Abstract
Transcription and pre-mRNA splicing are coupled to promote gene expression and regulation. However, mechanisms by which transcription and splicing influence each other are still under investigation. The ATPase Prp5p is required for pre-spliceosome assembly and splicing proofreading at the branch-point region. From an open UV mutagenesis screen for genetic suppressors of prp5 defects and subsequent targeted testing, we identify components of the TBP-binding module of the Spt–Ada–Gcn5 Acetyltransferase (SAGA) complex, Spt8p and Spt3p. Spt8Δ and spt3Δ rescue the cold-sensitivity of prp5-GAR allele, and prp5 mutants restore growth of spt8Δ and spt3Δ strains on 6-azauracil. By chromatin immunoprecipitation (ChIP), we find that prp5 alleles decrease recruitment of RNA polymerase II (Pol II) to an intron-containing gene, which is rescued by spt8Δ. Further ChIP-seq reveals that global effects on Pol II-binding are mutually rescued by prp5-GAR and spt8Δ. Inhibited splicing caused by prp5-GAR is also restored by spt8Δ. In vitro assays indicate that Prp5p directly interacts with Spt8p, but not Spt3p. We demonstrate that Prp5p's splicing proofreading is modulated by Spt8p and Spt3p. Therefore, this study reveals that interactions between the TBP-binding module of SAGA and the spliceosomal ATPase Prp5p mediate a balance between transcription initiation/elongation and pre-spliceosome assembly.
INTRODUCTION
Eukaryotic mRNA processing consists of multiple steps that occur in the nucleus after transcription, including 5′-capping, pre-mRNA splicing, 3′-polyadenylation, and RNA modifications, which occur mostly co-transcriptionally, and subsequent post-transcriptional steps of mRNA export and RNA surveillance (1,2). Although each of these steps can be investigated independently in vitro, much evidence in the past two decades demonstrated that these processes affect each other extensively and that such ‘coupling’ contributes to gene expression and regulation (reviewed in 3,4–6).
Transcription is central to the coupling of RNA processing events, primarily through the catalytic component, RNA polymerase II (Pol II) (6,reviewed in 7). Removal of introns from nascent transcripts by pre-mRNA splicing is essential in all eukaryotes. Coupling between transcription and splicing has been extensively studied. In one direction, components of the transcription machinery associate with splicing factors and regulate pre-mRNA splicing. For example, RNA Pol II has extensive association with SR proteins and other factors that promote efficient spliceosome assembly (8). The C-terminal domain (CTD) of RNA Pol II recruits SRp20, promotes exon skipping, and regulates alternative splicing (9). RNA Pol II and emergent splice sites in the nascent pre-mRNA are tethered together (10), with spliceosomal components being recruited in part by the Ser5-phosphorylated CTD of Pol II during transcription elongation (11). In the other direction, splicing factors also promote transcription elongation. Depletion of splicing factor SC35 induces RNA Pol II accumulation within the body of specific genes and attenuates transcription elongation, correlating with defective recruitment of transcription factor P-TEFb and a dramatic reduction of Ser2 phosphorylation of the CTD (12). Recruitment of SR proteins to nascent FOS transcript is RNase sensitive and transcription dependent, indicating that SR proteins are not pre-assembled with Pol II (13). A dual-function factor, Tat-SF1, identified as a transcription elongation factor in humans, interacts with snRNPs and strongly stimulates both polymerase elongation in vivo and splicing in vitro (14,15).
Two models have been proposed to explain the co-transcriptionality of splicing. The first is ‘recruitment coupling’, in which splicing factors are recruited by the transcription machinery. For example, the CTD of RNA Pol II directly interacts with a human spliceosomal U2AF65–Prp19 complex (16), and the yeast SR-like protein Npl3p facilitates co-transcriptional recruitment of splicing factors and thereby promotes splicing (17). The second model is ‘kinetic coupling’, which is achieved by coordinating the rates of transcription and splicing—i.e. the relative rates of sequential events are coordinated to optimize their function. In general, the transcription rate is hindered by chromatin structure, and the splicing rate is dependent on the strength of splice sites and binding of splicing regulators (18). However, transcription rates influence the outcome of splicing (19–21) and splice events also could be transcription checkpoints. Factors such as SC35, SAM68, and the DBC1–ZIRD (DBIRD) complex, which can modulate transcription and splicing rates, are important in the kinetic co-transcriptional model (12,22–25).
Evidence has pointed to U2 snRNP components and its related event, pre-spliceosome assembly, as critical in the process of co-transcriptional splicing. First, two core components of U2 snRNP, Lea1p/U2A′ and Msl1p/U2B″ (yeast/mammalian names), exhibit genetic interactions with GCN5; deletion of GCN5 rescues yeast lethality caused by double deletion of LEA1 and MSL1 (26). Gcn5p is a catalytic component of the Spt–Ada–Gcn5 Acetyltransferase (SAGA) complex, an evolutionary conserved, multifunctional transcription co-activator comprising two distinct enzymatic activities, acetylation and deubiquitination of histone residues (27,28). The histone acetyltransferase (HAT) activity of Gcn5p is required for co-transcriptional recruitment of the U2 snRNP (26). Second, Cus2p, a yeast U2 snRNP component and putative orthologue of human Tat-SF1 (29), has been proposed as a potential checkpoint factor in transcription elongation (30). Cus2p has been investigated as a functional target of Prp5p (31,32). Prp5p is a spliceosomal RNA-dependent ATPase required for stable binding of U2 snRNP to the branch site region (BS) and consequent pre-spliceosome assembly (33–36). PRP5 mutant allele prp5–1 causes transcriptional defects of intron-containing genes, in which accumulation of RNA Pol II across introns was observed, but this accumulation was relieved in the absence of Cus2p (30). This led to a model wherein Cus2p is a potential transcription elongation checkpoint factor during cotranscriptional spliceosome assembly.
Physical interactions have been reported between components of the SAGA complex and U2 snRNP, each complex containing around 20 subunits, conserved from yeast to mammals. The SAGA complex is important in transcription initiation and elongation, and in mRNA export (37,38). It is organized into four functional modules, including a HAT (histone acetyltransferase) module, a DUB (deubiquitinase) module, a Core structure module, and a TBP-binding module. 17S U2 snRNP is composed of U2A′, U2B'', Sm proteins, SF3a, and the SF3b complex (39). Human SAGA component SPT3 was found to interact with SF3B3/SAP130 (40,41), a subunit in the SF3b complex that is critical for pre-spliceosome assembly (42,43). In Drosophila, two SF3b subunits, SF3B3 and SF3B5, are proposed as components of the SAGA complex and interact with multiple SAGA subunits, including Sgf29 and Spt7, in a yeast two-hybrid assay (44).
Multiple genetic and physical interactions between the SAGA complex and U2 snRNP strongly suggest that these two complexes are important for regulation of co-transcriptional splicing. However, the details and mechanisms for how coupling between transcription and splicing is mediated remain unclear. In this study, we performed a genetic screen in yeast and found that a component of transcriptional complex SAGA, SPT8, genetically interacts with PRP5. Further genetic studies revealed that such interactions are limited to the TBP-binding module of the SAGA complex. We propose that the TBP-binding module (Spt8p/Spt3p) of SAGA, together with the spliceosomal RNA helicase Prp5p, mediate a balance between transcription initiation/elongation and pre-spliceosome assembly/proofreading.
