (MIC) <1 µg/mL collected before DAP treatment and development of the resistant phenotype. In addition, we provide evidence for the potential use of MinION to detect mutations known to be associated with DAP-NS phenotype within 5 hours after initiation of sequencing.
CASES REPORT
Case 1
A 63-year-old Caucasian male with chronic lymphocytic leukemia, which had recently transformed to AML, was admitted from the clinic to start induction chemotherapy in June 2018. The treatment started with a regimen of clofarabine and cytarabine, along with standard antibacterial and antifungal prophylaxis with levofloxacin and fluconazole. The patient became neutropenic after chemotherapy (absolute neutrophil count [ANC], 0). On hospital day 7, the patient developed diarrhea and fever up to 101.8ºF (38.8ºC), with left foot cellulitis noted on exam. Blood cultures were drawn, and the patient was started on vancomycin, cefepime, and micafungin. Cefepime treatment was changed the following day into meropenem because of suspicion of typhlitis. He developed acute kidney injury requiring hemodialysis after placement of a Quinton catheter. Blood cultures remained negative. The patient seemed to be gradually improving on this regimen but remained neutropenic. On day 13 of neutropenia (ANC, 0), the patient relapsed again with fevers of up to 102.8ºF (39.3ºC) and chills. Blood cultures were drawn, and the micafungin regimen was changed to liposomal amphotericin B (AmBisome). Vancomycin and meropenem were continued. Blood cultures grew VREfm (vancomycin MIC ≥ 64 µg/mL, DAP MIC ≤ 1 µg/mL). Vancomycin treatment was immediately discontinued, and DAP 6 mg/kg (dosing adjusted with renal function) was started. All central lines and hemodialysis catheters were removed. The patient, however, did not improve and had persistent blood cultures positive for VREfm. The DAP dose was increased to 8 mg/kg on day 3 and then 10 mg/kg on day 6 of bacteremia. Blood cultures drawn during DAP therapy showed that DAP MIC had increased to 8 µg/mL (reaching the nonsusceptibility breakpoint according to the Clinical and Laboratory Standards Institute [CLSI]) [7]. DAP was discontinued, and a linezolid regiment was started. The family decided to withdraw life-sustaining treatment, and antibiotics were stopped the next day. The patient was discharged home to inpatient hospice care (Supplementary Figure 1).
Case 2
A 64-year-old Caucasian male with high-risk myelodysplastic syndrome (MDS) was admitted for a haploidentical stem cell transplant in August 2018. He had a stem cell transplant on hospital day 8, with the post-transplant course complicated by delayed engraftment. The patient developed neutropenic fever up to 101.5ºF (38.6ºC) on day 21 after transplant (ANC, 100). Blood cultures drawn at that time grew VREfm (vancomycin MIC ≥ 64 µg/mL, DAP MIC ≤ 1 µg/mL). The patient was started on DAP 8 mg/kg with cefepime. Cefepime was discontinued once neutrophils recovered and all central venous catheters were removed. Despite initiation of DAP, the patient remained persistently bacteremic. Ceftaroline was added to DAP for synergistic effect; the dose of DAP was increased to 12 mg/kg on day 9 in view of persistent bacteremia and rising MICs. Transthoracic and transesophageal echocardiograms did not show any valvular vegetations. Magnetic resonance imaging of the spine showed no evidence of diskitis or osteomyelitis. DAP MIC increased from 4 µg/mL to 8 µg/mL during therapy with DAP on sequential blood cultures. On the 12th day of bacteremia, DAP and ceftaroline regimens were discontinued, and treatment with linezolid (MIC = 2 µg/mL) was started. Blood cultures obtained within 24 hours were negative. The patient was continued on linezolid treatment for an additional 4 weeks. Repeat bone marrow biopsy showed persistence of MDS. The family decided to focus on comfort measures, and life-sustaining treatment was stopped within 1 week (Supplementary Figure 1).
METHODS
Bacterial Isolates
The 20 VREfm isolates were identified from positive blood cultures from 2 inpatients with prolonged bacteremia from the University of Arkansas for Medical Sciences (UAMS). Samples collected at different time points from the 2 inpatients were grown on blood cultures and processed on the BacT/ALERT 3D (bioMérieux) system. The Accelerate Pheno system (Accelerate Diagnostics, Tucson, AZ, USA) was used for identification and susceptibility testing of initial positive blood cultures. For repeat positives from the same patient, the Vitek 2 MS and Vitek 2 systems were also used for identification and susceptibility testing.