MATERIALS AND METHODS
Yeast strains and plasmids
Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table S1. They were derived from strain 46ΔCUP (45), yYZX02 (MATa, ade2 cup1Δ::ura3 his3 leu2 lys2 prp5Δ::loxP trp1, pRS316-PRP5 [PRP5 URA3 CEN ARS]), or yYZX06 (MATa, ade2 cup1Δ::ura3 his3 leu2 lys2 prp5Δ::loxP trp1, cus2Δ::loxP pRS316-PRP5 [PRP5 URA3 CEN ARS]) (36). Strains with deletions of SAGA components or other factors were constructed by homologous recombination using PCR products of KAN-MX4 cassettes and G418 selection. Plasmids of prp5, prp28, spt8, gcn5 and ACT1-CUP1 reporter mutants were prepared by either in vivo gap repair cloning or traditional cloning.
Screen for suppressors of prp5-GAR cold-sensitivity
yYZX-02 cells (0.02 OD600) carrying the cold-sensitive prp5-GAR allele were spread on 10 cm -Trp plates and grown overnight at 30°C. To generate random mutations, cell plates were irradiated with 10, 25 or 50 mJ/cm2 ultraviolet light (Stratalinker, 254 nm) and then incubated at 16°C until colonies appeared (∼2 weeks). Suppressor colonies were then patched and cultured at 30°C. To exclude cus2 mutations, the CUS2 gene in each colony was amplified and Sanger sequenced. The remaining suppressor colonies were then transformed with yeast cDNA library plasmids. Plasmids in the yeast strains that reverted to 16°C cold sensitivity were isolated and sequenced to find suppressor candidates, which were further identified by sequencing of their chromosomal counterpart genes.
6-Azauracil and copper assays
Yeast strains were first grown overnight in synthetic complete (SC) media without indicated amino acid(s) depending on the plasmid back-bone. Cells were then diluted to OD 0.2 at 600 nm when the culture reached mid-log phase. For 6-azauracil (6-AU) assays, yeast cells in 10-fold serial dilutions were spotted on plates containing 75 μg/ml of 6-azauracil (Sigma-Aldrich) and cultured at various temperatures. Plates were photographed each day to monitor growth. Copper assays were carried out as described (36,45). Plates were scored by the maximum copper tolerance of strain growth after 4 days culture at 30°C.
Co-Immunoprecipitation, western blotting, and RT-PCR
To perform in vivo co-IP, Prp5p was tagged at the N-terminus with FLAG through overlapping PCR and cloning on pRS314-TRP plasmid, Spt8p was tagged at C-terminus with 3xHA on pRS313-HIS plasmid. Both plasmids were then transformed into a spt8 strain (yYZX15 with WT PRP5 on a URA plasmid), and transformants were further selected to lose the URA plasmid using 5-FOA plates. Tag-containing strains were grown into mid-log phase and harvested, and the cell pellets were washed once in TBS buffer and the cell lysates were prepared by glass beads using lysis buffer (50 mM Tris–HCl, pH 7.0, 0.1% Triton, 150 mM NaCl) with protease inhibitors. Cleared lysates were then tumbled with anti-HA beads (Invitrogen) for 4 h at 4°C, followed by washing three times using the lysis buffer. Bead-bound proteins were separated on SDS-PAGE gels and probed with anti-HA (Invitrogen) or anti-Flag (Sigma) antibodies.
Purification of 6xHis-tagged full-length and truncated Prp5p proteins was described previously (46). GST-tagged Spt8p and Spt3p were generated by cloning coding sequences of SPT8 and SPT3 into pGEX-4T-1 and expressed in Escherichia coli (Rosetta) for 20 h at 16°C with 1.0 mM IPTG followed by purification using glutathione-sepharose (GE) under standard conditions. All purified proteins were dialyzed and stored in buffer D (20 mM HEPES–KOH at pH 7.9, 0.2 mM EDTA, 100 mM KCl, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonylfluoride, 20% glycerol). For in vitro protein–protein interaction assays, Prp5p proteins (40 pmol) were incubated with Spt8p or Spt3p (10 pmol) in 500 μl binding buffer (20 mM Tris–Cl at pH 8.0, 150 mM NaCl, 1mM EDTA, 0.2% Triton X-100, 0.5 mM PMSF) for 4 h at 4°C with Ni-NTA agarose. Bead pellets were washed four times with wash buffer (50 mM Tris–Cl at pH 8.0, 140 mM NaCl, 1 mM EDTA, 0.1% Triton X-100) and then resuspended in 50 μl of 1× sample loading buffer for SDS-PAGE electrophoresis. Spt8p, Spt3p and Prp5p were detected by western blotting using anti-GST (Sigma) or anti-His (Sigma) monoclonal antibodies.
Total RNA was isolated from yeast strains grown under various conditions, and RT-PCR was then performed as described (36,47). All related primers are listed in Supplementary Table S2.
Chromatin immunoprecipitation and Hiseq-PE150 sequencing
Chromatin Immunoprecipitation (ChIP) was performed as described (17). Yeast strains were grown overnight from a starting culture and diluted in fresh medium to reach mid-log phase. Cells were then either shifted to 16°C for an additional 30 min culture, or added to 6-AU (final 75 μg/ml) for additional 1 h culture at 30°C. For cross-linking, cells were treated with 1% formaldehyde for 15 min and then quenched by addition of glycine to 125 mM. After centrifugation, cell pellets were washed with cold TBS and lysed by glass beads in FA lysis buffer (50 mM HEPES–KOH at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 0.1% SDS, 1 mM PMSF) containing protease inhibitors. Chromatin was sheared using a Diagenode Bioruptor. Immunoprecipitations were performed using anti-RNA Pol II antibody (8WG16; Covance MMS-126R), anti-HA-agarose beads (Sigma), and anti-Flag M2 Affinity Agarose (Sigma). Associated DNAs were then purified using a PCR clean-up kit (Axygen) and analyzed by QuantStudio Real-Time PCR (Thermo Fisher) and Hiseq-PE150 (Novogene). For Real-Time PCR, primers used are as described (30,48), and the PCR signals from the immunoprecipitated samples were normalized to input.
The standard framework of ChIP-seq was used from nf-core/ChIP-seq version 1.1.0 pipeline (49): reads were aligned to S. cerevisiae genome (version R64–1-1) using BWA version 0.7.17-r1188; reads that were marked as duplicates and mapped to multiple locations were removed by Samtools version 1.9; read counts per bin were normalized to one million mapped reads (RPM) by deepTools version 3.2.1; peak calling used MACS2 version 2.1.2. We used DESeq2 version 1.22.2 to make differential binding analysis of different IP samples. IP enrichment signals were calculated using ratio of RPMIP and RPMinput. Then, deepTools was used to draw the different value coverage of IP enrichment signal from two IP samples in exons, introns and 200 bp regions before the TSS. The gene annotation was obtained from the NCBI database.
RESULTS
spt8 mutants rescue the cs phenotype of the prp5-GAR allele
To identify additional interactions with or functions of Prp5p, we performed a yeast genetic screen using UV irradiation-induced mutagenesis and searched for suppressors that rescued the cs phenotype of the prp5-GAR allele (Figure 1A), which has mutations in the Prp5p SAT motif, has severely reduced ATPase activity, and is defective in splicing of branch site (BS) region mutant introns—i.e. introns with reduced base-pairing to U2 snRNA (36). The WT background S. cerevisiae strain with prp5-GAR allele cannot grow at the non-permissive temperature 16°C (Figure 1B); however, in the genetic screen we obtained dozens of clones that could grow at 16°C after UV-irradiation.
Figure 1.

Reciprocal genetic interactions between transcription factor SPT8 and splicing factor PRP5. (A) Strategy for genetic screen for suppressors of the cold sensitive (cs) prp5-GAR allele. One spt8 mutant allele was obtained. (B) Deletion of SPT8 gene rescues the cs phenotype of prp5-GAR allele at 16°C. (C) At the permissive temperature (30°C), prp5 alleles (-SAG, -TAG and -GAG) rescue growth defects of spt8Δ allele on 6-AU.
It was known that deletion of CUS2, encoding a protein component of U2 snRNP and an assumed functional target of Prp5p, rescued growth defects of the ts prp5–1 allele (31,32) and of the cs prp5-GAR allele (36). To eliminate cus2-mutated clones, we first amplified and sequenced the CUS2 gene in all suppressor clones, identifying more than 70% of clones that contained various mutations in CUS2, all of which introduced early stop codons thereby truncating Cus2p. We then focused on the non-cus2-mutated clones. We transformed cDNA library plasmids of S. cerevisiae (50) into the non-cus2 suppressor-containing clones to search for plasmids that could restore the cs phenotype, and identified a library plasmid carrying full-length transcription factor SPT8 that restored the yeast clone back to cold sensitivity. Further sequencing of this clone revealed that it had a mutation in the coding region of chromosomal SPT8, in which guanosine (position 980) was mutated to adenosine, introducing a stop codon (TAG) that terminated translation of Spt8p within its WD40 domain (Supplementary Figure S1A).
To confirm that loss of Spt8p, which is not essential for cell viability (51), suppressed prp5-GAR defects, we deleted the chromosomal SPT8 gene (spt8Δ) in the prp5-GAR strain and validated that spt8Δ also rescued the cs phenotype of the prp5-GAR allele (Figure 1B, cf. columns 2 to 1). Deletion of SPT8 did not change the growth of strains containing other prp5-SAT alleles (-SAG, -TAG, -GAG), all of which grew similarly to the WT PRP5-SAT strain at 16°C and 30°C (Figure 1B). Thus, inactivation of SAGA component Spt8p, either by mutation or deletion, partly rescues the severely ATPase-defective prp5-GAR allele.
prp5 alleles rescue growth defects due to SPT8 deletion
Spt8p is a component of SAGA, a chromatin-modifying complex that contains two distinct enzymatic activities, acetylation and deubiquitination of histone residues, and is involved in transcription initiation and elongation (27,28,52). We surmised from the SPT8−PRP5 genetic interaction that prp5 alleles might have previously-undetected effects on transcription. To address this possibility, we assayed growth in the presence of 6-azauracil (6-AU), which exacerbates transcription defects through inhibition of IMPDH, an enzyme catalyzing the first committed step in GMP biosynthesis (53,54). A series of prp5 alleles, including prp5-SAG, -TAG, -GAG and -GAR that have decreased ATPase activities (51%, 42%, 22% and 4%, respectively, relative to WT PRP5-SAT, 36), were tested at the permissive temperature (30°C). In comparison to the WT PRP5-SAT allele, strains with prp5-GAR allele grew mildly worse on 6-AU, but other prp5 mutant alleles did not show obvious defects in growth (Figure 1C, column 3). Consistent with Spt8p's primary function in transcription, growth of the spt8Δ strain on 6-AU was significantly inhibited (55); however, this inhibition was rescued by the presence of prp5-SAG, -TAG or -GAG alleles (Figure 1C column 4).
We further tested the 6-AU sensitivity at two other temperatures, 16°C and 37°C. At 16°C, all the yeast strains grew slower on 6-AU medium, but their patterns of inhibition and rescue were similar to that described above at 30°C (Supplementary Figure S1B). In contrast, at 37°C, the pattern was reversed: in the presence of all tested prp5 alleles, cells grown on 6-AU were significantly inhibited; however, spt8Δ rescued growth on 6-AU of the WT PRP5-SAT and prp5-GAR alleles, but not -SAG, -TAG, or -GAG alleles (Supplementary Figure S1C). Taken together, these data suggest a reciprocal genetic interaction between Prp5p and Spt8p that impacts transcriptional elongation.
SPT3, another SAGA component, also genetically interacts with PRP5
The yeast SAGA complex is composed of 19 protein components, which are roughly divided into four modules according to their functions, TBP-binding (Spt8p and Spt3p), core structural (Ada1p, Spt7p, Spt20p, Taf5p, Taf6p, Taf9p, Taf10p, and Taf12p), deubiquitinating (Sgf11p, Sgf73p, Sus1p and Ubp8p), and HAT (Ada2p, Ada3p, and Gcn5p) (37,56,57). To address whether other components of the SAGA complex are also involved in a reciprocal genetic interaction with Prp5p, we further tested deletion of the SPT3 gene, the other component of the TBP-binding module, and deletion of two genes selected from each of the other three SAGA modules, including spt7Δ, spt20Δ, ubp8Δ, sgf11Δ, gcn5Δ and ada2Δ (Figure 2A). None of the tested deletions could rescue the cs phenotype of prp5-GAR allele except for spt3Δ, which allowed yeast with prp5-GAR allele to grow at 16°C (Figure 2B, column 2), similar to the above findings with spt8Δ; in addition, the spt3Δ yeast strain grew slowly at 30°C, which was partially rescued by prp5-GAR allele (Figure 2B, cf. columns 4 to 3). Similarly, inhibition of growth on 6-AU of the spt3Δ strain could be rescued by prp5-TAG and -GAG alleles, but not by the prp5-GAR allele (Figure 2C, column 2). We note that prp5-GAR mildly reduces the growth at 30°C in the background of spt7Δ and gcn5Δ (Figure 2B, cf. columns 4 to 3).
Figure 2.
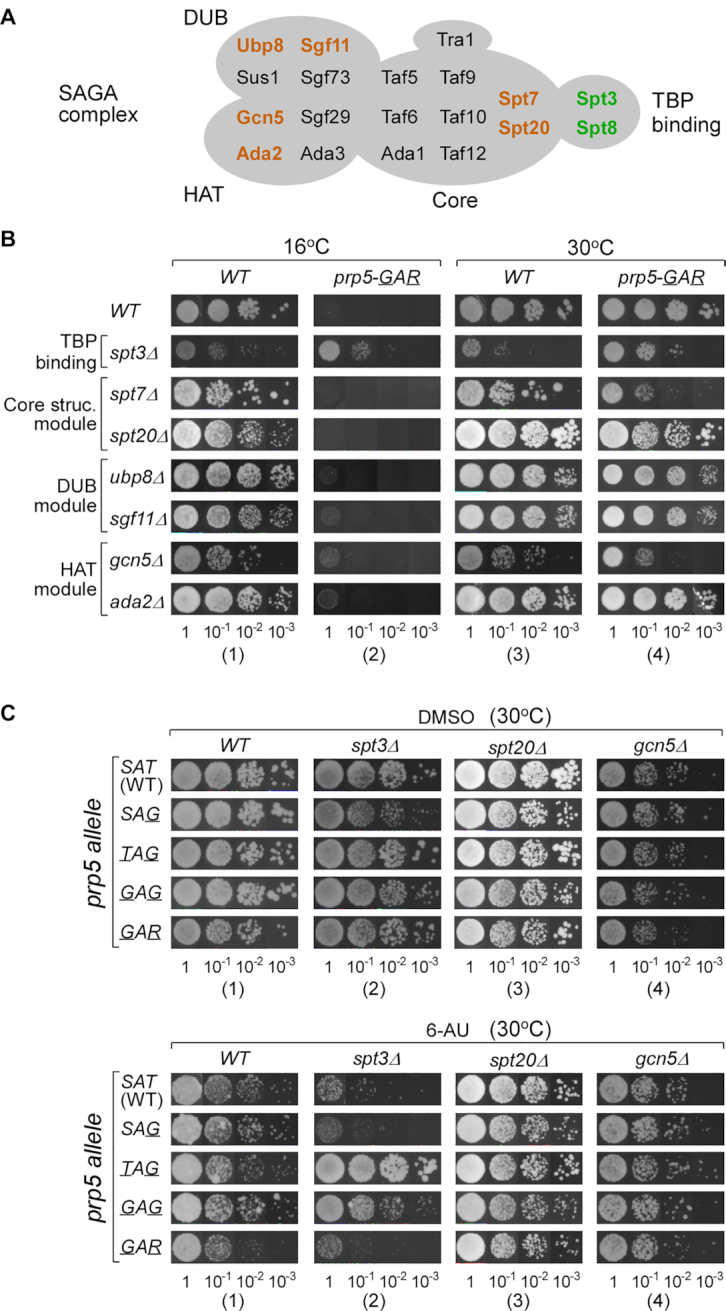
SPT3, another SAGA component, also genetically interacts with PRP5. (A) Illustration of the yeast SAGA complex. Components are classified into four groups (57), in which colored factors are tested in this study. (B) Deletion of SPT3 rescues the cs phenotype of the prp5-GAR allele, whereas deletion of other tested SAGA components cannot. (C) prp5 alleles (-TAG and -GAG) rescue growth defects of the spt3Δ allele on 6-AU.