DNA Extraction and Quantification
Isolated colonies on the blood agar plates were picked and re-suspended into a DNA/RNA Shield Collection and Lysis Tube (Zymo Research, Irvine, CA, USA). Genomic DNA was extracted from the tube using the Quick-DNA Fungal/Bacterial kit (Zymo Research) from colonies of VREfm. The purity of extracted DNA was determined using a NanoDrop Spectrophotometer by measuring the A260/280 and A260/230 ratios. DNA integrity and quantity were determined using an Agilent 2200 TapeStation and Qubit 3.0 Assay, respectively. Purified DNA was aliquoted into 2 tubes for MinION and Illumina sequencing.
Vancomycin and Daptomycin Susceptibility Testing
Vancomycin resistance was confirmed using E-tests (bioMérieux). Antimicrobial susceptibility test results were interpreted using the M100 CLSI standards [7]. Daptomycin MIC results were confirmed by the clinical microbiology lab using the E-test method.
Whole-Genome Sequencing Methods
MinION Sequencing
Nanopore sequencing libraries were prepared using a PCR-free method of multiplexing samples with the Rapid Barcoding kit (SQK-RBK004, ONT), and sequencing of the barcoded DNA was performed on a single R9.4/FLO-MIN106 ONT flow cell on the MinION (version Mk1B, ONT) for 48 hours. Base calling was carried out with Albacore, version 2.3.4, using standard default configuration. Reads were then filtered using a mean quality score of 9, and only reads >2000 bases were included in downstream analysis. To identify the presence of resistance genes, each data set was analyzed by ARMA workflow (version 2.2.14) via EPI2ME (https://epi2me.nanoporetech.com).
Illumina Sequencing
Paired-end 150-bp libraries were constructed using the KAPA HyperPlus kit (Roche Diagnostics Corporation, Indiana, USA) with enzymatic fragmentation for 10 minutes. The resulting genomic libraries of the remaining 20 E. faecium isolates were sequenced using the Illumina NextSeq 550 platform at the UAMS Myeloma Center. Adapters were trimmed using fastp, version 0.19.5 [8], with default settings. Trimmomatic, version 0.38 [9], was used to remove poor-quality reads,withthefollowingparameters:HEADCROP:15 LEADING:20 TRAILING:20 SLIDINGWINDOW:5:20 MINLEN:50. The quality of pre- and postprocessed reads was assessed with the FastQC tool, version 0.11.8 [10]. The resulting high-quality reads were assembled de novo using SPAdes, version 3.13.0 [11], with the settings “error-correction” and “careful,” k-mer of lengths 21, 33, 55, and 77, and minimum contig size of 50 bp. Draft genomes were quality-checked using default settings in QUAST, version 5.0.2 [12].
Single Nucleotide Variation Identification
Single nucleotide variation (SNV) calling was performed on sequenced reads (from 20 isolates) against liaS, liaR, and cls genes as reference using Snippy, version 4.4.0 (https://github.com/tseemann/snippy). DNA sequences of liaS (ANQ34009.1), liaR (ANQ34010.1), and cls (TEA56807.1) genes from the species Enterococcus faecium were downloaded from the CARD database. To perform SNVs calling, Snippy was run with the “--fbopt ‘--max-complex-gap 0’ --minfrac 0.7” parameter to map the short reads and with “--fbopt ‘--max-complex-gap 0’ --minfrac 0.7 --bwaopt ‘-x ont2d’” as the recommended parameter for Nanopore reads. The allele frequency of missense variants of all isolates was recalculated using pysam [13] and plotted using the ggplot2 package from R [14].
Comparative Analysis of Multilocus Sequence Typing, Core Genome Multilocus Sequence Typing, and Core Genome Single Nucleotide Polymorphism Typing
Multilocus sequence typing (MLST) of the 20 isolates from the 2 inpatients was obtained using “mlst” software (https://github.com/tseemann/mlst), which incorporates components of the PubMLST database (https://pubmlst.org/) [15]. To set up core genome MLST (cgMLST) schema for clinical 20 E. faecium isolates, we used the chewBBACA suite, version 2.0.16 [16]. The schema was defined using the parameter -p 1, removing the genes suggested by the previous genome quality test that was carried out with parameters -n 10 -t 50 -s 1. The final number of genes in the scheme for our isolates was 2519. The minimum spanning tree was plotted using PHYLOVIZ, version 2.0, applying the goeBURST clustering algorithm [17]. Snippy software (Snippy, version 4.4.0; https://github.com/tseemann/snippy) was used to identify core genome concatenated single nucleotide polymorphisms (SNPs) where the reference genome E. faecium UAMSEF_1 was used.