GCN5 has genetic interactions with U2 snRNP protein component genes LEA1/U2A′ and MSL1/U2B'', and gcn5Δ rescues the lethality of LEA1 and MSL1 double deletion (26). Therefore, we further used two gcn5 mutant alleles, gcn5-AAA(KQL), which eliminates the HAT activity of Gcn5p, and gcn5-AAA(RGY), which retains intermediate HAT function (26), and tested their genetic interaction with prp5 alleles. As with gcn5Δ above, these two gcn5 mutant alleles could not rescue the cs phenotype of prp5-GAR allele (Supplementary Figure S2), indicating no detectable genetic interaction between GCN5 and PRP5 under the conditions tested here.
Therefore, these data reveal a previously unknown reciprocal genetic interaction between the TBP-binding module (SPT8 and SPT3) of the SAGA complex and the spliceosomal ATPase gene PRP5. In one direction, spt8Δ or spt3Δ rescues the cs phenotype of the prp5-GAR allele, indicating that impaired splicing can be improved by deletion of transcription factors; in the other direction, prp5 mutant alleles rescue the growth defects on 6-AU of spt8Δ and spt3Δ, indicating that impaired transcription can be improved by the slower spliceosomal ATPase.
Decreased RNA Pol II binding in the presence of prp5 mutant alleles is reversed by spt8Δ
To further explore the mechanism of this genetic interaction between PRP5 and SPT8/SPT3, we performed ChIP using Rbp1p antibody (8WG16) to detect occupancy of RNA Polymerase II (Pol II) on regions of two frequently tested genes: DBP2, an intron-containing gene, and PDR5, an intronless gene (48). At the permissive temperature (30°C), binding of Pol II on the intron-containing gene DBP2 in the presence of prp5-TAG or -GAR allele was decreased across gene regions in comparison to the WT PRP5-SAT allele (Figure 3A upper left). Deletion of SPT8 significantly decreased Pol II binding in the presence of all prp5 alleles (Figure 3A upper right). After shift to the non-permissive temperature (16°C), binding of Pol II on DBP2 was generally reduced and the inhibition by prp5-TAG allele was not as strong as at 30°C (Figure 3A lower left). In contrast, spt8Δ significantly improved Pol II binding on DBP2 in the presence of prp5-GAR allele at 16°C, with a >4-fold enhancement on the promoter region in the presence of prp5-GAR allele compared to WT-PRP5 (Figure 3A, lower right). However, on the intronless gene PDR5, we did not observe decreased Pol II binding in the presence of prp5 mutant alleles, or the reverse effects by spt8Δ (Figure 3A′). Taken together, these results demonstrate that rescue of the prp5-GAR cs phenotype by spt8Δ correlates with the increased binding of RNA Pol II in an intron-dependent manner.
Figure 3.
In the absence of SPT8, prp5 mutant alleles stimulate RNA polymerase II binding to an intron-containing gene. ChIP assays on intron-containing gene DBP2 and intronless gene PDR5 were performed using RNA Pol II antibody 8WG16 (A and A’), anti-HA agarose for the HA-tagged Spt8 strain (B) and for the HA-tagged Gcn5 strain (C), and anti-Flag agarose for the Flag-tagged Prp5 strains with WT SPT8 (D) or spt8Δ (E). Temperatures for yeast cultures and location of PCR primers are indicated. Bar graphs represent the co-immunoprecipitated signals and were normalized to input. All the data were analyzed from triplicates. Error bars represent standard deviation from the mean.
We next performed the same ChIP assays on yeast that grew in media with 6-AU, and found that 6-AU dramatically inhibited the binding of Pol II on both DBP2 and PDR5 genes (Supplementary Figure S3), as previously described (58). Similar to the above data (Figure 3A, lower right), in the presence of 6-AU, mutant prp5 alleles inhibit Pol II binding on intron-containing gene DBP2, whereas spt8Δ allows slow prp5 alleles to increase Pol II binding. These results are consistent with our genetic results (Figure 1), that prp5-TAG allele rescues cell growth on 6-AU when SPT8 is deleted.
Spt8p and Prp5p reciprocally affect each other's recruitment to transcribed genes
To further address the role of Spt8–Prp5 interaction, we performed additional ChIP assays using antibodies against epitope tags on Spt8p, Gcn5p, and Prp5p (Figure 3B–E). First, we observed that Spt8p binds to promoter regions of both the intron-containing gene DBP2 and the intronless gene PDR5; this binding was decreased after the transcription initiation site and then restored at the elongation stage, as previously described (59). In comparison to WT PRP5-SAT, prp5-TAG, and -GAR mutant alleles significantly enhanced the binding of Spt8p at the elongation stage (Figure 3B). Second, Gcn5p, a component of the HAT module, exhibited overall less binding and no obvious changes during transcription on both intron-containing and intronless genes; and the prp5 alleles did not alter binding of Gcn5p (Figure 3C). These data again suggested that the function of Gcn5p is not coupled with Prp5p's activity. Third, we performed Prp5p ChIP in yeast strains with or without SPT8. In the presence of SPT8, recruitment of WT Prp5p to the intron-containing gene DBP2 was steadily increased from the transcription initiation site to the elongation stage, and prp5 mutant proteins exhibited enhanced recruitment to DBP2 (Figure 3D). In contrast, spt8Δ significantly changed the recruitment of Prp5p on DBP2: WT Prp5p was increased, whereas Prp5p mutants were significantly inhibited (cf. Figure 3D to E). However, for the intronless gene PDR5, recruitment of WT Prp5p and its mutants was not changed across the transcribed regions, and spt8Δ had no detectable effects (Figure 3D and E).
Taken together, these data demonstrate that slow prp5 mutants enhance the recruitment of Spt8p to transcribing genes, especially at the elongation stage, and the presence of Spt8p facilitates the recruitment of Prp5p to the transcribing intron-containing gene. These results are globally consistent with the above-identified reciprocal genetic interactions.
Inhibited splicing caused by prp5-GAR is rescued by SPT8 deletion
We previously found that prp5-GAR significantly inhibited splicing of the WT ACT1-CUP1 reporter (36). Here, we investigated splicing changes of endogenous genes in the mutant strains by RT-PCR. In comparison to the WT strain, splicing of all eight tested intron-containing genes was inhibited in the prp5-GAR strain, exhibiting increased levels of pre-mRNA, but not in the spt8Δ strain (Figure 4A). Although the prp5-GAR allele is cs, splicing inhibition was already obvious at 30°C. Interestingly, the inhibited splicing caused by prp5-GAR was rescued by the spt8Δ, evidenced by a decrease of pre-mRNA level to a similar level as in the WT strain (Figure 4A). These data demonstrate that Spt8p contributes to the modulation of spliceosome assembly at the stage of U2 snRNP binding that is facilitated by the ATPase/RNA helicase Prp5p.
Figure 4.
ChIP-seq reveals that global RNA Pol II binding is mutually rescued by prp5-GAR and spt8Δ. (A) Inhibited splicing by the prp5-GAR allele is restored by SPT8 deletion. RT-PCRs were performed and the inhibited splicing ratios were semi-quantitated by signals of pre-mRNA / (pre-mRNA + mRNA) from triplicates. Analyses of genome-wide RNA Pol II binding on yeast genes in the presence of prp5-GAR, spt8Δ, and the double-mutant alleles at 30°C (B) and 16°C (B'). Intronless and intron-containing genes were separately analyzed, and genes were grouped into increased, no change and decreased RNA Pol II binding according to the total mapped reads that are normalized by input. Individual analyses of three genes with RNA Pol II pausing in the intronic regions suggested the restored pausing in the double-mutant strain (C).
Genome-wide analysis of RNA Pol II binding shows that prp5 mutation and spt8Δ globally rescue each other's effects
To further investigate the role of Prp5p–Spt8p interaction on genome-wide transcription, we performed ChIP-seq of yeast strains with spt8Δ, prp5-GAR, and both mutant alleles using the RNA Pol II antibody. At the permissive temperature, deletion of SPT8 resulted in significantly decreased RNA Pol II binding over 661 genes (568 intronless genes plus 93 intron-containing genes), while 459 genes (442 plus 17) showed relatively mild increased Pol II binding (Figure 4B, left), consistent with the function of Spt8p being to facilitate transcription of a subset genes (60,61). The prp5-GAR allele decreased RNA Pol II binding over 327 genes (249 plus 78) and increased Pol II binding over 455 genes (438 plus 17), but with no significant direction preference as was observed for spt8Δ (Figure 4B middle). Interestingly, in the double mutant strain (prp5-GAR, spt8Δ), only 2 (decreased) and 132 (increased) genes showed significantly changed Pol II binding, suggesting a transcriptional effect of mutual rescue between the prp5 mutation and spt8Δ (Figure 4B, right). In contrast, we did not observe this mutual rescue effect at the non-permissive temperature, because the prp5-GAR allele changed RNA Pol II binding of far fewer genes at 16°C (Figure 4B′).
To further address co-transcriptional splicing, we looked at the transcription profile of all intron-containing genes, and found that three of them had restored pausing of RNA Pol II in their intron regions in the double-mutant strain. For example, the well-studied co-transcriptional splicing gene, DBP2, has a high peak of RNA Pol II binding in the intronic region after the 5′SS in the WT strain, and this peak was decreased in the prp5-GAR strain but significantly restored with additional spt8Δ, especially at the 16°C (Figure 4C). A similar pattern was also found for two other intron-containing genes, NOG2 and RPL6B, but most intron-containing genes did not exhibit an obvious pause of RNA Pol II around the intron region.
Prp5p physically interacts with Spt8p, but not with Spt3p
One possible basis of the Spt8p and Spt3p specific genetic interactions with Prp5p could be direct and physical interactions between the transcription and splicing factors. To address this, we first generated yeast strains in which Spt8p was tagged with 3xHA at its C-terminus and/or Prp5p was tagged with FLAG at its N-terminus; these tags did not change the cs phenotype of prp5-GAR allele (Supplementary Figure S4A). We found that Prp5p was co-immunoprecipitated with Spt8p using HA antibody in lysate from the double-tagged yeast strain, whereas no signal was detectable from the single-tagged yeast strain (Figure 5A, cf. lanes 4 to 3), suggesting that Spt8p and Prp5p form a stable interaction in vivo. Second, we tested in vitro protein-protein interactions using purified recombinant proteins and Ni-NTA beads, in which Prp5p was fused with a 6xHis tag, and Spt8p and Spt3p were fused to a GST tag. Pull-down assays showed that Prp5p directly interacted with Spt8p, but not Spt3p (Figure 5B). Furthermore, we also investigated the region of Prp5p required to interact with Spt8p. In contrast to the full-length Prp5p, two truncated Prp5p proteins without its N-terminal region (aa 1–206), ΔN and ΔN&ΔC, could not pull-down Spt8p. However, truncated Prp5p that lacked the C-terminal region, ΔC, still efficiently pulled-down Spt8p (Figure 5C). Although we were unable to express and test the N-terminal domain directly, these results imply that the N-terminal region of Prp5p is required for interaction with Spt8p.
Figure 5.
Prp5p physically interacts with Spt8p, but not Spt3p, in vivo and in vitro. (A) Spt8p interacts with Prp5p in cell lysate. In the cell lysate from a double-tagged strain, Spt8p-HA pulls down FLAG-Prp5p. Single-tagged strains are used as controls. (B) Prp5p directly interacts with Spt8p, but not Spt3p. Direct in vitro protein-protein interactions were tested using purified recombinant proteins and Ni-NTA agarose. Spt8p and Spt3p were GST-tagged, and Prp5p was fused to a 6xHis tag. No-bait pulldowns (i.e. beads alone) were used as controls. (C) The N-terminus of Prp5p is required for Spt8p–Prp5p interaction. Upper, schematic representation of the full-length and truncated Prp5p proteins. Lower, in vitro protein-protein interactions were tested as in panel B. (D) CUS2 is required for maintaining the genetic interaction between PRP5 and SPT8. cus2Δ abrogates the ability of all tested prp5 alleles to rescue the growth defects of spt8Δ on 6-AU medium at 16°C and 30°C.
We previously found that the N-terminal region of Prp5p also directly interacts with two different HEAT motif regions of Hsh155p/SF3B1, a U2 snRNP component (46). To address whether the Prp5p interaction with Spt8p and Hsh155p are mutually exclusive, an in vitro competition assay was performed. We found that Prp5p pulled down similar levels of Spt8p in the presence or absence of excess amounts of HEAT-motif fragments of Hsh155p (Supplementary Figure S4B), indicating that these two interactions might not be mutually exclusive. In addition, pull-down assays using yeast lysates showed that Spt8p-3HA could not co-IP Hsh155p (Supplementary Figure S4C). Taken together, these results suggest that Prp5p has different functional motifs within its N-terminal domain for these two interactions, but that the Prp5p-Spt8p and the Prp5p-Hsh155p interactions are for the most part at different stages/times. However, this does not rule out the possibility of transient simultaneous interactions (see Discussion).
As mentioned previously, another critical splicing factor for the function of Prp5p is Cus2p, which binds to Hsh155p through a UHM-ULM interaction (62). Depletion of Cus2p allows U2-intron binding in vitro in the absence of ATP, although Prp5p is still physically required (31,63,64); in vivo, cus2Δ suppresses a transcriptional elongation defect wherein RNA Pol II accumulates on introns (30). To address the role of CUS2 in the Prp5p−Spt8p interaction, we performed 6-AU assays in the cus2Δ background. In the absence of CUS2, the prp5-GAR allele is no longer cs (Figure 5D upper), as previously described (36). Deletion of CUS2 resulted in sensitivity to 6-AU and abrogated the rescue of spt8Δ growth defects on 6-AU by prp5 alleles under all tested temperatures (Figure 5D and Supplementary Figure S5A). Thus, CUS2 is required for maintaining the Spt8p−Prp5p interaction-mediated coupling between transcription and splicing.