RESULTS
To verify the presence of nucleotide substitutions in the LiaFSR 3-component system and cls gene, known to be associated with DAP-NS phenotype and clinical failures of therapy [6] in VREfm strains, we sequenced all isolates obtained from blood cultures drawn from both inpatients throughout the duration of their DAP treatment using Illumina paired-end sequencing. Furthermore, to evaluate the use of real-time sequencing in detection of variants for antibiotic stewardship, additionally obtaining high-quality complete genomes for in-depth analysis, 4 samples collected on the first and last days of bacteremia from both cases were sequenced using MinION long-read sequencing (Table 1).
Table 1.
Clinical Metadata and Resistance Phenotype Information for the Sequenced Isolates From Cases 1 and 2
| Isolate | Case | MLST | cgMLST | cgSNP | Vancomycin | Unit | Daptomycin MIC | Daptomycin Susceptibility | Collection Date | Sequencing Method | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UAMSEF_1 | 1 | 80 | 1 | 1 | vanA | E7 | ≤1 μg/mL | S | 6/28/2018 | MinION + Illumina | Blood |
| UAMSEF_2 | 1 | 80 | 2 | 2 | vanA | E7 | ≤1 μg/mL | S | 6/28/2018 | Illumina | Blood |
| UAMSEF_3 | 1 | 80 | 1 | 1 | vanA | E7 | 2 μg/mL | S | 6/29/2018 | Illumina | Blood |
| UAMSEF_4 | 1 | 80 | 2 | 3 | vanA | E7 | 4 μg/mL | SDD | 7/1/2018 | Illumina | Blood |
| UAMSEF_5 | 1 | 80 | 3 | 4 | vanA | E7 | 4 μg/mL | SDD | 7/2/2018 | Illumina | Blood |
| UAMSEF_6 | 1 | 80 | 4 | 5 | vanA | E7 | 4 μg/mL | SDD | 7/3/2018 | Illumina | Blood |
| UAMSEF_7 | 1 | 80 | 1 | 6 | vanA | E7 | 8 μg/mL | R | 7/5/2018 | Illumina | Blood |
| UAMSEF_8 | 1 | 80 | 2 | 3 | vanA | E7 | 8 μg/mL | R | 7/7/2018 | MinION + Illumina | Blood |
| UAMSEF_9 | 2 | 80 | 5 | 7 | vanA | E7 | ≤1 μg/mL | S | 9/18/2018 | MinION + Illumina | Blood |
| UAMSEF_10 | 2 | 80 | 5 | 8 | vanA | E7 | ≤1 μg/mL | S | 9/18/2018 | Illumina | Blood |
| UAMSEF_11 | 2 | 80 | 6 | 9 | vanA | E7 | 4 μg/mL | SDD | 9/19/2018 | Illumina | Blood |
| UAMSEF_12 | 2 | 80 | 6 | 9 | vanA | E7 | 4 μg/mL | SDD | 9/19/2018 | Illumina | Blood |
| UAMSEF_13 | 2 | 80 | 5 | 7 | vanA | E7 | 4 μg/mL | SDD | 9/20/2018 | Illumina | Blood |
| UAMSEF_14 | 2 | 80 | 5 | 10 | vanA | E7 | 4 μg/mL | SDD | 9/20/2018 | Illumina | Blood |
| UAMSEF_15 | 2 | 80 | 6 | 11 | vanA | E7 | 4 μg/mL | SDD | 9/21/2018 | Illumina | Blood |
| UAMSEF_16 | 2 | 80 | 5 | 7 | vanA | E7 | 4 μg/mL | SDD | 9/21/2018 | Illumina | Blood |
| UAMSEF_17 | 2 | 80 | 6 | 12 | vanA | E7 | 8 μg/mL | R | 9/24/2018 | Illumina | Blood |
| UAMSEF_18 | 2 | 80 | 6 | 11 | vanA | E7 | 8 μg/mL | R | 9/24/2018 | Illumina | Blood |
| UAMSEF_19 | 2 | 80 | 5 | 7 | vanA | E7 | 8 μg/mL | R | 9/26/2018 | Illumina | Blood |
| UAMSEF_20 | 2 | 80 | 6 | 13 | vanA | E7 | 8 μg/mL | R | 9/26/2018 | MinION + Illumina | Blood |
MLST, cgMLST, and cgSNP were performed using 20 isolates (8 from patient 1 and 12 from patient 2).