SPT8 and SPT3 contribute to BS proofreading by PRP5
Prp5p alleles, depending on their relative ATPase activities, can modulate the use of weak BS:U2 snRNA duplexes in splicing (36,47,65). We addressed whether disrupting the above-identified interactions would alter the BS region selectivity using the well-characterized splicing reporter gene ACT1-CUP1 in S. cerevisiae (45), where the tolerance to copper in the media is proportional to the splicing of the reporter intron (Figure 6A). In the WT SPT8 background, prp5-SAG, -TAG and -GAG alleles that are slow ATPases relative to WT PRP5-SAT improved splicing of suboptimal BS region substrates, such as U257C and A258U, whereas the WT reporter was unchanged, as previously reported (Figure 6B, left; 36). However, in the absence of SPT8, prp5-SAT mutant alleles no longer improved splicing of suboptimal BS substrates; instead, their splicing activities were decreased (Figure 6B, right); spt3Δ showed the same loss of improved splicing due to slow-ATPase prp5 alleles (Figure 6C). In contrast, deletion of other transcription factors, such as GCN5, DST1, or SPT4, did not alter the improved splicing of suboptimal BS substrate by SAT motif mutant allele prp5-TAG (Figure 6C). The exacerbation of BS region mutants in the absence of SPT8 is likely due to the failure of prp5 mutants to be recruited. These results indicate that the Prp5–Spt8/Spt3 interaction modulates splicing proofreading at the BS region.
Figure 6.
Deletion of the TBP-binding module components of SAGA complex, SPT8 or SPT3, reverses PRP5-dependent proofreading at branch site region. (A) Schematic of the ACT1-CUP1 reporter used for copper tolerance assay to monitor the efficiency of in vivo splicing. Mutant sites used in this study are indicated in red. (B) Deletion of SPT8 reverses the improvement of splicing activity of suboptimal branch site region substrates by prp5 alleles (SAT mutants). Growth of each strain on three representative copper concentrations was selected to illustrate altered splicing activities. (C) Deletion of SPT3, but not other transcription factors, also reverses proofreading at the branch site region by PRP5.
DISCUSSION
Prp5p is an RNA-dependent ATPase required for early spliceosome assembly and has been proposed to contribute to a splicing-dependent transcriptional checkpoint associated with pre-spliceosome formation (U1–U2–intron complexes) (30). In this study, we find partner proteins for Prp5p's coupling to the transcriptional machinery in the TBP-binding module of the SAGA complex. This is supported by several lines of evidence: (i) reciprocal genetic interactions between PRP5 and SAGA components, limited to SPT8 and SPT3; (ii) direct physical interaction between Prp5p and Spt8p; (iii) contribution of Spt8p and Prp5p to each other's recruitment to transcribed genes; (iv) global restoration of affected transcription in the prp5-GAR and spt8Δ double-mutant strain; (v) inhibited splicing by a prp5 mutant allele is rescued by spt8Δ and (vi) influence of Spt8p and Spt3p on Prp5p-dependent splicing proofreading at the BS region.
Reciprocal genetic interactions
Our data reveal a reciprocal genetic interaction between the TBP-binding module (SPT8 and SPT3) of SAGA complex and the spliceosomal ATPase gene PRP5. In one direction, spt8Δ or spt3Δ rescues the cs phenotype of the prp5-GAR allele, indicating that impaired splicing can be improved by deletion of transcription factors; in the other direction, prp5 mutant alleles with reduced ATPase activities rescue the growth defects on 6-AU of spt8Δ and spt3Δ, indicating that impaired transcription can be improved by the slower spliceosomal ATPase. This reciprocal relationship suggests that in WT cells there is a balance of activity of Prp5p and Spt8p/Spt3p. Mutation of either of these results in defects due to imbalance; in contrast, mutation of both restores this balance.
The genetic interaction of PRP5 with both SPT8 and SPT3 is consistent with previous findings that SPT8 and SPT3 are functionally similar to each other, form the TBP-binding module (51,57,60), and are structurally separated from other SAGA components (27). Null mutations in SPT8 are suppressed by several spt3 mutations (51). Along with Spt3p, one proposed role of Spt8p is to control the TBP–TATA interaction at the promoter region, identified by chemical crosslinking (66).
Direct Prp5p–Spt8p interaction
The reciprocal genetic effects of prp5 mutants and spt8Δ/spt3Δ suggested that they might be direct binding partners. We found that Prp5p and Spt8p co-immunoprecipitate from yeast lysates, indicating association within a complex. We directly tested this by making recombinant His-Prp5p, GST-Spt8p, and GST-Spt3p proteins and performed in vitro binding and pull-down assays (Figure 5). Prp5p directly interacts with Spt8p, but not Spt3p; this is consistent with newly revealed cryo-EM structures of the yeast SAGA complexes, in which Spt3p is located within the central core module (relatively inaccessible), while Spt8p is positioned outside on the adjacent surface (67,68).
Other described interactions between SAGA transcription components and splicing factors include U2 snRNP components, Lea1p/U2A′ and Msl1p/U2B'', with GCN5 in yeast (26), the co-purification of U2 snRNP SF3B3 (SAP130) with human GCN5-containing STAGA complex (41), and the co-purification of both U2 snRNP SF3B3 and SF3B5 with Drosophila SAGA (44). Recently, transcription elongation factor Spt5p has been shown to contribute to splicing efficiency by recruitment of U5 snRNP to intron-containing genes (69).
We previously demonstrated direct protein-protein interaction between Prp5p and Hsh155p, orthologue of human SF3B1 (46). Both Prp5p–Hsh155p and Prp5p–Spt8p interactions require the N-terminal domain of Prp5p, but we could not detect any mutual exclusivity of these interactions (Supplementary Figure S4). It is tempting to suggest that these interactions may occur simultaneously. This may help to first recruit Prp5p and U2 snRNPs to SAGA at the promoter of transcribed genes, and then subsequently to pass off Prp5p from SAGA to the Hsh155p component of U2 snRNP as U2 snRNA recognizes the branch site sequence in the transcript (Figure 7). Such simultaneous interaction would likely be transient, during pre-spliceosome formation itself, as pull-down assays using yeast lysates showed that Spt8p-3HA did not detectably co-IP Hsh155p. Further work is needed to address these possibilities.
Figure 7.
Coupling between transcription and splicing mediated by Spt8p/Spt3p and Prp5p. (A) Schematic depicting recruitment of Prp5p to the promoter region by the TBP-binding module of SAGA (Spt8p and Spt3p) and subsequent modulation of splicing proofreading at the BS region by Prp5p. This recruitment relies on direct interaction between Prp5p and Spt8p. (B) Deletion of SPT8 results in altered transcription of a subset of intronless and intron-containing genes, most are down-regulated. Loss of SPT8/SPT3 increases splicing proofreading at the BS region. (C) The prp5 mutant allele affects transcription of a subset of intronless and intron-containing genes and inhibits pre-mRNA splicing. (D) The double mutant, prp5 allele and deletion of SPT8, restores both affected transcription and inhibited pre-mRNA splicing. Cus2p plays a critical role in this coupling, deletion of CUS2 abrogates the coupling between transcription and splicing. Lack of Spt8p/Spt3p results in increased splicing proofreading on suboptimal BS substrates. For clarity, intronless genes are not depicted.
Prp5p–Spt8p interaction guides proper transcription and splicing
The physical interaction between splicing factor Prp5p and SAGA component Spt8p is important for the coupling between transcription and splicing (Figure 7A). Spt8p and the TBP-binding module of SAGA recruit Prp5p to the promoter region. The Spt8p−Prp5p interaction ensures proper initiation and elongation of transcription by RNA Pol II (Figures 3 and 4) and modulation of splicing proofreading of intron-containing gene at the BS region by Prp5p (Figure 6). In the presence of the WT Prp5p, lack of Spt8p significantly alters transcription of a subset of both intronless and intron-containing genes, in which most are down-regulated, but does not change the splicing efficiency of intron-containing genes (Figure 7B). On the other hand, in the presence of Spt8p, prp5 allele (-GAR) also affects transcription of a subset of intronless and intron-containing genes and inhibits pre-mRNA splicing of the intron-containing genes (Figure 7C). However, the double-mutant (prp5-GAR and spt8Δ) restores the affected transcription and the inhibited pre-mRNA splicing (Figure 7D).
The meaning of changes in BS fidelity
In the above model, Prp5p–Spt8p interaction promotes pre-spliceosome formation (Complex A). These findings lead us to propose that Prp5p–Spt8p interactions accelerate pre-spliceosome formation; this specifically suppresses splicing defects caused by suboptimal branch site region substrates. Conversely, loss of Prp5p–Spt8p interaction slows pre-spliceosome formation; this exacerbates splicing defects of suboptimal BS substrates (Figure 6). That a lack of Spt8p/Spt3p results in increased splicing proofreading on suboptimal BS substrates suggests that the Spt8p−Prp5p interaction contributes to both the efficiency of transcription and the fidelity of splicing. These data suggest that Spt8p/Spt3p have a direct effect on U2 snRNP function at the time of branch site recognition; alternatively, they may alter the quality (i.e. some feature) of U2 snRNPs that are recruited to nascent transcripts.
We note that the prp5-GAR mutant also changes the RNA Pol II occupancy on hundreds of intronless genes; this was unanticipated, suggesting that the recruitment of Prp5p to transcribed genes by Spt8p might not be intron- or splicing-dependent. Although the change in Pol II occupancy on intronless genes could be due to indirect effects, this possibility is less likely due to the short time interval (30 min) at the non-permissive temperature. The possible role of Prp5p on non-intron-containing genes will require further investigation.
As a splicing factor, deletion of CUS2 causes strong growth defects on 6-AU (Figure 5D), suggesting that it has a critical role in yeast transcription, and this is consistent with previous reports (30,70). The homolog of the yeast Cus2p in humans is Tat-SF1 (29), first identified as a Tat-dependent elongation factor important for HIV transcription (71). Removal of Cus2p is a powerful suppressor of the defect of ATPase deficient mutant Prp5p proteins (72). In the absence of CUS2, the Prp5p-Spt8p interaction-mediated coupling is no longer detectable, indicating a dynamic relationship between the transcription subcomplex SAGA and the spliceosome subcomplex U2 snRNP, and its mechanism needs further investigation. It is intriguing that all three identified suppressors of mutant prp5 (CUS2, SPT8, and SPT3) encode transcription factors, which underscores a role for Prp5p in mediating a connection between transcription and spliceosome assembly.
We considered the possibility that the Prp5p–Spt8p interaction represents non-canonical roles for these proteins. However, because prp5 mutants alter Spt8p-dependent pol II occupancy, and spt8Δ alters the Prp5p-dependent use of sub-optimal branch site sequences, we conclude that the interactions between Spt8p and Prp5p contribute to their respective canonical roles. We also performed 6-AU assays of spt8Δ with mutants of another ATPase Prp28p, which is required for a later stage of spliceosome assembly, facilitating the transition from 5'SS:U1 snRNA base-pairing to 5'SS:U6 snRNA base-pairing and resulting in the release of U1 snRNP (73,74). Using a series of prp28 mutants (-E404K, -E404V, -E404L, -E404A) previously identified (75), we did not observe any detectable genetic interaction between SPT8 and PRP28 (Supplementary Figure S5). These data suggest that the effects on splicing of the interaction between Spt8p and splicing factors is limited to pre-spliceosome assembly.
DATA AVAILABILITY
All NGS data have been deposited to NCBI under accession number GSE147179.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31400653, 31525022, 31472045, 31570821, 31971225]; National Institutes of Health [GM57829]; Hundred Talents program of Anhui Province [KJ2017A190], Scientific Research of BSKY [XJ201601]; Anhui Medical University Translational Medicine Program [2017ZHYX09]. Funding for open access charge: Scientific Research of BSKY from Anhui Medical University [XJ201601].
Conflict of interest statement. None declared.
REFERENCES
- 1. Darnell J.E., Jr. Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA. 2013; 19:443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pai A.A., Luca F.. Environmental influences on RNA processing: Biochemical, molecular and genetic regulators of cellular response. Wiley Interdiscip Rev. RNA. 2018; 10:e1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alpert T., Herzel L., Neugebauer K.M.. Perfect timing: splicing and transcription rates in living cells. Wiley Interdiscip Rev. RNA. 2017; 8:e1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saldi T., Cortazar M.A., Sheridan R.M., Bentley D.L.. Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 2016; 428:2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maniatis T., Reed R.. An extensive network of coupling among gene expression machines. Nature. 2002; 416:499–506. [DOI] [PubMed] [Google Scholar]
- 6. Pandit S., Wang D., Fu X.D.. Functional integration of transcriptional and RNA processing machineries. Curr. Opin. Cell. Biol. 2008; 20:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haberle V., Stark A.. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell. Biol. 2018; 19:621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Das R., Yu J., Zhang Z., Gygi M.P., Krainer A.R., Gygi S.P., Reed R.. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007; 26:867–881. [DOI] [PubMed] [Google Scholar]
- 9. de la Mata M., Kornblihtt A.R.. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat. Struct. Mol. Biol. 2006; 13:973–980. [DOI] [PubMed] [Google Scholar]
- 10. Dye M.J., Gromak N., Proudfoot N.J.. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006; 21:849–859. [DOI] [PubMed] [Google Scholar]
- 11. Nojima T., Rebelo K., Gomes T., Grosso A.R., Proudfoot N.J., Carmo-Fonseca M.. RNA Polymerase II Phosphorylated on CTD Serine 5 Interacts with the Spliceosome during Co-transcriptional Splicing. Mol. Cell. 2018; 72:369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin S., Coutinho-Mansfield G., Wang D., Pandit S., Fu X.D.. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008; 15:819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sapra A.K., Anko M.L., Grishina I., Lorenz M., Pabis M., Poser I., Rollins J., Weiland E.M., Neugebauer K.M.. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol. Cell. 2009; 34:179–190. [DOI] [PubMed] [Google Scholar]
- 14. Fong Y.W., Zhou Q.. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001; 414:929–933. [DOI] [PubMed] [Google Scholar]
- 15. Loerch S., Leach J.R., Horner S.W., Maji D., Jenkins J.L., Pulvino M.J., Kielkopf C.L.. The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface. J. Biol. Chem. 2019; 294:2892–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. David C.J., Boyne A.R., Millhouse S.R., Manley J.L.. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes. Dev. 2011; 25:972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kress T.L., Krogan N.J., Guthrie C.. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell. 2008; 32:727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wallace E.W.J., Beggs J.D.. Extremely fast and incredibly close: cotranscriptional splicing in budding yeast. RNA. 2017; 23:601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de la Mata M., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R.. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003; 12:525–532. [DOI] [PubMed] [Google Scholar]
- 20. Dujardin G., Lafaille C., Petrillo E., Buggiano V., Gomez Acuna L.I., Fiszbein A., Godoy Herz M.A., Nieto Moreno N., Munoz M.J., Allo M. et al.. Transcriptional elongation and alternative splicing. Biochim. Biophys. Acta. 2013; 1829:134–140. [DOI] [PubMed] [Google Scholar]
- 21. Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X.D., Bentley D.L.. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes. Dev. 2014; 28:2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014; 15:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C.Y., Xiao R., Burge C.B., Fu X.D.. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013; 153:855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Close P., East P., Dirac-Svejstrup A.B., Hartmann H., Heron M., Maslen S., Chariot A., Soding J., Skehel M., Svejstrup J.Q.. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012; 484:386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Batsche E., Yaniv M., Muchardt C.. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006; 13:22–29. [DOI] [PubMed] [Google Scholar]
- 26. Gunderson F.Q., Johnson T.L.. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS. Genet. 2009; 5:e1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu P.Y., Ruhlmann C., Winston F., Schultz P.. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell. 2004; 15:199–208. [DOI] [PubMed] [Google Scholar]