Abbreviations: cgMLST, core genome multilocus sequence typing; cgSNP, core genome single nucleotide polymorphism; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; S, Susceptible; R, Resistant; SDD, Susceptible-Dose Dependent.
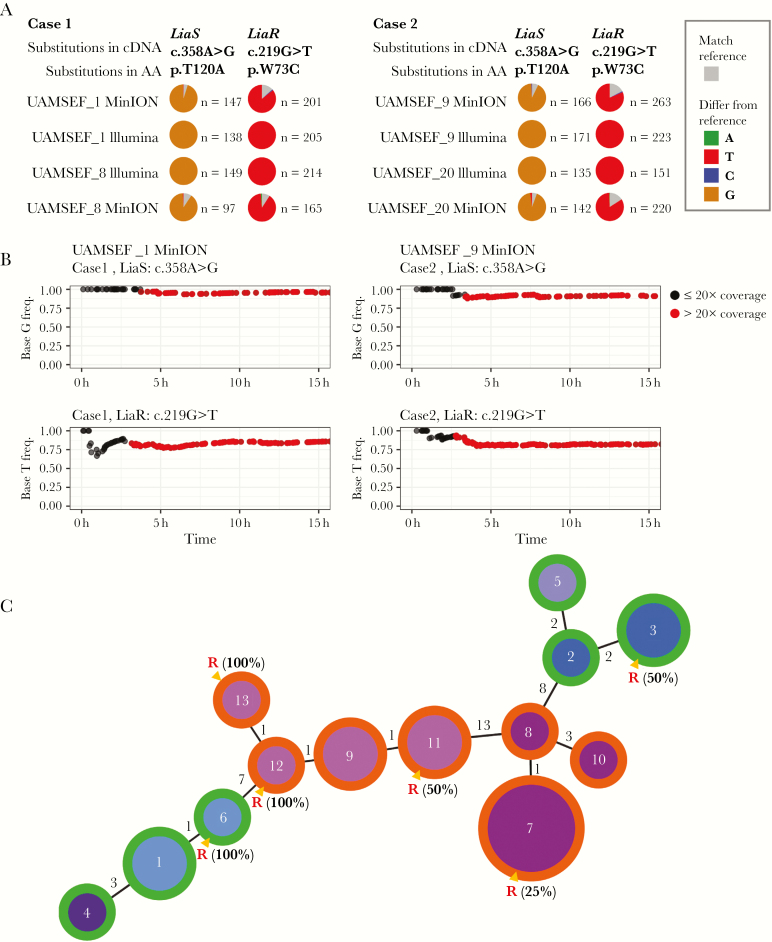
The real-time Antimicrobial Resistance Mapping Application (ARMA) workflow provided by Oxford Nanopore Technologies allows the detection of genes responsible for an antimicrobial resistance phenotype by aligning input reads against all reference sequences in the Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca/download) [18]. During the MinION sequencing, the integrated ARMA workflow detected reads for the vancomycin resistance vanA gene, the response regulator liaR gene, the histidine kinase liaS gene, and the cardiolipine synthetase cls gene in all 4 isolates from both cases. After whole-genome sequencing was completed, single nucleotide variant (SNV) calling was performed using the variant allele frequency threshold of 70% for mutations and a sequencing depth >20X. We identified the most common nonsynonymous co-mutations related to DAP resistance in liaS and liaR coding genes (protein variants LiaST120A and LiaRW73C) (Figure 1A) together with other missense variations. The presence of all variants previously mentioned was validated using Illumina reads (Supplementary Figure 2, Supplementary Table 1). To assess to the feasibility of rapid detection, we investigated the variant allele frequencies of the co-mutations LiaST120A and LiaRW73C in the liaS and liaR genes using Nanopore sequencing time stamps. Within 5 hours, co-mutations of both cases were observed with a sequencing depth of >20X (Figure 1B).
Figure 1.