- 28. Wu P.Y., Winston F.. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 2002; 22:5367–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan D., Perriman R., Igel H., Howe K.J., Neville M., Ares M. Jr.. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell Biol. 1998; 18:5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chathoth K.T., Barrass J.D., Webb S., Beggs J.D.. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol. Cell. 2014; 53:779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perriman R., Barta I., Voeltz G.K., Abelson J., Ares M. Jr.. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13857–13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perriman R.J., Ares M. Jr.. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes. Dev. 2007; 21:811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abu Dayyeh B.K., Quan T.K., Castro M., Ruby S.W.. Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J. Biol. Chem. 2002; 277:20221–20233. [DOI] [PubMed] [Google Scholar]
- 34. Ruby S.W., Chang T.H., Abelson J.. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes. Dev. 1993; 7:1909–1925. [DOI] [PubMed] [Google Scholar]
- 35. Xu Y.Z., Newnham C.M., Kameoka S., Huang T., Konarska M.M., Query C.C.. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO. J. 2004; 23:376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu Y.Z., Query C.C.. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell. 2007; 28:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO. Rep. 2009; 10:843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodriguez-Navarro S., Fischer T., Luo M.J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J.E., Reed R., Hurt E.. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004; 116:75–86. [DOI] [PubMed] [Google Scholar]
- 39. Will C.L., Luhrmann R.. Spliceosome structure and function. Cold Spring Harbor Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brand M., Moggs J.G., Oulad-Abdelghani M., Lejeune F., Dilworth F.J., Stevenin J., Almouzni G., Tora L.. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO. J. 2001; 20:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez E., Palhan V.B., Tjernberg A., Lymar E.S., Gamper A.M., Kundu T.K., Chait B.T., Roeder R.G.. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell Biol. 2001; 21:6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bai R., Wan R., Yan C., Lei J., Shi Y.. Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. Science. 2018; 360:1423–1429. [DOI] [PubMed] [Google Scholar]
- 43. Plaschka C., Lin P.C., Charenton C., Nagai K.. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature. 2018; 559:419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stegeman R., Spreacker P.J., Swanson S.K., Stephenson R., Florens L., Washburn M.P., Weake V.M.. The spliceosomal protein SF3B5 is a novel component of Drosophila SAGA that functions in gene expression independent of splicing. J. Mol. Biol. 2016; 428:3632–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lesser C.F., Guthrie C.. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993; 133:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang Q., Rodriguez-Santiago S., Wang J., Pu J., Yuste A., Gupta V., Moldon A., Xu Y.Z., Query C.C.. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes. Dev. 2016; 30:2710–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shao W., Kim H.S., Cao Y., Xu Y.Z., Query C.C.. A U1-U2 snRNP interaction network during intron definition. Mol. Cell. Biol. 2012; 32:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gornemann J., Kotovic K.M., Hujer K., Neugebauer K.M.. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005; 19:53–63. [DOI] [PubMed] [Google Scholar]
- 49. Ewels P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso P., Nahnsen S.. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020; 38:276–278. [DOI] [PubMed] [Google Scholar]
- 50. Hvorecny K.L., Prelich G.. A systematic CEN library of the Saccharomyces cerevisiae genome. Yeast. 2010; 27:861–865. [DOI] [PubMed] [Google Scholar]
- 51. Eisenmann D.M., Chapon C., Roberts S.M., Dollard C., Winston F.. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994; 137:647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koutelou E., Hirsch C.L., Dent S.Y.. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 2010; 22:374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Exinger F., Lacroute F.. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 1992; 22:9–11. [DOI] [PubMed] [Google Scholar]
- 54. Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W.. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 2002; 10:1139–1150. [DOI] [PubMed] [Google Scholar]
- 55. Desmoucelles C., Pinson B., Saint-Marc C., Daignan-Fornier B.. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 2002; 277:27036–27044. [DOI] [PubMed] [Google Scholar]
- 56. Samara N.L., Wolberger C.. A new chapter in the transcription SAGA. Curr. Opin. Struct. Biol. 2011; 21:767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Helmlinger D., Tora L.. Sharing the SAGA. Trends Biochem. Sci. 2017; 42:850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou K., Kuo W.H., Fillingham J., Greenblatt J.F.. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:6956–6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bian C., Xu C., Ruan J., Lee K.K., Burke T.L., Tempel W., Barsyte D., Li J., Wu M., Zhou B.O et al.. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO. J. 2011; 30:2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Belotserkovskaya R., Sterner D.E., Deng M., Sayre M.H., Lieberman P.M., Berger S.L.. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 2000; 20:634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baptista T., Grunberg S., Minoungou N., Koster M.J.E., Timmers H.T.M., Hahn S., Devys D., Tora L.. SAGA Is a General Cofactor for RNA Polymerase II Transcription. Mol. Cell. 2017; 68:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Talkish J., Igel H., Hunter O., Horner S.W., Jeffery N.N., Leach J.R., Jenkins J.L., Kielkopf C.L., Ares M.. Cus2 enforces the first ATP-dependent step of splicing by binding to yeast SF3b1 through a UHM-ULM interaction. RNA. 2019; 25:1020–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Perriman R., Ares M. Jr.. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol. Cell. 2010; 38:416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rodgers M.L., Tretbar U.S., Dehaven A., Alwan A.A., Luo G., Mast H.M., Hoskins A.A.. Conformational dynamics of stem II of the U2 snRNA. RNA. 2016; 22:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang Z.M., Yang F., Zhang J., Tang Q., Li J., Gu J., Zhou J., Xu Y.Z.. Crystal Structure of Prp5p Reveals Interdomain Interactions that Impact Spliceosome Assembly. Cell. Rep. 2013; 5:1269–1278. [DOI] [PubMed] [Google Scholar]
- 66. Sermwittayawong D., Tan S.. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO. J. 2006; 25:3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Papai G., Frechard A., Kolesnikova O., Crucifix C., Schultz P., Ben-Shem A.. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature. 2020; 577:711–716. [DOI] [PubMed] [Google Scholar]
- 68. Wang H., Dienemann C., Stutzer A., Urlaub H., Cheung A.C.M., Cramer P.. Structure of the transcription coactivator SAGA. Nature. 2020; 577:717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maudlin I.E., Beggs J.D.. Spt5 modulates co-transcriptional spliceosome assembly in Saccharomyces cerevisiae. RNA. 2019; 25:1298–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alexander R.D., Innocente S.A., Barrass J.D., Beggs J.D.. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010; 40:582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou Q., Sharp P.A.. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996; 274:605–610. [DOI] [PubMed] [Google Scholar]
- 72. Perriman R., Ares M. Jr.. ATP can be dispensable for prespliceosome formation in yeast. Genes. Dev. 2000; 14:97–107. [PMC free article] [PubMed] [Google Scholar]
- 73. Staley J.P., Guthrie C.. An RNA switch at the 5' splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999; 3:55–64. [DOI] [PubMed] [Google Scholar]
- 74. Murray H.L., Jarrell K.A.. Flipping the switch to an active spliceosome. Cell. 1999; 96:599–602. [DOI] [PubMed] [Google Scholar]
- 75. Yang F., Wang X.Y., Zhang Z.M., Pu J., Fan Y.J., Zhou J., Query C.C., Xu Y.Z.. Splicing proofreading at 5' splice sites by ATPase Prp28p. Nucleic Acids Res. 2013; 41:4660–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All NGS data have been deposited to NCBI under accession number GSE147179.