A, The concordance of MinION and Illumina in the co-mutations LiaST120A and LiaRW73C proteins in different time points and patients. We observed that the co-mutations of A to G at position 358 (c.358A > G) of liaS and G to T at position 219 (c.219G > T) of liaR in cDNA were responsible for substitutions in LiaS (T120A) and LiaR (W73C) proteins that are known to be associated with the daptomycin-nonsusceptible (DAP-NS) phenotype. The base frequency at each locus is represented with a gray color in the pie chart. If a nucleotide differs from the reference sequence, the pie chart was colored in proportion to the frequency of each base (A = green, C = blue, T = red, G = brown). On the right panel of the pie chart, “n” indicates the total number mapped reads on a given position. B, Detection of the co-mutations liaS (c.358A > G) and liaR (c.219G > T) in cDNA using MinION reads. The upper 2 plots represent the substitution frequency of G base in LiaS (c.358A > G) with information on sequencing depth, and the bottom 2 plots represent the substitution frequency of T base in LiaR (c.219G > T) with information on sequencing depth. Dots colored in black have a sequencing depth ≤20X, and dots in red have a sequencing depth >20X in coverage at a given time point. We observed that the co-mutations in cDNA responsible for co-mutations in protein known to be associated with the DAP-NS phenotype were already present in MinION data sequenced up to 5 hours with 20X coverage. C, Comparative analysis of core genome multilocus sequence typing (cgMLST) and core genome single nucleotide polymorphism (cgSNP) typing of 20 isolates from the 2 patients. The minimum spanning tree was built by goeBURST using single linkage cluster analysis based on the core genome SNP information obtained from Snippy, version 4.4.0, of 20 isolates. The numbers inside nodes indicate the cgSNP types (13 in total). The numbers on the edges connecting the nodes indicate allelic variations between isolates according to the goeBURST hierarchical clustering algorithm. The size of the nodes represents the number of isolates with the same cgSNP typing. The color of the nodes indicates cgMLST information obtained using chewBBACA, version 2.0.16. The outer ring of the nodes colored in green indicates nodes that are comprised of isolates from case 1, and nodes with the outer ring colored in orange contain isolates from case 2. Nodes marked with a red R contain DAP-NS strains with minimum inhibitory concentration values of 8 μg/mL. The percentage of DAP-NS strains in each cluster is also shown.
Multilocus sequence typing analyses of the 20 isolates of this work showed that all isolates from both cases belong to the same sequence type (ST80). Additional analyses of the SNPs found in all 20 isolates (core genome SNP [cgSNP]: core genome SNP analysis), using the first isolate of case 1 (E. faecium UAMSEF_1) as reference, revealed that isolates from case 1 represent a more divergent population than isolates from case 2. Isolates from case 1 differ in 0 to 17 core SNPs, whereas isolates from case 2 differ in 0 to 15 core SNPs. Only 28 SNPs were observed when comparing the 20 isolates together. Further, cgMLST analysis using our own schema with 2519 coding sequences confirmed that the number of allelic differences found between isolates from case 2 were smaller than those found in case 1, in agreement with cgSNP analysis results. Isolates from case 1 were resolved into 4 clonal complexes (CCs), whereas isolates from case 2 were clustered into 2 different CCs (Figure 1C, Table 1).
CONCLUSIONS
According to the results of the cgSNP analysis and the allelic variations from genes present in 20 isolates (cgMLST), we conclude that isolates from both cases may belong to the same genetic lineage, implying that both patients were colonized by the same strain. It is worth noting that the 2 inpatients occupied the same unit at the hospital but with a time lapse of 2 months between admission dates with no overlap (Table 1).
As shown in Table 1, the usage of DAP as treatment for VREfm bacteremia was strongly correlated to an increase in DAP MIC values in both cases, which increased as soon as the first DAP doses were administrated. Our data therefore support previous studies that argue that DAP exposure is a key driver of resistance development [19, 20] and that DAP-NS genotypes can be present in VREfm strains exhibiting DAP MICs <4 μg/mL [19, 21]. Changes in resistance levels to DAP during the course of antibiotic treatment in both cases could therefore be due to additional changes at the transcriptomic level as a result of exposure to the DAP antibiotic. The existence of these variants in genes linked to the DAP-NS phenotype, observed in samples collected before DAP therapy in both patients, with DAP MIC values ≤1 μg/mL, underscores the lack of correlation between the clinical breakpoints for DAP-NS (MIC > 8 μg/mL) and the development of the resistant phenotype.
Lastly, we observed that co-mutations in liaSR genes related to the DAP-NS phenotype were detected in MinION reads within 5 hours. This evidence supports the proof of concept that rapid mutation detection using the MinION platform could enable the early detection of antibiotic resistance genes and mutations. Real-time information on antibiotic resistance genotypes complementary to conventional clinical analyses could help accelerate the selection of the optimal drug for antibiotic therapy of multidrug-resistant strains.
Supplementary Material
Acknowledgments
Data processing for this work was performed in part using high-performance computing facilities, in particular the Grace cluster, provided by the UAMS, and managed by the Department of Biomedical Informatics.
Financial support. This work was supported by the UAMS College of Medicine Barton Pilot Grant FY19 program (AWD00052801) given to S.J. P.J. and T.W. are partially supported by the National Institute of General Medical Sciences of the National Institutes of Health award No. P20GM125503. Z.U., T.W., P.J., D.U., and A.K. are supported in part by the Helen Adams & Arkansas Research Alliance. D.U. is also supported by NIH/NIGMS grant 1P20GM121293.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. S.J., A.K., D.U., and B.W. conceived and designed the study. P.J. and Z.U. performed the bioinformatics analyses. P.J., Z.U., A.K., and S.J. were responsible for data interpretation. Z.U., P.J., A.K., and S.J. wrote the first draft of the paper. Z.U., P.J., A.K., S.J., E.M., D.U., and T.W. revised subsequent versions. C.A. carried out the experimental works in the clinical microbiology laboratory. J.L., M.M., and E.M. reviewed clinical metadata. T.W. performed Nanopore sequencing. R.T. performed Illumina sequencing. All authors have read and approved the final manuscript.
Ethical approval. Ethics approval and consent to participate. This study was approved by the Institutional Review Board of University of Arkansas for Medical Sciences (IRB No. 228137).
Data availability. Raw sequencing data and assembly and functional annotations for the isolates used in this study are available under the BioProject accession numbers PRJNA518133 and PRJNA520878 in the NCBI database.
References
- 1. Guzman Prieto AM, van Schaik W, Rogers MR, et al. . Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front Microbiol 2016; 7:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bender JK, Cattoir V, Hegstad K, et al. . Update on prevalence and mechanisms of resistance to linezolid, tigecycline and DAP in enterococci in Europe: towards a common nomenclature. Drug Resistance Updates 2018; 40:25–39. [DOI] [PubMed] [Google Scholar]
- 3. Britt NS, Potter EM, Patel N, Steed ME. Comparison of the effectiveness and safety of linezolid and daptomycin in vancomycin-resistant enterococcal bloodstream infection: a national cohort study of Veterans Affairs patients. Clin Infect Dis 2015; 61:871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Munita JM, Bayer AS, Arias CA. Evolving resistance among gram-positive pathogens. Clin Infect Dis 2015; 61(Suppl 2:S48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quick J, Loman NJ, Duraffour S, et al. . Real-time, portable genome sequencing for Ebola surveillance. Nature 2016; 530:228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xia Y, Li AD, Deng Y, et al. . MinION nanopore sequencing enables correlation between resistome phenotype and genotype of coliform bacteria in municipal sewage. Front Microbiol 2017; 8:2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 8. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018; 34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andrews S. FASTQC. A quality control tool for high throughput sequence data 2010. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (October 4, 2018, released v0.11.8).
- 11. Bankevich A, Nurk S, Antipov D, et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013; 29:1072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 15. Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010; 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silva M, Machado MP, Silva DN, et al. . chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom 2018; 4:e000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Francisco AP, Bugalho M, Ramirez M, Carriço JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 2009; 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McArthur AG, Waglechner N, Nizam F, et al. . The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 2013; 57:3348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munita JM, Panesso D, Diaz L, et al. . Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob Agents Chemother 2012; 56:4354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Egli A, Schmid H, Kuenzli E, et al. . Association of daptomycin use with resistance development in Enterococcus faecium bacteraemia—a 7-year individual and population-based analysis. Clin Microbiol Infect 2017; 23:118.e1–7. [DOI] [PubMed] [Google Scholar]
- 21. Diaz L, Tran TT, Munita JM, et al. . Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 2014; 58:4527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.